Introduction
Lung adenocarcinoma (LUAD) is the most common
primary lung cancer, accounting for ~40% of cases (1). Patients with early-stage LUAD mainly
receive surgical therapy, while advanced and metastatic LUAD often
require systemic therapy. These treatments improve symptoms and
prolong their overall survival to a certain extent (2). However, the anti-tumor effect in
patients with LUAD is often limited by chemotherapy or radiotherapy
resistance and immune escape, leading to tumor recurrence and poor
prognosis (3). For patients with
LUAD, individualized and comprehensive treatments should be
developed based on tumor progression. Molecular targeted therapy
targeting oncogenic driver genes, such as Kirsten rat sarcoma viral
oncogene homologue (KRAS), epidermal growth factor receptor (EGFR),
v-Raf murine sarcoma viral oncogene homolog B (BRAF) and anaplastic
lymphoma kinase (ALK) has greatly advanced the field of treatment
for non-small cell lung cancer, especially LUAD (4). Immune checkpoint inhibitors have been
approved by the US Food and Drug Administration for monotherapy or
combination with other agents in LUAD, which provide hope for
patients with negative expression of driver oncogenes or resistance
to molecular-targeted therapy (5,6).
However, clinical trials have shown that only a small number of
patients benefit from immunotherapy (7,8).
Therefore, it is necessary to identify more biomarkers to ensure
that more patients can benefit from them.
The tumor immune microenvironment is widely
recognized as the key regulator of tumor progression and clinical
response to therapy (9,10). Components of the tumor
microenvironment include cancer and inflammatory cells, and
suppressive cytokines that contribute to the transition of T cells
into ‘exhausted’ T cells by regulating T cell phenotypes and
functions (11,12). T cell exhaustion (TEX) is defined as
a decline in proliferation capacity, weakened effector function,
loss of cytokine production and sustained high expression of
inhibitory receptors (13,14). Exhausted T cells demonstrate
upregulation of immune checkpoint proteins such as cytotoxic T
lymphocyte antigen 4 (CTLA-4) and programmed cell death protein-1
(PD-1). Transcription and epigenetic reprogramming lead to
mitochondrial dysfunction and decreased metabolic activity, thereby
triggering immune evasion (15).
Immune checkpoint blockade (ICB) therapy can inhibit negative
regulatory signals and release T cells from exhaustion (16), however not all TEX cells respond to
ICB therapy. Recent studies have revealed heterogeneous
subpopulations within the exhausted T cell lineage including
ICB-sensitive and -resistant subpopulations, which lead to
differences in treatment responses (17,18).
TEX is closely related to tumor-associated macrophage (TAM)
polarization. Metabolites secreted by cancer cells promote cell
escape from M1 TAM polarization towards M2 TAM, contributing
further towards TEX development (19). M2 TAMs promote TEX, resulting in
anti-PD-1 blockade resistance (20). Therefore, inducing the switching of
TAM from M2 to M1 may reverse TEX and restore T cells from an
exhausted state, providing potential targets in tumor
immunotherapy.
The present study performed clustering analysis of
TEX-related genes in patients with LUAD patients by analyzing a
public database; subsequently the present study aimed to construct
a prognostic signature and nomogram to predict prognosis for LUAD.
Then, the study aimed to explored the potential of B cell antigen
receptor complex-associated protein β chain (CD79B) as a prognostic
marker and the role of immune function in LUAD, which may play a
key role in tumor immunotherapy.
Materials and methods
Data collection
RNA-sequencing expression profiles and corresponding
clinical information of 516 LUAD samples and 59 para-tumor tissue
samples were downloaded from The Cancer Genome Atlas (TCGA)
database (portal.gdc.cancer.gov/; age range: 33–88 year; female:
278, male: 238). STAR-counts data, mutation annotation format (MAF)
data, and corresponding clinical information were downloaded from
the TCGA database (portal.gdc.cancer.gov) (TCGA-LUAD dataset). Data
was then extracted in TPM format and performed normalization using
the log2 (TPM+1) transformation. After retaining samples that
included both RNAseq data and clinical information, 516 LUAD
samples and 59 para-tumor tissue samples were selected for further
analysis (21). Based on previous
research, 12 TEX-related genes that may have prognostic value were
obtained (22). The protein-protein
interaction network was obtained from the Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) database
(cn.string-db.org/) (23).
Consensus clustering analysis
To evaluate consistency in TCGA-LUAD database, R
package ConsensusClusterPlus (v1.72.0;
bioconductor.org/packages/release/bioc/html/ConsensusClusterPlus.html)
was used (24), with a maximum of 6
clusters and 80% of the samples were extracted 100 times with
clusterAlg=‘hc’, innerLinkage=‘ward. D2’. Patients were clustered
according to the optimal k-value based on the cumulative
distribution function (CDF) curve and consensus matrix. Principal
component analysis (PCA) was used to analyze the distribution
differences between clusters.
Survival analysis and construction of
prognostic signature
Survival analysis was conducted between clusters in
TCGA-LUAD database using the ‘survival’ and the ‘survminer’ R
packages (v0.4.9;
rdocumentation.org/packages/survminer/versions/0.4.9). The
appropriate conditions for the construction of the nomogram
prediction model were determined using univariate and multivariate
Cox regression analysis. Using the ‘forestplot’ R package (v3.1.7)
(https://github.com/cran/forestplot),
the P-value and hazard ratio (HR) with 95% confidence interval (CI)
of each variable were calculated. To predict the overall recurrence
rate over the next × years, a nomogram was created based on the
results of multivariate Cox proportional hazards analysis using the
‘rms’ R package (v8.0.0;
cran.r-project.org/web/packages/rms/index.html); the nomogram
represented factors which can be used to determine the likelihood
of recurrence for an individual patient (25).
The least absolute shrinkage and selection operator
(LASSO) regression used a feature selection algorithm,
incorporating 10-fold cross-validation, and was examined using the
‘glmnet’ R package (v4.1-10;
cran.r-project.org/web/packages/glmnet/index.html) (26). Using the ‘survival’ R package, a
novel prognostic model of TEX-related genes was constructed based
on multivariate Cox regression analysis. The time-receiver
operating characteristic (ROC) (v0.4) (huppertlab.net/home/training/knowledge-base/nirs_toolbox/fulldemos/receiver-operating-characteristic-roc/)
analysis was performed on TEX-related genes to determine the
accuracy and risk score (27).
Functional enrichment analysis
The ‘limma’ R package (v3.40.2)
(bioconductor.org/packages/release/bioc/html/limma.html) was used
to analyze the differential expression of mRNA between clusters
(28). The criteria for statistical
significance were adjusted P<0.05 and log2 (fold-change) >1
or <-1. Using the ClusterProfiler package in R software
(v3.18.0;
bioconductor.org/packages/release/bioc/html/clusterProfiler.htm)
(29), gene functions of enrichment
analysis were investigated in Gene Ontology (GO)
(geneontology.org/) (30) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg/) (31). The enrichment scores were calculated
using the Gene Set Variation Analysis package of R software
(v.1.3.0) (https://www.gsea-msigdb.org/gsea/index.jsp) based on
the absolute enrichment scores of gene sets in multiple
publications and gene signatures validated by previous experiments
(32–36), with the parameter chosen as
method=‘ssGESA’ (37). Spearman's
correlation was calculated to determine the relationship between
genes and pathway scores.
Tumor immune microenvironment
analysis
The ‘immuneeconv’ R package (v2.0.3;
rdocumentation.org/packages/immunedeconv/versions/2.0.3) was used
to predict 22 types of immune infiltrating cell based on the
CIBERSORT algorithm (a gene expression-based deconvolution
algorithm) (http://cibersort.stanford.edu/) (38). The mRNA levels of immune
checkpoint-related genes were compared between clusters, including
Programmed Cell Death 1 Ligand 1 (PD-L1/CD274), Cytotoxic
T-Lymphocyte Associated Protein 4 (CTLA4), Hepatitis A Virus
Cellular Receptor 2 (HAVCR2), Lymphocyte Activating 3 (LAG3),
Programmed Cell Death 1 (PDCD1), Programmed Cell Death 1 Ligand 2
(PDCD1LG2), T Cell Immunoreceptor With Ig And ITIM Domains (TIGIT),
and Sialic Acid Binding Ig Like Lectin 15 (SIGLEC15). To predict
the ICB response, the Tumor Immune Dysfunction and Exclusion (TIDE)
algorithm (v6.5.1) (http://tide.dfci.harvard.edu/) was used to simulate
computational approaches to tumor immune evasion, based on two
molecular mechanisms, including dysfunction of tumor infiltrating
cytotoxic T lymphocytes (CTLs) and rejection of CTLs by
immunosuppressive factors (7). The
correlation between gene expression and immune score was analyzed
by ‘ggstatsplot’ (v0.13.1;
cran.r-project.org/web/packages/ggstatsplot/index.html) and
‘pheatmap’ R package (v1.0.13) (https://www.rdocumentation.org/packages/pheatmap/versions/1.0.13/topics/pheatmap)
(39).
Prediction of therapeutic drug
sensitivity
The prediction of sensitivity to various therapeutic
agents were performed by the ‘pRRophetic’ R package (v4.0.3)
(github.com/paulgeeleher/pRRophetic), based on the Genomics of Drug
Sensitivity in Cancer (GDSC) dataset (cancerrxgene.org/) (40). The ridge regression method was used
to evaluate the half-maximal inhibitory concentration (IC50) of the
samples, and 10-fold cross-validation was performed based on the
GDSC training set to evaluate the prediction accuracy (41). The IC50 for each sample in TCGA-LUAD
database was estimated from the predictive model evaluated on GDSC
cell line data.
Cell culture and plasmid
transfection
Human normal lung epithelial HBE, macrophage THP-1
and LUAD A549, H358, PC-9, 95C, 95D cell lines were obtained from
the Cell Center of Central South University (Changsha, China). All
cells were cultured in RPMI-1640 medium (Gibo; Thermo Fisher
Scientific, Inc.) containing 10% fetal bovine serum (FBS; HyClone;
Cytiva). The pDONR223-CD79B overexpression plasmid (cat. no.
G104684) and empty vector control pDONR223 were purchased from
Youbio. Lipofectamine™ 3000 Transfection Reagent (cat. no.
L3000150, Thermo Fisher Scientific, Inc.) was used to transfect
cells with 6 µg plasmid transfection for 24 h at 37°C, followed by
48 h culture before downstream experiments.
Coculture assay
THP-1 monocytes (2×107 cells) were
differentiated into M0 macrophages by treatment with 100 ng/ml
phorbol 12-myristate 13-acetate (PMA; cat. no. P8139,
Sigma-Aldrich, Inc.) for 48 h in RPMI-1640 medium containing 10%
FBS (37°C). M0 macrophages (5×104 cells/insert) were
seeded in the upper chamber (0.4 µm pore size; Corning, Inc.) while
A549 cells (2×105 cells/well) were plated in the lower
chamber. Cells were co-cultured for 48 h under standard conditions
(37°C, 5% CO2). Following coculture, upper-chamber cells
were collected for downstream analysis by qPCR.
Quantitative (q)PCR
After 48 h co-culture with CD79B-overexpressing A549
cells in a humidified incubator (37°C, 5% CO2), total
RNA was extracted from M0 macrophage THP-1 cells using the RNA
extraction kit (cat. no. 15596026). Reverse transcription kit (cat.
no. K1621; both Thermo Fisher Scientific, Inc.) was used to obtain
cDNA, according to the manufacturer's instructions. SYBR Green kit
(cat. no. 4309155; Thermo Fisher Scientific, Inc.) was used for
qPCR analysis by ABI 7500. Thermocycling conditions were as
follows: 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec
and 60°C for 60 sec, with a melting-curve stage to confirm amplicon
specificity. β-actin was used as the reference gene for
normalization, and relative gene expression was calculated using
the 2−ΔΔCq method (42).
The primers are list in Table
SI.
Western blotting
The cells (HBE, A549, H358, PC9, 95C, 95D, A549-Vec,
A549-OE-CD79B cells) were lysed in immunoprecipitation buffer (cat.
no. 87787, Thermo Fisher Scientific, Inc.) containing inhibitors
cocktail (cat. no. 4693116001) and phosphatase inhibitor (cat. no.
4906845001, both Roche Diagnostics). The protein concentration was
measured with BCA reagent (cat. no. AR0197, Wuhan Boster Biological
Technology Ltd.). The samples (50 µg/lane) were separated by 10%
SDS-PAGE and transferred to a PVDF membrane (Merck KGaA). The
membranes were blocked with 5% non-fat dry milk in TBST (0.1%
Tween-20) at room temperature for 1 h, and then incubated at 4°C
overnight with rabbit anti-CD79B (1:400, cat. no. ab134147, Abcam)
or mouse anti-actin (1:20,000; cat. no. AC026, ABclonal, Biotech
Co., Ltd.), followed by incubation with HRP-conjugated goat
anti-rabbit secondary antibody (1:6,000; cat. no. AS014, ABclonal
Biotech Co., Ltd.) for 2 h at room temperature. Finally, the bands
were imaged on a ChemiDoc™ imaging system (Bio-Rad) after detection
with an enhanced chemiluminescence detection kit (cat. no. 36208-A,
Shanghai Yeasen Biotechnology Co., Ltd.). Densitometric analysis
was performed using Image Lab™ 6.1 software (Bio-Rad).
