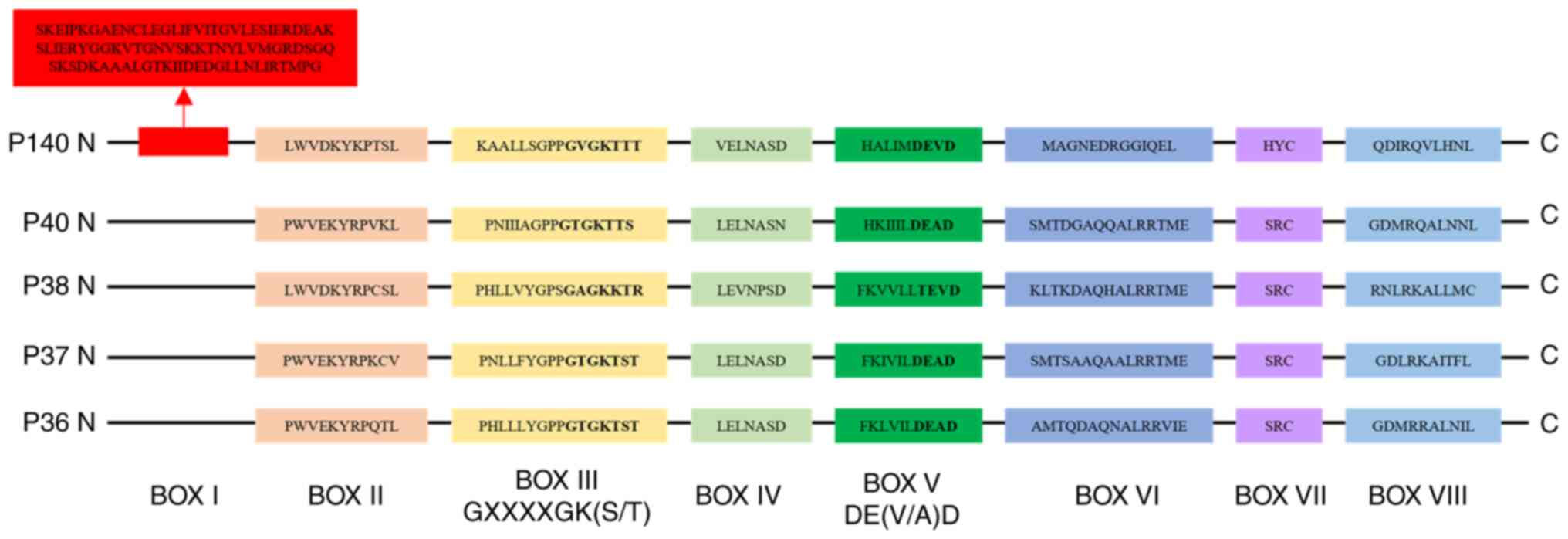
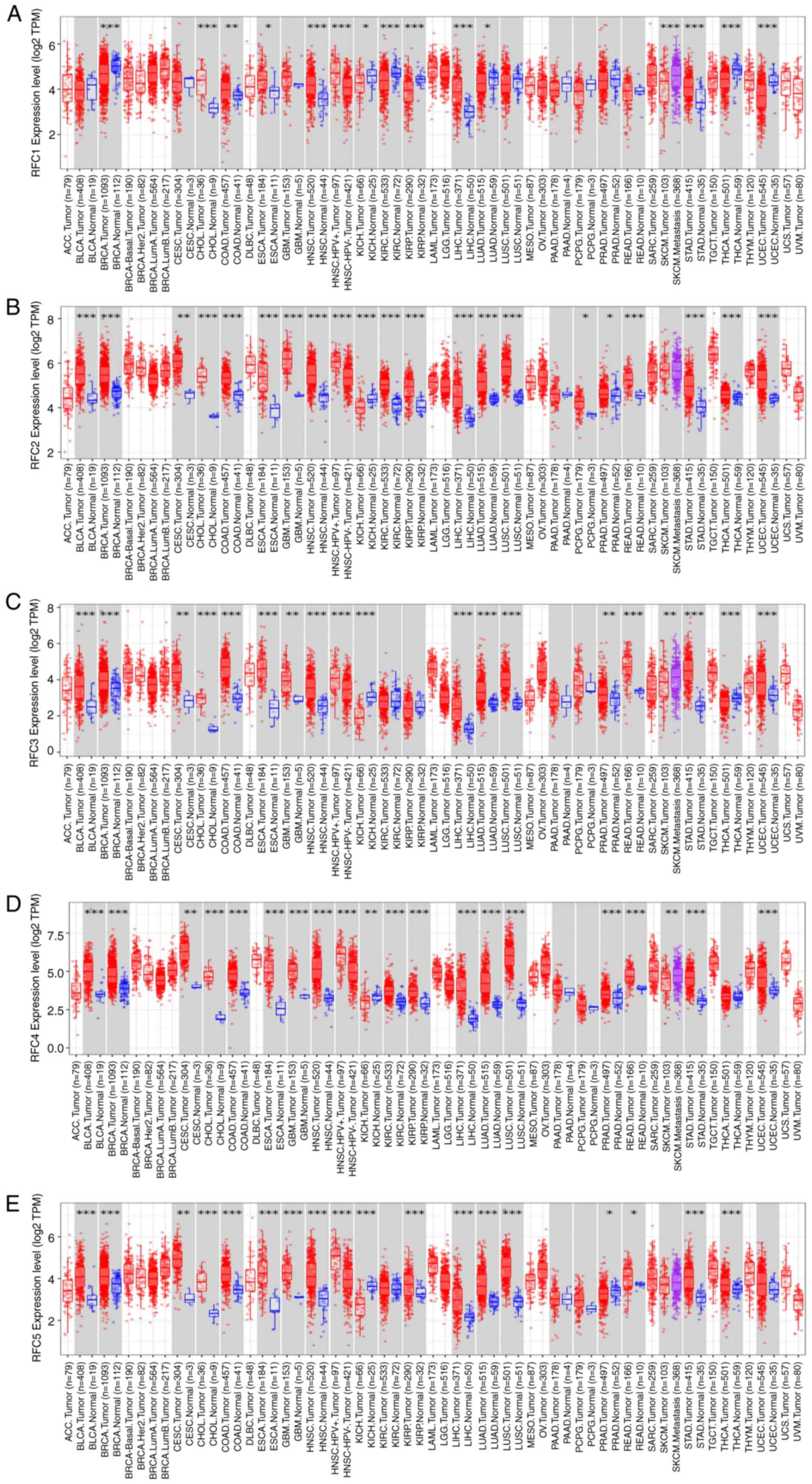
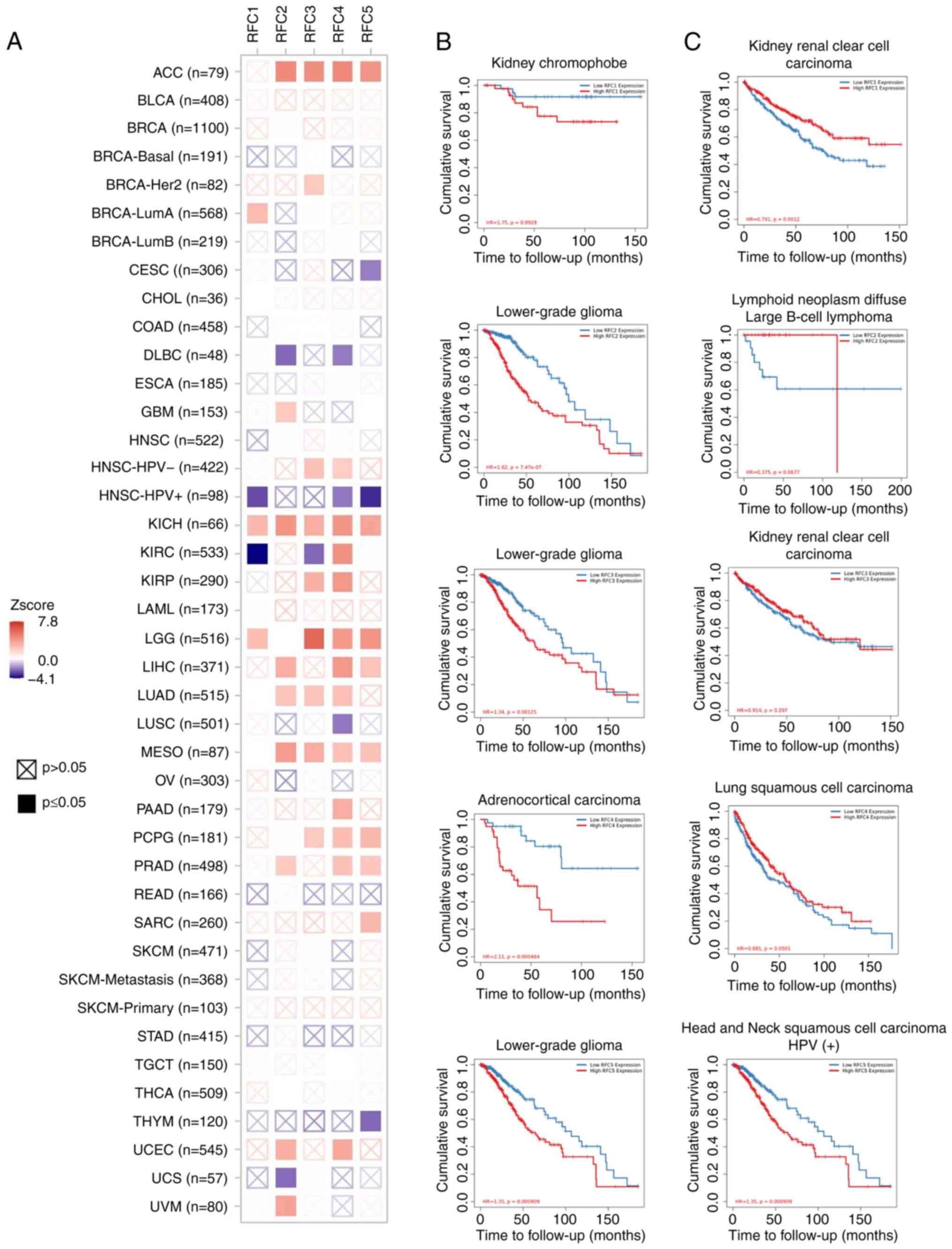
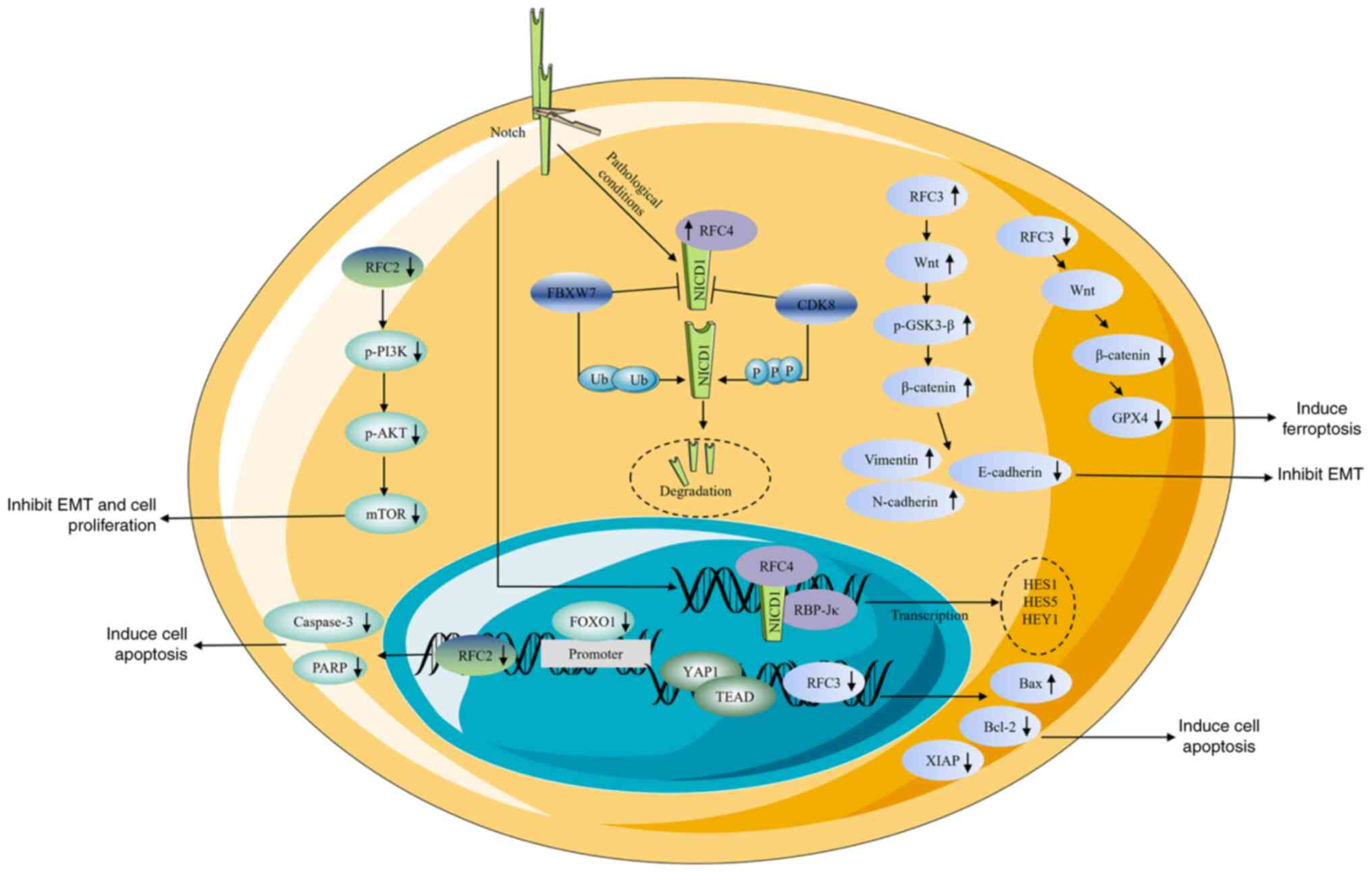
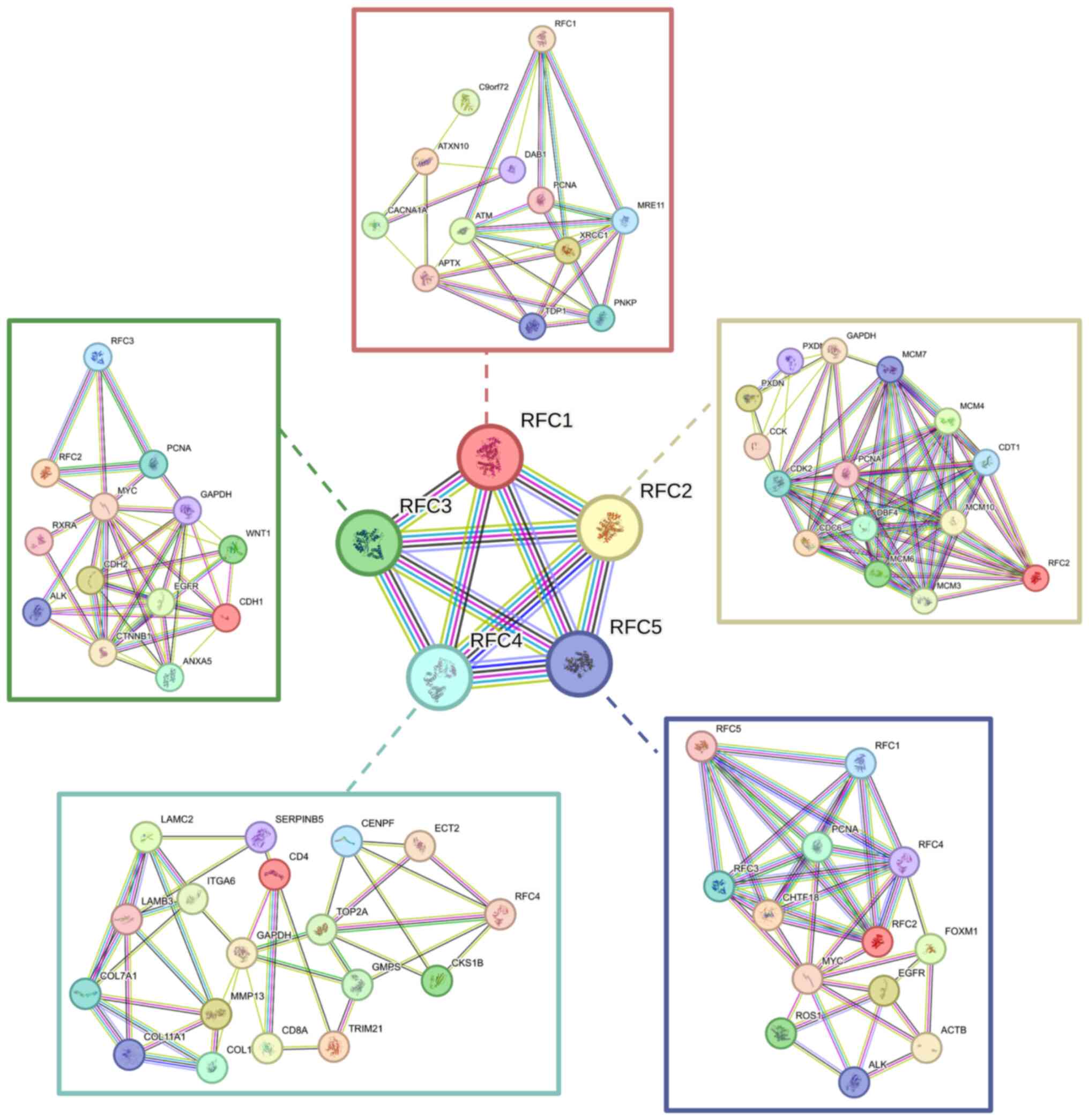
