Radiotherapy remains a major treatment option for
numerous patients, serving both as a radical therapy and as
palliative care (1,2). Since the discovery of X-rays by
Wilhelm Conrad Röntgen in 1895, the field of radiotherapy has
undergone marked evolution (3).
This advancement has been driven by the pioneering contributions of
renowned scientists such as Nikola Tesla, Mihajlo Idvorski Pupin
and Maria Sklodowska-Curie (4).
Their innovative efforts have established a solid foundation for
modern clinical applications of radiotherapy, which can be employed
individually or in combination with other therapies for both
curative and palliative purposes at all stages of cancer (5,6). More
than half of all patients with cancer undergo radiotherapy;
however, its effectiveness is often diminished by the emergence of
radioresistance, which can profoundly impact the quality of life of
patients (7,8). Radiation therapy exerts its effects by
destroying cancer cells either directly or indirectly through the
induction of DNA damage. Ionizing radiation (IR) can also activate
several prosurvival signaling pathways, including the p53, ataxia
telangiectasia mutated and NF-κB pathways, which enhance DNA damage
checkpoint activation, promote DNA repair and stimulate autophagy
(9–11). Together, these signaling pathways
provide protection to cancer cells against radiation-induced
damage, thereby contributing to their resistance to treatment
(12).
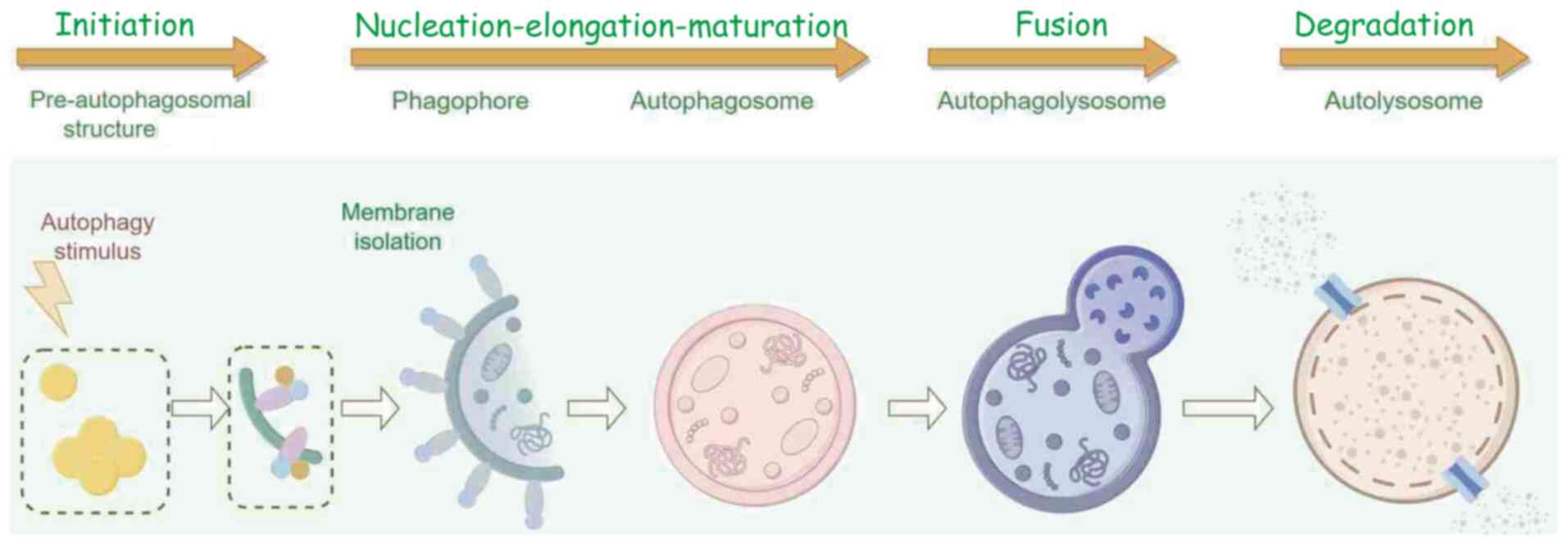
Autophagy is a vital protein degradation pathway in
cells, and is essential for maintaining cellular homeostasis and
overall health (13). Autophagy,
which was first identified in human liver cells, is categorized
into three forms in mammalian cells: Macroautophagy, microautophagy
and chaperone-mediated autophagy (14). The autophagy process typically
includes four main stages: Initiation, formation and maturation of
the autophagosome, precise fusion of the autophagosome with the
lysosome, and finally content degradation (15) (Fig.
1). During the autophagy process activated by radiotherapy, a
cellular mechanism directs old, damaged or malfunctioning
biomolecules and organelles to lysosomes for degradation (16,17).
This process is preserved through evolution and is triggered by
different internal and external stressors, including nutrient
shortage, absence of growth factors, IR or low oxygen levels
(18). Under such circumstances,
autophagy functions primarily as a survival tactic by clearing out
dysfunctional organelles or toxic aggregates that could otherwise
lead to apoptosis (19).
Furthermore, the lysosomal decomposition of excess cellular parts
supplies crucial raw materials and nutrients that can be reused to
create important biomolecules (20). Therefore, regulating tumor autophagy
levels to reduce radiotherapy resistance represents a novel
approach.
Long non-coding RNAs (lncRNAs) are defined as
transcripts longer than 200 nucleotides that do not encode proteins
(21). Despite their low
conservation across species, lncRNAs function as molecular switches
that influence cell survival, inflammation and lipid metabolism in
the vasculature by regulating autophagy (22). Research has established a
relationship between lncRNAs and resistance to tumor radiotherapy
(23,24). Additionally, lncRNAs serve various
roles, including acting as molecular signals, decoys, guides and
scaffolds (25). Therefore,
reviewing the role of autophagy-modulating lncRNAs in tumor
radioresistance can enhance the understanding of these regulatory
mechanisms and may lead to the identification of novel therapeutic
strategies aimed at increasing tumor radiosensitivity through the
modulation of autophagy.
The present review aims to present the current
understanding of the role of autophagy in cancer and its impact on
tumor radioresistance through interactions with lncRNAs. By
elucidating the function of autophagy-modulating lncRNAs in tumor
radioresistance, the present review aims to provide valuable
insights into the context-dependent applications of therapeutic
strategies.
The role of autophagy in cancer is complex and can
be viewed as a double-edged sword; it has the potential to inhibit
tumor development, while also being implicated in tumor initiation
(26). Autophagy operates at a
basal level in all cell types and actively contributes to tumor
prevention (27). As a critical
intracellular mechanism, autophagy effectively responds to
genotoxic stress through pathways involving reactive oxygen species
(ROS), helping to prevent the accumulation of mutations and
maintain genetic stability (28).
Furthermore, autophagy is responsible for the removal of damaged
organelles, ensuring that cellular damage does not accumulate,
thereby preserving homeostasis and normal cellular function
(29). This underscores its
precise, standardized and essential role in maintaining cellular
health and stability (30).
Among the numerous regulators of autophagy that have
been identified, beclin 1 (BECN1) is recognized as one of the most
crucial (31). As a vital
regulatory protein, BECN1 is essential for initiating autophagy and
serves a role in maintaining the delicate balance between cell
survival and death (32). Research
has indicated that heterozygous loss of the BECN1 gene enhances
tumor development in mice, highlighting its importance in
understanding the mechanisms underlying tumorigenesis (33). Notably, the deletion of the BECN1
gene is frequently observed in various malignant tumors, including
breast, ovarian and prostate cancer, underscoring its critical role
in cancer pathology (34–36). Furthermore, a study has demonstrated
that overexpression of BECN1 in breast cancer cells effectively
inhibited cell proliferation and colony formation, and reduced
tumor growth in nude mice (37). In
addition to BECN1, research has shown that the knockout of the
autophagy-related gene (ATG)5 and ATG7 genes in mice disrupts the
autophagy process, leading to exacerbated oxidative stress
responses and an increased risk of developing liver cancer
(38). These findings emphasize the
crucial role of autophagy in tumor prevention.
