Introduction
Colorectal cancer (CRC) is one of the most prevalent
malignancies worldwide, representing ~1/4 of all cancer diagnoses
and contributing to nearly one-third of cancer-related deaths
(1). New cases of CRC are expected
to increase to 3.2 million and related deaths are expected to reach
1.6 million by 2040. Although most CRC cases occur in developed
countries, the incidence continues to increase in developing
regions (2). Despite advancements
in non-invasive biomarkers for CRC diagnosis and improvements in
surgery, radiotherapy and other treatments, ~50% of CRC cases are
diagnosed at advanced stages, resulting in a 10% 5-year survival
rate (3). Thus, identifying novel
early screening markers and therapeutic targets for CRC is critical
for improving early diagnosis and treatment outcomes.
Leucine-rich repeat-containing protein 59 (LRRC59)
is a 244-amino acid endoplasmic reticulum (ER) membrane protein,
weighing 34 kDa, that was initially identified as a ribosomal
receptor in the ER (4–6). This protein is involved in the
progression of various tumors. Pallai et al (7) observed that LRRC59 translocate from
the ER to the nuclear membrane, facilitating nuclear import of
CIP2A in a PP2A-independent manner, thereby promoting tumorigenesis
in prostate cancer. In lung adenocarcinoma, LRRC59 is significantly
overexpressed and contributes to cancer cell proliferation and
invasion, which is correlated with a poor prognosis (8). Furthermore, LRRC59 is a prognostic
biomarker for risk assessment in patients with urothelial
carcinoma, and its elevated expression is associated with higher
pathological grades. LRRC59 also regulates signaling pathways
during ER stress (ERS). Depletion of LRRC59 potentiates
HYOU1/XBP1-driven ERS signaling, leading to marked inhibition of
malignant cell proliferation and migration (9). These findings suggested that LRRC59 is
broadly involved in tumor development and influences both apoptosis
and ERS. However, the biological functions and molecular mechanisms
of LRRC59 in CRC remain largely uncharacterized, and studies are
needed to determine its role in CRC progression and evaluate its
potential as a therapeutic target.
The ER is a specialized organelle that plays a
critical role in regulating key cellular processes such as protein
synthesis, folding, assembly and transport. ERS arises when
misfolded or unfolded proteins accumulate within the ER, a
condition triggered by factors such as ischemia, oxidative damage,
and disrupted in calcium homeostasis. ERS disrupts ER homeostasis
and affects cell proliferation, differentiation and apoptosis
(10). A total of 3 critical
sensors of ERS have been identified: protein kinase RNA-like ER
kinase (PERK), inositol-requiring enzyme 1α (IRE1α), and activating
transcription factor 6 (ATF6), which are all embedded in the ER
membrane. Upon activation of ERS, GRP78 dissociates from these
sensors and binds to misfolded or unfolded proteins, initiating the
unfolded protein response (UPR) to restore ER homeostasis (11,12).
However, severe or prolonged ERS can amplify protein misfolding and
initiate both caspase-dependent and independent apoptotic pathways
(13). In various tumor types,
oncogenic factors, and metabolic and transcriptional abnormalities
synergize to maintain a state of chronic ERS in tumor cells
(10,14,15).
Emerging research highlights a pivotal role of ERS in CRC
progression, where ERS-induced activation of PERK or IRE1α through
phosphorylation affects cell fate via the PERK pathway or IRE1α
pathway (16,17). Therefore, the targeted modulation of
ERS is a potential therapeutic approach for CRC. As LRRC59 may
regulate ERS, the starBase database was screened and it was found
that LRRC59 is significantly associated with key genes in the ERS
PERK signaling pathway. Thus, it was hypothesized that LRRC59
affects CRC progression by regulating the PERK signaling
pathway.
The present results demonstrated that LRRC59 was
overexpressed in CRC, leading to the promotion of cell
proliferation, migration and invasion and inhibition of apoptosis.
Additionally, LRRC59 inhibited apoptosis by mitigating PERK
signaling.
Materials and methods
Tissue sample collection
CRC tissues and their corresponding adjacent normal
tissue samples were obtained from six patients who underwent
surgical procedures (July 2024-December 2024) at the Second
Affiliated Hospital of Dalian Medical University (Dalian, China).
This was an opportunistic cohort. Detailed clinical and
pathological data for these patients are summarized in Table I [five males and one female, with a
median age of 69.5 years (range 65–80)]. Written informed consent
was obtained for the use of the tissue samples, and the study was
approved by the Ethics Committee of the Second Affiliated Hospital
of Dalian Medical University (approval no. KY2024-136-01; Dalian,
China). The limited specimen availability reflects the stringent
inclusion criteria: i) Signed patient informed consent, ii)
confirmed CRC diagnosis and iii) tissue quality sufficient for
immunohistochemical (IHC) and western blot analysis. While it is
recognized that this precluded formal power calculations, the
present sample size is consistent with exploratory translational
studies (18,19) and yielded statistically significant
results across key endpoints: Animal model outcomes, clinical
specimen IHC analysis and protein expression profiling. It is
acknowledged that larger samples would strengthen these findings
and will address this in future validation studies.
 | Table I.Specific details of the patients. |
Table I.
Specific details of the patients.
| Number | Age, years | Sex | Clinical
information | Pathological
information |
|---|
| 1 | 68 | Male | pT2N0M0 | Colonic moderately
differentiated adenocarcinoma |
| 2 | 67 | Male | pT3N0M0 | Moderately
differentiated adenocarcinoma of the rectum |
| 3 | 75 | Male | pT4N0M0 | Moderately
differentiated adenocarcinoma of the rectum |
| 4 | 71 | Male | pT2N0M0 | Colonic moderately
differentiated adenocarcinoma |
| 5 | 65 | Female | pT3N0M0 | Colonic moderately
differentiated adenocarcinoma |
| 6 | 80 | Male | pT2N0M0 | Colonic moderately
differentiated adenocarcinoma |
Cell culture and transfection
The CRC cell lines HCT116 and LoVo, purchased from
Suzhou HyCyte™ Biotechnology Co., Ltd., were cultured in RPMI-1640
medium (cat. no. 11875119; Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (cat. no. A5670701; Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin
(cat. no. SC118; Seven Innovations Biological Technology Co., Ltd).
The cells were maintained at 37°C in a 5% CO2
atmosphere. The PERK inhibitor GSK (cat. no. HY-13820;
MedChemExpress) was added to HCT116 and LoVo cells for 12 h (1 µM).
After the cells reached ~50% confluency, LRRC59-small interfering
RNA (siRNA) (20 µM) was transfected (37°C) with Lipofectamine 3000
(cat. no. L3000015; Thermo Fisher Scientific, Inc.), and the cells
were harvested after 24 or 48 h for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blotting. LRRC59-siRNA constructs were designed by Suzhou
GenePharma Co., Ltd., and siNC was used as a negative control. The
sequences were as follows: si-LRRC59-1:
5′-GACUACUCUACCGUCGGAUTT-3′; si-LRRC59-2:
5′-GUGCAAACAAGGUGUUACATT-3′; and si-NC:
5′-UUCUCCGAACGUGUCACGUTT-3′. The 293T cell line used in the present
study was purchased from the Cell Bank of the Chinese Academy of
Sciences. The cells were cultured in DMEM medium (cat. no.
PM150210; Procell Life Science & Technology Co., Ltd.)
supplemented with 10% fetal bovine serum (cat. no. A5670701; Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin
(cat. no. SC118; Seven Innovations Biological Technology Co., Ltd).
All cells were maintained at 37°C in a humidified incubator with 5%
CO2 and were used for subsequent lentiviral
transduction.
Stable transgenic cells via short
hairpin RNA lentiviral transduction
Stable LRRC59-knockdown LoVo cells were constructed
using 3rd-generation lentiviral vectors. The short hairpin
RNA-expressing plasmid pLV3-H1/GFP&Puro-shLRRC59 (target
sequence: 5′-CTGGATCTGTCTTGTAATAAA-3′) and non-targeting control
sh-NC (5′-TTCTCCGAACGTGTCACGT-3′) were obtained from Suzhou
GenePharma Co., Ltd. Viral particles were packaged in 293T cells
(Cell Bank of the Chinese Academy of Sciences) by co-transfecting
the transfer plasmid with packaging plasmids pGag/Pol, pRev, and
pVSV-G (GenePharma Co., Ltd.) at a mass ratio of 4:2:1:1 using
RNAi-Mate transfection reagent (cat. no. G04001; GenePharma Co.,
Ltd.). Transfection complexes were incubated with the cells in
15-cm dishes for 6h at 37°C in a 5% CO2 atmosphere,
followed by replacement of the medium with complete medium. Viral
supernatants were harvested 72 h post-transfection, filtered
through 0.45-µm membranes, and concentrated via ultracentrifugation
(32,000 × g, 2 h, 4°C). LoVo cells were transduced with viral
stocks at a multiplicity of infection of 10 in the presence of 5
µg/ml Polybrene (cat. no. H9268; MilliporeSigma) for 24 h at 37°C,
after which the medium was refreshed. Stable populations were
selected with 2 µg/ml puromycin (cat. no. HY-B1743; MedChemExpress)
for 7 days, followed by maintenance in 1 µg/ml puromycin. Knockdown
efficiency was validated by western blotting.
RT-qPCR
Total RNA was isolated using RNAkey™ Total RNA
Extraction Reagent (cat. no. SM139-02; Seven Innovations Biological
Technology Co., Ltd.). cDNA was synthesized using a two-step
RT-qPCR kit (cat. no. SRQ-01; Seven Innovations Biological
Technology Co., Ltd.) as follows: 50°C for 8 min and 85°C for 5
sec. Real-time PCR amplification of the cDNA templates was
performed using a Two-Step RT and qPCR Kit (cat. no. SRQ-01; Seven
Innovations Biological Technology Co., Ltd.) on a
QuantStudio™ 5 system (Thermo Fisher Scientific, Inc.).
