Introduction
IFN-stimulated genes (ISGs) are a group of genes
upregulated in response to IFN stimulation (1). IFNs are cytokines classified into
three main types: Type I, II and III (2). Type I IFNs have a broad range of
biological functions including modulation of innate and adaptive
immune responses, anti-proliferative function and antiviral
activity (3). Type II IFN is IFN-γ,
which is primarily produced by immune cells (activated T cells,
natural killer cells and macrophages) and has been described as an
immunomodulatory cytokine, but it also shows responses to viruses,
bacteria, and parasites (4,5). Type III IFNs are uniquely chemotactic,
with signaling and function largely restricted to epithelial cells,
and these interferons are as key players in mucosal immunity
(6,7). Following viral infection, pathogenic
invasion or other stimuli, pattern recognition receptors(PRR) in
host cells detect pathogen-associated molecular patterns,
triggering an immune response. This induces IFN production, which
then binds to specific cell surface receptors and activates
intracellular signaling cascades, leading to the transcriptional
induction of ISGs (8).
ISG20 is an RNA exonuclease belonging to the Rex4
subfamily, a homolog of yeast RNA exonuclease 4 and a member of the
DEDDh exonuclease family, comprising four conserved acidic
residues, three aspartates (D), and one glutamate(E), distributed
among three separate sequence motifs (Exo I–III) (9,10) and
with a fifth conserved residue of histidine (H). It has the ability
to degrade single-stranded (ss)RNA and DNA, and encodes a 20 kDa
protein (11). Under physiological
conditions, ISG20 primarily contributes to the antiviral defense of
the host. It directly degrades viral RNA and modulates
intracellular antiviral signaling pathways, such as inducing type I
IFN responses, thereby inhibiting viral replication and spread
(12). In addition, ISG20 may
regulate immune cell functions and cytokine signaling (13). Previous advances have further
revealed the complex role of ISG20 in tumorigenesis, attracting
increasing interest (14–18).
A growing body of research has demonstrated that
ISG20 expression is significantly dysregulated in various types of
cancer (19–22). In clear cell renal cell carcinoma
(ccRCC), ISG20 is markedly upregulated at both the mRNA and protein
levels, and is positively associated with advanced clinical stage.
Elevated ISG20 levels are associated with worse overall survival
(OS) and disease-free survival (DFS), suggesting its potential as a
diagnostic and prognostic biomarker (23). In glioma, ISG20 mRNA expression is
also significantly upregulated compared with that in normal brain
tissue, with differential expression observed across clinical
subgroups. High ISG20 expression is associated with poor prognosis,
and immunohistochemical and immunofluorescence analyses have
revealed stronger expression in high-grade glioma, predominantly
localized to M2 macrophages (24).
Furthermore, aberrant ISG20 expression has been reported in other
malignancies, including cervical and oral cancer, hepatocellular
carcinoma, breast cancer and acute myeloid leukemia, where it may
be upregulated, associated with specific tumor-promoting factors or
predictive of unfavorable outcomes (19,25,26).
Although considerable progress has been made in
elucidating the role of ISG20 in cancer, the number of publications
remains limited compared with other ISGs (13,27,28).
Therefore, a comprehensive summary of the functions of ISG20 in
tumorigenesis is key. The present review outlines the historical
discovery of ISG20, and elaborates on its molecular structure and
physiological functions, as well as the role of ISG20 in cancer and
its therapeutic potential.
Discovery and molecular characterization of
ISG20
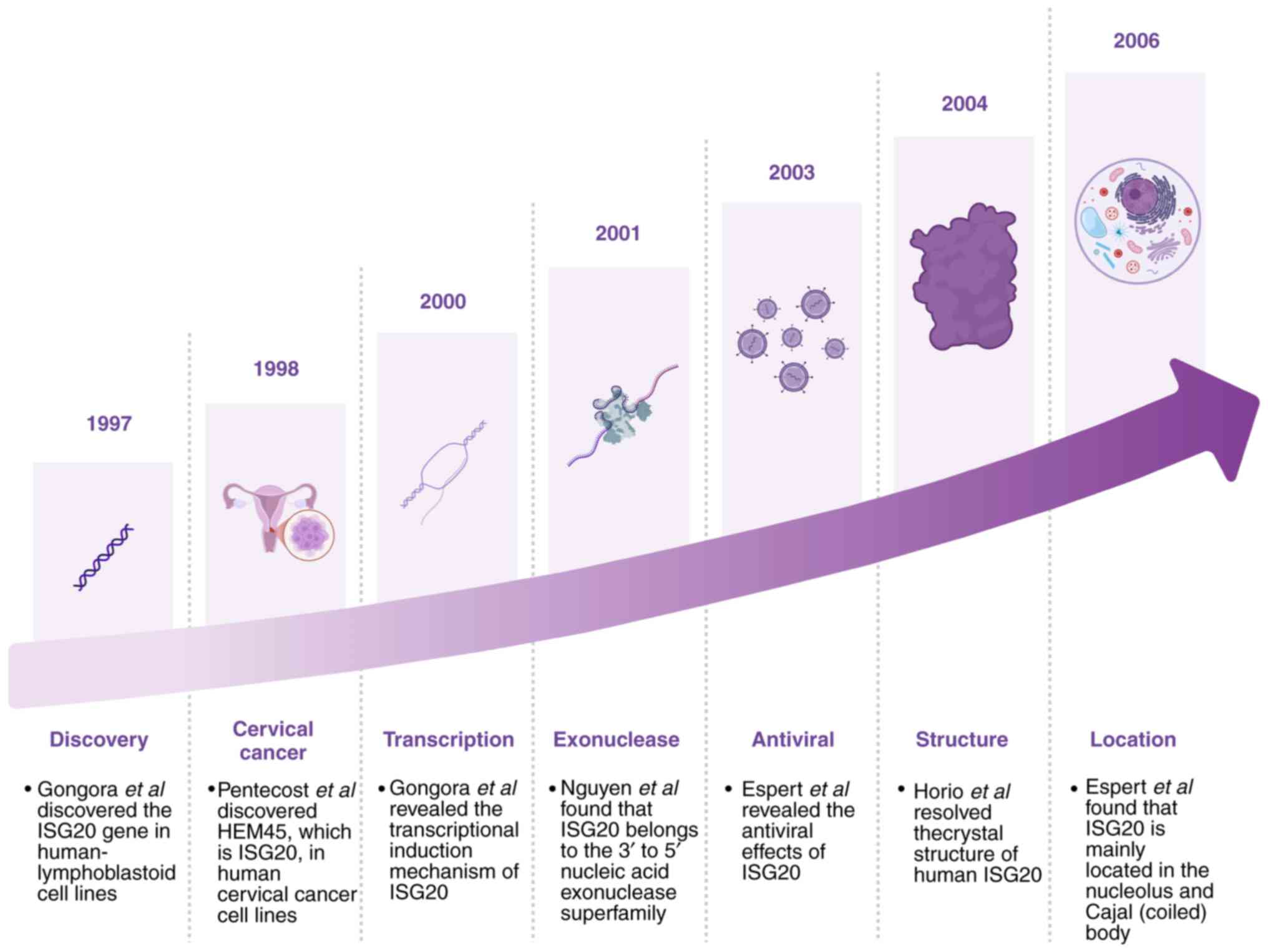
ISG20 was first identified in 1997 by Gongora et
al (29); this study employed
differential display techniques to screen IFN-regulated genes in
human Daudi lymphoblastoid cells treated with IFN-α/β (29). Among these genes, one was notably
inducible by both type I and II IFNs, and was designated as ISG20,
encoding a 20 kDa protein. In 1998, Pentecost (30) identified a transcript with low
baseline expression in multiple cell lines that was significantly
upregulated by estradiol in estrogen receptor (ER)-positive breast
cancer cells. This transcript, discovered via differential display
PCR in ER-transfected human cervical carcinoma cells (UP1), was
named HeLa estrogen-modulated 45 kDa band (HEM45). Subsequent
studies have confirmed that HEM45 and ISG20 are the same gene
product; as a result, ISG20 is also referred to as HEM45 (29,30).
In 2000, Gongora et al (31) demonstrated that ISG20 transcription
is induced by both type I (α/β) and II (γ) IFNs through a unique
IFN-stimulated response element located in its promoter, regulated
by IFN regulatory factor 1 (IRF1) (31). In 2001, Nguyen et al
(32) revealed that the ISG20
protein shares homology with members of the 3′-5; exonuclease
superfamily, including RNase T and D and the proofreading domain of
Escherichia coli DNA polymerase I. The catalytic site
consists of four conserved acidic residues arranged within three
characteristic exonuclease motifs, classifying it under the DEDDh
subfamily of 3′-5′exonucleases (32). In 2003, Espert et al
(33) revealed the antiviral
activity of ISG20; HeLa cells overexpressing ISG20 were resistant
to RNA viruses such as vesicular stomatitis virus (VSV), influenza
and encephalomyocarditis virus, even in the absence of IFN
treatment. This antiviral effect was attributed to the exonuclease
activity of ISG20, which also partially contributed to IFN-mediated
anti-VSV effects, demonstrating IFN-driven antiviral mechanisms
(33). ISG20 expression is directly
induced by synthetic double-stranded RNA (dsRNA) via activation of
NF-κB and IRF1; this induction is more rapid and robust than that
triggered by IFN (34). Moreover,
ISG20 translocates to the nuclear matrix upon induction. Its site
of action is different from the cytoplasmic functional region of
PKR (dsRNA-activated serine-threonine protein kinase, a member of
the IFN-induced antiviral signaling pathway, which together with
the 2′,5′-oligoadenylate synthetase/RNase L pathway constitutes the
core antiviral mechanism of the IFN system), suggesting its
involvement in the cell antiviral response, potentially through
aPKR-independent innate antiviral and anti-apoptotic pathway
(34). Horio et al (35) resolved the crystal structure of
human ISG20 in complex with two Mn2+ ions and uridine
5′-monophosphate (UMP) at a resolution of 1.9 Å. The structure
revealed marked similarity to DEDDh family DNases, suggesting a
shared catalytic mechanism. Unique residues within ISG20 may confer
its substrate preference for RNA, providing structural insights
into its antiviral function (35).
