Introduction
Esophageal cancer (EC) represents a major worldwide
health challenge, ranking seventh in incidence and sixth in terms
of cancer-related mortality (1).
There are two main histological variants of EC: Esophageal squamous
cell carcinoma (ESCC) and esophageal adenocarcinoma. ESCC is the
predominant variant, accounting for 85% of cases worldwide, with
Chinese patients representing up to 57% of all instances (2,3). Due
to non-specific symptoms in the early stages, a large proportion of
ESCC cases are identified at progressive stages (4). Among patients with locally advanced
ESCC, ~50% are eligible for R0 resection, while the remaining
patients may experience early recurrence post-surgery (5). Neoadjuvant chemotherapy or neoadjuvant
chemoradiotherapy in conjunction with transthoracic esophagectomy
has become the standard of care, enhancing the R0 resection rate
and survival outcomes (6). However,
the necessity of radiation therapy in neoadjuvant treatment
protocols in China remains a subject of debate (7,8).
Paclitaxel is recognized as an active chemotherapy
agent for ESCC, with paclitaxel-based regimens extensively utilized
in clinical settings. Nevertheless, response rates for both
paclitaxel and cisplatin have been reported to be as low as 33–40%
(9). The efficacy of paclitaxel is
notably compromised by resistance, which is associated with an
increase in postoperative complications and mortality, consequently
contributing to chemotherapy failure and unfavorable prognoses
(10,11). Therefore, further elucidation of the
molecular mechanisms underlying paclitaxel resistance in ESCC is
imperative. In this context, intrinsic resistance to paclitaxel in
ESCC is attributed to particular genetic, epigenetic and metabolic
characteristics (12).
N6-methyladenosine (m6A) represents the most
abundant post-transcriptional alteration in eukaryotic messenger
RNA (mRNA), serving a crucial function in determining RNA fate,
which encompasses mRNA splicing, export, stability, localization
and translation (13). The
biological functions of m6A modification undergo dynamic and
reversible control by three distinct groups of proteins: RNA m6A
methyltransferases (writers), RNA demethylases (erasers) and
m6A-binding proteins (readers). The ‘readers’ specifically identify
and engage with the m6A modification, influencing the destiny of
target transcripts that exhibit m6A methylation. Among this
significant group of readers, insulin-like growth factor 2 mRNA
binding proteins (IGF2BPs) function to suppress mRNA degradation
and enhance mRNA translation (14).
Notably, insulin-like growth factor 2 mRNA binding protein 2
(IGF2BP2) emerges as a prominent member. Studies indicated that
IGF2BP2 participates in the metabolism of target mRNAs,
encompassing Sirtuin 1 and 5-Hydroxytryptamine Receptor 3A in ESCC
(15,16). Nevertheless, the role and mechanisms
through which IGF2BP2 contributes to paclitaxel resistance in ESCC
remain unclear.
The investigation into the relationship between
tumor metabolic characteristics and cancer progression has been
gaining increasing attention within the realm of cancer research.
Cancer cells exhibit a preference for elevated rates of aerobic
glycolysis as their primary source of energy, even when sufficient
oxygen is present, and mitochondrial function remains normal
(referred to as the Warburg effect). This phenomenon enhances the
resistance of cancer cells to apoptotic cell death (17). The aberrantly increased glycolytic
rate in tumors is linked to both intrinsic and acquired resistance
to standard anticancer therapies (17–19).
Therefore, the modulation of tumor metabolism is anticipated to
function as a viable therapeutic strategy for multiple cancer
types, including ESCC.
In the current study, it was revealed that IGF2BP2
mediated paclitaxel resistance in ESCC and explored the mechanism
and identified the IGF2BP2-FOXM1 signaling as a potential biomarker
for predicting paclitaxel resistance in ESCC.
Materials and methods
Cell culture
The ESCC cell lines (KYSE30, KYSE150 and KYSE510)
utilized in the present study were procured from the Cell Resource
Center at the Shanghai Institutes for Biological Sciences, Chinese
Academy of Sciences. All cells were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum and maintained in an incubator at 37°C with 5%
CO2. The cells were passaged a maximum of 15 times.
Gene expression in ESCC datasets
The gene expression profiles were obtained from The
Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) and the Gene
Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/gds/). RNA sequencing
(RNA-seq) data from 82 ESCC tumor samples and 11 adjacent normal
tissues were retrieved from the TCGA database. Concurrently, gene
expression profiles GSE23400 and GSE75241 were downloaded from the
GEO database. Differential gene analysis was conducted on the
GSE20347 dataset from the GEO database. To ascertain differentially
expressed genes, the criteria |logFC|>1 and adjusted P<0.05
were employed and the differences were visualized using volcano
plots.
Plasmids
To generate KYSE30 and KYSE150 cells characterized
by FOXM1 overexpression, ESCC cells were transfected with the
FOXM1-overexpression plasmid (GV-FOXM1) or negative-control plasmid
(GV–Vector) using Lipofectamine™ 3000 (Thermo Fisher Scientific,
Inc.) at 37°C for 6 h, with 2 µg DNA and 5 µg HG-TransGene per well
in a 6-well plate, according to the supplier's protocols. The
overexpression plasmids utilized in the current study were
engineered and constructed by Genomeditech (Shanghai) Co., Ltd.
After the culture medium was replaced with fresh complete medium,
the efficiency of the FOXM1 overexpression was confirmed. ESCC
cells exhibiting FOXM1 overexpression were employed for subsequent
experiments 24 h post-medium change.
Cell transfection and lentiviral
infection
The IGF2BP2 RNA interference lentiviral vector and
the corresponding negative control lentiviral vectors were obtained
from Genomeditech (Shanghai) Co., Ltd. The shRNA was synthesized,
cloned and inserted into pLKO.1-puro vector. The generation system
is the third system. Subsequently, pLKO.1-puro-shRNA plasmid (20
µg), virus packaging plasmids (psPAX2, 15 µg) and envelope plasmids
(pMD2.G, 5 µg) were cotransfected into 293T cells (China Center for
Type Culture Collection) using Lipofectamine™ 3000 (Thermo Fisher
Scientific, Inc.) at 37°C for 6 h. Then medium was replaced with
fresh DMEM (Thermo Fisher Scientific, Inc.) containing 10% FBS and
incubated at 37°C with 5% CO2. Supernatants were
collected at 48 and 72 h post-transfection, filtered through 0.45
µm PVDF membrane and concentrated by ultracentrifugation (100,000 ×
g). MOI for lentivirus transfection was 10. KYSE30 and KYSE150
cells in the exponential growth phase were seeded in 6-well plates
for 24 h. The IGF2BP2 RNA interference and negative control
lentivirus were infected into KYSE30 and KYSE150 cells,
respectively. Infection was enhanced with 6 µg/ml Polybrene.
Following a 24-h infection, the medium was replaced with complete
medium, and the cells were cultured. Puromycin (cat. no. P8230;
Beijing Solarbio Science & Technology Co., Ltd.) at 5 µg/ml was
utilized to select stably transduced cell populations at 72 h
post-infection. The subsequent experiment was performed 10 days
post-medium change. The sequences of all shRNAs were as follows:
shRNA-IGF2BP2-1: 5′-GCCAGAACACUUCCAAGAUAU-3′; shRNA-IGF2BP2-2:
5′-CAGUUACUGGAGAUGAUUAAU-3′; shRNA-IGF2BP2-3:
5′-GCUAUCCACAAGGUCAGUAUU-3′; and non-targeting control shRNA:
5′-CCUAAGGUUAAGUCGCCCUCG-3′.
Small interfering RNA (siRNA) of FOXM1 and negative
control were obtained from Genomeditech (Shanghai) Co., Ltd. 5 µl
siRNA (20 µM) was diluted in 50 µl Opti-MEM (Gibco; Thermo Fisher
Scientific, Inc.) and 5 µl of Lipofectamine™ 3000 (Thermo Fisher
Scientific, Inc.) was diluted in 250 µl Opti-MEM. After incubating
separately for 5 min, the mixtures were combined and further
incubated for 20 min. The transfection complex was then added to
ESCC cells seeded in 6-well plates for transfection at 37°C for 6
h. Knockdown efficiency was assessed 24 h after transfection,
followed by subsequent experiments. The sequences of all siRNAs
were as follows: siRNA-FOXM1-1 sense, 5′-GAAGCGGACCUCAAUGUGAATT-3′
and antisense, 5′-UUCACAUUGAGGUCCGCUUCTT-3′; siRNA-FOXM1-2 sense,
5′-GGAGCUCAAGAAUCUGACUAUTT-3′ and antisense,
5′-UAGUCAGAUUCUUGAGCUCCTT-3′; and siRNA-Ctrl sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′.