Cell Counting Kit (CCK)8 assay
Cell viability was determined by CCK8 assay (cat.
no. C0005, TargetMol Chemicals, Inc.) according to the
manufacturer's instructions. A549-Vec and A549-OE-CD79B cells were
inoculated in 96-well plates, and incubated with CCK8 for 2 h. The
optical density at 450 nm was then determined by microplate reader
(BioTek ELx800; Agilent Technologies, Inc.).
Apoptosis assay
A549-Vec and A549-OE-CD79B cells were collected and
stained with annexin V-FITC and propidium iodide using an apoptosis
kit (cat. no. KGA103, Nanjing Keygen Biotech Co., Ltd.) according
to the manufacturer's instructions. Cells were analyzed on a BD
LSRFortessa™ X-20 flow cytometer (BD Biosciences). Data were
processed using FlowJo software (version 10.8.1; BD Biosciences),
with apoptotic cells defined as annexin
V+/PI− (early apoptotic) + annexin
V+/PI+ (late apoptotic) populations.
Scratch assay
A549-Vec and A549-OE-CD79B cells were seeded in
6-well culture plates (2×105 cells per well) and
incubated overnight to a density of 70–80% confluence (37°C, 5%
CO2). The medium was replaced with serum-free RPMI-1640,
a straight scratch was made with a sterile 100-µl pipette tip and
floating cells were rinsed away. Plates were incubated at 37°C for
12 h. Phase-contrast images were acquired at 0 and 12 h using an
inverted light microscope (AMEX1200, Thermo Fisher Scientific,
Inc.; magnification, ×100), and wound areas were quantified with
ImageJ (v1.53t, NIH, USA) (43).
Cell migration and invasion assay
Cell invasion assays were performed using 24-well
plates and Transwell invasion chamber (8 µm pore size, BD
Biosciences). For invasion assays, Matrigel (cat. no. 356234,
Corning, Inc.) was added at 37°C for 1 h before cell seeding.
Migration assays were performed without matrix gel. A549-Vec and
A549-OE-CD79B cells cultured at 37°C with 5% CO2 were
trypsinized, resuspended in serum-free RPMI-1640, and
5×104 cells in 200 µl were seeded into the upper
chamber. The lower chamber received 800 µl RPMI-1640 containing 10%
FBS as chemoattractant. After 24 h of incubation at 37°C with 5%
CO2, non-invading/migrating cells on the upper surface
were removed with a cotton swab. Cells on the lower surface were
fixed at room temperature with 4% paraformaldehyde for 20 min,
stained with 0.1% crystal violet at room temperature for 10 min,
washed three times with PBS), and air-dried. Invaded/migrated cells
in five randomly selected fields per insert were counted manually
under a light microscope.
Statistical analysis
The bioinformatics data were analyzed using R
software v4.1.3 (R Foundation for Statistical Computing, 2022), and
the sample test data were processed using SPSS24.0 (IBM Corp.) and
GraphPad Prism8.0.1 (Dotmatics) software. Paired Student t-test and
Wilcoxon test were applied to normally and non-normally distributed
data, respectively. One-way ANOVA followed by Tukey test was used
to determine the differences among multiple comparative groups. The
sample size for each group was ≥3. All data are presented as the
mean ± SD of three individual experiments. P<0.05 was considered
to indicate a statistically significant difference.
Results
Cluster and survival analysis of
TEX-related genes in patients with LUAD
A total of 12 TEX-related genes were collected from
a previous study (22). To explore
the TEX heterogeneity in LUAD, consensus clustering was performed
based on the expression of these 12 TEX-related genes in TCGA-LUAD.
Based on the consensus CDF curve (Fig.
S1A) and relative change in area under the CDF curve (Fig. S1B), k=2 was selected as the best
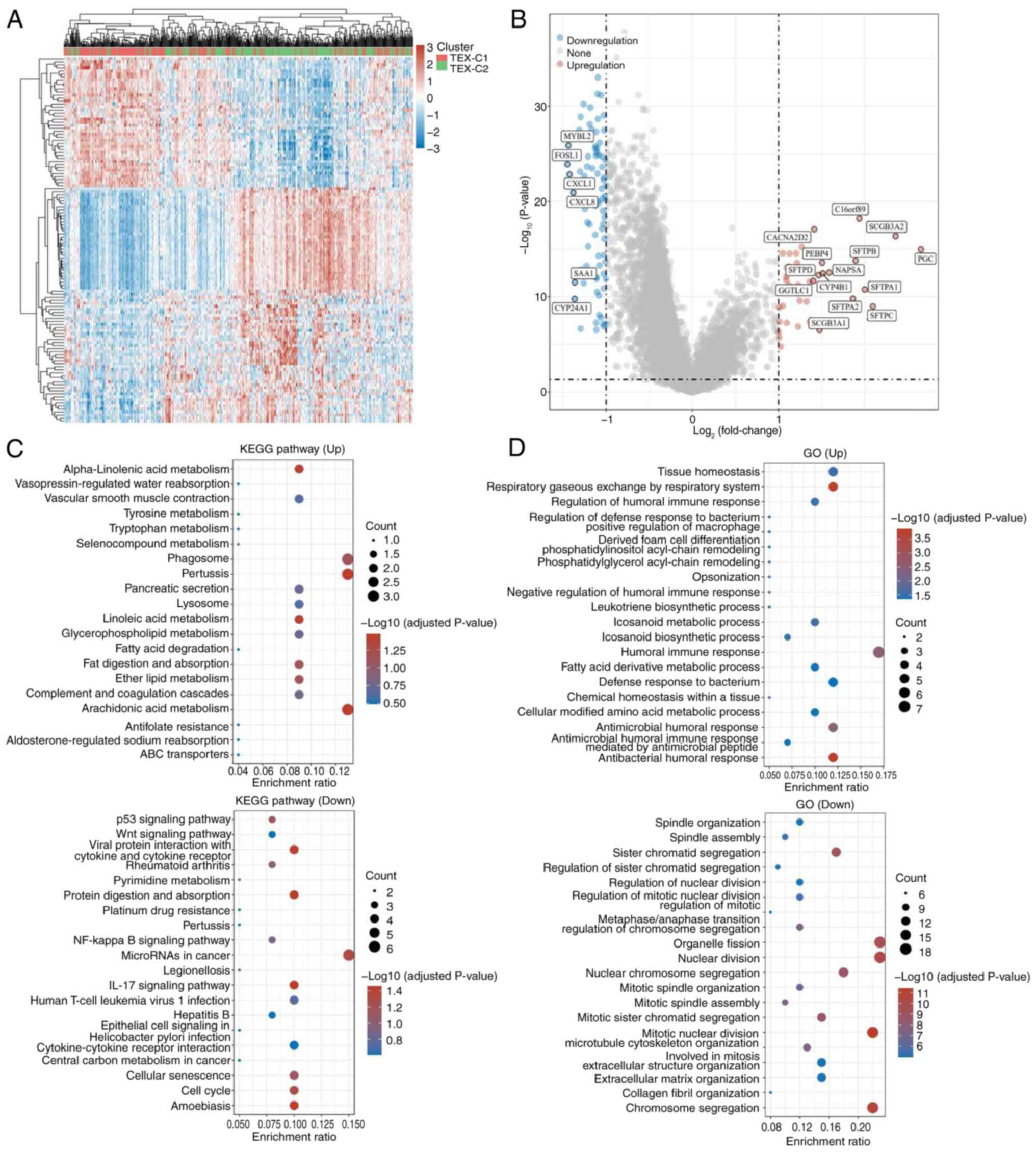
cluster and two clusters (TEX-C1 and -C2) were identified (Fig. 1A and B). There were 242 patients in
the TEX-C1 and 274 patients in the TEX-C2 cluster. The PCA results
confirmed significant differences between the two clusters
(Fig. S1C). The differences in
clinical characteristics between the two clusters are shown in
Table I. mRNA expression of eight
TEX-related genes was higher in the TEX-C2 than that in the TEX-C1
cluster, including cytoskeleton Associated Protein 4 (CKAP4), Dual
Specificity Phosphatase 5 (DUSP5), methylenetetrahydrofolate
dehydrogenase (NADP+ Dependent) 2 (MTHFD2), Opioid
Growth Factor Receptor (OGFR), Spondin 2 (SPON2), C-X-C Motif
Chemokine ligand 1 (CXCL1), C-C Motif Chemokine ligand 2 (CCL2) and
CXCL6. The expression of three TEX-related genes [CD79B, B Cell
Scaffold Protein With Ankyrin Repeats 1 (BANK1) and DNA Damage
Regulated Autophagy Modulator 1 (DRAM1)] was lower in the TEX-C2
cluster, however, the expression of Lipopolysaccharide Induced TNF
Factor (LITAF) was not significantly different between clusters
(Fig. 1C). Patients in the TEX-C2
cluster had worse prognosis with shorter overall survival (OS) and
progression-free survival (PFS), compared with those in the TEX-C1
cluster (Fig. 1D and E).
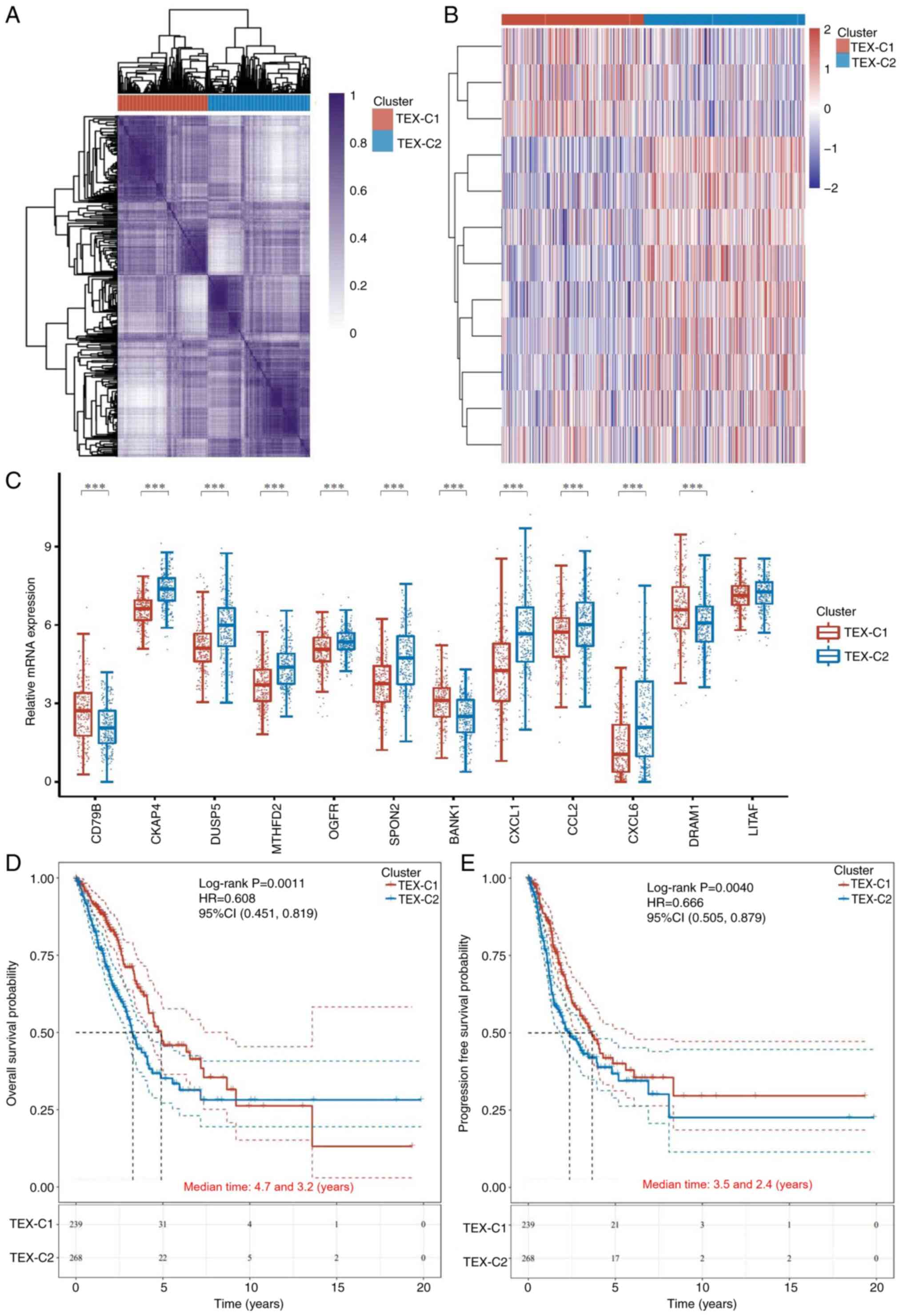 | Figure 1.Cluster and survival analysis of
TEX-related genes in patients with lung adenocarcinoma. (A)
Unsupervised consensus cluster analysis identifying two clusters
(k=2). (B) Heatmap of TEX-related genes in the two clusters. (C)
mRNA expression of TEX-related genes between clusters. Kaplan-Meier
survival analysis of (D) overall and (E) progression-free survival
between TEX clusters. ***P<0.001. TEX-C, T cell exhaustion
cluster; CD79B, B cell Antigen Receptor Complex-Associated Protein
Beta Chain; DUSP, dual Specificity Phosphatase; MTHFD,
Methylenetetrahydrofolate Dehydrogenase; OGFR, Opioid Growth Factor
Receptor; SPON, Spondin; BANK, B Cell Scaffold Protein With Ankyrin
Repeats; CXCL, C-X-C Motif Chemokine Ligand; CCL, C-C Motif
Chemokine Ligand; DRAM, DNA Damage Regulated Autophagy Modulator;
LITAF, Lipopolysaccharide Induced TNF Factor;, HR, Hazard Ratio;
CI, Confidence Interval. |
 | Table I.Clinical characteristics between
TEX-C1 and TEX-C2 in lung adenocarcinoma. |
Table I.