Espert et al (36) further
demonstrated, using immunofluorescence, that ISG20 primarily
localizes to the nucleolus and Cajal bodies, and associates with
complexes containing nuclear survival of motor neuron protein. As
its structure and regulatory mechanisms have become clearer,
investigations into the physiological roles of ISG20 under various
conditions have expanded (Fig. 1)
(37–39).
Structural features and physiological
functions of ISG20
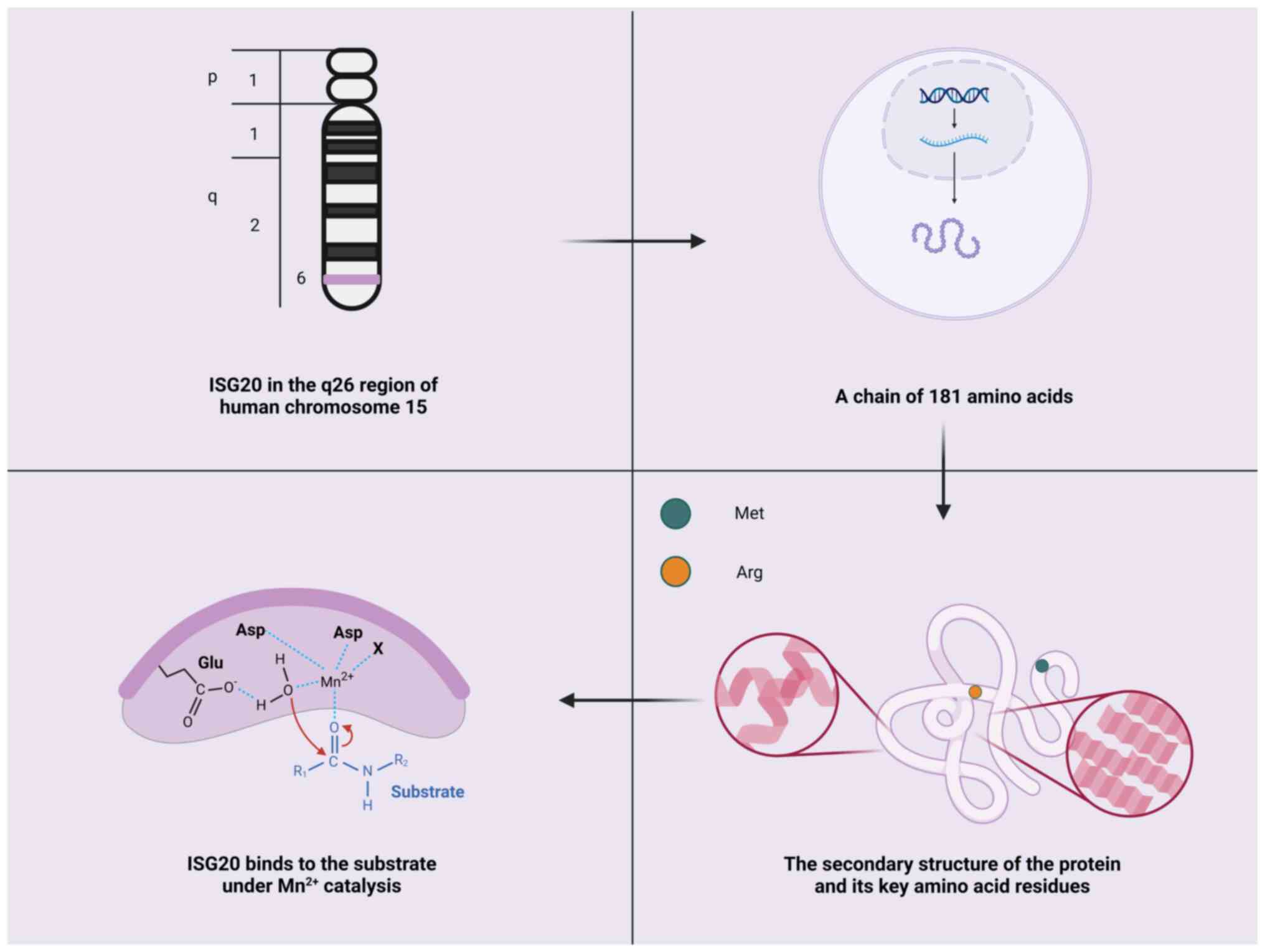
ISG20 is located on chromosome 15q26 in humans and
encodes a protein consisting of 181 amino acids with a molecular
weight of ~20.4 kDa (29). ISG20
belongs to the 3′-5′exonuclease superfamily (32). The catalytic site of this
superfamily consists of four conserved acidic residues
(Asp-Glu-Asp-Asp) distributed across three exonuclease motifs
(ExoI, ExoII and ExoIII) (10,40).
These residues coordinate two metal ions necessary for hydrolyzing
the terminal phosphodiester bond of RNA or DNA, thereby
facilitating an enzymatic reaction. Crystal structure analysis has
revealed that ISG20 contains a five-stranded β-sheet core flanked
by two clusters of α-helices (a3, a4 and a1, a2, a5, a7) (35). At the active site, specific amino
acid residues, such as Asp11, Glu13 and Asp154, participate in
coordinating metal ions (such as Mn2+), enabling
catalytic activity (35). In
vitro, ISG20 exhibits exonucleolytic activity against ssRNA and
ssDNA, with a marked preference for RNA substrates. This substrate
preference may be due to residues such as Met14 and Arg53, which
directly interact with the ribose 2′-OH group of UMP (35). However, the precise molecular
mechanism underlying this specificity remains to be elucidated
(Fig. 2).
Although purified ISG20 primarily cleaves ssRNA
in vitro, it may exhibit resistance to substrates with
3′structures. For example, Liu et al (41) found that the encapsidation signal
εRNA of hepatitis B virus (HBV) exhibits resistance to ISG20
(41). The εRNA is located near the
5′ end of the HBV pre-genomic RNA and features a unique secondary
structure, including a long 13-bp basal stem, a 6-nt bulge region
(initiator loop), an 11-bp upper stem and a terminal hexaloop
(42). It has been shown that ISG20
specifically binds the lower stem of εRNA, with binding stability
requiring a lower stem of >9 RNA bps (41). However, despite tight binding, ISG20
is unable to effectively cleave the εRNA substrate. Notably,
deletion of the C-terminal ExoIII motif of ISG20 results in
complete loss of RNA binding, suggesting that the ExoIII motif may
harbor an RNA-binding domain, or that the coordination of MnA by
Asp154 is key for RNA binding (41). In addition to its role in nucleic
acid degradation, ISG20 is involved in other key cellular
processes. ISG20 has been reported to mediate estrogen signaling,
serving a role in regulating cell proliferation and differentiation
(41). Its expression is
significantly upregulated in response to IFN or estrogen, further
highlighting its key role in hormone-regulated cell networks
(43–45).
The antiviral effects of ISG20 have been extensively
studied (12,45,47).
These include direct degradation of viral RNA, deamination of viral
DNA, IFN-induced and IFIT1 (IFN-induced protein with
tetratricopeptide repeats1)-mediated suppression of viral RNA
translation and non-IFN-induced inhibition of non-self RNA
translation (Fig. 3) (48). ISG20 exhibits RNase activity,
capable of degrading viral ssRNA and ssDNA. As aforementioned,
ISG20 can degrade HBV RNA by directly binding the ε-stem-loop
structure of viral RNA, thereby inhibiting HBV replication.
Additionally, Kang et al (49) studied the inhibitory mechanism of
ISG20 on bluetongue virus (BTV), and found that mutating Asp94 to
Gly in ISG20 to create a nuclease-deficient mutant significantly
decreased its inhibitory effect on BTV replication, suggesting that
ISG20 may directly degrade viral RNA through its exonuclease
activity during BTV infection (49). However, there remains controversy
regarding whether ISG20 directly degrades viral RNA and its
antiviral mechanism and specificity are not fully understood
(11,41,50).
