The biological hallmarks of cancer include
uncontrolled cell proliferation and differentiation, dysregulated
signaling pathways, genomic instability and metabolic reprogramming
(1). These characteristics make the
development of more effective cancer therapies a critical research
priority. In recent years, the emergence of immunotherapy
represents a major breakthrough in cancer treatment. As the fifth
treatment modality following surgery, radiotherapy, chemotherapy
and targeted therapy, immunotherapy works by re-establishing the
tumor-immune cycle and restoring the body's normal anti-tumor
immune response, thereby achieving tumor control and eradication
(2,3). Immune checkpoint blockade therapy has
been widely proven to be effective against various human tumors,
while cell therapy has shown notable efficacy in hematological
malignancies and remains in the clinical research stage for solid
tumors. In the field of immune checkpoint inhibitors, the main
approaches include: i) Monoclonal antibody therapy targeting
programmed cell death protein 1 (PD-1) and its ligand PD-L1; ii)
monoclonal antibody therapy targeting cytotoxic
T-lymphocyte-associated protein 4 (CTLA-4); and iii) monoclonal
antibody therapy targeting lymphocyte-activation gene 3. As for
adoptive cell therapy, it primarily encompasses: i)
Tumor-infiltrating lymphocyte therapy; ii) T cell
receptor-engineered T cell therapy; iii) chimeric antigen receptor
T cell (CAR-T) therapy; and iv) natural killer (NK) cell therapy
(4).
PD-1/PD-L1 monoclonal antibody therapy has emerged
as the most widely used immune checkpoint inhibitor in clinical
practice, demonstrating notable efficacy across various
malignancies (5). PD-1, also known
as CD279, is a pivotal immune checkpoint molecule that plays a
central role in maintaining immune homeostasis and regulating tumor
immune evasion. PD-1 is predominantly expressed on the surface of B
cells, T cells and NK cells, where it can specifically recognize
and bind to two ligands expressed on tumor cells: i) PD-L1 (CD274);
and ii) PD-L2 (CD273) (6). Unlike
CTLA-4 which primarily regulates immune responses during the early
stage of T cell activation, PD-1 predominantly suppresses effector
T cell function in peripheral tissues and the tumor
microenvironment during the effector phase (7). In the tumor microenvironment, the
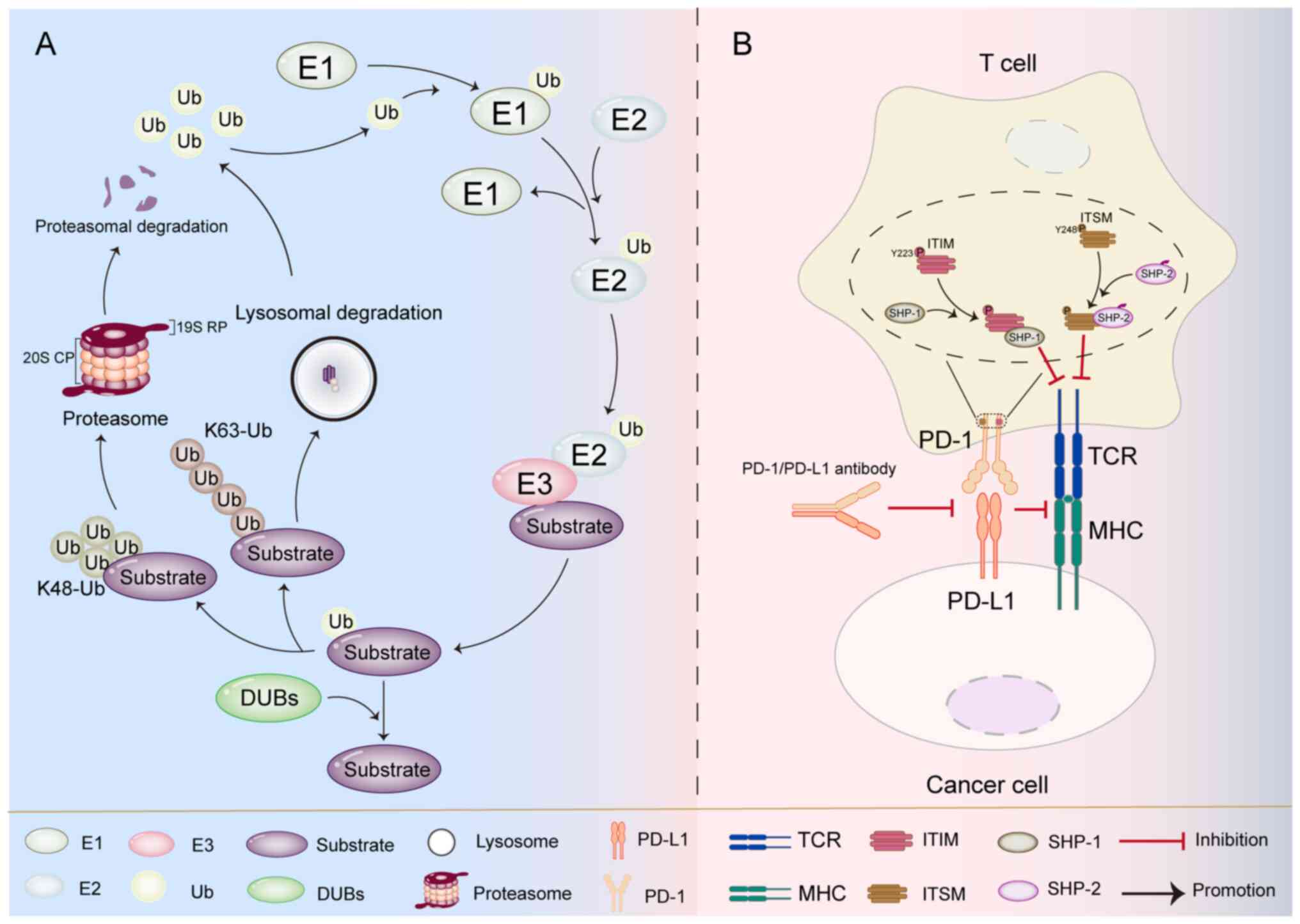
binding of PD-L1 expressed at high level on tumor cells to PD-1 on
T cell surfaces induces conformational changes in PD-1. This leads
to the exposure and phosphorylation of the immunoreceptor
tyrosine-based inhibitory motif (ITIM) at Y223 and the
immunoreceptor tyrosine-based switch motif (ITSM) at Y248. The
phosphorylated ITSM preferentially recruits the SHP-2 protein
tyrosine phosphatase, while the phosphorylated ITIM forms dimers
with the SHP-1 protein tyrosine phosphatase. These molecular events
collectively attenuate T cell activation signals, suppress T cell
cytotoxic function and consequently mediate negative regulation of
immune responses to maintain immune homeostasis (8–11).
PD-L1 can also interact with the costimulatory molecule CD80,
transmitting inhibitory signals to activated T cells (12). Currently, several PD-1/PD-L1
monoclonal antibodies such as nivolumab (13), pembrolizumab (14) and avelumab (15) have been widely used in clinical
cancer treatment. Although these immune checkpoint inhibitors
demonstrate notable efficacy against various solid tumors and
hematological malignancies, acquired resistance remains a major
clinical challenge (16).
Research has demonstrated that dysregulation of the
ubiquitin-dependent protein degradation pathway represents a
crucial molecular mechanism in cancer pathogenesis (17,18).
The UPS is an essential protein degradation mechanism in cells.
This system primarily works by ubiquitinating damaged, abnormal, or
functionally completed regulatory proteins and directing their
degradation by the proteasome. (19). The UPS consists of a series of
enzymes: i) Ubiquitin-activating enzyme E1 activates ubiquitin
molecules; ii) ubiquitin-conjugating enzyme E2 mediates ubiquitin
transfer; and iii) ubiquitin ligase E3 specifically recognizes
substrate proteins and completes ubiquitin tagging (Fig. 1). These enzymes work cooperatively
to ultimately achieve targeted protein degradation via the
proteasome pathway (20,21). Ubiquitinated substrates are
classified into different ubiquitination pathways based on the
types of polyubiquitin chains, with K48 and K63 being the two most
widely studied ubiquitination forms (22). Among these, K48-linked
ubiquitination is recognized to direct target proteins for
degradation via the proteasome pathway (23,24).
K63-linked ubiquitination is primarily involved in
proteasome-independent signaling pathways, typically associated
with positive regulatory processes such as protein stabilization,
subcellular localization and functional activation, including
critical biological processes such as endocytic trafficking, DNA
replication and signal transduction (25). Moreover, this modification can also
facilitate substrate protein degradation through the
autophagy-lysosome pathway (26).
