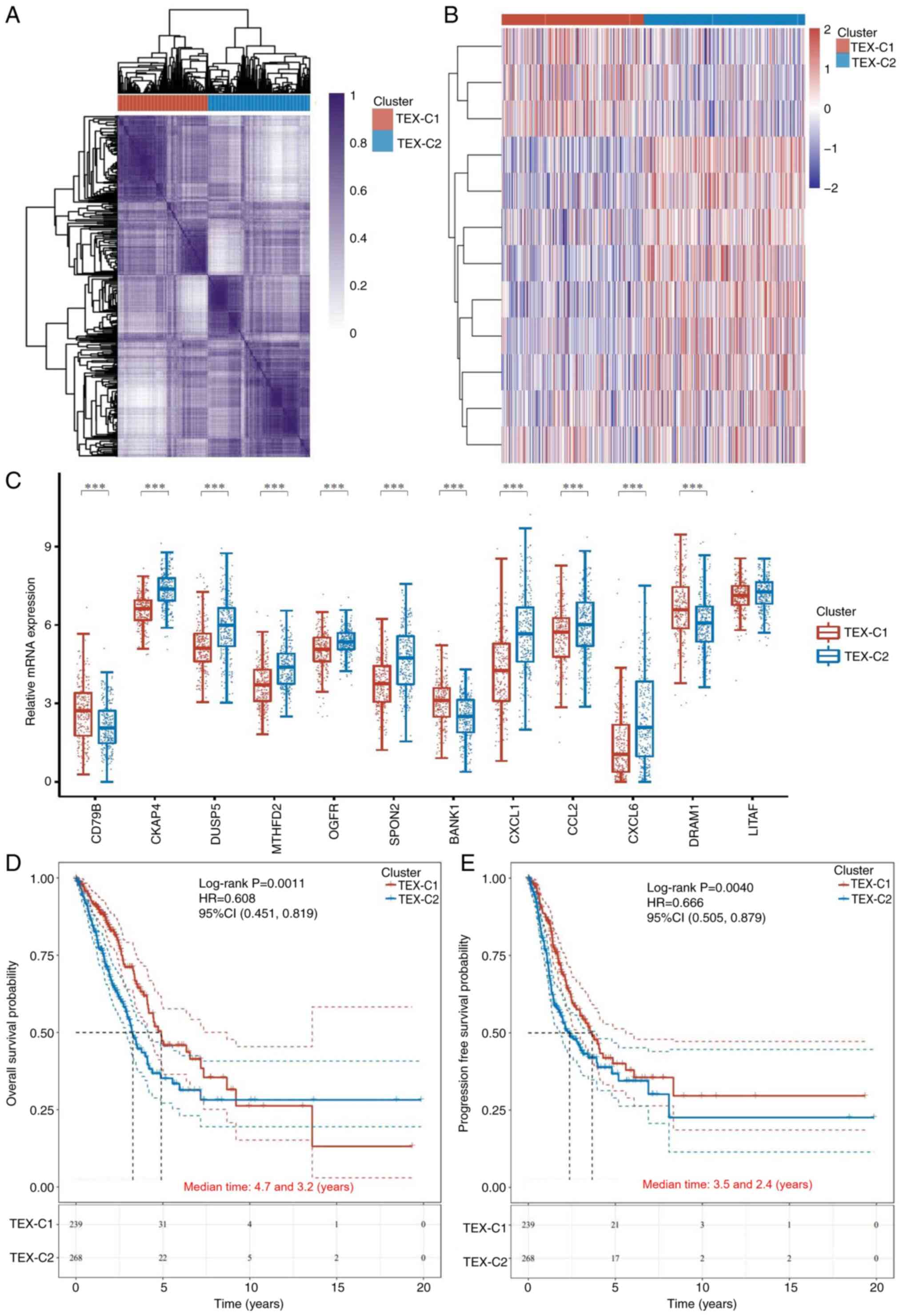
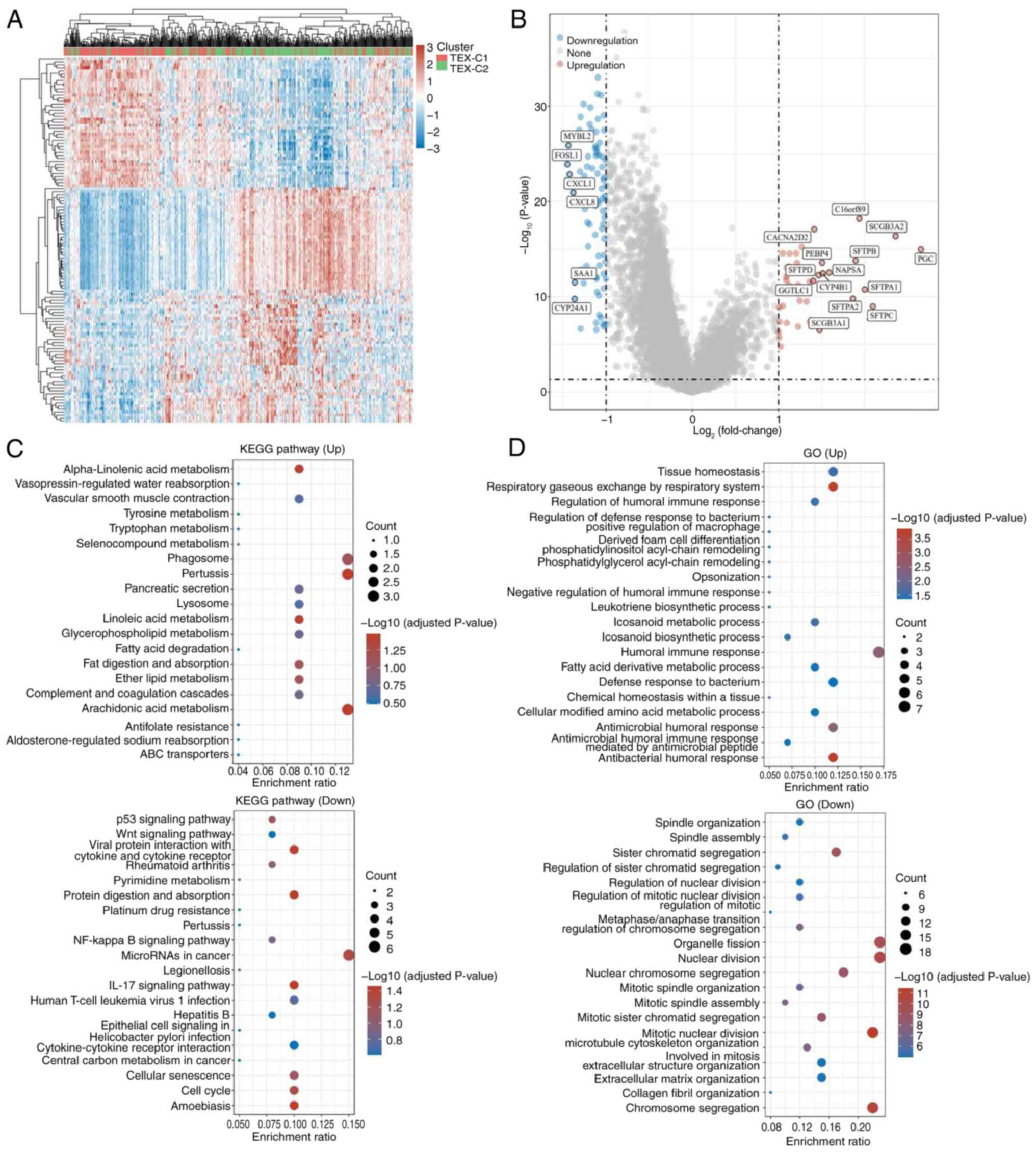
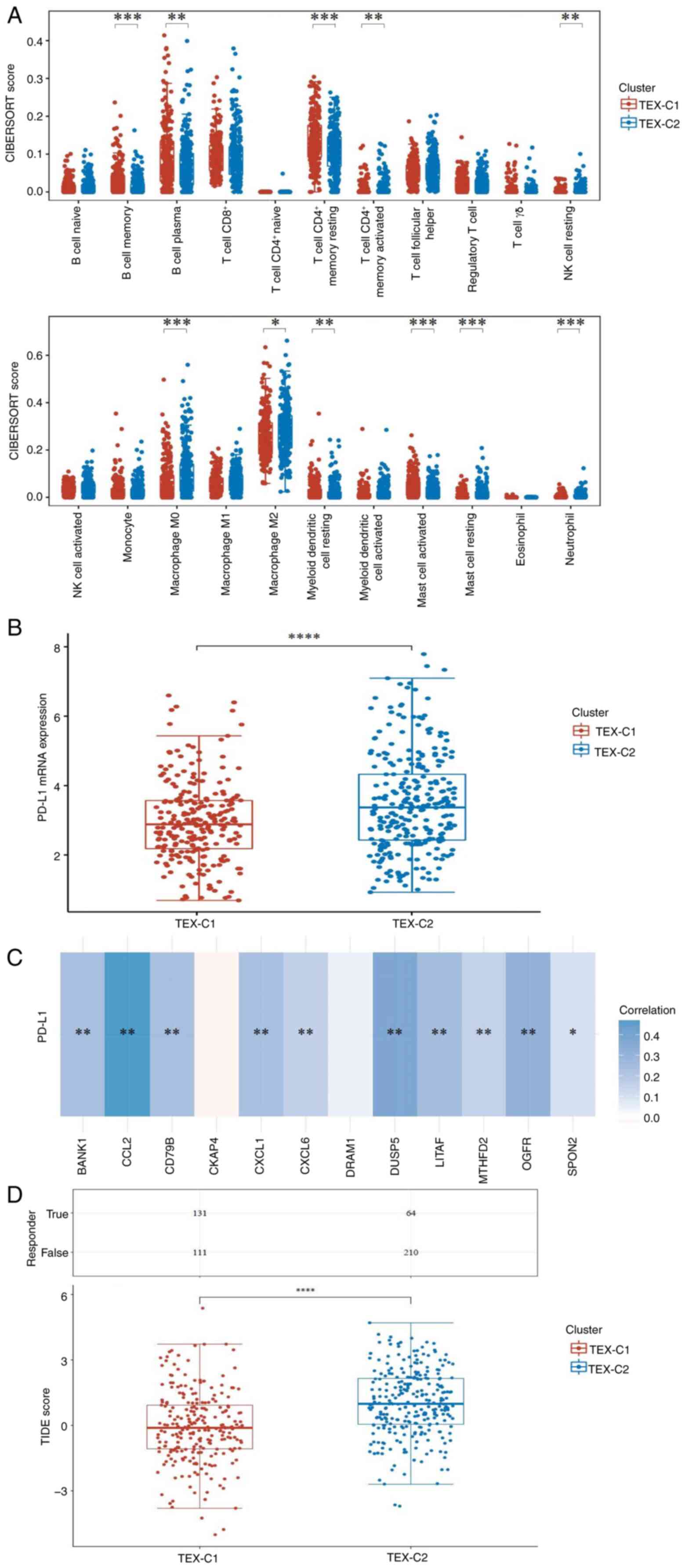
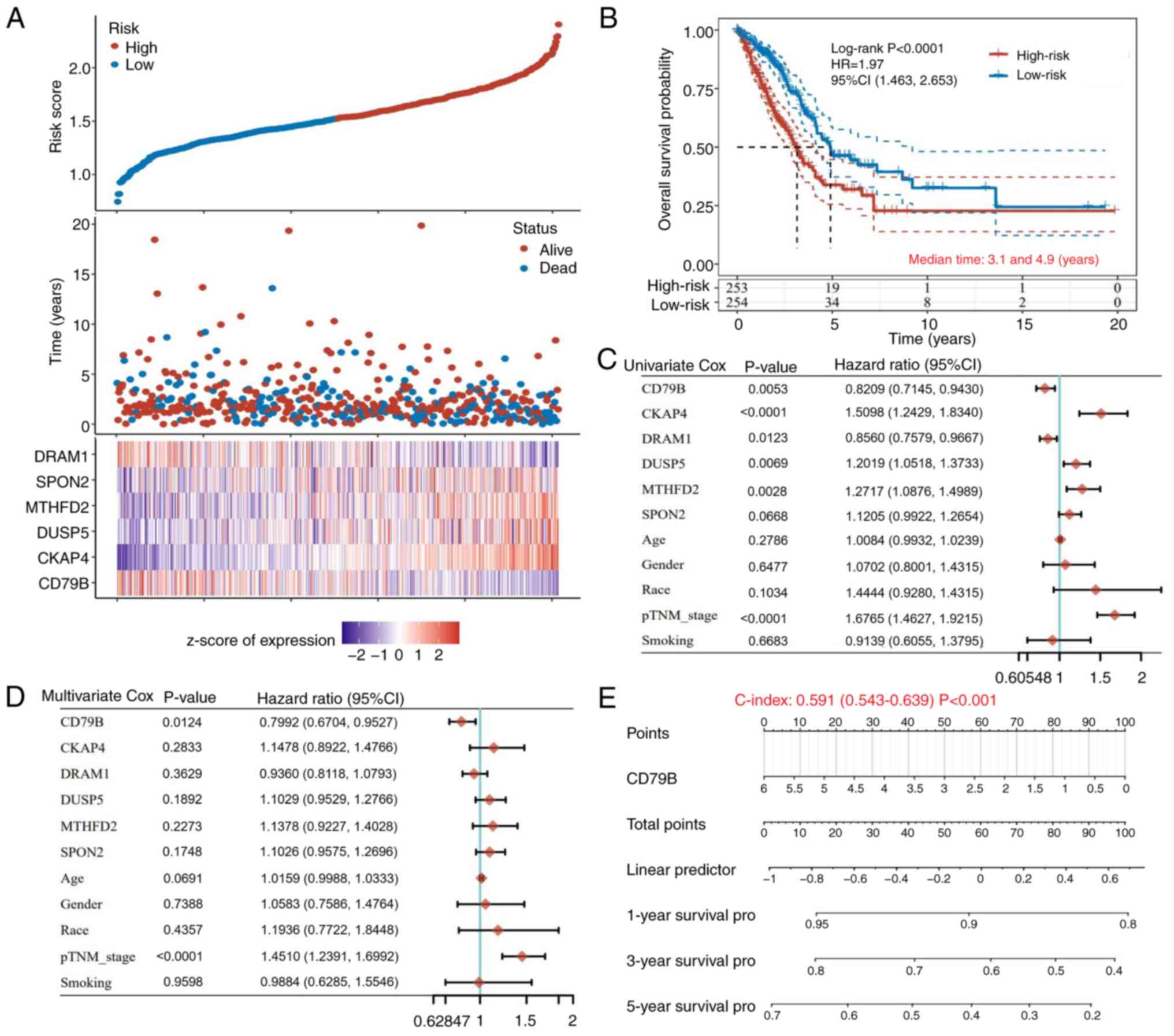
|
1
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Succony L, Rassl DM, Barker AP, McCaughan
FM and Rintoul RC: Adenocarcinoma spectrum lesions of the lung:
Detection, pathology and treatment strategies. Cancer Treat Rev.
99:1022372021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hughes PE, Caenepeel S and Wu LC: Targeted
therapy and checkpoint immunotherapy combinations for the treatment