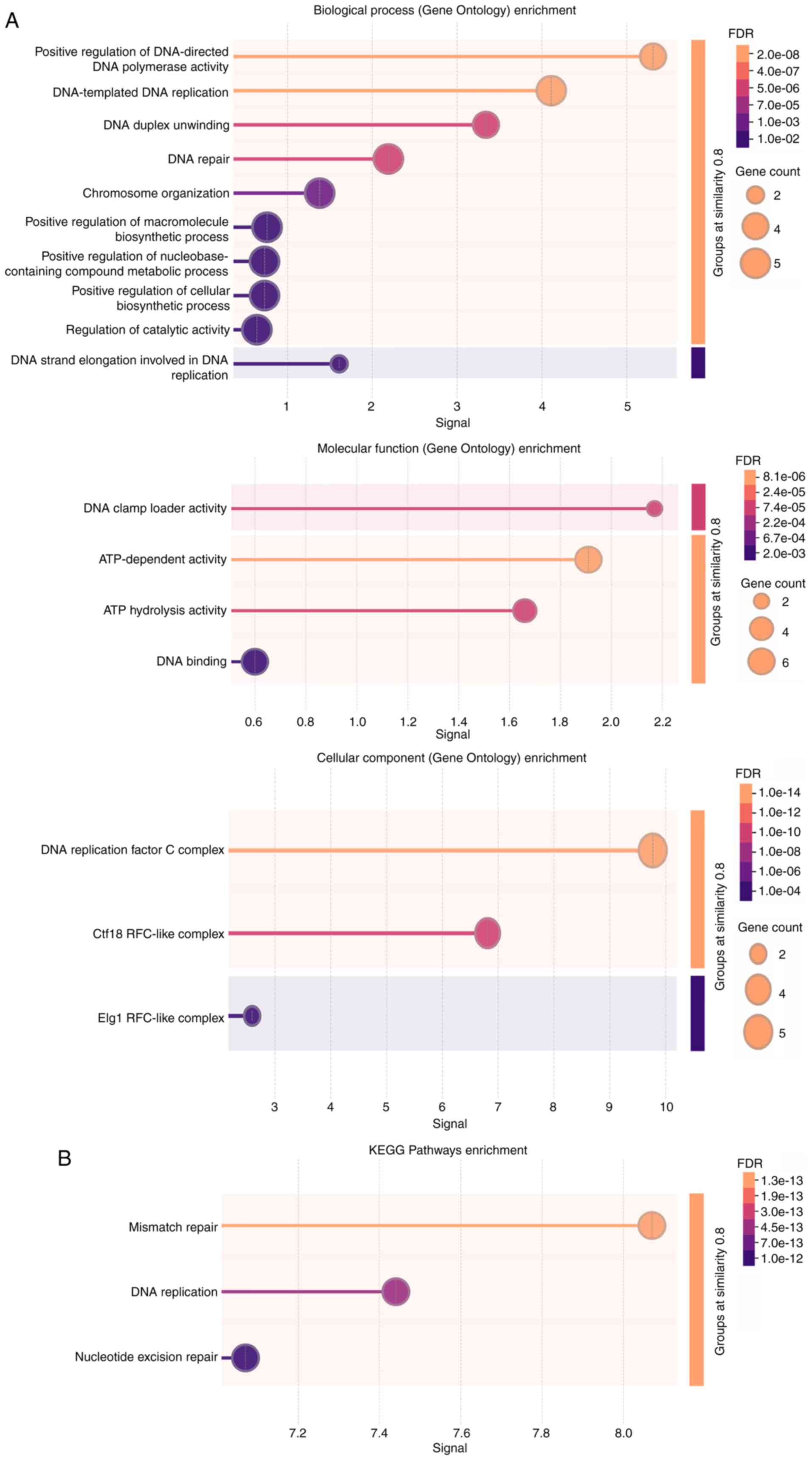
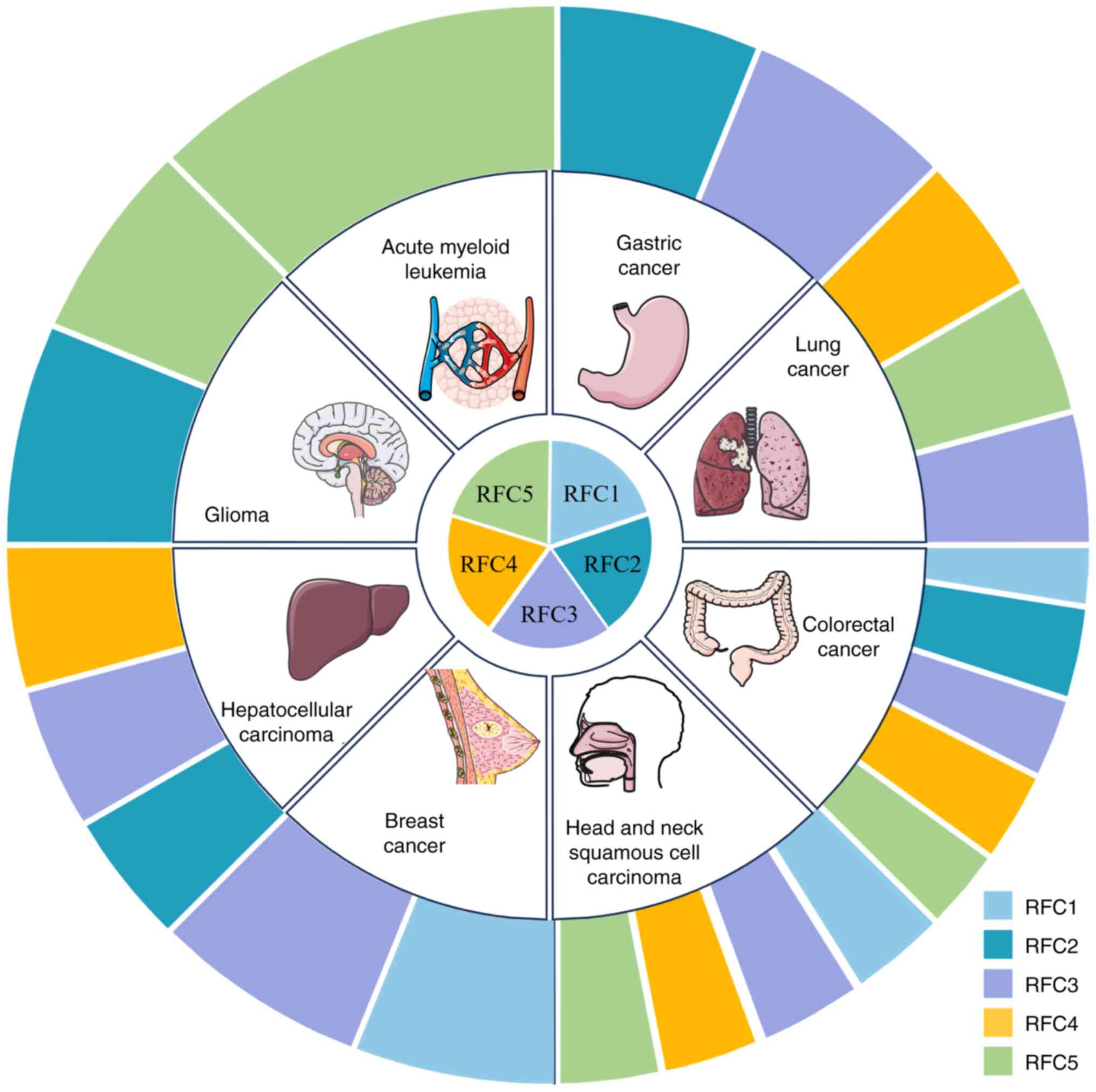
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.
|
|
2
|
Uhlmann F, Gibbs E, Cai J, O'Donnell M and
Hurwitz J: Identification of regions within the four small subunits
of human replication factor C required for complex formation and
DNA replication. J Biol Chem. 272:10065–10071. 1997. View Article : Google Scholar
|
|
3
|
Zhao X, Wang Y, Li J, Qu F, Fu X, Liu S,
Wang X, Xie Y and Zhang X: RFC2: A prognosis biomarker correlated
with the immune signature in diffuse lower-grade gliomas. Sci Rep.
