As one of the most prevalent and deadly malignancies
worldwide, non-small cell lung cancer (NSCLC) accounts for >85%
of LC cases (1–3). NSCLC exhibits notable molecular
heterogeneity, with key driver gene mutations including epidermal
growth factor receptor (EGFR), kirsten ratsarcoma viral oncogene
homolog (KRAS) and anaplastic lymphoma kinase. These mutations play
a crucial role in determining treatment strategies and prognosis
for patients (4,5). Although targeted therapies and
immunotherapies have notably improved survival in some patients,
the 5-year survival rate for NSCLC remains <20%, particularly in
advanced-stage cases, where tumor heterogeneity, metastatic
potential and drug resistance pose notable therapeutic challenges
(6,7). Therefore, investigating the mechanisms
underlying NSCLC development and progression, as well as
identifying novel therapeutic targets, remains a key focus.
Members of the SOX family are evolutionarily
conserved transcription factors that regulate gene expression by
binding DNA, playing a central role in embryonic development, cell
fate determination and the maintenance of stem cell pluripotency
(8,9). Studies have revealed the abnormal
expression of SOX family members such as SOX2, SOX4 and SOX9 in
various solid tumors, including colorectal, breast and liver
cancers (10–12). These factors contribute to tumor
malignancy by regulating cancer stemness, epithelial-mesenchymal
transition (EMT), metabolic reprogramming and microenvironment
remodeling (13,14). Particularly in NSCLC, SOX proteins
exhibit dual regulatory functions: Certain members (SOX2) may
promote chemotherapy resistance by maintaining cancer stem cell
properties, whereas others (SOX17) serve as tumor suppressors by
inhibiting oncogenic signaling pathways (15). This functional diversity suggests
the SOX family forms a complex regulatory network in NSCLC, the
precise mechanisms of which require systematic elucidation.
However, expression patterns of different SOX members across NSCLC
subtypes and molecular classifications, as well as their clinical
significance, remain incompletely understood. Moreover, the
interplay between their downstream regulatory networks and
epigenetic modifications requires further investigation.
Furthermore, translating the molecular functions of SOX proteins
into clinical applications, such as their development as biomarkers
or therapeutic targets, remains a notable challenge.
With advancements in high-throughput sequencing
technologies, researchers can gain a more detailed understanding of
genomic alterations in SOX family members, offering insight into
the complexity of NSCLC. Single-cell RNA sequencing provides an
opportunity to analyze intratumoral heterogeneity more precisely,
which is key for identifying the specific functions and roles of
SOX family members (16). Notably,
intervention strategies targeting SOX family members are also
evolving. Techniques based on small interfering (si)RNA or
clustered regularly interspaced short palindromic
repeats/CRISPR-associated protein 9 (CRISPR/Cas9) have been
explored to suppress the expression or function of SOX proteins
with the aim of achieving therapeutic effects (17). These emerging therapeutic approaches
not only expand understanding of potential NSCLC treatments but
also offer new perspectives for future personalized medicine.
The present study aimed to summarize the expression
patterns, molecular mechanism and clinical relevance of SOX protein
in NSCLC, with emphasis on SOX2, SOX4 and SOX9 (implicated in
critical oncogenic processes, including cell proliferation,
epithelial-mesenchymal transition, stemness maintenance, and
chemoresistance (18–21), their role in tumor progression, stem
cells and drug resistance, as well as their potential as diagnostic
biomarkers and therapeutic targets. Comprehensive understanding of
the SOX family in NSCLC may drive the development of more
personalized and effective treatments, ultimately improving patient
outcomes and quality of life.