of cancer. Trends Immunol. 37:462–476. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smit EF and Baas P: Lung cancer in 2015:
Bypassing checkpoints, overcoming resistance, and honing in on new
targets. Nat Rev Clin Oncol. 13:75–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Garon EB, Rizvi NA, Hui R, Leighl N,
Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L,
et al: Pembrolizumab for the treatment of non-small-cell lung
cancer. N Engl J Med. 372:2018–2028. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parra ER, Zhang J, Duose DY,
Gonzalez-Kozlova E, Redman MW, Chen H, Manyam GC, Kumar G, Zhang J,
Song X, et al: Multi-omics analysis reveals immune features
associated with immunotherapy benefit in patients with squamous
cell lung cancer from phase III Lung-MAP S1400I trial. Clin Cancer
Res. 30:1655–1668. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mao X, Xu J, Wang W, Liang C, Hua J, Liu
J, Zhang B, Meng Q, Yu X and Shi S: Crosstalk between
cancer-associated fibroblasts and immune cells in the tumor
microenvironment: New findings and future perspectives. Mol Cancer.
20:1312021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yuan Z, Li Y, Zhang S, Wang X, Dou H, Yu
X, Zhang Z, Yang S and Xiao M: Extracellular matrix remodeling in
tumor progression and immune escape: From mechanisms to treatments.
Mol Cancer. 22:482023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McLane LM, Abdel-Hakeem MS and Wherry EJ:
CD8 T cell exhaustion during chronic viral infection and cancer.
Annu Rev Immunol. 37:457–495. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cai MC, Zhao X, Cao M, Ma P, Chen M, Wu J,
Jia C, He C, Fu Y, Tan L, et al: T-cell exhaustion interrelates
with immune cytolytic activity to shape the inflamed tumor
microenvironment. J Pathol. 251:147–159. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wherry EJ: T cell exhaustion. Nat Immunol.