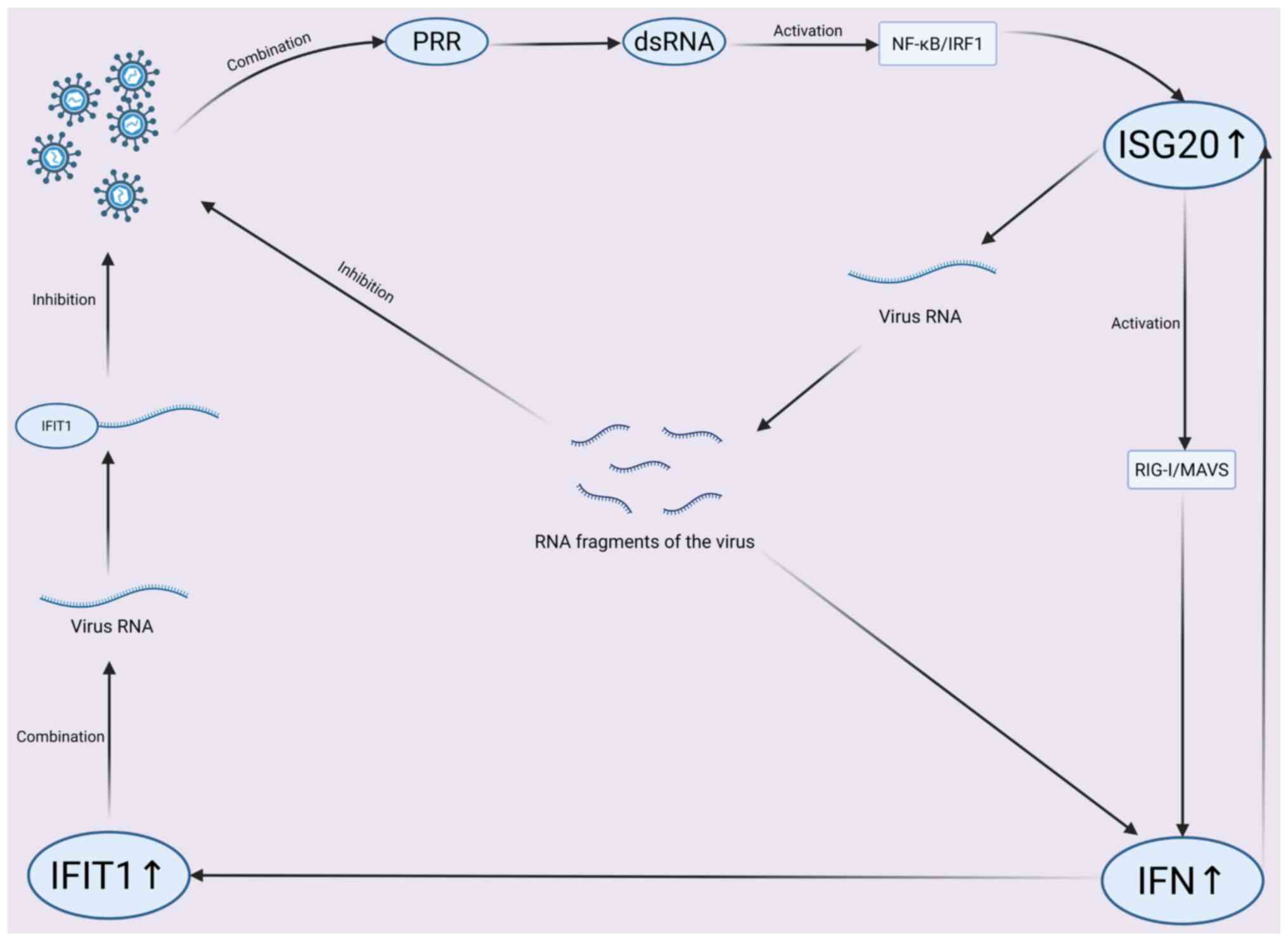 | Figure 3.Mechanistic pathway of ISG20
antiviral activity. Viral dsRNA is recognized by PRR, activating
NF-κB/IRF1 to upregulate ISG20. ISG20 degrades viral RNA and
activates RIG-I/MAVS for IFN production. IFN-induced IFIT1 binds
viral RNA, inhibiting replication and viral-PRR interaction. Viral
RNA fragments modulate the response, forming a coordinated innate
immune network. ISG, IFN-stimulated gene; IFIT, Interferon-induced
protein with tetratricopeptide repeats; IRF, IFN regulatory factor;
PRR, Pattern recognition receptor; ds, double-stranded; RIG-I,
Retinoic Acid-Inducible Gene I; MAVS, Mitochondrial Antiviral
Signaling Protein. |
Some studies have suggested that ISG20 may exert its
effects more through translational inhibition than RNA degradation
(12,46). For example, in mouse embryonic
fibroblasts, overexpression of mouse ISG20 indirectly suppresses
viral translation by inducing an IFN response and IFIT1, without
involving viral RNA degradation (12). However, ISG20 is an IFN regulatory
protein, and the mechanism by which it induces IFN, as well as how
it mediates suppression of a range of viruses, remains unclear.
Additionally, the source of the RNA (cellular or viral) targeted by
ISG20 during IFN induction has yet to be determined. Furthermore,
it has been demonstrated that the subcellular localization and
dynamic changes of ISG20 may influence its antiviral function
(36,46,51).
For example, under uninfected conditions, ISG20 is predominantly
localized in the nucleus, associated with nuclear structures such
as promyelocytic leukemia bodies, nucleoli and Cajal bodies, and
potentially involved in RNA processing and transcriptional
regulation (48). After viral
infection, a fraction of ISG20 translocates to processing bodies (P
bodies) in the cytoplasm, which serve a crucial role in RNA
degradation and translational regulation. The localization of ISG20
in P bodies may aid degradation of viral RNA or the suppression of
viral RNA translation, thereby contributing to its antiviral
activity (11).
ISG20 not only serves a key role in antiviral
responses but also participates in the immune regulation processes,
affecting numerous aspects of the immune system (52,53).
In innate immunity, ISG20 may serve as an important effector
molecule, participating in the responses of immune cells such as
macrophages and dendritic cells to pathogens (54). ISG20 serves a key role in
macrophages resisting Listeria infection, and in dendritic
cells responding to Mycobacterium tuberculosis and
Toxoplasma gondii infection, suggesting it may enhance
innate immune defense by regulating immune cell function (54,55).
ISG20 may also play a role in adaptive immunity. For example,
Rodríguez-Galán et al (56)
reported that the ISG20 family member ISG20L2 specifically binds
uridylated microRNAs (miRs), is upregulated in T lymphocytes under
T cell receptor and type I IFN stimulation, and participates in
regulating T cell function. Knockout of this gene may lead to
delayed T cell proliferation, increased apoptosis, abnormal
expression of activation-associated molecules, increased IL-2
secretion and impaired immunological synapse formation and
dynamics, indicating that ISG20 may influence T cell activation and
function, thereby indirectly affecting the adaptive immune response
(56).
The role of ISG20 in immune regulation may also be
associated with its impact on the interaction between immune and
tumor cells. In the tumor microenvironment (TME), the functional
state of immune cells markedly influences tumor initiation,
progression and the effectiveness of immunotherapy. Studies have
indicated that ISG20 may affect the immune balance in the TME by
regulating immune cell infiltration, modulating immune signaling
pathways and adjusting chemokines, thereby influencing tumor cell
proliferation and metastasis (14,43).
Role of ISG20 in cancer
ISG20 exhibits opposing mechanisms of action in
tumors, necessitating further research. From an immunological
perspective, when tumor cells successfully evade immune responses,
cancer progression occurs (57).
IFNs are a family of secreted proteins, also known as key drivers
of inflammation in the TME, that have a crucial role in triggering
immune responses in immune cells (57–59).
As the second protein induced by IFNs or dsRNA, ISG20 exhibits
differential expression in healthy individuals and certain types of
tumors, but its precise mechanism remains to be explored (Fig. 4).
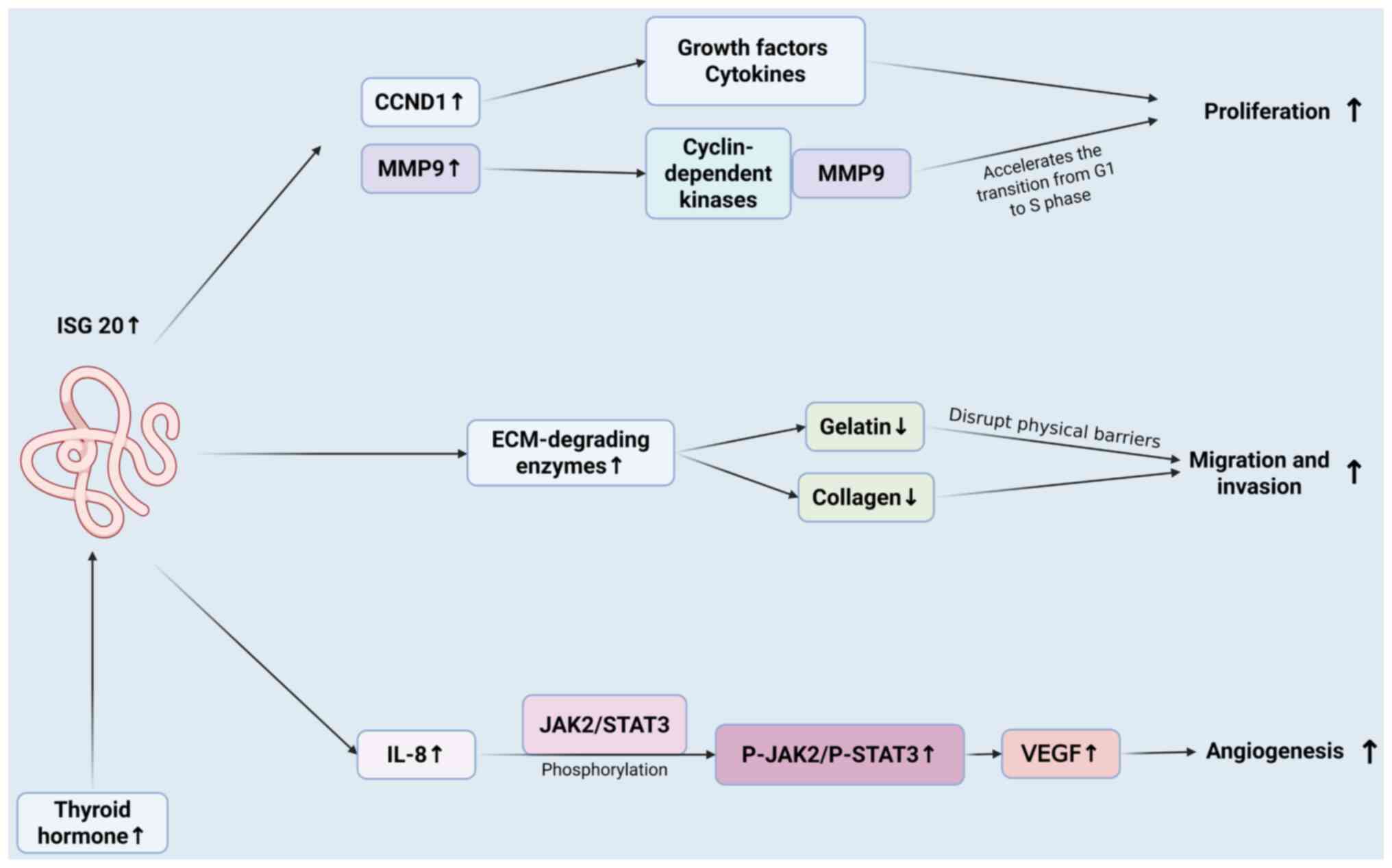 | Figure 4.Mechanism of ISG20 in promoting tumor
cell proliferation, migration, invasion and angiogenesis. ISG20,
induced by thyroid hormone, promotes cancer cell proliferation,
migration, invasion and angiogenesis. It acts via pathways like
CCND1/MMP9 for proliferation, ECM-degrading enzymes for
migration/invasion, and IL-8-JAK2/STAT3-VEGF for angiogenesis. ISG,
IFN-stimulated gene; CCND, cyclin D1; P, phosphorylation. |
Impact on tumor cell
proliferation
In certain types of cancer, such as RCC, breast and
cervical cancer, glioblastoma (GBM) and hepatocellular carcinoma,
ISG20 promotes tumor cell proliferation (Fig. 5). In ccRCC, Xu et al
(23) conducted Gene Set Enrichment
Analysis and revealed that high ISG20 expression notably
contributes to the ‘cell cycle pathway’, promoting tumor cell
proliferation. The aforementioned study also revealed that
silencing ISG20 leads to significant downregulation of MMP9 and
CCND1 expression in ccRCC, both of which are typically upregulated
in ccRCC (23). Therefore, ISG20
may accelerate the cell cycle progression and promote rapid tumor
cell proliferation by positively regulating the expression of the
cell cycle-associated proteins MMP9 and CCND1. Furthermore, ISG20
has been identified as a potential prognostic target in ccRCC.