The 26S proteasome is a multi-subunit proteolytic
complex composed of a 20S core particle (CP) and a 19S regulatory
particle (RP), which specifically recognizes polyubiquitin-tagged
proteins and degrades them into short peptides (27). The 19S RP performs three key
functions: i) Recognizing ubiquitinated substrate proteins; ii)
regulating the deubiquitination process; and iii) delivering
ubiquitinated proteins to the 20S CP. The 20S CP is a barrel-shaped
proteolytic core containing active catalytic sites, where the final
protein degradation occurs (28,29).
Deubiquitinating enzymes (DUBs) can reverse protein ubiquitination,
with their primary functions including: i) Maintaining cellular
free ubiquitin levels; ii) releasing substrate proteins from the
ubiquitin-proteasome degradation pathway; and iii) protecting
target proteins from degradation (22). Research has revealed an increasingly
clear connection between ubiquitination and cancer immunotherapy.
Tumor cells can modulate the UPS to stabilize immune checkpoint
protein expression and suppress immune-related protein function,
thereby evading immune surveillance and promoting tumor
progression.
In recent years, the regulatory role of the UPS in
cancer immunotherapy has received growing attention. The present
review summarized the key molecular mechanisms of UPS involvement
in tumor immune regulation and discusses potential therapeutic
strategies to enhance immunotherapeutic efficacy.
Ubiquitination is a series of biochemical reactions
mediated by ubiquitin-activating enzyme (E1), ubiquitin-conjugating
enzyme (E2) and ubiquitin ligase (E3) (20,21).
E3 ubiquitin ligase is a key component of this system, which can
recognize and target specific ubiquitinated substrate proteins
(25). The UPS, through dynamically
regulating the surface expression of PD-1/PD-L1, has become a
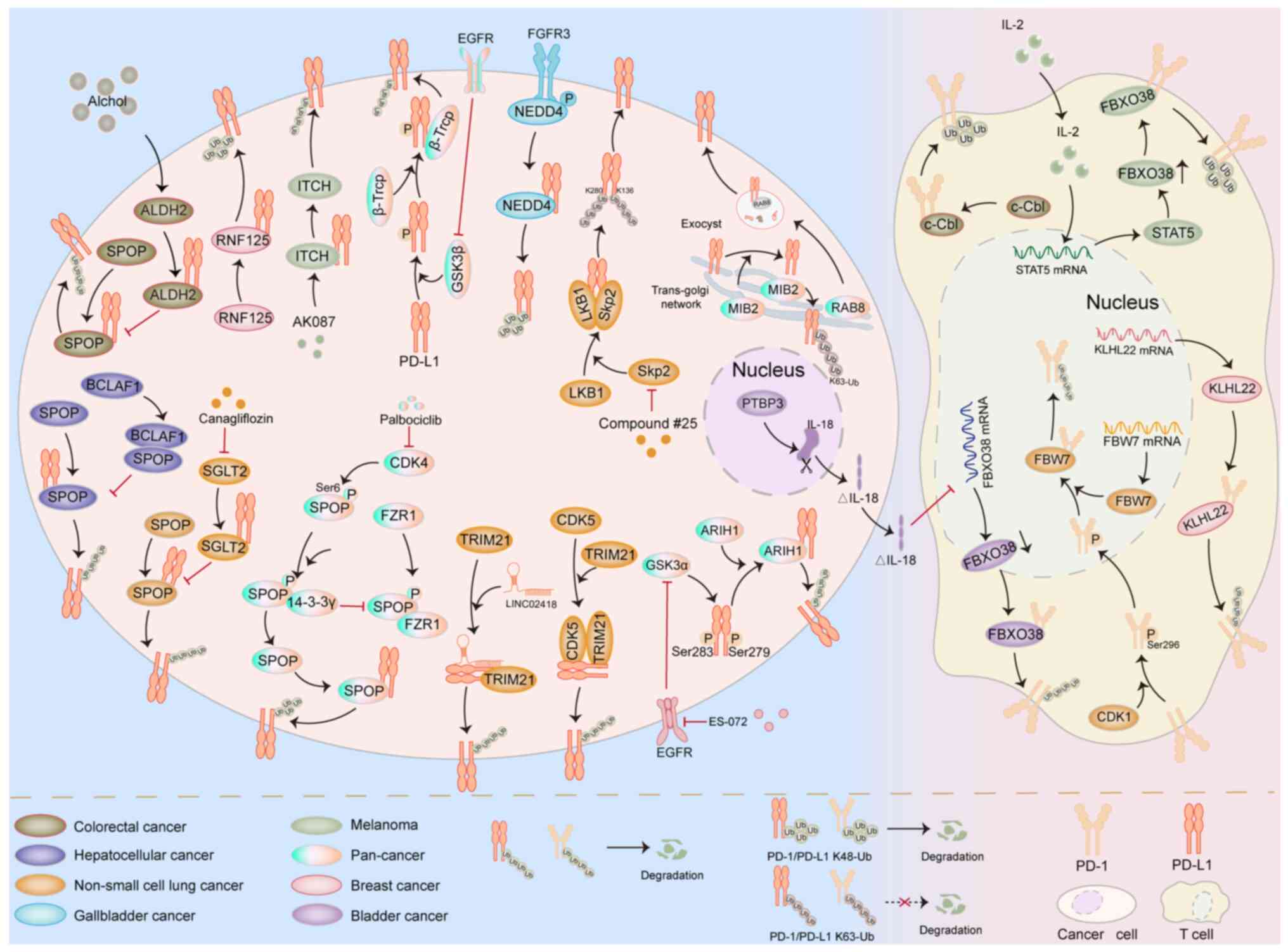
crucial link in tumor immune evasion (Fig. 2) (30,31).
Multiple studies have shown that speckle-type POZ
protein (SPOP) plays a crucial role in mediating the ubiquitination
process of PD-L1 and the mechanism of tumor immune evasion
(31–33). Zhang et al (31) found that in colorectal cancer cells,
the E3 ubiquitin ligase SPOP can promote the ubiquitination and
degradation of PD-L1. Meanwhile, the ALDH2 expressed at high levels
in cancer cells can competitively bind to PD-L1 with SPOP, thereby
inhibiting the ubiquitination process of PD-L1 mediated by SPOP and
ultimately weakening the antitumor effect of T cells. In addition,
in hepatocellular carcinoma, the transcription factor BCLAF1 can
inhibit the ubiquitination of PD-L1 by SPOP by targeting and
binding to SPOP. This mechanism enhances the stability of PD-L1 and
promotes tumor immune evasion (32). Ding et al (34) found that SGLT2 can competitively
bind to PD-L1 with the E3 ubiquitin ligase SPOP, thereby preventing
PD-L1 from being degraded through the proteasome pathway. The
small-molecule SGLT2 inhibitor canagliflozin can disrupt the
interaction between SGLT2 and PD-L1, prompting SPOP to recognize
PD-L1 and promote its ubiquitination, followed by degradation via
the proteasome pathway, thereby enhancing the antitumor activity of
T cells. Zhang et al (33)
found that in various types of cancer, CDK4 can directly promote
the phosphorylation of SPOP at Ser6. The phosphorylated SPOP can
bind to the scaffold protein 14-3-3γ, thereby blocking the binding
of SPOP to the complex activator FZR1, stabilizing the expression
level of SPOP, enabling it to recognize and promote the K48
ubiquitination of PD-L1 and eventually leading to its degradation
via the proteasome pathway.
While SPOP serves as a well-characterized E3 ligase
regulating PD-L1 stability, other ubiquitin ligases also contribute
markedly to this regulatory network. In non-small cell lung cancer,
the E3 ubiquitin ligase TRIM21 can ubiquitinate PD-L1 and promote
its degradation. LINC02418, acting as a molecular sponge, can form
a ternary complex with TRIM21 and PD-L1. This complex enhances the
ubiquitination of PD-L1 by TRIM21, ultimately leading to resistance
to anti-PD-L1-based immunotherapy in non-small cell lung cancer
(35). Gao et al (36) found that CDK5 can promote the
ubiquitination process of PD-L1 by TRIM21 by interacting with
TRIM21 and PD-L1. This mechanism notably exacerbates the resistance
of patients with non-small cell lung cancer to anti-PD-L1-based
immunotherapy. Wu et al (37) showed that in lymphosarcoma and
non-small cell lung cancer cells, glycogen synthase kinase 3 alpha
(GSK3α) can enhance the recognition and ubiquitination of PD-L1 by
the E3 ubiquitin ligase ARIH1 by promoting the phosphorylation of
PD-L1 at Ser279 and Ser283. However, the inhibitory effect of
epidermal growth factor (EGF) receptor (EGFR) on GSK3α in cancer
cells mediates tumor immune treatment resistance. The combined
application of the EGFR inhibitor ES-072 and immunotherapy can
effectively enhance the efficacy of immunotherapy (37). Wei et al (38) found that RNF125 can directly
interact with PD-L1, promote the K48 ubiquitination of PD-L1,
thereby accelerating its degradation and ultimately inhibiting the
immune evasion of tumor cells. Yang et al (39) found that in melanoma cells, ITCH can
promote the ubiquitination and degradation process of PD-L1. The
small-molecule ITCH agonist AK087 can effectively reduce the
accumulation of PD-L1 induced by MAPK inhibitors, thereby weakening
the tumor immune evasion and acquired resistance to anti-PD-L1
treatment induced by MAPK inhibitors. Glycogen synthase kinase 3β
(GSK3β), a serine/threonine kinase, has the function of catalyzing
the phosphorylation of substrates. Moreover, the phosphorylation
mediated by GSK3β usually promotes the recognition of substrates by
E3 ubiquitin ligases (40). Li
et al (41) showed that
GSK3β can interact with the E3 ligase β-TrCP and PD-L1, prompting
β-TrCP to ubiquitinate PD-L1 in a phosphorylation-dependent manner,
thereby accelerating the degradation of PD-L1. In basal-like breast
cancer, the EGF/EGFR signaling can stabilize PD-L1 by inhibiting
the activity of GSK3β, thereby mediating tumor immune treatment
resistance. The combination of the EGFR-targeted inhibitor
gefitinib and anti-PD-L1 treatment markedly enhances the efficacy
of tumor immunotherapy. In bladder cancer, the E3 ubiquitin ligase
NEDD4 can target PD-L1 and promote its K48 ubiquitination.