Actinomycin D treatment assays
ESCC cells were placed in a 6-well plate at
1×105 cells per well and exposed to Actinomycin D (2
µg/ml; Abcam) for 12 h. Subsequently, the measurement of the FOXM1
mRNA level was conducted using a reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay.
RT-qPCR
According to the manufacturer's instructions, total
RNA was extracted from tissues using TRIzol (cat. no. 15596026;
Thermo Fisher Scientific, Inc.). The extracted RNA was subsequently
converted to cDNA employing the Evo M-MLV RT Premix (cat. no.
AG11706; Accurate Biotechnology Co., Ltd.) according to the
supplier's instructions. The SYBR® Green Premix Pro Taq
HS qPCR kit (cat. no. AG11701; Accurate Biotechnology Co., Ltd.)
was used for qPCR, according to the manufacturer's instructions,
and the target sequence was amplified using a LightCycler 480II
device (Roche Diagnostics). The thermocycling parameters were set
as follows: Preincubation for 5 min at 95°C, followed by 45 cycles
of 10 sec at 95°C, 10 sec at 60°C, and 10 sec at 72°C. β-actin was
utilized as an internal control, and relative RNA expression was
analyzed using the 2−ΔΔCq method (20). The primer sequences were as follows:
FOXM1 forward, 5′-GCAGCGACAGGTTAAGGTTGAG-3′ and reverse,
5′-AGTGCTGTTGATGGCGAATTGTAT-3′; IGF2BP2 forward,
5′-TCGTCAGAATTATCGGGCACTTC-3′ and reverse,
5′-CCTGCTGCTTCACCTGTTGTA-3′; and β-actin forward
5′-TGGCACCCAGCACAATGAA-3′ and reverse,
5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′.
Western blot (WB) analysis
To extract proteins, ESCC cells were lysed in
PMSF-containing RIPA buffer solution (cat. no. R0010; Beijing
Solarbio Science & Technology Co., Ltd.) and centrifuged at
12,000 × g at 4°C for 20 min. The protein concentration was
determined using the Bicinchoninic acid reagent test kit (Beijing
Solarbio Science & Technology Co., Ltd.), according to the
manufacturer's instructions. Protein samples (30 µg) were separated
by SDS-PAGE on 10% gels (cat. no. PG112; Shanghai EpiZyme Co.,
Ltd.) and then transferred onto PVDF membranes (cat. no. WGPVDF22;
Servicebio Co., Ltd.). Subsequently, the membranes were blocked
with high-efficiency western blocking reagent (cat. no. GF1815;
Shanghai Genefist Co., Ltd.) for 15 min at room temperature and
maintained overnight at 4°C with the primary antibodies, including
anti-IGF2BP2 (1:2,000; cat. no. 11601-1-AP; Proteintech Group,
Inc.), anti-FOXM1 (1:2,000; cat. no. 13147-1-AP; Proteintech Group,
Inc.) and anti-GAPDH (1:5,000; cat. no. 10494-1-AP; Proteintech
Group, Inc.). After rinsing the membrane three times with TBST
(cat. no. G2150-1L; Wuhan Servicebio Technology Co., Ltd.), the
PVDF membranes were exposed to secondary rabbit antibodies
(1:5,000; cat. no. SA00001-2; Proteintech Group, Inc.) for 1 h at
ambient temperature. An enhanced chemiluminescence reagent (cat.
no. SQ201; Shanghai EpiZyme Co., Ltd.) was employed to detect
protein signals. Band intensities were analyzed utilizing ImageJ
software (V1.54p; National Institutes of Health), with GAPDH
serving as the internal control for normalizing target protein
expression.
Cell Counting Kit-8 (CCK-8)
ESCC cells in the exponential growth phase from both
the control and treated groups were inoculated into a 96-well plate
at 3,000 cells per well and maintained for 24, 48, 72 and 96 h.
After removing the culture medium, each well was washed with
phosphate-buffered saline (PBS), and the CCK-8 reagent
(MedChemExpress) was combined with serum-free medium to achieve a
concentration of 10%. Subsequently, a volume of 100 µl of the
prepared solution was dispensed into each well. Following
incubation of the 96-well plate at 37°C for 1 h, the absorbance of
each well was measured at 450 nm.
Colony formation assays
Following the counting of ESCC cells in both the
control and treated groups, the cells were placed into 6-well
plates at 1,000 cells per well and subsequently cultured in an
environment maintained at 37°C with 5% CO2. When the
count of cells within a single clone colony exceeded 50 or the
culture duration surpassed 14 days, the culture medium was
aspirated, and the plate underwent PBS washing. Following fixation
in 4% paraformaldehyde (Beyotime Institute of Biotechnology) for 30
min, staining was performed using 0.1% crystal violet (Beyotime
Institute of Biotechnology) at ambient temperature for 15 min.
Colonies were counted utilizing ImageJ software.
5-ethynyl-2′-deoxyuridine (EdU) cell
proliferation assays
Exponential growth ESCC cells from both the control
and treated groups were inoculated into 96-well plates at 5,000
cells per well and maintained for 24 h. The prepared EdU solution
(cat. no. C10310-1; Guangzhou RiboBio Co., Ltd.) was incorporated
into the culture medium and incubated for 2 h for EdU labeling.
Following the removal of the medium and washing with PBS, 4%
paraformaldehyde (Beyotime Institute of Biotechnology) was
introduced, and the plates were incubated for 30 min at ambient
conditions. Subsequently, glycine solution (MedChemExpress), PBS,
and an osmotic agent (Beyotime Institute of Biotechnology) were
sequentially added to immobilize the cells. The wells were then
subjected to Apollo and DNA staining for 30 min at ambient
temperature respectively, with images acquired and analyzed using a
fluorescence microscope (magnification, ×200).
Flow cytometry
The Annexin V-FITC Cell Apoptosis Detection Kit
(cat. no. E-CK-A211; Wuhan Elabscience Biotechnology Co., Ltd.) was
employed to evaluate the level of cell apoptosis. ESCC cells
underwent trypsin digestion to obtain a single-cell suspension (the
density of cells was 5×105/ml). Following a 15-min dual
staining procedure with V-FITC and PI in the 200-µl single-cell
suspension, the cell specimens were examined via flow cytometry
using BD LSRFortessa (BD Biosciences), BD FACSDivaTM
software (V7.0; BD Biosciences) and FlowJo software (V10.8.1;
FlowJo LLC).
Glycolysis assays
A glycolysis assay was performed utilizing a lactate
assay kit (cat. no. 600450; Cayman Chemical Company). Exponentially
growing ESCC cells from both the control and treated groups were
inoculated into 96-well plates at 5,000 cells per well. After 24 h,
the culture medium was harvested for lactate content assessment.
The specimens were examined following the instructions outlined in
the reagent kit, the values were noted, and calculations were
performed.
RNA-seq
Following transfection of IGF2BP2-knockdown KYSE30
and control cells, the cell samples were collected by scraping and
subsequently lysed in TRIzol (cat. no. 15596026; Thermo Fisher
Scientific, Inc.) for total RNA extraction. DNase (cat. no.
AG12001; Accurate Biotechnology Co., Ltd.) was used to remove
genomic DNA contamination. The purity of the sample was determined
by NanoPhotometer® (IMPLEN, USA). The concentration and
integrity of RNA samples were detected by Agilent 2100 RNA nano
6000 assay kit (cat. no. 5067-1511; Agilent Technology Co., Ltd.).
Sequencing libraries were generated using VAHTS Universal V6
RNA-seq Library Prep Kit for Illumina® (cat. no.
NR604-01/02; Vazyme Co., Ltd.) following the manufacturer's
recommendations. Poly-T magnetic beads were used to isolate mRNA,
which was then fragmented. First- and second-strand cDNA were
synthesized, purified, and subjected to end repair, adenylation,
and adapter ligation. The library was size-selected and enriched by
PCR prior to sequencing. The library was quantified using the
Bio-RAD CFX 96 fluorescence quantitative PCR instrument with the
iQ™ SYBR® Green kit (cat. no. 1708880; Bio-Rad
Laboratories, Inc.) to the effective concentration of >10 nM.
Sequencing was performed on the Illumina NovaSeq 6000 platform
(Illumina, Inc.) with the NovaSeq 6000 S4 Reagent Kit V1.5 (cat.
no. 20028312; Illumina, Inc.), generating 150 bp paired-end reads.
