Introduction
Due to the high incidence rates and mortality
burden, cancer is a notable societal, public health and economic
problem. Although a variety of treatments have been proposed, the
therapeutic efficiency and safety remain to be improved. There is
an urgent need to develop more effective treatment strategies
(1). Consequently, understanding
the functional mechanisms of key genes during tumorigenesis holds
notable clinical implications.
The replication factor C (RFC) family consists of
five paralogous members (RFC1-5) characterized by distinct
structural domains and conserved functional motifs. The sequence
homology arises from the strong similarity in the sequences of
these family members. RFC1 exhibits unique structural features
including the exclusive RFC box I domain, while boxes II–VIII are
structurally conserved across all RFC paralogs (2).
Accumulating evidence highlights the critical role
of RFC family members in tumorigenesis and cancer progression.
Functional experimental studies have reported the antitumor or
pro-tumor effects of RFC family members, such as promoting cell
apoptosis (3), inducing cell cycle
arrest (4) and inhibiting
epithelial-mesenchymal transition (EMT) (5). Numerous studies have also demonstrated
the regulatory function of RFC family members, including microRNA
(miR)-mediated translational suppression (6), post-translational modification via
cascade effects (7) and epigenetic
modification (8). In addition,
substantial evidence suggests some RFC family members also
participate in several key signaling pathways, including the
Wnt/β-catenin (9), Notch pathway
(10) and p53 pathways (11).
Moreover, RFC-like complexes (RLCs) also are
involved in tumorigenic processes, particularly in DNA-related
damage and repair. Notably, RLCs have emerged as integral
components of tumorigenic processes, particularly in maintaining
genomic stability through coordinated participation in DNA damage
response pathways and repair mechanisms (12). Due to the multifaceted roles of RFC
family in the context of cancer, its members have the potential to
be biomarkers for precision oncology. However, comprehensive
analyses of their contributions to tumor development and
progression remain limited.
In the present review, the differential expression
of RFC family members in pan-cancer was systematically evaluated
using Tumor IMmune Estimation Resource (TIMER2.0) (13). As potential biomarkers for patients,
the relationship between expression levels and clinical importance
were investigated. Furthermore, the present review summarized
current research on RFC family members in various malignancies with
particular emphasis on their specific contributions to signaling
pathway regulation. The present article aimed to systematically
summarize the expression characteristics, functional heterogeneity
and molecular mechanisms of the members of the RFC family in
various cancers, and to explore their potential as diagnostic
markers and therapeutic targets.
Overview of RFC
The replication factor C (RFC) protein, first
purified from 293T cells, has been shown to interact with SV40
large T antigen during the initial phases of viral DNA replication
in vitro (14–16). Subsequently, RFC was also purified
from HeLa cells. RFC exhibits multiple activities, including its
ability to participate in DNA replication in an ATP- and
proliferating cell nuclear antigen (PCNA)-dependent reaction
(17). Moreover, it is also
involved in pleiotropic biological activities, including DNA
repair, transcriptional regulation, cell cycle, apoptosis, cellular
differentiation and telomere-length regulation (14).
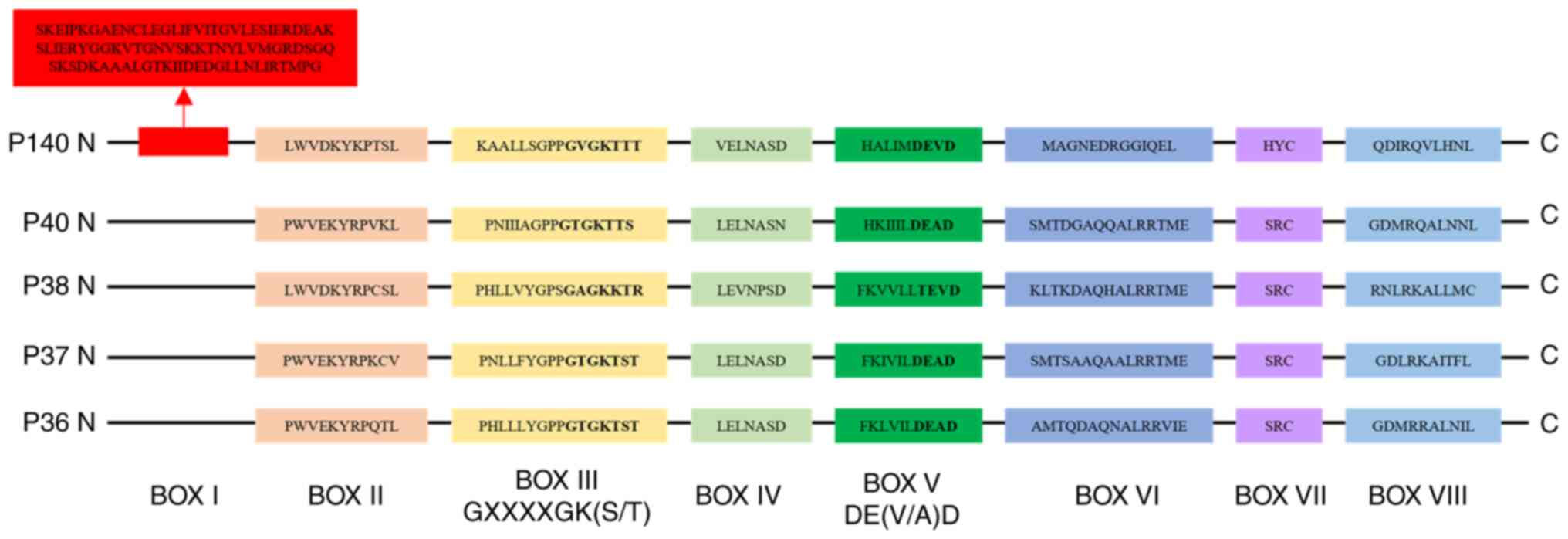
Structure and expression of RFC family
members in pan-cancer
The RFC family comprises of five distinct members
(RFC1, RFC2, RFC3, RFC4 and RFC5) with molecular weights of 140,
40, 38, 37 and 36 kDa respectively. These proteins have been
discovered in Saccharomyces cerevisiae, Mus musculus, Homo
sapiens, Calf thymus and Escherichia coli (18–22).
It has been reported that p140 (RFC1), p40 (RFC2),
p38 (RFC3), p37 (RFC4) and p36 (RFC5) are
located on human chromosomes at 4p13-p14, 7q11.23, 13q12.3-q13,
3q27 and 12q24.2-q24.3, respectively (23,24).
In humans, the five subunits of RFC complex exhibit
sequence conservation across conserved regions known as RFC boxes
II–VIII, which are critical for their functional interactions
(2,20). The RFC box I, which is located on
the large subunit p140 and consists of 89 amino acids, demonstrates
sequence homology with prokaryotic DNA ligases (2). Additionally, it has been demonstrated
that deletion of RFC box II domain markedly impairs DNA synthesis,
which requires the RFC complex for its function (25). The highly conserved RFC box III
GXXXXGK(S/T) motif contains the phosphate-binding loop (P-loop),
the most evolutionarily conserved structural element within this
motif. By contrast, RFC box IV exhibits limited conservation in
prokaryotic proteins. RFC box V DE(V/A)D and DEAD-box proteins have
similar features. Within the large subunit p140, RFC box VI is
designated VIa, whereas it is referred to as VIb in the other four
small subunits (14,26). RFC box VII (SRC) is conserved within
the small subunits and the prokaryotic accessory proteins, but only
the cysteine is present in the large RFC subunits. Notably, the HYC
motif in RFC1 corresponds to box VII (27). In the human RFC family, RFC box VIII
displays subunit-specific amino acid sequence variations. The
characteristic sequences of RFC boxes are shown in Fig. 1.
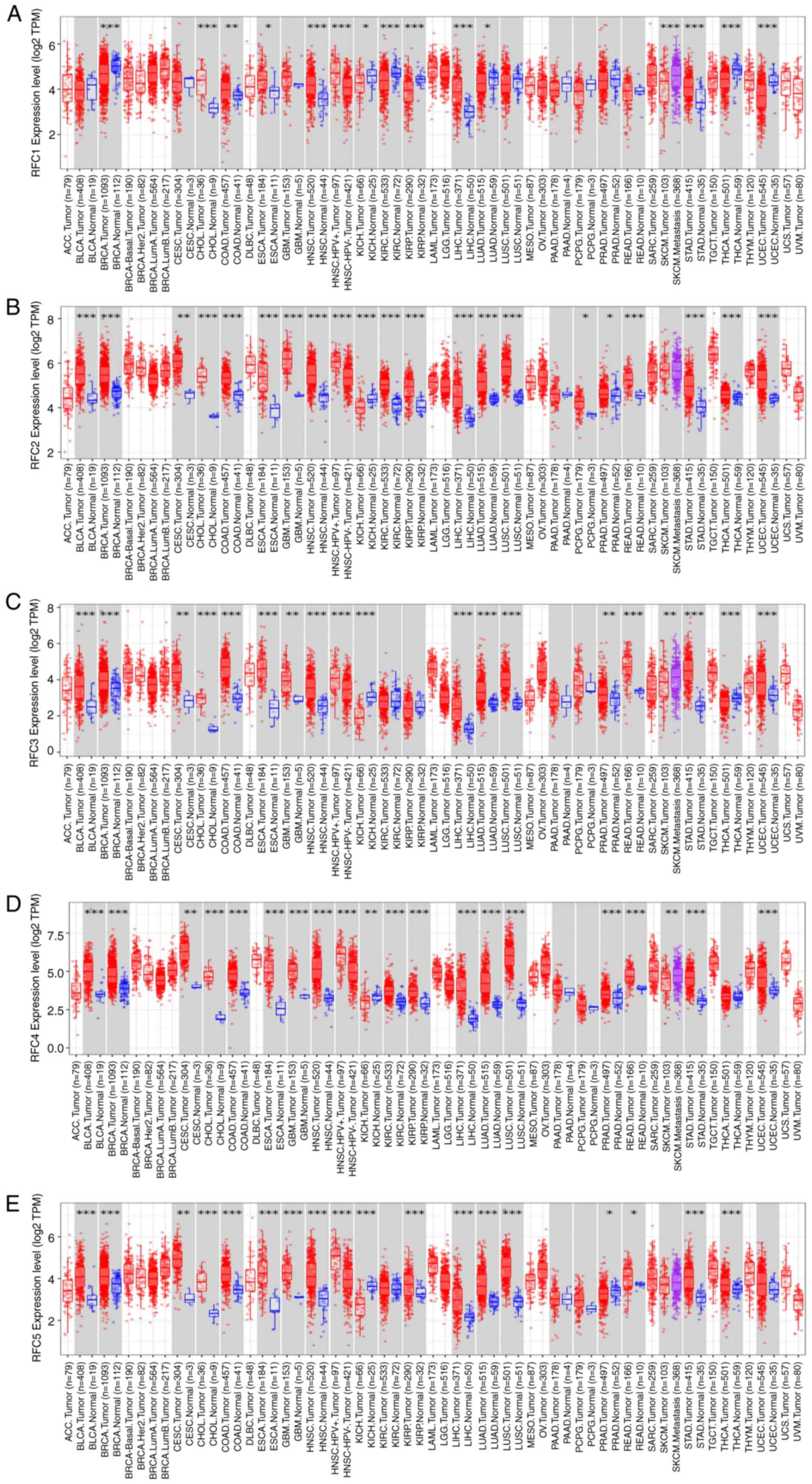
The mRNA expression of RFC family members was
evaluated in pan-cancer using TIMER 2.0 (Fig. 2) (13). According to the results, in the most
types of tumors, the RFC family members are expressed at higher
levels in tumor tissues compared with in normal tissues. Notably,
in kidney chromophobe, the expression levels of all RFC family
members are markedly lower in tumor tissues compared with in normal
tissues. The underlying mechanisms warrant further
investigation.