Clinical characteristics between
TEX-C1 and TEX-C2 in lung adenocarcinoma.
| Characteristic | TEX-C1 | TEX-C2 | P-value |
|---|
| Status |
|
|
|
|
Alive | 170 | 159 |
|
|
Dead | 72 | 115 | 0.005 |
| Mean age,
years | 66.4 (9.8) | 64.3 (10.1) |
|
| Median age, years
(range) | 67 (33–88) | 65 (39–87) | 0.021 |
| Sex |
|
|
|
|
Female | 128 | 150 |
|
|
Male | 114 | 124 | 0.739 |
| Ethnicity |
|
|
|
|
Asian | 3 | 5 |
|
|
Black | 24 | 28 |
|
|
White | 189 | 200 |
|
| Native
American | 0 | 1 | 0.788 |
| pT stage |
|
|
|
| T1 | 41 | 26 |
|
|
T1a | 27 | 20 |
|
|
T1b | 34 | 21 |
|
| T2 | 74 | 95 |
|
|
T2a | 32 | 50 |
|
|
T2b | 10 | 17 |
|
| T3 | 15 | 32 |
|
| T4 | 8 | 11 |
|
| TX | 1 | 2 | 0.007 |
| pN stage |
|
|
|
| N0 | 173 | 159 |
|
| N1 | 35 | 61 |
|
| N2 | 25 | 49 |
|
| NX | 8 | 3 |
|
| N3 | 0 | 2 | 0.001 |
| pM stage |
|
|
|
| M0 | 159 | 188 |
|
| M1 | 6 | 12 |
|
|
M1a | 1 | 1 |
|
|
M1b | 1 | 4 |
|
| MX | 73 | 67 | 0.343 |
| pTNM stage |
|
|
|
| I | 1 | 4 |
|
| IA | 86 | 45 |
|
| IB | 68 | 72 |
|
| II | 1 | 0 |
|
|
IIA | 20 | 30 |
|
|
IIB | 24 | 47 |
|
|
IIIA | 25 | 48 |
|
|
IIIB | 4 | 7 |
|
| IV | 8 | 18 | <0.001 |
| New tumor
event |
|
|
|
|
Metastasis (NOS) | 24 | 37 |
|
|
Metastasis (recurrence) | 3 | 5 |
|
|
Primary | 7 | 4 |
|
|
Recurrence | 22 | 26 |
|
|
Metastasis (primary) | 0 | 1 | 0.482 |
| Smoking status |
|
|
|
|
Non-smoking | 40 | 35 |
|
|
Smoking | 194 | 233 | 0.255 |
| Therapy |
|
|
|
|
Ancillary chemotherapy | 2 | 1 |
|
|
Chemotherapy | 70 | 85 |
|
|
Chemotherapy; targeted
molecular therapy | 1 | 5 |
|
|
Targeted molecular
therapy | 2 | 0 |
|
|
Chemotherapy;
immunotherapy | 4 | 0 |
|
|
Immunotherapy | 1 | 0 |
|
|
Vaccine | 0 | 1 | 0.378 |
Differential gene expression and gene
enrichment analysis of TEX-based clusters in patients with
LUAD
TEX-C1 and TEX-C2 clusters demonstrated 121
differentially expressed genes. Compared with the TEX-C1 cluster,
43 genes were upregulated in the TEX-C2 cluster, such as the
pepsinogen C and Surfactant Protein B (SFTP) families. SFTP is a
pulmonary surface-active substance synthesized by alveolar
epithelial type II cells, which is involved in the development and
progression of LUAD by regulating multiple immune-associated
pathways (44). In the TEX-C2
cluster, 78 genes were downregulated, including CXCL1 and CXCL8,
which are ligands for the chemokine receptor CXCR2 and involved in
the recruitment of tumor immune cells such as TAMs and regulatory T
cells (Tregs; Fig. 2A and B)
(45). Compared with the TEX-C1
cluster, the differentially expressed genes in the TEX-C2 cluster
were mainly involved in immune-related pathways. The KEGG analysis
of upregulated genes in the TEX-C2 cluster demonstrated enrichment
of metabolism-related pathways, while downregulated genes were
associated with ‘p53 signaling pathway’, ‘Wnt signaling pathways’,
‘NF-κB signaling pathways’, ‘IL-17 signaling pathways’ and other
signaling pathways (Fig. 2C). In
addition, upregulated genes in the TEX-C2 cluster were enriched in
metabolism-related pathways according to GO enrichment analysis;
downregulated genes were enriched in mitosis (Fig. 2D). Therefore, worse prognosis for
patients in the TEX-C2 cluster may be related to tumor immune- and
metabolism-associated pathways.
Tumor immune microenvironment analysis
of TEX-based clusters in patients with LUAD
To investigate the heterogeneity of the tumor immune
microenvironment between clusters, tumor immune cell infiltration
was calculated using the CIBERSORT algorithm (Fig. 3A). Compared with the TEX-C1 cluster,
the TEX-C2 cluster had a significantly higher abundance of M0 and
M2 macrophage cells, while the abundance of CD4 memory T and B
cells was lower. PD-L1 is used as an indicator to predict responses
to immunotherapy (46). The present
study investigated the association between PD-L1 and TEX in LUAD;
expression of PD-L1 in the TEX-C2 cluster was significantly higher
than that in the TEX-C1 cluster (Fig.
3B). Moreover, PD-L1 expression was positively correlated with
the expression of TEX-related genes such as BANK1, CCL2, CD79B,
CXCL1 and CXCL6 (Fig. 3C). TIDE
method was used to predict response to ICB. The TIDE score of the
TEX-C2 cluster was significantly higher than that of the TEX-C1
cluster, suggesting this cluster had a poor ICB response and
shorter survival time following ICB treatment (Fig. 3D).
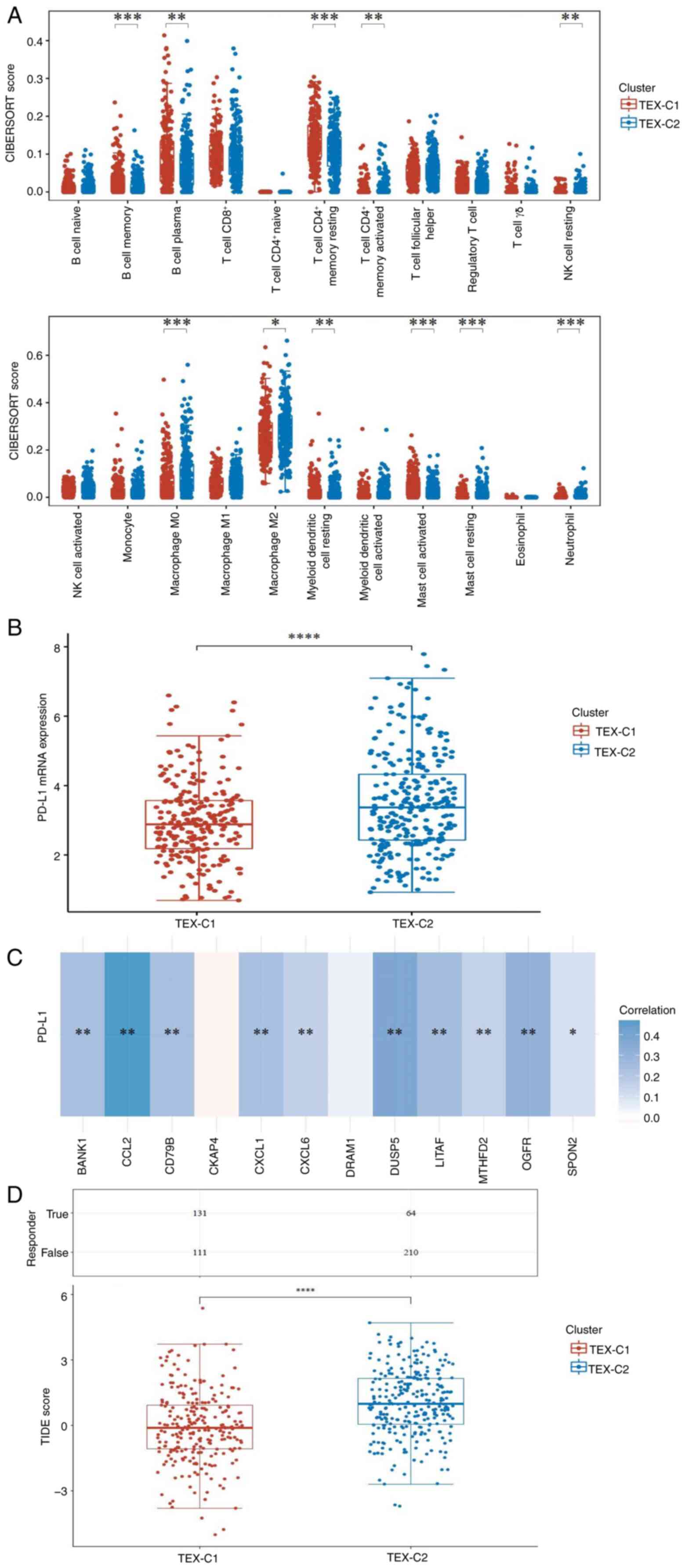 | Figure 3.Tumor immune microenvironment
analysis of TEX-based clusters in patients with lung
adenocarcinoma. (A) Infiltration of immune cells between TEX
clusters was analyzed using the CIBERSORT algorithm. (B) mRNA
expression of PD-L1 between TEX clusters. (C) Heatmap of
correlation between PD-L1 and TEX-related genes. Blue, positive;
red, negative; the darker the color, the stronger the association.
(D) TIDE scores in TEX clusters. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001. TEX-C, T cell exhaustion
cluster; TIDE, Tumor Immune Dysfunction and Exclusion; CD79B, B
cell Antigen Receptor Complex-Associated Protein Beta Chain; DUSP,
Dual Specificity Phosphatase; MTHFD, Methylenetetrahydrofolate
Dehydrogenase; OGFR, Opioid Growth Factor Receptor; SPON, Spondin;
BANK, B Cell Scaffold Protein With Ankyrin Repeats; CXCL, C-X-C
Motif Chemokine Ligand; CCL, C-C motif chemokine ligand; DRAM, DNA
Damage Regulated Autophagy Modulator; LITAF, Lipopolysaccharide
Induced TNF Factor; PD-L1, Programmed Cell Death 1 Ligand 1; NK,
natural killer cell. |
Construction of the prognostic
TEX-related gene signature in patients with LUAD
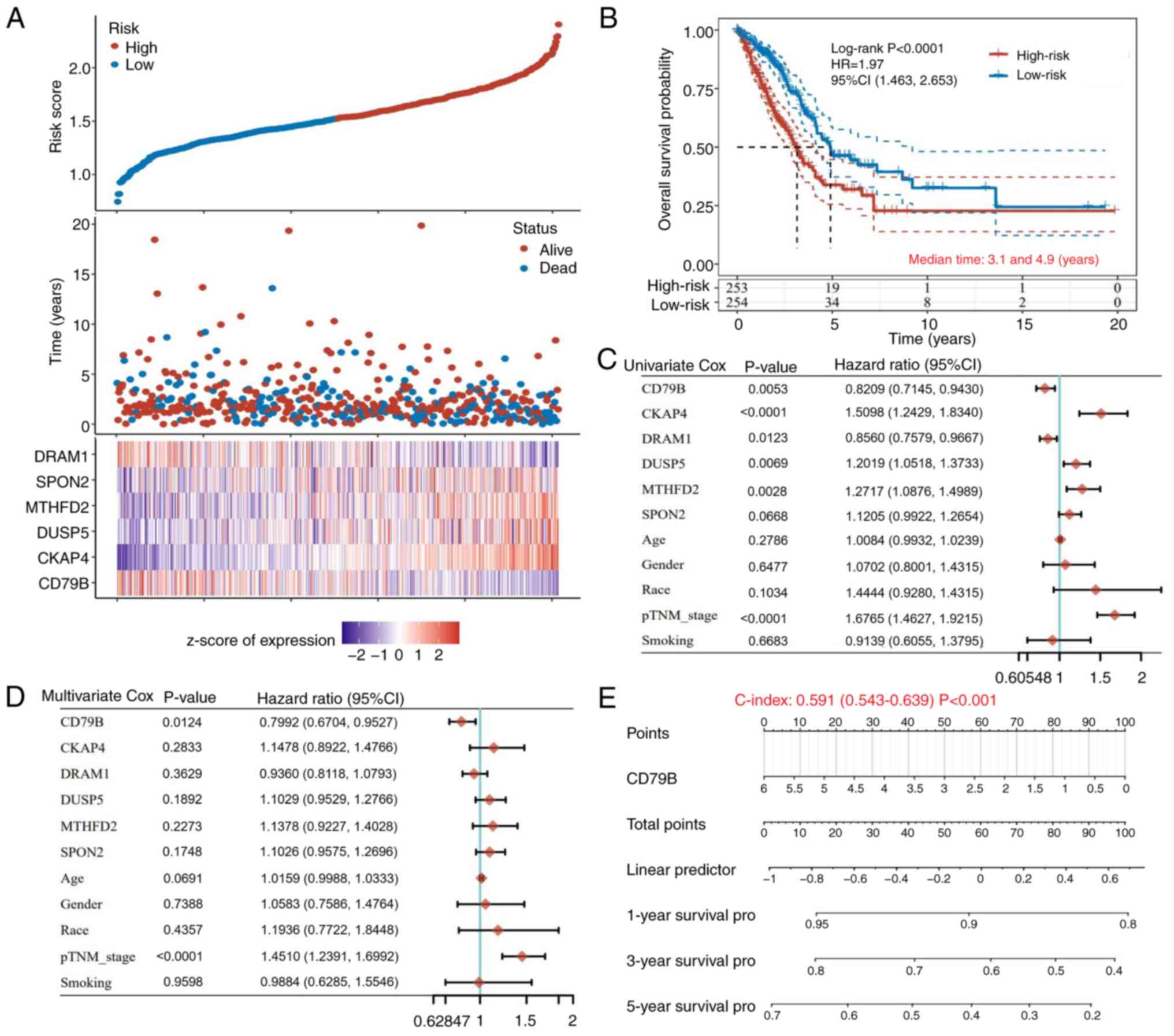
A risk prediction model for patients with LUAD was
constructed based on TEX-related genes and prognosis-related genes
were selected using the LASSO algorithm combined with 10-fold
cross-validation for further analysis in the best subset
regression. A total of six genes (CD79B, CKAP4, DUSP5, MTHFD2,
SPON2 and DRAM1) were identified as the best candidates, with a λ
value of 0.027. The risk score for prognosis prediction was
calculated as follows: Risk score=(−0.0863) × CD79B + (0.2314) ×
CKAP4 + (0.068) × DUSP5 + (0.0249) × MTHFD2 + (0.0203) × SPON2 +
(−0.0694) × DRAM1. The best subset regression model and coefficient
and deviance profiles are shown in Figs. 4A and S2A and B. Patients were divided into low-
and high-risk groups using median risk score (1.487) as the cutoff
value. Kaplan-Meier survival curves indicated that the median OS of
the low-risk group was longer than that of the high-risk group
(Fig. 4B). ROC analysis showed that
the 1-, 3- and 5-year prediction accuracies of the TEX-related gene
prediction model were 0.695, 0.668 and 0.645, respectively
(Fig. S2C). Cox analysis was
performed to analyze the association between CD79B, CKAP4, DUSP5,
MTHFD2, SPON2, DRAM1 and clinicopathological factors such as age,
sex, ethnicity and grade in the OS of patients with LUAD.
