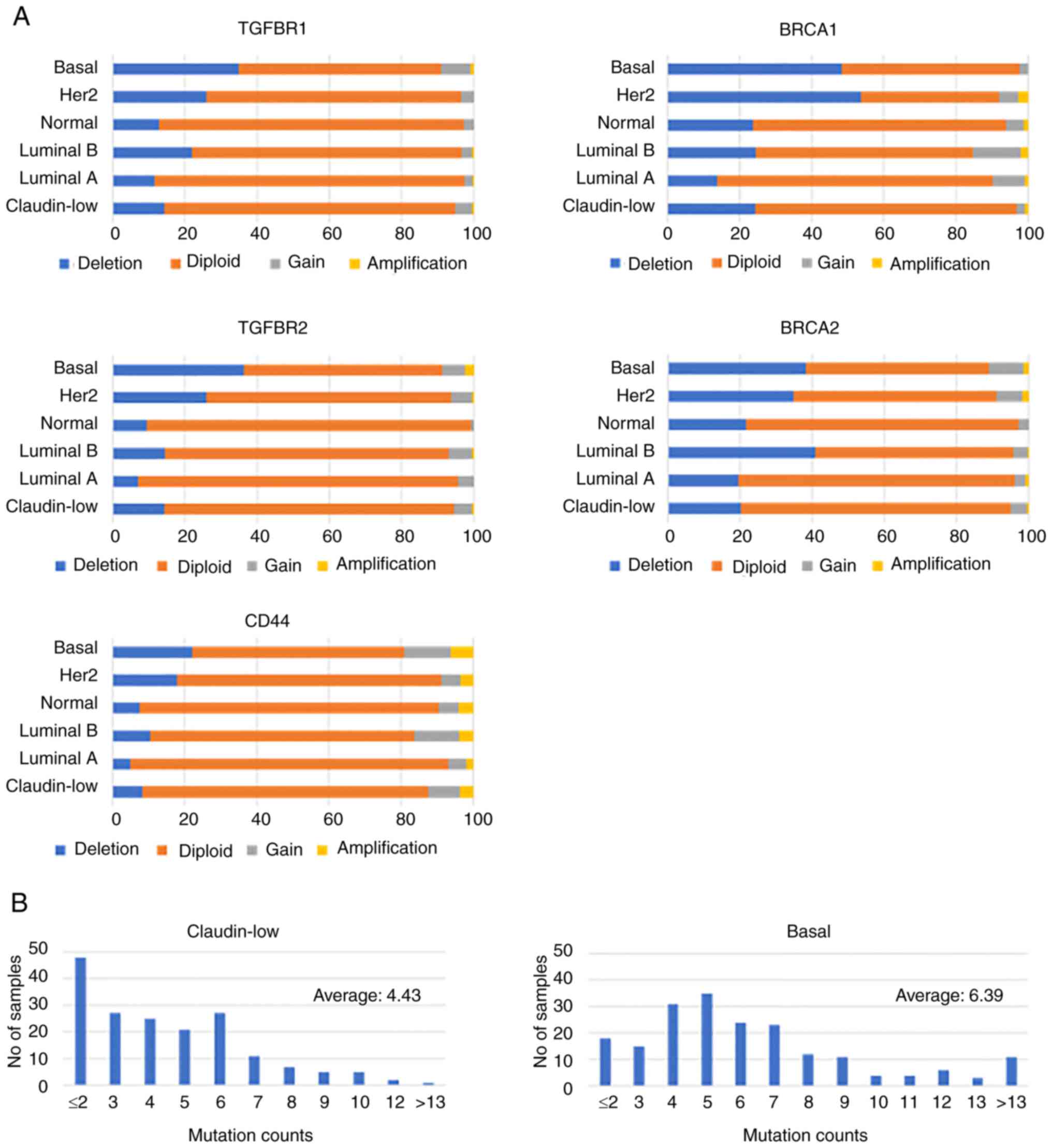
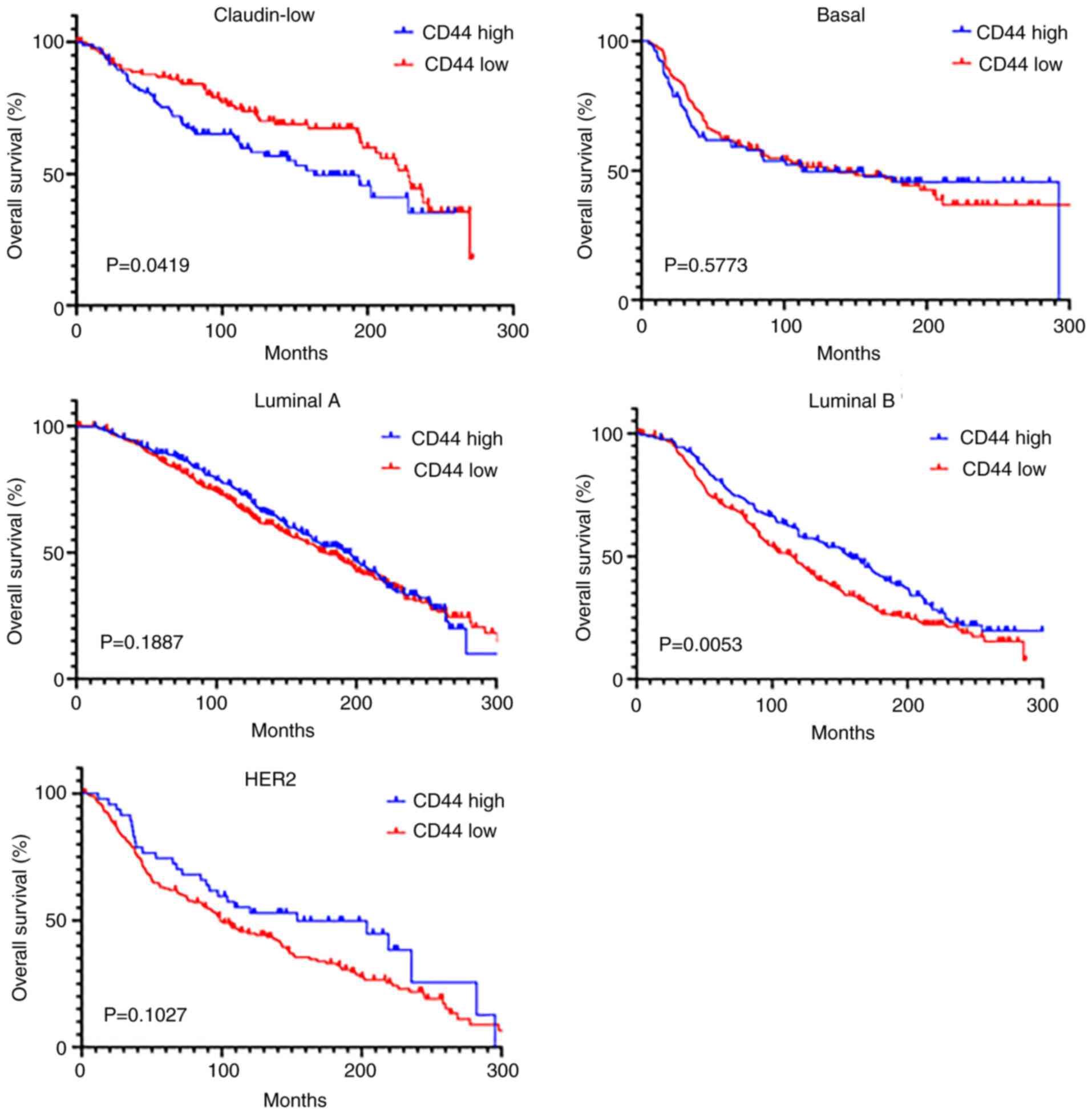