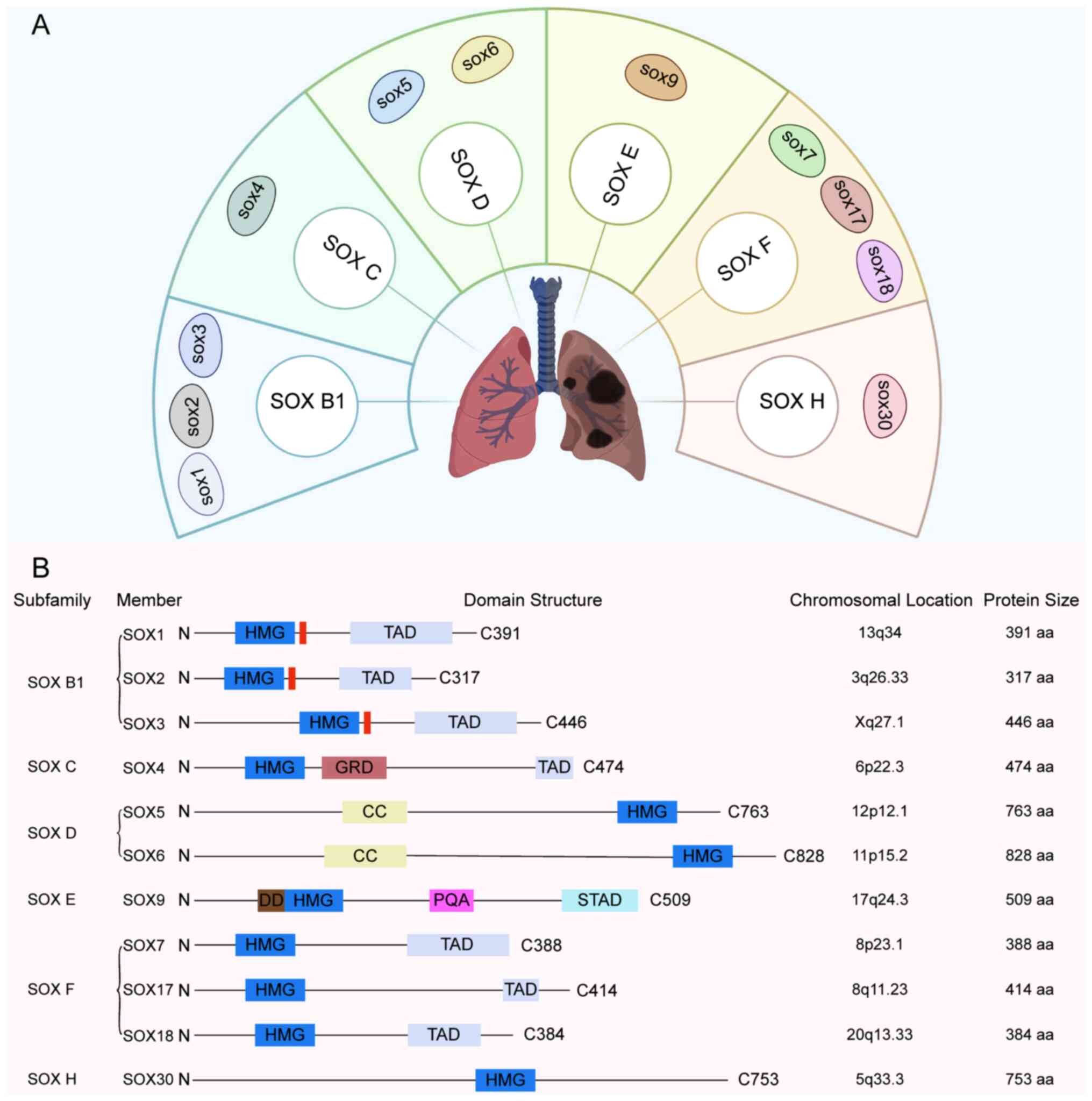
The SOX gene family encodes a series of
transcription factors characterized by a highly conserved domain
and is classified into ten subgroups (A-J) based on protein
homology (22). In LC research, the
subgroups most frequently involved include B1, C, D, E, F and H
(Fig. 1). These transcription
factors bind DNA through their high mobility group box domains and
regulate gene expression, thereby serving pivotal roles in
physiological processes such as embryonic development, cell fate
determination and differentiation. For example, SOX4, a member of
the C subgroup, serves as a key mediator of TNF-α-induced
transformation of fibroblast-like synoviocytes in the pathological
progression of arthritis. It exerts its regulatory effects by
interacting with the transcription factor v-rel
reticuloendotheliosis viral oncogene homolog A/p65) in the NF-κB
signaling pathway, thereby cooperatively modulating the expression
of downstream genes under TNF signaling (23). Similarly, SOX3, a member of the
SOXB1 subfamily, is broadly expressed during embryogenesis,
neurogenesis and gonadal development in Misgurnus (loach),
and this expression pattern is conserved throughout vertebrate
evolution (24). In addition, the
expression of multiple SOX genes is observed in stem,
undifferentiated progenitor and differentiated cells with
neuro-sensory characteristics in cnidarians, further supporting the
ancient evolutionary conservation of the SOX gene family in
developmental biology (22).