12:492–499. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Z, Chen L, Chen H, Zhao J, Li K, Sun
J and Zhou M: Pan-cancer landscape of T-cell exhaustion
heterogeneity within the tumor microenvironment revealed a
progressive roadmap of hierarchical dysfunction associated with
prognosis and therapeutic efficacy. EBioMedicine. 83:104–207. 2022.
View Article : Google Scholar
|
|
15
|
Liu J, Li M, Wu J, Qi Q, Li Y, Wang S,
Liang S, Zhang Y, Zhu Z, Huang R, et al: Identification of ST3GAL5
as a prognostic biomarker correlating with CD8+ T cell exhaustion
in clear cell renal cell carcinoma. Front Immunol. 13:9796052022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu-Monette ZY, Zhang M, Li J and Young KH:
PD-1/PD-L1 Blockade: Have we found the key to unleash the antitumor
immune response? Front Immunol. 8:15972017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Z, Zhang Y, Ma N, Yang Y, Ma Y, Wang
F, Wang Y, Wei J, Chen H, Tartarone A, et al: Progenitor-like
exhausted SPRY1+CD8+ T cells potentiate responsiveness to
neoadjuvant PD-1 blockade in esophageal squamous cell carcinoma.
Cancer Cell. 41:1852–1870.e9. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tanoue K, Ohmura H, Uehara K, Ito M,
Yamaguchi K, Tsuchihashi K, Shinohara Y, Lu P, Tamura S, Shimokawa
H, et al: Spatial dynamics of CD39+CD8+ exhausted T cell reveal
tertiary lymphoid structures-mediated response to PD-1 blockade in
esophageal cancer. Nat Commun. 15:90332024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang H, Wu C, Tong X and Chen S: A
Biomimetic Metal-organic framework nanosystem modulates
immunosuppressive tumor microenvironment metabolism to amplify
immunotherapy. J Control Release. 353:727–737. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li W, Wu F, Zhao S, Shi P, Wang S and Cui
D: Correlation between PD-1/PD-L1 expression and polarization in
tumor-associated macrophages: A key player in tumor immunotherapy.
Cytokine Growth Factor Rev. 67:49–57. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang T, Hao L, Cui R, Liu H, Chen J, An J,
Qi S and Li Z: Identification of an immune prognostic 11-gene
signature for lung adenocarcinoma. PeerJ. 9:e107492021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu YH, Jin HQ and Liu HP: Identification
of T-cell exhaustion-related gene signature for predicting
prognosis in glioblastoma multiforme. J Cell Mol Med. 27:3503–3513.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szklarczyk D, Kirsch R, Koutrouli M,
Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT,
Pyysalo S, et al: The STRING database in 2023: Protein-protein
association networks and functional enrichment analyses for any
sequenced genome of interest. Nucleic Acids Res. 51:D638–D646.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wilkerson MD and Hayes DN:
ConsensusClusterPlus: A class discovery tool with confidence
assessments and item tracking. Bioinformatics. 26:1572–1573. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Balachandran VP, Gonen M, Smith JJ and
DeMatteo RP: Nomograms in oncology: More than meets the eye. Lancet
Oncol. 16:e173–e180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xi LJ, Guo ZY, Yang XK and Ping ZG:
Application of LASSO and its extended method in variable selection
of regression analysis. Zhonghua Yu Fang Yi Xue Za Zhi. 57:107–111.
2023.(In Chinese). PubMed/NCBI
|
|
27
|
Li Y, Lu F and Yin Y: Applying logistic
LASSO regression for the diagnosis of atypical Crohn's disease. Sci
Rep. 12:113402022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ashburner M, Ball CA, Blake JA. Botstein
D. Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene Ontology: Tool for the unification of biology. Nat
Genet. 25:25–29. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanehisa M, Goto S, Furumichi M, Tanabe M
and Hirakawa M: KEGG for representation and analysis of molecular
networks involving diseases and drugs. Nucleic Acids Res.
38:D355–D360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hung JH, Yang TH, Hu Z, Weng Z and DeLisi
C: Gene set enrichment analysis: Performance evaluation and usage
guidelines. Brief Bioinform. 13:281–291. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Edelman E, Porrello A, Guinney J,
Balakumaran B, Bild A, Febbo PG and Mukherjee S: Analysis of sample
set enrichment scores: Assaying the enrichment of sets of genes for
individual samples in genome-wide expression profiles.