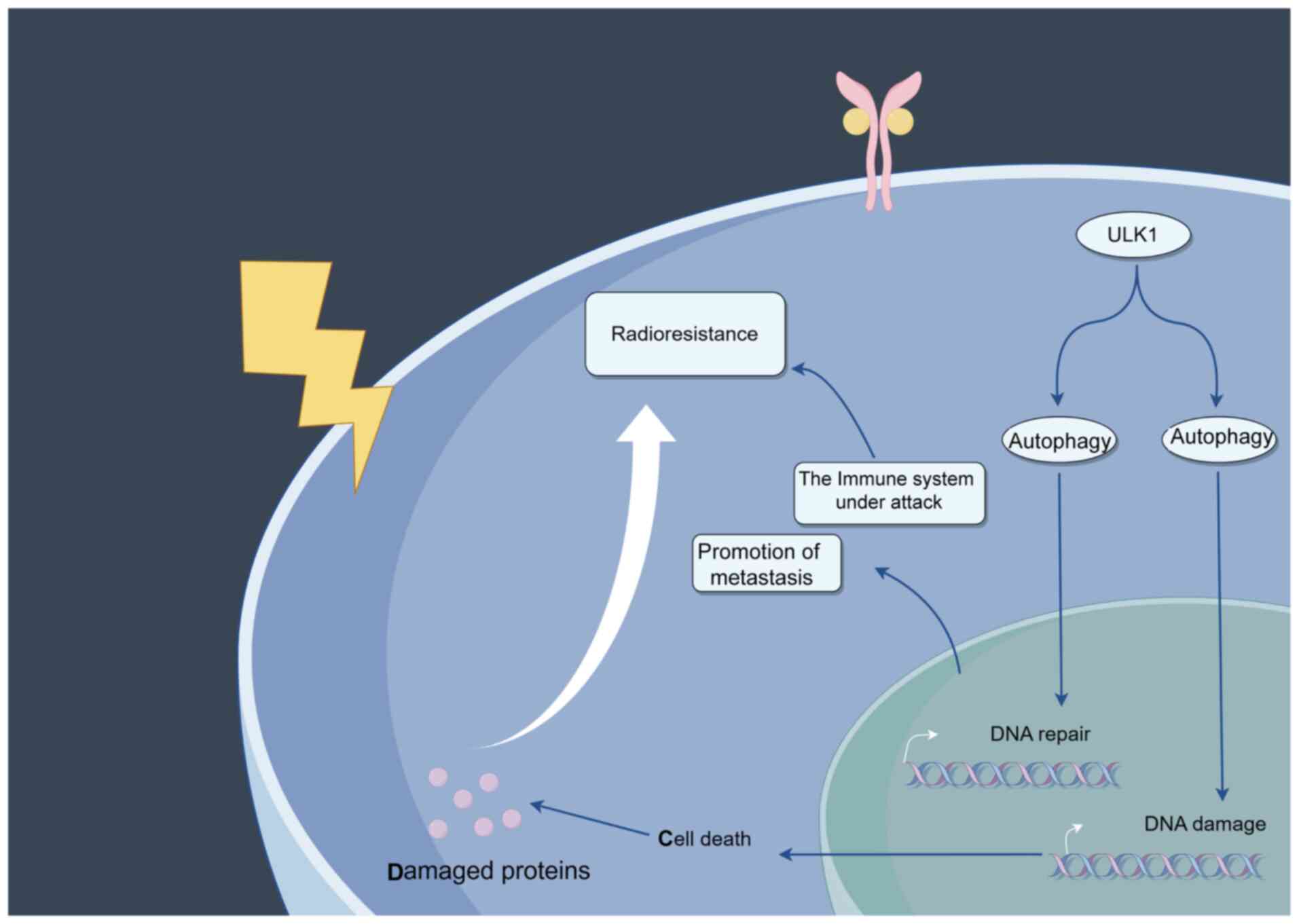
Although radiotherapy is a main approach for the
treatment of cancer, resistance frequently arises, resulting in
unsuccessful treatments (44). This
radioresistance arises from the capacity of tumors to disrupt
stress responses, such as DNA damage and repair processes, which
either promote or inhibit autophagy and aid in nutrient recycling
(45) (Fig. 2). For example, by modulating Unc-51
like autophagy activating kinase 1 (ULK1)-mediated autophagy,
butyrophilin subfamily 3 member A1 contributes to tumor progression
and radiation resistance in esophageal squamous cell carcinoma
(46). Xu et al (47) reported that blocking autophagic flux
and DNA repair in tumor cells could enhance the effectiveness of
radiotherapy for orthotopic glioblastoma. In addition to damaged
proteins, various intermediate molecules, their complexes, and
organelles such as mitochondria and micronuclei can serve as cargo
for the autophagy process (48).
Previous studies have revealed both pro-survival and anti-survival
characteristics of autophagic pathways (49,50).
Cancer cells frequently take advantage of the dual role of
autophagy to survive in metabolically difficult conditions, escape
immune system attacks, prevent cell death and induce metastasis
(51,52). Research has revealed that using both
radiation and berberine together suppresses autophagy and
alternative end-joining DNA repair, which are linked to
radioresistance, thereby enhancing sensitivity to radiation
(53).
The sensitivity of tumor cells to radiation is
frequently affected by autophagic processes (54). BECN1 moves to the nucleus after IR
exposure, resulting in G2/M cell cycle arrest (55). Furthermore, autophagy driven by ATG5
has been demonstrated to increase the radiosensitivity of prostate
cancer cells, especially when nutrients are scarce or glutamine is
depleted, and also when MYC is silenced (56). Chen et al (57) proposed that inhibiting the nuclear
factor erythroid 2-related factor 2 (NRF2) antioxidative pathway
could make U-2 osteosarcoma cells more sensitive to radiation, with
the NRF2 antioxidative response being controlled by
autophagy-driven activation of ERK 1/2 kinases. Consequently,
reducing the expression levels of BECN1, ATG5 or NRF2 has been
shown to decrease the IR sensitivity of cancer cells. Furthermore,
the inhibition of ATG7 by lncRNA HOTAIR, which is notably
upregulated in prostate cancer cell lines exposed to radiation, has
been associated with radioresistance in these cells (58). These results emphasize the essential
function of autophagy in influencing radiosensitivity and suggest
the possibility of targeting autophagic pathways to enhance
radiotherapy effectiveness (58).
Research on breast cancer cells, which naturally
resist apoptosis, further underscores the link between IR and
autophagic pathways (59). After IR
exposure, these cells show increased autophagic characteristics,
resulting in more iron accumulation. This accumulation, in
conjunction with subsequent ROS generation, oxidative stress and
DNA damage, may trigger cell death through a mechanism known as
ferroptosis (60,61). These insights have sparked a rising
interest in pairing autophagic inducers with IR to improve the
radiosensitivity of cancer cells. For instance, in a similar
approach, the treatment of non-small cell lung carcinoma (NSCLC)
cells with rapamycin alongside a histone deacetylase inhibitor has
been found to promote radiosensitization (62). This combined treatment has two
effects: It boosts autophagy and at the same time hinders DNA
damage repair processes. Observations in cultured cells and tumor
xenograft mouse models have demonstrated the effectiveness of the
strategy, pointing to a potential improvement in radiotherapy
outcomes (63).
Autophagy affects cancer cell survival in complex
ways, acting both as a supporter and an opponent of
radiosensitization (62,64). Various cancer cell types have been
shown to exploit enhanced autophagy as a survival strategy. For
instance, the application of autophagy inhibitors renders otherwise
radio-resistant bladder cancer (BLCA) cells more sensitive to
chemotherapy (65). Similarly,
silencing of ATG5, a key component of the autophagy process, has
been found to increase IR-induced cell death in nasopharyngeal
carcinoma (66). Furthermore, the
use of autophagy inhibitors in combination with IR has emerged as a
factor influencing the bystander and abscopal effects associated
with chemotherapy and radiotherapy (67). These findings underscore the
importance of context in the role of autophagy in cancer treatment
and highlight the potential of targeting autophagy pathways to
enhance treatment efficacy.
Radiotherapy is a common cancer treatment that
induces different forms of cell death, including autophagy
(74). Besides directly causing DNA
strand breaks, IR can lead to the production of significant amounts
of ROS in cells, resulting in oxidative stress damage (75). According to Yang et al
(76), mitophagy triggered by IR
boosts ferroptosis by raising the levels of free fatty acids inside
cells. Lysosomes serve a crucial role in autophagy, a cellular
degradation process (77). A
conserved protein associated with autophagy, Atgs/P62/LC3II,
controls the process (78). In
cancer cells, autophagy is a double-edged sword. In the initial
phases, it might restrict the development of tumors. Nevertheless,
it might also offer a survival advantage for adaptation and
detoxification in challenging conditions such as starvation, low
oxygen levels, and chemotherapy or radiotherapy (79). Investigators have reported that
autophagy activity rises following IR. This acts as a mechanism for
cells to survive when faced with cytotoxic agents, including IR and
temozolomide, in the treatment of glioblastoma (80,81).
Furthermore, researchers have revealed that IR promotes autophagy
via the Wnt/β-Catenin pathway (82). In conclusion, IR has an important
effect on tumor autophagy.