DESeq2 (V1.44.0) and EdgeR (V3.44.0) were used for Difference
analysis (21,22), and the Gene Ontology (GO; http://geneontology.org/) was used for function
enrichment analysis.
m6A-qPCR and m6A sequencing
(m6A-seq)
Total RNA was obtained and purified from ESCC
samples with TRIzol (cat. no. 15596026; Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions. Following
extraction, RNA was incubated with m6A antibodies utilizing the
Magna Methylated RNA Immunoprecipitation m6A Kit (MilliporeSigma)
for immunoprecipitation. The concentration of m6A-modified mRNA
underwent examination through m6A-qPCR or m6A-seq methodologies
(Haplox Biotechnology Co., Ltd.). Primers targeting the negative
region of FOXM1 m6A served as negative controls, whereas primers
targeting the positive region of FOXM1 m6A were employed as
positive controls. For m6A-seq, the enriched m6A RNA underwent
reverse transcription to generate cDNA, and the library was
prepared per the instructions provided by Illumina's NEBNext Ultra
RNA Library Preparation Kit (New England Biolabs, Inc.).
High-throughput library sequencing was conducted to acquire
sequence information about m6A-modified RNA using the Illumina
HiSeq X™ Ten platform (Illumina, Inc.).
Statistical analysis
Statistical analysis was performed utilizing
GraphPad Prism 9 (Dotmatics). Data are presented as the mean ±
standard deviation (SD). Comparisons among several groups were
performed using one-way ANOVA followed by Least Significance
Difference test was used for the post hoc test, whereas evaluations
between paired groups employed paired Student's t-test. P<0.05
was considered to indicate a statistically significant
difference.
Results
IGF2BP2 is highly expressed in
ESCC
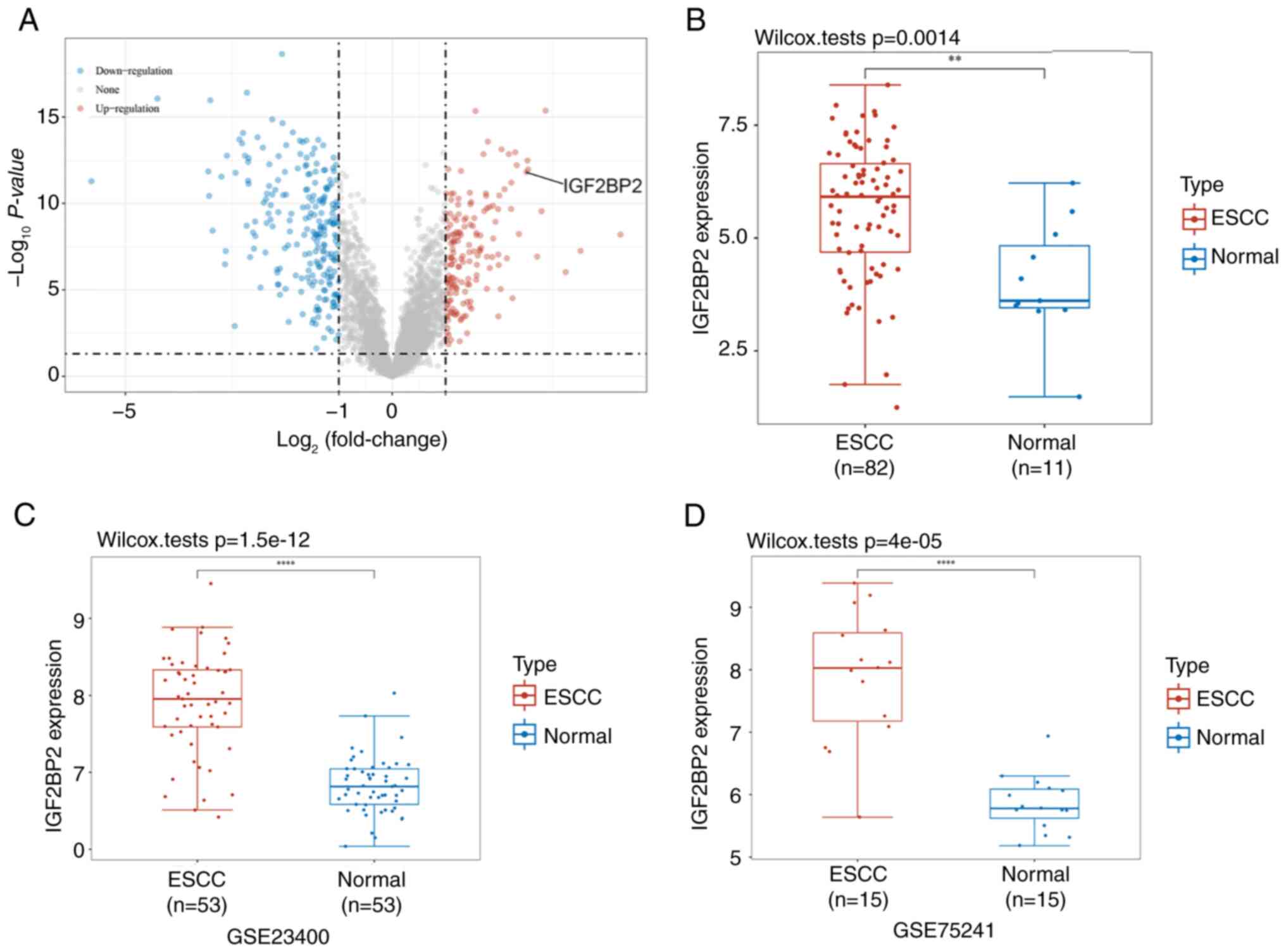
Differential gene analysis was performed utilizing
the GSE20347 dataset, which revealed a total of 389 differentially
expressed genes, encompassing 226 downregulated and 163 upregulated
genes (Fig. 1A). Among these, the
10 most markedly upregulated genes were identified as COL11A1,
ZIC1, COL1A2, ECT2, ACKR3, LUM, MFHAS1, FNDC3B, IGF2BP2 and KIF23.
Furthermore, a comparison of IGF2BP2 expression levels between ESCC
tumor tissues and normal tissue samples from the TCGA database
indicated a significant increase in IGF2BP2 expression (P=0.0014;
Fig. 1B). Data obtained from
GSE23400 and GSE75241 confirmed the upregulation of IGF2BP2 in ESCC
(P<0.0001; Fig. 1C and D).
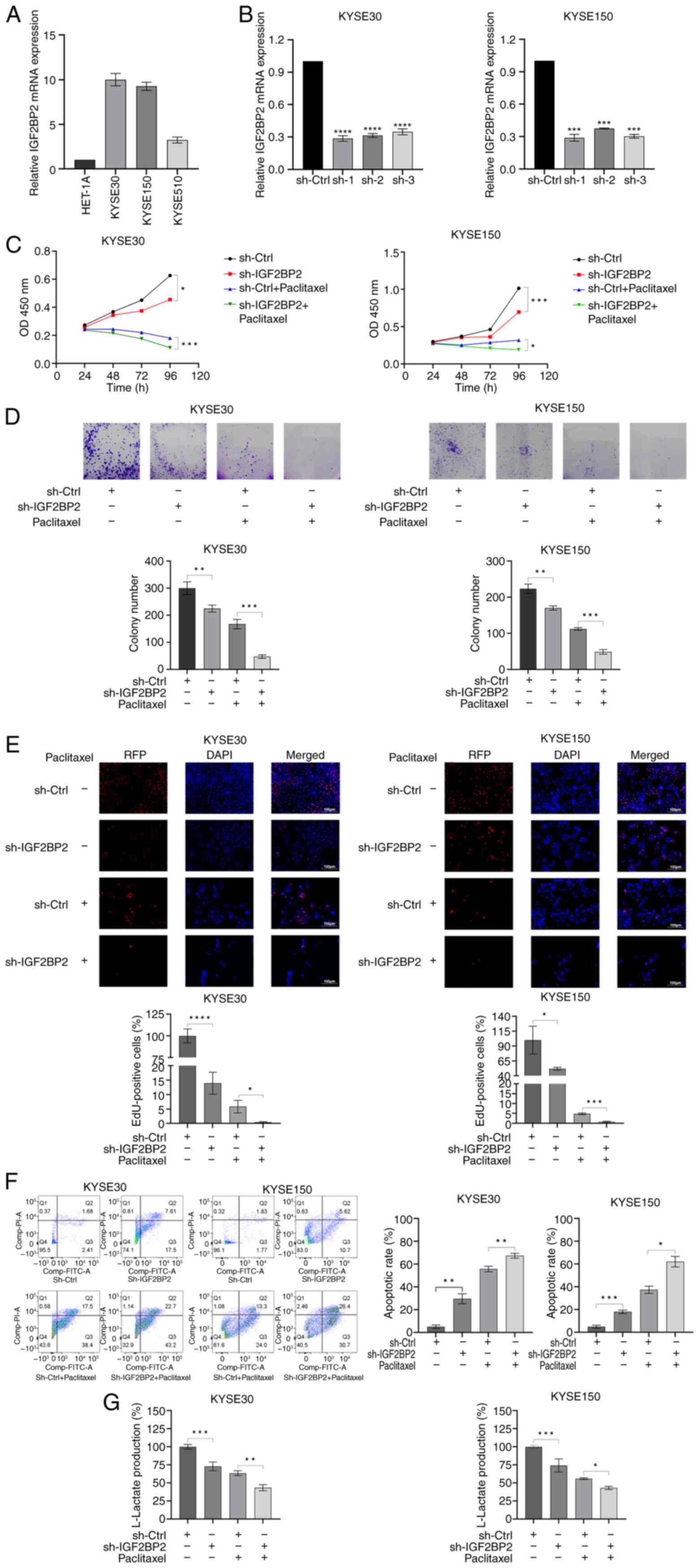
Regarding the ESCC cell lines, RT-qPCR (Fig. 2A) demonstrated a significant
upregulation of IGF2BP2 in KYSE30 and KYSE150 relative to HET-1A
(the normal esophageal cell line). Consequently, both cell lines
were chosen for the following investigations.