Different SOX subgroups demonstrate functional specificity. For
example, SOX5 and SOX6 from the SOXD subgroup are involved in
transcriptional regulation during embryonic development, neural
growth and chondrogenesis (25). A
member of the SOXE subgroup, SOX9, serves a key role in sex
determination and gonadal development in various species (26). Notably, the majority of SOX family
members exhibit aberrant expression in NSCLC, influenced by
mechanisms including gene mutations, DNA methylation and regulation
by microRNAs (miRs; Table I)
(27). These dysregulated SOX genes
have notable effects on the initiation and progression of NSCLC.
Elucidating the functions and regulatory mechanisms of these SOX
genes will not only deepen understanding of NSCLC pathogenesis but
may provide potential targets for the development of novel targeted
therapeutic strategies.
SOX4 plays a pivotal role in the initiation and
progression of NSCLC. Studies have demonstrated that SOX4 is
significantly upregulated in NSCLC tissue and serves as an
independent prognostic marker, underlining its key role in
malignant tumor progression (33,34).
At the molecular level, SOX4 promotes NSCLC cell migration,
invasion and EMT by upregulating B cell-specific MLV integration
site-1 (BMI1) expression (35).
BMI1 induces the ubiquitination of histone H2A, which suppresses
the expression of zinc finger protein 24 and decreases the
secretion of vascular endothelial growth factor A, thereby
promoting tumor angiogenesis and providing support for tumor cell
proliferation and metastasis. Moreover, SOX4 regulates cell
processes, including proliferation, survival and migration of NSCLC
cells, through interactions with signaling pathways such as
PI3K/AKT (36,37). Furthermore, the hypoxia-sensitive
lncRNA CASC15 can promote the occurrence of LC by regulating the
SOX4/β-catenin axis. Under hypoxic conditions, CASC15 transcription
is activated, promoting the expression of SOX4, stabilizing the
β-catenin protein and ultimately enhancing NSCLC cell proliferation
and migration abilities (38).
Meanwhile, circular (circ)RNA circ_0020714 serves as an endogenous
miR-30a-5p sponge, enhancing the expression of SOX4 and promoting
immune escape and anti-PD-1 resistance in patients with NSCLC
(39). These findings highlight the
multifaceted role of SOX4 in NSCLC pathogenesis and progression,
making it a potential target for therapeutic interventions.
EGFR and KRAS mutations are common oncogenic drivers
in lung adenocarcinoma and serve a crucial role in tumorigenesis
(40). SOX9 expression is elevated
in lung adenocarcinoma, particularly those with KRAS mutations, and
mediates Notch1-induced EMT. A previous study demonstrated higher
SOX9 mRNA in KRAS-compared with EGFR-mutant tumors (41). This suggests SOX9 acts downstream of
Notch in KRAS-driven NSCLC, promoting invasion and metastasis, with
potential indirect antagonism to EGFR signaling due to mutual
exclusivity of KRAS/EGFR mutations (41). Furthermore, SOX9 is essential for
KRAS-driven lung adenocarcinoma progression. Loss of SOX9 decreases
tumor burden, extends survival and enhances anti-tumor immunity by
increasing levels of tumor-infiltrating dendritic cells and
CD8+ T cells (42).
Although direct evidence regarding the interaction between SOX9 and
the EGFR/KRAS pathways in lung adenocarcinoma remains limited,
studies in other types of cancer suggest that SOX9 may regulate the
biological behavior of lung adenocarcinoma cells through crosstalk
with these signaling pathways (42–44).
For example, aberrant activation of EGFR or KRAS may regulate SOX9
expression or activity via downstream signaling cascades, thereby
affecting cell processes such as proliferation, differentiation,
migration and invasion (42–44).