Fibroblast growth factor receptor 3 (FGFR3) can activate the
enzymatic activity of NEDD4 by phosphorylating it, thereby
promoting the degradation of PD-L1. Therefore, the use of FGFR3
inhibitors to treat bladder cancer may lead to the occurrence of
immune evasion in bladder cancer (42).
K63 ubiquitination is a type of non-proteolytic
ubiquitination, which is usually closely associated with positive
regulatory processes such as the maintenance of protein stability,
subcellular localization and functional activation (25). It has been shown that MIB2 can
promote the K63 ubiquitination of PD-L1 and stabilize the
expression of PD-L1. This mechanism drives the RAB8-mediated
exocytosis, facilitating the transportation of PD-L1 from the
trans-Golgi network to the plasma membrane and ultimately mediating
tumor immune evasion (43). In
non-small cell lung cancer, S-phase kinase-associated protein 2
(Skp2), as a key linker molecule between LKB1 and PD-L1, has been
shown to be able to stabilize the expression level of PD-L1 by
promoting the K63 ubiquitination of K136 and K280 residues on
PD-L1. In addition, LKB1 can promote the expression of Skp2 and
PD-L1. This series of actions ultimately mediates the phenomenon of
immune evasion in non-small cell lung cancer (44).
Studies have shown that FBXO38 can promote the K48
ubiquitination of PD-1 and accelerate its degradation process.
Exogenous interleukin (IL)-2 can enhance the transcriptional
activity of signal transducer and activator of transcription 5,
upregulate the expression level of FBXO38 and thereby enhance the
anti-tumor ability of T cells (30). In gallbladder cancer (GBC), PTBP3
which is expressed at high levels can promote the production of the
IL-18 splice variant ΔIL-18. ΔIL-18 can downregulate the
transcriptional level of FBXO38. This mechanism inhibits the
ubiquitination process of PD-1 mediated by FBXO38, ultimately
promoting the immune treatment evasion phenomenon in GBC (45). Zhou et al (46) demonstrated that the E3 ubiquitin
ligase KLHL22 can recognize PD-1 and promote its ubiquitination.
This process reduces the expression level of PD-1 on the surface of
breast cancer cells and ultimately enhances the immune function of
T cells. Liu et al (47)
demonstrated that CDK1 can promote the nuclear translocation of
PD-1 by enhancing the phosphorylation of PD-1 at Ser296. This
mechanism facilitates the interaction between the E3 ubiquitin
ligase F-box and WD repeat domain-containing 7 in the nucleus and
PD-1, thereby mediating the ubiquitination and degradation process
of PD-1 in non-small cell lung cancer and ultimately enhancing the
anti-tumor ability of T cells. In colorectal cancer, c-Cbl binds to
and interacts with PD-1, leading to its degradation via the
ubiquitin-proteasome pathway. This reduces PD-1 expression levels,
enhances the anti-tumor activity of T cells and promotes
immunotherapy efficacy (48).
DUBs can regulate the metabolic level of substrate
proteins by cleaving monoubiquitin or polyubiquitin molecules,
thereby modulating a variety of cellular activities, such as gene
transcription, tumorigenesis and inflammatory immune responses
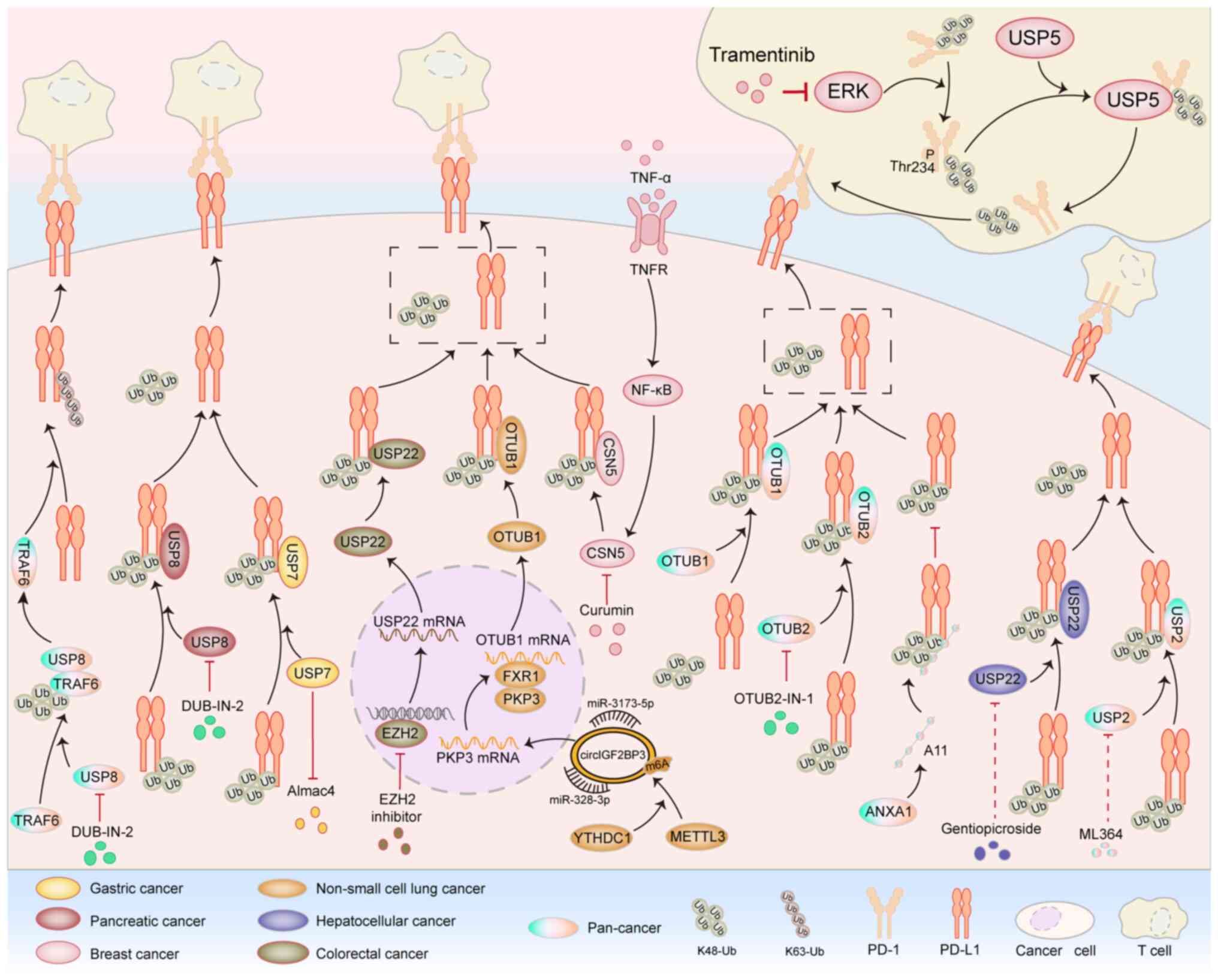
(Fig. 3) (49).
The ubiquitin-specific proteases (USP) family is a
group of enzymes that specifically participate in protein
deubiquitination modification and belongs to the DUB family.