Univariate prognostic analysis indicated that CD79B, CKAP4, DUSP5,
DRAM1, MTHFD2 and p-TNM grades were associated with OS in patients
with LUAD (Fig. 4C). Multivariate
prognostic analysis further demonstrated that CD79B and pTNM grade
were reliable independent prognostic factors for predicting the
prognosis of patients with LUAD. In survival analysis, CD79B
indicated a decreased risk while pTNM grade indicated an increased
risk (Fig. 4D). CD79B was marked
with a scale indicating the value range; the length of the line
reflected contribution to the prognostic analysis; consistency
between the prognosis predicted by CD79B and the actual occurrence
was statistically significant. (Fig.
4E). Based on the score of these factors, the nomogram
predicted the 1-, 3- and 5-year OS in patients with LUAD (Fig. S2D).
Prognosis analysis between CD79B
expression and clinical data in patients with LUAD
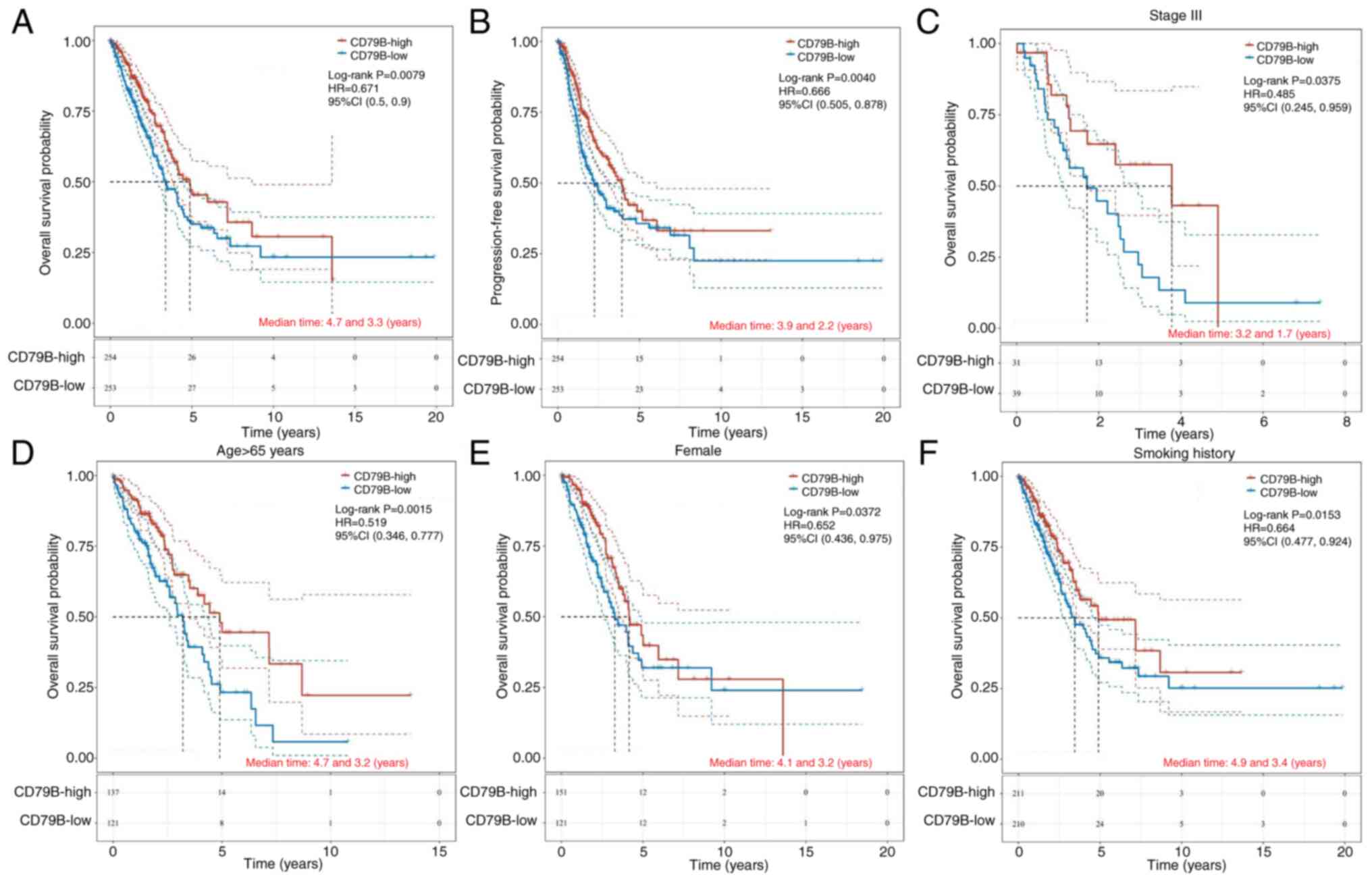
To confirm the prognostic role of CD79B in LUAD,
TCGA-LUAD patients were divided into a CD79B-low and a -high group
based on the median CD79B mRNA expression (2.391). Low CD79B
expression in patients with LUAD was associated with shorter OS and
PFS, compared with those with high expression of CD79B (Fig. 5A and B). Survival analysis showed
that patients with higher CD79B expression had better prognosis in
advanced LUAD (stage III; Fig. 5C),
while there was no significant difference in the prognostic effect
of CD79B in early-stage lung cancer (stages I and II; Fig. S3A). Similarly, elderly patients
(aged ≥65 years) with high CD79B expression had longer OS, compared
with low CD79B expression (Fig.
5D), while there was no significant difference in the
prognostic role of CD79B for patients <65 years (Fig. S3B). Female patients with high CD79B
expression had better prognosis than male patients, however, there
was no significant difference in prognosis among male patients
based on CD79B levels (P=0.109; Fig.
S3C). High expression of CD79B in smokers indicated better
prognosis (Fig. 5F), however, there
was no significant difference in prognosis for non-smokers based on
CD79B levels (Fig. S3D). These
results suggested that CD79B can be a potential prognostic
biomarker for LUAD, especially in advanced cases, elderly
individuals, female patients and those with a history of
smoking.
Bioinformatics and tumor immune
microenvironment analysis of CD79B expression in patients with
LUAD
Tumor proliferation signature and G2M checkpoint
were negatively associated with CD79B expression by Spearman's
correlation analysis based on the Gene Set Variation Analysis
(Fig. S4A). However, the p53
pathway, apoptosis, inflammatory response and IL-10
anti-inflammatory signaling pathway were positively correlated with
CD79B expression (Fig. S4A). The
IL-10 signaling pathway inhibits activation of pro-inflammatory
macrophages by downregulating mTOR and promoting mitochondrial
autophagy (47). IL-10 is secreted
by Tregs, which synergistically promote TEX in tumors by regulating
the expression of numerous inhibitory receptors (48). In KEGG enrichment analysis, genes
upregulated by CD79B expression were associated with immune-related
pathways such as ‘T cell receptor signaling pathway’, ‘B cell
receptor signaling pathway’, ‘Th1 and Th2 cell differentiation’,
‘Th17 cell differentiation’, ‘PD-L1 expression and PD-1’ and
‘primary immunodeficiency’ (Fig.
S4B). GO analysis of CD79B expression demonstrated enrichment
of innate and adaptive immune-related pathways, including
‘mononuclear cell proliferation’ and ‘leukocyte proliferation’ as
well as ‘T cell activation’, ‘T cell proliferation’ and ‘B cell
activation’ (Fig. S4C). The
protein-protein interaction network of CD79B was obtained from the
STRING database. Most of the proteins interacting with CD79B were B
cell markers (CD19 and CD79A) as well as components of B cell
receptor (BCR)-dependent kinase signaling pathways (Spleen
Associated Tyrosine Kinase and Bruton Tyrosine Kinase, BTK), which
are common in diffuse large B cell lymphoma (DLBCL; Fig. S4D) (49). These results indicated that CD79B
may affect prognosis in patients with LUAD patients through
immune-related pathways.
The correlation between CD79B expression and immune
cell levels in patients with LUAD was analyzed; six types of immune
cells (B and CD4+ and CD8+ T cells,
neutrophils, macrophages and dendritic cells) were all positively
correlated with CD79B expression (Fig.
6A). CIBERSORT algorithm was used to investigate the
association between CD79B expression and selected immune cells to
determine their effect on the tumor microenvironment. Compared with
the CD79B-low group, there was more infiltration of CD8+
T cells and M1 macrophages (classically activated type 1,
pro-inflammatory type), but less infiltration of M0 (the
undifferentiated) and M2 macrophages (the alternatively activated
type 2, anti-inflammatory type), which may be relevant to the
inhibition of TEX (Fig. 6B). In
addition, the association between CD79B and immune markers of
infiltrating immune cells in LUAD was investigated. The expression
of CD79B in LUAD was strongly correlated with innate immune cell
markers, including monocyte [CD14, CD86 and Fc γ receptor IIIa
(FCGR3A)], TAM (CD68, CCL2 and CCL5), M1 [CXCL10 and Tumor Necrosis
Factor (TNF)] and M2 macrophage [Mannose Receptor C-Type 1 (MRC1)
and CD163], neutrophil [Integrin Subunit Alpha M (ITGAM) and C-C
Motif Chemokine Receptor 7 (CCR7)] and natural killer [Killer Cell
Immunoglobulin Like Receptor, Two Ig Domains And long cytoplasmic
tail 1 (KIR2DL1), KIR2DL3, KIR2DL4, KIR3DL1, KIR3DL2 and KIR2DS4]
and dendritic cell markers [Major Histocompatibility Complex, Class
II, DP Beta 1 (HLA-DPB1), HLA-DQB1, HLA-DRA, HLA-DPA1 and CD1C;
Table II]. The CD79B expression
was significantly correlated with adaptive immunity cell markers,
including CD3D, CD3E and CD2 of T cells, CD19, Membrane Spanning
4-Domains A1 (MS4A1) and Fc Epsilon Receptor II (FCER2) of B cells,
T-Box Transcription Factor 21 (TBX21), Signal Transducer And
Activator Of Transcription 4 (STAT4), STAT1 and Interferon Gamma
(IFNG) of T helper 1 (Th1) cells, Trans-Acting T-Cell-Specific
Transcription Factor GATA-3 (GATA3), STAT6, STAT5A and Interleukin
13 (IL13) of Th2 cells, B Cell CLL/Lymphoma 6 (BCL6), IL21,
Inducible T Cell Costimulator (ICOS) and CXCL13 of T follicular
helper (Tfh) cells, STAT3 and IL17A of Th17 cells, as well as
Forkhead Box P3 (FOXP3), CCR8, STAT5B, Transforming Growth Factor
Beta 1 (TGFB1) and Interleukin 2 Receptor Subunit Alpha (IL2RA) of
Tregs (Table III). The expression
of CD79B was significantly correlated with gene markers for both
innate and adaptive immune cells; most correlations were positive,
CD79B negatively correlated with BCL6, STAT6 and STAT3 (Fig. 6C and D). mRNA expression of eight
immune checkpoint genes was significantly increased in the
CD79B-high group (Fig. 6E). Based
on these results, the high expression of CD79B was closely
associated with immune cell infiltration in LUAD, which may serve a
key role in reprogramming T cells from TEX states.