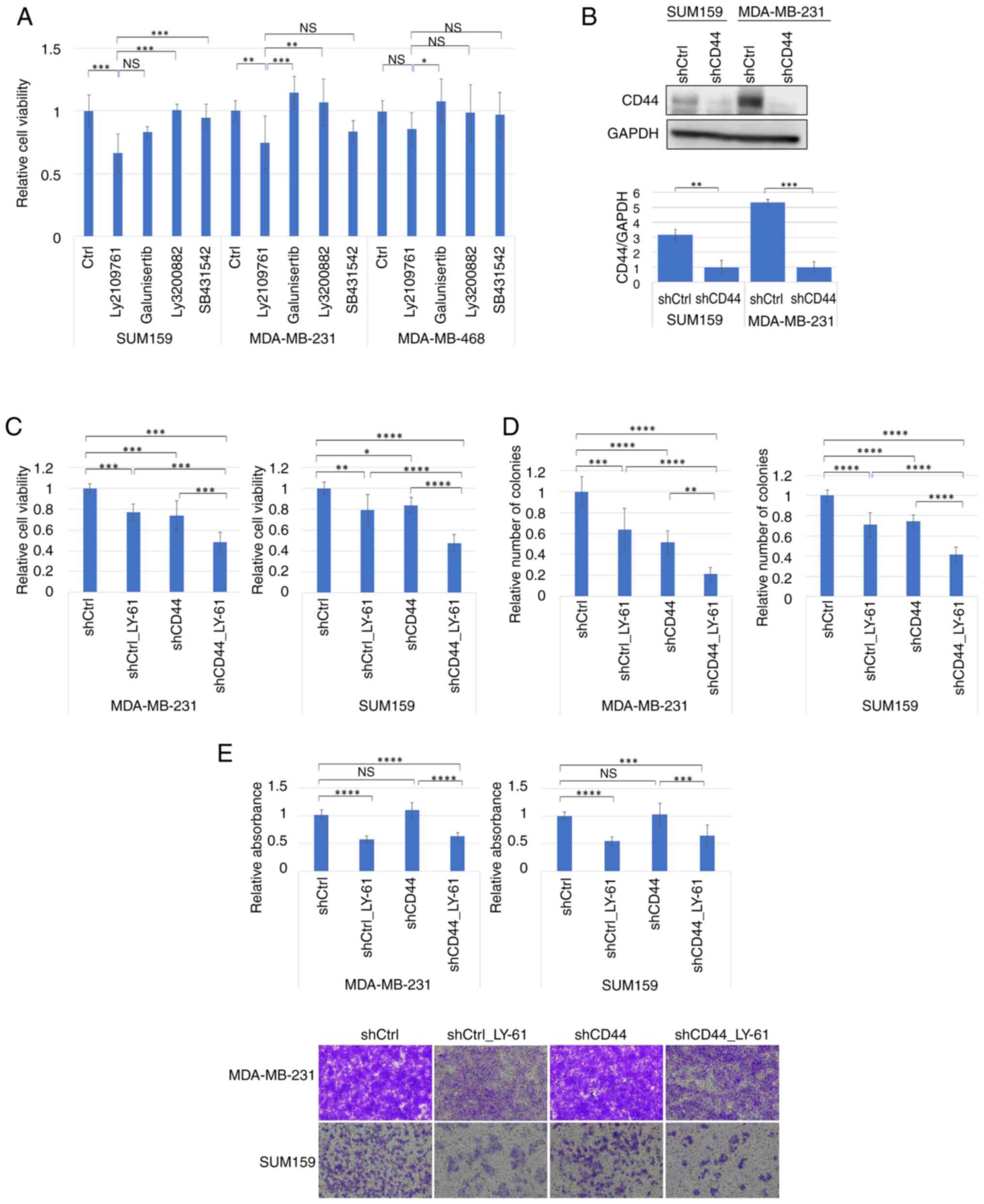
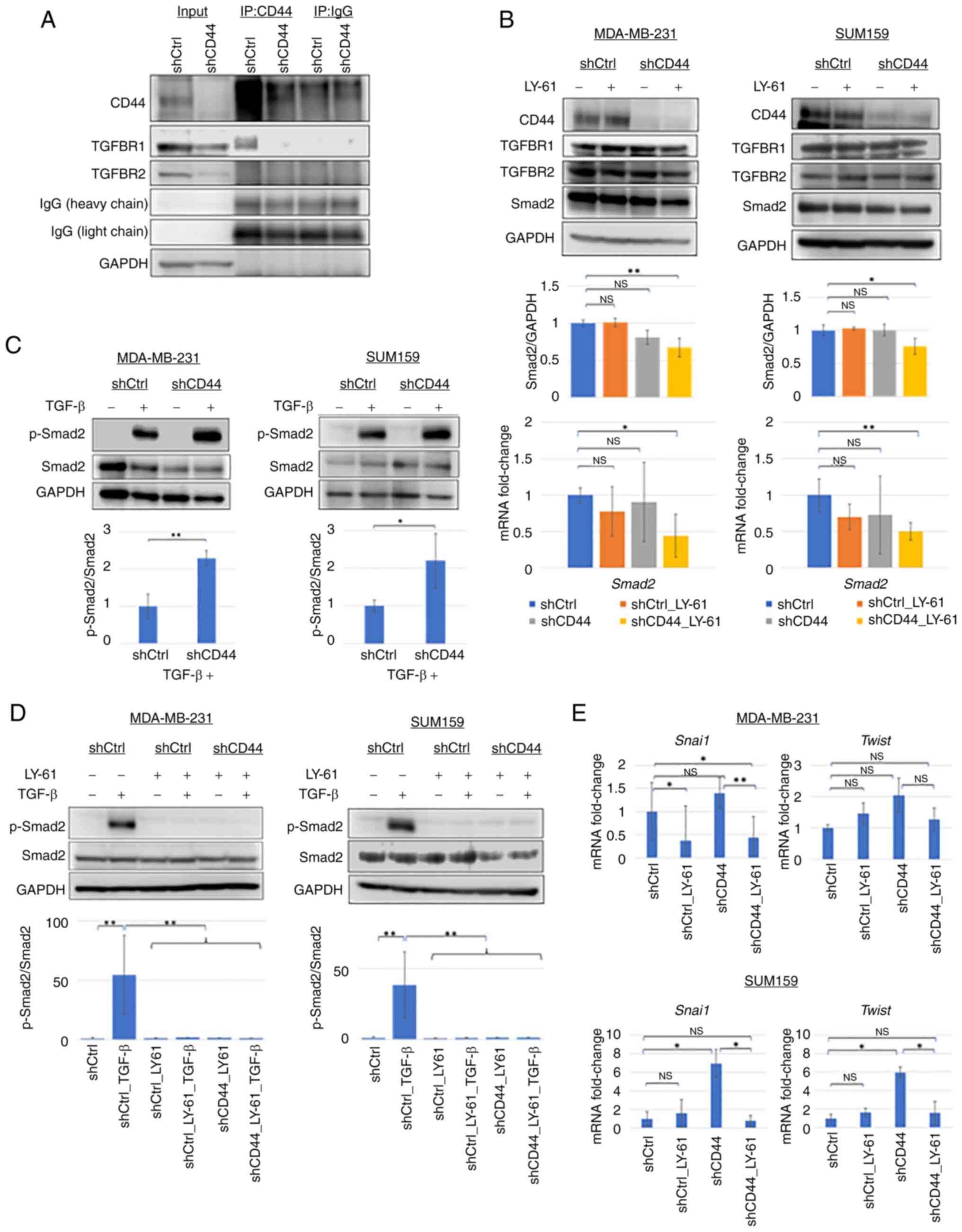
|
1
|
Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J
and Shi B: Breast cancer intrinsic subtype classification, clinical
use and future trends. Am J Cancer Res. 5:2929–2943.