12:31222022. View Article : Google Scholar
|
|
4
|
Yao Z, Hu K, Huang HE, Xu S, Wang Q, Zhang
P, Yang P and Liu B: shRNA-mediated silencing of the RFC3 gene
suppresses hepatocellular carcinoma cell proliferation. Int J Mol
Med. 36:1393–1399. 2015. View Article : Google Scholar
|
|
5
|
Guo Z and Guo L: Abnormal activation of
RFC3, A YAP1/TEAD downstream target, promotes gastric cancer
progression. Int J Clin Oncol. 29:442–455. 2024. View Article : Google Scholar
|
|
6
|
Misbah M, Kumar M, Najmi AK and Akhtar M:
Identification of expression profiles and prognostic value of RFCs
in colorectal cancer. Sci Rep. 14:66072024. View Article : Google Scholar
|
|
7
|
Lou F and Zhang M: RFC2 promotes aerobic
glycolysis and progression of colorectal cancer. BMC Gastroenterol.
23:3532023. View Article : Google Scholar
|
|
8
|
Li H, Chai L, Ding Z and He H: CircCOL1A2
sponges MiR-1286 to promote cell invasion and migration of gastric
cancer by elevating expression of USP10 to downregulate RFC2
ubiquitination level. J Microbiol Biotechn. 32:938–948. 2022.
View Article : Google Scholar
|
|
9
|
Wu Y, Chen T, Wu S, Huang Y and Li F:
Knockdown of RFC3 enhances the sensitivity of colon cancer cells to
oxaliplatin by inducing ferroptosis. Fund Clin Pharmacol.
39:e130442025. View Article : Google Scholar
|
|
10
|
Liu L, Tao T, Liu S, Yang X, Chen X, Liang
J, Hong R, Wang W, Yang Y, Li X, et al: An RFC4/Notch1 signaling
feedback loop promotes NSCLC metastasis and stemness. Nat Commun.
12:26932021. View Article : Google Scholar
|
|
11
|
Yang T, Fan Y, Bai G and Huang Y: RFC4
confers radioresistance of esophagus squamous cell carcinoma
through regulating DNA damage response. Am J Physiol Cell Physiol.
328:C367–C380. 2025. View Article : Google Scholar
|
|
12
|
Lee K and Park SH: Eukaryotic clamp
loaders and unloaders in the maintenance of genome stability. Exp
Mol Med. 52:1948–1958. 2020. View Article : Google Scholar
|
|
13
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020. View Article : Google Scholar
|
|
14
|
Mossi R and Hubscher U: Clamping down on
clamps and clamp Loaders-the eukaryotic replication factor C. Eur J
Biochem. 254:209–216. 1998. View Article : Google Scholar
|
|
15
|
Virshup DM, Kauffman MG and Kelly TJ:
Activation of SV40 DNA replication in vitro by cellular protein
phosphatase 2A. EMBO J. 8:3891–3898. 1989. View Article : Google Scholar
|
|
16
|
Virshup DM and Kelly TJ: Purification of
replication protein C, a cellular protein involved in the initial
stages of simian virus 40 DNA replication in vitro. Proc Natl Acad
Sci USA. 86:3584–3588. 1989. View Article : Google Scholar
|
|
17
|
Yoder BL and Burgers PM: Saccharomyces
cerevisiae replication factor C. I. Purification and
characterization of its ATPase activity. J Biol Chem.
266:22689–22697. 1991. View Article : Google Scholar
|
|
18
|
Burbelo PD, Utani A, Pan ZQ and Yamada Y:
Cloning of the large subunit of activator 1 (replication factor C)
reveals homology with bacterial DNA ligases. Proc Natl Acad Sci
USA. 90:11543–11547. 1993. View Article : Google Scholar
|
|
19
|
Li X and Burgers PM: Cloning and
characterization of the essential Saccharomyces cerevisiae RFC4
gene encoding the 37-kDa subunit of replication factor C. J Biol
Chem. 269:21880–21884. 1994. View Article : Google Scholar
|
|
20
|
Li X and Burgers PM: Molecular cloning and
expression of the Saccharomyces cerevisiae RFC3 gene, an essential
component of replication factor C. Proc Natl Acad Sci USA.
91:868–872. 1994. View Article : Google Scholar
|
|
21
|
Noskov V, Maki S, Kawasaki Y, Leem SH, Ono
B, Araki H, Pavlov Y and Sugino A: The RFC2 gene encoding a subunit
of replication factor C of Saccharomyces cerevisiae. Nucleic Acids
Res. 22:1527–1535. 1994. View Article : Google Scholar
|
|
22
|
Podust VN, Georgaki A, Strack B and
Hubscher U: Calf thymus RF-C as an essential component for DNA
polymerase delta and epsilon holoenzymes function. Nucleic Acids
Res. 20:4159–4165. 1992. View Article : Google Scholar
|
|
23
|
Li Y, Gan S, Ren L, Yuan L, Liu J, Wang W,
Wang X, Zhang Y, Jiang J, Zhang F and Qi X: Multifaceted regulation
and functions of replication factor C family in human cancers. Am J
Cancer Res. 8:1343–1355. 2018.
|
|
24
|
Okumura K, Nogami M, Taguchi H, Dean FB,
Chen M, Pan ZQ, Hurwitz J, Shiratori A, Murakami Y and Ozawa K:
Assignment of the 36.5-kDa (RFC5), 37-kDa (RFC4), 38-kDa (RFC3),
and 40-kDa (RFC2) subunit genes of human replication factor C to
chromosome bands 12q24.2-q24.3, 3q27, 13q12.3-q13, and 7q11.23.