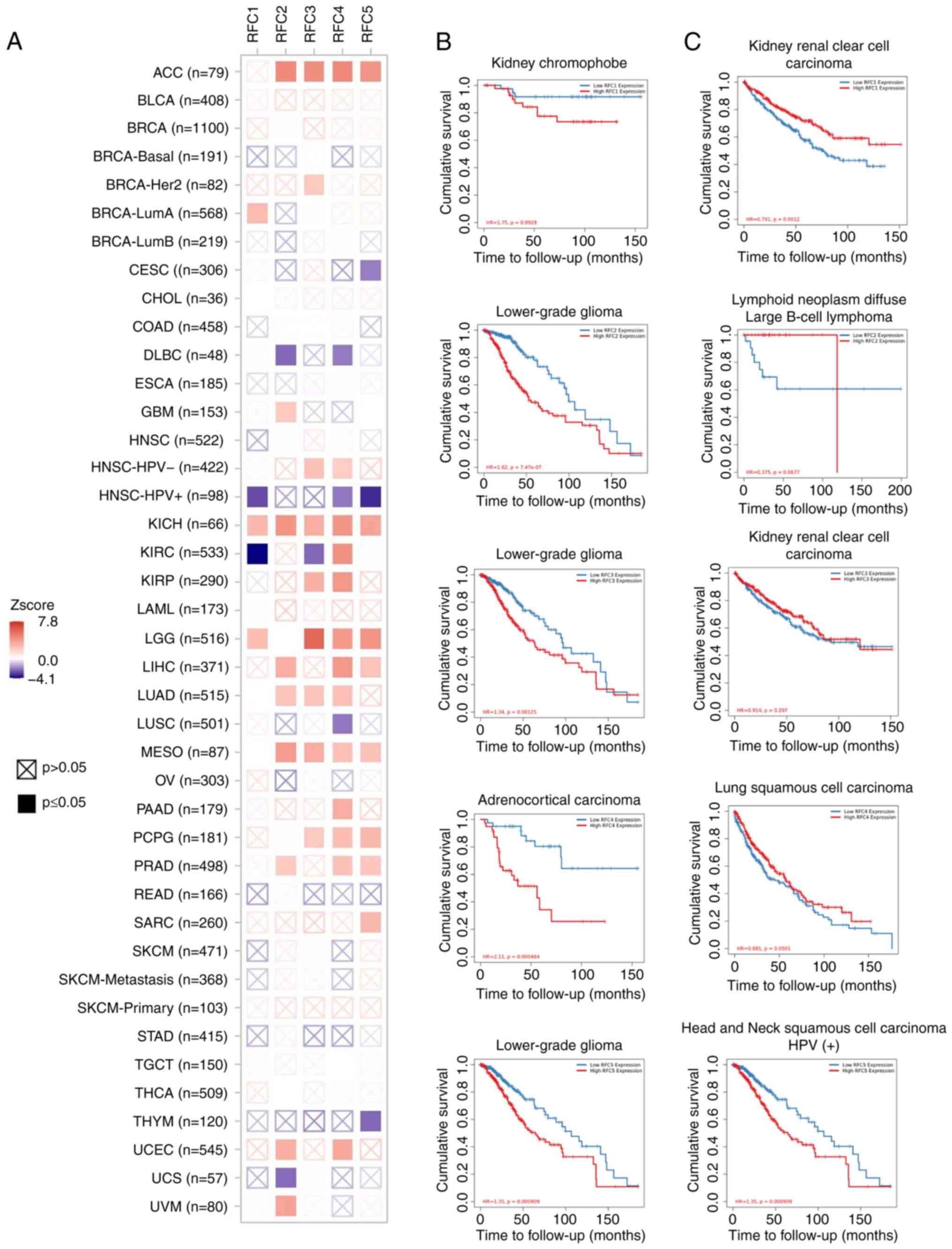
The clinical relevance of RFC gene expression across
various cancer types was assessed using TIMER 2.0 (Fig. 3). For each RFC subunit, the cancer
type with the highest and lowest Z score were summarized. The
results are presented in Table I.
Kaplan-Meier analysis was used to evaluate the overall survival
(OS). The potential relationship between the mRNA expression of RFC
subunits and overall survival (OS) in patients with various cancers
was analyzed (Fig. 3) (28). Elevated expression levels of RFC2,
RFC3 and RFC5 were associated with worse outcomes in brain
lower-grade glioma (LGG). Similarly, high RFC4 expression was
associated with poor survival in adrenocortical carcinoma (ACC). By
contrast, reduced RFC1 expression was associated with worse
outcomes in kidney renal clear cell carcinoma, while decreased RFC5
expression was associated with poor prognosis in human
papillomavirus (HPV)-positive head and neck squamous cell carcinoma
(HNSCC). These results indicate that RFC family members have the
potential to be biomarkers for precision oncology.
 | Table I.Association between cancer type and
the mRNA expression of RFC subunits. |
Table I.
Association between cancer type and
the mRNA expression of RFC subunits.
| Gene | Cancer | Z score | adj.p |
|---|
| RFC1 | KICH (n=66) | 2.711109947 | 0.047403342 |
|
| KIRC (n=533) | −4.07798685 | 0.000924303 |
| RFC2 | LGG (n=516) | 7.83870425 |
0.000000000000933 |
|
| DLBC (n=48) | −2.37849173 | 0.091375475 |
| RFC3 | LGG (n=516) | 5.892615212 | 0.00000039 |
|
| KIRC (n=533) | −2.39697366 | 0.091375475 |
| RFC4 | ACC (n=79) | 4.644463897 | 0.00023299 |
|
| LUSC (n=501) | −2.14599312 | 0.142045069 |
| RFC5 | LGG (n=516) | 4.114157942 | 0.000924303 |
|
| HNSC-HPV+
(n=98) | −3.47315808 | 0.005858129 |
Function of RFC subunits in human
cancers
RFC1
The RFC1 gene, localized on chromosome 4, encodes
the ubiquitously expressed RFC1 protein which serves as the largest
subunit of the RFC family. This protein serves a critical role in
DNA replication and repair processes (29). In humans, three functional homologs
of RFC1 have been identified including RAD17 checkpoint clamp
loader component (RAD17), chromosome transmission fidelity factor
18 (CTF18) and ATPase family AAA domain containing 5 (ATAD5). These
homologs can associate with other subunits through protein-protein
interactions, which facilitates the assembly of RLCs (30).
The expression of RFC1 is overexpressed in malignant
nasopharyngeal carcinoma (NPC) cells (31). In breast cancer (BC), the
curcumin-analog PAC markedly upregulates the expression of RFC1
which is involved in the nucleotide excision repair (NER) pathway
(32). A study on estrogen receptor
(ER)-negative MDA-MB-231 BC cells revealed that 17β-estradiol
exposure downregulated RFC1 expression, leading to re-expression of
ERα (33). Emerging evidence in
colorectal cancer (CRC) research suggests that reduced RFC1
expression may function as a tumor suppressor mechanism.
Mechanistic investigation identified post-transcriptional
regulation of RFC1 via microRNA-26a-5p targeting of its
3′-untranslated region, which modulates critical DNA maintenance
processes, including mismatch repair, DNA replication fidelity and
NER pathway functionality (6). RFC1
demonstrates more distinctive features than the other four RFC
family members, making it a promising candidate for further cancer
research. Now that the notable effects of RFC1 have drawn attention
in cancer research, similar focus has been directed to other
members of the RFC family. Evidently, a deeper understanding of the
roles of RFC family members in cancer biology is crucial, as it may
facilitate the discovery of novel signaling pathways and potential
therapeutic targets.
RFC2
The RFC2 gene, located on chromosome 7, encodes the
unique RFC subunit that can independently unload PCNA and suppress
DNA polymerase activity.
In diffuse LGG, bioinformation analysis has shown
that overexpressed RFC2 is strongly associated with pivotal immune
checkpoint genes [including programmed cell death protein 1 (PD-1),
programmed death ligand 1 (PD-L1), PD-L2, B7-H2 and cytotoxic
T-lymphocyte associated protein 4 (CTLA4)] and serves as an
independent predictor of adverse prognosis. In cell models,
functional experiments demonstrate that RFC2 serves an anticancer
role by promoting cell apoptosis, inhibiting proliferation and
inducing cell G2 phase arrest (3). In gastric cancer (GC), circular
RNA-collagen type Iα2 chain functions as a competing endogenous RNA
by sponging miR-1286, upregulating ubiquitin specific peptidase 10
expression and attenuating RFC2 ubiquitination, ultimately
enhancing cell invasion and migration (8). In hepatocellular carcinoma (HCC), as a
novel biomarker for the prognosis, high expression of RFC2 was
associated with worse OS and disease-free survival. Functional
experiments demonstrate that RFC2 knockdown inhibits malignant
behaviors including proliferation and migration of HCC cell lines
(34). In CRC, upregulated RFC2
expression is associated with poor clinical outcomes.
Functional studies have demonstrated that RFC2
silencing notably attenuates malignant phenotypes, including tumor
cell proliferation, migration and invasion. Notably, knockdown of
CAMP responsive element binding protein 5 which is the
transcription factor of RFC2 markedly suppresses RFC2 expression.
Moreover, RFC2 promotes aerobic glycolysis and MET/PI3K/AKT/mTOR
pathway, thereby driving tumorigenesis (7). In castration-resistant prostate cancer
(CRPC) cell lines, RFC2 downregulation markedly inhibits cell
proliferation, induces apoptosis and aggravates DNA damage
(35). Furthermore, clinical
evidence demonstrates that elevated RFC2 protein expression is
associated with unfavorable prognosis in patients with prostate
cancer (PCa). Notably, RFC2 expression levels are markedly higher
in CRPC tissues compared with localized PCa, suggesting a potential
role in disease progression (35).
In metastatic Ewing's sarcoma (ES), RFC2 mRNA expression is
markedly upregulated. Elevated RFC2 levels are inversely associated
with poor OS and event-free survival in patients with ES (36). In cervical cancer (CC), RFC2 serves
a pivotal role in promoting cervical cancer progression by
enhancing cell growth, regulating the cell cycle and promoting
metastatic behavior (37).