IGF2BP2 inhibition decreases anaerobic
glycolysis and increases the sensitivity to paclitaxel in ESCC
cells
To explore the role of IGF2BP2 in ESCC, alterations
in cellular phenotypes were assessed following the inhibition of
IGF2BP2 expression in cell lines. Efficient knockdown of IGF2BP2
was achieved using two short-hairpin (sh)RNAs (sh-IGF2BP2-1 and
sh-IGF2BP2-2) in both KYSE30 and KYSE150 cells (Fig. 2B). Given that sh-IGF2BP2-1 exhibited
the highest efficiency, this sequence was selected for subsequent
depletion experiments.
CCK-8 assays were performed to evaluate the
influence of IGF2BP2 on the proliferation of ESCC cells. The
results demonstrated that IGF2BP2 knockdown resulted in
significantly decreased cell proliferation (Fig. 2C). Images obtained from colony
formation assays illustrated that silencing IGF2BP2 significantly
impaired the formation of colonies of ESCC cells (Fig. 2D). Moreover, EdU assays validated
the substantial suppressive effect of IGF2BP2 knockdown on ESCC
cell proliferation (Fig. 2E).
Additionally, flow cytometry assays indicated that the knockdown of
IGF2BP2 markedly enhanced the apoptosis of ESCC cells (Fig. 2F). Notably, it was observed that
silencing IGF2BP2 resulted in a reduction of anaerobic glycolysis
in KYSE30 and KYSE150 cells (Fig.
2G). Moreover, the depletion of IGF2BP2 significantly enhanced
ESCC cell responsiveness to paclitaxel (Fig. 2C-F). Collectively, these findings
indicated that IGF2BP2 serves a crucial function in anaerobic
glycolysis and resistance to paclitaxel in ESCC cells.
FOXM1 is the m6A modification target
of IGF2BP2
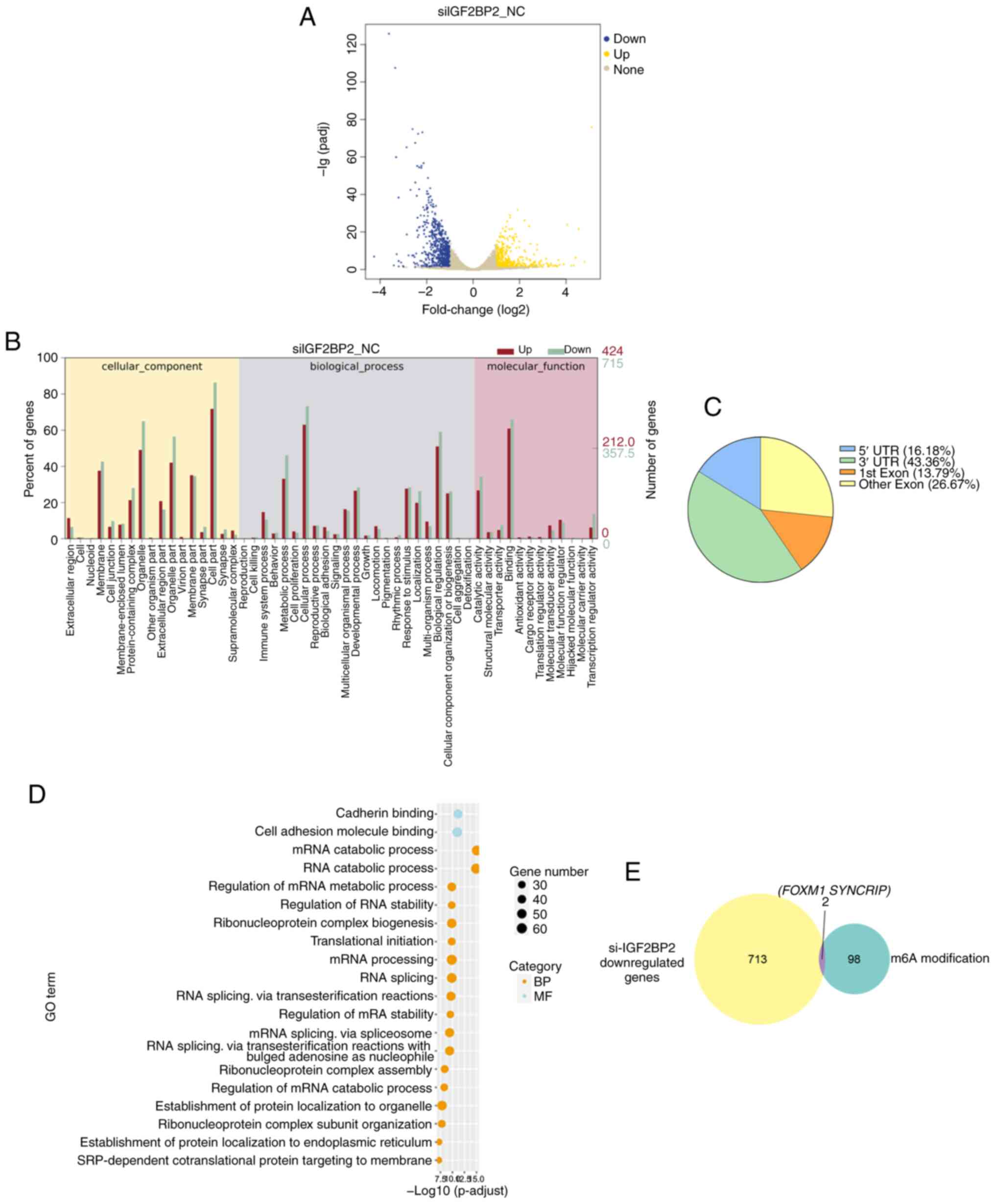
To elucidate the mechanism through which IGF2BP2
contributes to paclitaxel resistance in ESCC, RNA-seq was conducted
using both IGF2BP2-knockdown KYSE30 and control cells. It was
demonstrated that the expression of 1,139 genes was globally
modified following IGF2BP2 knockdown, with 424 genes exhibiting
upregulation and 715 genes displaying downregulation (Fig. 3A). GO analysis suggested that
several pathways, encompassing the MAPK signaling pathway, TGF-β
signaling pathway, and protein processing in the endoplasmic
reticulum, were markedly enriched (Fig.
3B). These findings collectively highlight the oncogenic role
of IGF2BP2 in ESCC.
It is widely acknowledged that IGF2BP2 functions as
an m6A reader, capable of recognizing and binding to m6A-methylated
transcripts to regulate target genes. Consequently, m6A-seq was
conducted in KYSE30 cells, resulting in the identification of 8,104
m6A modification peaks across 3,990 genes. The majority of these
m6A modifications were located within the 3′-untranslated regions
(3′-UTRs; 43.36%; Fig. 3C).
Functional annotation indicated that these mRNAs were associated
with several distinct gene clusters, including the regulation of
RNA stability and RNA splicing (Fig.