2015.PubMed/NCBI
|
|
2
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xiong X, Zheng LW, Ding Y, Chen YF, Cai
YW, Wang LP, Huang L, Liu CC, Shao ZM and Yu KD: Breast cancer:
Pathogenesis and treatments. Signal Transduct Target Ther.
10:492025. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Herschkowitz JI, Simin K, Weigman VJ,
Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S,
Chandrasekharan S, et al: Identification of conserved gene
expression features between murine mammary carcinoma models and
human breast tumors. Genome Biol. 8:R762007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Prat A, Parker JS, Karginova O, Fan C,
Livasy C, Herschkowitz JI, He X and Perou CM: Phenotypic and
molecular characterization of the claudin-low intrinsic subtype of
breast cancer. Breast Cancer Res. 12:R682010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sabatier R, Finetti P, Guille A, Adelaide
J, Chaffanet M, Viens P, Birnbaum D and Bertucci F: Claudin-low
breast cancers: Clinical, pathological, molecular and prognostic
characterization. Mol Cancer. 13:2282014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fougner C, Bergholtz H, Norum JH and
Sørlie T: Re-definition of claudin-low as a breast cancer
phenotype. Nat Commun. 11:17872020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garrido-Castro AC, Lin NU and Polyak K:
Insights into molecular classifications of triple-negative breast
cancer: Improving patient selection for treatment. Cancer Discov.
9:176–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hassn Mesrati M, Syafruddin SE, Mohtar MA
and Syahir A: CD44: A multifunctional mediator of cancer
progression. Biomolecules. 11:18502021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim KJ, Godarova A, Seedle K, Kim MH, Ince
TA, Wells SI, Driscoll JJ and Godar S: Rb suppresses collective
invasion, circulation and metastasis of breast cancer cells in
CD44-dependent manner. PLoS One. 8:e805902013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dehbokri SG, Noorolyai S, Baghbani E,
Moghaddamneshat N, Javaheri T and Baradaran B: Effects of CD44
siRNA on inhibition, survival, and apoptosis of breast cancer cell
lines (MDA-MB-231 and 4T1). Mol Biol Rep. 51:6462024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nam K, Oh S, Lee KM, Yoo SA and Shin I:
CD44 regulates cell proliferation, migration, and invasion via
modulation of c-Src transcription in human breast cancer cells.
Cell Signal. 27:1882–1894. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang Z, Chen D, Nie J, Zhou S, Wang J,
Tang Q and Yang X: MicroRNA-143 targets CD44 to inhibit breast
cancer progression and stem cell-like properties. Mol Med Rep.
13:5193–5199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Montgomery N, Hill A, McFarlane S, Neisen
J, O'Grady A, Conlon S, Jirstrom K, Kay EW and Waugh DJ: CD44
enhances invasion of basal-like breast cancer cells by upregulating
serine protease and collagen-degrading enzymatic expression and
activity. Breast Cancer Res. 14:R842012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ouhtit A, Madani S, Gupta I,
Shanmuganathan S, Abdraboh ME, Al-Riyami H, Al-Farsi YM and Raj MH:
TGF-β2: A novel target of CD44-promoted breast cancer invasion. J
Cancer. 4:566–572. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu H, Niu M, Yuan X, Wu K and Liu A: CD44
as a tumor biomarker and therapeutic target. Exp Hematol Oncol.
9:362020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou L, Sheng D, Deng Q, Wang D and Liu S:
Development of a novel method for rapid cloning of shRNA vectors,
which successfully knocked down CD44 in mesenchymal triple-negative
breast cancer cells. Cancer Commun (Lond). 38:572018.PubMed/NCBI
|
|
19
|
Huang P, Chen A, He W, Li Z, Zhang G, Liu
Z, Liu G, Liu X, He S, Xiao G, et al: BMP-2 induces EMT and breast
cancer stemness through Rb and CD44. Cell Death Discov.