Rajkumar et al (18) used
microarray technology to analyze samples from various stages of
cervical cancer, cervical intraepithelial neoplasia and normal
cervical tissue; the findings initially indicated a potential
expression difference for ISG20, which was validated by reverse
transcription-quantitative PCR and calibrated to normal samples,
confirming high ISG20 expression in cervical cancer tissue. These
results suggested that ISG20 may serve an important role in the
progression of cervical cancer (18).
By contrast, in ovarian cancer, ISG20 inhibits tumor
cell proliferation both in vivo and in vitro
(45). ISG20 overexpression can
significantly inhibit the proliferation of Caov3 cells in
vivo. Additionally, xenograft tumor assays in nude mice
revealed that stable overexpression of ISG20 in Caov3 cells using
pLVX–ISG20-IRES-Neo slows tumor growth. These results suggested
that ISG20 may suppress tumorigenesis in vivo (45).
Effect on tumor cell migration and
invasion
Tumor migration and invasion are regulated by
multiple factors, including genetic mutation, signaling pathway
activation, TME remodeling and phenotypic plasticity. ISG20exhibits
heterogeneity in its effect on tumor migration and invasion.
Alsheikh et al (43)
reported that the NMI (N-Myc and STAT interactor protein)-STAT5A
signaling axis inhibits ISG20 expression via miR-20a, maintaining
normal breast epithelial differentiation and suppressing tumor
metastasis. Clinical samples revealed that in 70% of patients with
metastatic breast cancer, NMI protein levels are lower than those
in primary tumor samples, whereas ISG20 expression is significantly
higher in metastatic than in primary sites. Functional assays,
including Transwell, Matrigel and lung metastasis experiments,
confirmed that ISG20 overexpression enhances migration, invasion
and lung metastasis potential in breast cancer cells (43).
Additionally, ISG20 may promote the expression of
certain extracellular matrix (ECM)-degrading enzymes, leading to
the breakdown of collagen and gelatin components in the ECM,
thereby disrupting physical barriers around tumor cells and
facilitating migration and invasion (23). Notably, in ccRCC, high ISG20
expression may promote MMP9 activity, thereby enhancing tumor
migration and invasion (23). MMP9,
a key ECM-degrading enzyme, disrupts the basement membrane and ECM,
providing a physical passage for tumor cell migration (60). ISG20 knockdown inhibits cell
invasion, confirming its role in ECM remodeling through an
MMP9-dependent mechanism (23).
However, in ovarian cancer, Yu et al
(45) employed Transwell assays to
assess the invasion and migration of Caov3 cells in vitro,
finding that ISG20 overexpression significantly inhibited cell
invasion and migration compared with the control group, however,
the underlying mechanism remains unclear (45).
ISG20 upregulation induces tumor
angiogenesis
Tumor formation is typically driven by the
proliferation and metastasis of tumor cells, which depend on the
ability of tumor cells to recruit their own blood supply.
Angiogenesis is the key pathway by which tumors acquire nutrients
and oxygen and achieve distant dissemination, and is therefore key
for the development of human cancer (61). In the intricate regulatory network
of angiogenesis, the positive regulator vascular endothelial growth
factor and the negative regulator thrombospondin-1 form a dynamic
balance (62,63).
Previous studies have shown that ISG20
overexpression does not inhibit angiogenesis, whereas its dominant
negative mutation ExoII (lacking exonuclease activity) inhibits
angiogenesis by 43% (64,65). In addition, fibrin gel assay
suggested that the exonuclease activity of ISG20 may be essential
for angiogenesis (64). In
hepatocellular carcinoma, Lin et al (65) reported that thyroid hormone
(triiodothyronine) upregulates ISG20, which promotes hepatocellular
carcinoma angiogenesis by regulating IL-8 and activating the
phosphorylated (p-)JAK2/p-STAT3 signaling pathway, providing
therapeutic targets and a theoretical basis for liver cancer
treatment. In vitro, conditioned medium from
ISG20-overexpressing hepatocellular carcinoma cells significantly
promotes the formation of lumen-like structures by human umbilical
vein endothelial cells (HUVECs), whereas ISG20 knockdown may
inhibit thyroid hormone-induced HUVEC tube formation (65). In vivo, experiments using the
Matrigel plug and chicken embryo chorioallantoic membrane (CAM)
models have confirmed that ISG20-overexpressing cells enhance
angiogenesis, as evidenced by increased hemoglobin content in the
Matrigel plugs, more CAM blood vessel branches and elevated
expression of the endothelial marker CD31 (65). Knockdown of ISG20 decreases these
effects. Clinical analysis has also revealed that ISG20 expression
in liver cancer tissue is positively associated with
angiogenesis-associated clinical parameters, such as vascular
invasion and tumor size (65).
Additionally, patients with high ISG20 expression have shorter
recurrence-free survival, providing evidence that ISG20 promotes
angiogenesis in hepatocellular carcinoma (65,66).
Furthermore, human angiogenesis chip assays have revealed that
ISG20 overexpression significantly upregulate the angiogenic factor
IL-8 and activates the p-JAK2/p-STAT3 signaling pathway in HUVECs
(65). By contrast, knockdown of
IL-8 or the use of IL-8 neutralizing antibodies inhibits
ISG20-induced tube formation in vitro and angiogenesis in
vivo. Treatment with JAK2/STAT3 inhibitors also significantly
blocks ISG20-mediated angiogenesis, confirming the key role of this
pathway (65).
Regulation of TME
The TME refers to the local environment that
supports the survival and development of tumor cells, encompassing
various cells, molecules and the ECM, including tumor and immune
cells (67). These components form
a complex ecosystem that serves a key role in tumor initiation,
growth, metastasis and response to treatment (68). The regulatory mechanisms of ISG20
expression in the TME involve numerous signaling pathways and
molecular mechanisms, with its effect on tumor cells varying
depending on tumor type and microenvironment. In some tumors, it
may suppress tumor progression by enhancing immune responses,
whereas in others, it may promote tumor progression by fostering
immune suppression or enhancing tumor invasiveness (Fig. 6) (14,22,23).