Bioinformatics. 22:e108–e116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee E, Chuang HY, Kim JW, Ideker T and Lee
D: Inferring pathway activity toward precise disease
classification. PLoS Comput Biol. 4:e10002172008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tomfohr J, Lu J and Kepler TB: Pathway
level analysis of gene expression using singular value
decomposition. BMC Bioinformatics. 6:2252005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al: The
prognostic landscape of genes and infiltrating immune cells across
human cancers. Nat Med. 21:938–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hu W, Wang G, Chen Y, Yarmus LB, Liu B and
Wan Y: Coupled immune stratification and identification of
therapeutic candidates in patients with lung adenocarcinoma. Aging
(Albany NY). 12:16514–16538. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Geeleher P, Cox NJ and Huang RS: Clinical
drug response can be predicted using baseline gene expression
levels and in vitro drug sensitivity in cell lines. Genome Biol.
15:R472014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Geeleher P, Cox N and Huang RS:
pRRophetic: An R package for prediction of clinical
chemotherapeutic response from tumor gene expression levels. PLoS
One. 9:e1074682014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Suarez-Arnedo A, Torres Figueroa F,
Clavijo C, Arbeláez P, Cruz JC and Muñoz-Camargo C: An image J
plugin for the high throughput image analysis of in vitro scratch
wound healing assays. PLoS One. 15:e02325652020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yuan L, Wu X, Zhang L, Yang M, Wang X,
Huang W, Pan H, Wu Y, Huang J, Liang W, et al: SFTPA1 is a
potential prognostic biomarker correlated with immune cell
infltration and response to immunotherapy in lung adenocarcinoma.
Cancer Immunol Immunother. 71:399–415. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Korbecki J, Kupnicka P, Chlubek M, Gorący
J, Gutowska I and Baranowska-Bosiacka I: CXCR2 receptor: Regulation
of expression, signal transduction, and involvement in cancer. Int
J Mol Sci. 23:21682022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee D, Cho M, Kim E, Seo Y and Cha JH:
PD-L1: From cancer immunotherapy to therapeutic implications in
multiple disorders. Mol Ther. 32:4235–4255. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ip EWK, Hoshi N, Shouval DS, Snapper S and
Medzhitov R: Anti-inflammatory effect of IL-10 mediated by
metabolic reprogramming of macrophages. Science. 356:513–519. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sawant DV, Yano H, Chikina M, Zhang Q,
Liao M, Liu C, Callahan DJ, Sun Z, Sun T, Tabib T, et al: Adaptive
plasticity of IL-10+ and IL-35+ Treg cells cooperatively promotes
tumor T cell exhaustion. Nat Immunol. 20:724–735. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Phelan JD, Young RM, Webster DE, Roulland
S, Wright GW, Kasbekar M, Shaffer AL, Ceribelli M, Wang JQ, Schmitz
R, et al: A multiprotein supercomplex controlling oncogenic
signalling in lymphoma. Nature. 560:387–391. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shukla M and Sarkar RR: Differential
cellular communication in tumor immune microenvironment during
early and advanced stages of lung adenocarcinoma. Mol Genet
Genomics. 299:1002024. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chi X, Luo S, Ye P, Hwang WL, Cha JH, Yan
X and Yang WH: T-cell exhaustion and stemness in antitumor
immunity: Characteristics, mechanisms, and implications. Front
Immunol. 14:11047712023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pichler AC, Carrié N, Cuisinier M, Ghazali
S, Voisin A, Axisa PP, Tosolini M, Mazzotti C, Golec DP, Maheo S,
et al: TCR-independent CD137 (4-1BB) signaling promotes
CD8+-exhausted T cell proliferation and terminal differentiation.