3D). Given that IGF2BP2 has been recognized for its role in
maintaining RNA stability (14),
the attention was directed towards transcripts that were
downregulated following IGF2BP2 knockdown and exhibited the top 100
highest peak m6A modifications. By overlapping the results from
RNA-seq and m6A modification analyses, two candidate genes, FOXM1
and SYNCRIP, were identified as meeting the criteria. Among these,
FOXM1 mRNA levels declined more markedly upon IGF2BP2 knockdown in
ESCC cell lines. Furthermore, emerging evidence suggested that
FOXM1 contributes to paclitaxel resistance (23). These findings prompted a shift in
focus towards FOXM1, which is considered a critical factor in the
mediation of paclitaxel resistance through IGF2BP2 in ESCC
(Fig. 3E).
IGF2BP2 sustains the stability of
FOXM1 mRNA
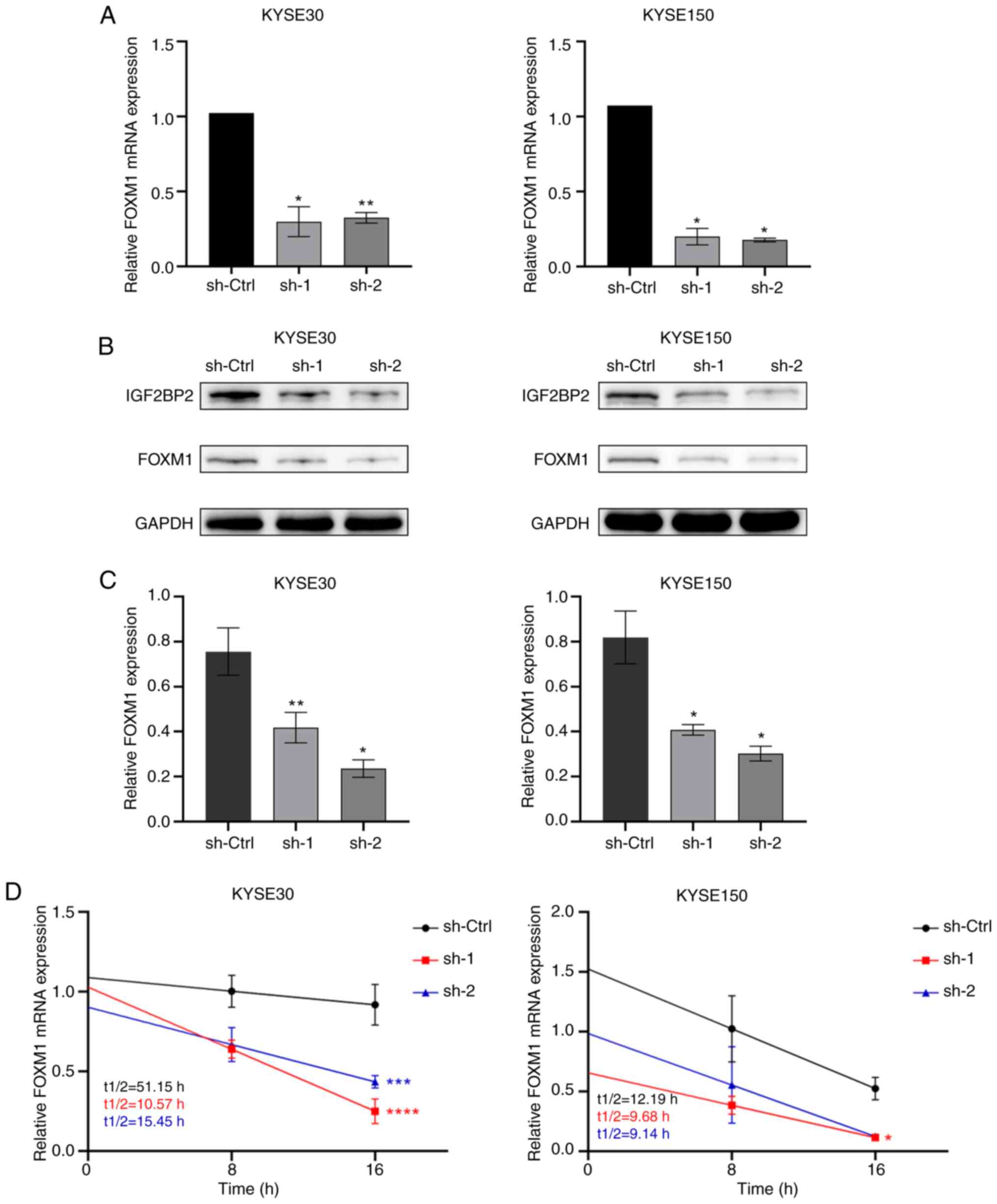
To ascertain whether IGF2BP2 modulates FOXM1
expression in ESCC cells, FOXM1 expression was assessed at both
transcriptional and translational levels following IGF2BP2
knockdown, utilizing RT-qPCR and WB analysis, respectively. As
anticipated, both the RNA expression levels (Fig. 4A) and protein levels (Fig. 4B and C) of FOXM1 exhibited a
significant decrease correlating with IGF2BP2 depletion. Given that
IGF2BP2 deficiency impaired the levels of FOXM1 mRNA, it was
hypothesized that IGF2BP2 may regulate FOXM1 by maintaining the
stability of FOXM1 mRNA. Subsequently, 2 g/ml Actinomycin D was
administered to treat KYSE30 cells transfected with IGF2BP2-shRNA
or control shRNA at various time points. As illustrated in Fig. 4D, a significantly accelerated decay
of FOXM1 mRNA was observed in the absence of IGF2BP2 (Fig. 4D), indicating that IGF2BP2 can
sustain the stability of FOXM1 mRNA.
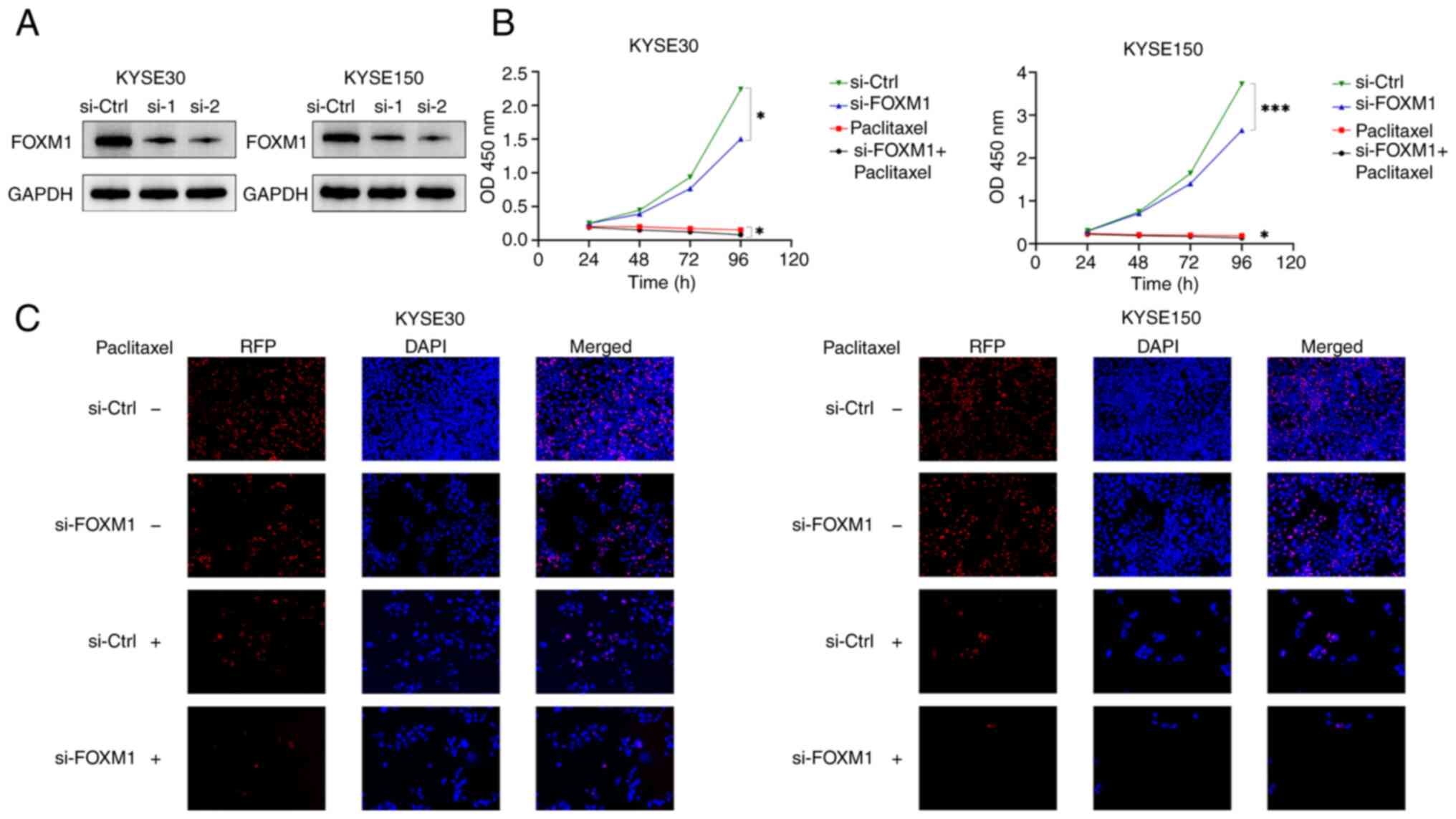
FOXM1 serves an oncogenic function in
ESCC cells
The oncogenic function of FOXM1 has been documented
in ESCC (21–23). To further validate the function of
FOXM1 in ESCC, the phenotypes of ESCC cells, encompassing cell
growth and proliferation, were examined through gene-loss
experiments following FOXM1 depletion (Fig. 5A). Notably, a significant reduction
in cell growth (Fig. 5B) was
observed through the CCK-8 assays, alongside a diminished formation
of colonies (Fig. 5C) of the KYSE30
and KYSE150 cells observed in the EdU assays. Consequently, it was
hypothesized that FOXM1 facilitates oncogenesis in ESCC cells.