Therefore, RFC2 exerts pro-tumorigenic effects. Designing a
targeted drug tailored to RFC2 may be a promising precision
medicine approach.
RFC3
The RFC3 gene, located on chromosome 13 in humans,
is involved in cell proliferation and DNA damage repair, and its
abnormal activation is associated with various malignant
tumors.
In HNSCC, bioinformation analysis has demonstrated
that the high expression of RFC3 is notably associated with
clinicopathological features, including tumor stage, grade,
metastasis and patient survival (38). Additionally, it is also associated
with immune cell infiltration and well-known oncogenic signaling
pathways, such as MYC/MYCN, Hippo and mTOR (38). In the ER-positive BC cell line
MCF-7, upregulated RFC3 is notably associated with poor prognosis,
and downregulated RFC3 induces S phase arrest and attenuated cell
proliferation, migration and invasion (39). The downregulation of
hsa_circ_0011946 inhibits the migration and invasion of MCF-7 cells
by targeting RFC3 (40).
Upregulation of RFC3 is associated with poor prognostic phenotype
in BC (41). In lung adenocarcinoma
(LUAD), RFC3 overexpression is not only associated with poor
prognosis but also facilitates cell cycle progression from
G1 to S phase, potentially contributing to tumor growth.
Mechanistically, RFC3 promotes EMT through the Wnt/β-catenin
signaling pathway (42). Similarly,
RFC3 silencing induces cell cycle arrest at S phase, leading to
proliferation inhibition in HCC (4)
and CC (43). In GC, the
yes-associated protein 1 (YAP1)/TEA domain family member 1 (TEAD)
pathway transcriptionally activates RFC3, driving tumor
progression; conversely, RFC3 depletion abolishes malignant
behaviors in vitro and in vivo (5). In CRC, kinesin family member 14
upregulation counteracts the suppressive effects of RFC3 depletion
on aggressive phenotypes, highlighting its functional role
(44).
RFC4
The RFC4 gene, localized to chromosome 3q27 in
humans, serves a critical role in DNA replication and repair. RFC4
is indispensable for the initiation of DNA template expansion by
DNA polymerase δ (Pol δ) and Pol ε, essential components of the DNA
replication machinery (45).
In NPC, RFC4 expression is markedly upregulated in
tumor tissues compared with adjacent normal tissues. Functional
studies have revealed that RFC4 knockdown induces G2/M
cell cycle arrest and inhibits cellular proliferation both in
vitro and in vivo. Notably, HOXA10 has been identified
as a downstream effector of RFC4, and overexpression of HOXA10
partially rescues the suppressive effects of RFC4 silencing,
including impaired cell proliferation, inhibited colony formation
and cell cycle arrest (46). In
oral squamous cell carcinoma (OSCC), bioinformatics analysis
identified high RFC4 expression is an independent prognostic factor
for poor survival and associated with increased levels of MET,
along with reduced levels of CD274 and CD160 (47). Knockdown of RFC4 led to
G2/M phase cell cycle arrest and inhibited the
proliferation of OSCC cells both in vitro and in vivo
(47).
In esophageal squamous cell carcinoma (ESCC), RFC4
overexpression may be driven by copy number alterations, which
contribute to genomic instability and oncogenic activation.
Notably, RFC4 is upregulated not only in early-stage tumors but
also during early nodal metastasis, implying that it has a role in
early dissemination. Clinically, high RFC4 expression predicts poor
prognosis, independent of the presence of abundant
tumor-infiltrating immune cells (such as CD4+ T cells,
CD8+ T cells, B cells, dendritic cells and monocytes),
suggesting that RFC4-mediated immune evasion or resistance may
bypass immune surveillance (48).
In addition, Zeng et al (49). collected a small sample database
which comprised seven patients and the data showed that patients
with low expression of RFC4 exhibited a higher tumor shrinkage rate
compared with patients with high expression of RFC4. Further
validation in larger cohorts is required to confirm whether RFC4
expression is indeed associated with radiotherapy response in
esophageal cancer.
In CRC, RFC4 functions as a radiation resistance
factor, protecting CRC cells from radiation-induced DNA
double-strand breaks (DSBs) and apoptosis in vitro as well
as in nude mouse xenograft models (50). RFC4 promotes non-homologous end
joining-mediated DNA DSB repair by interacting with the Ku70/Ku80
complex. Moreover, RFC4 upregulation in tumor cells predicts
reduced tumor regression and poor prognosis in patients with
locally advanced rectal cancer (LARC) treated with neoadjuvant
radiotherapy. Therefore, RFC4 levels might serve as an effective
predictive biomarker of radiation sensitivity and a target for
radio-sensitization in patients with LARC (50). Additionally, RFC4 is frequently
overexpressed in CRC, driving tumor progression and poor survival
outcomes. Downregulated RFC4 inhibits cell proliferation and
induces S phase arrest (51). In
the HCC cell line HepG2, RFC4 downregulation enhances the cytotoxic
effects of doxorubicin and camptothecin, suggesting a role in
chemoresistance (52). In the lung
cancer cell line A549, RFC4 exhibits notable co-expression with
protein kinase C-like (PKCι) and PKCι knockdown markedly suppresses
RFC4 expression, suggesting a potential regulatory axis between
PKCι and RFC4 (53).
Moreover, Zheng et al (54) reported that high expression of RFC4
was not only associated with poor prognosis but also indicated a
stronger therapeutic response to immunotherapy in patients with
LUAD using bioinformatics methods. Possibly because RFC4
participated in reshaping the tumor immune microenvironment (TIME),
CD8+ T cells and macrophages M1 were positively
associated with RFC4 gene expression. In BC cell lines, silencing
RFC4 inhibited cell proliferation, induced G1 cell cycle
arrest and reduced cell migratory and invasive ability (55). Moreover, knockdown of RFC4
attenuated stemness and downregulated the expression of CD44 and
SOX2. RFC4 silencing inhibited migration and invasion, promoted
apoptosis and improved sensitivity to radiotherapy. Regarding the
mechanism, insulin like growth factor 2 mRNA binding protein 2
(IGF2BP2) can bind to the RFC4 mRNA coding sequence, and knockdown
of RFC4 eliminates the effects of overexpressed IGF2BP2 on
increasing cell viability, invasion, expression of stemness markers
and radio resistance (56).
Therefore, it also has potential to be a therapeutic target for
BC.
In patients with CC, a higher expression of RFC4 is
associated with an improved prognosis, possibly because RFC4 is
positively associated with the expression of three
immunostimulatory factors (UL16 binding protein 1, TNF receptor
superfamily member 13C/18/25 and inducible T cell costimulator
ligand) and three immunosuppressive factors (IL10 receptor subunit
b, V-set domain-containing T-cell activation inhibitor 1 and
galectin 9) (57).
RFC5
The RFC5 gene, located on chromosome 12 in humans,
serves vital roles in DSB repair, DNA excision and cell cycle
regulation, thereby maintaining genomic stability and influencing
tumorigenesis.
Mechanistic studies have demonstrated that astrocyte
elevated gene-1 (AEG-1) knockdown markedly downregulates RFC5
expression, impairing homologous recombination (HR)-mediated DNA
repair following radiation exposure. This effect is associated with
enhanced radiosensitivity in glioma cells, highlighting RFC5 as a
critical mediator of DNA damage response (58). Clinically, high levels of AEG-1 and
RFC5 are associated with poor prognosis specifically in patients
with glioma undergoing radiotherapy, further supporting their roles
as therapeutic targets (58). RFC5
is upregulated in HPV-negative HNSCC, where it serves a critical
role in DNA damage repair pathways. This upregulation enhances the
tumor cell capacity to withstand genotoxic stress, contributing to
therapy resistance and poor clinical outcomes (59). RFC5 functions as an oncogene in lung
cancer, where its overexpression is notably associated with poor
patient prognosis. Elevated RFC5 levels are associated with
aggressive clinicopathological features, including advanced tumor
stage, lymph node metastasis and enhanced tumor cell proliferation,
thereby promoting tumorigenesis and disease progression (60).
Bioinformatics analyses reveal that RFC5 harbors
putative binding sites for serine/arginine-rich splicing factor 10
(SRSF10). Functional studies have demonstrated that RFC5
overexpression promotes CRC cell proliferation and metastasis, even
when SRSF10 is silenced, suggesting an oncogenic dependency
(61). Mechanistically, SRSF10
regulates alternative splicing of RFC5, preferentially excluding
exon 2-AS1 (alternative splicing isoform 1 of exon 2) (61). Notably, Yao et al (62) confirmed that RFC5 promotes a
malignant phenotype in vitro, and downregulation of RFC5
markedly suppresses tumor growth in vivo. Collectively, the
researchers demonstrate that the circ_0038985/miR-3614-5p/RFC5 axis
serves a critical role in the progression of CRC, and RFC5 may
promote CRC progression via modulation of the VEGFA/VEGFR2/ERK
signaling pathway.
In acute myeloid leukemia (AML), bioinformatics
analysis showed that high RFC5 expression served as an independent
prognostic factor for the poor OS of patients with AML.
Additionally, elevated RFC5 mRNA levels were positively associated
with plasma cell and M2 macrophage infiltration, implicating TIME
remodeling in RFC5-driven leukemic progression (63). Zhang et al (64) demonstrated that basic transcription
factor 3 directly binds to the RFC5 promoter and upregulates RFC5
expression in PCa cells.
Role of the RFC family in different
signaling pathways
The RFC family members mainly participate in DNA
associated regulation. Here, the molecular mechanism associated
with the RFC family in cancers are discussed (Fig. 4).