Additionally, in colorectal cancer, SOX9 activates the canonical
Wnt/β-catenin pathway by promoting β-catenin stability, nuclear
translocation, and transcription of Wnt components like FZD7 and
LRP6, which drives tumor growth, metastasis, and stem cell-like
properties (45,46). In pancreatic cancer, the oncogene
KRAS induces the expression of SOX9 at both the mRNA and protein
levels, including its phosphorylated form, thereby promoting SOX9
nuclear translocation and transcriptional activity (47,48).
The transforming growth factor-β-activated kinase 1/IκBα/NF-κB
signaling pathway is involved in the regulation of SOX9 by KRAS. In
addition, SOX9 modulates the expression of mediator of DNA damage
checkpoint 1and minichromosome maintenance complex components,
which are associated with tumor cell proliferation, invasion and
metastasis (47). Furthermore, in
the context of KRAS mutations, ectopic expression of SOX9 in acinar
cells synergizes with oncogenic KRAS to markedly accelerate the
formation of precancerous lesions (48). In urothelial carcinoma, the
activation of EGFR can upregulate the expression of SOX9 via the
ERK signaling pathway, thereby promoting tumor occurrence; the
EGFR-ERK-SOX9 signaling cascade mechanism suggests that a similar
regulatory pathway may also exist in EGFR-mutated NSCLC (44). Elucidating the mechanistic interplay
between SOX9 and the EGFR/KRAS pathways in lung adenocarcinoma
holds promise for identifying novel therapeutic targets and
developing precision medicine strategies.
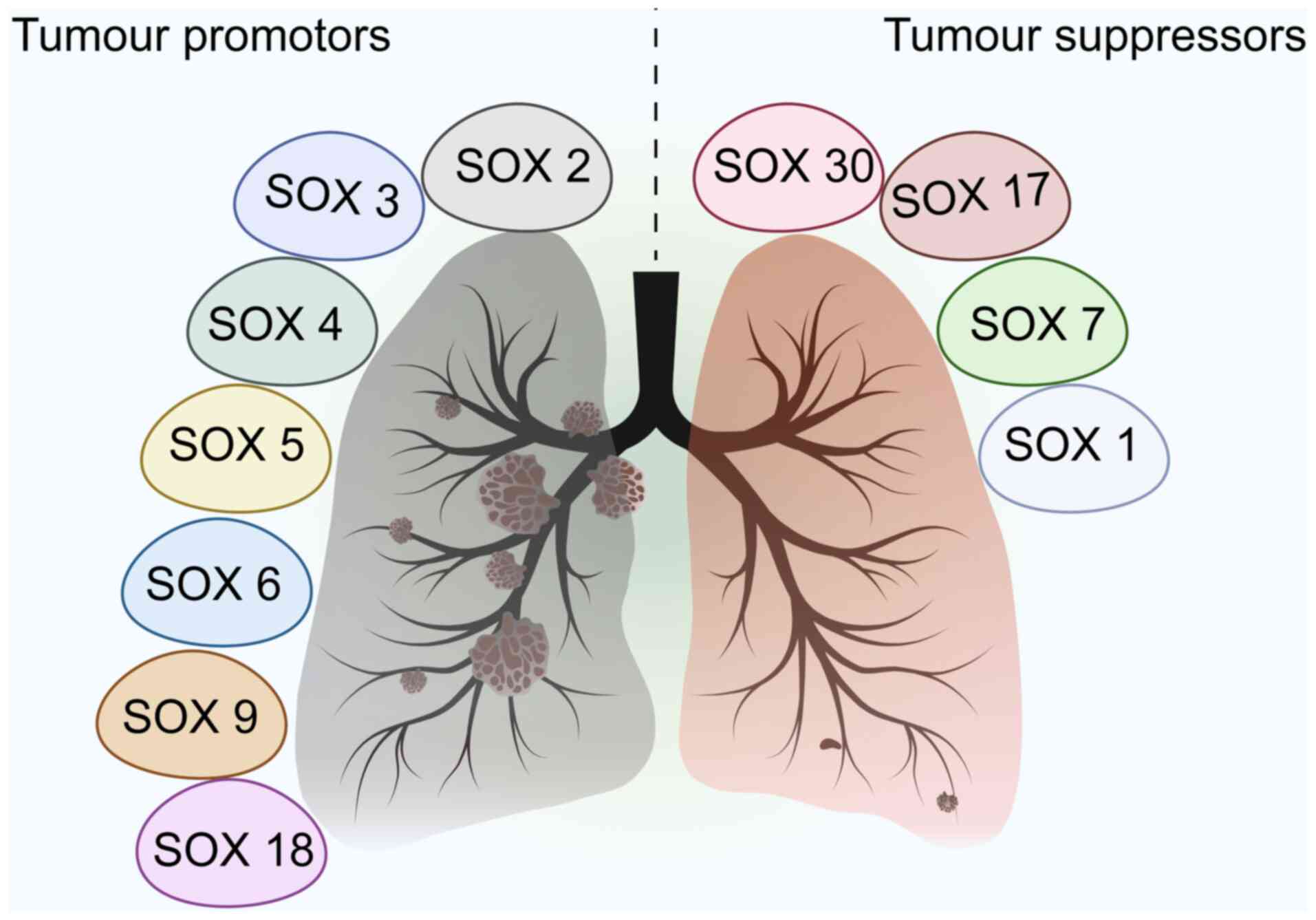
In addition to SOX2, SOX4, and SOX9, other members
of the SOX family regulate the initiation and progression of NSCLC
(Fig. 2). For example, SOX5 is
highly expressed in NSCLC and may serve as a supportive prognostic
marker for the diagnosis of NSCLC (49,50).
In a cohort of 90 patients with lung adenocarcinoma,
immunohistochemical analysis revealed elevated SOX5 expression in
tumor compared with adjacent non-tumor tissues. High SOX5 levels
were significantly associated with advanced clinical stage, lymph
node metastasis and reduced overall survival. Multivariate Cox
regression confirmed SOX5 as an independent prognostic factor for
poor survival (51). Functional