3:170392017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao F, Zhang G, Liu Y, He Y, Sheng Y, Sun
X, Du Y and Yang C: Activation of CD44 signaling in leader cells
induced by tumor-associated macrophages drives collective
detachment in luminal breast carcinomas. Cell Death Dis.
13:5402022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pan C, Xu A, Ma X, Yao Y, Zhao Y, Wang C
and Chen C: Research progress of Claudin-low breast cancer. Front
Oncol. 13:12261182023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pommier RM, Sanlaville A, Tonon L,
Kielbassa J, Thomas E, Ferrari A, Sertier AS, Hollande F, Martinez
P, Tissier A, et al: Comprehensive characterization of claudin-low
breast tumors reflects the impact of the cell-of-origin on cancer
evolution. Nature Commun. 11:34312020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gooding AJ and Schiemann WP:
Epithelial-mesenchymal transition programs and cancer stem cell
phenotypes: Mediators of breast cancer therapy resistance. Mol
Cancer Res. 18:1257–1270. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Akhurst RJ and Hata A: Targeting the TGFβ
signalling pathway in disease. Nat Revi Drug Discov. 11:790–811.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Matsunuma R, Chan DW, Kim BJ, Singh P, Han
A, Saltzman AB, Cheng C, Lei JT, Wang J, Roberto da Silva L, et al:
DPYSL3 modulates mitosis, migration, and epithelial-to-mesenchymal
transition in claudin-low breast cancer. Proc Natl Acad Sci USA.
115:E11978–E11987. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Porsch H, Mehić M, Olofsson B, Heldin P
and Heldin CH: Platelet-derived growth factor β-receptor,
transforming growth factor β type I receptor, and CD44 protein
modulate each other's signaling and stability. J Biol Chem.
289:19747–19757. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang K and Yi T: Tumor cell stemness in
gastrointestinal cancer: Regulation and targeted therapy. Front Mol
Biosci. 10:12976112023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen C, Zhao S, Karnad A and Freeman JW:
The biology and role of CD44 in cancer progression: Therapeutic
implications. J Hematol Oncol. 11:642018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Flanagan L, Van Weelden K, Ammerman C,
Ethier SP and Welsh J: SUM-159PT cells: A novel estrogen
independent human breast cancer model system. Breast Cancer Res
Treat. 58:193–204. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chan DW, Mody CH, Ting NS and Lees-Miller
SP: Purification and characterization of the double-stranded
DNA-activated protein kinase, DNA-PK, from human placenta. Biochem
Cell Biol. 74:67–73. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Voutsadakis IA: Molecular characteristics
and therapeutic vulnerabilities of claudin-low breast cancers
derived from cell line models. Cancer Genomics Proteomics.
20:5392023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Donovan J and Slingerland J: Transforming
growth factor-beta and breast cancer: Cell cycle arrest by
transforming growth factor-beta and its disruption in cancer.
Breast Cancer Res. 2:116–124. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang X, Eichhorn PJA and Thiery JP: TGF-β,
EMT, and resistance to anti-cancer treatment. Semin Cancer Biol.
97:1–11. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fougner C, Bergholtz H, Kuiper R, Norum JH
and Sørlie T: Claudin-low-like mouse mammary tumors show distinct
transcriptomic patterns uncoupled from genomic drivers. Breast
Cancer Res. 21:852019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Barcellos-Hoff MH and Gulley JL: Molecular
pathways and mechanisms of TGFβ in cancer therapy. Clin Cancer Res.
29:2025–2033. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kwei KA, Kung Y, Salari K, Holcomb IN and
Pollack JR: Genomic instability in breast cancer: Pathogenesis and
clinical implications. Mol Oncol. 4:255–266. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu H, Tian Y, Yuan X, Wu H, Liu Q, Pestell
RG and Wu K: The role of CD44 in epithelial-mesenchymal transition
and cancer development. Onco Targets Ther. 8:3783–3792.