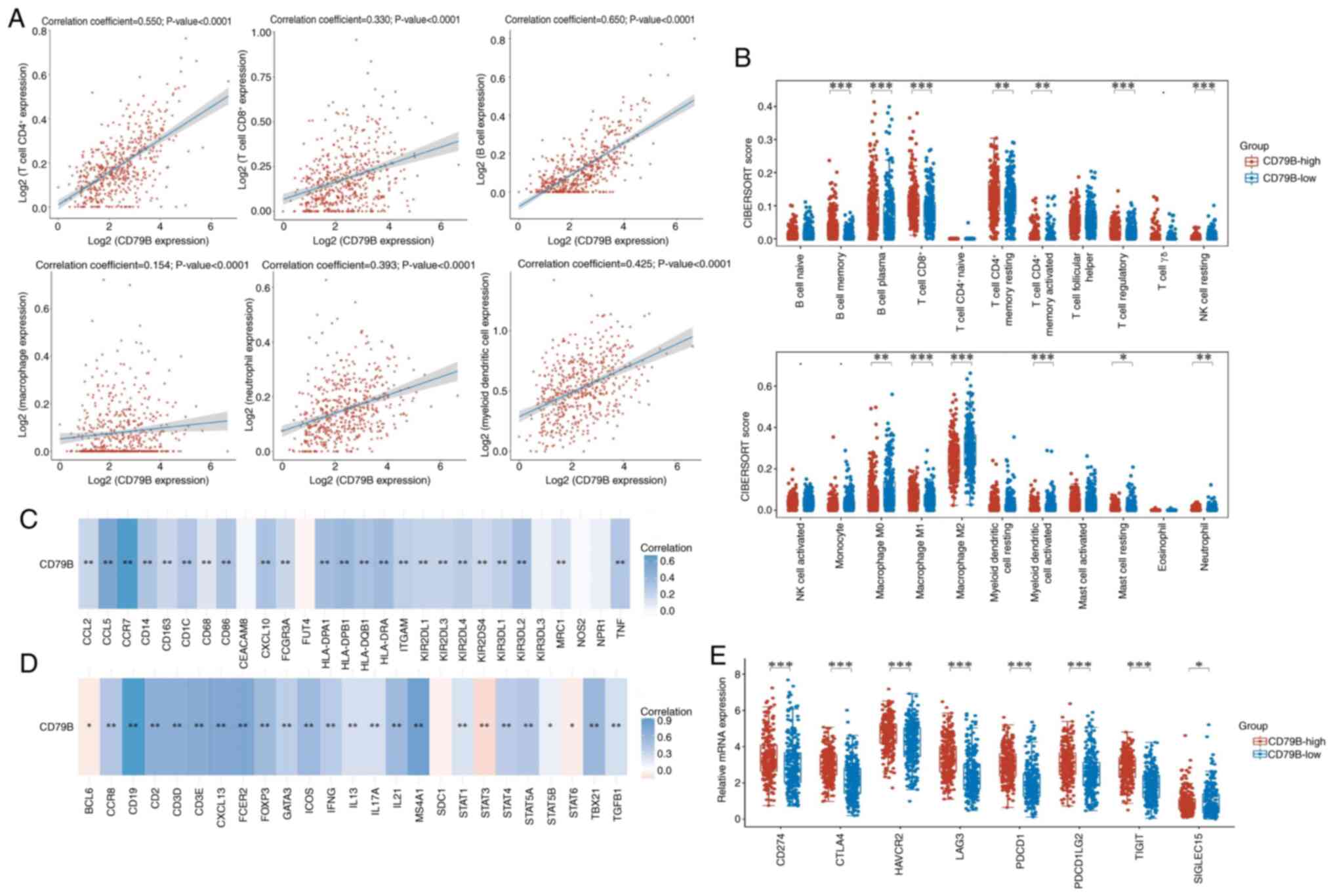 | Figure 6.Tumor immune microenvironment
analysis of CD79B expression in patients with lung adenocarcinoma.
(A) Correlation between CD79B expression and immune cell levels was
analyzed using Spearman's correlation. (B) Infiltration of immune
cells between CD79B-low and -high groups was analyzed using the
CIBERSORT algorithm. Correlation between CD79B and markers of (C)
innate and (D) adaptive immune cells. (E) mRNA expression levels of
immune checkpoint genes between CD79B-low and -high groups.
*P<0.05, **P<0.01, ***P<0.001. CD79B, B cell Antigen
Receptor Complex-Associated Protein Beta Chain; NK, natural killer
cell; CTLA, Cytotoxic T-Lymphocyte Associated Protein 4; HAVCR,
Hepatitis A Virus cellular receptor 1; LAG, Lymphocyte Activating
3; PDCD1LG2, Programmed Cell Death 1 Ligand 2; TIGIT, T Cell
Immunoreceptor with Ig and ITIM Domains; SIGLEC, Sialic Acid
Binding Ig Like Lectin. |
 | Table II.Correlation analysis between B cell
Antigen Receptor Complex-Associated Protein Beta Chain and markers
of innate immunity cells. |
Table II.
Correlation analysis between B cell
Antigen Receptor Complex-Associated Protein Beta Chain and markers
of innate immunity cells.
| Cell | Gene markers | Cor | P-value |
|---|
| Monocyte | CD14 | 0.306 | <0.001 |
|
| CD86 | 0.313 | <0.001 |
|
| FCGR3A | 0.192 | <0.001 |
| TAM | CD68 | 0.124 | 0.005 |
|
| CCL2 | 0.200 | <0.001 |
|
| CCL5 | 0.544 | <0.001 |
| M1 macrophage | NOS2 | 0.029 | 0.515 |
|
| CXCL10 | 0.319 | <0.001 |
|
| TNF | 0.300 | <0.001 |
| M2 macrophage | MRC1 | 0.145 | <0.001 |
|
| CD163 | 0.192 | <0.001 |
| Neutrophils | CEACAM8 | 0.042 | 0.338 |
|
| ITGAM | 0.240 | <0.001 |
|
| CCR7 | 0.666 | <0.001 |
|
| FUT4 | −0.342 | 0.438 |
| Natural killer | KIR2DL1 | 0.214 | <0.001 |
|
| KIR2DL3 | 0.209 | <0.001 |
|
| KIR2DL4 | 0.250 | <0.001 |
|
| KIR3DL1 | 0.232 | <0.001 |
|
| KIR3DL2 | 0.353 | <0.001 |
|
| KIR3DL3 | 0.063 | 0.154 |
|
| KIR2DS4 | 0.185 | <0.001 |
| Dendritic | HLA-DPB1 | 0.394 | <0.001 |
|
| HLA-DQB1 | 0.324 | <0.001 |
|
| HLA-DRA | 0.367 | <0.001 |
|
| HLA-DPA1 | 0.349 | <0.001 |
|
| CD1C | 0.298 | <0.001 |
|
| NPR1 | 0.068 | 0.123 |
 | Table III.Correlation analysis between CD79B
and markers of adaptive immunity cells. |
Table III.
Correlation analysis between CD79B
and markers of adaptive immunity cells.
| Cell | Gene markers | Cor | P-value |
|---|
| T | CD3D | 0.641 | <0.001 |
|
| CD3E | 0.672 | <0.001 |
|
| CD2 | 0.631 | <0.001 |
| B | CD19 | 0.902 | <0.001 |
|
| MS4A1 | 0.816 | <0.001 |
|
| SDC1 | −0.075 | 0.088 |
|
| FCER2 | 0.722 | <0.001 |
| Th1 | TBX21 | 0.541 | <0.001 |
|
| STAT4 | 0.346 | <0.001 |
|
| STAT1 | 0.187 | <0.001 |
|
| IFNG | 0.357 | <0.001 |
| Th2 | GATA3 | 0.334 | <0.001 |
|
| STAT6 | −0.090 | 0.041 |
|
| STAT5A | 0.378 | <0.001 |
|
| IL13 | 0.197 | <0.001 |
| Tfh | BCL6 | −0.098 | 0.026 |
|
| IL21 | 0.516 | <0.001 |
|
| ICOS | 0.521 | <0.001 |
|
| CXCL13 | 0.701 | <0.001 |
| Th17 | STAT3 | −0.179 | <0.001 |
|
| IL17A | 0.208 | <0.001 |
| Regulatory T | FOXP3 | 0.515 | <0.001 |
|
| CCR8 | 0.397 | <0.001 |
|
| STAT5B | 0.095 | 0.031 |
|
| TGFB1 | 0.212 |
|
|
| IL2RA | 0.369 | <0.001 |
Survival analysis between CD79B
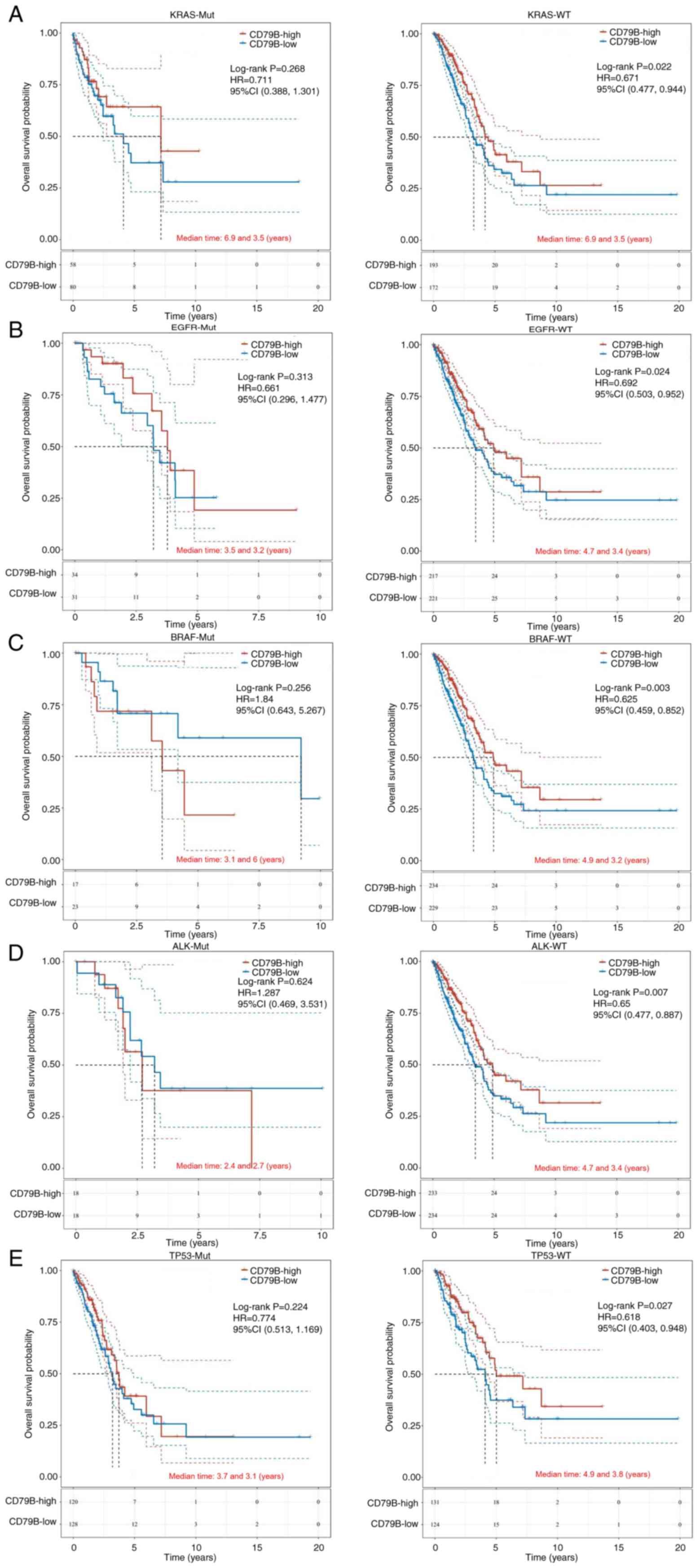
expression and driver gene mutations in patients with LUAD
MAF files from TCGA database were used to show the
mutation rate in LUAD patients, and the top 20 gene landscape of
driver genes with mutation frequency is illustrated in Fig. S5. Patients with LUAD with high
CD79B expression and wild-type KRAS, EGFR, BRAF, ALK or TP53 had
better OS compared with the CD79B-low group (Fig. 7). However, there was no significant
difference in OS between patients with KRAS, EGFR, BRAF, ALK or
TP53 mutations.