The thermocycling conditions were as follows: 95°C for 30 sec; 40
cycles at 95°C for 20 sec, and 60°C for 20 sec. The
2−ΔΔCq method was used to calculate the relative mRNA
expression (20). GAPDH was used as
an internal control. The primer sequences were as follows: LRRC59
forward, 5′-CCTTGCCTGTCAGCTTTGC-3′ and reverse,
5′-TCATGTGCTGTAACACCTTGTTTG-3′; and GAPDH forward,
5′-CATGTTCGTCATGGGTGTGAA-3′ and reverse,
5′-GGCATGGACTGTGGTCATGAG-3′.
Cell Counting Kit-8 (CCK-8) assay
CRC cells were transfected with either si-LRRC59 or
si-NC. After 24 h incubation cell counts were determined, and the
cells were seeded into 96-well plates (5×103
cells/well). Following incubation for 24, 48 and 72 h, fresh medium
containing 10% CCK-8 reagent (cat. no. SC119; Seven Innovations
Biological Technology Co., Ltd.) was added. After 2 h, optical
density was measured at 450 nm.
Colony formation assay
CRC cells were transfected with either si-LRRC59 or
si-NC for 24 h. After counting, the cells were seeded into 6-well
plates (1×103 cells/well). The plates were maintained in
culture for one week. Colonies (contains at least 50 cells) were
fixed with 10% methanol (15 min at room temperature) and stained
with 0.1% crystal violet (30 min at room temperature) to enhance
visualization.
Scratch wound healing assay
CRC cells were transfected with si-LRRC59 or si-NC,
seeded into 6-well plates, and cultured to 90% confluence. Scratch
wounds were created using a sterile 10-µl pipette tip, and cellular
debris was removed by washing with phosphate-buffered saline (PBS).
Basal medium (without serum) was added, and images of the scratch
areas were captured and measured at both 0 h and 48 h using a Leica
microscope (inverted light microscope). Wound closure percentages
were analyzed using ImageJ 1.54d software (National Institutes of
Health).
Migration and invasion assays
Cells (1×105 cells/well) were plated into
24-well 8-µm pore size Transwell chambers (cat. no. 3422; Corning,
Inc.), with or without Matrigel (cat. no. 354230; Corning, Inc.)
coating (37°C, 6 h). The lower chambers contained basal medium with
10% fetal bovine serum as a chemoattractant. After 48 h, migratory
cells were fixed in methanol (1 h at room temperature) and stained
with 0.1% crystal violet (20 min at room temperature). A total of 5
random fields were selected for analyses.
IHC analysis
CRC tissues, adjacent normal tissues, and xenograft
tumor sections (4-µm thickness) underwent sequential
deparaffinization in xylene and rehydration in a graded ethanol
series at room temperature. Following antigen retrieval in
Tris/EDTA buffer (pH 9.0), endogenous peroxidase activity was
quenched by adding 3% H2O2 incubation (10 min
incubation, room temperature). The sections were rinsed three times
with distilled water (5 min/wash) and equilibrated in PBS for 5
min. After blocking with 5–10% normal goat serum (cat. no.
G1208-5ML; Servicebio Technology Co., Ltd.) for 10 min at room
temperature, the sections were incubated overnight at 4°C with the
following primary antibodies: anti-LRRC59 (1:200; cat. no.
27208-1-AP; Proteintech Group, Inc.), anti-GRP78 (1:200; cat. no.
11587-1-AP; Proteintech Group, Inc.), anti-p-PERK (1:300; cat. no.
PA5-40294; Invitrogen), anti-ATF4 (1:200; cat. no. 28657-1-AP;
Proteintech Group, Inc.), and anti-CHOP (1:200; cat. no.
15204-1-AP; Proteintech). Next, horseradish peroxidase-conjugated
secondary antibody (1:200; cat. no. G1213; Servicebio Technology
Co., Ltd.) was incubated with the sections at 37°C for 1 h. A DAB
Chromogenic Kit cat. no. G1212; Wuhan Servicebio Technology Co.,
Ltd.) was used to treat the sections for 15 min, followed by
hematoxylin counterstaining (cat. no. G1004; Servicebio Technology
Co., Ltd.) for 3 min at room temperature and bright-field
microscopy using a Leica DMI1 system. The staining intensity and
percentage were quantified using the H-score formula: H-score=∑(pi
× i), where pi is the percentage of positive pixels and i is the
intensity grade.
Western blotting and antibodies
Proteins were extracted from tissues and cells using
cell lysis buffer for western blotting and immunoprecipitation (IP;
cat. no. P0013; Beyotime Institute of Biotechnology) containing
phenyl-methyl-sulphonyl-fluoride (cat. no. SW107; Seven Innovations
Biological Technology Co., Ltd.) and phosphatase inhibitors (cat.
no. SW106; Seven Innovations Biological Technology Co., Ltd.). The
protein concentration was quantified using the BCA assay (cat. no.
KGB2101; Nanjing KeyGen Biotech Co., Ltd.). Precast polyacrylamide
gels (7.5–12.5%; cat. no. SW145; Seven Innovations Biological
Technology Co., Ltd) were loaded with 20 µg protein per lane along
with pre-stained protein molecular weight markers (cat. no. SW176;
Seven Innovations Biological Technology Co., Ltd.). Electrophoresis
was performed at 80 V for 30 min followed by 120 V for 60–90 min,
followed by wet transfer (300 mA, 30 min) onto polyvinylidene
fluoride membranes (cat. no. ISEQ00005; MilliporeSigma). The
membranes were blocked with 5% non-fat milk in Tris-buffered saline
containing 0.1% Tween-20 at room temperature for 1 h, followed by
incubation with primary antibodies overnight at 4°C. The following
primary antibodies were used: β-tubulin (1:10,000; cat. no. AP0064;
Bioworld Technology, Inc.), LRRC59 (1:10,000; cat. no. 27208-1-AP;
Proteintech Group, Inc.), GRP78 (1:10,000; cat. no. 11587-1-AP;
Proteintech Group, Inc.), PERK (1:1,000; cat. no. 24390-1-AP;
Proteintech Group, Inc.), phosphorylated PERK (p-PERK) (1:1,000;
cat. no. AP1501; ABclonal Biotech Co., Ltd.), ATF4 (1:10,000; cat.
no. 81798-1-RR; Proteintech Group, Inc.), p-EIF2α (1:1,000; cat.
no. AP0692; ABclonal Biotech Co., Ltd.), EIF2α (1:10,000; cat. no.
82936-1-PBS; Proteintech Group, Inc.), CHOP (1:1,000; cat. no.
15204-1-AP; Proteintech Group, Inc.), Caspase 3 (1:1,0000; cat. no.
82202-1-RR; Proteintech Group, Inc.), active-caspase-3 (1:1,000;
cat. no. A11021; ABclonal Biotech Co., Ltd.), Bax (1:1,000; cat.
no. A19684; ABclonal Biotech Co., Ltd.), Bcl-2 (1:1,000; cat. no.
A19693; ABclonal Biotech Co., Ltd.). Horseradish
peroxidase-conjugated rabbit secondary antibodies (1:2,000; cat.
no. AS014; Abcam) and mouse secondary antibodies (1:2,000; cat. no.
AS003; ABclonal Biotech Co., Ltd.) were applied for 1 h at room
temperature. Protein signals were detected using Super ECL Reagent
(cat. no. 36208ES60; Shanghai Yeasen Biotechnology Co., Ltd.) and
visualized using a ChemiDoc Imaging System (Bio-Rad Laboratories,
Inc.). Quantitative analysis was performed using ImageJ 1.54d
software. All experiments were conducted in triplicate.
Flow cytometry
HCT116 and LoVo cells (5×104) were
stained using an Annexin V-FITC/PI Apoptosis Detection Kit (cat.
no. 40302ES50; Shanghai Yeasen Biotechnology Co., Ltd.). The cells
were trypsinized using 0.25% ethylene-diamine-tetraacetic acid-free
trypsin (cat. no. SC108, Seven Innovations Biological Technology
Co., Ltd.) and pelleted by centrifugation at 300 × g for 5 min at
4°C. After two washes with ice-cold PBS (centrifugation: 300 × g,
4°C, 5 min), 5×105 cells were resuspended in 100 µl 1X
Annexin V Binding Buffer. Subsequently, 5 µl FITC-conjugated
Annexin V and 10 µl propidium iodide working solution (20 µg/ml)
were added, followed by gentle vortexing and 15 min incubation in
light-protected conditions at room temperature. The reaction was
terminated by adding 400 µl ice-cold binding buffer. The samples
were maintained on ice and analyzed within 60 min using a NovoCyte
Advanteon flow cytometer (Agilent Technologies, Inc.). Apoptosis
levels were assessed using NovoExpress software (version 1.6.2;
Agilent). All experiments were conducted in triplicate.
Xenograft tumor formation
Female BALB/c female mice (aged 5 weeks; n=10;
weight, 18.23±0.74 g) were acquired from Beijing Vital River
Laboratory Animal Technology Co., Ltd. The mice were housed in a
specific pathogen-free grade animal room in individually ventilated
cages within the barrier at a rate of five mice per cage. The mice
were housed under controlled environmental conditions (26–28°C,
40–60% humidity) with a standardized 10 h light/14 h dark
photoperiod. All mice had unrestricted access to standard
laboratory chow and water throughout the study period, along with
environmental enrichment (for example, nesting materials and
shelters). The specific humane endpoint criteria implemented in the
experiment were as follows: i) The tumor volume of any single tumor
did not exceed 2,000 mm3; ii) Tumor ulceration or
necrosis: Progressive ulceration, severe necrosis, or infection of
the tumor surface was expected to be irreversible or cause
significant pain; iii) weight loss: Mice lost more than 20% of
their initial weight within three consecutive days; iv) activity
status and behavioral changes: Mice exhibited severely reduced
activity, hunchbacks, coarse and messy fur, difficulty breathing,
inability to access food or water normally, and other significant
signs of pain or impending death; v) Functional impairment caused
by tumors: Tumor growth significantly impaired the mobility (such
as walking) and normal physiological functions of mice; and vi)
monitoring and execution: Trained researchers observed the
experimental animals at least once daily. Mice were monitored for
any potential signs of humane endpoints twice a day.
When a mouse met any of the humane endpoint criteria
aforementioned, it was euthanized immediately. The maximum tumor
volume was 1372.24 mm3 and the maximum tumor diameter
was 16.14 mm.