2015.PubMed/NCBI
|
|
41
|
Li L, Qi L, Liang Z, Song W, Liu Y, Wang
Y, Sun B, Zhang B and Cao W: Transforming growth factor-β1 induces
EMT by the transactivation of epidermal growth factor signaling
through HA/CD44 in lung and breast cancer cells. Int J Mol Med.
36:113–122. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gao Y, Ruan B, Liu W, Wang J, Yang X,
Zhang Z, Li X, Duan J, Zhang F, Ding R, et al: Knockdown of CD44
inhibits the invasion and metastasis of hepatocellular carcinoma
both in vitro and in vivo by reversing epithelial-mesenchymal
transition. Oncotarget. 6:7828–7837. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Deng Z, Fan T, Xiao C, Tian H, Zheng Y, Li
C and He J: TGF-β signaling in health, disease and therapeutics.
Signal Transduct Target Ther. 9:612024. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chanmee T, Ontong P, Mochizuki N,
Kongtawelert P, Konno K and Itano N: Excessive hyaluronan
production promotes acquisition of cancer stem cell signatures
through the coordinated regulation of Twist and the transforming
growth factor β (TGF-β)-Snail signaling axis. J Biol Chem.
289:26038–26056. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang K, Liu X, Hao F, Dong A and Chen D:
Targeting TGF-β1 inhibits invasion of anaplastic thyroid carcinoma
cell through SMAD2-dependent S100A4-MMP-2/9 signalling. Am J Transl
Res. 8:2196–2209. 2016.PubMed/NCBI
|
|
46
|
Katsuno Y, Meyer DS, Zhang Z, Shokat KM,
Akhurst RJ, Miyazono K and Derynck R: Chronic TGF-β exposure drives
stabilized EMT, tumor stemness, and cancer drug resistance with
vulnerability to bitopic mTOR inhibition. Sci Signal.
12:eaau85442019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ouhtit A, Madani S, Gupta I,
Shanmuganathan S, Abdraboh ME, Al-Riyami H, Al-Farsi YM and Raj MH:
TGF-β2: A novel target of CD44-promoted breast cancer invasion. J
Cancer. 4:566–572. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Park NR, Cha JH, Jang JW, Bae SH, Jang B,
Kim JH, Hur W, Choi JY and Yoon SK: Synergistic effects of CD44 and
TGF-β1 through AKT/GSK-3β/β-catenin signaling during
epithelial-mesenchymal transition in liver cancer cells. Biochem
Biophys Res Commun. 477:568–574. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hao Y, Baker D and Ten Dijke P:
TGF-β-mediated epithelial-mesenchymal transition and cancer
metastasis. Int J Mol Sci. 20:27672019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Voutsadakis IA: EMT Features in
Claudin-Low versus Claudin-Non-Suppressed Breast Cancers and the
Role of Epigenetic Modifications. Curr Issues Mol Biol.
45:6040–6054. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Werden SJ, Sphyris N, Sarkar TR, Paranjape
AN, LaBaff AM, Taube JH, Hollier BG, Ramirez-Peña EQ, Soundararajan
R, den Hollander P, et al: Phosphorylation of serine 367 of FOXC2
by p38 regulates ZEB1 and breast cancer metastasis, without
impacting primary tumor growth. Oncogene. 35:5977–5988. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Taube JH, Herschkowitz JI, Komurov K, Zhou
AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW, et
al: Core epithelial-to-mesenchymal transition interactome
gene-expression signature is associated with claudin-low and
metaplastic breast cancer subtypes. Proc Natl Acad Sci USA.
107:15449–15454. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liang H, Benard O, Kumar V, Griffen A, Ren
Z, Sivalingam K, Wang J, de Simone Benito E, Zhang X, Zhang J, et
al: Wnt/ERK/CDK4/6 activation in the partial EMT state coordinates
mammary cancer stemness with self-renewal and inhibition of
differentiation. Br J Cancer. 133:986–1002. 2025. View Article : Google Scholar : PubMed/NCBI
|