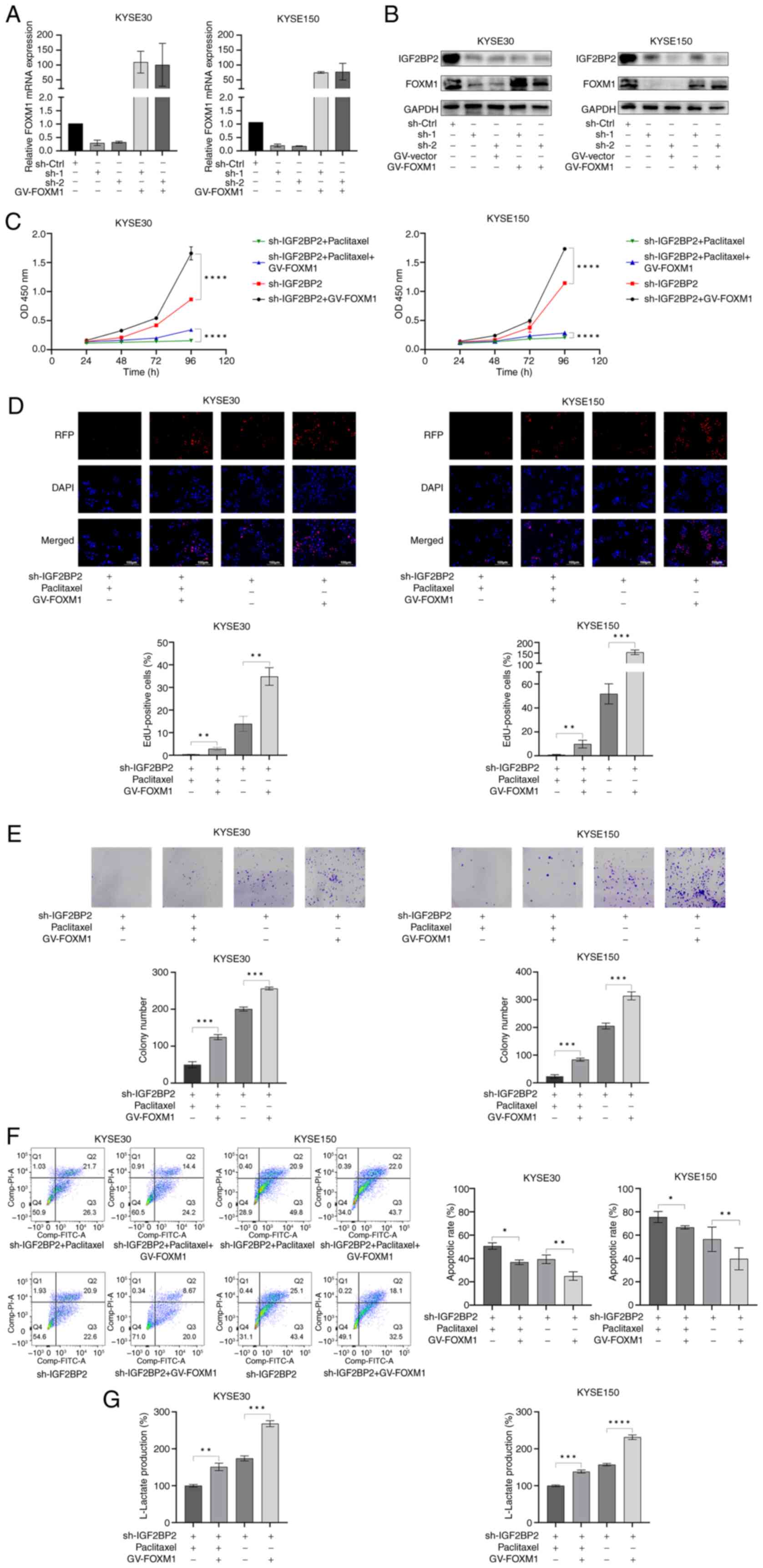
Ectopic expression of FOXM1
ameliorates the impact of IGF2BP2 deficiency in ESCC cells
To further investigate whether FOXM1 mediates the
oncogenic effects of IGF2BP2, FOXM1 expression was upregulated in
ESCC cells that were silenced for IGF2BP2. The efficacy of FOXM1
overexpression was validated through WB analysis (Fig. S1A and B). Furthermore, the efficacy
of ectopic FOXM1 expression in conjunction with silenced IGF2BP2
was assessed at both transcriptional and translational levels
through RT-qPCR and WB analysis, respectively. The results
indicated that both RNA expression (Fig. 6A) and protein abundance (Fig. 6B) of ectopically expressed FOXM1
were markedly increased in IGF2BP2-silenced ESCC cells.