All procedures were approved by the Institutional
Animal Care and Use Committee of Wuhan Servicebio Technology Co.,
Ltd. (approval no. 2024167). The experiments strictly adhered to
the institution's guidelines for animal welfare and use. The mice
were randomly assigned to two groups with five animals per group.
Each mouse received a subcutaneous injection of LoVo cells
(1×107 cells) that had been transfected with either
sh-NC or shLRRC59, resuspended in 100 µl of PBS. Tumor volumes were
measured at 3-day intervals, and the tumors were excised and
weighed after 34 days. Mice were anesthetized by intraperitoneal
injection of 2% tribromoethanol (20 ml/kg), and deep anesthesia was
confirmed by the absence of a pedal reflex. Euthanasia was induced
via cervical dislocation. No experimental procedures were performed
on the mice after tribromoethanol anesthesia.
Our sample size decisions were guided by both
field-specific standards and ethical considerations. Animal studies
(n=5/group): this sample size aligns with established norms for
xenograft tumor models in published literature (21,22).
Consistent with the 3R principles (Replacement, Reduction,
Refinement) under international animal research guidelines, group
sizes were optimized to minimize animal use while maintaining
experimental validity.
The choice of the LoVo cell line for in vivo
studies was based on: Literature-established protocols:
Subcutaneous xenograft models using LoVo are well-documented for
CRC studies (23,24). Ethical optimization: This approach
minimized animal usage while maintaining experimental validity,
consistent with 3R principles. Biological rationale: Complementary
in vitro experiments demonstrated consistent LRRC59 effects
across both HCT116 and LoVo cell lines. Critically, our
dual-cell-line validation confirmed that LRRC59′s modulation of the
PERK pathway occurs independently of cellular context. LoVo cells
were therefore selected for in vivo validation as they
represent a mechanistically conserved model of LRRC59 function in
CRC progression.
Mice were euthanized on Day 34 for two reasons: i)
Tumor size limits: IACUC protocol requires euthanasia before tumor
volume exceeds 2,000 mm3. Projected growth curves
indicated most mice would reach this threshold by Day 34. ii)
Humane endpoints: Mice were monitored daily for distress signs (for
example, >20% weight loss, lethargy, ulceration). Any mouse
meeting these criteria was euthanized immediately; none required
early euthanasia before Day 34. This schedule preempted welfare
violations while allowing meaningful cross-group comparison at a
standardized endpoint.
Blinding method
Blinding was applied to evaluate the results during
the measurement phase of all experiments involving subjective
evaluation.
Statistical analysis
Data are expressed as the mean values accompanied by
standard deviations calculated from three independent experiments.
Statistical analyses were conducted using GraphPad Prism 8.0
software (GraphPad, Inc.; Dotmatics). For comparisons between two
groups, two-tailed, paired Student's t-test was used, whereas
one-way analysis of variance was used to analyze multiple groups.
Normality was assessed using the Shapiro-Wilk test for all
experimental groups, with results indicating P>0.05 for each
dataset, confirming adherence to normality assumptions. Prior to
ANOVA, homogeneity of variance was further verified using Levene's
test (P>0.05), confirming that all ANOVA prerequisites were
satisfied. For comparisons across the three groups (including
controls), Dunnett's post hoc test was applied. All pairwise
comparisons against the control group showed statistically
significant differences (P<0.05). Statistical significance
thresholds were set at *P<0.05, **P<0.01 and
***P<0.001.
Results
Elevated expression of LRRC59 in CRC
tissues
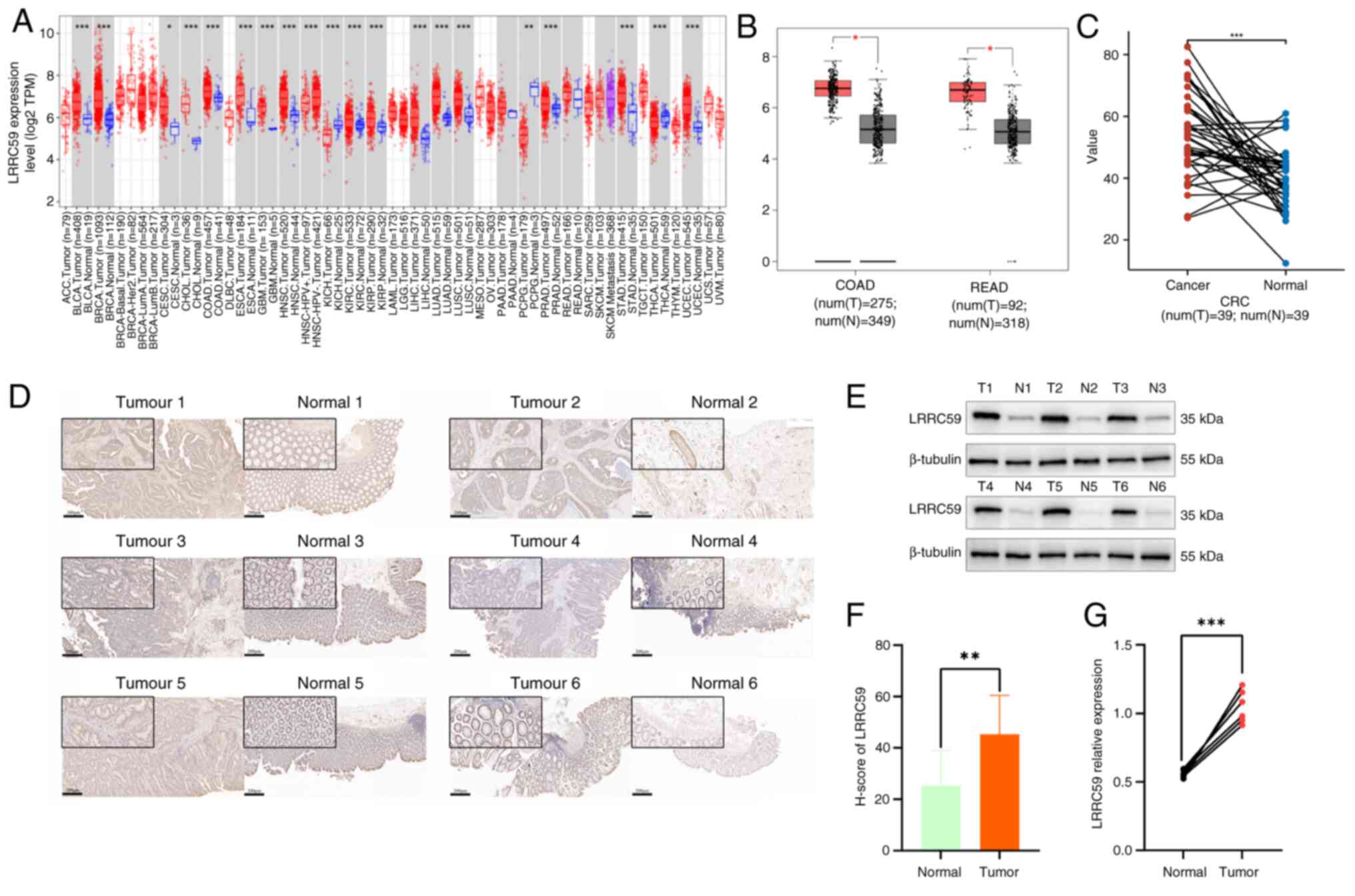
To evaluate the LRRC59 expression in CRC tissues in
compared with that in normal tissues, mRNA expression levels were
initially analyzed using the TIMER2.0 database (http://timer.comp-genomics.org/) (Fig. 1A). The results revealed
significantly higher LRRC59 mRNA levels in colon adenocarcinoma
tissues than in normal tissues. Further data were acquired from
both paired and unpaired CRC samples using the GEPIA (http://gepia.cancer-pku.cn/index.html)
(Fig. 1B) and TCGA databases
(https://www.tcpaportal.org/) (Fig. 1C), respectively. Statistical
analyses showed that LRRC59 mRNA expression was markedly
upregulated in CRC tissues compared with that in corresponding
adjacent normal tissues. To substantiate these results, CRC and
adjacent normal tissues from six patients were analyzed using IHC
(Fig. 1D and F) and western
blotting (Fig. 1E and G). The
results revealed a significant increase in LRRC59 protein
expression in CRC tissues compared with that in their normal tissue
counterparts. Collectively, these findings indicated that LRRC59 is
substantially overexpressed in CRC tissues compared with in normal
paracancerous tissues.