Studies have shown that the USP family plays a crucial role in
regulating the deubiquitination of PD-L1 to mediate tumor immune
evasion (50–52). Wang et al (51) found that that USP7 can directly
target PD-L1 and deubiquitinate it, thereby stabilizing the
expression level of the PD-L1 protein and ultimately promoting the
process of immune evasion in gastric cancer. Another study showed
that USP8 can directly bind to PD-L1 and remove its ubiquitination
modification, thereby stabilizing the protein expression level of
PD-L1 and ultimately promoting the process of immune evasion in
pancreatic cancer (52). USP8 can
not only directly mediate the deubiquitination of PD-L1, but has
also been proved to stabilize the expression of TRAF6 by
deubiquitinating TRAF6 in various types of cancer. Once TRAF6 is
stably expressed, it will further promote the K63 ubiquitination of
PD-L1 mediated by itself, ultimately stabilizing PD-L1 and
promoting tumor immune evasion. The inhibition of USP8 by DUBs-IN-2
can effectively enhance the antitumor activity of T cells (53). In colorectal cancer and prostate
cancer cells, USP2 can directly interact with PD-L1 and promote the
K48 deubiquitination of PD-L1. This process stabilizes the
expression of PD-L1 and then mediates tumor immune evasion
(54). In liver cancer, USP22 can
deubiquitinate PD-L1 and thereby mediate the antitumor immune
resistance in liver cancer (55).
In colorectal cancer, the inhibition of enhancer of zeste homolog 2
upregulates the expression of USP22 at the transcriptional level.
The upregulated USP22 further deubiquitinates and stabilizes PD-L1,
ultimately promoting the process of tumor immune evasion (56). In breast cancer, lung cancer and
melanoma, the derivative peptide A11 of annexin A1 can
competitively bind to PD-L1 with USP7, which is a deubiquitinase of
PD-L1. This process inhibits the deubiquitination process of PD-L1
mediated by USP7, thereby promoting the degradation of PD-L1 and
ultimately leading to the phenomenon of immune treatment resistance
in tumors (57).
The OTUB family is a part of the DUB family. Studies
have shown that the OTUB family also plays an important role in
mediating the ubiquitination of PD-L1 (58–60).
Zhu et al (59) found that
in various types of cancer, OTUB1 can directly bind to PD-L1 and
remove its K48 ubiquitination modification. This process inhibits
the degradation of PD-L1 through the endoplasmic
reticulum-associated degradation pathway, ultimately promoting
tumor immune evasion. In addition, in non-small cell lung cancer,
METTL3 mediates m6A modification in a YTHDC1-dependent manner,
thereby promoting the circularization of circIGF2BP3. Acting as a
molecular sponge for microRNA (miR)-328-3p and miR-3173-5p,
circIGF2BP3 upregulates the expression of PKP3. Subsequently, PKP3
stabilizes OTUB1 mRNA through fragile × mental retardation
syndrome-related protein 1 and ultimately, through the
OTUB1-mediated deubiquitination of PD-L1, inhibits the function of
CD8+ T cells (61).
OTUB2 has also been proven to be able to directly interact with
PD-L1 and deubiquitinate it. This process inhibits the
ubiquitination and degradation of PD-L1 in the endoplasmic
reticulum. The OTUB2 inhibitor OTUB2-IN-1 can effectively interfere
with the DUB activity of OTUB2, thereby markedly enhancing the
antitumor immune effect (60). COP9
signalosome 5 (CSN5), as a subunit of the COP9 signalosome,
possesses DUB activity and can remove ubiquitin chains from
substrate proteins, thereby preventing substrate proteins from
being degraded by the proteasome. In triple-negative breast cancer
(TNBC), tumor necrosis factor-α upregulates the expression level
and activity of CSN5 through the nuclear factor-κB signaling
pathway, thereby promoting the deubiquitination process of PD-L1
mediated by CSN5 and ultimately enhancing the resistance of cancer
cells to PD-1/PD-L1 immunotherapy. The CSN5-targeting inhibitor
curcumin can promote the degradation of PD-L1 and thus enhance the
efficacy of tumor immunotherapy (62).
The UPS is the core mechanism for maintaining
protein homeostasis within cells. During tumor immune evasion, UPS
mainly regulates the dynamic balance of immune checkpoint
molecules, antigen presentation processes and immunosuppressive
cells through ubiquitination tagging and proteasomal degradation
(64). Studies have confirmed that
small-molecule drugs targeting the UPS play an important role in
improving the therapeutic effects of tumor immune checkpoint
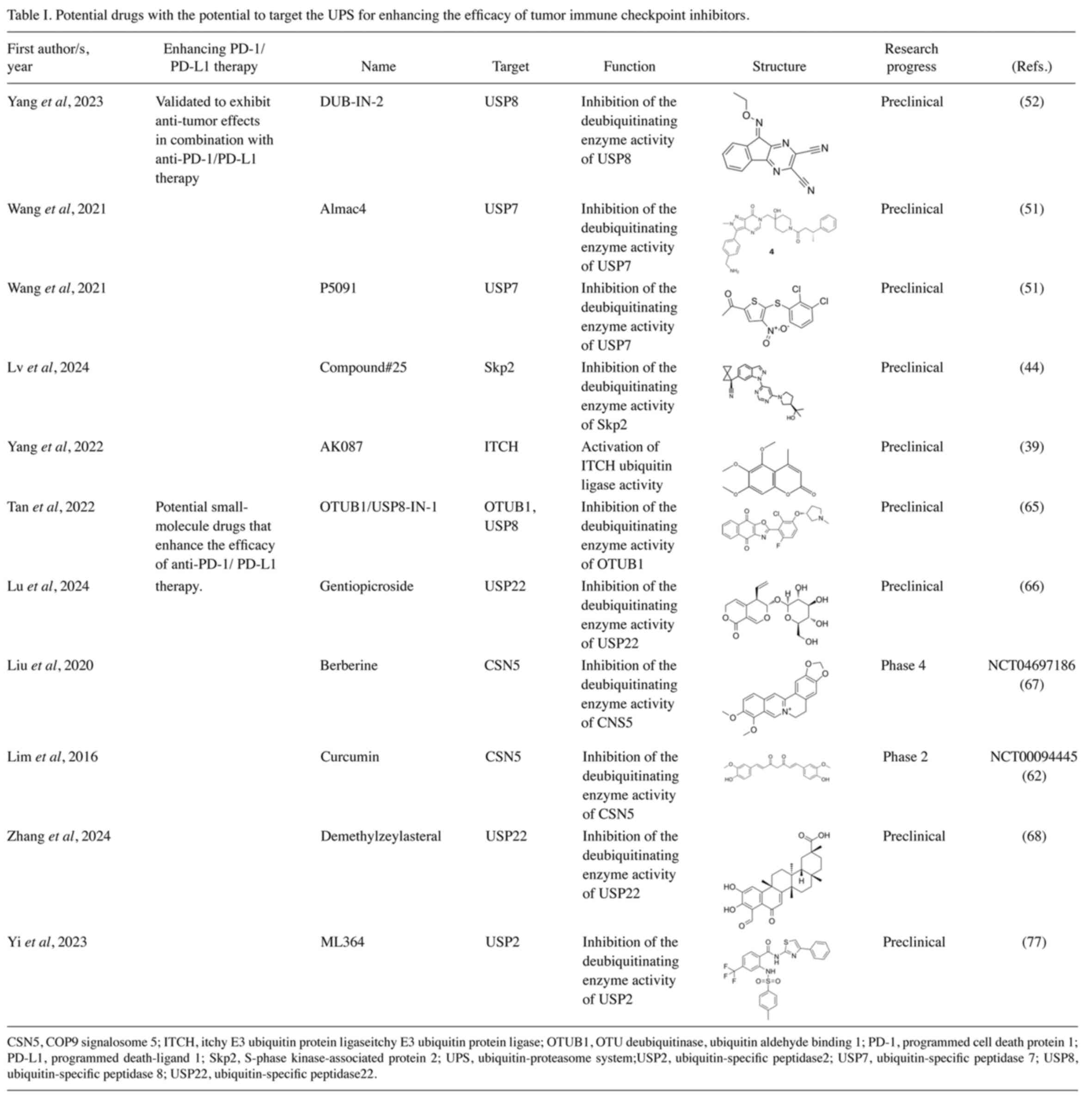
inhibitors (Table I) (51,52,60).
In pancreatic cancer, DUB-IN-2 inhibits the deubiquitination
function of USP8, reduces the expression level of PD-L1 and thereby
reverses the immunosuppressive state in the tumor microenvironment
(52). In gastric cancer, the use
of the small-molecule inhibitors Almac4 and P5091 targeting USP7
can inhibit the deubiquitination activity of USP7, thereby
suppressing the proliferation of cancer cells. In addition, these
two inhibitors can also downregulate the expression level of PD-L1
and enhance the anti-tumor immune response (51). In melanoma and colorectal cancer,
the small-molecule inhibitor OTUB2-IN-1 can notably inhibit the DUB
activity of OTUB2 and reduce the expression level of PD-L1 in tumor
cells in a dose-dependent manner, thereby promoting antitumor
immune function (60).
Recent studies have further revealed that specific
small-molecule compounds can modulate the stability of PD-L1
through ubiquitination pathways. In non-small cell lung cancer,
compound #25 can enhance the anti-tumor immune response by
inhibiting the Skp2-mediated K63 ubiquitination process of PD-L1
(44). The small-molecule agonist
AK087 of ITCH can effectively promote the ubiquitination and
degradation process of PD-L1 mediated by ITCH and notably inhibit
the resistance of tumors to PD-1/PD-L1 treatment (39).