Subsequently, a series of functional assays were conducted to
evaluate ESCC cell activity. CCK-8 assays suggested that the
proliferation rate of ESCC cells exhibiting ectopic FOXM1
expression was significantly higher than that of cells lacking
ectopic FOXM1, irrespective of the presence of paclitaxel (Fig. 6C). Furthermore, EdU assays
illustrated that the reduced cell proliferation activity due to
IGF2BP2 depletion was significantly enhanced through ectopic FOXM1
expression (Fig. 6D). The results
of colony formation assays corroborated the findings observed in
the EdU assays (Fig. 6E). Moreover,
flow cytometric analysis revealed that ectopic FOXM1 expression
mitigated the apoptosis induced through IGF2BP2 silencing (Fig. 6F). These findings demonstrated that
ectopic FOXM1 expression improved cell proliferation, enhanced the
formation of colonies and alleviated apoptosis associated with
IGF2BP2 deficiency in ESCC cells. Notably, it was observed that the
decrease in anaerobic glycolysis resulting from IGF2BP2 silencing
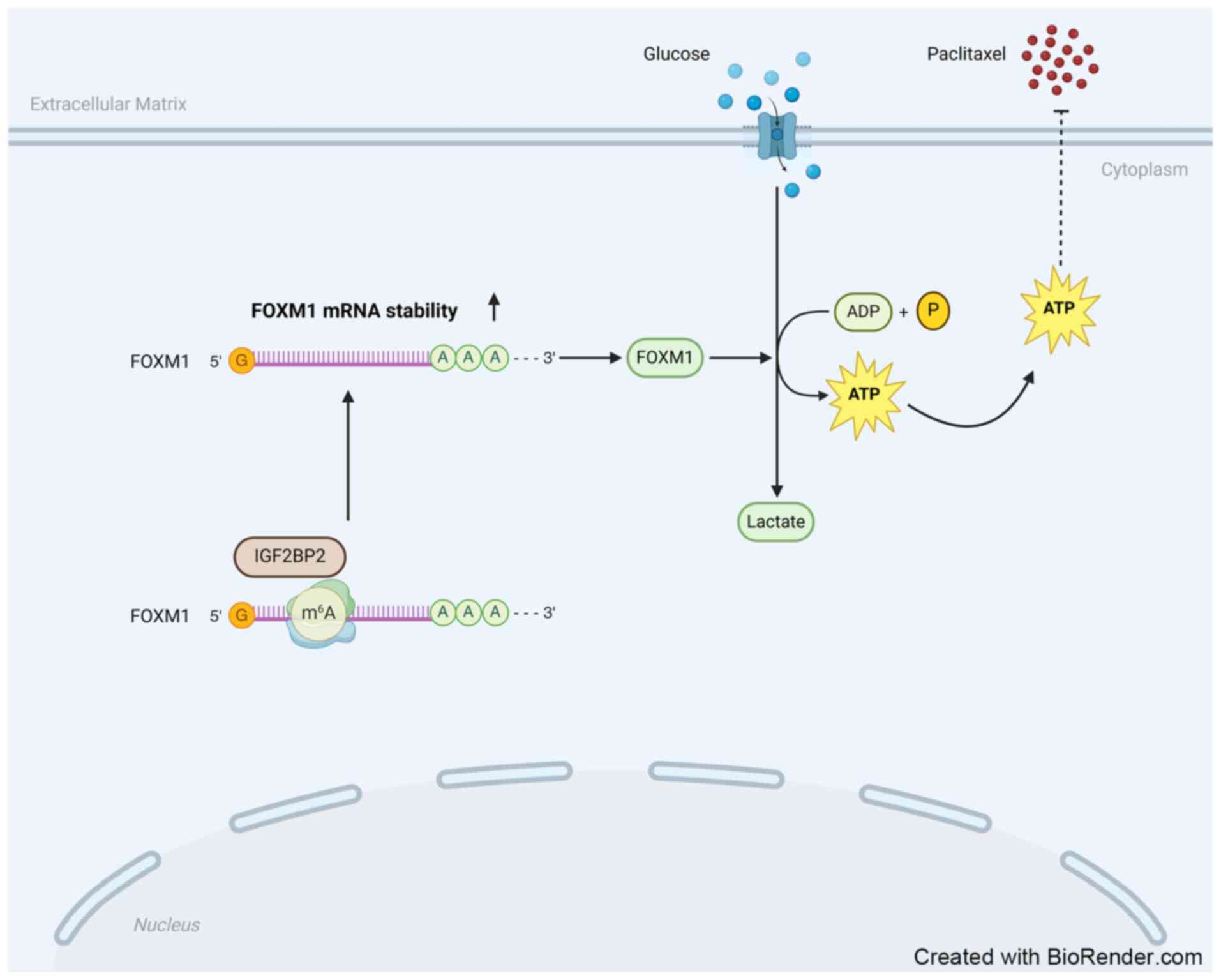
in ESCC cells was reversed upon FOXM1 upregulation (Fig. 6G). Therefore, it is proposed that
IGF2BP2 promotes anaerobic glycolysis and diminishes sensitivity to
paclitaxel in ESCC cells by regulating FOXM1 (Fig. 7).
Discussion
In the current investigation, the overexpression of
IGF2BP2 was confirmed, along with its association with paclitaxel
resistance and anaerobic glycolysis in ESCC in vitro.
Mechanistically, it was demonstrated that IGF2BP2 stabilizes FOXM1
mRNA.
Paclitaxel is extensively utilized in combination
chemotherapy for ESCC. However, tumor resistance to paclitaxel
frequently results in treatment failure, prompting considerable
attention towards underlying the mechanisms. Wu et al
(12) conducted single-cell RNA-seq
experiments to elucidate the heterogeneity of gene expression in
paclitaxel-resistant ESCC cells, which may contribute to the
observed resistance. The findings were validated through drug
toxicity assays, colony formation assays and rodent xenograft model
experiments. It was concluded that intrinsic paclitaxel resistance
exists in KYSE30 ESCC cells, with potential association with KRT19
expression. In the current study, KYSE30 ESCC cells were similarly
selected to focus on the role of the m6A reader IGF2BP2 in
paclitaxel resistance within ESCC. RNA m6A modification has been
reported to mediate paclitaxel resistance in various malignancies.
Liu et al (27) demonstrated
that lncRNA RFPL1S-202 was capable of boosting paclitaxel
cytotoxicity in ovarian cancer by physically interacting with
DEAD-Box Helicase 3 X-linked (DDX3X) protein, thereby increasing
the m6A modification of IFNB1 to reduce the expression of
IFN-inducible genes. Additionally, Xie et al (28) reported that LINC02489 with m6A
modification linked to PKNOX2 via the PTEN/mTOR axis, thereby
enhancing the sensitivity of ovarian cancer to paclitaxel and
reducing the migration and invasion of chemotherapy-resistant
ovarian cancer cells.
A recent study reported that m6A reader IGF2BP2 is
upregulated in ESCC tissues in comparison with normal esophageal
mucosa. Li et al (29)
demonstrated a significant upregulation of IGF2BP2 in ESCC tissues
through epi-transcriptomics. Additionally, Wang et al
(30) suggested that IGF2BP2
exhibits elevated expression in ESCC than those in normal
esophageal tissues, with its overexpression being closely linked to
lymph node metastasis. In the present study, the upregulation of
IGF2BP2 in ESCC was confirmed, consistent with previous findings
(26–28).
Furthermore, it was demonstrated for the first time
that IGF2BP2 expression is positively linked to paclitaxel
resistance in ESCC. The current study serves as a valuable
supplement to the understanding of m6A modification in the
regulation of paclitaxel resistance across various malignancies.
One of the hallmarks of cancer is the observation that, even in the
presence of normal oxygen concentrations, cancer cells
predominantly generate energy through glycolysis at elevated rates,
a phenomenon referred to as the Warburg effect. In addition to its
tumor-promoting properties, aerobic glycolysis is capable of
enhancing drug resistance in cancer cells by creating a supportive
microenvironment (32).
Consequently, this may present a novel clinical approach,
suggesting that the inhibition of glycolysis could partially
reverse chemotherapy resistance.
Enhanced sensitivity of KYSE150 ESCC cells to
cisplatin, both in vitro and in vivo, was reported to
be associated with inhibited glycolysis, as evidenced by reduced
glucose consumption and diminished lactate production. This
observation suggests that enhancing the sensitivity of ESCC to
cisplatin could be achieved through the reduction of glycolysis
(33). Wu et al (34) demonstrated that disrupted glycolysis
was positively correlated with paclitaxel resistance in epithelial
ovarian cancer. In the present study, it was revealed for the first
time, to the best of our knowledge, that the inhibition of IGF2BP2
markedly reversed paclitaxel resistance while concurrently
decreasing anaerobic glycolysis in ESCC. Further study will be
implemented to investigate whether IGF2BP2 mediates paclitaxel
resistance in ESCC by modulating glycolytic pathways.
As an m6A reader, the biological function of IGF2BP2
is contingent upon its interaction with m6A-modified RNA. In the
present study, m6A modifications in ESCC were systematically
screened, revealing a prevalence of such modifications in the UTRs
of mRNAs, aligning with previous findings (35). Concurrently, transcriptional
analysis following IGF2BP2 knockdown in ESCC cells indicated that
the majority of transcripts exhibiting m6A modification were
downregulated, suggesting that IGF2BP2 maintains mRNA stability in
an m6A-dependent manner within ESCC. Among these genes, it was
confirmed that FOXM1 serves as a direct target of IGF2BP2 in ESCC.
A previous study indicated that the Akt/FOXM1 signaling cascade
inhibits paclitaxel-induced cell death in ESCC (23). In the present investigation, it was
established that FOXM1 is modulated by IGF2BP2 at both
transcriptional and translational levels in ESCC. Further
experiments demonstrated that FOXM1 represents a pivotal factor
through which IGF2BP2 mediates paclitaxel resistance and aberrant
anaerobic glycolysis in ESCC. The results showed that the FOXM1
transcript is m6A-modified and its expression decreases upon
IGF2BP2 knockdown, indicating that IGF2BP2 regulates FOXM1
expression in an m6A-dependent manner.
While the current findings yielded several
significant conclusions, certain limitations persist. The mechanism
by which IGF2BP2 regulates FOXM1 in an m6A-dependent manner remains
to be experimentally substantiated due to the complexity of the
associated experiments. IGF2BP2 comprises six canonical RNA-binding
domains, encompassing two RNA recognition motifs and four K
homology domains. It has been established that the KH3-4 di-domains
are critical for IGF2BP2 to bind to m6A-modified mRNAs and modulate
the expression of target genes. In forthcoming research, basic
experiments will be undertaken to elucidate the mechanism by which
IGF2BP2 regulates FOXM1 through the creation of an IGF2BP2 mutant
featuring alterations in the KH domains, alongside an assessment of
the interaction between IGF2BP2 and FOXM1 mRNA.