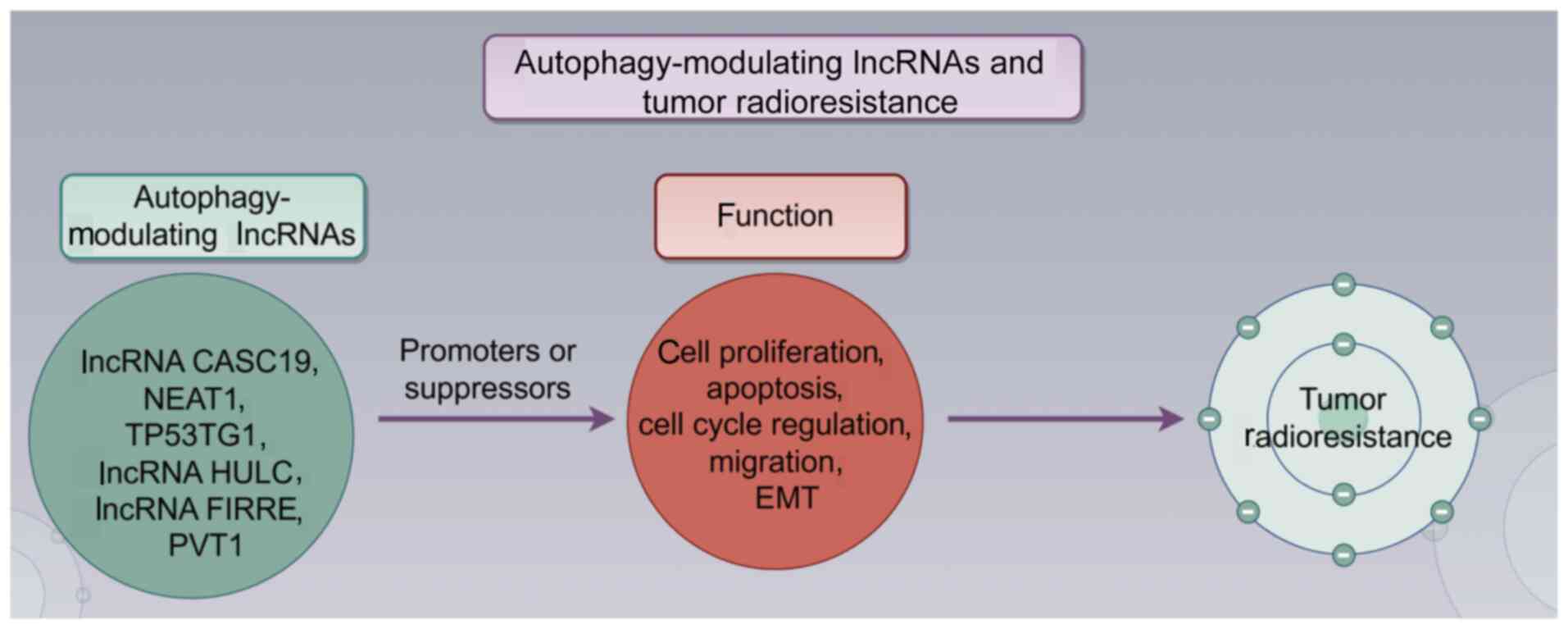
Numerous cancer types have been found to exhibit
abnormal expression of lncRNAs, and the relationship between
lncRNAs and autophagy has garnered interest across various cancer
types (Table I), including lung,
gastric, breast and prostate cancer (83). The majority of research suggests
that lncRNAs regulate autophagy mainly through mechanisms such as
microRNA (miRNA/miR) sponging (functioning as competing endogenous
RNAs), interactions between RNAs, RNA-protein regulation and
various other pathways (84,85).
Studies have indicated that lncRNAs are involved in various phases
of the autophagy process from the beginning to the maturation stage
(86,87). They assist in starting autophagic
phagocytosis by controlling essential proteins such as ULK1, mTOR
and BECN1, and they also affect the extension of autophagic
vesicles by regulating ATG3, ATG5, ATG4, ATG12 and ATG7 (88). The present review highlights the
complex and diverse functions of lncRNAs in regulating autophagy,
and their possible impact on cancer biology and treatment.
Proliferation is one of the most critical malignant
phenotypes observed in cancer, while apoptosis is equally
significant in the context of neoplastic development (89). Although their functions are
fundamentally opposite (proliferation promotes cell growth and
survival, while apoptosis leads to programmed cell death), their
interactions work synergistically to regulate cancer development.
This delicate balance between cell proliferation and apoptosis
serves a crucial role in tumor progression and resistance to
therapy (90). In digestive system
cancers, various lncRNAs have been identified to serve roles in
tumor progression and autophagy regulation (91). For instance, lncRNA SNHG11 is
upregulated in gastric cancer and is associated with poor patient
prognosis. lncRNA SNHG11 post-transcriptionally enhances ATG12
expression through miR-1276, thereby promoting autophagy, cell
proliferation and activation of the Wnt/β-catenin signaling pathway
(92). Similarly, in HCC, lncRNA
MALAT1 exhibits elevated expression levels compared with those in
normal tissues. Silencing MALAT1 has been shown to increase HCC
autophagy by promoting the conversion of LC3-I to LC3-II and
simultaneously suppressing HCC cell proliferation (93). Conversely, lncRNA NBR2 acts as a
tumor suppressor in HCC by inhibiting BECN1 and autophagy through
the ERK/JNK pathways, thereby restraining HCC cell proliferation
(94). In colorectal cancer (CRC),
lncRNA SLCO4A1-AS1 acts as an oncogenic element, exhibiting a
positive association with par-3 family cell polarity regulator
(PARD3) and sponging miR-508-3p. This lncRNA enhances the
proliferation of CRC cells and initiates autophagy through the
miR-508-3p/PARD3 pathway. Nonetheless, the regulatory effect is
interrupted by the application of the autophagy inhibitor
3-methyladenine (95). Another
lncRNA in CRC, FIRRE, directly interacts with polypyrimidine tract
binding protein 1 to enhance BECN1 mRNA stability, thus reducing
autophagy and promoting CRC cell proliferation (96). In the context of oral squamous cell
carcinoma (OSCC) and cholangiocarcinoma (CCA), several upregulated
lncRNAs have been shown to suppress autophagy. For instance,
LINC01207 is highly expressed in OSCC, where its upregulation
promotes cell proliferation but inhibits both apoptosis and
autophagy via the miR-1301-3p/lactate dehydrogenase A axis
(97). By contrast, overexpression
of LINC00958 reduces apoptosis while promoting autophagy by
upregulating autophagy-related proteins such as BECN1 and ATG5, and
increasing the LC3-II/LC3-I ratio, driven by p53 mediated by
sirtuin 1 (SIRT1) (98). In CCA,
lncRNA HOTAIR inhibits both apoptotic and autophagic processes,
thereby promoting the proliferation of CCA cells by targeting the
miR-204-5p/high mobility group box 1 axis (99). Pancreatic cancer (PANC), known for
its poor prognosis, remains a complex malignancy with unclear
progression mechanisms (100). A
study by Wu et al (101)
revealed that LZTS1-AS1, a highly expressed lncRNA in PANC cells
and tissues, promoted PANC cell proliferation while inhibiting
apoptosis and autophagy via the miR-532/twist family bHLH
transcription factor 1 axis. Overall, these findings highlight the
diverse and critical roles of lncRNAs in modulating autophagy and
influencing the malignant phenotypes of various cancer types.
The downregulation of lncRNA CASC2 enhances
apoptosis in NSCLC and diminishes ATG5-mediated autophagy by
regulating the miR-214/tripartite motif containing 16 axis in A549
and H1299 NSCLC cell lines (105).
Conversely, a reversed autophagic state has been noted in papillary
thyroid carcinoma (PTC), where overexpression of lncRNA SLC26A4-AS1
inhibited the proliferation of PTC cells and stimulated autophagy
by recruiting the transcription factor ETS1 and elevating inositol
1,4,5-trisphosphate receptor type 1 expression (106). Additionally, Qin et al
(107) revealed that lncRNA SNHG5
was stabilized by RNA binding motif protein 47 (RBM47) and directed
to FOXO3, resulting in decreased proliferation and the activation
of autophagy in PTC cells through the RBM47/small nucleolar RNA
host gene 5/FOXO3 pathway.
CASC19, a newly identified lncRNA located on
chromosome 8q24.21, consists of 324 nucleotides (110). A study has demonstrated that
CASC19 expression is upregulated in a variety of human cancer
types, including NSCLC, gastric cancer, CRC, PANC, ccRCC, glioma,
cervical cancer and nasopharyngeal carcinoma (111). Notably, some research has
indicated that CASC19 can enhance radioresistance in nasopharyngeal
carcinoma by modulating autophagy signaling pathways (112,113). Furthermore, the dysregulation of
CASC19 has been closely linked to numerous clinicopathological
characteristics (prognosis and pathological staging) and cancer
progression, particularly in HCC and gastric cancer (114,115). In summary, CASC19 influences
various cellular phenotypes, including cell proliferation,
apoptosis, cell cycle regulation, migration, invasion, EMT,
autophagy and therapeutic resistance (116).
NEAT1 is predominantly located in the nucleus,
although a smaller fraction is present in the cytoplasm (117). NEAT1 functions as a structural
component of nuclear paraspeckles and is involved in regulating
gene expression at both transcriptional and post-transcriptional
levels (118). Research has
demonstrated that NEAT1 contributes to radioresistance in HCC cells
by inducing autophagy through GABA type A receptor-associated
protein (119). The role of NEAT1
in carcinogenesis is multifaceted, encompassing interactions with
miRNAs, modulation of gene expression, regulation of epigenetic
factors and participation in various signaling pathways, such as
the PI3K-AKT and TGF-β1 pathways (120). Additionally, the involvement of
NEAT1 in cancer is complicated by its impact on CSCs and the tumor
microenvironment (121). The
interaction of NEAT1 with autophagy further adds to the complexity
of its functions in cancer biology (122). Understanding the interactions
between NEAT1, autophagy and radioresistance is crucial for
researchers as it may lead to the identification of novel
therapeutic targets and strategies aimed at disrupting oncogenic
processes, reducing treatment resistance and improving patient
survival (123). Consequently,
this understanding could facilitate the development of specialized
treatment approaches and regimens.
lncRNA TP53TG1 (National Center for Biotechnology
Information reference sequence, NR_015381.1), located on chromosome
12, is transcribed as an ~0.7-kilobase lncRNA molecule (124). Its expression can be induced under
conditions of cellular stress in a wild-type TP53-dependent manner
(125). As a relatively recent
discovery in the field of lncRNAs, TP53TG1 has been implicated to
serve oncogenic and anti-oncogenic roles across various cancer
types, such as gastric and breast cancer (126). For example, overexpression of
TP53TG1 promotes proliferation of colon cancer cells (127). In addition, Cheng et al
(128) found that lncRNA TP53TG1
exerted anticancer effects in cervical cancer by comprehensively
regulating the transcriptome profile of HeLa cells. Research has
indicated a close association between TP53TG1 and autophagy
(129). Furthermore, it has been
demonstrated that lncRNA TP53TG1 contributes to the radioresistance
of glioma cells through the miR-524-5p/RAB5A axis (130). Given these findings, TP53TG1
represents a promising target to enhance the sensitivity of tumors
to radiotherapy.