In addition, some small-molecule drugs have shown
great potential in the field of tumor immunotherapy.
OTUB1/USP8-IN-1 is a dual inhibitor targeting OTUB1 and USP8, which
can effectively inhibit the functions of these two DUBs and is a
potential small-molecule drug for tumor immunotherapy (65). Natural compounds also play a marked
role in the field of tumor immunotherapy. Lu et al (66) found that the natural compound
gentiopicroside can inhibit the DUB activity of USP22, thereby
reducing the expression level of PD-L1 in lung adenocarcinoma and
enhancing the body's anti-tumor immune ability. Curcumin, as a
natural dietary supplement, has been proven to have the potential
for anti-tumor immunity. In TNBC, curcumin can inhibit the DUB
activity of CSN5, reduce the stability of PD-L1, induce the
ubiquitination and degradation of PD-L1 and thereby enhance the
immune system's ability to attack tumor cells (62). In lung cancer, CSN5 can promote the
deubiquitination process of PD-L1, thereby inducing tumor immune
evasion. The natural compound berberine can inhibit the DUB
activity of CSN5, reduce the expression level of PD-L1 and thus
enhance the body's anti-tumor immune ability (67). In colorectal and lung cancers,
demethylzeylasteral specifically binds to USP22 and induces its
degradation, thereby promoting ubiquitin-dependent proteasomal
degradation of PD-L1 and ultimately enhancing T cell-mediated
antitumor immune responses (68).
Small-molecule inhibitors targeting the UPS have
shown great potential in the field of tumor immunotherapy and are
very likely to be one of the important strategies for cancer
treatment in the future.
During the occurrence and development of tumors,
tumor cells can evade the strict surveillance of the immune system
through a variety of complex mechanisms. Among these mechanisms,
the activation of immune checkpoint pathways is one of the core
mechanisms by which tumors achieve immune evasion. At present,
anti-PD-1/PD-L1 therapy, as a representative of immune checkpoint
inhibitor therapy, is one of the most widely used tumor
immunotherapy strategies in clinical practice (5). Although anti-PD-1/PD-L1 therapy has
shown good efficacy in some patients, a considerable number of
patients still do not respond to the treatment after receiving it
and even develop acquired resistance due to anti-PD-1/PD-L1 therapy
(69). Therefore, elucidating
further the potential molecular mechanisms underlying tumor
resistance to anti-PD-1/PD-L1 and exploring effective combination
therapy strategies are of great significance for improving the
therapeutic efficacy of anti-PD-1/PD-L1 and prolonging the survival
of patients (Table II).
The UPS, as a crucial molecular mechanism
responsible for protein degradation and stabilization within cells,
plays a pivotal role in regulating various biological processes
such as cell cycle progression, signal transduction networks and
immune response reactions. It is one of the main pathways mediating
protein degradation or stabilization inside cells (70,71).
The UPS has the ability to precisely recognize and selectively tag
damaged, abnormal, or function-completed proteins with ubiquitin
‘tags’. Subsequently, the proteasome recognizes these tagged
proteins and precisely regulates the degradation process of the
proteins (19). In recent years,
with the continuous deepening of research, an increasing number of
studies have shown that the UPS plays a crucial role in the
pathological and physiological processes of tumor proliferation,
invasion, metastasis, immune regulation and drug resistance
(72–74). It has been confirmed that the UPS
can mediate tumor immune evasion and drug resistance induced by
immune checkpoint inhibitor therapy by regulating the
ubiquitination level of PD-1/PD-L1 (31,51,52).
In-depth exploration of the potential molecular mechanisms by which
the UPS regulates PD-1/PD-L1 will not only help improve the
efficacy of tumor immunotherapy, but also provide a solid
theoretical basis for the development of novel therapeutic
strategies.
In recent years, targeting the UPS for disease
treatment has become an important direction in drug development,
showing great therapeutic potential in a number of fields such as
autoimmune diseases and neurodegenerative diseases and cancer
(75,76). Studies have shown that targeting the
ubiquitination process of PD-1/PD-L1 mediated by the UPS has
achieved notable effects in inhibiting tumor immune evasion and
improving tumor resistance to anti-PD-1/PD-L1 therapy. For example,
the small-molecule agonist AK087 of ITCH can effectively promote
the ubiquitination and degradation of PD-L1 mediated by ITCH and
markedly inhibit the resistance of tumors to anti-PD-1/PD-L1
treatment (39). The USP8 inhibitor
DUB-IN-2 can effectively inhibit the deubiquitination process of
PD-L1 mediated by USP8. This action notably inhibits tumor immune
evasion and can effectively improve the efficacy of tumor
immunotherapy (53). The
small-molecule compound ML364 directly binds to USP2 and inhibits
its deubiquitinase activity (77).
The UPS not only determines the protein stability of PD-L1, but
also forms a ‘positive-negative feedback’ loop with the
IFN-γ/JAK-STAT signaling pathway through key nodes including
TRIM25/SOCS1/USP18. Targeting this regulatory loop can amplify or
suppress IFN-γ signaling across different cancer types, thereby
guiding combination strategies between UPS inhibitors and immune
checkpoint inhibitors (78,79).
This therapeutic model targeting the UPS has a
unique mechanism of action. Instead of directly blocking protein
functions, it regulates the stability and degradation of proteins
from the source, thereby affecting the levels of abnormal proteins.
Meanwhile, this therapeutic model has precise targeting ability,
which can reduce the biological toxicity caused by broad-spectrum
inhibition. Due to tissue specificity, the functions of E3
ubiquitin ligases also vary. The ‘functional switch’ of the same E3
ubiquitin ligase in different cancers is collectively determined by
protein expression in the tumor microenvironment, signaling
pathways and post-translational modifications. For instance, the
mutational inactivation and dysregulated expression of SPOP in
various types of cancer can affect its E3 ubiquitin ligase
activity. Notably, Skp2-mediated PD-L1 K63 ubiquitination in
non-small cell lung cancer depends on LKB1 inactivation (Fig. 2), whereas BRAF inhibitors in
melanoma suppress PD-L1 ubiquitination by inhibiting the
ERK-GSK3β-β-TrCP axis (41,80). This explains the reason Skp2
inhibitors (compound #25) can enhance the efficacy of PD-1
antibodies in non-small cell lung cancer, while melanoma requires
combined MAPK inhibition to relieve β-TrCP suppression. For
example, preclinical data on the USP7 inhibitor P5091 in gastric
cancer showed that H. pylori-positive patients (with
concomitant USP7 overexpression) had a 3.2-fold higher response
rate compared with negative patients (P<0.01), suggesting that
future trials should stratify patients based on microbiome-UPS
co-mutation status (51). By
contrast, a phase II trial of the CSN5 inhibitor curcumin in TNBC
(trial no. NCT00094445) demonstrated limited efficacy due to the
lack of screening for CSN5-high populations, highlighting the
necessity of biomarker-guided therapy. Although UPS-targeted drugs
such as USP8 inhibitors may develop resistance due to mutations or
compensatory pathways, current evidence suggests that such
resistance mechanisms are independent of PD-1/PD-L1 ubiquitination
regulation. Future studies should explore whether resistance to
UPS-targeted drugs upregulates PD-L1 through non-UPS pathways such
as through transcriptional reprogramming or exosome release,
thereby indirectly leading to immunotherapy failure. It is
recommend that future studies employ cancer-specific organoid
models to validate UPS-targeting drugs, thereby mimicking the
effect of stromal cells on ubiquitination regulation within the
tumor microenvironment.
Although some molecular drugs targeting the UPS have
achieved preliminary progress in improving tumor immunotherapy, the
number of those in clinical verification stage is still limited. As
of November 2024, <20 UPS-targeting agents have entered clinical
stages globally, with most concentrated in the
proteolysis-targeting chimera (PROTAC). Notably, UPS modulators
specifically targeting PD-1/PD-L1 remain in Phase I or earlier
development (81). High expression
of E3 ubiquitin ligases such as CRBN and VHL in the liver and
kidneys often leads to hematological and renal toxicity, Excessive
PROTACs may form nonfunctional binary complexes, disrupting UPS
activity and causing drug ‘rebound effects’ (82). Dong et al (83) encapsulated VPS18/11 inhibitors such
as RD-N into lung-targeted nanoparticles, effectively reversing
tumor resistance and suppressing metastasis. Similar strategies
could enable tissue-specific delivery of deubiquitinase inhibitors,
minimizing off-target effects and systemic toxicity. UPS gene
mutations or compensatory pathway activation can result in adaptive
resistance during long-term treatment (84). Real-time drug concentration
tracking, combined with artificial intelligence and machine
learning-based predictive models, may optimize pharmacokinetic
profiles and address long-term adaptive resistance.