Prediction of therapeutic drug
sensitivity based on CD79B expression in patients with LUAD
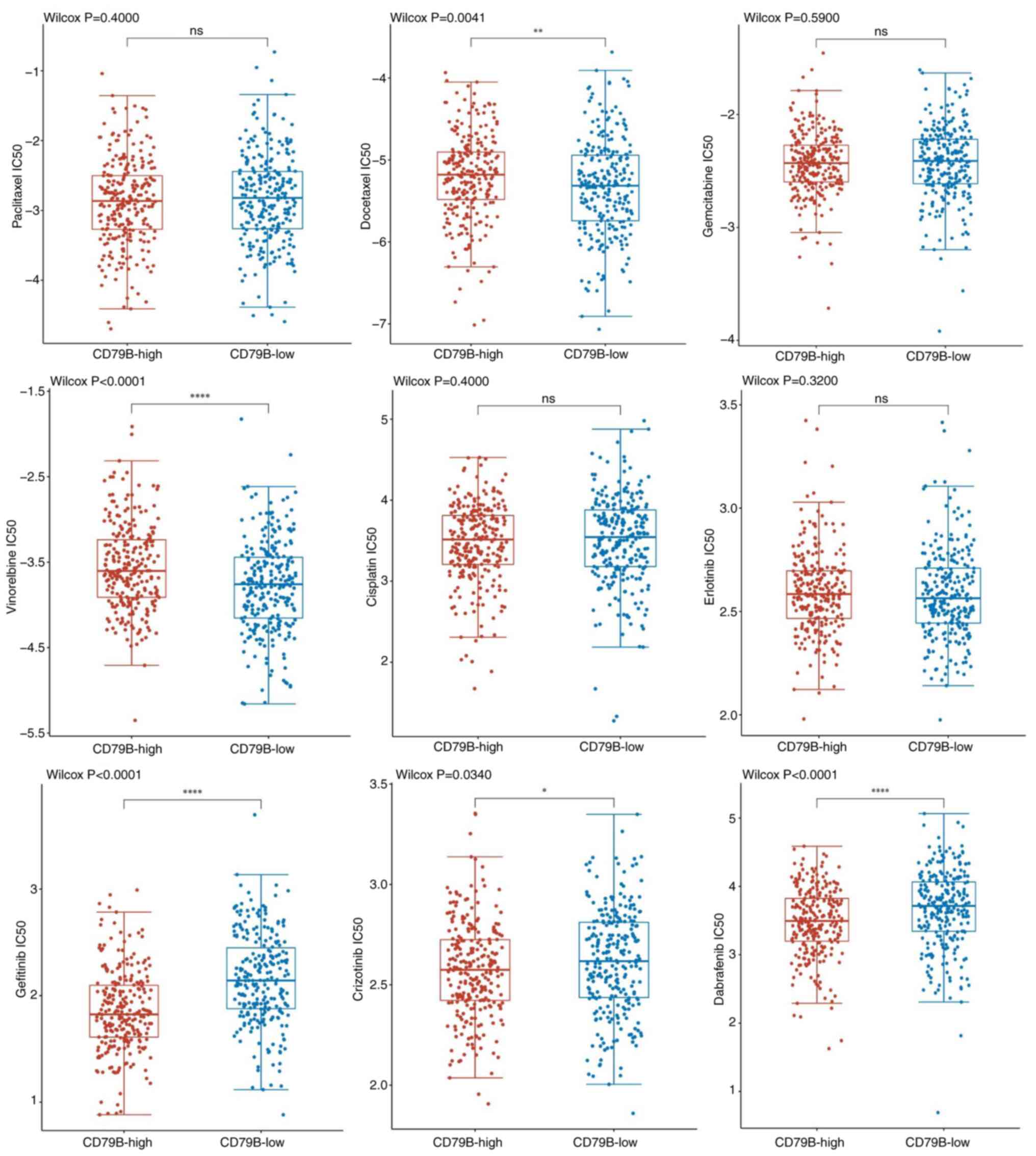
To explore the drug sensitivity between CD79B
expression subgroups, IC50 values of common chemotherapy and
molecular targeted therapeutic drugs for LUAD were calculated. The
CD79B-low group had higher sensitivity to chemotherapeutic drugs
such as docetaxel and vinorelbine, compared with CD79B-high group.
However, patients in the CD79B-high group had higher sensitivity to
targeted agents such as gefitinib, crizotinib and dabrafenib.
Additionally, there was no significant difference in the
sensitivity to paclitaxel, gemcitabine, cisplatin and erlotinib
(Fig. 8). Given the extensive
crosstalk between tumor and immune cells within the tumor
microenvironment (50), TEX-related
gene CD79B may be a potential target for combination therapy to
improve tumor immunotherapy.
CD79B inhibits proliferation,
migration and invasion and promotes apoptosis of LUAD cells, and
induces M1-like TAM polarization in vitro
qPCR and western blotting showed that the expression
of CD79B in LUAD cells was lower than that in normal lung
epithelial cells (Fig. 9A and B).
To investigate the role of CD79B in LUAD cells, A549 cells were
transfected with the CD79B-overexpression plasmid and verified by
qPCR and western blotting (Fig. 9C and
D). Compared with the control group, the viability of A549
cells overexpressing CD79B was significantly inhibited (Fig. 9E). Flow cytometry revealed that
overexpression of CD79B could induce apoptosis in A549 cells
(Fig. 9F). Scratch and Transwell
assay results indicated that the migration and invasion ability of
A549 cells overexpressing CD79B significantly weakened (Fig. 9G and H). M0 macrophage THP-1 cells
were co-culture with A549 cells overexpressing CD79B. Compared with
the vector group, macrophages co-cultured with A549 cells
overexpressing CD79B demonstrated upregulation of M1 marker genes
such as CD86, TNF-α and iNOS, while expression of M2 macrophage
marker genes such as CD206 and IL-10 was significantly decreased
(Fig. 9I). Thus, the TEX-related
gene CD79B is not only involved in proliferation, invasion,
migration and apoptosis of cancer cells but also contributed to
M1-like TAM polarization in LUAD.
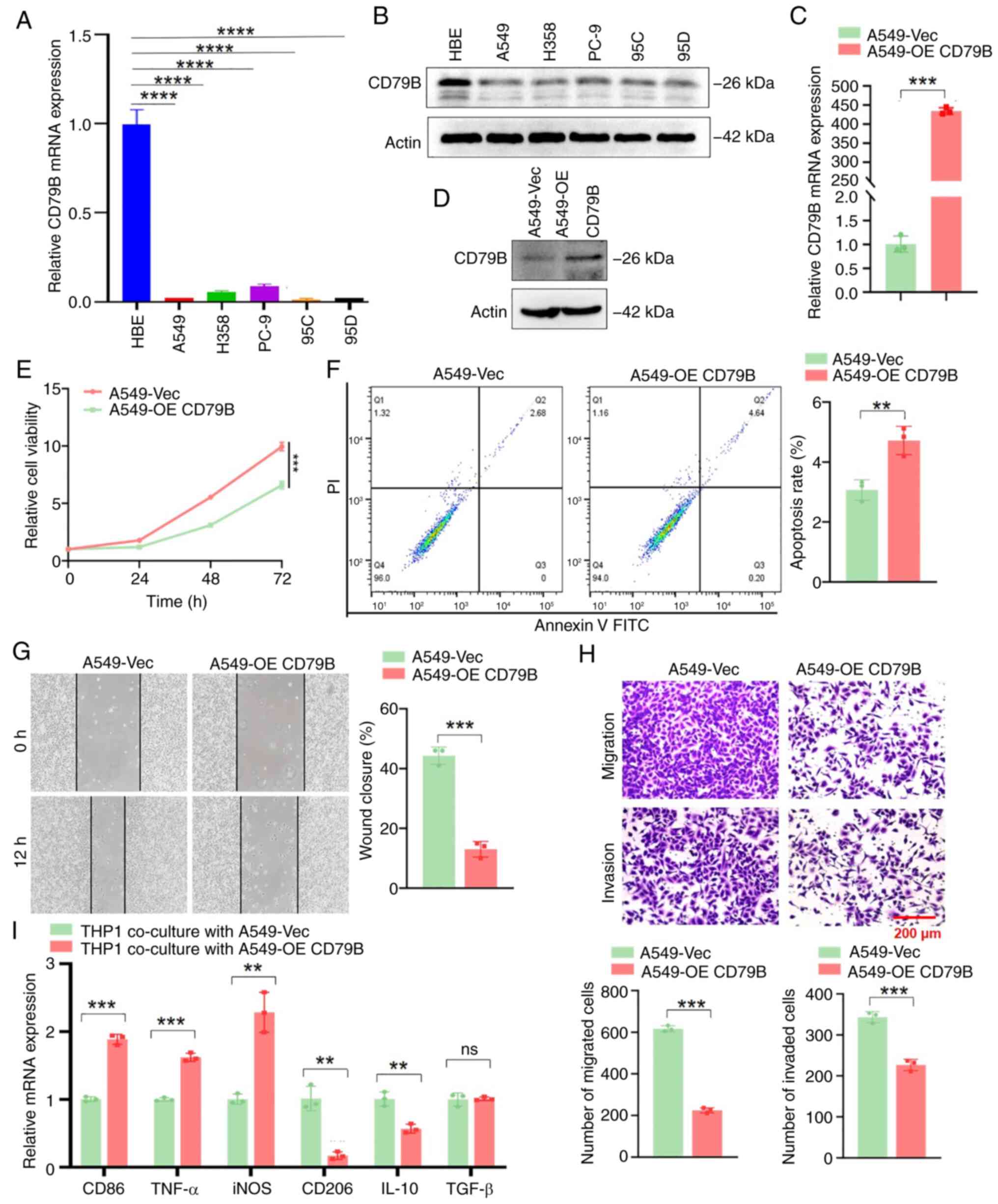 | Figure 9.CD79B inhibits proliferation,
migration and invasion and promotes apoptosis of LUAD cells, and
induces M1-like tumor-associated macrophage polarization in
vitro. mRNA and protein expression of CD79B were detected by
(A) qPCR and (B) western blotting in normal lung epithelial HBE and
LUAD cells. A549 cells were transfected with the CD79B-OE plasmid
and mRNA and protein expression of CD79B were detected by (C) qPCR
and (D) western blotting. (E) Cell viability was detected by Cell
Counting Kit 8 assay. (F) Flow cytometry was used to analyze cell
apoptosis. (G) Scratch assay was used to detect cell migration. (H)
Transwell assay was used to analyze cell migration and invasion
ability. (I) M0 macrophage THP-1 cells were co-culture with A549
cells, and the mRNA levels of M1/M2 marker genes were detected by
qPCR after 48 h. **P<0.01, ***P<0.001 and ****P<0.0001.
CD79B, B-Cell Antigen Receptor Complex-Associated Protein Beta
Chain; LUAD, lung adenocarcinoma; q, quantitative; OE,
Over-Expression; ns, not significant; Vec, Vector; iNOS. Nitric
Oxide Synthase, Inducible. |
Discussion
TEX, a state of effector T cell dysfunction, is a
notable obstacle to developing effective cancer immunotherapy
(51). Therefore, exploring the
mechanism of TEX and the effect of immunotherapy has become a
target in anti-tumor research (17,52).
The present study performed consensus clustering analysis of
TCGA-LUAD patients based on the expression of 12 TEX-related genes,
and the patients were divided into two clusters. Patients in the
TEX-C2 cluster had worse OS and PFS compared with those in the
TEX-C1 cluster. Upregulated genes in the TEX-C2 cluster were
enriched in metabolism-related pathways, while downregulated genes
were enriched in p53, NF-κB and IL-17 signaling pathways. TEX-C2
cluster had a poor ICB response and shorter survival following ICB
treatment. T cells are prone to exhaustion in an environment
characterized by mitochondrial capacity loss, glucose metabolism
defects and exposure to hypoxia, particularly intolerable levels of
reactive oxygen species (53). TEX
may further reduce metabolic fitness, resulting in anti-tumor
immune dysfunction and suppression (54). In acute myeloid leukemia with TP53
mutation, Tregs show gene expression signatures suggestive of
metabolic adaptation to their environment, whereas CTLs exhibit
features of exhaustion/dysfunction with stronger expression of T
cell Immunoglobulin and mucin domain 3 (55). Inhibition of NF-κB activity due to
loss of Interleukin 1 Receptor Associated Kinase 4 leads to T cell
dysfunction through production of immunosuppressive factors and
checkpoint ligands in pancreatic ductal adenocarcinoma (56). CD4+ T cells trigger the
recruitment of CD11b+Gr-1+ Myeloid-derived suppressor cells (MDSCs)
to promote terminal exhaustion of CD8+ T cells and tumor
progression in vivo by secreting IL-17 (57). ICB therapy is used to inhibit TEX by
blocking antibodies against immune checkpoints, including CTLA4,
PD-1 and its ligand PD-L1. However, the efficacy of ICB therapy
depends on the stage of T cell differentiation and is most
successful when antigen-specific T cells are functional,
proliferate and produce effective responses to maintain antitumor
function (58). Additionally, ICB
therapy alone does not reprogram T cells out of their exhausted
state (59). Current studies aim to
develop new strategies to target terminally exhausted T cells to
reactivate and restore their immune function for immunotherapy
(60,61). The present results indicate that the
poor prognosis of TEX-C2 cluster patients may be associated with
higher susceptibility to TEX due to tumor immune metabolism-related
pathways, which makes it more difficult to benefit from ICB
therapy.
A prognostic TEX-related gene signature model was
constructed to predict the prognosis of patients with LUAD. Among
the prognosis-related genes, CD79B was considered an independent
prognostic biomarker. Survival analysis showed that the high
expression of CD79B in patients with LUAD was associated with a
good prognosis, especially in advanced lung cancer, the elderly,
female patients and patients with a history of smoking. The results
of qPCR and western blot showed that CD79B was expressed at low
levels in LUAD cells. Previous studies have shown that CD79B
expression is also downregulated in cervical cancer tissues and
hepatocellular carcinoma cell lines (62,63).