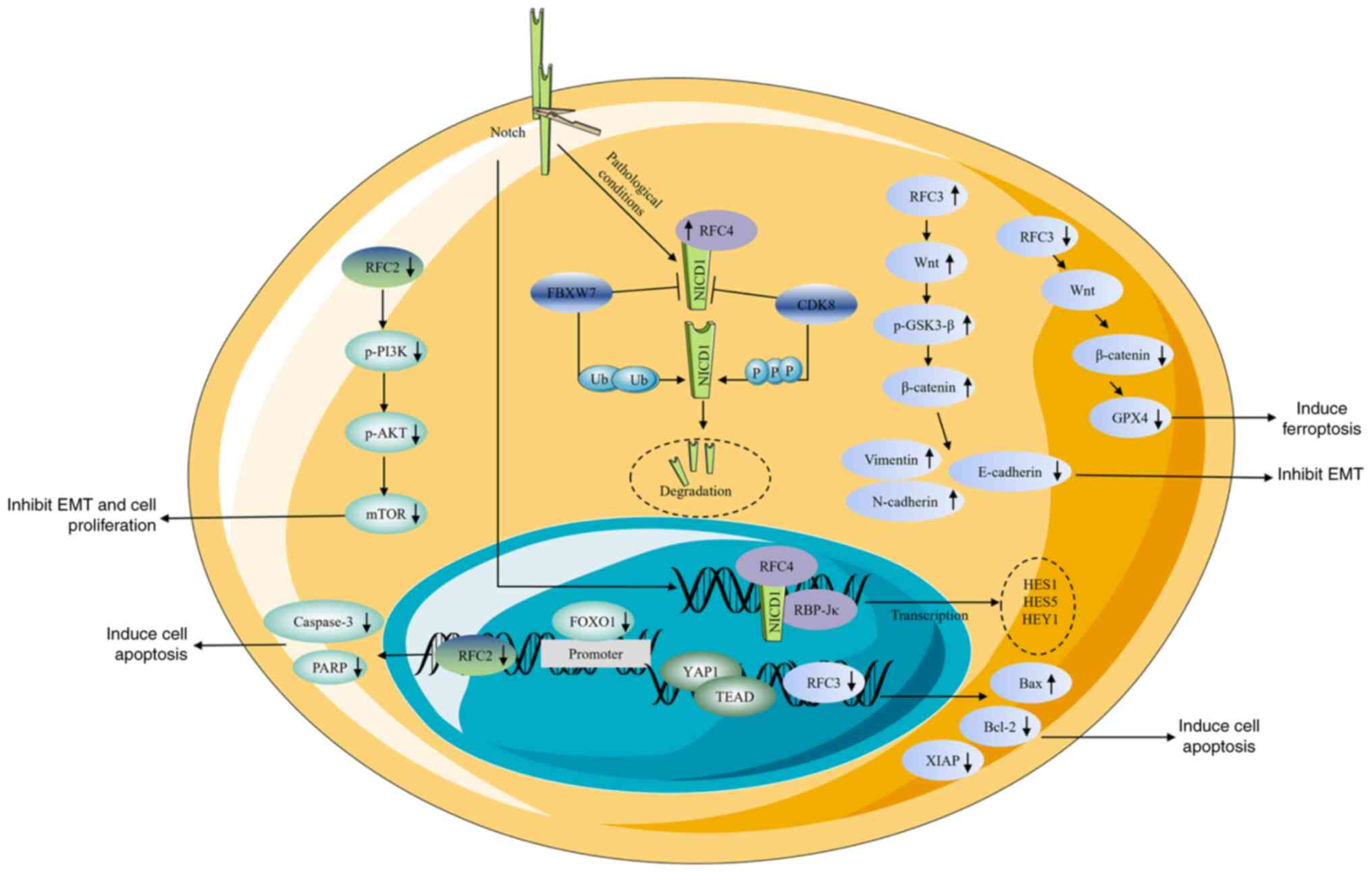 | Figure 4.Roles of RFC subunits in different
signaling pathways. RFC2 participates in the FOXO1 and PI3K/AKT
signal pathways. RFC3 participates in the YAP1/TEAD1 and
Wnt/β-catenin signal pathways. RFC4 participates in Notch1
signaling pathway. ↑, increase; ↓, decrease; RFC, replication
factor C; FOXO1, Forkhead box protein O1; YAP1, yes-associated
protein 1; TEAD, TEA domain family member 1; p-, phosphorylated;
PARP, poly(ADP-ribose) polymerase; EMT, epithelial-mesenchymal
transition GSK3β, glycogen synthase kinase-3 β; GPX4, glutathione
peroxidase 4. |
It has been demonstrated that Forkhead box O1
(FOXO1), a nuclear transcription factor, is able to directly
activate RFC2 expression in a transcriptional process by binding to
the promoter of RFC2. Knockdown of the FOXO1/RFC2 gene regulatory
network increases caspase-3 and poly(ADP-ribose) polymerase (PARP)
in temozolomide resistance a glioma cell line (65). In CRC cells, RFC2 activates the
MET/PI3K/AKT/mTOR pathway, and RFC2 knockdown decreases the levels
of phosphorylated (p-)PI3K, p-AKT, p-mTOR and p-70S6K in both
HCT116 and SW480 cell lines (7). In
CRC chemo-resistant cells, knockdown of RFC3 inhibited the
expression of β-catenin and glutathione peroxidase 4 (GPX4).
Immunostaining assays further demonstrated that the loss of RFC3
diminished the nuclear localization of β-catenin in these cells and
rescue experiments also supported these results. These findings
indicate that RFC3 knockdown disrupts the Wnt/β-catenin/GPX4 axis
(9).
In GC, transcriptional coactivator YAP1 binds to the
transcriptional factor TEAD and induces the expression of RFC3.
RFC3 silencing increased the expression of Bax and decreased the
expression of Bcl-2 (5). In
addition, RFC3 knockdown also reduces X-linked inhibitor of
apoptosis protein expression (5).
Overexpression of RFC3 increased Wnt expression, the
p-GSK3β(Ser9)/GSK3β ratio, and β-catenin protein levels. Since
β-catenin has a critical function in EMT, this results in
downregulated E-cadherin and upregulated N-cadherin. These results
suggest that RFC3 may trigger the Wnt/β-catenin signaling pathway
and promote LUAD migration and invasion through EMT (42).
Upon canonical Notch signaling activation, the
Notch1 intracellular domain (NICD1) undergoes nuclear translocation
and binds to the transcription factor recombination signal binding
protein for immunoglobulin kJ, initiating transcription of
downstream targets such as hairy and enhancer of split (HES) 1,
HES5 and hes related family bHLH transcription factor with YRPW
motif 1. These transcriptional repressors suppress
differentiation-promoting genes, maintaining progenitor cell
states. Under pathological conditions, RFC4 directly interacts with
NICD1 via high-affinity binding. This interaction competitively
abrogates CDK8-induced phosphorylation and F-box and WD repeat
domain containing 7-induced ubiquitination-dependent degradation of
NICD1. A feedforward loop between elevated RFC4 and NICD1 levels
drives sustained overactivation of Notch signaling, promoting NSCLC
tumorigenesis and metastasis (10).
In ESCC cells, overexpression of RFC4 increases cyclin D1 and Rad51
levels, whereas p53 and p21 levels are notably decreased.
Conversely, RFC4 knockdown has the opposite effect (11). However, the precise nature of the
regulation of the p53 signaling pathway by RFC4 remains somewhat
unclear.
Function and regulation of RLCs in
humans
Rad17-RFC complex
RAD17 interacts with the small subunits RFC2-5 and
forms a sophisticated complex, which is composed of two effective
layers: i) The lower part is an open and slightly twisted C-shaped
ring formed by the N-terminal Rossmann fold domains and central
helical bundle domains of RAD17, RFC2, RFC3, RFC4 and RFC5; and ii)
the upper ring, which forms a collar over the central cavity, is
offset from the central of the lower C-shaped ring, lying
predominantly over RFC3, RFC4 and RFC5 (66).
The Rad17-RFC complex functions as DNA damage sensor
(67). When DNA damage occurs, it
recruits the 9-1-1 (RAD9-RAD1-HUS1 checkpoint clamp component)
complex in an ATP-dependent manner to the damage site and initiates
the phosphorylation of ATM serine/threonine kinase and ataxia
telangiectasia and Rad3 related (ATR) (68). Additionally, it loads the 9-1-1
complex at the double stranded DNA/single stranded DNA junctions of
DNA damage sites in an replication protein-dependent manner and
promotes the activation of ATR and checkpoint kinase 1 (CHK1)
(12). Wang et al (69) demonstrated that RAD17 knockout
HCT116 cells are unable to form colonies, and the protein level of
Chk1phospho-Ser 345 is also downregulated. These results
indicate that the RAD17 complex is required for ATR-mediated
phosphorylation of Chk1 (69). The
Rad17-RFC complex and 9-1-1 complex also bind with DNA
topoisomerase II binding protein 1 which participates in the
activation of ATR (12). Moreover,
the Rad17-RFC complex is involved in ATP hydrolysis, but the
precise role of this process remains unclear.
CTF18-RFC
The multifunctional factor CTF18 exhibits two
distinct binding modalities within the cellular context: i) CTF18
stably associates with DNA replication and sister chromatid
cohesion 1 (DCC1) and CTF8, and the three proteins form a
heptameric complex with RFC2-5; and ii) CTF18 can also combine with
RFC2-5 to form a pentamer. The heptameric CTF18-DCC1-CTF8-RLC
complex (CTF18-RLC) has emerged as a central regulator of DNA
metabolism, orchestrating pivotal processes such as PCNA loading at
replication forks, establishment of sister chromatid cohesion and
activation of the DNA replication checkpoint (12).
In human cells, CTF18-RLC interacts with Pol ε by
binding to the CTF18-DCC1-CTF8 (CTF18-1-8) module's core component
DCC1 and loads PCNA; this combination is more efficient compared
with the pentamer. Meanwhile, Terret et al (70) further underscore the critical role
of CTF18-RLC in sister chromatid cohesion and maintenance of
processive DNA replication fork. The authors studies using
DCC1-knockout hTERT-RPE1 cells revealed severe defects in these
pathways, highlighting the indispensable nature of the intact
heptameric complex for genomic stability (70). Mounting evidence demonstrates that
patients with CRC and high DCC1 expression have a lower survival
rate compared with the lower-expression group. It has also been
demonstrated that HCT116 cells with DCC1 knockdown grow more slowly
and exhibit lower invasiveness compared with the control group. The
low protein expression levels of cyclin D1 and E-cadherin in the
DCC1 knockdown group also support these phenotypic changes.
Furthermore, DCC1 knockdown induces the proteolysis
of CTF18, whereas CTF18 knockdown does not affect cell invasion.
This differential effect establishes DCC1 as the dominant
functional component within the CTF18-1-8 module for promoting CRC
progression. It may be a promising molecular target for therapeutic
intervention in CRC (71).
ATAD5-RLC
Once DNA synthesis has ceased, PCNA must be
unloaded, a process primarily mediated by the ATAD5-RLC complex
(12). The C-terminal region of
ATAD5 contains both an ATPase domain and an RFC2-5 binding motif,
which are essential for efficient PCNA unloading. Experimental
evidence demonstrates that ATAD5 depletion leads to S phase arrest,
impaired PCNA unloading and markedly reduced DNA replication rates
(72). The N-terminal domain of
ATAD5 facilitates interactions with multiple protein partners
involved in regulating various aspects of DNA metabolism (72). With the depletion of ATAD5,
spontaneous HR increases and the DSB-induced HR decreases (73).
In U2OS cells, the ATAD5 knockdown group showed
higher sensitivity to olaparib compared with the control group.
This result was caused by trapping of PARP1 on chromatin (30). This observation was further
validated in both HeLa and 293T cell lines, where ATAD5 depletion
resulted in the accumulation of replication factors on chromatin in
a PCNA-dependent manner (74).
These findings collectively suggest that ATAD5 serves a crucial
role in regulating interactions between proteins and chromatin
during DNA replication and repair processes.