assays show SOX5 promotes EMT, enhancing invasion and migration,
while knockdown inhibits these processes in vitro and in
vivo (52). SOX5 is also
aberrantly upregulated in NSCLC cell lines, where it promotes
proliferation, migration, invasion and EMT via interaction with
YAP1. Knockdown of SOX5 suppresses tumor growth and metastasis, and
ncRNAs such as miR-143-3p that target SOX5 inhibit cancer
progression, underscoring its prognostic significance (51,53).
Additionally, elevated SOX18 expression is associated with poor
prognosis, and knockdown of SOX18 notably impairs cellular
migratory capacity (54,55). In a cohort of 198 NSCLC cases, SOX18
was expressed in the nuclei and cytoplasm of cancer cells in 94.4
and 47% of the NSCLC cases, respectively (54). SOX18 mRNA levels are lower in NSCLC
than in non-malignant lung tissue, but protein levels are higher.
Cytoplasmic SOX18 expression is associated with poor patient
outcome, and nuclear SOX18 is positively associated with Ki-67
proliferation index, suggesting its role in tumor progression and
potential as a prognostic biomarker (54). Although the specific role of SOX3 in
NSCLC remains largely unexplored, aberrant expression of SOX3 has
been reported in other malignancies such as osteosarcoma and breast
cancer (56,57). In these contexts, SOX3 dysregulation
is implicated in tumor development and progression, potentially via
its influence on apoptosis, migration and proliferation (56,58,59).
To elucidate its potential role in NSCLC, machine learning
approaches may be employed to integrate multi-omics datasets (such
as The Cancer Genome Atlas/Gene Expression Omnibus) for predicting
SOX3 interactions with pathways such as Wnt or EGFR. Furthermore,
artificial intelligence-driven clustering of SOX expression with
tumor mutational burden (TMB) and immune signatures may help
identify resistance patterns or novel biomarkers, while
bioinformatics analysis of SOX3 methylation may demonstrate
diagnostic value (60,61). Furthermore, SOX7 and SOX17 are
notably associated with prognosis in lung adenocarcinoma (60,62,63).
Patients with high expression of SOX7 and SOX17 exhibit better
overall survival, suggesting these genes may serve as potential
prognostic biomarkers (60,62,63).