HULC, found on chromosome 6p24.3, has been
identified to be upregulated in HCC and is linked to cancer
advancement (131). The expression
of this transcript is regulated by miRNAs and transcriptionally
modulated by Sp1 family factors (132). HULC has been identified as a novel
biomarker in different types of cancer, such as prostate cancer and
CRC, serving a role as an oncogenic factor (133). Research conducted previously
indicated that inhibiting HULC could reduce angiogenesis in human
gliomas (134). Another study
demonstrated that HULC facilitated the advancement of CRC (135). A study has shown that lncRNA HULC
serves a role in radioresistance by reducing autophagy in prostate
cancer cells (136). Thus, lncRNA
HULC could serve as a therapeutic target to combat tumor resistance
to radiotherapy.
PVT1, a lncRNA, was initially identified as a driver
of variant translocations in mouse plasmacytomas (142). The human lncPVT1 gene is situated
on chromosome 8q24.21, sharing the same location as the MYC
oncogene (143). lncPVT1 has been
identified as a factor that accelerates the development of several
cancer types, such as nasopharyngeal cancer and lung cancer
(144,145). Radioresistance poses a major
challenge to tumor treatment success, and is connected to the
abnormal regulation of physiological functions in cancer cells,
including apoptosis, autophagy, stemness (for CSCs), hypoxia, EMT
and DNA damage repair (146,147). Recently, lncPVT1 has been linked
to the modulation of radioresistance in certain cancer types,
including nasopharyngeal cancer and lung cancer (148).
Research interest in lncRNA therapeutics has only
emerged in the past decade, and so far, no treatments targeting
lncRNAs have reached clinical development (149). SP100-AS1, a lncRNA that targets
the antisense sequence of the SP100 gene, is upregulated in the
tissues of patients with CRC who are resistant to radiation,
according to RNA-sequencing analysis (150). Notably, reducing SP100-AS1
expression lowered radioresistance and decreased cell proliferation
and tumor growth in both laboratory and live models. To identify
the proteins and miRNAs interacting with SP100-AS1, mass
spectrometry and bioinformatics analyses were utilized. SP100-AS1
was shown to interact with and stabilize the ATG3 protein via the
ubiquitination-dependent proteasome mechanism (150). This interaction highlights the
possible involvement of SP100-AS1 in regulating autophagy-related
mechanisms and enhancing radioresistance in CRC. SP100-AS1 holds
potential as a therapeutic target to enhance the response of CRC to
radiation therapy (150). As
aforementioned, autophagy affects tumor cell proliferation,
survival and the response to treatment in intricate ways across
different types of cancer, such as nasopharyngeal cancer and CRC.
Methods to target autophagy might involve aiming at specific
proteins or pathways associated with autophagy, as well as using
agents that promote or block autophagy.
Scientists seek to improve treatment effectiveness
by utilizing the interaction between autophagy and processes
regulated by lncRNAs. They aim to develop more effective treatments
by integrating autophagy modulation with lncRNA-targeted methods
(151). Furthermore, these
combination therapies provide a comprehensive strategy to boost
patient outcomes (152).
There are both obstacles and prospects in the future
research of radiation oncology and lncRNAomics (153). For precise tumor targeting,
technological advancements such as hybrid MRI and positron emission
tomography scans are vital, although they entail significant costs
and demand specialized expertise (154). One major obstacle in lncRNA
research is the restricted knowledge gained from examining just a
small portion of lncRNAs, which limits the understanding of their
mechanisms, roles and structures (155). Despite significant progress in the
field, delivering oligonucleotides to solid tumors continues to be
a major challenge. Advancements in artificial intelligence and
sophisticated statistical modeling have enabled molecular-guided
treatment plans customized for each patient, representing a
significant achievement in precision medicine. Autophagy-modulating
lncRNAs represent a vast yet largely unexplored reservoir of
oncogenes, tumor suppressors and radioresponse modifiers.
Integrating this knowledge into treatment strategies, from
diagnosis and response prediction to the design of targeted
therapies, could greatly enhance management and outcomes for
patients with cancer (156). In
conclusion, the present review summarizes the effects of
autophagy-modulating lncRNAs on tumor radioresistance, providing
novel ideas for tumor radiosensitization.
lncRNAs that modulate autophagy are crucial elements
in the radioresistance of tumors. These lncRNAs are crucial in
different stages of cancer development and advancement, such as
tumor expansion, spread and resistance to treatment. Studies are
concentrating on using autophagy-based treatment methods to tackle
radioresistance in patients with cancer (157,158). Modifying autophagy can influence
the survival of tumor cells, their response to therapy and the
overall success of treatment. Creating targeted cancer treatments
requires a deep comprehension of the interaction between lncRNAs
and autophagy. Targeting specific lncRNAs could potentially adjust
autophagy and increase tumor radiosensitivity. Current clinical
trials and preclinical research are investigating the potential of
targeting autophagy and lncRNAs to enhance treatment outcomes for
patients with cancers resistant to radiation (159,160).
Not applicable.
The present study was supported by The Wings Scientific and
Technological Foundation of The First People's Hospital of Changde
City (grant no. Z2025ZC04) and Interdisciplinary Research Program
in Medicine and Engineering, The First Affiliated Hospital of
University of South China (grant no. IRP-M&E-2025-05)
Not applicable.
HL wrote and reviewed the drafts of the paper, while
ZH created the figures and tables, and revised the paper. Data
authentication is not applicable. All authors have read and
approved the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Schaue D and McBride WH: Opportunities and
challenges of radiotherapy for treating cancer. Nat Rev Clin Oncol.
12:527–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yerolatsite M, Torounidou N, Amylidi AL,
Rapti IC, Zarkavelis G, Kampletsas E and Voulgari PV: A Systematic
review of pneumonitis following treatment with immune checkpoint
inhibitors and radiotherapy. Biomedicines. 13:9462025. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tubiana M: Wilhelm conrad röntgen and the
discovery of X-rays. Bull Acad Natl Med. 180:97–108. 1996.(In
French). PubMed/NCBI
|
|
4
|
Babic RR, Stankovic Babic G, Babic SR and
Babic NR: 120 years since the discovery of X-Rays. Med Pregl.
69:323–330. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Klucznik KA, Ravkilde T, Skouboe S, Møller
DS, Hokland S, Keall P, Buus S, Bentzen L and Poulsen PR: Cone-beam
CT-based estimations of prostate motion and dose distortion during
radiotherapy. Phys Imaging Radiat Oncol. 35:1007982025. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lam MB, Landrum MB, McWilliams JM, Buzzee
B, Wright AA, Keating NL and Landon BE: Practice-level spending
variation for radiation treatment episodes among older adults with
cancer. JAMA Health Forum. 6:e2519522025. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hill RM, Fok M, Grundy G, Parsons JL and
Rocha S: The role of autophagy in hypoxia-induced radioresistance.
Radiother Oncol. 189:1099512023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang C, Song R, Yuan J, Hou G, Chu AL,
Huang Y, Xiao C, Chai T, Sun C and Liu Z: Exosome-Shuttled METTL14
From AML-Derived mesenchymal stem cells promotes the proliferation
and radioresistance in AML cells by stabilizing ROCK1 expression
via an m6A-IGF2BP3-dependent mechanism. Drug Dev Res.
86:e700252025. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo H, Huang MF, Xu A, Wang D, Gingold JA,
Tu J, Wang R, Huo Z, Chiang YT, Tsai KL, et al: Mutant p53 confers
chemoresistance by activating KMT5B-mediated DNA repair pathway in
nasopharyngeal carcinoma. Cancer Lett. 625:2177362025. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feng Y, Zhang L, Zhang Y, Xu Y, Zhou K,
Yang Z, Zhu W, Zhang Q, Cao J, Wang L and Jiao Y: NEDD4-mediated
endothelial-mesenchymal transition participates in
radiation-induced lung injury through the ATM signaling pathway.
Dose Response. 23:155932582513527262025. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qu Z, Shi L, Wang P, Zhao A, Zheng X and
Yin Q: Dual targeting of inflammatory and immune checkpoint
pathways to overcome radiotherapy resistance in esophageal squamous
cell carcinoma. J Inflamm Res. 18:9091–9106. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Porrazzo A, Cassandri M, D'Alessandro A,
Morciano P, Rota R, Marampon F and Cenci G: DNA repair in tumor
radioresistance: Insights from fruit flies genetics. Cell Oncol.