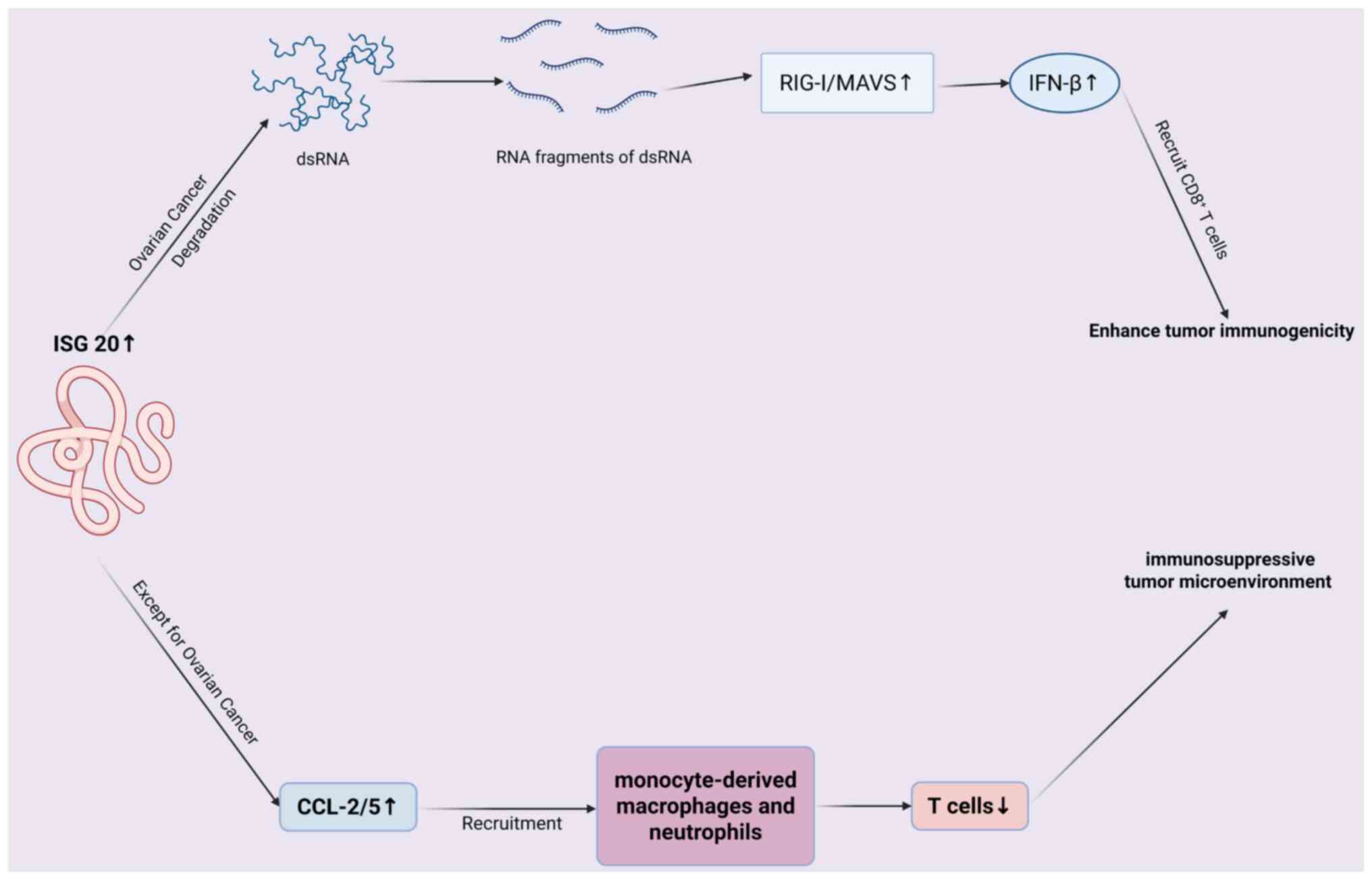 | Figure 6.Mechanism of ISG20 regulating tumor
immune microenvironment. In ovarian cancer, ISG20 degrades dsRNA
into fragments, activating RIG-I/MAVS to induce IFN-β, which
recruits CD8+ T cells and enhances immunogenicity. In
other cancers, ISG20 upregulates CCL-2/5, recruiting
monocyte-derived macrophages/neutrophils, reducing T cells and
forming an immunosuppressive tumor microenvironment. ISG,
IFN-stimulated gene; ds, double-stranded; RIG-I, retinoic
Acid-Inducible Gene I; MAVS, Mitochondrial Antiviral Signaling
Protein. |
Chen et al (22) revealed that ISG20 stimulates
antitumor immunity in ovarian cancer through a dsRNA-induced IFN
response (22). Specifically,
through protein-protein interactions and survival analysis, ISG20
was identified as the only gene significantly associated with OS in
patients with ovarian cancer. Comparison of the high and low ISG20
subgroups in The Cancer Genome Atlas (TCGA) ovarian cancer dataset
revealed that high ISG20 expression is associated with increased
CD8+ T-cell infiltration and enhanced tumor
immunogenicity. Functional experiments demonstrated that ISG20
degrades endogenous long dsRNA into shorter fragments through its
exonuclease activity, activating the RIG-I/MAVS (retinoic
acid-inducible gene I/Mitochondrial antiviral-signaling protein)
signaling pathway and promoting IFN-β secretion. IFN-β upregulates
ISGs, such as CXCL10 and CXCL11, enhancing tumor cell
immunogenicity and CD8+ T cell infiltration. The
aforementioned study identified the critical role of the RIG-I/MAVS
signaling pathway, dependent on ISG20 exonuclease activity, in
ovarian cancer. Enhancing the ISG20/RIG-I pathway (through dsRNA
agonists) or use of immune checkpoint inhibitors (such as
anti-PD-1) may offer novel therapeutic strategies for ovarian
cancer immunotherapy.
In glioma, ISG20 has been shown to promote local
tumor immunity (14). Gao et
al (14) used functional
enrichment and correlation analysis to reveal that ISG20 recruits
monocyte-derived macrophages and neutrophils by upregulating
chemokines such as CCL2/5, while inhibiting the infiltration of
antitumor T cell subsets, including central memory and follicular
helper T cells. This expression pattern is positively associated
with immune checkpoint molecules such as PD-1/PD-L1 and CTLA4 and
JAK/STAT pathway activation, thus shaping an immune-suppressive
microenvironment (14). Although
the aforementioned study revealed the key role of ISG20 in the
glioma immune microenvironment, its specific molecular mechanisms,
such as direct target interactions and signaling pathway
regulation, require further investigation. Another study using
large TCGA cohorts and meta-analysis demonstrated that higher ISG20
expression is associated with poor prognosis in patients with
glioma (24). The aforementioned
study further revealed that ISG20 is expressed in tumor-associated
macrophages, influencing immune cell infiltration and immune
checkpoint regulation, thereby participating in the regulation of
the glioma immune microenvironment (24). ISG20 is primarily expressed in
M2-type tumor-associated macrophages, with high expression
positively associated with M2 macrophage and regulatory T cell
(Treg) infiltration, and negatively associated with plasma and
naïve T cells (24). It was also
positively associated with immune checkpoint molecules such as PD-1
and CTLA4, suggesting that it promotes tumor immune escape by
shaping an immune-suppressive microenvironment (24). Immunofluorescence confirmed the
co-localization of ISG20 with CD163, indicating its predominant
expression in M2-type tumor-associated macrophages. Furthermore,
enrichment analysis showed that ISG20-associated genes are directly
involved in pathways such as IL6/JAK/STAT3 (cell proliferation),
PI3K/AKT/mTOR (survival signaling) and ECM remodeling (invasion and
metastasis) (24). This suggested
that ISG20 may serve as a novel biomarker for malignant phenotype
and prognosis in glioma. It influences tumor progression by
regulating the immune microenvironment and pro-cancer pathways,
offering a target for immunotherapy aimed at M2 macrophages.
A previous study investigated the precise effect of
ISG20 on macrophage polarization in GBM (69). The results demonstrated that
FOSL2(Fos-like 2), a potential transcription factor, may activate
ISG20 transcription through DNA hypomethylation, inducing
macrophage polarization towards the M2 phenotype. This can lead to
the secretion of immunosuppressive cytokines, such as IL-10 and
TGF-β, which suppress T cell function and establish an
immune-suppressive microenvironment, thus promoting tumor
progression. The aforementioned study reveals the critical role of
the FOSL2/ISG20 pathway in GBM progression, providing new targets
for therapies aimed at M2 macrophage polarization and
FOSL2/ISG20-associated treatment strategies. However, the
aforementioned study did not elucidate the mechanism by which ISG20
induces M2 macrophage polarization in the GBM TME.
In ccRCC, ISG20 promote the infiltration of
suppressive immune cells, enhances immune checkpoint signaling and
regulates disulfide metabolism, thereby constructing an
immune-suppressive TME that enables tumor cells to evade host
immune surveillance and attack (70). Specifically, in the C2 subtype
(high-risk group) with high ISG20 expression, tumor-infiltrating
immune cells, including Tregs and myeloid-derived suppressor cells,
are significantly enriched (70).
Single-cell and spatial transcriptomic analyses
(70) have revealed that ISG20 is
highly expressed in Tregs and exhausted CD8+ T cells,
co-localizing with T cell exhaustion markers such as lymphocyte
activation Gene 3). This suggests that ISG20 may maintain the
immunosuppressive phenotype of Tregs and the functional decline of
exhausted T cells, thus inhibiting effector T cell activity
(70). Furthermore, high ISG20
expression is significantly associated with the upregulation of
immune checkpoint proteins, such as PD-1, CTLA-4 and T cell
Immunoglobulin and ITIM Domain), which transmit inhibitory signals
leading to T cell inactivation (70). The high-risk group exhibits
significantly higher T cell exhaustion and dysfunction scores,
further confirming that ISG20 contributes to the inhibitory TME,
hindering T cell attacks on tumors (70). ISG20 is also positively associated
with the disulfide death core gene SLC7A11 at the protein level,
and their co-expression may regulate intracellular disulfide bond
metabolism and redox homeostasis, influencing ferroptosis and
shaping a microenvironment that supports tumor survival (70).
In summary, ISG20 exhibits a dual role in the TME,
driving anti-tumor immunity while promoting immunosuppression and
tumor progression. This functional heterogeneity is shaped by a
series of key determinants of tumor type specificity. In-depth
investigation of these factors is essential for understanding the
biological role of ISG20 and developing precision targeting
strategies. Cellular localization of ISG20 action is a key factor.