The PROTAC technology, as an emerging therapeutic
strategy for targeted protein degradation in recent years, has
gradually attracted widespread attention (85,86).
The PROTAC consists of three parts: i) A target protein-binding
ligand; ii) an E3 ligase ligand; and iii) a linker. Its main
mechanism of action is to induce the binding of the E3 ligase to
the target protein, mediate the ubiquitination of the target
protein and subsequently promote the degradation of the target
protein (87). This technology has
overcome the limitation of the ‘undruggable’ status of certain
proteins and can reversibly regulate the expression levels of
proteins over time. However, PROTAC drugs usually have a relatively
large molecular weight, which leads to limited bioavailability.
Therefore, the drug design of PROTAC still needs further
optimization.
Conventional PROTACs suffer from high molecular
weight, poor membrane permeability and low oral bioavailability,
leading to weak in vitro-in vivo association and limited
clinical translation. To overcome these hurdles, Sun et al
(88) developed tumor
microenvironment (TME)-responsive enzyme-activated click-forming
PROTACs (ENCTACs). By exploiting cathepsin B overexpressed in
>90% of solid tumors as a biological trigger, an orthogonal
cleavage-click reaction assembles the active degrader in
situ, selectively eliminating the epigenetic regulator BRD4 and
consequently downregulating PD-L1 to remodel the immune
microenvironment. In the 4T1 TNBC mouse model, ENCTACs achieved a
65% tumor-growth inhibition, a three-fold deeper tissue penetration
and negligible systemic toxicity compared with traditional PROTACs.
In parallel, a recent study conjugated a CD47 antibody with a
folate ligand to create a Folate Receptor Targeting Chimera
(FRTAC). Leveraging the high folate-receptor expression on cancer
cells, FRTAC drives CD47 into lysosomal degradation via
receptor-mediated endocytosis, markedly potentiating
macrophage-mediated phagocytosis while sparing normal tissues
(89). At present, multiple PROTAC
drugs for tumor treatment have entered the clinical stage, such as
ARV-110 for prostate cancer (90)
and ARV-471 for ER+ breast cancer (91). Looking ahead, next-generation PROTAC
platforms that integrate TME-specific activation with
nanoparticle-based delivery are poised to surmount the dual
barriers of cellular permeability and off-target toxicity, offering
unprecedented opportunities for cancer immunotherapy.
In conclusion, the present review discussed the
potential molecular mechanisms by which the UPS plays a role in
tumor immune evasion and resistance to anti-PD-1/PD-L1 therapy and
has summarized the potential targeted drugs that can inhibit tumor
immune evasion and overcome resistance to immune checkpoint therapy
by targeting the UPS. Future research should focus on the following
key aspects: i) Further in-depth exploration of the regulatory
mechanisms of the UPS on various immune checkpoints in different
tumor types and immune microenvironments; ii) optimization of the
design of targeted UPS drugs to improve their targeting ability and
bioavailability; and iii) development of combined treatment
regimens of immune checkpoint inhibitors and targeted UPS drugs to
enhance the synergistic therapeutic effect. These research
directions will lay a solid theoretical foundation for the
development of the next-generation precision immunotherapy
strategies.
Not applicable.
The present study was supported by the National Natural Science
Foundation of China (grant no. 82373124 and 81872163), the Shandong
Provincial Natural Science Foundation (grant no. ZR2023MH073) and
the research startup fund of Shaoxing People's Hospital (grant nos.
PI202501 and YJ202402).
Not applicable.
LHG and WLD were involved in conceptualization. LHG,
AG, YYD, XJW, HXZ and WLD performed the literature search, data
collection and writing. WLD and BGZ reviewed and edited the
manuscript. Data authentication is not applicable. All authors read
and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Wang Q, Shao X, Zhang Y, Zhu M, Wang FXC,
Mu J, Li J, Yao H and Chen K: Role of tumor microenvironment in
cancer progression and therapeutic strategy. Cancer Med.
12:11149–11165. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang Y and Zhang Z: The history and
advances in cancer immunotherapy: Understanding the characteristics
of tumor-infiltrating immune cells and their therapeutic
implications. Cell Mol Immunol. 17:807–821. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Esfahani K, Roudaia L, Buhlaiga N, Del
Rincon SV, Papneja N and Miller WH Jr: A review of cancer
immunotherapy: From the past, to the present, to the future. Curr
Oncol. 27:S87–S97. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang H, Kaur G, Sankin AI, Chen F, Guan F
and Zang X: Immune checkpoint blockade and CAR-T cell therapy in
hematologic malignancies. J Hematol Oncol. 12:592019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Byun DJ, Wolchok JD, Rosenberg LM and
Girotra M: Cancer immunotherapy-immune checkpoint blockade and
associated endocrinopathies. Nat Rev Endocrinol. 13:195–207. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Naimi A, Mohammed RN, Raji A, Chupradit S,
Yumashev AV, Suksatan W, Shalaby MN, Thangavelu L, Kamrava S,
Shomali N, et al: Tumor immunotherapies by immune checkpoint
inhibitors (ICIs); the pros and cons. Cell Commun Signal.
20:442022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dong H, Strome SE, Salomao DR, Tamura H,
Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al:
Tumor-associated B7-H1 promotes T-cell apoptosis: A potential
mechanism of immune evasion. Nat Med. 8:793–800. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gauen LK, Zhu Y, Letourneur F, Hu Q, Bolen
JB, Matis LA, Klausner RD and Shaw AS: Interactions of p59fyn and
ZAP-70 with T-cell receptor activation motifs: Defining the nature
of a signalling motif. Mol Cell Biol. 14:3729–3741. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Straus DB and Weiss A: Genetic evidence
for the involvement of the lck tyrosine kinase in signal
transduction through the T cell antigen receptor. Cell. 70:585–593.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang H, Dai Z, Wu W, Wang Z, Zhang N,
Zhang L, Zeng WJ, Liu Z and Cheng Q: Regulatory mechanisms of
immune checkpoints PD-L1 and CTLA-4 in cancer. J Exp Clin Cancer
Res. 40:1842021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang Q, Chen Y, Li X, Long S, Shi Y, Yu Y,
Wu W, Han L and Wang S: The role of PD-1/PD-L1 and application of
immune-checkpoint inhibitors in human cancers. Front Immunol.
13:9644422022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Butte MJ, Keir ME, Phamduy TB, Sharpe AH
and Freeman GJ: Programmed death-1 ligand 1 interacts specifically
with the B7-1 costimulatory molecule to inhibit T cell responses.
Immunity. 27:111–122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paik J: Nivolumab plus relatlimab: First
approval. Drugs. 82:925–931. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Harrington KJ, Burtness B, Greil R,
Soulières D, Tahara M, de Castro G Jr, Psyrri A, Brana I, Basté N,
Bratland Å, et al: Pembrolizumab with or without chemotherapy in
recurrent or metastatic head and neck squamous cell carcinoma:
Updated results of the phase III KEYNOTE-048 study. J Clin Oncol.
41:790–802. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Powles T, Park SH, Voog E, Caserta C,
Valderrama BP, Gurney H, Kalofonos H, Radulović S, Demey W, Ullén
A, et al: Avelumab maintenance therapy for advanced or metastatic
urothelial carcinoma. N Engl J Med. 383:1218–1230. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kiasari BA, Abbasi A, Darestani NG, Adabi
N, Moradian A, Yazdani Y, Hosseini GS, Gholami N and Janati S:
Combination therapy with nivolumab (anti-PD-1 monoclonal antibody):
A new era in tumor immunotherapy. Int Immunopharmacol.
113:1093652022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Scheffner M, Werness BA, Huibregtse JM,
Levine AJ and Howley PM: The E6 oncoprotein encoded by human
papillomavirus types 16 and 18 promotes the degradation of p53.
Cell. 63:1129–1136. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Joazeiro CA, Wing SS, Huang H, Leverson
JD, Hunter T and Liu YC: The tyrosine kinase negative regulator
c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase.
Science. 286:309–312. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Han D, Wang L, Jiang S and Yang Q: The
ubiquitin-proteasome system in breast cancer. Trends Mol Med.
29:599–621. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Eldridge AG and O'Brien T: Therapeutic
strategies within the ubiquitin proteasome system. Cell Death
Differ. 17:4–13. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fang S and Weissman AM: A field guide to
ubiquitylation. Cell Mol Life Sci. 61:1546–1561. 2004.PubMed/NCBI
|
|
22
|
Pfoh R, Lacdao IK and Saridakis V:
Deubiquitinases and the new therapeutic opportunities offered to
cancer. Endocr Relat Cancer. 22:T35–T54. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hochstrasser M: Ubiquitin-dependent
protein degradation. Annu Rev Genet. 30:405–439. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hofmann K and Falquet L: A
ubiquitin-interacting motif conserved in components of the
proteasomal and lysosomal protein degradation systems. Trends
Biochem Sci. 26:347–350. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park J, Cho J and Song EJ:
Ubiquitin-proteasome system (UPS) as a target for anticancer
treatment. Arch Pharm Res. 43:1144–1161. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McKeon JE, Sha D, Li L and Chin LS:
Parkin-mediated K63-polyubiquitination targets ubiquitin C-terminal
hydrolase L1 for degradation by the autophagy-lysosome system. Cell
Mol Life Sci. 72:1811–1824. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pickart CM and Eddins MJ: Ubiquitin:
Structures, functions, mechanisms. Biochim Biophys Acta.