47:717–732. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mizushima N and Komatsu M: Autophagy:
Renovation of cells and tissues. Cell. 147:728–741. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cursaro I, Rossi S, Butini S, Gemma S,
Carullo G and Campiani G: A focus on natural autophagy modulators
as potential host-directed weapons against emerging and re-emerging
viruses. Med Res Rev. Jul 16–2025.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Parzych KR and Klionsky DJ: An overview of
autophagy: Morphology, mechanism, and regulation. Antioxid Redox
Signal. 20:460–473. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Debnath J, Gammoh N and Ryan KM: Autophagy
and autophagy-related pathways in cancer. Nat Rev Mol Cell Biol.
24:560–575. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vafadar A, Tajbakhsh A,
Hosseinpour-Soleimani F, Savardshtaki A and Hashempur MH:
Phytochemical-mediated efferocytosis and autophagy in inflammation
control. Cell Death Discov. 10:4932024. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feng X, Zhang H, Meng L, Song H, Zhou Q,
Qu C, Zhao P, Li Q, Zou C, Liu X and Zhang Z: Hypoxia-induced
acetylation of PAK1 enhances autophagy and promotes brain
tumorigenesis via phosphorylating ATG5. Autophagy. 17:723–742.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu S, Yao S, Yang H, Liu S and Wang Y:
Autophagy: Regulator of cell death. Cell Death Dis. 14:6482023.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Martini-Stoica H, Xu Y, Ballabio A and
Zheng H: The autophagy-lysosomal pathway in neurodegeneration: A
TFEB perspective. Trends Neurosci. 39:221–234. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin W, Zhou Q, Wang CQ, Zhu L, Bi C, Zhang
S, Wang X and Jin H: LncRNAs regulate metabolism in cancer. Int J
Biol Sci. 16:1194–1206. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu Y, Liu L, Wu H, Zheng Y, Zhan H and Li
L: LncRNA GAS5 regulated by FTO-mediated m6A demethylation promotes
autophagic cell death in NSCLC by targeting UPF1/BRD4 axis. Mol
Cell Biochem. 479:553–566. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou X, Tong Y, Yu C, Pu J, Zhu W, Zhou Y,
Wang Y, Xiong Y and Sun X: FAP positive cancer-associated
fibroblasts promote tumor progression and radioresistance in
esophageal squamous cell carcinoma by transferring exosomal lncRNA
AFAP1-AS1. Mol Carcinog. 63:1922–1937. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hjazi A, Jasim SA, Altalbawy FMA, Kaur H,
Hamzah HF, Kaur I, Deorari M, Kumar A, Elawady A and Fenjan MN:
Relationship between lncRNA MALAT1 and Chemo-radiotherapy
resistance of cancer cells: Uncovered truths. Cell Biochem Biophys.
82:1613–1627. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang H, Jin H, Lei R, He Z, He S, Chen J,
Saw PE, Qiu Z, Ren G and Nie Y: lncRNA-WAL promotes triple-negative
breast cancer aggression by inducing β-catenin nuclear
translocation. Mol Cancer Res. 22:1036–1050. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Russell RC and Guan KL: The multifaceted
role of autophagy in cancer. EMBO J. 41:e1100312022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bhol CS, Senapati PK, Kar RK, Chew G,
Mahapatra KK, Lee EHC, Kumar AP, Bhutia SK and Sethi G: Autophagy
paradox: Genetic and epigenetic control of autophagy in cancer
progression. Cancer Lett. 630:2179092025. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao L, Loveless J, Shay C and Teng Y:
Targeting ROS-Mediated crosstalk between autophagy and apoptosis in
cancer. Adv Exp Med Biol. 1260:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carretero-Fernández M, Cabrera-Serrano AJ,
Sánchez-Maldonado JM, Ruiz-Durán L, Jiménez-Romera F,
García-Verdejo FJ, González-Olmedo C, Cardús A, Díaz-Beltrán L,
Gutiérrez-Bautista JF, et al: Autophagy and oxidative stress in
solid tumors: Mechanisms and therapeutic opportunities. Crit Rev
Oncol Hematol. 212:1048202025. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hama Y, Ogasawara Y and Noda NN: Autophagy
and cancer: Basic mechanisms and inhibitor development. Cancer Sci.
114:2699–2708. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Z, Liu F, Li F, Zeng G, Wen X, Ding J
and Zhou J: DHX9-mediated epigenetic silencing of BECN1 contributes
to impaired autophagy and tumor progression in breast cancer via
recruitment of HDAC5. Cell Death Dis. 16:5242025. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao Z, Tian K, Ran Y, Zhou H, Zhou L, Ding
Y and Tang X: Beclin-1: A therapeutic target at the intersection of
autophagy, immunotherapy, and cancer treatment. Front Immunol.
15:15064262024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li X, Yang KB, Chen W, Mai J, Wu XQ, Sun
T, Wu RY, Jiao L, Li DD, Ji J, et al: CUL3 (cullin 3)-mediated
ubiquitination and degradation of BECN1 (beclin 1) inhibit
autophagy and promote tumor progression. Autophagy. 17:4323–4340.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen X, Sun Y, Wang B and Wang H:
Prognostic significance of autophagy-related genes BECN1 and LC3 in
ovarian cancer: A meta-analysis. J Int Med Res.
48:3000605209682992020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu C, Xu P, Chen D, Fan X, Xu Y, Li M,
Yang X and Wang C: Roles of autophagy-related genes Beclin-1 and
LC3 in the development and progression of prostate cancer and
benign prostatic hyperplasia. Biomed Rep. 1:855–860. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yao Q, Chen J, Lv Y, Wang T, Zhang J, Fan
J and Wang L: The significance of expression of autophagy-related
gene Beclin, Bcl-2, and Bax in breast cancer tissues. Tumour Biol.
32:1163–1171. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu CL, Liu JF, Liu Y, Wang YX, Fu KF, Yu
XJ, Pu Q, Chen XX and Zhou LJ: BECN1 inhibition enhances
paclitaxel-mediated cytotoxicity in breast cancer in vitro and in
vivo. Int J Mol Med. 43:1866–1878. 2019.PubMed/NCBI
|
|
38
|
Tran S, Juliani J, Harris TJ, Evangelista
M, Ratcliffe J, Ellis SL, Baloyan D, Reehorst CM, Nightingale R,
Luk IY, et al: BECN1 is essential for intestinal homeostasis
involving autophagy-independent mechanisms through its function in
endocytic trafficking. Commun Biol. 7:2092024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ishaq M, Ojha R, Sharma AP and Singh SK:
Autophagy in cancer: Recent advances and future directions. Semin
Cancer Biol Semin Cancer Biol. 66:171–181. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Senapati PK, Mahapatra KK, Singh A and
Bhutia SK: mTOR inhibitors in targeting autophagy and
autophagy-associated signaling for cancer cell death and therapy.
Biochim Biophys Acta Rev Cancer. 1880:1893422025. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ferro F, Servais S, Besson P, Roger S,
Dumas JF and Brisson L: Autophagy and mitophagy in cancer metabolic
remodelling. Semin Cell Dev Biol. 98:129–138. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang M, Liu S, Chua MS, Li H, Luo D, Wang
S, Zhang S, Han B and Sun C: SOCS5 inhibition induces autophagy to
impair metastasis in hepatocellular carcinoma cells via the
PI3K/Akt/mTOR pathway. Cell Death Dis. 10:6122019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Maycotte P, Jones KL, Goodall ML, Thorburn
J and Thorburn A: Autophagy supports breast cancer stem cell
maintenance by regulating IL6 secretion. Mol Cancer Res.
13:651–658. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Song J, Yang P, Chen C, Ding W, Tillement
O, Bai H and Zhang S: Targeting epigenetic regulators as a
promising avenue to overcome cancer therapy resistance. Signal
Transduct Target Ther. 10:2192025. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
ALKhemeiri N, Eljack S and Saber-Ayad MM:
Perspectives of targeting autophagy as an adjuvant to
anti-PD-1/PD-L1 therapy for colorectal cancer treatment. Cells.
14:7452025. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang W, Cheng B, Chen P, Sun X, Wen Z and
Cheng Y: BTN3A1 promotes tumor progression and radiation resistance
in esophageal squamous cell carcinoma by regulating ULK1-mediated
autophagy. Cell Death Dis. 13:9842022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu Q, Zhang H, Liu H, Han Y, Qiu W and Li
Z: Inhibiting autophagy flux and DNA repair of tumor cells to boost
radiotherapy of orthotopic glioblastoma. Biomaterials.
280:1212872022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chaurasia M, Bhatt AN, Das A, Dwarakanath
BS and Sharma K: Radiation-induced autophagy: Mechanisms and
consequences. Free Radic Res. 50:273–290. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yue T, Zheng D, Yang J, He J and Hou J:
Potential value and cardiovascular risks of programmed cell death
in cancer treatment. Front Pharmacol. 16:16159742025. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu S, Wang L, Zhu L, Zhao T, Han P, Yan
F, Wang X, Li C, Wang Z and Yang BF: Mechanism and regulation of
mitophagy in liver diseases: A review. Front Cell Dev Biol.
13:16149402025. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yun CW and Lee SH: The roles of autophagy
in cancer. Int J Mol Sci. 19:34662018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Singh SS, Vats S, Chia AY, Tan TZ, Deng S,
Ong MS, Arfuso F, Yap CT, Goh BC, Sethi G, et al: Dual role of
autophagy in hallmarks of cancer. Oncogene. 37:1142–1158. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Clark A, Villarreal MR, Huang SB,
Jayamohan S, Rivas P, Hussain SS, Ybarra M, Osmulski P, Gaczynska
ME, Shim EY, et al: Targeting S6K/NFκB/SQSTM1/Polθ signaling to
suppress radiation resistance in prostate cancer. Cancer Lett.