Immunity. 56:1631–1648.e10. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Scharping NE, Rivadeneira DB, Menk AV,
Vignali PDA, Ford BR, Rittenhouse NL, Peralta R, Wang Y, Wang Y,
DePeaux K, et al: Mitochondrial stress induced by continuous
stimulation under hypoxia rapidly drives T cell exhaustion. Nat
Immunol. 22:205–215. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Watowich MB, Gilbert MR and Larion M: T
cell exhaustion in malignant gliomas. Trends Cancer. 9:270–292.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Abolhalaj M, Sincic V, Lilljebjörn H,
Sandén C, Aab A, Hägerbrand K, Ellmark P, Borrebaeck CAK, Fioretos
T and Lundberg K: Transcriptional profiling demonstrates altered
characteristics of CD8+ cytotoxic T-cells and regulatory T-cells in
TP53-mutated acute myeloid leukemia. Cancer Med. 11:3023–3032.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Somani VK, Zhang D, Dodhiawala PB, Lander
VE, Liu X, Kang LI, Chen HP, Knolhoff BL, Li L, Grierson PM, et al:
IRAK4 signaling drives resistance to checkpoint immunotherapy in
pancreatic ductal adenocarcinoma. Gastroenterology. 162:2047–2062.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kim BS, Kuen DS, Koh CH, Kim HD, Chang SH,
Kim S, Jeon YK, Park YJ, Choi G, Kim J, et al: Type 17 immunity
promotes the exhaustion of CD8+ T cells in cancer. J Immunother
Cancer. 9:e0026032021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Huang Y, Jia A, Wang Y and Liu G: CD8+ T
cell exhaustion in anti-tumour immunity: The new insights for
cancer immunotherapy. Immunology. 168:30–48. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zebley CC and Youngblood B: Mechanisms of
T cell exhaustion guiding next generation immunotherapy. Trends
Cancer. 8:726–734. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ford BR, Vignali PDA, Rittenhouse NL,
Scharping NE, Peralta R, Lontos K, Frisch AT, Delgoffe GM and
Poholek AC: Tumor microenvironmental signals reshape chromatin
landscapes to limit the functional potential of exhausted T cells.
Sci Immunol. 7:eabj91232022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang L, Zhang B, Li L, Ye Y, Wu Y, Yuan
Q, Xu W, Wen X, Guo X and Nian S: Novel targets for immunotherapy
associated with exhausted CD8 + T cells in cancer. J Cancer Res
Clin Oncol. 149:2243–2258. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Pu D, Liu D, Li C, Chen C, Che Y, Lv J,
Yang Y and Wang X: A novel ten-gene prognostic signature for
cervical cancer based on CD79B-related immunomodulators. Front
Genet. 13:9337982022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang Y, Yang Y, Zhao Z, Sun H, Luo D,
Huttad L, Zhang B and Han B: A new nomogram model for prognosis of
hepatocellular carcinoma based on novel gene signature that
regulates cross-talk between immune and tumor cells. BMC Cancer.
22:3792022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huse K, Bai B, Hilden VI, Bollum LK,
Våtsveen TK, Munthe LA, Smeland EB, Irish JM, Wälchli S and
Myklebust JH: Mechanism of CD79A and CD79B support for IgM+ B cell
fitness through BCR surface expression. J Immunol. 209:2042–2053.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tkachenko A, Kupcova K and Havranek O:
B-cell receptor signaling and beyond: The role of Igα (CD79a)/Igβ
(CD79b) in normal and malignant B cells. Int J Mol Sci. 25:102023.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ramesh S, Go M, Call ME and Call MJ: Deep
mutational scanning reveals transmembrane features governing
surface expression of the B cell antigen receptor. Front Immunol.
15:14267952024. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xie Z, Qin Y, Chen X, Yang S, Yang J, Gui
L, Liu P, He X, Zhou S, Zhang C, et al: Deciphering the prognostic
significance of MYD88 and CD79B mutations in diffuse large B-Cell
lymphoma: Insights into treatment outcomes. Target Oncol.
19:383–400. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ushmorov A, Leithäuser F, Sakk O,
Weinhaüsel A, Popov SW, Möller P and Wirth T: Epigenetic processes
play a major role in B-cell-specific gene silencing in classical
Hodgkin lymphoma. Blood. 107:2493–2500. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bullerwell CE, Robichaud PP, Deprez PML,
Joy AP, Wajnberg G, D'Souza D, Chacko S, Fournier S, Crapoulet N,
Barnett DA, et al: EBF1 drives hallmark B cell gene expression by
enabling the interaction of PAX5 with the MLL H3K4
methyltransferase complex. Sci Rep. 11:15372021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Khasnis S, Veenstra H, McClellan MJ,
Ojeniyi O, Wood CD and West MJ: Regulation of B cell receptor
signalling by Epstein-Barr virus nuclear antigens. Biochem J.