Genomics. 25:274–278. 1995. View Article : Google Scholar
|
|
25
|
Uhlmann F, Cai J, Gibbs E, O'Donnell M and
Hurwitz J: Deletion analysis of the large subunit p140 in human
replication factor C reveals regions required for complex formation
and replication activities. J Biol Chem. 272:10058–10064. 1997.
View Article : Google Scholar
|
|
26
|
Cai J, Gibbs E, Uhlmann F, Phillips B, Yao
N, O'Donnell M and Hurwitz J: A complex consisting of human
replication Factor C p40, p37, and p36 subunits is a DNA-dependent
ATPase and an intermediate in the assembly of the holoenzyme. J
Biol Chem. 272:18974–18981. 1997. View Article : Google Scholar
|
|
27
|
Cullmann G, Fien K, Kobayashi R and
Stillman B: Characterization of the five replication factor C genes
of Saccharomyces cerevisiae. Mol Cell Biol. 15:4661–4671. 1995.
View Article : Google Scholar
|
|
28
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for Large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar
|
|
29
|
Delforge V, Tard C, Davion JB, Dujardin K,
Wissocq A, Dhaenens CM, Mutez E and Huin V: RFC1: Motifs and
phenotypes. Rev Neurol (Paris). 180:393–409. 2024. View Article : Google Scholar
|
|
30
|
Giovannini S, Weller M, Hanzlíková H,
Shiota T, Takeda S and Jiricny J: ATAD5 deficiency alters DNA
damage metabolism and sensitizes cells to PARP inhibition. Nucleic
Acids Res. 48:4928–4939. 2020. View Article : Google Scholar
|
|
31
|
Fung LF, Lo AK, Yuen PW, Liu Y, Wang XH
and Tsao SW: Differential gene expression in nasopharyngeal
carcinoma cells. Life Sci. 67:923–936. 2000. View Article : Google Scholar
|
|
32
|
Almalki E, Al-Amri A, Alrashed R,
AL-Zharani M and Semlali A: The curcumin analog PAC is a potential
solution for the treatment of Triple-negative breast cancer by
modulating the gene expression of DNA repair pathways. Int J Mol
Sci. 24:96492023. View Article : Google Scholar
|
|
33
|
Moggs JG, Murphy TC, Lim FL, Moore DJ,
Stuckey R, Antrobus K, Kimber I and Orphanides G:
Anti-proliferative effect of estrogen in breast cancer cells that
re-express ERalpha is mediated by aberrant regulation of cell cycle
genes. J Mol Endocrinol. 34:535–551. 2005. View Article : Google Scholar
|
|
34
|
Ji Z, Li J and Wang J: Up-regulated RFC2
predicts unfavorable progression in hepatocellular carcinoma.
Hereditas. 158:172021. View Article : Google Scholar
|
|
35
|
Oshima M, Takayama K, Yamada Y, Kimura N,
Kume H, Fujimura T and Inoue S: Identification of DNA damage
response-related genes as biomarkers for castration-resistant
prostate cancer. Sci Rep. 13:196022023. View Article : Google Scholar
|
|
36
|
Li G, Zhang P, Zhang W, Lei Z, He J, Meng
J, Di T and Yan W: Identification of key genes and pathways in
Ewing's sarcoma patients associated with metastasis and poor
prognosis. Onco Targets Ther. 12:4153–4165. 2019. View Article : Google Scholar
|
|
37
|
Koh JW and Park S: Knockdown of RFC2
prevents the proliferation, migration and invasion of cervical
cancer cells. Anticancer Res. 45:989–1000. 2025. View Article : Google Scholar
|
|
38
|
Avs KR, Pandi C, Kannan B, Pandi A,
Jayaseelan VP and Arumugam P: RFC3 serves as a novel prognostic
biomarker and target for head and neck squamous cell carcinoma.
Clin Oral Invest. 27:6961–6969. 2023. View Article : Google Scholar
|
|
39
|
Zhu J, Ye L, Sun S, Yuan J, Huang J and
Zeng Z: Involvement of RFC3 in tamoxifen resistance in ER-positive
breast cancer through the cell cycle. Aging. 15:13738–13752. 2023.
View Article : Google Scholar
|
|
40
|
Zhou J, Zhang W, Peng F, Sun J, He Z and
Wu S: Downregulation of hsa_circ_0011946 suppresses the migration
and invasion of the breast cancer cell line MCF-7 by targeting
RFC3. Cancer Manag Res. 10:535–544. 2018. View Article : Google Scholar
|
|
41
|
He Z, Wu S, Peng F, Zhang Q, Luo Y, Chen M
and Bao Y: Up-Regulation of RFC3 promotes triple negative breast
cancer metastasis and is associated with poor prognosis via EMT.
Transl Oncol. 10:1–9. 2017. View Article : Google Scholar
|
|
42
|
Gong S, Qu X, Yang S, Zhou S, Li P and
Zhang Q: RFC3 induces epithelial-mesenchymal transition in lung
adenocarcinoma cells through the Wnt/β-catenin pathway and
possesses prognostic value in lung adenocarcinoma. Int J Mol Med.
44:2276–2288. 2019.
|
|
43
|
Koh JW and Park S: RFC3 Knockdown
decreases cervical cancer cell proliferation, migration and
invasion. Cancer Genomics Proteomics. 22:127–135. 2024. View Article : Google Scholar
|
|
44
|
Yu R, Wu X, Qian F and Yang Q: RFC3 drives
the proliferation, migration, invasion and angiogenesis of
colorectal cancer cells by binding KIF14. Exp Ther Med. 27:2222024.
View Article : Google Scholar
|
|
45
|
Yu L, Li J, Zhang M, Li Y, Bai J, Liu P,
Yan J and Wang C: Identification of RFC4 as a potential biomarker
for pan-cancer involving prognosis, tumour immune microenvironment
and drugs. J Cell Mol Med. 28:e184782024. View Article : Google Scholar
|
|
46
|
Guan S, Feng L, Wei J, Wang G and Wu L:
Knockdown of RFC4 inhibits the cell proliferation of nasopharyngeal
carcinoma in vitro and in vivo. Front Med. 17:132–142. 2023.