597:2170632024. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sezen Us A, Dagsuyu E, Us H, Cöremen M,
Karabulut Bulan O and Yanardag R: Apocynin may alleviate side
effects of autophagy-blocked radiotherapy through antioxidant
effects. Biotech Histochem. Jun 30–2025.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ma S, Fu X, Liu L, Liu Y, Feng H, Jiang H,
Liu X, Liu R, Liang Z, Li M, et al: Iron-dependent autophagic cell
death induced by radiation in MDA-MB-231 breast cancer cells. Front
Cell Dev Biol. 9:7238012021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mukha A, Kahya U, Linge A, Chen O, Löck S,
Lukiyanchuk V, Richter S, Alves TC, Peitzsch M, Telychko V, et al:
GLS-driven glutamine catabolism contributes to prostate cancer
radiosensitivity by regulating the redox state, stemness and
ATG5-mediated autophagy. Theranostics. 11:7844–7868. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen N, Zhang R, Konishi T and Wang J:
Upregulation of NRF2 through autophagy/ERK 1/2 ameliorates ionizing
radiation induced cell death of human osteosarcoma U-2 OS. Mutat
Res Genet Toxicol Environ Mutagen. 813:10–17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wu C, Yang L, Qi X, Wang T, Li M and Xu K:
Inhibition of long non-coding RNA HOTAIR enhances radiosensitivity
via regulating autophagy in pancreatic cancer. Cancer Manag Res.
10:5261–5271. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cui L, Song Z, Liang B, Jia L, Ma S and
Liu X: Radiation induces autophagic cell death via the p53/DRAM
signaling pathway in breast cancer cells. Oncol Rep. 35:3639–3647.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lei G, Zhang Y, Koppula P, Liu X, Zhang J,
Lin SH, Ajani JA, Xiao Q, Liao Z, Wang H and Gan B: The role of
ferroptosis in ionizing radiation-induced cell death and tumor
suppression. Cell Res. 30:146–162. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chen X, Kang R, Kroemer G and Tang D:
Broadening horizons: The role of ferroptosis in cancer. Nat Rev
Clin Oncol. 18:280–296. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang Y, Liu F, Fang C, Xu L, Chen L, Xu Z,
Chen J, Peng W, Fu B and Li Y: Combination of rapamycin and SAHA
enhanced radiosensitization by inducing autophagy and acetylation
in NSCLC. Aging (Albany NY). 13:18223–18237. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen J, Luo H, Wu X, Dong M, Wang D, Ou Y,
Wang Y, Sun S, Liu Z, Yang Z, et al: Inhibition of Phosphoglycerate
Kinase 1 Enhances Radiosensitivity of Esophageal Squamous Cell
Carcinoma to X-rays and Carbon Ion Irradiation. Front Biosci
(Landmark Ed). 30:364302025. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Mukha A, Kahya U and Dubrovska A:
Targeting glutamine metabolism and autophagy: The combination for
prostate cancer radiosensitization. Autophagy. 17:3879–3881. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ma X, Mao G, Chang R, Wang F, Zhang X and
Kong Z: Down-regulation of autophagy-associated protein increased
acquired radio-resistance bladder cancer cells sensitivity to
taxol. Int J Radiat Biol. 97:507–516. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mo N, Lu YK, Xie WM, Liu Y, Zhou WX, Wang
HX, Nong L, Jia YX, Tan AH, Chen Y, et al: Inhibition of autophagy
enhances the radiosensitivity of nasopharyngeal carcinoma by
reducing Rad51 expression. Oncol Rep. 32:1905–1912. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Prise KM and O'Sullivan JM:
Radiation-induced bystander signalling in cancer therapy. Nat Rev
Cancer. 9:351–360. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Jia H, Wei J, Zheng W and Li Z: The dual
role of autophagy in cancer stem cells: Implications for tumor
progression and therapy resistance. J Transl Med. 23:5832025.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhou S, Wang X, Ding J, Yang H and Xie Y:
Increased ATG5 expression predicts poor prognosis and promotes EMT
in cervical carcinoma. Front Cell Dev Biol. 9:7571842021.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Pustovalova M, Alhaddad L, Blokhina T,
Smetanina N, Chigasova A, Chuprov-Netochin R, Eremin P,
Gilmutdinova I, Osipov AN and Leonov S: The CD44high subpopulation
of multifraction irradiation-surviving NSCLC cells exhibits partial
EMT-program activation and DNA damage response depending on their
p53 Status. Int J Mol Sci. 22:23692021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yazal T, Bailleul J, Ruan Y, Sung D, Chu
FI, Palomera D, Dao A, Sehgal A, Gurunathan V, Aryan L, et al:
Radiosensitizing pancreatic cancer via effective autophagy
inhibition. Mol Cancer Ther. 21:79–88. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Gajate C, Gayet O, Fraunhoffer NA, Iovanna
J, Dusetti N and Mollinedo F: Induction of apoptosis in human
pancreatic cancer stem cells by the endoplasmic reticulum-targeted
alkylphospholipid analog edelfosine and potentiation by autophagy
inhibition. Cancers (Basel). 13:61242021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Fischer P, Schmid M, Ohradanova-Repic A,
Schneeweiss R, Hadatsch J, Grünert O, Benedum J, Röhrer A,
Staudinger F, Schatzlmaier P, et al: Molecular features of TNBC
govern heterogeneity in the response to radiation and autophagy
inhibition. Cell Death Dis. 16:5402025. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Xiao S, Wang Y, Pan S, Mu M, Chen B, Li H,
Feng C, Fan R, Yu W, Han B, et al: Bismuth-functionalized
probiotics for enhanced antitumor radiotherapy and immune
activation. J Mater Chem B. Jul 14–2025.(Epub ahead of print).
View Article : Google Scholar
|
|
76
|
Yang P, Li J, Zhang T, Ren Y, Zhang Q, Liu
R, Li H, Hua J, Wang WA, Wang J and Zhou H: Ionizing
radiation-induced mitophagy promotes ferroptosis by increasing
intracellular free fatty acids. Cell Death Differ. 30:2432–2445.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Liu C, Liu X, Li Z, Wei Y, Liu B, Zhu P,
Liu Y and Zhao R: VPS37A activates the autophagy-lysosomal pathway
for TNFR1 degradation and induces NF-κB-Regulated cell death under
metabolic stress in colorectal cancer. Oncol Res. 33:2085–2105.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Azmat MA, Zaheer M, Shaban M, Arshad S,
Hasan M, Ashraf A, Naeem M, Ahmad A and Munawar N: Autophagy: A new
avenue and biochemical mechanisms to mitigate the climate change.
Scientifica (Cairo). 2024:99083232024. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Choi Y, Bowman JW and Jung JU: Autophagy
during viral infection-a double-edged sword. Nat Rev Microbiol.
16:341–354. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Daniel P, Sabri S, Chaddad A, Meehan B,
Jean-Claude B, Rak J and Abdulkarim BS: Temozolomide induced
hypermutation in glioma: Evolutionary mechanisms and therapeutic
opportunities. Front Oncol. 9:412019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Liu EK, Sulman EP, Wen PY and Kurz SC:
Novel therapies for glioblastoma. Curr Neurol Neurosci Rep.
20:192020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Tsai CY, Ko HJ, Huang CF, Lin CY, Chiou
SJ, Su YF, Lieu AS, Loh JK, Kwan AL, Chuang TH and Hong YR:
Ionizing radiation induces resistant glioblastoma stem-like cells
by promoting autophagy via the Wnt/β-catenin pathway. Life (Basel).
11:4512021.PubMed/NCBI
|
|
83
|
Wang Y, Fu Y, Lu Y, Chen S, Zhang J, Liu B
and Yuan Y: Unravelling the complexity of lncRNAs in autophagy to
improve potential cancer therapy. Biochim Biophys Acta Rev Cancer.
1878:1889322023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zheng J, Mat Ludin AF, Rajab NF, Shaolong
L and Jufri NF: The roles of lncMALAT1 in coronary artery disease
regulation and therapeutic perspective: A systematic review.
iScience. 28:1129452025. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang X, Shi Y, Wang C and Zhang K: LncRNA
MALAT1 knockdown inhibits apoptosis of mouse hippocampus neuron
cells with high glucose by Silencing autophagy. BMC Endocr Disord.
25:1732025. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Yang L, Wang H, Shen Q, Feng L and Jin H:
Long non-coding RNAs involved in autophagy regulation. Cell Death
Dis. 8:e30732017. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Xia N, Zhang P, Yang L, Yin X, Wu SQ, Yao
Y, Shang MY and Weng L: The lncRNA MIR503HG/miR-16-5p/FOSL1 pathway
mediates autophagy to promote esophageal epithelial cells
proliferation and EMT in esophageal restenosis. Arch Biochem
Biophys. 772:1105362025. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhu X, Sun Y, Yu Q, Wang X, Wang Y and
Zhao Y: Exosomal lncRNA GAS5 promotes M1 macrophage polarization in
allergic rhinitis via restraining mTORC1/ULK1/ATG13-mediated
autophagy and subsequently activating NF-кB signaling. Int
Immunopharmacol. 121:1104502023. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
DeBerardinis RJ, Lum JJ, Hatzivassiliou G
and Thompson CB: The biology of cancer: Metabolic reprogramming
fuels cell growth and proliferation. Cell Metab. 7:11–20. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhou X, Song Y, Wang Z, Fu L, Xu L, Feng
X, Zhang Z and Yuan K: Dietary sugar intervention: A promising
approach for cancer therapy. Biochim Biophys Acta Rev Cancer.