In conclusion, the present study demonstrated the
significant role of IGF2BP2-FOXM1 signaling in modulating
paclitaxel resistance in ESCC. The insights and conclusions
presented may offer novel directions for the effective treatment of
ESCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science
Foundation of Shandong (grant no. ZR2021MH155).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The data generated in the
present study may be found in the Figshare under accession number
10.6084/m9.figshare.29312819.v2 or at the following URL:
(https://doi.org/10.6084/m9.figshare.29312819.v2).
Authors' contributions
SR and WJ conceived and designed the study. SR and
JW performed the experiments. JW conducted statistical analysis
using GraphPad Prism. GG performed bioinformatics analysis. LZ
assisted in experiments, contributed to manuscript editing based on
reviewers' comments and revised figures. WJ secured funding,
supervised the project and provided critical revision of the
manuscript. All authors contributed to the writing and revision of
the article. All authors read and approved the final version of the
manuscript. SR and WJ confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Morgan E, Soerjomataram I, Rumgay H,
Coleman HG, Thrift AP, Vignat J, Laversanne M, Ferlay J and Arnold
M: The Global landscape of esophageal squamous cell carcinoma and
esophageal adenocarcinoma incidence and mortality in 2020 and
projections to 2040: New estimates from GLOBOCAN 2020.
Gastroenterology. 163:649–658.e2. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
An L, Zheng R, Zeng H, Zhang S, Chen R,
Wang S, Sun K, Li L, Wei W and He J: The survival of esophageal
cancer by subtype in China with comparison to the United States.
Int J Cancer. 152:151–161. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rustgi AK and El-Serag HB: Esophageal
carcinoma. N Engl J Med. 371:2499–2509. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Leng XF, Daiko H, Han YT and Mao YS:
Optimal preoperative neoadjuvant therapy for resectable locally
advanced esophageal squamous cell carcinoma. Ann N Y Acad Sci.
1482:213–224. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu H, Ma X, Ye T, Wang H, Wang Z, Liu Q
and Zhao K: Esophageal cancer in China: Practice and research in
the new era. Int J Cancer. 152:1741–1751. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang H, Liu H, Chen Y, Zhu C, Fang W, Yu
Z, Mao W, Xiang J, Han Y, Chen Z, et al: Neoadjuvant
Chemoradiotherapy followed by surgery versus surgery alone for
locally advanced squamous cell carcinoma of the esophagus
(NEOCRTEC5010): A phase III multicenter, randomized, open-label
clinical trial. J Clin Oncol. 36:2796–2803. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kotecki N, Hiret S, Etienne PL, Penel N,
Tresch E, François E, Galais MP, Ben Abdelghani M, Michel P, Dahan
L, et al: First-line chemotherapy for metastatic esophageal
squamous cell carcinoma: Clinico-biological predictors of disease
control. Oncology. 90:88–96. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mariette C, Dahan L, Mornex F, Maillard E,
Thomas PA, Meunier B, Boige V, Pezet D, Robb WB, Le Brun-Ly V, et
al: Surgery alone versus chemoradiotherapy followed by surgery for
stage I and II esophageal cancer: Final analysis of randomized
controlled phase III trial FFCD 9901. J Clin Oncol. 32:2416–2422.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chan KKW, Saluja R, Delos Santos K, Lien
K, Shah K, Cramarossa G, Zhu X and Wong RKS: Neoadjuvant treatments
for locally advanced, resectable esophageal cancer: A network
meta-analysis. Int J Cancer. 143:430–437. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu H, Chen S, Yu J, Li Y, Zhang XY, Yang
L, Zhang H, Hou Q, Jiang M, Brunicardi FC, et al: Single-cell
Transcriptome analyses reveal molecular signals to intrinsic and
acquired paclitaxel resistance in esophageal squamous cancer cells.
Cancer Lett. 420:156–167. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meyer KD and Jaffrey SR: The dynamic
epitranscriptome: N6-methyladenosine and gene expression control.
Nat Rev Mol Cell Biol. 15:313–326. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA
stability and translation. Nat Cell Biol. 20:285–295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang Z, Wan J, Ma L, Li Z, Yang R, Yang H,
Li J, Zhou F and Ming L: Long non-coding RNA HOXC-AS1 exerts its
oncogenic effects in esophageal squamous cell carcinoma by
interaction with IGF2BP2 to stabilize SIRT1 expression. J Clin Lab
Anal. 37:e248012023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang GW, Chen QQ, Ma CC, Xie LH and Gu J:
Linc01305 promotes metastasis and proliferation of esophageal
squamous cell carcinoma through interacting with IGF2BP2 and
IGF2BP3 to stabilize HTR3A mRNA. Int J Biochem Cell Biol.
136:1060152021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liberti MV and Locasale JW: The warburg
effect: How does it benefit cancer cells? Trends Biochem Sci.
41:211–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Haque MM and Desai KV: Pathways to
endocrine therapy resistance in breast cancer. Front Endocrinol
(Lausanne). 10:5732019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma L and Zong X: Metabolic symbiosis in
Chemoresistance: Refocusing the role of aerobic glycolysis. Front
Oncol. 10:52020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Meng RY, Jin H, Nguyen TV, Chai OH, Park
BH and Kim SM: Ursolic acid accelerates paclitaxel-induced cell
death in esophageal cancer cells by suppressing Akt/FOXM1 signaling
cascade. Int J Mol Sci. 22:114862021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu C, Liu T, Han C, Xuan Y, Jiang D, Sun
Y, Zhang X, Zhang W, Xu Y, Liu Y, et al: HPV E6/E7 promotes aerobic
glycolysis in cervical cancer by regulating IGF2BP2 to stabilize
m6A-MYC expression. Int J Biol Sci. 18:507–521. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pu J, Wang J, Qin Z, Wang A, Zhang Y, Wu
X, Wu Y, Li W, Xu Z, Lu Y, et al: IGF2BP2 promotes liver cancer
growth through an m6A-FEN1-dependent mechanism. Front Oncol.
10:5788162020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen
YX, Liu J, Luo XJ, Meng Q, Pu HY, et al: LncRNA LINRIS stabilizes
IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer.
Mol Cancer. 18:1742019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu S, Chen X, Huang K, Xiong X, Shi Y,
Wang X, Pan X, Cong Y, Sun Y, Ge L, et al: Long noncoding RNA
RFPL1S-202 inhibits ovarian cancer progression by downregulating
the IFN-β/STAT1 signaling. Exp Cell Res. 422:1134382023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xie Y, Wang L, Luo Y, Chen H, Yang Y, Shen
Q and Cao G: LINC02489 with m6a modification increase paclitaxel
sensitivity by inhibiting migration and invasion of ovarian cancer
cells. Biotechnol Genet Eng Rev. 39:1128–1142. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Xiao Z, Wang Y, Zhang D and Chen Z:
The m6A reader IGF2BP2 promotes esophageal cell
carcinoma progression by enhancing EIF4A1 translation. Cancer Cell
Int. 24:1622024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang C, Zhou M, Zhu P, Ju C, Sheng J, Du
D, Wan J, Yin H, Xing Y, Li H, et al: IGF2BP2-induced circRUNX1
facilitates the growth and metastasis of esophageal squamous cell
carcinoma through miR-449b-5p/FOXP3 axis. J Exp Clin Cancer Res.
41:3472022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiao Y, Tang J, Yang D, Zhang B, Wu J, Wu
Z, Liao Q, Wang H, Wang W and Su M: Long noncoding RNA LIPH-4
promotes esophageal squamous cell carcinoma progression by
regulating the miR-216b/IGF2BP2 axis. Biomark Res. 10:602022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bhattacharya B, Mohd Omar MF and Soong R:
The Warburg effect and drug resistance. Br J Pharmacol.
173:970–979. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fang J, Ma Y, Li Y, Li J, Zhang X, Han X,
Ma S and Guan F: CCT4 knockdown enhances the sensitivity of
cisplatin by inhibiting glycolysis in human esophageal squamous
cell carcinomas. Mol Carcinog. 61:1043–1055. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu X, Qiu L, Feng H, Zhang H, Yu H, Du Y,
Wu H, Zhu S, Ruan Y and Jiang H: KHDRBS3 promotes paclitaxel
resistance and induces glycolysis through modulated MIR17HG/CLDN6
signaling in epithelial ovarian cancer. Life Sci. 293:1203282022.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zaccara S, Ries RJ and Jaffrey SR:
Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell
Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI
|