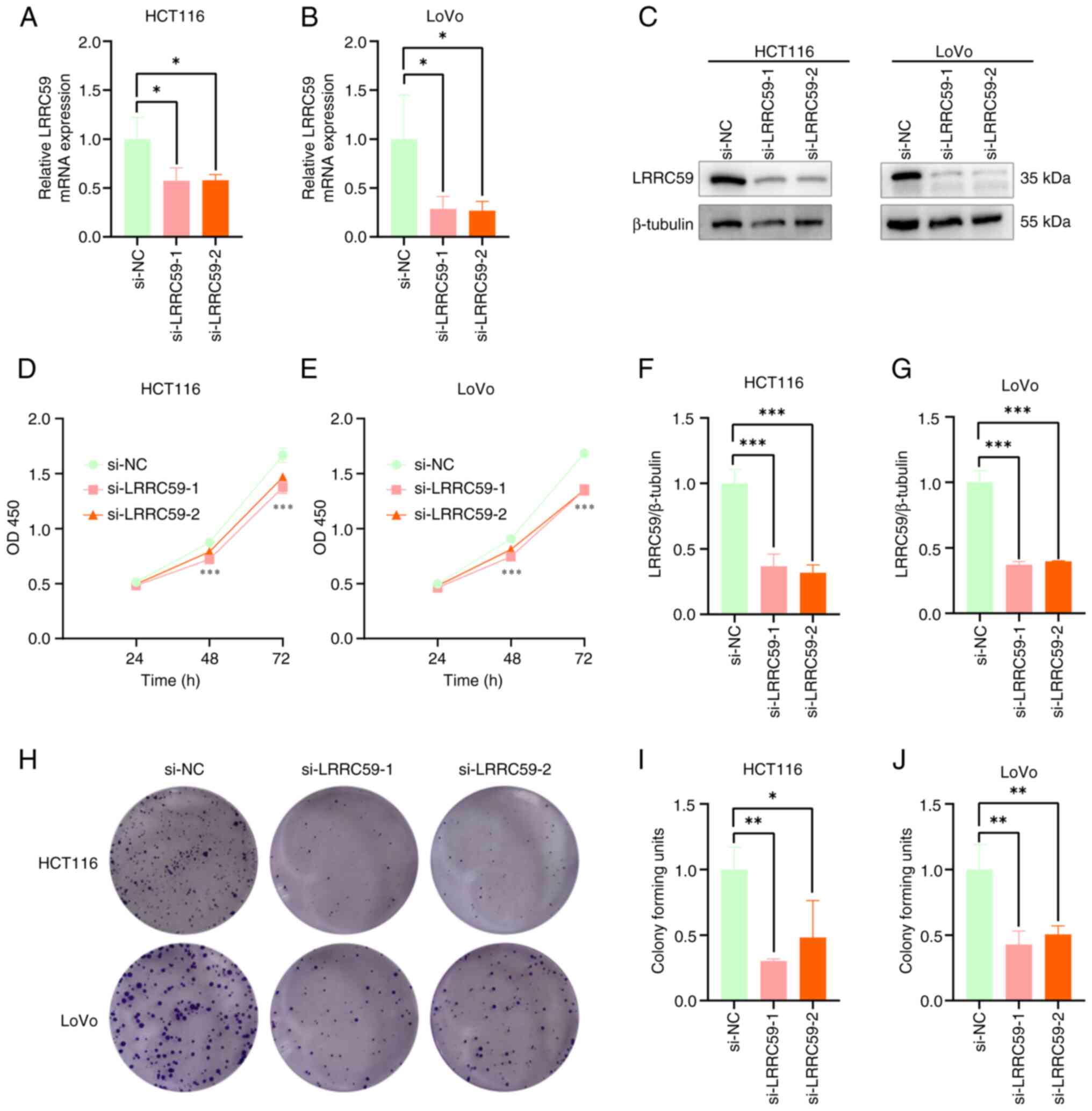
LRRC59 knockdown suppresses CRC cell
proliferation
The function of LRRC59 in promoting CRC cell
proliferation was examined by silencing its expression in HCT116
and LoVo cell lines using siRNA. RT-qPCR (Fig. 2A and B) and western blot analysis
(Fig. 2C-E) confirmed the
successful silencing of LRRC59. Cell viability and proliferation
potential were detected using the CCK-8 (Fig. 2F and G) and colony formation assays
(Fig. 2H-J). These results revealed
reduced colony formation and inhibition of cell proliferation
following LRRC59 knockdown. These findings suggested that LRRC59
promotes CRC cell proliferation.
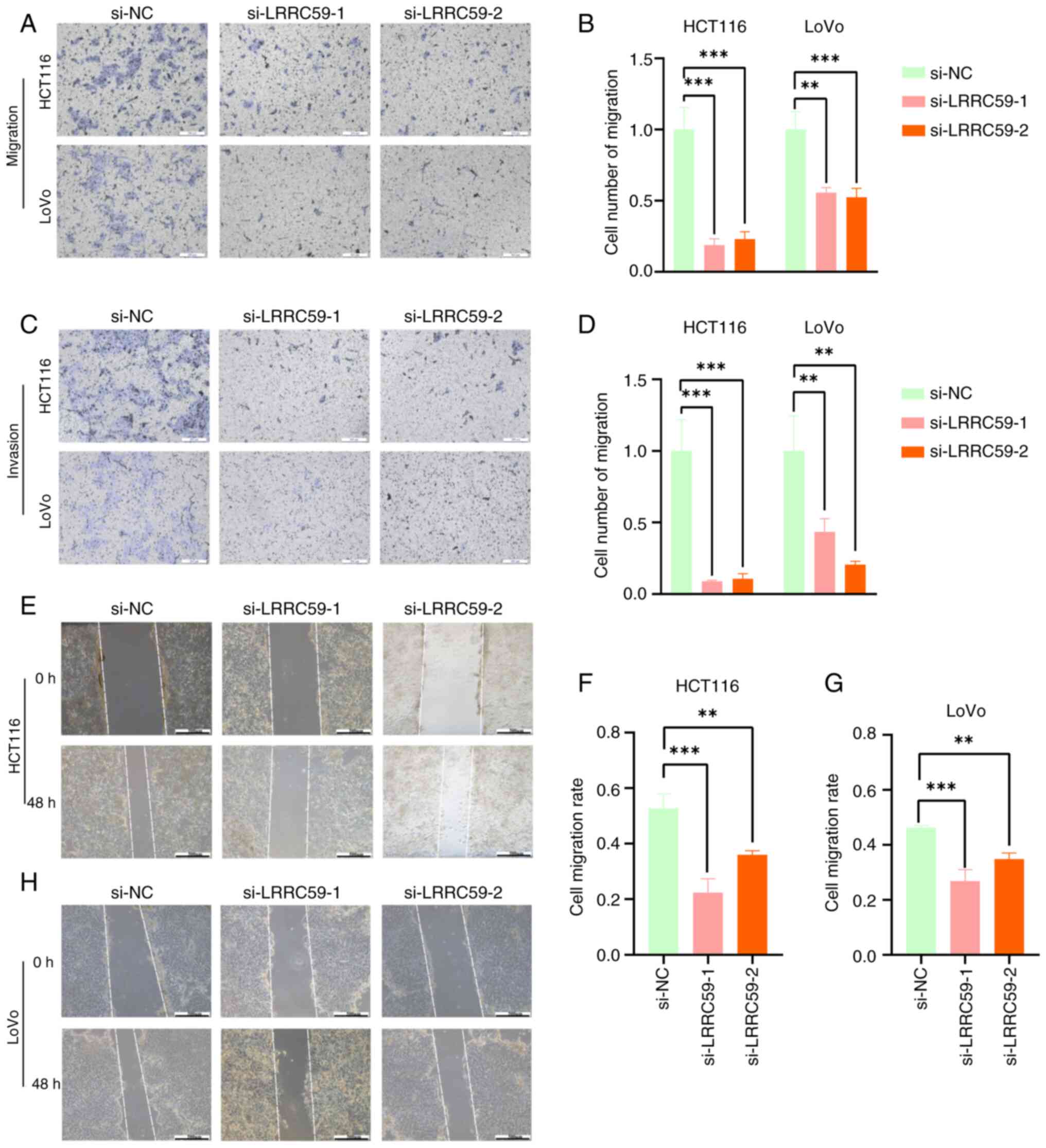
Silencing LRRC59 reduces CRC cell
migration and invasion
The effect of LRRC59 on CRC cell migration and
invasion was evaluated using Transwell and wound healing assays.
LRRC59 silencing significantly reduced the number of cells that
migrated through the porous membrane or Matrigel-coated inserts
(Fig. 3A-D). Similarly, wound
healing assays revealed diminished motility in cells with reduced
LRRC59 expression (Fig. 3E-H).
These results highlighted the role of LRRC59 in promoting CRC cell
migration and invasion.
Knockdown of LRRC59 induces activation
of the PERK pathway in CRC cells
Emerging evidence suggests that LRRC59 is involved
in ERS regulation (9).
Bioinformatics analysis using the starBase database (http://starbase.sysu.edu.cn/) revealed a significant
positive correlation between LRRC59 and GRP78 (HSPA5), a master
chaperone of ERS. These results suggested that LRRC59 modulates ERS
signaling, implicating a regulatory role in CRC pathogenesis. The
potential associations between LRRC59 and key regulatory components
of the three UPR branches were subsequently examined: IRE1α, PERK
and ATF6 pathways. The results revealed a significant association
between LRRC59 and PERK signaling pathways (ATF4 and CHOP)
(Fig. 4A). To validate these
findings, LRRC59 in HCT116 and LoVo cells was knocked down,
followed by western blot analysis of PERK pathway components.
Altered expression of PERK pathway-related proteins was observed,
including increased levels of GRP78, p-PERK, p-EIF2α, ATF4 and
CHOP, in LRRC59-silenced cells (Fig.
4B-D). These data demonstrated that LRRC59 is a negative
regulator of PERK-mediated ERS signaling in CRC cells.