CD79B, as a component of the B cell antigen receptor, has a notable
impact on immune synapse formation, antigen affinity and cell
migration and serves a crucial role in the maturation and
maintenance of B cells (64). CD79B
is highly expressed on the surface of cells in almost all patients
with non-Hodgkin lymphoma and chronic lymphocytic leukemia
(65). There generally exists a
positive correlation between the intensity of BCR signaling and the
expression of CD79B (66). Another
mechanism through which mutated CD79B increases BCR signaling is
its inability to properly activate the Src-family kinase Lyn, which
triggers a negative feedback loop-based inhibition of BCR signaling
(67). However, in tumor cells from
classical Hodgkin lymphoma, many B cell lineage-specific genes are
downregulated, including CD79B. Silencing of CD79B is associated
with its promoter methylation (68). In CD30-positive diffuse large B cell
lymphoma, low CD79B expression is related to paired box 5
interacting with complexes of histones that modify enhancers and
promoters for target genes, thereby influencing transcriptional
activation (69). In Epstein-Barr
Virus-negative Burkitt lymphoma cells, Epstein-Barr nuclear antigen
2 inhibits the expression of CD79B by binding its promoter and
interfering with the binding and transcriptional activation of
Early B cell factor 1 (70).
However, the molecular mechanisms underlying downregulation of
CD79B expression remain to be elucidated in LUAD. These may involve
complex alterations in gene regulatory networks, abnormal signaling
pathways and other factors such as promoter methylation.
In the present study, high CD79B expression was
associated with immune-related pathways. Moreover, CD79B was
strongly associated with a large number of immune cell populations
and most immune genetic markers, including immune
checkpoint-related genes, which also supported the poor OS of the
CD79B-low group. In addition, CD79B was positively correlated with
infiltrating CD8+ T cells and M1 macrophages and
negatively correlated with M0 and M2 macrophages. Regardless of the
expression of CD79B, targeted therapy could affect the prognosis of
LUAD patients with driver gene mutations. EGFR, ALK, BRAF, KRAS and
TP53 can provide targeted therapy for eligible patients with LUAD
(71). In vitro,
upregulation of CD79B could inhibit proliferation, migration and
invasion while promoting apoptosis of LUAD cells and inducing
M1-like TAM polarization. Expansion of IFN gene numbers can be
found at the CD79B locus (72). IFN
is associated with numerous B cell receptor pathway genes and genes
involved in adaptive immune responses. Among them, CD79B has also
been validated as an IFN-γ response gene in multiple sclerosis
(73). CD79B lymphocyte cells
produce notable amounts of IL-2, granulocyte-macrophage
colony-stimulating factor (CSF), IFN-γ, and IL-17A in response to
tumor cells in B cell lymphomas (74). M1 macrophages are generated
following stimulation by lipopolysaccharide and IFN-γ while M2
macrophages are generated by IL-4, IL-10 and IL-13 stimulation
(75). Therefore, it was
hypothesized that the upregulation of CD79B in tumor cells may
promote the production of cytokines such as IFN-γ, thereby
facilitating the polarization of M0 to M1 macrophages and
reprogramming TEX to inhibit malignant progression of LUAD cells.
Exhausted T cells could contribute to the recruitment and
polarization of macrophages by secreting CSF1 (76). TAMs present cancer cell antigens to
CD8+ T cells via interferon regulatory factor 8, leading
to the induction of PD-1 expression, a decrease in effector
cytokines and TEX (77). M0 TAMs
significantly increase the expression of PD-L2 via the PD-1
signaling pathway, which promotes the polarization of
immunosuppressive M2 type macrophages and decreases anti-tumor
effector T cells, leading to immune evasion and tumor promotion
(78). Xanthine oxidoreductase loss
in TAMs promotes M2-like polarization by increasing Isocitrate
Dehydrogenase [NADP(+)] 3 activity and α-ketoglutarate production.
It can also promote CD8+ TEX by upregulating
immunosuppressive metabolites, thereby exacerbating hepatocellular
carcinoma (HCC) progression (79).
Inhibition of 3-Oxoacid CoA-Transferase 1 (OXCT1) expression in
TAMs promotes reprogramming to an M1 phenotype via the
succinyl-H3K4me3-Arg1 axis, which may also decrease CD8+
TEX and enhance the anti-tumor effect of T cells (80). In breast cancer model mice,
Interleukin 1 Receptor Type 2 (IL1R2) blockade could decrease tumor
burden and prolong survival by decreasing PD-L1 transcriptional
activators and macrophage recruitment while inhibiting TAM
polarization and CD8+ TEX (81). Hence, TAM M2 polarization results in
CD8+ TEX, whereas TAM M1 polarization restores migration
and infiltration of CD8+ T cells. This suggests that TAM
M1 polarization may be an immunotherapeutic target for
reprogramming TEX.
TAM serves key functions in promoting a suppressive
tumor immune microenvironment and immune evasion, which limits the
treatment effects of ICB therapy in different types of cancer. By
using TAM modulators, the suppressive tumor immune microenvironment
may be reversed, thereby polarizing M2 into M1 TAMs releasing
IL-12, IL-6 and TNF-α, enhancing the activation of CD8+
T and natural killer cells, and possibly synergizing with
PD-1/PD-L1 checkpoint inhibition to enhance anti-cancer immune
responses (82). In glioblastoma
multiforme, combination therapy with rapamycin and
hydroxychloroquine downregulates the CD47-signal regulatory protein
α) axis in tumor cells, promoting the polarization of TAMs to a
pro-inflammatory M1-like phenotype and enhancing the therapeutic
response to anti-PD-1 antibody (83). Humanized modified macrophage-derived
microparticles loaded with methionine induce M2-like TAMs to
repolarize into the M1 phenotype, promote the infiltration of
CD8+ T cells into tumor tissue, and thereby enhance the
anti-cancer activity of anti-PD-1 antibody (84). In the treatment of oral cancer, the
combination of selective receptor tyrosine kinase inhibitor and ICB
immunotherapy increases the ratio of M1 to M2 macrophages in the
tumor microenvironment while promoting polarization of TAMs towards
an immunostimulatory state, thereby prolonging survival time
(85). The aforementioned studies
indicate that TAMs may serve as a potential target for combination
therapy to enhance the efficacy of tumor immunotherapy. Patients in
CD79B-high group had higher sensitivity to targeted agents such as
gefitinib, crizotinib and dabrafenib. The upregulation of CD79B
induced M1-like TAM polarization to inhibit malignant progression
of LUAD cells in vitro. It was hypothesized that patients
with high levels of CD79B may exhibit a better response to combined
use of immune checkpoint inhibitors and TAM polarizing agents.
However, there are certain limitations in the
present study. Firstly, the present retrospective study was based
on TCGA database, which may have potential selection bias.
Therefore, it is necessary to conduct prospective studies to verify
the conclusions. Secondly, there was a lack of molecular mechanism
studies to investigate the functional role of CD79B in TAM
polarization toward the M1 phenotype. Further in vitro and
in vivo experiments are required to elucidate the potential
regulatory mechanisms. Thirdly, given the absence of relevant data
from patients with LUAD, the results require further validation
through clinical studies involving larger sample sizes.
In summary, the present study performed clustering
analysis using TEX-related genes and developed a prognostic
TEX-related gene signature model to predict the prognosis of
patients with LUAD. Among the prognosis-related genes, CD79B was
considered an independent prognostic marker. Upregulation of CD79B
inhibited proliferation, migration and invasion while promoting
apoptosis in LUAD cells and inducing M1-like TAM polarization.
These results demonstrated the clinical value of TEX-related genes,
suggesting CD79B may be a potential prognostic indicator and
therapeutic target for improving tumor immunotherapy.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science
Foundation of Hunan Province, China (grant no. 2025JJ50551) and the
Fundamental Research Funds for the Central Universities of Central
South University (grant no. 2024ZZTS0912).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XW constructed figures, analyzed data, performed
experiments and wrote the manuscript. CQ performed experiments. YO
designed and performed the experiments. LY analyzed data, designed
and performed the experiments and edited the manuscript. WJ
conceived the study, designed and performed the experiments,
analyzed data and edited the manuscript. XW and CQ confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Succony L, Rassl DM, Barker AP, McCaughan
FM and Rintoul RC: Adenocarcinoma spectrum lesions of the lung:
Detection, pathology and treatment strategies. Cancer Treat Rev.
99:1022372021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hughes PE, Caenepeel S and Wu LC: Targeted
therapy and checkpoint immunotherapy combinations for the treatment
of cancer. Trends Immunol. 37:462–476. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smit EF and Baas P: Lung cancer in 2015:
Bypassing checkpoints, overcoming resistance, and honing in on new
targets. Nat Rev Clin Oncol. 13:75–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Garon EB, Rizvi NA, Hui R, Leighl N,
Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L,
et al: Pembrolizumab for the treatment of non-small-cell lung
cancer. N Engl J Med. 372:2018–2028. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parra ER, Zhang J, Duose DY,
Gonzalez-Kozlova E, Redman MW, Chen H, Manyam GC, Kumar G, Zhang J,
Song X, et al: Multi-omics analysis reveals immune features
associated with immunotherapy benefit in patients with squamous
cell lung cancer from phase III Lung-MAP S1400I trial. Clin Cancer
Res. 30:1655–1668. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mao X, Xu J, Wang W, Liang C, Hua J, Liu
J, Zhang B, Meng Q, Yu X and Shi S: Crosstalk between
cancer-associated fibroblasts and immune cells in the tumor
microenvironment: New findings and future perspectives. Mol Cancer.
20:1312021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yuan Z, Li Y, Zhang S, Wang X, Dou H, Yu
X, Zhang Z, Yang S and Xiao M: Extracellular matrix remodeling in
tumor progression and immune escape: From mechanisms to treatments.
Mol Cancer. 22:482023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McLane LM, Abdel-Hakeem MS and Wherry EJ:
CD8 T cell exhaustion during chronic viral infection and cancer.
Annu Rev Immunol. 37:457–495. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cai MC, Zhao X, Cao M, Ma P, Chen M, Wu J,
Jia C, He C, Fu Y, Tan L, et al: T-cell exhaustion interrelates
with immune cytolytic activity to shape the inflamed tumor
microenvironment. J Pathol. 251:147–159. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wherry EJ: T cell exhaustion. Nat Immunol.
12:492–499. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Z, Chen L, Chen H, Zhao J, Li K, Sun
J and Zhou M: Pan-cancer landscape of T-cell exhaustion
heterogeneity within the tumor microenvironment revealed a
progressive roadmap of hierarchical dysfunction associated with
prognosis and therapeutic efficacy. EBioMedicine. 83:104–207. 2022.
View Article : Google Scholar
|
|
15
|
Liu J, Li M, Wu J, Qi Q, Li Y, Wang S,
Liang S, Zhang Y, Zhu Z, Huang R, et al: Identification of ST3GAL5
as a prognostic biomarker correlating with CD8+ T cell exhaustion
in clear cell renal cell carcinoma. Front Immunol. 13:9796052022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu-Monette ZY, Zhang M, Li J and Young KH:
PD-1/PD-L1 Blockade: Have we found the key to unleash the antitumor
immune response? Front Immunol. 8:15972017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Z, Zhang Y, Ma N, Yang Y, Ma Y, Wang
F, Wang Y, Wei J, Chen H, Tartarone A, et al: Progenitor-like
exhausted SPRY1+CD8+ T cells potentiate responsiveness to
neoadjuvant PD-1 blockade in esophageal squamous cell carcinoma.
Cancer Cell. 41:1852–1870.e9. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tanoue K, Ohmura H, Uehara K, Ito M,
Yamaguchi K, Tsuchihashi K, Shinohara Y, Lu P, Tamura S, Shimokawa
H, et al: Spatial dynamics of CD39+CD8+ exhausted T cell reveal
tertiary lymphoid structures-mediated response to PD-1 blockade in
esophageal cancer. Nat Commun. 15:90332024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang H, Wu C, Tong X and Chen S: A
Biomimetic Metal-organic framework nanosystem modulates
immunosuppressive tumor microenvironment metabolism to amplify
immunotherapy. J Control Release. 353:727–737. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li W, Wu F, Zhao S, Shi P, Wang S and Cui
D: Correlation between PD-1/PD-L1 expression and polarization in
tumor-associated macrophages: A key player in tumor immunotherapy.
Cytokine Growth Factor Rev. 67:49–57. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang T, Hao L, Cui R, Liu H, Chen J, An J,
Qi S and Li Z: Identification of an immune prognostic 11-gene
signature for lung adenocarcinoma. PeerJ. 9:e107492021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu YH, Jin HQ and Liu HP: Identification
of T-cell exhaustion-related gene signature for predicting
prognosis in glioblastoma multiforme. J Cell Mol Med. 27:3503–3513.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szklarczyk D, Kirsch R, Koutrouli M,
Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT,
Pyysalo S, et al: The STRING database in 2023: Protein-protein
association networks and functional enrichment analyses for any
sequenced genome of interest. Nucleic Acids Res. 51:D638–D646.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wilkerson MD and Hayes DN:
ConsensusClusterPlus: A class discovery tool with confidence
assessments and item tracking. Bioinformatics. 26:1572–1573. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Balachandran VP, Gonen M, Smith JJ and
DeMatteo RP: Nomograms in oncology: More than meets the eye. Lancet
Oncol. 16:e173–e180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xi LJ, Guo ZY, Yang XK and Ping ZG:
Application of LASSO and its extended method in variable selection
of regression analysis. Zhonghua Yu Fang Yi Xue Za Zhi. 57:107–111.