In ovarian cancer, Chen et al (22) clarified that its function mainly
originates from the expression of tumor cells, which activates the
RIG-I/MAVS/IFN-β pathway through degradation of endogenous dsRNA in
tumor cells, which in turn recruits CD8+ T cells. By
contrast, in glioma and ccRCC, key evidence (co-localization with
CD163, expression in Tregs/depleted CD8+ T cells)
suggests that the pro-tumorigenic effect of ISG20 stems primarily
from its expression in specific immune cell subsets (especially
M2-type tumor-associated macrophages and Tregs), which are the key
components of the immune-suppressive TME. Downstream pathways and
effector molecules activated by ISG20 show functional shifts due to
differences in the molecular composition of the TME. The abundant
endogenous dsRNA in ovarian cancer provides ISG20 with a substrate
to activate the RIG-I pathway, which produces chemokines such as
CXCL10/11 that recruit effector T cells. In glioma, high expression
of ISG20 (potentially activated by FOSL2 transcription) is
associated with the sustained activation of the JAK/STAT pathway,
leading to the upregulation of chemokines such as CCL2/5 that
recruit suppressor myeloid cells and promoting the expression of
immune checkpoints such as PD-1/PD-L1. Renal cancer studies
(22,23). have further revealed ISG20 synergy
with disulfide death-related genes (such as SLC7A11) and its
spatial co-localization with T cell depletion markers, suggesting
that it may shape the suppressive TME by regulating cell metabolism
and directly participating in the maintenance of the depletion
phenotype. Finally, the ‘functional output’ of ISG20 is dependent
on the composition and activation status of the immune cell network
in which it operates. CCL2/5 may recruit different cell types in
different TMEs; ISG20-induced production of IFN-β may enhance
immunogenicity in ovarian cancer, whereas it may promote immune
tolerance or depletion in the context of chronic inflammation or in
the presence of other suppressive signals. The association of high
ISG20 expression with decreased infiltration of specific T cell
subsets (central memory and follicular helper T) observed in glioma
studies (14,24) and with Tregs/myeloid-Derived
Suppressor Cells) enrichment and elevated T cell dysfunction score
in renal carcinoma (70) is a
manifestation of this network dependence. Thus, ISG20 functions are
affected by the cellular ecological niche, molecular signaling
network and immune context of specific tumor types. Understanding
the determinants of tumor specificity, such as cellular
localization, microenvironmental molecular context (substrate,
pathway coupling) and immune cell network composition and status,
is a fundamental prerequisite for resolving its dual roles,
evaluating its value as a biomarker and designing effective
targeted intervention strategies. Furthermore, in terms of
potential molecular mechanisms between ISG20 and immune checkpoint
regulation, existing studies (14,24)
have primarily provided correlation evidence (co-expression,
co-localization) and pathway associations (JAK/STAT), but lack
conclusive molecular mechanistic models (key transcription factor
complexes, direct substrate-product association, specific protein
interactions) and in-depth experimental validation such as
chromatin and RNA immunoprecipitation followed by Sequencing,
reporter gene analysis, effects of conditional
knockout/overexpression in specific cell types. In glioblastoma
(69), the transcription factor
FOSL2 activates ISG20 transcription through DNA hypomethylation,
induces macrophage polarization towards M2 type and secretes
inhibitory factors such as IL-10 and TGF-β. M2 type macrophages may
further upregulate PD-L1 expression through the JAK/STAT pathway,
creating an immunosuppressive microenvironment. In ccRCC (70), high ISG20 expression was positively
associated with the phosphorylation of STAT3, a key transcriptional
activator of PD-L1, which is enhanced by direct binding to the
PD-L1 promoter. In addition, ISG20 is positively correlated with
the disulfide death core gene SLC7A11 in renal cancer, which
regulates intracellular redox homeostasis. SLC7A11 enhances PD-L1
transcription by stabilizing hypoxia-inducible factor 1α, and ISG20
may indirectly support this process through metabolic reprogramming
(70). Future studies are needed to
characterize these factors in more tumor types and explore the
precise molecular mechanisms of their interaction.
Potential application of ISG20 in cancer
therapy
As a tumor-associated antigen
Due to marked dysregulation of the IFN signaling
pathway in various tumors, ISG20 is upregulated in tumors of the
brain, uterus, breast, cervix, esophagus, kidney, liver, pancreas,
skin and testes (24). In ovarian
cancer, ISG20 enhances type I IFN signaling, increasing the
immunogenicity of tumor cells and promoting immune cell
infiltration and antitumor immune responses (22). This suggests that ISG20 gene
delivery via viral vectors (including lentiviruses) may directly
increase ISG20 expression in tumor cells, promoting dsRNA
fragmentation and IFN-β secretion, thereby activating both innate
and adaptive immune responses. Alternatively, drugs specifically
activating ISG20 RNase activity, or alleviating ISG20 inhibitors
(protein modification regulation), may be developed to induce
endogenous dsRNA accumulation and IFN pathway activation for
ovarian cancer treatment. However, ISG20 has a tumor-promoting role
in most types of cancer, including glioma and ccRCC (23,24).
In the future, ISG20 could be used as a tumor-associated antigen to
design vaccines (such as peptide, DNA/RNA or dendritic cell
vaccines) to activate specific immune responses targeting tumor
cells for antitumor effects. However, due to the complex and
potentially risky mechanisms of ISG20 in different tumors, the
development of ISG20-based vaccines requires further research and
validation. Future research should focus on clarifying the specific
mechanisms of ISG20 in different types of disease, optimizing
vaccine design and evaluating their safety and efficacy.
As a potential biomarker
Biomarkers are typically defined as objective
indicators that can be measured to reflect physiological or
pathological processes, as well as biological effects following
exposure to treatment or intervention. As aforementioned, ISG20 is
upregulated in various tumors, and is significantly associated with
disease progression and prognosis. Therefore, ISG20 is increasingly
recognized as a potential biomarker for certain types of tumor
(14,23,71).
For example, Xu et al (23)
reported that ISG20 is highly upregulated in ccRCC tumors, where it
enhances tumor cell proliferation and metastasis by modulating the
MMP9/CCND1 signaling pathway. ISG20 has been identified as a
potential predictive target for ccRCC, with high ISG20 expression
associated with poorer OS and DFS (23). In cervical cancer tissue, Rajkumar
et al (29) observed high
expression of ISG20, which may be associated with the onset and
progression of cervical cancer (18). In ovarian cancer (72), a significant decrease in ISG20
levels has been observed, which may be associated with ethnicity,
advanced clinical stage, higher pathological grade and poor
prognosis. Lower ISG20 mRNA expression is detected in
cisplatin-resistant ovarian cancer compared with in a
cisplatin-sensitive group (25).
Furthermore, Lin et al (65)
reported that ISG20 upregulation in patients with hepatocellular
carcinoma is positively associated with clinical factors such as
vascular invasion and tumor size, and poorer relapse-free survival
(65). Additionally, in
radioresistant oral cancer cells, the mRNA and protein expression
levels of ISG20 are higher than those in corresponding parental
cells, suggesting that ISG20 upregulation may be essential for the
radioresistant phenotype in oral cancer (26). In summary, ISG20 may serve as a
potential biomarker for early tumor diagnosis, prognostic
assessment and treatment response prediction.
As an immunological adjuvant
An immunoadjuvant is a non-specific immune
enhancer, typically administered with an antigen to enhance the
immune response to a specific antigen or alter the type of immune
response. Its primary function is to improve vaccine efficacy and
immune response strength by altering the physical properties of the
antigen, promoting antigen processing and presentation and
stimulating immune cell activation (73). Tumor neoantigen vaccines and PD-L1
inhibitors are promising immunotherapeutic approaches to the
clinical treatment of various types of tumor, but show limited
efficacy in patients with tumors lacking functional T cell
infiltration (74,75). As aforementioned, ISG20 has been
shown to enhance tumor immunogenicity in ovarian cancer, recruiting
various immune cells, including CD8+ T cells, to
infiltrate tumor tissue and enhance antitumor immune responses
(22). This suggests that ISG20 may
serve as an immunoadjuvant when combined with tumor antigens to
achieve antitumor immune effects. However, studies have shown that
ISG20 promotes tumor immune evasion in glioma, indicating that its
use as an immunoadjuvant should be selected based on the specific
tumor type and microenvironment (14,24).
Conclusion
ISG20 is an insufficiently explored molecular
target in cancer. It is upregulated in various types of tumor
tissue, and influences processes such as tumor cell proliferation,
migration, invasion, angiogenesis and immune modulation in the TME
through complex mechanisms. ISG20 may serve as a tumor antigen,
biomarker and immunoadjuvant. However, prior to clinical
application, further research is needed to clarify its structure
and functional sites as a tumor antigen, evaluate its role as a
biomarker and explore its precise application as an immunoadjuvant
across different tumor types and microenvironments.
Future research should focus on elucidating the
molecular mechanisms underlying the roles of ISG20 in various
tumors and further investigating its regulatory network in the TME,
particularly its impact on immune cells. Leveraging advanced
technology to analyze its three-dimensional structure and
protein-protein interaction sites may accelerate the development of
small-molecule drugs or biologics targeting ISG20. Additionally,
large-scale, multicenter clinical studies should be conducted to
validate its value in tumor precision diagnosis and personalized
treatment, facilitating its efficient translation from basic
research to clinical application, and providing novel strategies
and approaches for cancer treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 82260596), the Natural Science
Foundation of Jiangxi Province (grant nos. 20242BAB25506 and
20242BAB20406), the Science and Technology Program of Jiangxi
Provincial Health and Family Planning Commission (grant no.
202410246), the China Postdoctoral Science Foundation (grant no.