Exploration of RFC family molecular
functions and regulation pathways using bioinformation tools
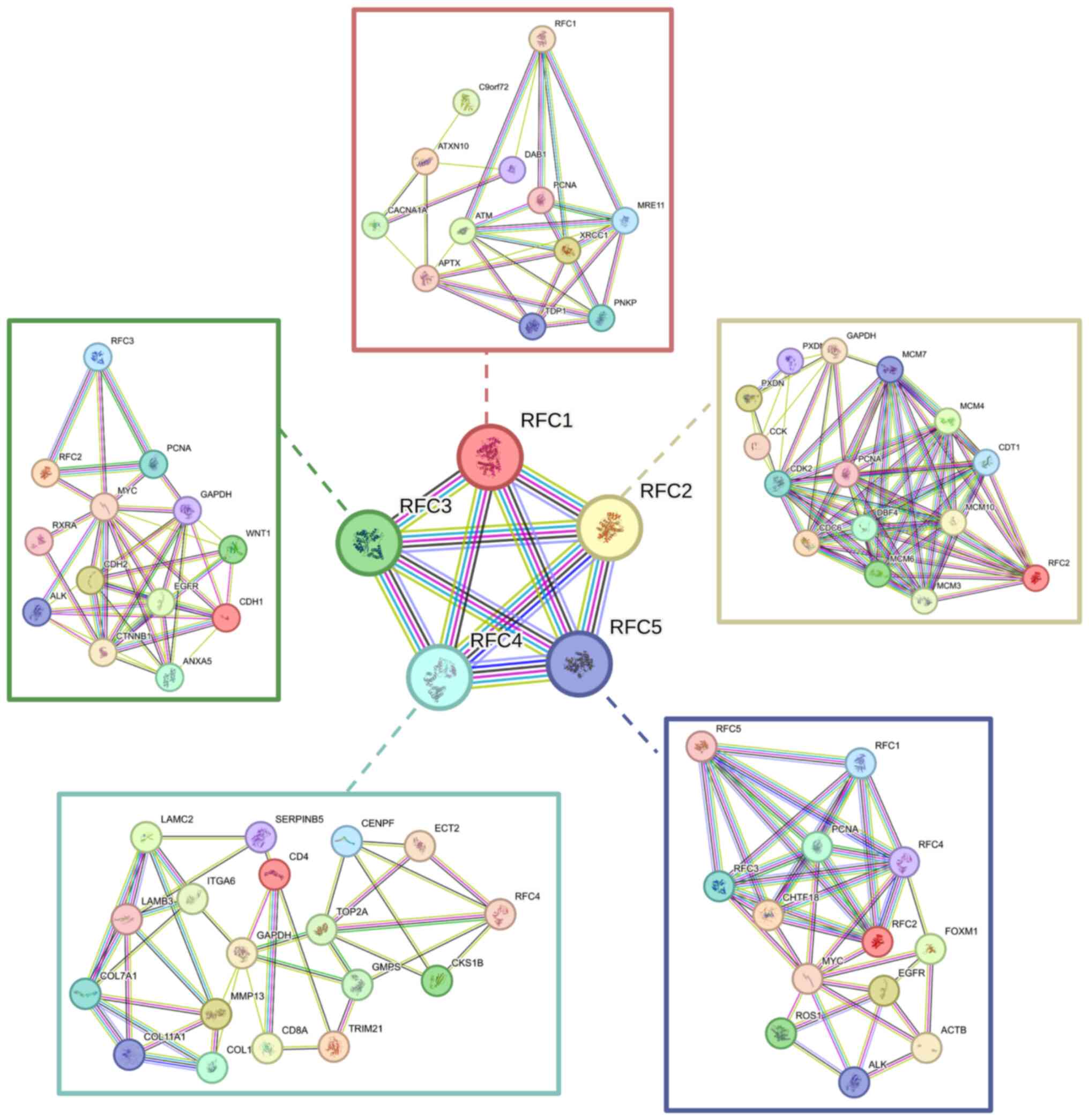
Using bioinformatics tools, the potential molecular
functions involved in interactions and regulatory pathways of the
RFC family have been explored. First, a co-expression network among
RFC family members was constructing using the STRING database
(75) and their potential
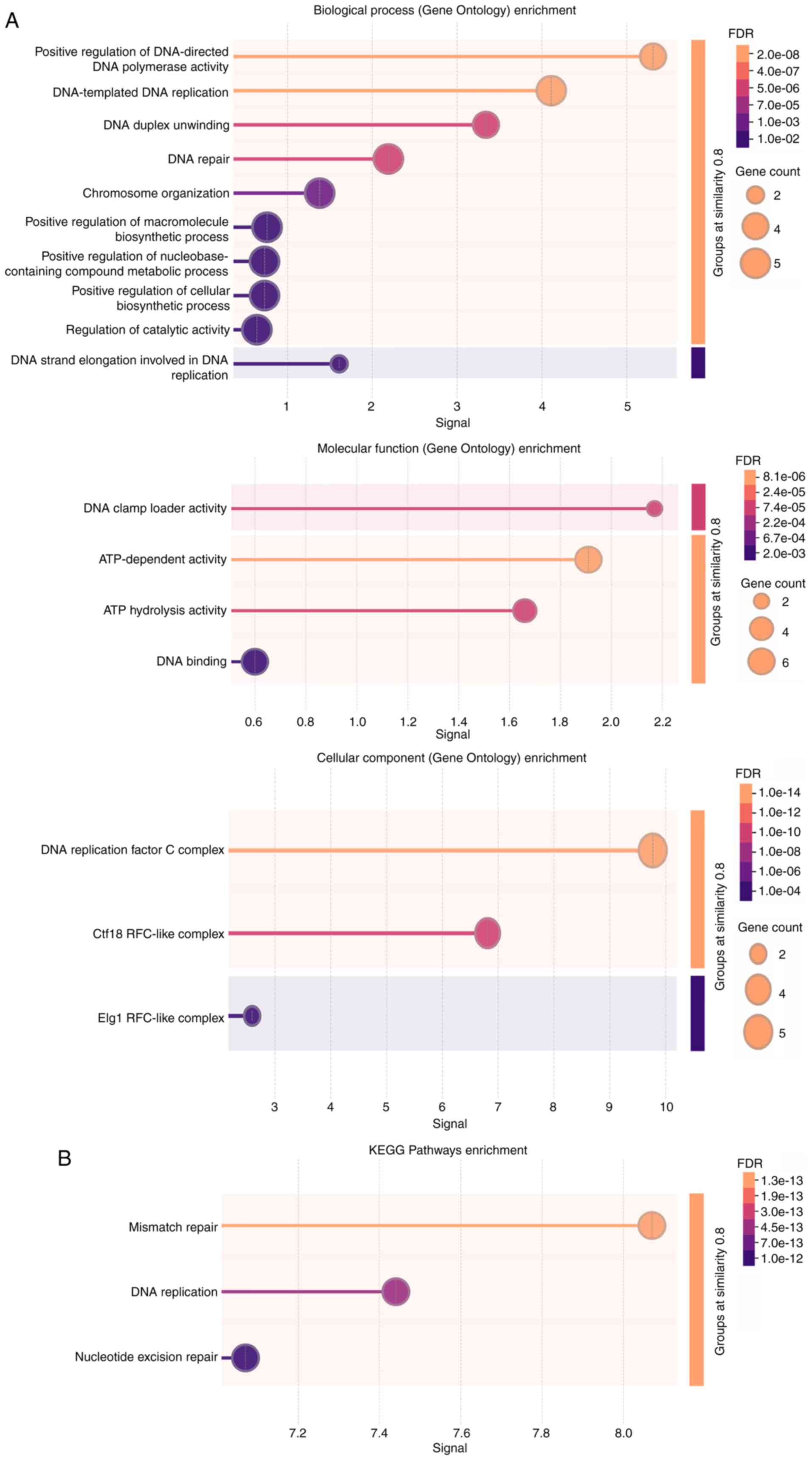
connections with other genes were investigated (Fig. 5). Additionally, Gene Ontology
analysis was performed on the RFC family (Fig. 6A) to identify their biological
processes, molecular functions and cellular components. Kyoto
Encyclopedia of Genes and Genomes pathway analysis was performed to
identify molecular pathways in which RFC family members were
involved (Fig. 6B). In summary, RFC
family members primarily participate in DNA damage and repair.
Since DNA damage acts as an initiator of innate immunity (76), future investigations should focus on
the connection between RFC family members and immune
regulation.
Clinical relevance of the RFC family
In recent years, research on biomarkers has drawn
increasing attention due to its ability to improve diagnostic
accuracy, guide individualized treatment and evaluate long-term
prognosis. Moreover, biomarkers also assist doctors in assessing
the potential effectiveness of treatments and prognosis (77). The rise of bioinformatics methods
contributes to identification of new biomarkers using tumor data
platforms. In CRC, sarcoma, HCC and LGG, patients with high RFC2
expression had worse OS (3,7,78,79).
Similarly, increased expression of RFC3 predicts worsened OS in
HNSCC, BC and LC (38,41,42).
Higher RFC4 expression is associated with worse prognosis in ESCC
and HCC (3,48), whereas it is associated with
improved OS in patients with CC (57,80).
For patients with CRC and CC, higher RFC5 expression is associated
with a worse OS (62,81). These results provide new insights
for cancer research; however, to a certain extent, they reflect the
association between clinical prognosis and potential biomarker
expression level. Further experimental validation and larger
databases are needed to draw more definitive conclusions in the
future.
Numerous patients have benefited from multiple
treatments, including chemotherapy, radiotherapy and immunotherapy;
however, the benefits remain limited. Chemoresistance and
radioresistance are the main causes of treatment failure; thus,
combination therapies may overcome the limitations of therapeutic
outcomes. Oshima et al (35)
proposed that RFC2 inhibition could be a potential therapeutic
strategy for CRPC resistant to PARP inhibitor (PARPi) by inhibiting
the NER pathway. In glioma, knockdown of RFC2 suppressed
temozolomide resistance (65,82).
Wu et al (9) demonstrated
that depletion of RFC3 enhances the sensitivity of CRC cells to
oxaliplatin by inducing ferroptosis. In BC, RFC3 knockdown enhances
tamoxifen sensitivity by inducing S-phase arrest (39). RFC4 deficiency enhances
radiosensitivity by promoting DNA damage in ESCC, BC and LARC
(11,50,56).
Nelarabine, in combination with other chemotherapies, is able to
reduce RFC4 gene expression (83).
In addition, RFC5 knockdown enhances radiation sensitivity by
impaired homologous recombination repair activity (58). The high RFC4 expression indicates a
stronger therapeutic response to immunotherapy in LUAD (54). Notably, most of these studies are
based on model organisms; therefore, more clinical trials are
needed in the future.
Due to the poor selectivity and off-target effects,
traditional treatments have notable clinical limitations. Thus, the
specific target therapy focusing on the molecular targets has
become a potential alternative. Previously, research by Alaa et
al (83) and colleagues
demonstrated that vorinostat and trichostatin A act as RFC4
inhibitors using molecular docking and molecular dynamics
simulation.