1880:1894022025. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Li C, Wang K, Zhao L, Liu J, Jin Y, Zhang
C, Xu M, Wang M, Kuang Y, Liu J, et al: LINC00622 transcriptionally
promotes RRAGD to repress mTORC1-modulated autophagic cell death by
associating with BTF3 in cutaneous melanoma. Cell Death Dis.
16:5152025. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wu Q, Ma J, Wei J, Meng W, Wang Y and Shi
M: lncRNA SNHG11 promotes gastric cancer progression by activating
the Wnt/β-Catenin pathway and oncogenic autophagy. Mol Ther.
29:1258–1278. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Chen F, Zhong Z, Tan HY, Guo W, Zhang C,
Cheng CS, Wang N, Ren J and Feng Y: Suppression of lncRNA MALAT1 by
betulinic acid inhibits hepatocellular carcinoma progression by
targeting IAPs via miR-22-3p. Clin Transl Med. 10:e1902020.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Sheng JQ, Wang MR, Fang D, Liu L, Huang
WJ, Tian DA, He XX and Li PY: LncRNA NBR2 inhibits tumorigenesis by
regulating autophagy in hepatocellular carcinoma. Biomed
Pharmacother. 133:1110232021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Wang Z and Jin J: LncRNA SLCO4A1-AS1
promotes colorectal cancer cell proliferation by enhancing
autophagy via miR-508-3p/PARD3 axis. Aging (Albany NY).
11:4876–4889. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wang Y, Li Z, Xu S, Li W, Chen M, Jiang M
and Fan X: LncRNA FIRRE functions as a tumor promoter by
interaction with PTBP1 to stabilize BECN1 mRNA and facilitate
autophagy. Cell Death Dis. 13:982022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Lu X, Chen L, Li Y, Huang R, Meng X and
Sun F: Long non-coding RNA LINC01207 promotes cell proliferation
and migration but suppresses apoptosis and autophagy in oral
squamous cell carcinoma by the microRNA-1301-3p/lactate
dehydrogenase isoform A axis. Bioengineered. 12:7780–7793. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Jiang N, Zhang X, Gu X, Li X and Shang L:
Progress in understanding the role of lncRNA in programmed cell
death. Cell Death Discov. 7:302021. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lu M, Qin X, Zhou Y, Li G, Liu Z, Yue H
and Geng X: LncRNA HOTAIR suppresses cell apoptosis, autophagy and
induces cell proliferation in cholangiocarcinoma by modulating the
miR-204-5p/HMGB1 axis. Biomed Pharmacother. 130:1105662020.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Murakami K, Yokoi Y, Hirane N, Otaki M and
Nishimura SI: Nanosomal irinotecan targeting pancreatic cancer cell
surface neuraminidase-1 sialidase. Adv Healthc Mater. Jul
22–2025.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wu H, Li A, Zheng Q, Gu J and Zhou W:
LncRNA LZTS1-AS1 induces proliferation, metastasis and inhibits
autophagy of pancreatic cancer cells through the miR-532/TWIST1
signaling pathway. Cancer Cell Int. 23:1302023. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Lv D, Xiang Y, Yang Q, Yao J and Dong Q:
Long Non-Coding RNA TUG1 promotes cell proliferation and inhibits
cell apoptosis, autophagy in clear cell renal cell carcinoma via
MiR-31-5p/FLOT1 axis. Onco Targets Ther. 13:5857–5868. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Shao Q, Wang Q and Wang J: LncRNA SCAMP1
regulates ZEB1/JUN and autophagy to promote pediatric renal cell
carcinoma under oxidative stress via miR-429. Biomed Pharmacother.
120:1094602019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhang Z, Jia JP, Zhang YJ, Liu G, Zhou F
and Zhang BC: Long Noncoding RNA ADAMTS9-AS2 inhibits the
proliferation, migration, and invasion in bladder tumor cells. Onco
Targets Ther. 13:7089–7100. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Li Q, Chen K, Dong R and Lu H: LncRNA
CASC2 inhibits autophagy and promotes apoptosis in non-small cell
lung cancer cells via regulating the miR-214/TRIM16 axis. RSC Adv.
8:40846–40855. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Peng D, Li W, Zhang B and Liu X:
Overexpression of lncRNA SLC26A4-AS1 inhibits papillary thyroid
carcinoma progression through recruiting ETS1 to promote
ITPR1-mediated autophagy. J Cell Mol Med. 25:8148–8158. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Qin Y, Sun W, Wang Z, Dong W, He L, Zhang
T, Lv C and Zhang H: RBM47/SNHG5/FOXO3 axis activates autophagy and
inhibits cell proliferation in papillary thyroid carcinoma. Cell
Death Dis. 13:2702022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Wang Y, Liu Z, Xu Z, Shao W, Hu D, Zhong H
and Zhang J: Introduction of long non-coding RNAs to regulate
autophagy-associated therapy resistance in cancer. Mol Biol Rep.
49:10761–10773. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Shaw A and Gullerova M: Home and away: The
role of non-coding RNA in intracellular and intercellular DNA
damage response. Genes (Basel). 12:14752021. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Wang S, Qiao C, Fang R, Yang S, Zhao G,
Liu S and Li P: LncRNA CASC19: A novel oncogene involved in human
cancer. Clin Transl Oncol. 25:2841–2851. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Wu Y, Mou J, Zhou G and Yuan C: CASC19: An
oncogenic long non-coding RNA in different cancers. Curr Pharm Des.
30:1157–1166. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Liu H, Zheng W, Chen Q, Zhou Y, Pan Y,
Zhang J, Bai Y and Shao C: lncRNA CASC19 contributes to
radioresistance of nasopharyngeal carcinoma by promoting autophagy
via AMPK-mTOR pathway. Int J Mol Sci. 22:14072021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Liu H, Chen Q, Zheng W, Zhou Y, Bai Y, Pan
Y, Zhang J and Shao C: LncRNA CASC19 enhances the radioresistance
of nasopharyngeal carcinoma by regulating the miR-340-3p/FKBP5
axis. Int J Mol Sci. 24:30472023. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Wang WJ, Guo CA, Li R, Xu ZP, Yu JP, Ye Y,
Zhao J, Wang J, Wang WA, Zhang A, et al: Long non-coding RNA CASC19
is associated with the progression and prognosis of advanced
gastric cancer. Aging (Albany NY). 11:5829–5847. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Hou Y, Tang Y, Ma C, Yu J and Jia Y:
Overexpression of CASC19 contributes to tumor progression and
predicts poor prognosis after radical resection in hepatocellular
carcinoma. Dig Liver Dis. 55:799–806. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Teng S, Ge J, Yang Y, Cui Z, Min L, Li W,
Yang G, Liu K and Wu J: M1 macrophages deliver CASC19 via exosomes
to inhibit the proliferation and migration of colon cancer cells.
Med Oncol. 41:2862024. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Chiu HS, Somvanshi S, Patel E, Chen TW,
Singh VP, Zorman B, Patil SL, Pan Y, Chatterjee SS; Cancer Genome
Atlas Research Network;, ; et al: Pan-cancer analysis of lncRNA
regulation supports their targeting of cancer genes in each tumor
context. Cell Rep. 23:297–312.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Huang Z, Li X, Wang X, Wu J, Gong Z, Kõks
S and Wang M: Nuclear paraspeckle assembly transcript 1 promotes
photophobia behavior in mice via miR-196a-5p/Trpm3 coupling. J
Headache Pain. 26:1182025. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Sakaguchi H, Tsuchiya H, Kitagawa Y,
Tanino T, Yoshida K, Uchida N and Shiota G: NEAT1 Confers
radioresistance to hepatocellular carcinoma cells by inducing
autophagy through GABARAP. Int J Mol Sci. 23:7112022. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Mahmoud HS, Eldesoky NA, Shaker OG and
El-Husseiny AA: The diagnostic value of LncRNA NEAT1 targeting
miR-129-5p in pancreatic cancer patients. Sci Rep. 15:276382025.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Dong P, Xiong Y, Yue J, Xu D, Ihira K,
Konno Y, Kobayashi N, Todo Y and Watari H: Long noncoding RNA NEAT1
drives aggressive endometrial cancer progression via
miR-361-regulated networks involving STAT3 and tumor
microenvironment-related genes. J Exp Clin Cancer Res. 38:2952019.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Almujri SS and Almalki WH: The paradox of
autophagy in cancer: NEAT1′s role in tumorigenesis and therapeutic
resistance. Pathol Res Pract. 262:1555232024. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Wen J, Zheng W, Zeng L, Wang B, Chen D,
Chen Y, Lu X, Shao C, Chen J and Fan M: LTF Induces Radioresistance
by Promoting Autophagy and Forms an AMPK/SP2/NEAT1/miR-214-5p
feedback loop in lung squamous cell carcinoma. Int J Biol Sci.