View Article : Google Scholar
|
|
47
|
You P, Wang D, Liu Z, Guan S, Xiao N, Chen
H, Zhang X, Wu L, Wang G and Dong H: Knockdown of RFC 4 inhibits
cell proliferation of oral squamous cell carcinoma in vitro and in
vivo. FEBS Open Bio. 15:346–358. 2025. View Article : Google Scholar
|
|
48
|
Wang J, Luo F, Huang T, Mei Y, Peng L,
Qian C and Huang B: The upregulated expression of RFC4 and GMPS
mediated by DNA copy number alteration is associated with the early
diagnosis and immune escape of ESCC based on a bioinformatic
analysis. Aging (Albany NY). 13:21758–21777. 2021. View Article : Google Scholar
|
|
49
|
Zeng B, Liu X, Liu J, Qiu H, Zhang T, Chen
X and Wang L: Roles of MARCKSL1, MCM6, RFC4, and PLAU genes in
esophageal cancer and their association with radiotherapy response.
Sci Rep. 15:235592025. View Article : Google Scholar
|
|
50
|
Wang X, Yue X, Zhang R, Liu T, Pan Z, Yang
M, Lu Z, Wang Z, Peng J, Le L, et al: Genome-wide RNAi screening
identifies RFC4 as a factor that mediates radioresistance in
colorectal cancer by facilitating nonhomologous end joining repair.
Clin Cancer Res. 25:4567–4579. 2019. View Article : Google Scholar
|
|
51
|
Xiang J, Fang L, Luo Y, Yang Z, Liao Y,
Cui J, Huang M, Yang Z, Huang Y, Fan X, et al: Levels of human
replication factor C4, a clamp loader, correlate with tumor
progression and predict the prognosis for colorectal cancer. J
Transl Med. 12:3202014. View Article : Google Scholar
|
|
52
|
Arai M, Kondoh N, Imazeki N, Hada A,
Hatsuse K, Matsubara O and Yamamoto M: The knockdown of endogenous
replication factor C4 decreases the growth and enhances the
chemosensitivity of hepatocellular carcinoma cells. Liver Int.
29:55–62. 2009. View Article : Google Scholar
|
|
53
|
Erdogan E, Klee EW, Thompson EA and Fields
AP: Meta-analysis of oncogenic protein kinase Ciota signaling in
lung adenocarcinoma. Clin Cancer Res. 15:1527–1533. 2009.
View Article : Google Scholar
|
|
54
|
Zheng J, Lin N, Huang B, Wu M, Xiao L and
Zeng B: Comprehensive analysis of RFC4 as a potential biomarker for
regulating the immune microenvironment and predicting immune
therapy response in lung adenocarcinoma. Front Immunol.
16:15782432025. View Article : Google Scholar
|
|
55
|
Koh JW and Park S: Replication Factor C
Subunit 4 plays a role in human breast cancer cell progression.
Cancer Genomics Proteomics. 22:478–490. 2025. View Article : Google Scholar
|
|
56
|
Zhu XY, Li PS, Qu H, Ai X, Zhao ZT and He
JB: Replication factor C4, which is regulated by insulin-like
growth factor 2 mRNA binding protein 2, enhances the
radioresistance of breast cancer by promoting the stemness of tumor
cells. Hum Cell. 38:652025. View Article : Google Scholar
|
|
57
|
Huang B, Zheng J, Chen B, Wu M and Xiao L:
Analysis of the correlation between RFC4 expression and tumor
immune microenvironment and prognosis in patients with cervical
cancer. Front Genet. 16:15143832025. View Article : Google Scholar
|
|
58
|
Zhao X, Sun Y, Sun X, Li J, Shi X, Liang
Z, Ma Y and Zhang X: AEG-1 Knockdown sensitizes glioma cells to
radiation through impairing homologous recombination via targeting
RFC5. DNA Cell Biol. 40:895–905. 2021. View Article : Google Scholar
|
|
59
|
Martinez I, Wang J, Hobson KF, Ferris RL
and Khan SA: Identification of differentially expressed genes in
HPV-positive and HPV-negative oropharyngeal squamous cell
carcinomas. Eur J Cancer. 43:415–432. 2007. View Article : Google Scholar
|
|
60
|
Wang M, Xie T, Wu Y, Yin Q, Xie S, Yao Q,
Xiong J and Zhang Q: Identification of RFC5 as a novel potential
prognostic biomarker in lung cancer through bioinformatics
analysis. Oncol Lett. 16:4201–4210. 2018.
|
|
61
|
Xu S, Zhong F, Jiang J, Yao F, Li M, Tang
M, Cheng Y, Yang Y, Wen W, Zhang X, et al: High Expression of
SRSF10 promotes colorectal cancer progression by aberrant
alternative splicing of RFC5. Technol Cancer Res.
23:153303382412719062024. View Article : Google Scholar
|
|
62
|
Yao H, Zhou X, Zhou A, Chen J, Chen G, Shi
X, Shi B, Tai Q, Mi X, Zhou G, et al: RFC5, regulated by
circ_0038985/miR-3614-5p, functions as an oncogene in the
progression of colorectal cancer. Mol Carcinog. 62:771–785. 2023.
View Article : Google Scholar
|
|
63
|
Li W, Wu D, Zhou Y, Zhang C and Liao X:
Prognostic biomarker replication factor C subunit 5 and its
correlation with immune infiltrates in acute myeloid leukemia.
Hematology. 27:555–564. 2022. View Article : Google Scholar
|
|
64
|
Zhang Y, Gao X, Yi J, Sang X, Dai Z, Tao
Z, Wang M, Shen L, Jia Y, Xie D, et al: BTF3 confers oncogenic
activity in prostate cancer through transcriptional upregulation of
Replication Factor C. Cell Death Dis. 12:122021. View Article : Google Scholar
|
|
65
|
Qiu X, Tan G, Wen H, Lian L and Xiao S:
Forkhead box O1 targeting replication factor C subunit 2 expression
promotes glioma temozolomide resistance and survival. Ann Transl
Med. 9:6922021. View Article : Google Scholar
|
|
66
|
Day M, Oliver AW and Pearl LH: Structure
of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp
bound to a dsDNA-ssDNA junction. Nucleic Acids Res. 50:8279–8289.