Moreover, the potential application of RLCs in
clinical treatment has also drawn more attention. ATAD5-depleted
cells show high sensitivity to antitumor drugs including methyl
methanesulphonate, camptothecin, mitomycin C and PARPi (30). Rohde et al (84) discovered a new compound ML367 which
inhibits ATAD5 stabilization. ML367 could block DNA repair pathways
including RPA32-phosphorylation and CHK1-phosphorylation in
response to UV irradiation. It serves as a sensitizer of PARPi to
enhance the antitumor effects synergistically. For inhibitors
intended for clinical use, further experimental validation is
essential. In ESCC, negative elongation factor complex member A
(NELFA) mRNA promotes cell apoptosis, partially by inhibiting the
interaction between Rad17 and the Rad17-RFC2-5 complex.
Furthermore, higher NELFA expression is associated with worse OS.
As a key regulator of the Rad17-RFC2-5 complex, NELFA holds promise
as a novel therapeutic target in cancer (85).
Conclusions and future perspectives
The RFC family members serve critical roles in
tumorigenesis and cancer progression. Specifically, individual RFC
subunits serve as crucial regulators of malignant phenotypes
including aberrant cell proliferation, EMT and tumor metastasis.
Clinically, elevated expression levels of RFC members are
associated with poor OS across multiple malignancies, establishing
them as potent diagnostic and prognostic biomarkers. Although
current research emphasizes the effects of RFC family member on
particular signaling pathways, there are relatively few studies
regarding RFC1 and RFC5. In 2019, the (AAAGGG)n motif expansions of
RFC1 were first shown to be a main cause late-onset ataxia
(86). To date, the majority of
research into RFC1 are concentrated on genopathy (29). Moreover, research on the regulatory
mechanisms of RFC5 in cancers has not been sufficiently in-depth.
This may be a new direction in cancer biology.
In the present review, the map of RFC family members
which are associated with cancers is shown in Fig. 7 and the regulation mechanisms are
shown in Table II. The present
review had some limitations. Much of the analysis remains at basic
experimental level, lacking forward-looking perspective.
Additionally, there is insufficient discussion on the connection
between RFC family members. With the rapid progression of
understanding of RFC family members in human cancers, their dual
potential as either oncogenes or tumor suppressors have been
demonstrated. It was hypothesized that the contradictory effects
observed in previous studies arise from variations in sample
sources and tumor heterogeneity. Cancer cells and the TIME can
exhibit molecular characteristic variations; these changes may
further contribute to the distinct roles of specific factors across
different tumors (87). Functional
experiments often rely on immortalized cell lines and standardized
animal models, which may overlook other potential influencing
factors. Furthermore, some results derived from publicly available
datasets could be affected by multiple confounding variables,
including variations in sample composition, batch effects,
differences in sequencing technologies, data processing pipelines
and normalization methods (88).
 | Table II.Expression and function of
replication factor C family members in human cancers. |
Table II.
Expression and function of
replication factor C family members in human cancers.
| RFC family
members | Cancer type | Roles in human
cancers |
|---|
| RFC1 | Nasopharyngeal
carcinoma | Overexpressed |
|
| Breast cancer | Repressed by E2 in
ERα-negative breast cancer cells in which ERα has been
re-expressed. |
|
| Colorectal
cancer | Targeted by
microRNA-26a-5p. |
| RFC2 | Lower-grade
glioma | RFC2 overexpression
correlates with immune checkpoint genes. An independent predictor
of adverse prognosis. An anti-tumor factor. |
|
| Gastric cancer | RFC2 ubiquitination
enhances cell invasion and migration. |
|
| Hepatocellular
carcinoma | Severed as a novel
biomarker for the prognosis. RFC2 silencing inhibits proliferation
and migration. |
|
| Colorectal
cancer | Upregulated RFC2
expression correlates with poor clinical outcomes. RFC2 promotes
aerobic glycolysis and MET/PI3K/AKT/mTOR pathway. |
|
|
Castration-resistant prostate cancer | Overexpressed. RFC2
downregulation inhibits cell proliferation, induces apoptosis and
aggravates DNA damage. Elevated RFC2 protein expression is
associated with unfavorable prognosis |
|
| Ewing's
sarcoma | Overexpressed. High
expression predicted poor OS and EFS. |
| RFC3 | Head and neck
squamous cell carcinoma | High expression
predicts poor clinicopathological features and associates with
oncogenic signaling pathways, such as MYC/MYCN, HIPPO, and
mTOR. |
|
| ER positive breast
cancer | High expression
predicts poor prognosis, downregulated RFC3 induces S phase arrest
and attenuated cell. |
|
| Lung
adenocarcinoma | High expression
predicts poor prognosis, overexpressed RFC3 facilitates cell cycle
progression from G1 to S phase, RFC3 promoted EMT
through the Wnt/β-catenin signaling pathway. |
|
| Hepatocellular
carcinoma | RFC3 silencing
induces cell cycle arrest at S phase. |
|
| Gastric cancer | The YAP1/TEAD
pathway transcriptionally activated RFC3, driving tumor
progression. |
|
| Colorectal
cancer | KIF14 upregulation
counteracts the suppressive effects of RFC3 depletion on aggressive
phenotypes. |
| RFC4 | Nasopharyngeal
carcinoma | Overexpressed. RFC4
knockdown induces G2/M cell cycle arrest and inhibits
cellular proliferation. Overexpression of HOXA10 partially rescues
the suppressive effects of RFC4 silencing. |
|
| Esophageal squamous
cell carcinoma | Overexpressed. high
RFC4 expression predicts poor prognosis. |
|
| Locally advanced
rectal cancer | A radiation
resistance factor. Served as an effective predictive biomarker of
radiation sensitivity and a target for radio-sensitization. |
|
| Colorectal
cancer | Overexpressed,
Downregulated RFC4 inhibits cell proliferation and induces S phase
arrest. |
|
| Hepatocellular
carcinoma | RFC4 downregulation
enhances the cytotoxic effects of doxorubicin and
camptothecin. |
|
| Lung
adenocarcinoma | RFC4 exhibits
significant co-expression with PKCι, and PKCι knockdown markedly
suppresses RFC4 expression. |
| RFC5 | Glioma | AEG-1 knockdown
downregulates RFC5 expression. RFC5 serves as a critical mediator
of DNA damage response. High levels of RFC5 is associated with poor
prognosis. |
|
| HPV negative
squamous cell carcinoma of the head and neck | Overexpressed RFC5
enhances the tumor cells' capacity to withstand genotoxic
stress. |
|
| Lung cancer | RFC5 functions as
an oncogene in lung cancer. Elevated RFC5 levels correlate with
aggressive clinicopathological features. |
|
| Colorectal
cancer | RFC5 overexpression
promotes CRC cell proliferation and metastasis. RFC5 may promote
CRC progression via modulation of the VEGFA/VEGFR2/ERK signaling
pathway. |
|
| Acute myeloid
leukemia | High RFC5
expression served as an independent prognostic factor for the poor
OS. Elevated RFC5 mRNA levels correlated positively with plasma
cell and M2 macrophage infiltration. |
While individual members of the RFC have been
characterized across various cancer types, their underlying
mechanisms require further investigation. Several critical
questions emerge from the current research: First, do the family
members have more notable oncogenic effects collectively compared
with individually, potentially through synergistic interactions?
Second, are there novel mechanisms being explored to elucidate
tumorigenesis? Finally, from a clinical perspective, could the RFC
family members serve as valuable diagnostic and prognostic
biomarkers? Addressing these questions will require comprehensive
studies to elucidate the complex roles of RFC proteins in cancer
biology and their potential translational applications. Future
research should focus on characterizing these molecular
relationships and evaluating their clinical utility for improved
cancer management.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of Hubei Province (grant no. 2022CFB808) and the Open
Project of Minda Hospital Affiliated to Hubei University for
Nationalities (grant no. OIR202302Q).
Availability of data and materials
Not applicable.
Authors' contributions
DL wrote the original draft. DL and BL prepared the
figures. XJ reviewed and revised the manuscript. All authors
reviewed the manuscript. Data authentication is not applicable. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263.
2024.
|
|
2
|
Uhlmann F, Gibbs E, Cai J, O'Donnell M and
Hurwitz J: Identification of regions within the four small subunits
of human replication factor C required for complex formation and
DNA replication. J Biol Chem. 272:10065–10071. 1997. View Article : Google Scholar
|
|
3
|
Zhao X, Wang Y, Li J, Qu F, Fu X, Liu S,
Wang X, Xie Y and Zhang X: RFC2: A prognosis biomarker correlated
with the immune signature in diffuse lower-grade gliomas. Sci Rep.
12:31222022. View Article : Google Scholar
|
|
4
|
Yao Z, Hu K, Huang HE, Xu S, Wang Q, Zhang
P, Yang P and Liu B: shRNA-mediated silencing of the RFC3 gene
suppresses hepatocellular carcinoma cell proliferation. Int J Mol
Med. 36:1393–1399. 2015. View Article : Google Scholar
|
|
5
|
Guo Z and Guo L: Abnormal activation of
RFC3, A YAP1/TEAD downstream target, promotes gastric cancer
progression. Int J Clin Oncol. 29:442–455. 2024. View Article : Google Scholar
|
|
6
|
Misbah M, Kumar M, Najmi AK and Akhtar M:
Identification of expression profiles and prognostic value of RFCs
in colorectal cancer. Sci Rep. 14:66072024. View Article : Google Scholar
|
|
7
|
Lou F and Zhang M: RFC2 promotes aerobic
glycolysis and progression of colorectal cancer. BMC Gastroenterol.
23:3532023. View Article : Google Scholar
|
|
8
|
Li H, Chai L, Ding Z and He H: CircCOL1A2
sponges MiR-1286 to promote cell invasion and migration of gastric
cancer by elevating expression of USP10 to downregulate RFC2
ubiquitination level. J Microbiol Biotechn. 32:938–948. 2022.
View Article : Google Scholar
|
|
9
|
Wu Y, Chen T, Wu S, Huang Y and Li F:
Knockdown of RFC3 enhances the sensitivity of colon cancer cells to
oxaliplatin by inducing ferroptosis. Fund Clin Pharmacol.
39:e130442025. View Article : Google Scholar
|
|
10
|
Liu L, Tao T, Liu S, Yang X, Chen X, Liang
J, Hong R, Wang W, Yang Y, Li X, et al: An RFC4/Notch1 signaling
feedback loop promotes NSCLC metastasis and stemness. Nat Commun.
12:26932021. View Article : Google Scholar
|
|
11
|
Yang T, Fan Y, Bai G and Huang Y: RFC4
confers radioresistance of esophagus squamous cell carcinoma
through regulating DNA damage response. Am J Physiol Cell Physiol.