19:1509–1527. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
He H: Targeting TP53TG1: A promising
prognostic biomarker and therapeutic target for personalized cancer
therapy. Discov Oncol. 16:12712025. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Li J, Liu R, Hu H, Huang Y, Shi Y, Li H,
Chen H, Cai M, Wang N, Yan T, et al: Methionine deprivation
inhibits glioma proliferation and EMT via the
TP53TG1/miR-96-5p/STK17B ceRNA pathway. NPJ Precis Oncol.
8:2702024. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Liao D, Liu X, Yuan X, Feng P, Ouyang Z,
Liu Y and Li C: Long non-coding RNA tumor protein 53 target gene 1
promotes cervical cancer development via regulating microRNA-33a-5p
to target forkhead box K2. Cell Cycle. 21:572–584. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Ji W, Wang W and Wei Y: TP53TG1/STAT axis
is involved in the development of colon cancer through affecting
PD-L1 expression and immune escape mechanism of tumor cells. Am J
Cancer Res. 13:5218–5235. 2023.PubMed/NCBI
|
|
128
|
Cheng Y, Huang N, Yin Q, Cheng C, Chen D,
Gong C, Xiong H, Zhao J, Wang J, Li X, et al: LncRNA TP53TG1 plays
an anti-oncogenic role in cervical cancer by synthetically
regulating transcriptome profile in HeLa cells. Front Genet.
13:9810302022. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Luan F, Chen W, Chen M, Yan J, Chen H, Yu
H, Liu T and Mo L: An autophagy-related long non-coding RNA
signature for glioma. FEBS Open Bio. 9:653–667. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Gao W, Qiao M and Luo K: Long noncoding
RNA TP53TG1 contributes to radioresistance of glioma cells Via
miR-524-5p/RAB5A Axis. Cancer Biother Radiopharm. 36:600–612.
2021.PubMed/NCBI
|
|
131
|
Wang C, Li Y, Yan S, Wang H, Shao X, Xiao
M, Yang B, Qin G, Kong R, Chen R and Zhang N: Interactome analysis
reveals that lncRNA HULC promotes aerobic glycolysis through LDHA
and PKM2. Nat Commun. 11:31622020. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Gandhy SU, Imanirad P, Jin UH, Nair V,
Hedrick E, Cheng Y, Corton JC, Kim K and Safe S: Specificity
protein (Sp) transcription factors and metformin regulate
expression of the long non-coding RNA HULC. Oncotarget.
6:26359–26372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Chen X, Lin J, Liu Y, Peng J, Cao Y, Su Z,
Wang T, Cheng J and Hu D: AlncRNA HULC as an effective biomarker
for surveillance of the outcome of cancer: A meta-analysis. PLoS
One. 12:e01712102017. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Li YP, Liu Y, Xiao LM, Chen LK, Tao EX,
Zeng EM and Xu CH: Induction of cancer cell stemness in glioma
through glycolysis and the long noncoding RNA HULC-activated
FOXM1/AGR2/HIF-1α axis. Lab Invest. 102:691–701. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Li S, Chang M, Tong L, Wang Y, Wang M and
Wang F: Screening potential lncRNA biomarkers for breast cancer and
colorectal cancer combining random walk and logistic matrix
factorization. Front Genet. 13:10236152023. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Chen C, Wang K, Wang Q and Wang X: LncRNA
HULC mediates radioresistance via autophagy in prostate cancer
cells. Braz J Med Biol Res. 51:e70802018. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Hacisuleyman E, Goff LA, Trapnell C,
Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG,
Sauvageau M, Kelley DR, et al: Topological organization of
multichromosomal regions by the long intergenic noncoding RNA
Firre. Nat Struct Mol Biol. 21:198–206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Meng X, Fang E, Zhao X and Feng J:
Identification of prognostic long noncoding RNAs associated with
spontaneous regression of neuroblastoma. Cancer Med. 9:3800–3815.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Shi X, Cui Z, Liu X, Wu S, Wu Y, Fang F
and Zhao H: LncRNA FIRRE is activated by MYC and promotes the
development of diffuse large B-cell lymphoma via Wnt/β-catenin
signaling pathway. Biochem Biophys Res Commun. 510:594–600. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Wang Z, Zhang J, Liu Y, Zhao R, Zhou X and
Wang H: An integrated autophagy-related long noncoding RNA
signature as a prognostic biomarker for human endometrial cancer: A
bioinformatics-based approach. Biomed Res Int. 2020:57174982020.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Cai J, Wang R, Chen Y, Zhang C, Fu L and
Fan C: LncRNA FIRRE regulated endometrial cancer radiotherapy
sensitivity via the miR-199b-5p/SIRT1/BECN1 axis-mediated
autophagy. Genomics. 116:1107502024. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Cory S, Graham M, Webb E, Corcoran L and
Adams JM: Variant (6;15) translocations in murine plasmacytomas
involve a chromosome 15 locus at least 72 kb from the c-myc
oncogene. EMBO J. 4:675–681. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Tseng YY, Moriarity BS, Gong W, Akiyama R,
Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell
TC, et al: PVT1 dependence in cancer with MYC copy-number increase.
Nature. 512:82–86. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Boloix A, Masanas M, Jiménez C, Antonelli
R, Soriano A, Roma J, Sánchez de Toledo J, Gallego S and Segura MF:
Long Non-coding RNA PVT1 as a prognostic and therapeutic target in
pediatric cancer. Front Oncol. 9:11732019. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Ogunwobi OO and Segura MF: Editorial: PVT1
in cancer. Front Oncol. 10:5887862020. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Lai SW, Chen MY, Bamodu OA, Hsieh MS,
Huang TY, Yeh CT, Lee WH and Cherng YG: Exosomal lncRNA PVT1/VEGFA
axis promotes colon cancer metastasis and stemness by
downregulation of tumor suppressor miR-152-3p. Oxid Med Cell
Longev. 2021:99598072021. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Zhu Y, Wu F, Gui W, Zhang N, Matro E, Zhu
L, Eserberg DT and Lin X: A positive feedback regulatory loop
involving the lncRNA PVT1 and HIF-1α in pancreatic cancer. J Mol
Cell Biol. 13:676–689. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Yao W, Li S, Liu R, Jiang M, Gao L, Lu Y,
Liang X and Zhang H: Long non-coding RNA PVT1: A promising
chemotherapy and radiotherapy sensitizer. Front Oncol.
12:9592082022. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Tan YT, Lin JF, Li T, Li JJ, Xu RH and Ju
HQ: LncRNA-mediated posttranslational modifications and
reprogramming of energy metabolism in cancer. Cancer Commun (Lond).
41:109–120. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zhou Y, Shao Y, Hu W, Zhang J, Shi Y, Kong
X and Jiang J: A novel long noncoding RNA SP100-AS1 induces
radioresistance of colorectal cancer via sponging miR-622 and
stabilizing ATG3. Cell Death Differ. 30:111–124. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Guo Z, Zhang X, Zhu H, Zhong N, Luo X,
Zhang Y, Tu F, Zhong J, Wang X, He J and Huang L: TELO2 induced
progression of colorectal cancer by binding with RICTOR through
mTORC2. Oncol Rep. 45:523–534. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Chang S, Zhang M, Liu C, Li M, Lou Y and
Tan H: Redox mechanism of glycerophospholipids and relevant
targeted therapy in ferroptosis. Cell Death Discov. 11:3582025.
View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Zhang L, Yu S, Wang C, Jia C, Lu Z and
Chen J: Establishment of a non-coding RNAomics screening platform
for the regulation of KRAS in pancreatic cancer by RNA sequencing.
Int J Oncol. 53:2659–2670. 2018.PubMed/NCBI
|
|
154
|
Muthoni A, Anyango A and V Wanjala H:
Mucilage-based nanocarriers for targeted cancer therapy-design,
functionalization, and therapeutic potential. Drug Dev Ind Pharm.
Aug 4–2025.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Wu T and Du Y: LncRNAs: From basic
research to medical application. Int J Biol Sci. 13:295–307. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Zhang L, Chen H, Yang Y, Zhao L, Xie H, Li
P, Lv X, He L, Liu N and Liu B: Targeting LINC02320 prevents
colorectal cancer growth via GRB7-dependent inhibition of MAPK
signaling pathway. Cell Mol Biol Lett. 30:862025. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Taylor MA, Das BC and Ray SK: Targeting
autophagy for combating chemoresistance and radioresistance in
glioblastoma. Apoptosis. 23:563–575. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Han F, Chen S, Zhang K, Zhang K, Wang M
and Wang P: Targeting the HNRNPA2B1/HDGF/PTN axis to overcome
radioresistance in non-small cell lung cancer. Antioxid Redox
Signal. 43:189–214. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Liu Y, Chen X, Chen X, Liu J, Gu H, Fan R
and Ge H: Long non-coding RNA HOTAIR knockdown enhances
radiosensitivity through regulating microRNA-93/ATG12 axis in
colorectal cancer. Cell Death Dis. 11:1752020. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Yan Y, Chen X, Wang X, Zhao Z, Hu W, Zeng
S, Wei J, Yang X, Qian L, Zhou S, et al: The effects and the
mechanisms of autophagy on the cancer-associated fibroblasts in
cancer. J Exp Clin Cancer Res. 38:1712019. View Article : Google Scholar : PubMed/NCBI
|