2023.(In Chinese). PubMed/NCBI
|
|
27
|
Li Y, Lu F and Yin Y: Applying logistic
LASSO regression for the diagnosis of atypical Crohn's disease. Sci
Rep. 12:113402022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ashburner M, Ball CA, Blake JA. Botstein
D. Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene Ontology: Tool for the unification of biology. Nat
Genet. 25:25–29. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanehisa M, Goto S, Furumichi M, Tanabe M
and Hirakawa M: KEGG for representation and analysis of molecular
networks involving diseases and drugs. Nucleic Acids Res.
38:D355–D360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hung JH, Yang TH, Hu Z, Weng Z and DeLisi
C: Gene set enrichment analysis: Performance evaluation and usage
guidelines. Brief Bioinform. 13:281–291. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Edelman E, Porrello A, Guinney J,
Balakumaran B, Bild A, Febbo PG and Mukherjee S: Analysis of sample
set enrichment scores: Assaying the enrichment of sets of genes for
individual samples in genome-wide expression profiles.
Bioinformatics. 22:e108–e116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee E, Chuang HY, Kim JW, Ideker T and Lee
D: Inferring pathway activity toward precise disease
classification. PLoS Comput Biol. 4:e10002172008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tomfohr J, Lu J and Kepler TB: Pathway
level analysis of gene expression using singular value
decomposition. BMC Bioinformatics. 6:2252005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al: The
prognostic landscape of genes and infiltrating immune cells across
human cancers. Nat Med. 21:938–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hu W, Wang G, Chen Y, Yarmus LB, Liu B and
Wan Y: Coupled immune stratification and identification of
therapeutic candidates in patients with lung adenocarcinoma. Aging
(Albany NY). 12:16514–16538. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Geeleher P, Cox NJ and Huang RS: Clinical
drug response can be predicted using baseline gene expression
levels and in vitro drug sensitivity in cell lines. Genome Biol.
15:R472014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Geeleher P, Cox N and Huang RS:
pRRophetic: An R package for prediction of clinical
chemotherapeutic response from tumor gene expression levels. PLoS
One. 9:e1074682014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Suarez-Arnedo A, Torres Figueroa F,
Clavijo C, Arbeláez P, Cruz JC and Muñoz-Camargo C: An image J
plugin for the high throughput image analysis of in vitro scratch
wound healing assays. PLoS One. 15:e02325652020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yuan L, Wu X, Zhang L, Yang M, Wang X,
Huang W, Pan H, Wu Y, Huang J, Liang W, et al: SFTPA1 is a
potential prognostic biomarker correlated with immune cell
infltration and response to immunotherapy in lung adenocarcinoma.
Cancer Immunol Immunother. 71:399–415. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Korbecki J, Kupnicka P, Chlubek M, Gorący
J, Gutowska I and Baranowska-Bosiacka I: CXCR2 receptor: Regulation
of expression, signal transduction, and involvement in cancer. Int
J Mol Sci. 23:21682022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee D, Cho M, Kim E, Seo Y and Cha JH:
PD-L1: From cancer immunotherapy to therapeutic implications in
multiple disorders. Mol Ther. 32:4235–4255. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ip EWK, Hoshi N, Shouval DS, Snapper S and
Medzhitov R: Anti-inflammatory effect of IL-10 mediated by
metabolic reprogramming of macrophages. Science. 356:513–519. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sawant DV, Yano H, Chikina M, Zhang Q,
Liao M, Liu C, Callahan DJ, Sun Z, Sun T, Tabib T, et al: Adaptive
plasticity of IL-10+ and IL-35+ Treg cells cooperatively promotes
tumor T cell exhaustion. Nat Immunol. 20:724–735. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Phelan JD, Young RM, Webster DE, Roulland
S, Wright GW, Kasbekar M, Shaffer AL, Ceribelli M, Wang JQ, Schmitz
R, et al: A multiprotein supercomplex controlling oncogenic
signalling in lymphoma. Nature. 560:387–391. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shukla M and Sarkar RR: Differential
cellular communication in tumor immune microenvironment during
early and advanced stages of lung adenocarcinoma. Mol Genet
Genomics. 299:1002024. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chi X, Luo S, Ye P, Hwang WL, Cha JH, Yan
X and Yang WH: T-cell exhaustion and stemness in antitumor
immunity: Characteristics, mechanisms, and implications. Front
Immunol. 14:11047712023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pichler AC, Carrié N, Cuisinier M, Ghazali
S, Voisin A, Axisa PP, Tosolini M, Mazzotti C, Golec DP, Maheo S,
et al: TCR-independent CD137 (4-1BB) signaling promotes
CD8+-exhausted T cell proliferation and terminal differentiation.
Immunity. 56:1631–1648.e10. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Scharping NE, Rivadeneira DB, Menk AV,
Vignali PDA, Ford BR, Rittenhouse NL, Peralta R, Wang Y, Wang Y,
DePeaux K, et al: Mitochondrial stress induced by continuous
stimulation under hypoxia rapidly drives T cell exhaustion. Nat
Immunol. 22:205–215. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Watowich MB, Gilbert MR and Larion M: T
cell exhaustion in malignant gliomas. Trends Cancer. 9:270–292.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Abolhalaj M, Sincic V, Lilljebjörn H,
Sandén C, Aab A, Hägerbrand K, Ellmark P, Borrebaeck CAK, Fioretos
T and Lundberg K: Transcriptional profiling demonstrates altered
characteristics of CD8+ cytotoxic T-cells and regulatory T-cells in
TP53-mutated acute myeloid leukemia. Cancer Med. 11:3023–3032.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Somani VK, Zhang D, Dodhiawala PB, Lander
VE, Liu X, Kang LI, Chen HP, Knolhoff BL, Li L, Grierson PM, et al:
IRAK4 signaling drives resistance to checkpoint immunotherapy in
pancreatic ductal adenocarcinoma. Gastroenterology. 162:2047–2062.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kim BS, Kuen DS, Koh CH, Kim HD, Chang SH,
Kim S, Jeon YK, Park YJ, Choi G, Kim J, et al: Type 17 immunity
promotes the exhaustion of CD8+ T cells in cancer. J Immunother
Cancer. 9:e0026032021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Huang Y, Jia A, Wang Y and Liu G: CD8+ T
cell exhaustion in anti-tumour immunity: The new insights for
cancer immunotherapy. Immunology. 168:30–48. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zebley CC and Youngblood B: Mechanisms of
T cell exhaustion guiding next generation immunotherapy. Trends
Cancer. 8:726–734. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ford BR, Vignali PDA, Rittenhouse NL,
Scharping NE, Peralta R, Lontos K, Frisch AT, Delgoffe GM and
Poholek AC: Tumor microenvironmental signals reshape chromatin
landscapes to limit the functional potential of exhausted T cells.
Sci Immunol. 7:eabj91232022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang L, Zhang B, Li L, Ye Y, Wu Y, Yuan
Q, Xu W, Wen X, Guo X and Nian S: Novel targets for immunotherapy
associated with exhausted CD8 + T cells in cancer. J Cancer Res
Clin Oncol. 149:2243–2258. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Pu D, Liu D, Li C, Chen C, Che Y, Lv J,
Yang Y and Wang X: A novel ten-gene prognostic signature for
cervical cancer based on CD79B-related immunomodulators. Front
Genet. 13:9337982022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang Y, Yang Y, Zhao Z, Sun H, Luo D,
Huttad L, Zhang B and Han B: A new nomogram model for prognosis of
hepatocellular carcinoma based on novel gene signature that
regulates cross-talk between immune and tumor cells. BMC Cancer.
22:3792022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huse K, Bai B, Hilden VI, Bollum LK,
Våtsveen TK, Munthe LA, Smeland EB, Irish JM, Wälchli S and
Myklebust JH: Mechanism of CD79A and CD79B support for IgM+ B cell
fitness through BCR surface expression. J Immunol. 209:2042–2053.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tkachenko A, Kupcova K and Havranek O:
B-cell receptor signaling and beyond: The role of Igα (CD79a)/Igβ
(CD79b) in normal and malignant B cells. Int J Mol Sci. 25:102023.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ramesh S, Go M, Call ME and Call MJ: Deep
mutational scanning reveals transmembrane features governing
surface expression of the B cell antigen receptor. Front Immunol.
15:14267952024. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xie Z, Qin Y, Chen X, Yang S, Yang J, Gui
L, Liu P, He X, Zhou S, Zhang C, et al: Deciphering the prognostic
significance of MYD88 and CD79B mutations in diffuse large B-Cell
lymphoma: Insights into treatment outcomes. Target Oncol.
19:383–400. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ushmorov A, Leithäuser F, Sakk O,
Weinhaüsel A, Popov SW, Möller P and Wirth T: Epigenetic processes
play a major role in B-cell-specific gene silencing in classical
Hodgkin lymphoma. Blood. 107:2493–2500. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bullerwell CE, Robichaud PP, Deprez PML,
Joy AP, Wajnberg G, D'Souza D, Chacko S, Fournier S, Crapoulet N,
Barnett DA, et al: EBF1 drives hallmark B cell gene expression by
enabling the interaction of PAX5 with the MLL H3K4
methyltransferase complex. Sci Rep. 11:15372021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Khasnis S, Veenstra H, McClellan MJ,
Ojeniyi O, Wood CD and West MJ: Regulation of B cell receptor
signalling by Epstein-Barr virus nuclear antigens. Biochem J.
479:2395–2417. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Dantoing E, Piton N, Salaün M, Thiberville
L and Guisier F: Anti-PD1/PD-L1 immunotherapy for Non-small cell
lung cancer with actionable oncogenic driver mutations. Int J Mol
Sci. 22:62882021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu F, Wang T, Petit J, Forlenza M, Chen
X, Chen L, Zou J and Secombes CJ: Evolution of IFN subgroups in
bony fish-2. Analysis of subgroup appearance and expansion in
teleost fish with a focus on salmonids. Fish Shellfish Immunol.
98:564–573. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Khsheibun R, Paperna T, Volkowich A,
Lejbkowicz I, Avidan N and Miller A: Gene expression profiling of
the response to interferon beta in Epstein-Barr-transformed and
primary B cells of patients with multiple sclerosis. PLoS One.
9:e1023312014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chu F, Cao J, Liu J, Yang H, Davis TJ,
Kuang SQ, Cheng X, Zhang Z, Karri S, Vien LT, et al: Chimeric
antigen receptor T cells to target CD79b in B-cell lymphomas. J
Immunother Cancer. 11:e0075152023. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang S, Liu G, Li Y and Pan Y: Metabolic
reprogramming induces macrophage polarization in the tumor
microenvironment. Front Immunol. 13:8400292022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kersten K, Hu KH, Combes AJ, Samad B,
Harwin T, Ray A, Rao AA, Cai E, Marchuk K, Artichoker J, et al:
Spatiotemporal co-dependency between macrophages and exhausted CD8+
T cells in cancer. Cancer Cell. 40:624–638.e9. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Nixon BG, Kuo F, Ji L, Liu M, Capistrano
K, Do M, Franklin RA, Wu X, Kansler ER, Srivastava RM, et al:
Tumor-associated macrophages expressing the transcription factor
IRF8 promote T cell exhaustion in cancer. Immunity.
55:2044–2058.e5. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yang H, Zhang Q, Xu M, Wang L, Chen X,
Feng Y, Li Y, Zhang X, Cui W and Jia X: CCL2-CCR2 axis recruits
tumor associated macrophages to induce immune evasion through PD-1
signaling in esophageal carcinogenesis. Mol Cancer. 19:412020.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lu Y, Sun Q, Guan Q, Zhang Z, He Q, He J,
Ji Z, Tian W, Xu X, Liu Y, et al: The XOR-IDH3α axis controls
macrophage polarization in hepatocellular carcinoma. J Hepatol.
79:1172–1184. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhu CX, Yan K, Chen L, Huang RR, Bian ZH,
Wei HR, Gu XM, Zhao YY, Liu MC, Suo CX, et al: Targeting
OXCT1-mediated ketone metabolism reprograms macrophages to promote
antitumor immunity via CD8+ T cells in hepatocellular carcinoma. J
Hepatol. 81:690–703. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Xia J, Zhang L, Peng X, Tu J, Li S, He X,
Li F, Qiang J, Dong H, Deng Q, et al: IL1R2 blockade alleviates
immunosuppression and potentiates anti-PD-1 efficacy in
triple-negative breast cancer. Cancer Res. 84:2282–2296. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Peters S, Paz-Ares L, Herbst RS and Reck
M: Addressing CPI resistance in NSCLC: Targeting TAM receptors to
modulate the tumor microenvironment and future prospects. J
Immunother Cancer. 10:e0048632022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Hsu SPC, Chen YC, Chiang HC, Huang YC,
Huang CC, Wang HE, Wang YS and Chi KHL: Rapamycin and
hydroxychloroquine combination alters macrophage polarization and
sensitizes glioblastoma to immune checkpoint inhibitors. J
Neurooncol. 146:417–426. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Wei Z, Zhang X, Yong T, Bie N, Zhan G, Li
X, Liang Q, Li J, Yu J, Huang G, et al: Boosting anti-PD-1 therapy
with metformin-loaded macrophage-derived microparticles. Nat
Commun. 12:4402021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Oliva M, Chepeha D, Araujo DV, Diaz-Mejia
JJ, Olson P, Prawira A, Spreafico A, Bratman SV, Shek T, de Almeida
J, et al: Antitumor immune effects of preoperative sitravatinib and
nivolumab in oral cavity cancer: SNOW window-of-opportunity study.
J Immunother Cancer. 9:e0034762021. View Article : Google Scholar : PubMed/NCBI
|