479:2395–2417. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Dantoing E, Piton N, Salaün M, Thiberville
L and Guisier F: Anti-PD1/PD-L1 immunotherapy for Non-small cell
lung cancer with actionable oncogenic driver mutations. Int J Mol
Sci. 22:62882021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu F, Wang T, Petit J, Forlenza M, Chen
X, Chen L, Zou J and Secombes CJ: Evolution of IFN subgroups in
bony fish-2. Analysis of subgroup appearance and expansion in
teleost fish with a focus on salmonids. Fish Shellfish Immunol.
98:564–573. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Khsheibun R, Paperna T, Volkowich A,
Lejbkowicz I, Avidan N and Miller A: Gene expression profiling of
the response to interferon beta in Epstein-Barr-transformed and
primary B cells of patients with multiple sclerosis. PLoS One.
9:e1023312014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chu F, Cao J, Liu J, Yang H, Davis TJ,
Kuang SQ, Cheng X, Zhang Z, Karri S, Vien LT, et al: Chimeric
antigen receptor T cells to target CD79b in B-cell lymphomas. J
Immunother Cancer. 11:e0075152023. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang S, Liu G, Li Y and Pan Y: Metabolic
reprogramming induces macrophage polarization in the tumor
microenvironment. Front Immunol. 13:8400292022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kersten K, Hu KH, Combes AJ, Samad B,
Harwin T, Ray A, Rao AA, Cai E, Marchuk K, Artichoker J, et al:
Spatiotemporal co-dependency between macrophages and exhausted CD8+
T cells in cancer. Cancer Cell. 40:624–638.e9. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Nixon BG, Kuo F, Ji L, Liu M, Capistrano
K, Do M, Franklin RA, Wu X, Kansler ER, Srivastava RM, et al:
Tumor-associated macrophages expressing the transcription factor
IRF8 promote T cell exhaustion in cancer. Immunity.
55:2044–2058.e5. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yang H, Zhang Q, Xu M, Wang L, Chen X,
Feng Y, Li Y, Zhang X, Cui W and Jia X: CCL2-CCR2 axis recruits
tumor associated macrophages to induce immune evasion through PD-1
signaling in esophageal carcinogenesis. Mol Cancer. 19:412020.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lu Y, Sun Q, Guan Q, Zhang Z, He Q, He J,
Ji Z, Tian W, Xu X, Liu Y, et al: The XOR-IDH3α axis controls
macrophage polarization in hepatocellular carcinoma. J Hepatol.
79:1172–1184. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhu CX, Yan K, Chen L, Huang RR, Bian ZH,
Wei HR, Gu XM, Zhao YY, Liu MC, Suo CX, et al: Targeting
OXCT1-mediated ketone metabolism reprograms macrophages to promote
antitumor immunity via CD8+ T cells in hepatocellular carcinoma. J
Hepatol. 81:690–703. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Xia J, Zhang L, Peng X, Tu J, Li S, He X,
Li F, Qiang J, Dong H, Deng Q, et al: IL1R2 blockade alleviates
immunosuppression and potentiates anti-PD-1 efficacy in
triple-negative breast cancer. Cancer Res. 84:2282–2296. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Peters S, Paz-Ares L, Herbst RS and Reck
M: Addressing CPI resistance in NSCLC: Targeting TAM receptors to
modulate the tumor microenvironment and future prospects. J
Immunother Cancer. 10:e0048632022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Hsu SPC, Chen YC, Chiang HC, Huang YC,
Huang CC, Wang HE, Wang YS and Chi KHL: Rapamycin and
hydroxychloroquine combination alters macrophage polarization and
sensitizes glioblastoma to immune checkpoint inhibitors. J
Neurooncol. 146:417–426. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Wei Z, Zhang X, Yong T, Bie N, Zhan G, Li
X, Liang Q, Li J, Yu J, Huang G, et al: Boosting anti-PD-1 therapy
with metformin-loaded macrophage-derived microparticles. Nat
Commun. 12:4402021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Oliva M, Chepeha D, Araujo DV, Diaz-Mejia
JJ, Olson P, Prawira A, Spreafico A, Bratman SV, Shek T, de Almeida
J, et al: Antitumor immune effects of preoperative sitravatinib and
nivolumab in oral cavity cancer: SNOW window-of-opportunity study.
J Immunother Cancer. 9:e0034762021. View Article : Google Scholar : PubMed/NCBI
|