2023M741523), the Outstanding Youth Foundation of Jiangxi Province
(grant no. 20252BAC220050), the Graduate Student Innovation Special
Foundation of Jiangxi Province (grant no. YC2025-S245) and the
Science and Technology Program of Jiangxi Provincial Administration
of Traditional Chinese Medicine (grant no. 2024A0028).
Availability of data and materials
Not applicable.
Authors' contributions
XZ and SJ designed the study. LZ and SZ wrote the
manuscript. HZ, JZ and LC constructed the figures. HL and ZZ
performed the literature review. All authors have read and approved
the final manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang X, Yang W, Wang X, Zhang X, Tian H,
Deng H, Zhang L and Gao G: Identification of new type I
interferon-stimulated genes and investigation of their involvement
in IFN-β activation. Protein Cell. 9:799–807. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Negishi H, Taniguchi T and Yanai H: The
interferon (IFN) class of cytokines and the IFN regulatory factor
(IRF) transcription factor family. Cold Spring Harb Perspect Biol.
10:a0284232017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
LaFleur DW, Nardelli B, Tsareva T, Mather
D, Feng P, Semenuk M, Taylor K, Buergin M, Chinchilla D, Roshke V,
et al: Interferon-kappa, a novel type I interferon expressed in
human keratinocytes. J Biol Chem. 276:39765–39771. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bach EA, Aguet M and Schreiber RD: The IFN
gamma receptor: A paradigm for cytokine receptor signaling. Annu
Rev Immunol. 15:563–591. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schroder K, Hertzog PJ, Ravasi T and Hume
DA: Interferon-gamma: An overview of signals, mechanisms and
functions. J Leukoc Biol. 75:163–189. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kotenko SV, Gallagher G, Baurin VV,
Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H
and Donnelly RP: IFN-lambdas mediate antiviral protection through a
distinct class II cytokine receptor complex. Nat Immunol. 4:69–77.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ank N, Iversen MB, Bartholdy C, Staeheli
P, Hartmann R, Jensen UB, Dagnaes-Hansen F, Thomsen AR, Chen Z,
Haugen H, et al: An important role for type III interferon
(IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol.
180:2474–2485. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schoggins JW: Interferon-stimulated genes:
What do they all do? Annu Rev Virol. 6:567–584. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moser MJ, Holley WR, Chatterjee A and Mian
IS: The proofreading domain of Escherichia coli DNA polymerase I
and other DNA and/or RNA exonuclease domains. Nucleic Acids Res.
25:5110–5118. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zuo Y and Deutscher MP: Exoribonuclease
superfamilies: Structural analysis and phylogenetic distribution.
Nucleic Acids Res. 29:1017–1026. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zheng Z, Wang L and Pan J:
Interferon-stimulated gene 20-kDa protein (ISG20) in infection and
disease: Review and outlook. Intractable Rare Dis Res. 6:35–40.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weiss CM, Trobaugh DW, Sun C, Lucas TM,
Diamond MS, Ryman KD and Klimstra WB: The Interferon-Induced
exonuclease ISG20 exerts antiviral activity through upregulation of
type I interferon response proteins. mSphere. 3:e00209–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Suo J, Wang Z, Ran K, Tian Y, Han
W, Liu Y and Peng X: The PTPRZ1-MET/STAT3/ISG20 axis in glioma
stem-like cells modulates Tumor-associated macrophage polarization.
Cell Signal. 120:1111912024. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao M, Lin Y, Liu X, Li Y, Zhang C, Wang
Z, Wang Z, Wang Y and Guo Z: ISG20 promotes local tumor immunity
and contributes to poor survival in human glioma. Oncoimmunology.
8:e15340382018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhong R, Li JQ, Wu SW, He XM, Xuan JC,
Long H and Liu HQ: Transcriptome analysis reveals possible
antitumor mechanism of Chlorella exopolysaccharide. Gene.
779:1454942021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shou Y, Yang L, Yang Y, Zhu X, Li F and Xu
J: Determination of hypoxia signature to predict prognosis and the
tumor immune microenvironment in melanoma. Mol Omics. 17:307–316.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei C, Liu X, Wang Q, Li Q and Xie M:
Identification of hypoxia signature to assess the tumor immune
microenvironment and predict prognosis in patients with ovarian
cancer. Int J Endocrinol. 2021:41561872021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rajkumar T, Sabitha K, Vijayalakshmi N,
Shirley S, Bose MV, Gopal G and Selvaluxmy G: Identification and
validation of genes involved in cervical tumourigenesis. BMC
Cancer. 11:802011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiong H, Zhang X, Chen X, Liu Y, Duan J
and Huang C: High expression of ISG20 predicts a poor prognosis in
acute myeloid leukemia. Cancer Biomark. 31:255–261. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang Z, Xu J, Zhang S, Lan H and Bao Y: A
pairwise immune gene model for predicting overall survival and
stratifying subtypes of colon adenocarcinoma. J Cancer Res Clin
Oncol. 149:10813–10829. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bishara I, Liu X, Griffiths Jl, Cosgrove
PA, McQuerry JR, Liu J, Ihle KK, Bacon ER, Chi F, Wallet P, et al:
Abstract 5133: End-stage breast cancer metastases manifest as two
subtypes with distinct dissemination patterns, proliferation/EMT
signatures, and immune microenvironments. Cancer Res. 85 (Suppl
1):S51332025. View Article : Google Scholar
|
|
22
|
Chen Z, Yin M, Jia H, Chen Q and Zhang H:
ISG20 stimulates anti-tumor immunity via a double-stranded
RNA-induced interferon response in ovarian cancer. Front Immunol.
14:11761032023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu T, Ruan H, Gao S, Liu J, Liu Y, Song Z,
Cao Q, Wang K, Bao L, Liu D, et al: ISG20 serves as a potential
biomarker and drives tumor progression in clear cell renal cell
carcinoma. Aging (Albany NY). 12:1808–1827. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Peng Y, Liu H, Wu Q, Wang L, Yu Y, Yin F,
Feng C, Ren X, Liu T, Chen L and Zhu H: Integrated bioinformatics
analysis and experimental validation reveal ISG20 as a novel
prognostic indicator expressed on M2 macrophage in glioma. BMC
Cancer. 23:5962023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu J, Jiang L, Wang S, Peng L, Zhang R and
Liu Z: TGF β1 promotes the polarization of M2-type macrophages and
activates PI3K/mTOR signaling pathway by inhibiting ISG20 to
sensitize ovarian cancer to cisplatin. Int Immunopharmacol.
134:1122352024. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miyashita H and Fukumoto M, Kuwahara Y,
Takahashi T and Fukumoto M: ISG20 is overexpressed in clinically
relevant radioresistant oral cancer cells. Int J Clin Exp Pathol.
13:1633–1639. 2020.PubMed/NCBI
|
|
27
|
Lin H, Zhou Z, Sun H, Li C, Lu Y, Wu Z,
Zhou L, Wang Y, Pu Z, Mou L and Yang MM: Mapping the role of
cytokine signaling at single-cell and structural resolution in
uveal melanoma. Genes Immun. Jun 9–2025.doi:
10.1038/s41435-025-00337-3 (Epub ahead of print). View Article : Google Scholar
|
|
28
|
Wang T, Liu Y, Wu X, Wang X, Shi S, Song
X, Ma Y, Zhang Z, Gao J, Sun R and Song G: Multi-omics reveals
miR-181a-5p regulates PPAR-driven lipid metabolism in Oral squamous
cell carcinoma: Insights from CRISPR/Cas9 knockout models. J
Proteomics. 319:1054802025. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gongora C, David G, Pintard L, Tissot C,
Hua TD, Dejean A and Mechti N: Molecular cloning of a new
Interferon-induced PML nuclear Body-associated Protein. J Biol
Chem. 272:19457–19463. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pentecost BT: Expression and estrogen
regulation of the HEM45 MRNA in human tumor lines and in the rat
uterus. J Steroid Biochem Mol Biol. 64:25–33. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gongora C, Degols G, Espert L, Hua TD and
Mechti N: A unique ISRE, in the TATA-less human Isg20 promoter,
confers IRF-1-mediated responsiveness to both interferon type I and
type II. Nucleic Acids Res. 28:2333–2341. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nguyen LH, Espert L, Mechti N and Wilson
DM III: The human interferon- and estrogen-regulated ISG20/HEM45
gene product degrades single-stranded RNA and DNA in vitro.
Biochemistry. 40:7174–7179. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Espert L, Degols G, Gongora C, Blondel D,
Williams BR, Silverman RH and Mechti N: ISG20, a new
interferon-induced RNase specific for single-stranded RNA, defines
an alternative antiviral pathway against RNA genomic viruses. J
Biol Chem. 278:16151–16158. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Espert L, Rey C, Gonzalez L, Degols G,
Chelbi-Alix MK, Mechti N and Gongora C: The exonuclease ISG20 is
directly induced by synthetic dsRNA via NF-kappaB and IRF1
activation. Oncogene. 23:4636–4640. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Horio T, Murai M, Inoue T, Hamasaki T,
Tanaka T and Ohgi T: Crystal structure of human ISG20, an
interferon-induced antiviral ribonuclease. FEBS Lett. 577:111–116.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Espert L, Eldin P, Gongora C, Bayard B,
Harper F, Chelbi-Alix MK, Bertrand E, Degols G and Mechti N: The
exonuclease ISG20 mainly localizes in the nucleolus and the Cajal
(Coiled) bodies and is associated with nuclear SMN
protein-containing complexes. J Cell Biochem. 98:1320–1333. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Karasawa T, Sato R, Imaizumi T, Fujita M,
Aizawa T, Tsugawa K, Mattinzoli D, Kawaguchi S, Seya K, Terui K, et
al: Expression of interferon-stimulated gene 20 (ISG20), an
antiviral effector protein, in glomerular endothelial cells:
Possible involvement of ISG20 in lupus nephritis. Ren Fail.