328:C367–C380. 2025. View Article : Google Scholar
|
|
12
|
Lee K and Park SH: Eukaryotic clamp
loaders and unloaders in the maintenance of genome stability. Exp
Mol Med. 52:1948–1958. 2020. View Article : Google Scholar
|
|
13
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020. View Article : Google Scholar
|
|
14
|
Mossi R and Hubscher U: Clamping down on
clamps and clamp Loaders-the eukaryotic replication factor C. Eur J
Biochem. 254:209–216. 1998. View Article : Google Scholar
|
|
15
|
Virshup DM, Kauffman MG and Kelly TJ:
Activation of SV40 DNA replication in vitro by cellular protein
phosphatase 2A. EMBO J. 8:3891–3898. 1989. View Article : Google Scholar
|
|
16
|
Virshup DM and Kelly TJ: Purification of
replication protein C, a cellular protein involved in the initial
stages of simian virus 40 DNA replication in vitro. Proc Natl Acad
Sci USA. 86:3584–3588. 1989. View Article : Google Scholar
|
|
17
|
Yoder BL and Burgers PM: Saccharomyces
cerevisiae replication factor C. I. Purification and
characterization of its ATPase activity. J Biol Chem.
266:22689–22697. 1991. View Article : Google Scholar
|
|
18
|
Burbelo PD, Utani A, Pan ZQ and Yamada Y:
Cloning of the large subunit of activator 1 (replication factor C)
reveals homology with bacterial DNA ligases. Proc Natl Acad Sci
USA. 90:11543–11547. 1993. View Article : Google Scholar
|
|
19
|
Li X and Burgers PM: Cloning and
characterization of the essential Saccharomyces cerevisiae RFC4
gene encoding the 37-kDa subunit of replication factor C. J Biol
Chem. 269:21880–21884. 1994. View Article : Google Scholar
|
|
20
|
Li X and Burgers PM: Molecular cloning and
expression of the Saccharomyces cerevisiae RFC3 gene, an essential
component of replication factor C. Proc Natl Acad Sci USA.
91:868–872. 1994. View Article : Google Scholar
|
|
21
|
Noskov V, Maki S, Kawasaki Y, Leem SH, Ono
B, Araki H, Pavlov Y and Sugino A: The RFC2 gene encoding a subunit
of replication factor C of Saccharomyces cerevisiae. Nucleic Acids
Res. 22:1527–1535. 1994. View Article : Google Scholar
|
|
22
|
Podust VN, Georgaki A, Strack B and
Hubscher U: Calf thymus RF-C as an essential component for DNA
polymerase delta and epsilon holoenzymes function. Nucleic Acids
Res. 20:4159–4165. 1992. View Article : Google Scholar
|
|
23
|
Li Y, Gan S, Ren L, Yuan L, Liu J, Wang W,
Wang X, Zhang Y, Jiang J, Zhang F and Qi X: Multifaceted regulation
and functions of replication factor C family in human cancers. Am J
Cancer Res. 8:1343–1355. 2018.
|
|
24
|
Okumura K, Nogami M, Taguchi H, Dean FB,
Chen M, Pan ZQ, Hurwitz J, Shiratori A, Murakami Y and Ozawa K:
Assignment of the 36.5-kDa (RFC5), 37-kDa (RFC4), 38-kDa (RFC3),
and 40-kDa (RFC2) subunit genes of human replication factor C to
chromosome bands 12q24.2-q24.3, 3q27, 13q12.3-q13, and 7q11.23.
Genomics. 25:274–278. 1995. View Article : Google Scholar
|
|
25
|
Uhlmann F, Cai J, Gibbs E, O'Donnell M and
Hurwitz J: Deletion analysis of the large subunit p140 in human
replication factor C reveals regions required for complex formation
and replication activities. J Biol Chem. 272:10058–10064. 1997.
View Article : Google Scholar
|
|
26
|
Cai J, Gibbs E, Uhlmann F, Phillips B, Yao
N, O'Donnell M and Hurwitz J: A complex consisting of human
replication Factor C p40, p37, and p36 subunits is a DNA-dependent
ATPase and an intermediate in the assembly of the holoenzyme. J
Biol Chem. 272:18974–18981. 1997. View Article : Google Scholar
|
|
27
|
Cullmann G, Fien K, Kobayashi R and
Stillman B: Characterization of the five replication factor C genes
of Saccharomyces cerevisiae. Mol Cell Biol. 15:4661–4671. 1995.
View Article : Google Scholar
|
|
28
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for Large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar
|
|
29
|
Delforge V, Tard C, Davion JB, Dujardin K,
Wissocq A, Dhaenens CM, Mutez E and Huin V: RFC1: Motifs and
phenotypes. Rev Neurol (Paris). 180:393–409. 2024. View Article : Google Scholar
|
|
30
|
Giovannini S, Weller M, Hanzlíková H,
Shiota T, Takeda S and Jiricny J: ATAD5 deficiency alters DNA
damage metabolism and sensitizes cells to PARP inhibition. Nucleic
Acids Res. 48:4928–4939. 2020. View Article : Google Scholar
|
|
31
|
Fung LF, Lo AK, Yuen PW, Liu Y, Wang XH
and Tsao SW: Differential gene expression in nasopharyngeal
carcinoma cells. Life Sci. 67:923–936. 2000. View Article : Google Scholar
|
|
32
|
Almalki E, Al-Amri A, Alrashed R,
AL-Zharani M and Semlali A: The curcumin analog PAC is a potential
solution for the treatment of Triple-negative breast cancer by
modulating the gene expression of DNA repair pathways. Int J Mol
Sci. 24:96492023. View Article : Google Scholar
|
|
33
|
Moggs JG, Murphy TC, Lim FL, Moore DJ,
Stuckey R, Antrobus K, Kimber I and Orphanides G:
Anti-proliferative effect of estrogen in breast cancer cells that
re-express ERalpha is mediated by aberrant regulation of cell cycle
genes. J Mol Endocrinol. 34:535–551. 2005. View Article : Google Scholar
|
|
34
|
Ji Z, Li J and Wang J: Up-regulated RFC2
predicts unfavorable progression in hepatocellular carcinoma.
Hereditas. 158:172021. View Article : Google Scholar
|
|
35
|
Oshima M, Takayama K, Yamada Y, Kimura N,
Kume H, Fujimura T and Inoue S: Identification of DNA damage
response-related genes as biomarkers for castration-resistant
prostate cancer. Sci Rep. 13:196022023. View Article : Google Scholar
|
|
36
|
Li G, Zhang P, Zhang W, Lei Z, He J, Meng
J, Di T and Yan W: Identification of key genes and pathways in
Ewing's sarcoma patients associated with metastasis and poor
prognosis. Onco Targets Ther. 12:4153–4165. 2019. View Article : Google Scholar
|
|
37
|
Koh JW and Park S: Knockdown of RFC2
prevents the proliferation, migration and invasion of cervical
cancer cells. Anticancer Res. 45:989–1000. 2025. View Article : Google Scholar
|
|
38
|
Avs KR, Pandi C, Kannan B, Pandi A,
Jayaseelan VP and Arumugam P: RFC3 serves as a novel prognostic
biomarker and target for head and neck squamous cell carcinoma.
Clin Oral Invest. 27:6961–6969. 2023. View Article : Google Scholar
|
|
39
|
Zhu J, Ye L, Sun S, Yuan J, Huang J and
Zeng Z: Involvement of RFC3 in tamoxifen resistance in ER-positive
breast cancer through the cell cycle. Aging. 15:13738–13752. 2023.
View Article : Google Scholar
|
|
40
|
Zhou J, Zhang W, Peng F, Sun J, He Z and
Wu S: Downregulation of hsa_circ_0011946 suppresses the migration
and invasion of the breast cancer cell line MCF-7 by targeting
RFC3. Cancer Manag Res. 10:535–544. 2018. View Article : Google Scholar
|
|
41
|
He Z, Wu S, Peng F, Zhang Q, Luo Y, Chen M
and Bao Y: Up-Regulation of RFC3 promotes triple negative breast
cancer metastasis and is associated with poor prognosis via EMT.
Transl Oncol. 10:1–9. 2017. View Article : Google Scholar
|
|
42
|
Gong S, Qu X, Yang S, Zhou S, Li P and
Zhang Q: RFC3 induces epithelial-mesenchymal transition in lung
adenocarcinoma cells through the Wnt/β-catenin pathway and
possesses prognostic value in lung adenocarcinoma. Int J Mol Med.
44:2276–2288. 2019.
|
|
43
|
Koh JW and Park S: RFC3 Knockdown
decreases cervical cancer cell proliferation, migration and
invasion. Cancer Genomics Proteomics. 22:127–135. 2024. View Article : Google Scholar
|
|
44
|
Yu R, Wu X, Qian F and Yang Q: RFC3 drives
the proliferation, migration, invasion and angiogenesis of
colorectal cancer cells by binding KIF14. Exp Ther Med. 27:2222024.
View Article : Google Scholar
|
|
45
|
Yu L, Li J, Zhang M, Li Y, Bai J, Liu P,
Yan J and Wang C: Identification of RFC4 as a potential biomarker
for pan-cancer involving prognosis, tumour immune microenvironment
and drugs. J Cell Mol Med. 28:e184782024. View Article : Google Scholar
|
|
46
|
Guan S, Feng L, Wei J, Wang G and Wu L:
Knockdown of RFC4 inhibits the cell proliferation of nasopharyngeal
carcinoma in vitro and in vivo. Front Med. 17:132–142. 2023.
View Article : Google Scholar
|
|
47
|
You P, Wang D, Liu Z, Guan S, Xiao N, Chen
H, Zhang X, Wu L, Wang G and Dong H: Knockdown of RFC 4 inhibits
cell proliferation of oral squamous cell carcinoma in vitro and in
vivo. FEBS Open Bio. 15:346–358. 2025. View Article : Google Scholar
|
|
48
|
Wang J, Luo F, Huang T, Mei Y, Peng L,
Qian C and Huang B: The upregulated expression of RFC4 and GMPS
mediated by DNA copy number alteration is associated with the early
diagnosis and immune escape of ESCC based on a bioinformatic
analysis. Aging (Albany NY). 13:21758–21777. 2021. View Article : Google Scholar
|
|
49
|
Zeng B, Liu X, Liu J, Qiu H, Zhang T, Chen
X and Wang L: Roles of MARCKSL1, MCM6, RFC4, and PLAU genes in
esophageal cancer and their association with radiotherapy response.
Sci Rep. 15:235592025. View Article : Google Scholar
|
|
50
|
Wang X, Yue X, Zhang R, Liu T, Pan Z, Yang
M, Lu Z, Wang Z, Peng J, Le L, et al: Genome-wide RNAi screening
identifies RFC4 as a factor that mediates radioresistance in
colorectal cancer by facilitating nonhomologous end joining repair.
Clin Cancer Res. 25:4567–4579. 2019. View Article : Google Scholar
|
|
51
|
Xiang J, Fang L, Luo Y, Yang Z, Liao Y,
Cui J, Huang M, Yang Z, Huang Y, Fan X, et al: Levels of human
replication factor C4, a clamp loader, correlate with tumor
progression and predict the prognosis for colorectal cancer. J
Transl Med. 12:3202014. View Article : Google Scholar
|
|
52
|
Arai M, Kondoh N, Imazeki N, Hada A,
Hatsuse K, Matsubara O and Yamamoto M: The knockdown of endogenous
replication factor C4 decreases the growth and enhances the
chemosensitivity of hepatocellular carcinoma cells. Liver Int.