1695:55–72. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Groll M, Ditzel L, Löwe J, Stock D,
Bochtler M, Bartunik HD and Huber R: Structure of 20S proteasome
from yeast at 2.4 A resolution. Nature. 386:463–471. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bedford L, Paine S, Sheppard PW, Mayer RJ
and Roelofs J: Assembly, structure, and function of the 26S
proteasome. Trends Cell Biol. 20:391–401. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Meng X, Liu X, Guo X, Jiang S, Chen T, Hu
Z, Liu H, Bai Y, Xue M, Hu R, et al: FBXO38 mediates PD-1
ubiquitination and regulates anti-tumour immunity of T cells.
Nature. 564:130–135. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang H, Xia Y, Wang F, Luo M, Yang K,
Liang S, An S, Wu S, Yang C, Chen D, et al: Aldehyde dehydrogenase
2 mediates alcohol-induced colorectal cancer immune escape through
stabilizing PD-L1 expression. Adv Sci (Weinh). 8:20034042021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu Z, Wu X, Zhu J, Yan H, Li Y, Zhang H,
Zhong Y, Lin M, Ye G, Li X, et al: BCLAF1 binds SPOP to stabilize
PD-L1 and promotes the development and immune escape of
hepatocellular carcinoma. Cell Mol Life Sci. 81:822024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Bu X, Wang H, Zhu Y, Geng Y,
Nihira NT, Tan Y, Ci Y, Wu F, Dai X, et al: Cyclin D-CDK4 kinase
destabilizes PD-L1 via cullin 3-SPOP to control cancer immune
surveillance. Nature. 553:91–95. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ding L, Chen X, Zhang W, Dai X, Guo H, Pan
X, Xu Y, Feng J, Yuan M, Gao X, et al: Canagliflozin primes
antitumor immunity by triggering PD-L1 degradation in endocytic
recycling. J Clin Invest. 133:e1547542023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun Z, Mai H, Xue C, Fan Z, Li J, Chen H,
Huo N, Kang X, Tang C, Fang L, et al:
Hsa-LINC02418/mmu-4930573I07Rik regulated by METTL3 dictates
anti-PD-L1 immunotherapeutic efficacy via enhancement of
Trim21-mediated PD-L1 ubiquitination. J Immunother Cancer.
11:e0074152023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gao L, Xia L, Ji W, Zhang Y, Xia W and Lu
S: Knockdown of CDK5 down-regulates PD-L1 via the
ubiquitination-proteasome pathway and improves antitumor immunity
in lung adenocarcinoma. Transl Oncol. 14:1011482021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu Y, Zhang C, Liu X, He Z, Shan B, Zeng
Q, Zhao Q, Zhu H, Liao H, Cen X, et al: ARIH1 signaling promotes
anti-tumor immunity by targeting PD-L1 for proteasomal degradation.
Nat Commun. 12:23462021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wei M, Mo Y, Liu J, Zhai J, Li H, Xu Y,
Peng Y, Tang Z, Wei T, Yang X, et al: Ubiquitin ligase RNF125
targets PD-L1 for ubiquitination and degradation. Front Oncol.
12:8356032022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang Z, Wang Y, Liu S, Deng W, Lomeli SH,
Moriceau G, Wohlschlegel J, Piva M and Lo RS: Enhancing PD-L1
degradation by ITCH during MAPK inhibitor therapy suppresses
acquired resistance. Cancer Discov. 12:1942–1959. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Doble BW and Woodgett JR: GSK-3: Tricks of
the trade for a multi-tasking kinase. J Cell Sci. 116:1175–1186.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo
CW, Khoo KH, Chang SS, Cha JH, Kim T, et al: Glycosylation and
stabilization of programmed death ligand-1 suppresses T-cell
activity. Nat Commun. 7:126322016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jing W, Wang G, Cui Z, Xiong G, Jiang X,
Li Y, Li W, Han B, Chen S and Shi B: FGFR3 destabilizes PD-L1 via
NEDD4 to control T-cell-mediated bladder cancer immune
surveillance. Cancer Res. 82:114–129. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu X, Li W, Liu H, Wang X, Coarfa C, Cheng
C, Yu X, Zeng Z, Cao Y, Young KH and Li Y: PD-L1 translocation to
the plasma membrane enables tumor immune evasion through MIB2
ubiquitination. J Clin Invest. 133:e1604562023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lv L, Miao Q, Zhan S, Chen P, Liu W, Lv J,
Yan W, Wang D, Liu H, Yin J, et al: LKB1 dictates sensitivity to
immunotherapy through Skp2-mediated ubiquitination of PD-L1 protein
in non-small cell lung cancer. J Immunother Cancer. 12:e0094442024.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao C, Zhao JW, Zhang YH, Zhu YD, Yang
ZY, Liu SL, Tang QY, Yang Y, Wang HK, Shu YJ, et al: PTBP3 Mediates
IL-18 exon skipping to promote immune escape in gallbladder cancer.
Adv Sci (Weinh). 11:e24066332024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou XA, Zhou J, Zhao L, Yu G, Zhan J, Shi
C, Yuan R, Wang Y, Chen C, Zhang W, et al: KLHL22 maintains PD-1
homeostasis and prevents excessive T cell suppression. Proc Natl
Acad Sci USA. 117:28239–28250. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu J, Wei L, Hu N, Wang D, Ni J, Zhang S,
Liu H, Lv T, Yin J, Ye M and Song Y: FBW7-mediated ubiquitination
and destruction of PD-1 protein primes sensitivity to anti-PD-1
immunotherapy in non-small cell lung cancer. J Immunother Cancer.
10:e0051162022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lyle C, Richards S, Yasuda K, Napoleon MA,
Walker J, Arinze N, Belghasem M, Vellard I, Yin W, Ravid JD, et al:
c-Cbl targets PD-1 in immune cells for proteasomal degradation and
modulates colorectal tumor growth. Sci Rep. 9:202572019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Clague MJ, Urbé S and Komander D: Breaking
the chains: Deubiquitylating enzyme specificity begets function.
Nat Rev Mol Cell Biol. 20:338–352. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gao H, Yin J, Ji C, Yu X, Xue J, Guan X,
Zhang S, Liu X and Xing F: Targeting ubiquitin specific proteases
(USPs) in cancer immunotherapy: From basic research to preclinical
application. J Exp Clin Cancer Res. 42:2252023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang Z, Kang W, Li O, Qi F, Wang J, You Y,
He P, Suo Z, Zheng Y and Liu HM: Abrogation of USP7 is an
alternative strategy to downregulate PD-L1 and sensitize gastric
cancer cells to T cells killing. Acta Pharm Sin B. 11:694–707.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang H, Zhang X, Lao M, Sun K, He L, Xu J,
Duan Y, Chen Y, Ying H, Li M, et al: Targeting ubiquitin-specific
protease 8 sensitizes anti-programmed death-ligand 1 immunotherapy
of pancreatic cancer. Cell Death Differ. 30:560–575. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xiong W, Gao X, Zhang T, Jiang B, Hu MM,
Bu X, Gao Y, Zhang LZ, Xiao BL, He C, et al: USP8 inhibition
reshapes an inflamed tumor microenvironment that potentiates the
immunotherapy. Nat Commun. 13:17002022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kuang Z, Liu X, Zhang N, Dong J, Sun C,
Yin M, Wang Y, Liu L, Xiao D, Zhou X, et al: USP2 promotes tumor
immune evasion via deubiquitination and stabilization of PD-L1.
Cell Death Differ. 30:2249–2264. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Huang X, Zhang Q, Lou Y, Wang J, Zhao X,
Wang L, Zhang X, Li S, Zhao Y, Chen Q, et al: USP22 deubiquitinates
CD274 to suppress anticancer immunity. Cancer Immunol Res.
7:1580–1590. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Huang J, Yin Q, Wang Y, Zhou X, Guo Y,
Tang Y, Cheng R, Yu X, Zhang J, Huang C, et al: EZH2 inhibition
enhances PD-L1 protein stability through USP22-mediated
deubiquitination in colorectal cancer. Adv Sci (Weinh).