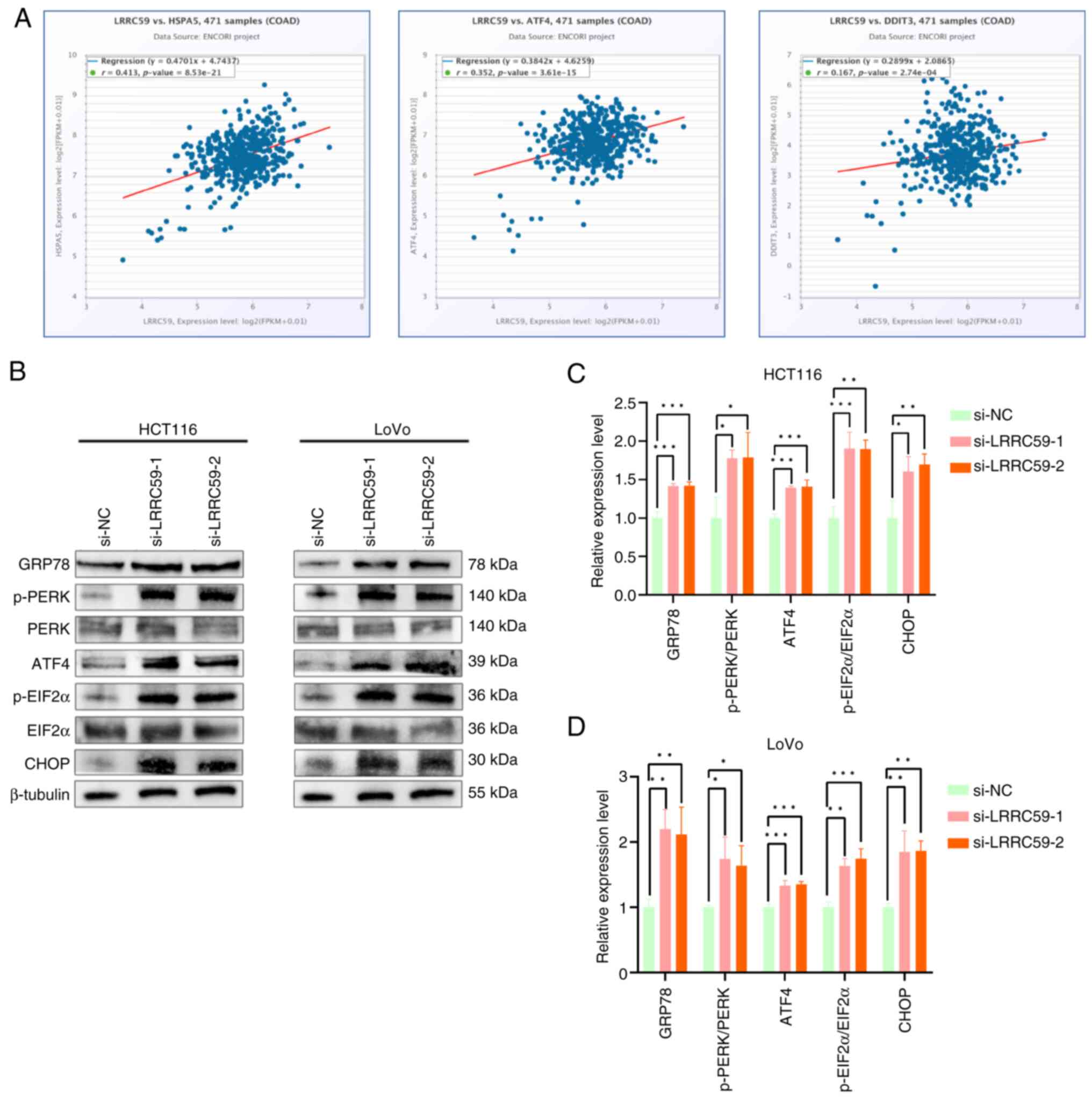 | Figure 4.LRRC59 regulates the PERK-mediated
ERS signaling pathway. (A) According to the starBase database,
LRRC59 is associated with ERS pathway key genes GRP78, ATF4, and
CHOP in colorectal cancer. (B-D) Western blot analysis of PERK
pathway-related proteins (GRP78, p-PERK, PERK, ATF4, p-EIF2α, EIF2α
and CHOP). *P<0.05, **P<0.01 and ***P<0.001. ER,
endoplasmic reticulum; PERK, protein kinase RNA-like ER kinase;
ERS, ER stress; p-, phosphorylated; si-, small interfering; NC,
negative control; COAD, colon adenocarcinoma. |
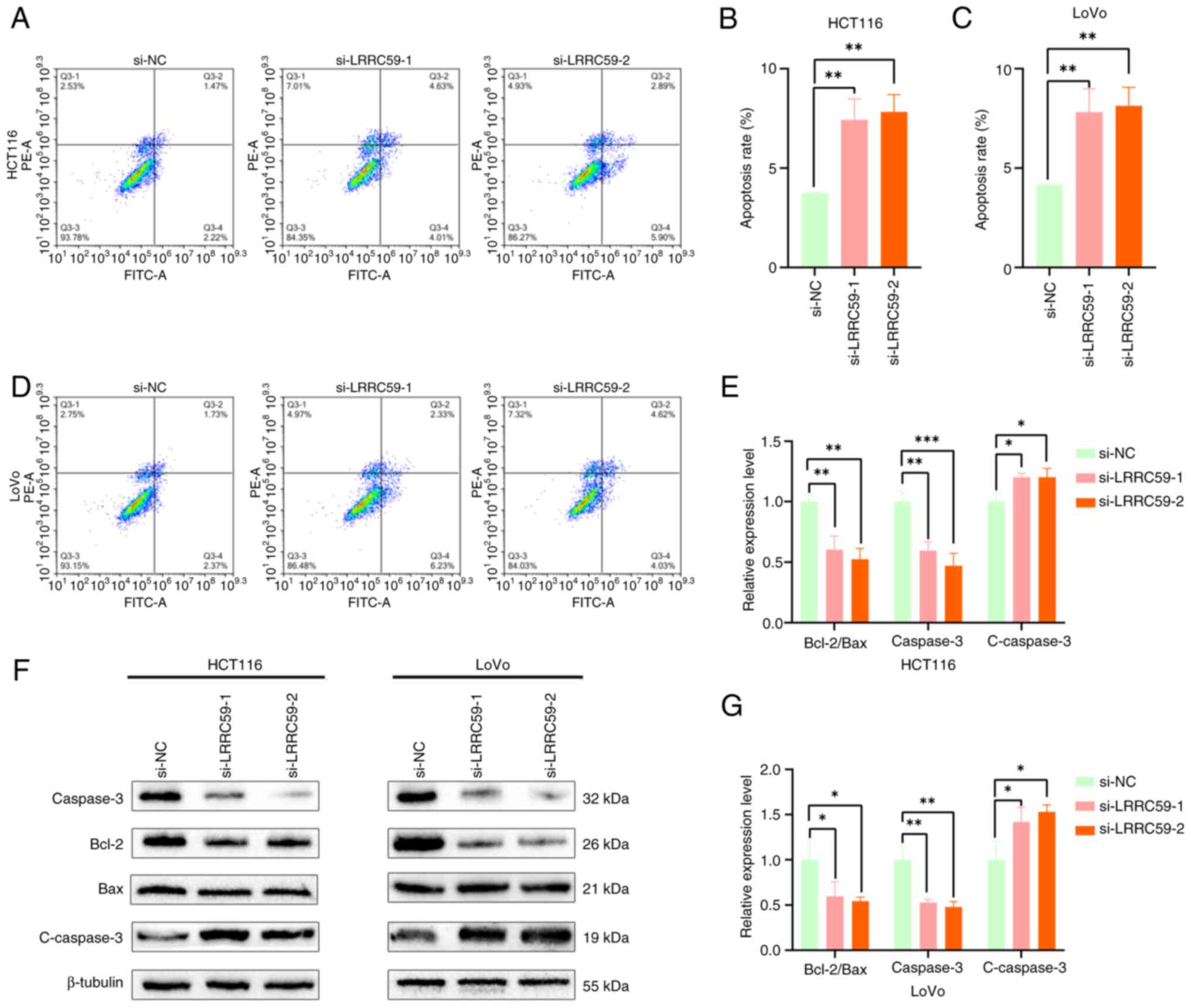
LRRC59 suppresses apoptosis in CRC
cells
ERS plays a critical role in modulating apoptosis
(25–27). To investigate whether LRRC59
regulates apoptosis through ERS pathways, apoptotic responses were
first examined following LRRC59 knockdown. Flow cytometric analysis
demonstrated that LRRC59 deficiency in HCT116 and LoVo cells
resulted in a substantial increase in apoptotic cells compared with
that in control cells (Fig. 5A-D).
Western blot analysis of apoptosis-related proteins (Fig. 5E-G) demonstrated upregulation of
active caspase-3 and decreased Bcl-2 and Caspase3 expression. These
findings provided further evidence that LRRC59 functions as an
anti-apoptotic factor in CRC cells.
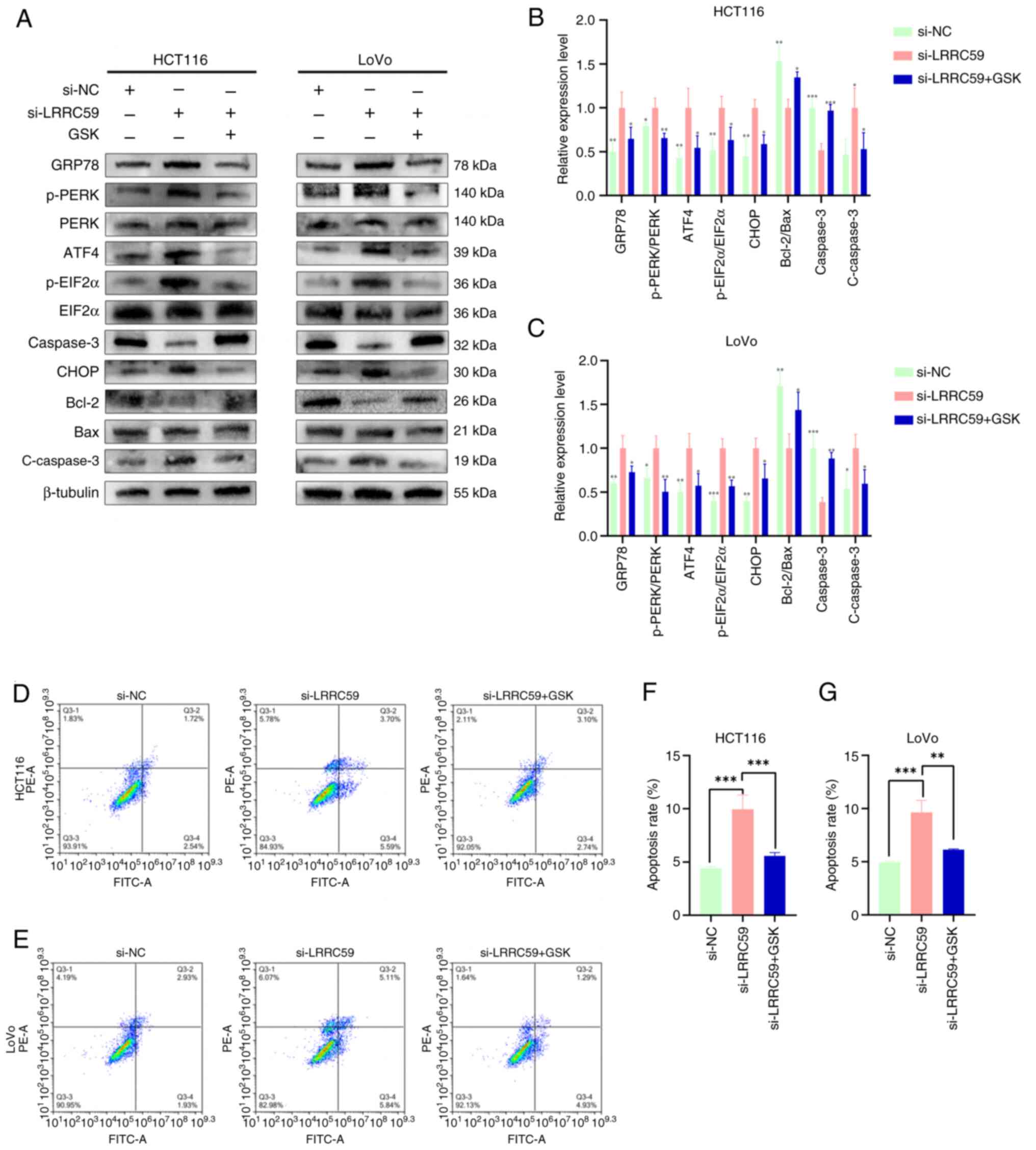
LRRC59 modulates apoptosis via PERK
signaling in CRC
Increased PERK phosphorylation is a hallmark of PERK
pathway activation. To assess whether LRRC59 influenced apoptosis
by regulating the PERK pathway, LRRC59 knockdown CRC cells were
treated with GSK, a PERK pathway inhibitor. The results showed that
GSK treatment significantly inhibited PERK phosphorylation. Western
blot analysis (Fig. 6A-C) revealed
decreased expression of ERS pathway proteins and a reduction in
apoptosis-related proteins, including active-caspase-3, whereas the
anti-apoptotic protein Bcl-2 was upregulated. Flow cytometry
(Fig. 6D-G) confirmed a significant
reduction in apoptosis following GSK treatment. These findings
suggested that LRRC59 regulates apoptosis by modulating the PERK
signaling pathway in CRC cells.