2022. View Article : Google Scholar
|
|
67
|
Niida H and Nakanishi M: DNA damage
checkpoints in mammals. Mutagenesis. 21:3–9. 2006. View Article : Google Scholar
|
|
68
|
Bermudez VP, Lindsey-Boltz LA, Cesare AJ,
Maniwa Y, Griffith JD, Hurwitz J and Sancar A: Loading of the human
9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader
hRad17-replication factor C complex in vitro. Proc Natl Acad Sci
USA. 100:1633–1638. 2003. View Article : Google Scholar
|
|
69
|
Wang X, Zou L, Zheng H, Wei Q, Elledge SJ
and Li L: Genomic instability and endoreduplication triggered by
RAD17 deletion. Gene Dev. 17:965–970. 2003. View Article : Google Scholar
|
|
70
|
Terret ME, Sherwood R, Rahman S, Qin J and
Jallepalli PV: Cohesin acetylation speeds the replication fork.
Nature. 462:231–234. 2009. View Article : Google Scholar
|
|
71
|
Kim JT, Cho HJ, Park SY, Oh BM, Hwang YS,
Baek KE, Lee YH, Kim HC and Lee HG: DNA replication and sister
chromatid cohesion 1 (DSCC1) of the replication factor complex
CTF18-RFC is critical for colon cancer cell growth. J Cancer.
10:6142–6153. 2019. View Article : Google Scholar
|
|
72
|
Kang M, Ryu E, Lee S, Park J, Ha NY, Ra
JS, Kim YJ, Kim J, Abdel-Rahman M, Park SH, et al: Regulation of
PCNA cycling on replicating DNA by RFC and RFC-like complexes. Nat
Commun. 10:24202019. View Article : Google Scholar
|
|
73
|
Sikdar N, Banerjee S, Lee KY, Wincovitch
S, Pak E, Nakanishi K, Jasin M, Dutra A and Myung K: DNA damage
responses by human ELG1 in S phase are important to maintain
genomic integrity. Cell Cycle. 8:3199–3207. 2009. View Article : Google Scholar
|
|
74
|
Lee KY, Fu H, Aladjem MI and Myung K:
ATAD5 regulates the lifespan of DNA replication factories by
modulating PCNA level on the chromatin. J Cell Biol. 200:31–44.
2013. View Article : Google Scholar
|
|
75
|
Szklarczyk D, Nastou K, Koutrouli M,
Kirsch R, Mehryary F, Hachilif R, Hu D, Peluso ME, Huang Q, Fang T,
et al: The STRING database in 2025: Protein networks with
directionality of regulation. Nucleic Acids Res. 53:D730–D737.
2025. View Article : Google Scholar
|
|
76
|
Hopfner KP and Hornung V: Molecular
mechanisms and cellular functions of cGAS-STING signalling. Nat Rev
Mol Cell Bio. 21:501–521. 2020. View Article : Google Scholar
|
|
77
|
Yuan Q, Cai S, Chang Y, Zhang J, Wang M,
Yang K and Jiang D: The multifaceted roles of mucins family in lung
cancer: From prognostic biomarkers to promising targets. Front
Immunol. 16:16081402025. View Article : Google Scholar
|
|
78
|
Wu G, Zhou J, Zhu X, Tang X, Liu J, Zhou
Q, Chen Z, Liu T, Wang W, Xiao X and Wu T: Integrative analysis of
expression, prognostic significance and immune infiltration of RFC
family genes in human sarcoma. Aging (Albany NY). 14:3705–3719.
2022. View Article : Google Scholar
|
|
79
|
Deng J, Zhong F, Gu W and Qiu F:
Exploration of prognostic biomarkers among replication factor C
family in the hepatocellular carcinoma. Evol Bioinform.
17:11769343219941092021. View Article : Google Scholar
|
|
80
|
Zhang J, Meng S, Wang X, Wang J, Fan X,
Sun H, Ning R, Xiao B, Li X, Jia Y, et al: Sequential gene
expression analysis of cervical malignant transformation identifies
RFC4 as a novel diagnostic and prognostic biomarker. BMC Med.
20:4372022. View Article : Google Scholar
|
|
81
|
Tu S, Zhang H, Yang X, Wen W, Song K, Yu X
and Qu X: Screening of cervical cancer-related hub genes based on
comprehensive bioinformatics analysis. Cancer Biomark. 32:303–315.
2021. View Article : Google Scholar
|
|
82
|
Ho KH, Kuo TC, Lee YT, Chen PH, Shih CM,
Cheng CH, Liu AJ, Lee CC and Chen KC: Xanthohumol regulates
miR-4749-5p-inhibited RFC2 signaling in enhancing temozolomide
cytotoxicity to glioblastoma. Life Sci. 254:1178072020. View Article : Google Scholar
|
|
83
|
Alaa Eldeen M, Mamdouh F, Abdulsahib WK,
Eid RA, Alhanshani AA, Shati AA, Alqahtani YA, Alshehri MA, Samir
A, Zaki M, et al: Oncogenic potential of replication factor C
subunit 4: Correlations with tumor progression and assessment of
potential inhibitors. Pharmaceuticals (Basel). 17:1522024.
View Article : Google Scholar
|
|
84
|
Rohde JM, Rai G, Choi YJ, Sakamuru S, Fox
JT, Huang R, Xia M, Myung K, Boxer MB and Maloney DJ: Discovery of
ML367, inhibitor of ATAD5 stabilization. Probe Reports from the NIH
Molecular Libraries Program [Internet] Bethesda (MD): National
Center for Biotechnology Information (US); 2010
|
|
85
|
Xu J, Wang G, Gong W, Guo S, Li D and Zhan
Q: The noncoding function of NELFA mRNA promotes the development of
oesophageal squamous cell carcinoma by regulating the Rad17-RFC2-5
complex. Mol Oncol. 14:611–624. 2020. View Article : Google Scholar
|
|
86
|
Cortese A, Simone R, Sullivan R,
Vandrovcova J, Tariq H, Yau WY, Humphrey J, Jaunmuktane Z,
Sivakumar P, Polke J, et al: Biallelic expansion of an intronic
repeat in RFC1 is a common cause of late-onsetataxia. Nat Genet.
51:649–658. 2019. View Article : Google Scholar
|
|
87
|
Haffner MC, Zwart W, Roudier MP, True LD,
Nelson WG, Epstein JI, De Marzo AM, Nelson PS and Yegnasubramanian
S: Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev
Urol. 18:79–92. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Huang S, Chen Y, Hu M, Fu S, Yao Z, He H,
Wang L, Chen Z and Liu X: The role of the SIX family in cancer
development and therapy: Insights from foundational and
cutting-edge research. Crit Rev Oncol Hemat. 214:1048602025.
View Article : Google Scholar : PubMed/NCBI
|