11:e23080452024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yu ZZ, Liu YY, Zhu W, Xiao D, Huang W, Lu
SS, Yi H, Zeng T, Feng XP, Yuan L, et al: ANXA1-derived peptide for
targeting PD-L1 degradation inhibits tumor immune evasion in
multiple cancers. J Immunother Cancer. 11:e0063452023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sivakumar D, Kumar V, Naumann M and Stein
M: Activation and selectivity of OTUB-1 and OTUB-2
deubiquitinylases. J Biol Chem. 295:6972–6982. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhu D, Xu R, Huang X, Tang Z, Tian Y,
Zhang J and Zheng X: Deubiquitinating enzyme OTUB1 promotes cancer
cell immunosuppression via preventing ER-associated degradation of
immune checkpoint protein PD-L1. Cell Death Differ. 28:1773–1789.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ren W, Xu Z, Chang Y, Ju F, Wu H, Liang Z,
Zhao M, Wang N, Lin Y, Xu C, et al: Pharmaceutical targeting of
OTUB2 sensitizes tumors to cytotoxic T cells via degradation of
PD-L1. Nat Commun. 15:92024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liu Z, Wang T, She Y, Wu K, Gu S, Li L,
Dong C, Chen C and Zhou Y: N6-methyladenosine-modified
circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor
immune evasion by promoting the deubiquitination of PD-L1 in
non-small cell lung cancer. Mol Cancer. 20:1052021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu
Y, Chang SS, Lin WC, Hsu JM, Hsu YH, et al: Deubiquitination and
stabilization of PD-L1 by CSN5. Cancer Cell. 30:925–939. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xiao X, Shi J, He C, Bu X, Sun Y, Gao M,
Xiang B, Xiong W, Dai P, Mao Q, et al: ERK and USP5 govern PD-1
homeostasis via deubiquitination to modulate tumor immunotherapy.
Nat Commun. 14:28592023. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ding P, Ma Z, Fan Y, Feng Y, Shao C, Pan
M, Zhang Y, Huang D, Han J, Hu Y and Yan X: Emerging role of
ubiquitination/deubiquitination modification of PD-1/PD-L1 in
cancer immunotherapy. Genes Dis. 10:848–863. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tan L, Shan H, Han C, Zhang Z, Shen J,
Zhang X, Xiang H, Lu K, Qi C, Li Y, et al: Discovery of potent
OTUB1/usp8 dual inhibitors targeting proteostasis in non-small-cell
lung cancer. J Med Chem. 65:13645–13659. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lu W, Chu P, Tang A, Si L and Fang D: The
secoiridoid glycoside Gentiopicroside is a USP22 inhibitor with
potent antitumor immunotherapeutic activity. Biomed Pharmacother.
177:1169742024. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu Y, Liu X, Zhang N, Yin M, Dong J, Zeng
Q, Mao G, Song D, Liu L and Deng H: Berberine diminishes cancer
cell PD-L1 expression and facilitates antitumor immunity via
inhibiting the deubiquitination activity of CSN5. Acta Pharm Sin B.
10:2299–2312. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang Y, Huang Y, Yu D, Xu M, Hu H, Zhang
Q, Cai M, Geng X, Zhang H, Xia J, et al: Demethylzeylasteral
induces PD-L1 ubiquitin-proteasome degradation and promotes
antitumor immunity via targeting USP22. Acta Pharm Sin B.
14:4312–4328. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Denis M, Grasselly C, Choffour PA,
Wierinckx A, Mathé D, Chettab K, Tourette A, Talhi N, Bourguignon
A, Birzele F, et al: In vivo syngeneic tumor models with acquired
resistance to anti-PD-1/PD-L1 therapies. Cancer Immunol Res.
10:1013–1027. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Laine A and Ronai Z: Ubiquitin chains in
the ladder of MAPK signaling. Sci STKE. 26:re52005.PubMed/NCBI
|
|
71
|
Çetin G, Klafack S, Studencka-Turski M,
Krüger E and Ebstein F: The ubiquitin-proteasome system in immune
cells. Biomolecules. 11:602021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Han M, Guo Y, Li Y, Zeng Q, Zhu W and
Jiang J: SMURF2 facilitates ubiquitin-mediated degradation of ID2
to attenuate lung cancer cell proliferation. Int J Biol Sci.
19:3324–3340. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Cui H, Wang Q, Lei Z, Feng M, Zhao Z, Wang
Y and Wei G: DTL promotes cancer progression by PDCD4
ubiquitin-dependent degradation. J Exp Clin Cancer Res. 38:3502019.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chen Y, Xian M, Ying W, Liu J, Bing S,
Wang X, Yu J, Xu X, Xiang S, Shao X, et al: Succinate dehydrogenase
deficiency-driven succinate accumulation induces drug resistance in
acute myeloid leukemia via ubiquitin-cullin regulation. Nat Commun.
15:98202024. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Reichelt J, Sachs W, Frömbling S, Fehlert
J, Studencka-Turski M, Betz A, Loreth D, Blume L, Witt S, Pohl S,
et al: Non-functional ubiquitin C-terminal hydrolase L1 drives
podocyte injury through impairing proteasomes in autoimmune
glomerulonephritis. Nat Commun. 14:21142023. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Fuseya Y, Kadoba K, Liu X, Suetsugu H,
Iwasaki T, Ohmura K, Sumida T, Kochi Y, Morinobu A, Terao C and
Iwai K: Attenuation of HOIL-1L ligase activity promotes systemic
autoimmune disorders by augmenting linear ubiquitin signaling. JCI
Insight. 9:e1711082024. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yi J, Tavana O, Li H, Wang D, Baer RJ and
Gu W: Targeting USP2 regulation of VPRBP-mediated degradation of
p53 and PD-L1 for cancer therapy. Nat Commun. 14:19412023.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhang XZ, Li FH and Wang XJ: Regulation of
tripartite motif-containing proteins on immune response and viral
evasion. Front Microbiol. 12:7948822021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Gao X, Cao Y, Li H, Yu F, Xi J, Zhang J,
Zhuang R, Xu Y and Xu L: Mechanisms underlying altered
ubiquitin-proteasome system activity during heart failure and
pharmacological interventions. Eur J Med Chem. 292:1177252025.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yamaguchi H, Hsu JM, Yang WH and Hung MC:
Mechanisms regulating PD-L1 expression in cancers and associated
opportunities for novel small-molecule therapeutics. Nat Rev Clin
Oncol. 19:287–305. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhong G, Chang X, Xie W and Zhou X:
Targeted protein degradation: Advances in drug discovery and
clinical practice. Signal Transduct Target Ther. 9:3082024.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang C, Zhang Y, Chen W, Wu Y and Xing D:
New-generation advanced PROTACs as potential therapeutic agents in
cancer therapy. Mol Cancer. 23:1102024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Dong T, Niu H, Chu Z, Zhou C, Gao Y, Jia
M, Sun B, Zheng X, Zhang W, Zhang J, et al: Targeting VPS18 hampers
retromer trafficking of PD-L1 and augments immunotherapy. Sci Adv.
10:eadp49172024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Shende S, Rathored J and Budhbaware T:
Role of metabolic transformation in cancer immunotherapy
resistance: Molecular mechanisms and therapeutic implications.
Discov Oncol. 16:4532025. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Raina K, Lu J, Qian Y, Altieri M, Gordon
D, Rossi AM, Wang J, Chen X, Dong H, Siu K, et al: PROTAC-induced
BET protein degradation as a therapy for castration-resistant
prostate cancer. Proc Natl Acad Sci USA. 113:7124–7129. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Xiao M, Zhao J, Wang Q, Liu J and Ma L:
Recent advances of degradation technologies based on PROTAC
mechanism. Biomolecules. 12:12572022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chen Y, Tandon I, Heelan W, Wang Y, Tang W
and Hu Q: Proteolysis-targeting chimera (PROTAC) delivery system:
Advancing protein degraders towards clinical translation. Chem Soc
Rev. 51:5330–5350. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Sun C, Liu S, Lau JW, Yang H, Chen Y and
Xing B: Enzyme-Activated orthogonal proteolysis chimeras for tumor
microenvironment-responsive immunomodulation. Angew Chem Int Ed
Engl. 64:e2024230572025. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zhou Y, Li C, Chen X, Zhao Y, Liao Y,
Huang P, Wu W, Nieto NS, Li L and Tang W: Development of folate
receptor targeting chimeras for cancer selective degradation of
extracellular proteins. Nat Commun. 15:86952024. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
He Y, Zheng Y, Zhu C, Lei P, Yu J, Tang C,
Chen H and Diao X: Radioactive ADME demonstrates ARV-110′s high
druggability despite low oral bioavailability. J Med Chem.
67:14277–14291. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Gough SM, Flanagan JJ, Teh J, Andreoli M,
Rousseau E, Pannone M, Bookbinder M, Willard R, Davenport K,
Bortolon E, et al: Oral estrogen receptor PROTAC vepdegestrant
(ARV-471) is highly efficacious as monotherapy and in combination
with CDK4/6 or PI3K/mTOR pathway inhibitors in preclinical ER+
breast cancer models. Clin Cancer Res. 30:3549–3563. 2024.
View Article : Google Scholar : PubMed/NCBI
|