29:55–62. 2009. View Article : Google Scholar
|
|
53
|
Erdogan E, Klee EW, Thompson EA and Fields
AP: Meta-analysis of oncogenic protein kinase Ciota signaling in
lung adenocarcinoma. Clin Cancer Res. 15:1527–1533. 2009.
View Article : Google Scholar
|
|
54
|
Zheng J, Lin N, Huang B, Wu M, Xiao L and
Zeng B: Comprehensive analysis of RFC4 as a potential biomarker for
regulating the immune microenvironment and predicting immune
therapy response in lung adenocarcinoma. Front Immunol.
16:15782432025. View Article : Google Scholar
|
|
55
|
Koh JW and Park S: Replication Factor C
Subunit 4 plays a role in human breast cancer cell progression.
Cancer Genomics Proteomics. 22:478–490. 2025. View Article : Google Scholar
|
|
56
|
Zhu XY, Li PS, Qu H, Ai X, Zhao ZT and He
JB: Replication factor C4, which is regulated by insulin-like
growth factor 2 mRNA binding protein 2, enhances the
radioresistance of breast cancer by promoting the stemness of tumor
cells. Hum Cell. 38:652025. View Article : Google Scholar
|
|
57
|
Huang B, Zheng J, Chen B, Wu M and Xiao L:
Analysis of the correlation between RFC4 expression and tumor
immune microenvironment and prognosis in patients with cervical
cancer. Front Genet. 16:15143832025. View Article : Google Scholar
|
|
58
|
Zhao X, Sun Y, Sun X, Li J, Shi X, Liang
Z, Ma Y and Zhang X: AEG-1 Knockdown sensitizes glioma cells to
radiation through impairing homologous recombination via targeting
RFC5. DNA Cell Biol. 40:895–905. 2021. View Article : Google Scholar
|
|
59
|
Martinez I, Wang J, Hobson KF, Ferris RL
and Khan SA: Identification of differentially expressed genes in
HPV-positive and HPV-negative oropharyngeal squamous cell
carcinomas. Eur J Cancer. 43:415–432. 2007. View Article : Google Scholar
|
|
60
|
Wang M, Xie T, Wu Y, Yin Q, Xie S, Yao Q,
Xiong J and Zhang Q: Identification of RFC5 as a novel potential
prognostic biomarker in lung cancer through bioinformatics
analysis. Oncol Lett. 16:4201–4210. 2018.
|
|
61
|
Xu S, Zhong F, Jiang J, Yao F, Li M, Tang
M, Cheng Y, Yang Y, Wen W, Zhang X, et al: High Expression of
SRSF10 promotes colorectal cancer progression by aberrant
alternative splicing of RFC5. Technol Cancer Res.
23:153303382412719062024. View Article : Google Scholar
|
|
62
|
Yao H, Zhou X, Zhou A, Chen J, Chen G, Shi
X, Shi B, Tai Q, Mi X, Zhou G, et al: RFC5, regulated by
circ_0038985/miR-3614-5p, functions as an oncogene in the
progression of colorectal cancer. Mol Carcinog. 62:771–785. 2023.
View Article : Google Scholar
|
|
63
|
Li W, Wu D, Zhou Y, Zhang C and Liao X:
Prognostic biomarker replication factor C subunit 5 and its
correlation with immune infiltrates in acute myeloid leukemia.
Hematology. 27:555–564. 2022. View Article : Google Scholar
|
|
64
|
Zhang Y, Gao X, Yi J, Sang X, Dai Z, Tao
Z, Wang M, Shen L, Jia Y, Xie D, et al: BTF3 confers oncogenic
activity in prostate cancer through transcriptional upregulation of
Replication Factor C. Cell Death Dis. 12:122021. View Article : Google Scholar
|
|
65
|
Qiu X, Tan G, Wen H, Lian L and Xiao S:
Forkhead box O1 targeting replication factor C subunit 2 expression
promotes glioma temozolomide resistance and survival. Ann Transl
Med. 9:6922021. View Article : Google Scholar
|
|
66
|
Day M, Oliver AW and Pearl LH: Structure
of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp
bound to a dsDNA-ssDNA junction. Nucleic Acids Res. 50:8279–8289.
2022. View Article : Google Scholar
|
|
67
|
Niida H and Nakanishi M: DNA damage
checkpoints in mammals. Mutagenesis. 21:3–9. 2006. View Article : Google Scholar
|
|
68
|
Bermudez VP, Lindsey-Boltz LA, Cesare AJ,
Maniwa Y, Griffith JD, Hurwitz J and Sancar A: Loading of the human
9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader
hRad17-replication factor C complex in vitro. Proc Natl Acad Sci
USA. 100:1633–1638. 2003. View Article : Google Scholar
|
|
69
|
Wang X, Zou L, Zheng H, Wei Q, Elledge SJ
and Li L: Genomic instability and endoreduplication triggered by
RAD17 deletion. Gene Dev. 17:965–970. 2003. View Article : Google Scholar
|
|
70
|
Terret ME, Sherwood R, Rahman S, Qin J and
Jallepalli PV: Cohesin acetylation speeds the replication fork.
Nature. 462:231–234. 2009. View Article : Google Scholar
|
|
71
|
Kim JT, Cho HJ, Park SY, Oh BM, Hwang YS,
Baek KE, Lee YH, Kim HC and Lee HG: DNA replication and sister
chromatid cohesion 1 (DSCC1) of the replication factor complex
CTF18-RFC is critical for colon cancer cell growth. J Cancer.
10:6142–6153. 2019. View Article : Google Scholar
|
|
72
|
Kang M, Ryu E, Lee S, Park J, Ha NY, Ra
JS, Kim YJ, Kim J, Abdel-Rahman M, Park SH, et al: Regulation of
PCNA cycling on replicating DNA by RFC and RFC-like complexes. Nat
Commun. 10:24202019. View Article : Google Scholar
|
|
73
|
Sikdar N, Banerjee S, Lee KY, Wincovitch
S, Pak E, Nakanishi K, Jasin M, Dutra A and Myung K: DNA damage
responses by human ELG1 in S phase are important to maintain
genomic integrity. Cell Cycle. 8:3199–3207. 2009. View Article : Google Scholar
|
|
74
|
Lee KY, Fu H, Aladjem MI and Myung K:
ATAD5 regulates the lifespan of DNA replication factories by
modulating PCNA level on the chromatin. J Cell Biol. 200:31–44.
2013. View Article : Google Scholar
|
|
75
|
Szklarczyk D, Nastou K, Koutrouli M,
Kirsch R, Mehryary F, Hachilif R, Hu D, Peluso ME, Huang Q, Fang T,
et al: The STRING database in 2025: Protein networks with
directionality of regulation. Nucleic Acids Res. 53:D730–D737.
2025. View Article : Google Scholar
|
|
76
|
Hopfner KP and Hornung V: Molecular
mechanisms and cellular functions of cGAS-STING signalling. Nat Rev
Mol Cell Bio. 21:501–521. 2020. View Article : Google Scholar
|
|
77
|
Yuan Q, Cai S, Chang Y, Zhang J, Wang M,
Yang K and Jiang D: The multifaceted roles of mucins family in lung
cancer: From prognostic biomarkers to promising targets. Front
Immunol. 16:16081402025. View Article : Google Scholar
|
|
78
|
Wu G, Zhou J, Zhu X, Tang X, Liu J, Zhou
Q, Chen Z, Liu T, Wang W, Xiao X and Wu T: Integrative analysis of
expression, prognostic significance and immune infiltration of RFC
family genes in human sarcoma. Aging (Albany NY). 14:3705–3719.
2022. View Article : Google Scholar
|
|
79
|
Deng J, Zhong F, Gu W and Qiu F:
Exploration of prognostic biomarkers among replication factor C
family in the hepatocellular carcinoma. Evol Bioinform.
17:11769343219941092021. View Article : Google Scholar
|
|
80
|
Zhang J, Meng S, Wang X, Wang J, Fan X,
Sun H, Ning R, Xiao B, Li X, Jia Y, et al: Sequential gene
expression analysis of cervical malignant transformation identifies
RFC4 as a novel diagnostic and prognostic biomarker. BMC Med.
20:4372022. View Article : Google Scholar
|
|
81
|
Tu S, Zhang H, Yang X, Wen W, Song K, Yu X
and Qu X: Screening of cervical cancer-related hub genes based on
comprehensive bioinformatics analysis. Cancer Biomark. 32:303–315.
2021. View Article : Google Scholar
|
|
82
|
Ho KH, Kuo TC, Lee YT, Chen PH, Shih CM,
Cheng CH, Liu AJ, Lee CC and Chen KC: Xanthohumol regulates
miR-4749-5p-inhibited RFC2 signaling in enhancing temozolomide
cytotoxicity to glioblastoma. Life Sci. 254:1178072020. View Article : Google Scholar
|
|
83
|
Alaa Eldeen M, Mamdouh F, Abdulsahib WK,
Eid RA, Alhanshani AA, Shati AA, Alqahtani YA, Alshehri MA, Samir
A, Zaki M, et al: Oncogenic potential of replication factor C
subunit 4: Correlations with tumor progression and assessment of
potential inhibitors. Pharmaceuticals (Basel). 17:1522024.
View Article : Google Scholar
|
|
84
|
Rohde JM, Rai G, Choi YJ, Sakamuru S, Fox
JT, Huang R, Xia M, Myung K, Boxer MB and Maloney DJ: Discovery of
ML367, inhibitor of ATAD5 stabilization. Probe Reports from the NIH
Molecular Libraries Program [Internet] Bethesda (MD): National
Center for Biotechnology Information (US); 2010
|
|
85
|
Xu J, Wang G, Gong W, Guo S, Li D and Zhan
Q: The noncoding function of NELFA mRNA promotes the development of
oesophageal squamous cell carcinoma by regulating the Rad17-RFC2-5
complex. Mol Oncol. 14:611–624. 2020. View Article : Google Scholar
|
|
86
|
Cortese A, Simone R, Sullivan R,
Vandrovcova J, Tariq H, Yau WY, Humphrey J, Jaunmuktane Z,
Sivakumar P, Polke J, et al: Biallelic expansion of an intronic
repeat in RFC1 is a common cause of late-onsetataxia. Nat Genet.
51:649–658. 2019. View Article : Google Scholar
|
|
87
|
Haffner MC, Zwart W, Roudier MP, True LD,
Nelson WG, Epstein JI, De Marzo AM, Nelson PS and Yegnasubramanian
S: Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev
Urol. 18:79–92. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Huang S, Chen Y, Hu M, Fu S, Yao Z, He H,
Wang L, Chen Z and Liu X: The role of the SIX family in cancer
development and therapy: Insights from foundational and
cutting-edge research. Crit Rev Oncol Hemat. 214:1048602025.
View Article : Google Scholar : PubMed/NCBI
|