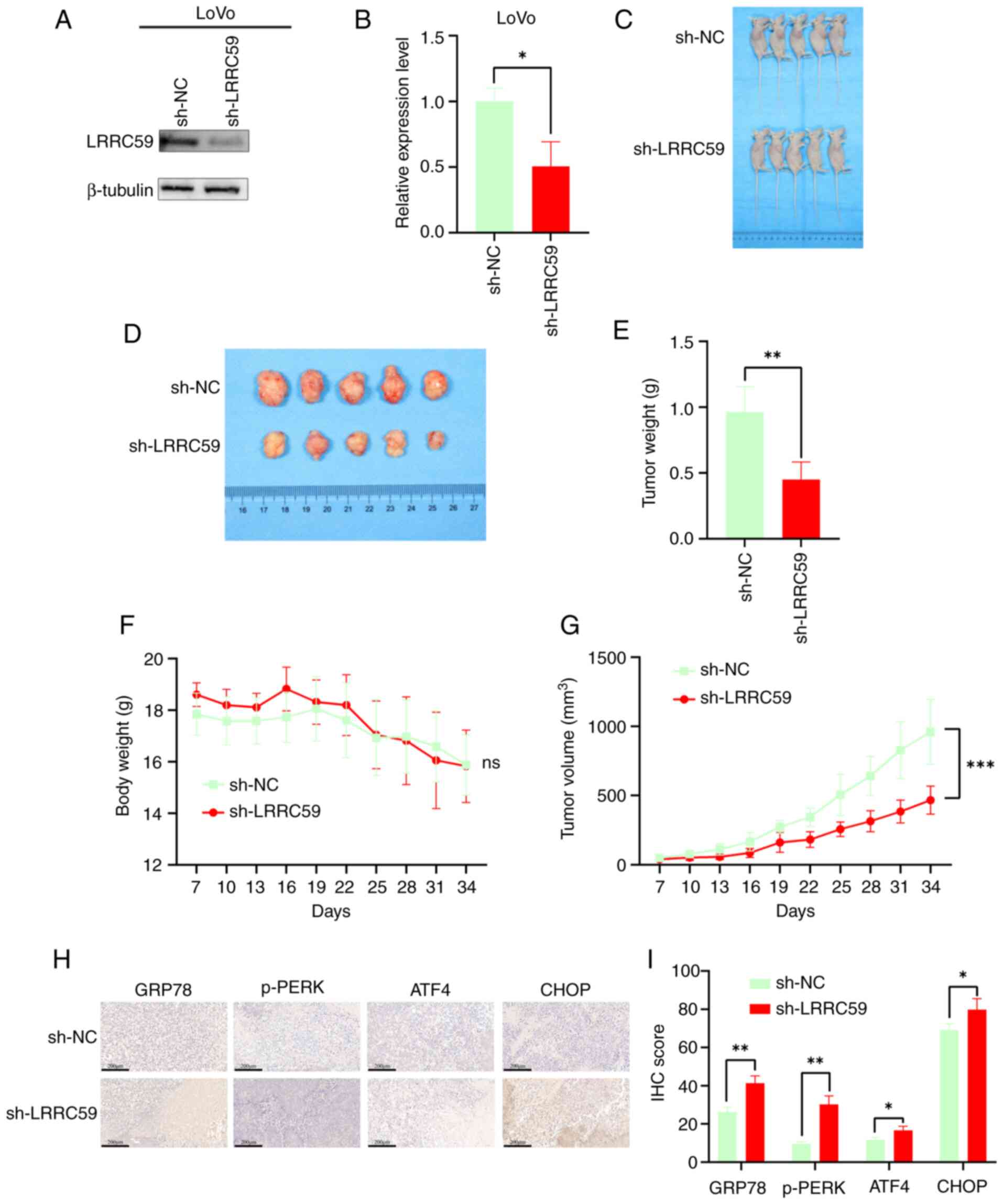
LRRC59 knockdown reduces tumor growth
in mice
The effect of LRRC59 on CRC tumorigenesis was
assessed in vivo in a mouse xenograft model. Western blot
analysis confirmed the successful establishment of stable
LRRC59-knockdown LoVo cells prior to implantation (Fig. 7A and B). Mice injected with
sh-LRRC59 transfected LoVo cells exhibited significantly smaller
tumor volumes and weights than the controls (Fig. 7C-E and G). Body weight remained
consistent across groups (Fig. 7F).
IHC analysis demonstrated increased activation of the PERK pathway
in sh-LRRC59 tumors (Fig. 7H and
I). These results indicated that LRRC59 knockdown hinders CRC
progression by activating the PERK pathway.
Discussion
The role of LRRC59 in CRC progression was
investigated. The present findings revealed notable upregulation of
LRRC59 expression in CRC tissues, leading to the promotion of cell
proliferation, migration and invasion. Additionally, LRRC59
inhibited apoptosis by modulating the PERK signaling. These results
suggested that LRRC59 can be used as target for the treatment of
CRC.
LRRC59 is a 244-amino acid protein localized to the
ER membrane that was initially identified as a ribosome-binding
protein (4–6). It is involved in various cellular
functions, including immune responses, anti-inflammatory
activities, and the translation and translocation of proteins
within the ER (28). Previous
studies highlighted the involvement of LRRC59 in multiple cancer
types (7–9,29,30).
However, its role in CRC remains unclear. A recent study suggested
that LRRC59 suppresses CRC progression (31), which contrast the current findings.
It was found that LRRC59 mRNA expression was significantly higher
in CRC tissues than in adjacent normal tissues. Subsequent analyses
confirmed its overexpression at the protein level in CRC tissues.
Furthermore, silencing of LRRC59 reduced CRC cell proliferation and
impaired cell migration and invasion. Additionally, LRRC59
knockdown significantly attenuated tumor growth in a mouse
xenograft model. These findings suggest that LRRC59 is
significantly overexpressed in CRC and plays a key role in
promoting CRC progression by inhibiting apoptosis.
The leucine-rich repeat domain of LRRC59 mediates
protein-protein interactions, which influence processes such as
apoptosis, autophagy and nuclear mRNA transport (32). Previous studies demonstrated that
LRRC59 interacts with FGF1 to promote its nuclear import; inhibit
apoptosis; and regulate tumor cell survival, proliferation and
migration (33,34). Additionally, LRRC59 knockdown
induced G1 cell cycle arrest, thereby inhibiting tumor progression.
In the present study, silencing of LRRC59 elevated apoptotic
markers such as active-caspase-3 in CRC cells and decreased
expression of the anti-apoptotic protein Bcl-2. These results
suggested that LRRC59 suppresses apoptosis in CRC cells.
Malignant tumor progression is characterized by
uncontrolled cell proliferation, which creates a highly metabolic,
hypoxic, nutrient-deficient and acidic microenvironment. This
environment activates the UPR, which amplifies the ERS (35,36).
ERS is a dynamic process that responds to protein imbalances in the
ER by activating the IRE1-, PERK- and ATF6-mediated ERS pathways
during CRC development, thus helping to restore ER homeostasis and
support cell survival (37). These
pathways initially reduce protein overload in the ER through
mechanisms such as PERK-mediated EIF2α phosphorylation,
IRE1-dependent mRNA decay, and autophagy activation via the
IRE1α-JNK axis. UPR transcription factors activate ATF6 and splice
XBP1 to promote adaptive responses, restore ER function, and
support cell survival (38–43). However, if ERS persists, the cells
shift from an adaptive response to a death-inducing response
(44,45). Numerous studies indicated that the
overactivation of the ERS signaling promotes apoptosis, autophagy
and ferroptosis in tumor cells (46–50).
When ERS conditions are prolonged, IRE1α recruits TRAF2, activating
ASK1 and JNK to promote apoptosis (51). Sustained PERK signaling induces
apoptosis by upregulating CHOP, a pro-apoptotic transcription
factor that downregulates BCL-2 and induces BH3-only protein
expression, leading to cell death (52). LRRC59 affects HYOU1- and
XBP1-mediated ERS signaling and promotes cancer cell proliferation
and migration. However, whether LRRC59 plays a role in CRC via the
ERS pathway remains unclear. An association between LRRC59 and the
expression of key proteins involved in the PERK-mediated ERS
pathway was identified. In CRC cell lines with LRRC59 knockdown,
expression of GRP78, p-PERK, p-EIF2α, ATF4 and CHOP was increased.