45:22248902023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jia M, Li L, Chen R, Du J, Qiao Z, Zhou D,
Liu M, Wang X, Wu J, Xie Y, et al: Targeting RNA oxidation by
ISG20-mediated degradation is a potential therapeutic strategy for
acute kidney injury. Mol Ther. 31:3034–3051. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Louvat C, Deymier S, Nguyen XN, Labaronne
E, Noy K, Cariou M, Corbin A, Mateo M, Ricci EP, Fiorini F and
Cimarelli A: Stable structures or PABP1 loading protects cellular
and viral RNAs against ISG20-mediated decay. Life Sci Alliance.
7:e2023022332024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Viswanathan M and Lovett ST: Exonuclease X
of Escherichia coli. A novel 3′-5′ DNase and Dnaq superfamily
member involved in DNA repair. J Biol Chem. 274:30094–31100. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu Y, Nie H, Mao R, Mitra B, Cai D, Yan
R, Guo JT, Block TM, Mechti N and Guo H: Interferon-inducible
ribonuclease ISG20 inhibits hepatitis B virus replication through
directly binding to the epsilon stem-loop structure of viral RNA.
PLoS Pathog. 13:e10062962017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pollack JR and Ganem D: An RNA stem-loop
structure directs hepatitis B virus genomic RNA encapsidation. J
Virol. 67:3254–3263. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Alsheikh HAM, Metge BJ, Pruitt HC,
Kammerud SC, Chen D, Wei S, Shevde LA and Samant RS: Disruption of
STAT5A and NMI signaling axis leads to ISG20-driven metastatic
mammary tumors. Oncogenesis. 10:452021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Qu X, Shi Z, Guo J, Guo C, Qiu J and Hua
K: Identification of a novel six-gene signature with potential
prognostic and therapeutic value in cervical cancer. Cancer Med.
10:6881–6896. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu J, Liu TT, Liang LL, Liu J, Cai HQ,
Zeng J, Wang TT, Li J, Xiu L, Li N and Wu LY: Identification and
validation of a novel glycolysis-related gene signature for
predicting the prognosis in ovarian cancer. Cancer Cell Int.
21:3532021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu N, Nguyen XN, Wang L, Appourchaux R,
Zhang C, Panthu B, Gruffat H, Journo C, Alais S, Qin J, et al: The
interferon stimulated gene 20 protein (ISG20) is an innate defense
antiviral factor that discriminates self versus Non-self
translation. PLoS Pathog. 15:e10080932019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
El Kazzi P, Rabah N, Chamontin C, Poulain
L, Ferron F, Debart F, Rossi S, Bribes I, Rouilly L, Simonin Y, et
al: Internal RNA 2′O-methylation in the HIV-1 genome counteracts
ISG20 nuclease-mediated antiviral effect. Nucleic Acids Res.
51:2501–2515. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Deymier S, Louvat C, Fiorini F and
Cimarelli A: ISG20: An enigmatic antiviral RNase targeting multiple
viruses. FEBS Open Bio. 12:1096–1111. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kang D, Gao S, Tian Z, Zhang G, Guan G,
Liu G, Luo J, Du J and Yin H: ISG20 inhibits bluetongue virus
replication. Virol Sin. 37:521–530. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Espert L, Degols G, Lin YL, Vincent T,
Benkirane M and Mechti N: Interferon-induced exonuclease ISG20
exhibits an antiviral activity against human immunodeficiency virus
type 1. J Gen Virol. 86:2221–2229. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Decker CJ and Parker R: P-bodies and
stress granules: possible roles in the control of translation and
mRNA degradation. Cold Spring Harb Perspect Biol. 4:a0122862012.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hehl M, Scherer M, Raubuch EM, Ploil C,
Rummel T, Kirchner P, Kottmann N, Reichel A, Katharina R, König AC,
et al: Exonuclease ISG20 inhibits human cytomegalovirus replication
by inducing an innate immune defense signature. bioRxiv. Feb
10–2025.doi: 10.1101/2025.02.10.637376.
|
|
53
|
Xing L, Meng G, Chen T, Zhang X, Bai D and
Xu H: Crosstalk between RNA-Binding proteins and immune
microenvironment revealed Two RBP regulatory patterns with distinct
immunophenotypes in periodontitis. J Immunol Res. 2021:55884292021.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Staege H, Brauchlin A, Schoedon G and
Schaffner A: Two novel genes FIND and LIND differentially expressed
in deactivated and Listeria-infected human macrophages.
Immunogenetics. 53:105–113. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chaussabel D, Semnani RT, McDowell MA,
Sacks D, Sher A and Nutman TB: Unique gene expression profiles of
human macrophages and dendritic cells to phylogenetically distinct
parasites. Blood. 102:672–681. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rodríguez-Galán A, Dosil SG, Hrčková A,
Fernández-Messina L, Feketová Z, Pokorná J, Fernández-Delgado I,
Camafeita E, Gómez MJ, Ramírez-Huesca M, et al: ISG20L2: An RNA
nuclease regulating T cell activation. Cell Mol Life Sci.
80:2732023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Budhwani M, Mazzieri R and Dolcetti R:
Plasticity of Type I Interferon-mediated responses in cancer
therapy: From Anti-tumor immunity to resistance. Front Oncol.
8:3222018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dunn GP, Koebel CM and Schreiber RD:
Interferons, immunity and cancer immunoediting. Nat Rev Immunol.
6:836–848. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hervas-Stubbs S, Perez-Gracia JL, Rouzaut
A, Sanmamed MF, Le Bon A and Melero I: Direct effects of type I
interferons on cells of the immune system. Clin Cancer Res.
17:2619–2627. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Brinckerhoff CE and Matrisian LM: Matrix
metalloproteinases: A tail of a frog that became a prince. Nat Rev
Mol Cell Biol. 3:207–214. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
61
|
Baeriswyl V and Christofori G: The
angiogenic switch in carcinogenesis. Semin Cancer Biol. 19:329–337.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhou L, Isenberg JS, Cao Z and Roberts DD:
Type I collagen is a molecular target for inhibition of
angiogenesis by endogenous thrombospondin-1. Oncogene. 25:536–545.
2005. View Article : Google Scholar
|
|
63
|
Herbert SP and Stainier DYR: Molecular
control of endothelial cell behaviour during blood vessel
morphogenesis. Nat Rev Mol Cell Biol. 12:551–564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Taylor KL, Leaman DW, Grane R, Mechti N,
Borden EC and Lindner DJ: Identification of
interferon-beta-stimulated genes that inhibit angiogenesis in
vitro. J Interferon Cytokine Res. 28:733–740. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lin SL, Wu SM, Chung IH, Lin YH, Chen CY,
Chi HC, Lin TK, Yeh CT and Lin KH: Stimulation of
Interferon-stimulated gene 20 by thyroid hormone enhances
angiogenesis in liver cancer. Neoplasia. 20:57–68. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Waugh DJ and Wilson C: The interleukin-8
pathway in cancer. Clin Cancer Res. 14:6735–6741. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Mun JY, Leem SH, Lee JH and Kim HS: Dual
relationship between stromal cells and immune cells in the tumor
microenvironment. Front Immunol. 13:8647392022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Shah DD, Chorawala MR, Raghani NR, Patel
R, Fareed M, Kashid VA and Prajapati BG: Tumor microenvironment:
Recent advances in understanding and its role in modulating cancer
therapies. Med Oncol. 42:1172025. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Du H, Sun J, Wang X, Zhao L, Liu X, Zhang
C, Wang F and Wu J: FOSL2-mediated transcription of ISG20 induces
M2 polarization of macrophages and enhances tumorigenic ability of
glioblastoma cells. J Neurooncol. 169:659–670. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Peng K, Wang N, Liu Q, Wang L, Duan X, Xie
G, Li J and Ding D: Identification of disulfidptosis-related
subtypes and development of a prognosis model based on stacking
framework in renal clear cell carcinoma. J Cancer Res Clin Oncol.
149:13793–13810. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Meng K, Li YY, Liu DY, Hu LL, Pan YL,
Zhang CZ and He QY: A five-protein prognostic signature with GBP2
functioning in immune cell infiltration of clear cell renal cell
carcinoma. Comput Struct Biotechnol J. 21:2621–2630. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Geng H, Zhang H, Cheng L and Dong S:
Corrigendum to ‘Sivelestat ameliorates sepsis-induced myocardial
dysfunction by activating the PI3K/AKT/mTOR signaling pathway’
[Int. Immunopharmacol. 128 (2024) https://doi.org/10.1016/j.intimp.2023.111466simplehttps://doi.org/10.1016/j.intimp.2023.111466].
Int Immunopharmacol. 131:1118732024. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Banstola A, Jeong JH and Yook S:
Immunoadjuvants for cancer immunotherapy: A review of recent
developments. Acta Biomater. 114:16–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Moehler M, Delic M, Goepfert K, Aust D,
Grabsch HI, Halama N, Heinrich B, Julie C, Lordick F, Lutz MP, et
al: Immunotherapy in gastrointestinal cancer: Recent results,
current studies and future perspectives. Eur J Cancer. 59:160–170.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yang M, Cui M, Sun Y, Liu S and Jiang W:
Mechanisms, combination therapy, and biomarkers in cancer
immunotherapy resistance. Cell Commun Signal. 22:3382024.
View Article : Google Scholar : PubMed/NCBI
|