To confirm whether LRRC59 affects apoptosis through the
PERK-mediated ERS pathway, the PERK pathway-specific inhibitor GSK
was used, which resulted in reduced apoptosis of CRC cells,
increased expression of Bcl-2, and decreased expression of
active-caspase-3. These findings suggested that LRRC59 inhibits
apoptosis and promotes CRC cell survival by suppressing excessive
activation of the PERK-mediated ERS signaling pathway.
Given that the PERK pathway plays a central role in
the ERS response in CRC cells, targeting the PERK pathway for the
treatment of CRC may have broad application prospects (36). Numerous drugs or compounds capable
of directly acting on ER-associated proteins have been discovered
(53). For instance, V8, a novel
derivative of the natural flavonoid wogonin and a potential
anticancer agent, has demonstrated significant antitumor activity
both in vitro and in vivo. In vitro studies using
acute myeloid leukemia cell lines showed that V8 selectively
activates the PERK pathway, ultimately triggering CHOP-mediated
apoptosis via an ERS-specific pathway (54). Terpenoids, one of the largest and
most structurally diverse classes of natural compounds, have been
shown in some studies to reduce the incidence of cervical cancer
and promote cancer cell apoptosis through the PERK pathway
(55). 2-Deoxyglucose, a glucose
analog, has been found to exert antitumor effects by promoting the
PERK pathway; when combined with HCQ, it can further inhibit the
viability and migration of breast tumor cells and induce apoptosis
(56). However, the vast majority
of currently identified compounds or drugs that may act against
tumors via the PERK pathway are still confined to preclinical
studies, and their translation into clinical therapies requires
further validation through clinical trials. Recent research
suggests that cancer treatment strategies employing nanoparticles
to target the ER and regulate programmed cell death may represent a
promising new direction for clinical translation (57). In the field of nasopharyngeal
carcinoma, relevant in vitro studies have been conducted: A
photothermal nano-vaccine based on phenylboronic acid-modified poly
dendrimers of generation 5 attached with indocyanine green can
serve as a carrier for the ERS-inducing drug toyocamycin,
demonstrating satisfactory antitumor efficacy in vitro
(58). However, direct targeting of
the PERK pathway in vivo may not be effective and could
result in systemic effects (59).
PERK inhibition destroys pancreatic cells and osteoblasts, leading
to diabetes and severe bone defects (60,61).
Therefore, indirectly affecting the PERK pathway through LRRC59 may
be a safe approach for treating CRC. Although LRRC59 shows
potential as a therapeutic target, the development of inhibitors
and related drugs targeting LRRC59 requires further analysis.
Although it was found that LRRC59 can affect CRC
cells through the PERK pathway, there were limitations to the
present study. The numbers of clinical specimens and animal model
samples used were relatively small, and only two CRC cell lines
were evaluated, limiting the generalizability of the results.
Further studies are needed to expand the number of clinical
samples, increase the number of CRC cell lines and animal models,
and explore the molecular mechanism of LRRC59 in greater depth to
obtain more universally applicable results. The main objective of
the present study was to preliminarily validate the effect of
LRRC59 knockdown on CRC cells in vivo using a mouse
xenograft model. Additional studies are necessary to establish
in situ transplantation and metastasis models to
comprehensively examine the effects of LRRC59 on CRC metastasis and
the tumor microenvironment. In the present study, focus was
addressed on the PERK pathway branch in ERS, and the literature
suggests that LRRC59 is related to other branches of ERS (62). Given the complexity of the UPR
pathway, future multi-omics analysis is needed to determine whether
LRRC59 can affect CRC through the other two main branches, the
IRE1α and ATF6 pathways. Although the PERK pathway was established
as the main target of LRRC59, the involvement of non-PERK pathways
in tumor progression cannot be ruled out. This is an important
topic for future research.
In conclusion, the present study demonstrated that
LRRC59 is significantly upregulated in CRC tissues, where it
enhances cell proliferation, migration and invasion. Moreover,
LRRC59 suppresses apoptosis by attenuating PERK-mediated activation
of the ERS pathway. Although research on LRRC59 remains in the
early stages, further mechanistic explorations and large-scale
clinical trials are required. The current findings indicate the
potential of LRRC59 as a therapeutic target for CRC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science
Foundation of Liaoning (grant no. 2024-MS-152).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
XH and YW contributed to the methodology,
validation, and writing of the original draft. YC participated in
data validation. PZ was involved in conceptualization of the study.
GW contributed to study conceptualization, writing, review and
editing of the manuscript. BLi conducted formal analyses. BLu
performed the experiments. HJ provided resources and performed the
experiments. SN was responsible for conceptualization, funding
acquisition, supervision, and writing, reviewing and editing the
manuscript. XH and SN confirm the authenticity of all the raw data.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the Declaration of Helsinki and was approved (approval no.
kY2024-136-01) by the Ethics Committee of the Second Affiliated
Hospital of Dalian Medical University (Dalian, China). Animal
experiments were conducted in accordance with the ARRIVE guidelines
and were approved (approval no. 2024167) by the Institutional
Animal Care and Use Committee of Wuhan Servicebio Technology Co.,
Ltd.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Wagle NS, Cercek A, Smith RA
and Jemal A: Colorectal cancer statistics, 2023. CA Cancer J Clin.
73:233–254. 2023.PubMed/NCBI
|
|
2
|
Morgan E, Arnold M, Gini A, Lorenzoni V,
Cabasag CJ, Laversanne M, Vignat J, Ferlay J, Murphy N and Bray F:
Global burden of colorectal cancer in 2020 and 2040: Incidence and
mortality estimates from GLOBOCAN. Gut. 72:338–344. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leowattana W, Leowattana P and Leowattana
T: Systemic treatment for metastatic colorectal cancer. World J
Gastroenterol. 29:1569–1588. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ichimura T, Shindo Y, Uda Y, Ohsumi T,
Omata S and Sugano H: Anti-(p34 protein) antibodies inhibit
ribosome binding to and protein translocation across the rough
microsomal membrane. FEBS Lett. 326:241–245. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ohsumi T, Ichimura T, Sugano H, Omata S,
Isobe T and Kuwano R: Ribosome-binding protein p34 is a member of
the leucine-rich-repeat-protein superfamily. Biochem J.
294:465–472. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ichimura T, Ohsumi T, Shindo Y, Ohwada T,
Yagame H, Momose Y, Omata S and Sugano H: Isolation and some
properties of a 34-kDa-membrane protein that may be responsible for
ribosome binding in rat liver rough microsomes. FEBS Lett.
296:7–10. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pallai R, Bhaskar A, Barnett-Bernodat N,
Gallo-Ebert C, Pusey M, Nickels JT Jr and Rice LM: Leucine-rich
repeat-containing protein 59 mediates nuclear import of cancerous
inhibitor of PP2A in prostate cancer cells. Tumour Biol.
36:6383–6390. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li D, Xing Y, Tian T, Guo Y and Qian J:
Overexpression of LRRC59 is associated with poor prognosis and
promotes cell proliferation and invasion in lung adenocarcinoma.
Onco Targets Ther. 13:6453–6463. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pei L, Zhu Q, Zhuang X, Ruan H, Zhao Z,
Qin H and Lin Q: Identification of leucine-rich repeat-containing
protein 59 (LRRC59) located in the endoplasmic reticulum as a novel
prognostic factor for urothelial carcinoma. Transl Oncol.
23:1014742022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen X and Cubillos-Ruiz JR: Endoplasmic
reticulum stress signals in the tumour and its microenvironment.
Nat Rev Cancer. 21:71–88. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Walter P and Ron D: The unfolded protein
response: From stress pathway to homeostatic regulation. Science.
334:1081–1086. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hetz C, Zhang K and Kaufman RJ:
Mechanisms, regulation and functions of the unfolded protein
response. Nat Rev Mol Cell Biol. 21:421–438. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shore GC, Papa FR and Oakes SA: Signaling
cell death from the endoplasmic reticulum stress response. Curr
Opin Cell Biol. 23:143–149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Long D, Chen K, Yang Y and Tian X:
Unfolded protein response activated by endoplasmic reticulum stress
in pancreatic cancer: Potential therapeutical target. Front Biosci
(Landmark Ed). 26:1689–1696. 2021. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song M and Cubillos-Ruiz JR: Endoplasmic
reticulum stress responses in intratumoral immune cells:
Implications for cancer immunotherapy. Trends Immunol. 40:128–141.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Geng J, Guo Y, Xie M, Li Z, Wang P, Zhu D,
Li J and Cui X: Characteristics of endoplasmic reticulum stress in
colorectal cancer for predicting prognosis and developing treatment
options. Cancer Med. 12:12000–12017. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou F, Gao H, Shang L, Li J, Zhang M,
Wang S, Li R, Ye L and Yang S: Oridonin promotes endoplasmic
reticulum stress via TP53-repressed TCF4 transactivation in
colorectal cancer. J Exp Clin Cancer Res. 42:1502023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li F, Dong Q, Kai Z, Pan Q and Liu C:
CYP8B1 is a prognostic biomarker with important functional
implications in hepatocellular carcinoma. Oncol Rep. 54:1042025.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma H, Suleman M, Zhang F, Cao T, Wen S,
Sun D, Chen L, Jiang B, Wang Y, Lin F, et al: Pirin inhibits
FAS-mediated apoptosis to support colorectal cancer survival. Adv
Sci (Weinh). 11:e23014762024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu H, Wang T, Nie H, Sun Q, Jin C, Yang S,
Chen Z, Wang X, Tang J, Feng Y and Sun Y: USP36 promotes colorectal
cancer progression through inhibition of p53 signaling pathway via
stabilizing RBM28. Oncogene. 43:3442–3455. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao L, Sun X, Hou C, Yang Y, Wang P, Xu
Z, Chen Z, Zhang X, Wu G, Chen H, et al: CPNE7 promotes colorectal
tumorigenesis by interacting with NONO to initiate ZFP42
transcription. Cell Death Dis. 15:8962024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim J, Jeong Y, Shin YM, Kim SE and Shin
SJ: FL118 enhances therapeutic efficacy in colorectal cancer by
inhibiting the homologous recombination repair pathway through
survivin-RAD51 downregulation. Cancers (Basel). 16:33852024.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Z, Wang L, Zhao L, Wang Q, Yang C,
Zhang M, Wang B, Jiang K, Ye Y, Wang S and Shen Z:
N6-methyladenosine demethylase ALKBH5 suppresses colorectal cancer
progression potentially by decreasing PHF20 mRNA methylation. Clin
Transl Med. 12:e9402022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu H, Zheng S, Zhang J, Xu S and Miao Z:
Cadmium induces endoplasmic reticulum stress-mediated apoptosis in
pig pancreas via the increase of Th1 cells. Toxicology.
457:1527902021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee H, Lee YS, Harenda Q, Pietrzak S,
Oktay HZ, Schreiber S, Liao Y, Sonthalia S, Ciecko AE, Chen YG, et
al: Beta cell dedifferentiation induced by IRE1α deletion prevents
type 1 diabetes. Cell Metab. 31:822–836.e825. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim TW, Lee SY, Kim M, Cheon C and Ko SG:
Kaempferol induces autophagic cell death via IRE1-JNK-CHOP pathway
and inhibition of G9a in gastric cancer cells. Cell Death Dis.
9:8752018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pallai R, Bhaskar A, Barnett-Bernodat N,
Gallo-Ebert C, Nickels JT Jr and Rice LM: Cancerous inhibitor of
protein phosphatase 2A promotes premature chromosome segregation
and aneuploidy in prostate cancer cells through association with
shugoshin. Tumour Biol. 36:6067–6074. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maurizio E, Wiśniewski JR, Ciani Y, Amato
A, Arnoldo L, Penzo C, Pegoraro S, Giancotti V, Zambelli A, Piazza
S, et al: Translating proteomic into functional data: An high
mobility group A1 (HMGA1) proteomic signature has prognostic value
in breast cancer. Mol Cell Proteomics. 15:109–123. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pan B, Cheng J, Tan W, Wu X, Fan Q, Fan L,
Jiang M, Yu R, Cheng X and Deng Y: Pan-cancer analysis of LRRC59
with a focus on prognostic and immunological roles in
hepatocellular carcinoma. Aging (Albany NY). 16:8171–8197.
2024.PubMed/NCBI
|
|
31
|
Chen H, Zhao T, Fan J, Yu Z, Ge Y, Zhu H,
Dong P, Zhang F, Zhang L, Xue X and Lin X: Construction of a
prognostic model for colorectal adenocarcinoma based on Zn
transport-related genes identified by single-cell sequencing and
weighted co-expression network analysis. Front Oncol.
13:12074992023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xian H, Yang S, Jin S, Zhang Y and Cui J:
LRRC59 modulates type I interferon signaling by restraining the
SQSTM1/p62-mediated autophagic degradation of pattern recognition
receptor DDX58/RIG-I. Autophagy. 16:408–418. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhen Y, Sørensen V, Skjerpen CS, Haugsten
EM, Jin Y, Wälchli S, Olsnes S and Wiedlocha A: Nuclear import of
exogenous FGF1 requires the ER-protein LRRC59 and the importins
Kpnα1 and Kpnβ1. Traffic. 13:650–664. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu N, Zhang J, Sun S, Yang L, Zhou Z, Sun
Q and Niu J: Expression and clinical significance of fibroblast
growth factor 1 in gastric adenocarcinoma. Onco Targets Ther.
8:615–621. 2015.PubMed/NCBI
|
|
35
|
Oakes SA: Endoplasmic reticulum stress
signaling in cancer cells. Am J Pathol. 190:934–946. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang J, Pan H, Wang J, Wang T, Huo X, Ma
Y, Lu Z, Sun B and Jiang H: Unfolded protein response in colorectal
cancer. Cell Biosci. 11:262021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Urra H, Dufey E, Avril T, Chevet E and
Hetz C: Endoplasmic reticulum stress and the hallmarks of cancer.
Trends Cancer. 2:252–262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Harding HP, Novoa I, Zhang Y, Zeng H, Wek
R, Schapira M and Ron D: Regulated translation initiation controls
stress-induced gene expression in mammalian cells. Mol Cell.
6:1099–1108. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Han D, Lerner AG, Vande Walle L, Upton JP,
Xu W, Hagen A, Backes BJ, Oakes SA and Papa FR: IRE1alpha kinase
activation modes control alternate endoribonuclease outputs to
determine divergent cell fates. Cell. 138:562–575. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
4Hollien J, Lin JH, Li H, Stevens N,
Walter P and Weissman JS: Regulated Ire1-dependent decay of
messenger RNAs in mammalian cells. J Cell Biol. 186:323–331. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hollien J and Weissman JS: Decay of
endoplasmic reticulum-localized mRNAs during the unfolded protein
response. Science. 313:104–107. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee AH, Iwakoshi NN and Glimcher LH: XBP-1
regulates a subset of endoplasmic reticulum resident chaperone
genes in the unfolded protein response. Mol Cell Biol.
23:7448–7459. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ameri K and Harris AL: Activating
transcription factor 4. Int J Biochem Cell Biol. 40:14–21. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shi Y, Jiang B and Zhao J: Induction
mechanisms of autophagy and endoplasmic reticulum stress in
intestinal ischemia-reperfusion injury, inflammatory bowel disease,
and colorectal cancer. Biomed Pharmacother. 170:1159842024.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sisinni L, Pietrafesa M, Lepore S,
Maddalena F, Condelli V, Esposito F and Landriscina M: Endoplasmic
reticulum stress and unfolded protein response in breast cancer:
The balance between apoptosis and autophagy and its role in drug
resistance. Int J Mol Sci. 20:8572019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ji X, Chen Z, Lin W, Wu Q, Wu Y, Hong Y,
Tong H, Wang C and Zhang Y: Esculin induces endoplasmic reticulum
stress and drives apoptosis and ferroptosis in colorectal cancer
via PERK regulating eIF2α/CHOP and Nrf2/HO-1 cascades. J
Ethnopharmacol. 328:1181392024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Qu J, Zeng C, Zou T, Chen X, Yang X and
Lin Z: Autophagy induction by trichodermic acid attenuates
endoplasmic reticulum stress-mediated apoptosis in colon cancer
cells. Int J Mol Sci. 22:55662021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang G, Han J, Wang G, Wu X, Huang Y, Wu M
and Chen Y: ERO1α mediates endoplasmic reticulum stress-induced
apoptosis via microRNA-101/EZH2 axis in colon cancer RKO and HT-29
cells. Hum Cell. 34:932–944. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Piao MJ, Han X, Kang KA, Fernando PDSM,
Herath HMUL and Hyun JW: The endoplasmic reticulum stress response
mediates shikonin-induced apoptosis of 5-fluorouracil-resistant
colorectal cancer cells. Biomol Ther (Seoul). 30:265–273. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Quan JH, Gao FF, Chu JQ, Cha GH, Yuk JM,
Wu W and Lee YH: Silver nanoparticles induce apoptosis via
NOX4-derived mitochondrial reactive oxygen species and
endoplasmic reticulum stress in colorectal cancer cells.
Nanomedicine (Lond). 16:1357–1375. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Urano F, Wang X, Bertolotti A, Zhang Y,
Chung P, Harding HP and Ron D: Coupling of stress in the ER to
activation of JNK protein kinases by transmembrane protein kinase
IRE1. Science. 287:664–666. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tabas I and Ron D: Integrating the
mechanisms of apoptosis induced by endoplasmic reticulum stress.
Nat Cell Biol. 13:184–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhou R, Wang W, Li B, Li Z, Huang J and Li
X: Endo-plasmic reticulum stress in cancer. MedComm (2020).
6:e702632025. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Guo YJ, Zhu MY, Wang ZY, Chen HY, Qing YJ,
Wang HZ, Xu JY, Hui H and Li H: Therapeutic effect of V8 affecting
mitophagy and endoplasmic reticulum stress in acute myeloid
leukemia mediated by ROS and CHOP signaling. FASEB J.
39:e706222025. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang P, Liu H, Yu Y, Peng S, Zeng A and
Song L: Terpenoids mediated cell apoptotsis in cervical cancer:
Mechanisms, advances and prospects. Fitoterapia. 180:1063232024.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhou N, Liu Q, Wang X, He L, Zhang T, Zhou
H, Zhu X, Zhou T, Deng G and Qiu C: The combination of
hydroxychloroquine and 2-deoxyglucose enhances apoptosis in breast
cancer cells by blocking protective autophagy and sustaining
endoplasmic reticulum stress. Cell Death Discov. 8:2862022.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liang L, Zhu Z, Jiang X, Tang Y, Li J,
Zhang Z, Ding B, Li X, Yu M and Gan Y: Endoplasmic
reticulum-targeted strategies for programmed cell death in cancer
therapy: Approaches and prospects. J Control Release.
385:1140592025. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liu Z, Deng X, Wang Z, Guo Y, Hameed MMA,
El-Newehy M, Zhang J, Shi X and Shen M: A biomimetic therapeutic
nanovaccine based on dendrimer-drug conjugates coated with
metal-phenolic networks for combination therapy of nasopharyngeal
carcinoma: An in vitro investigation. J Mater Chem B.
13:5440–5452. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yeap JW, Ali IAH, Ibrahim B and Tan ML:
Chronic obstructive pulmonary disease and emerging ER
stress-related therapeutic targets. Pulm Pharmacol Ther.
81:1022182023. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Harding HP, Zeng H, Zhang Y, Jungries R,
Chung P, Plesken H, Sabatini DD and Ron D: Diabetes mellitus and
exocrine pancreatic dysfunction in perk-/- mice reveals a role for
translational control in secretory cell survival. Mol Cell.
7:1153–1163. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang P, McGrath B, Li S, Frank A, Zambito
F, Reinert J, Gannon M, Ma K, McNaughton K and Cavener DR: The PERK
eukaryotic initiation factor 2 alpha kinase is required for the
development of the skeletal system, postnatal growth, and the
function and viability of the pancreas. Mol Cell Biol.
22:3864–3874. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yuan Z, Wang Y, Xu S, Zhang M and Tang J:
Construction of a prognostic model for colon cancer by combining
endoplasmic reticulum stress responsive genes. J Proteomics.
309:1052842024. View Article : Google Scholar : PubMed/NCBI
|