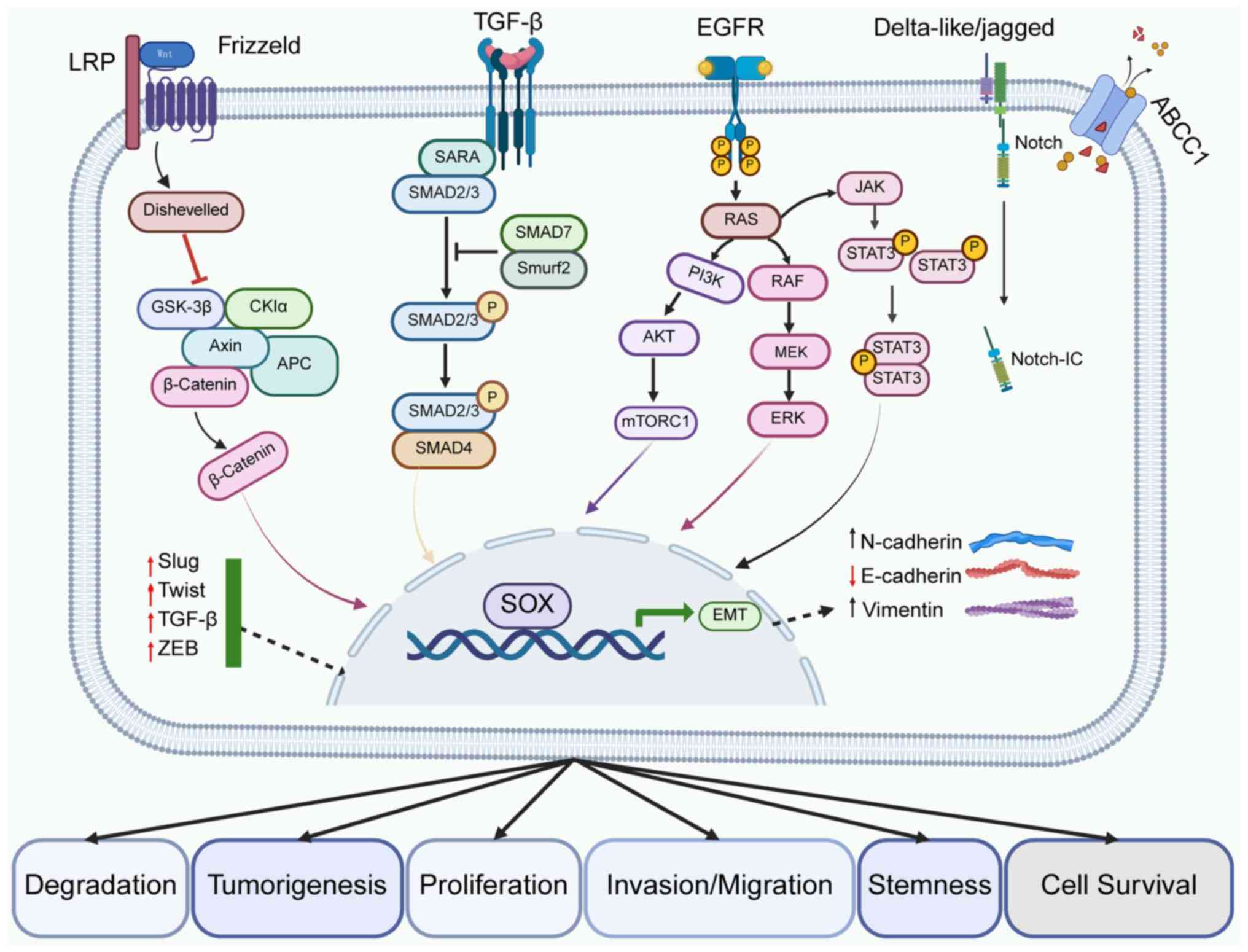
The SOX family is involved and development of NSCLC
by regulating biological behaviors, including cell proliferation,
apoptosis, invasion and metastasis, as well as the maintenance of
tumor stem cells (Fig. 3).
Studies have demonstrated that certain SOX members
actively promote NSCLC cell proliferation (35,63,64).
For example, SOX2 facilitates cell proliferation and survival by
regulating cell cycle progression and DNA damage repair (64). In models of radioresistant NSCLC
cell lines, SOX2 expression is markedly upregulated (65–67).
Overexpression of SOX2 enhances resistance to radiotherapy and
improves DNA repair capacity, thereby promoting proliferation
(68). Conversely, SOX2 knockdown
impairs these functions and suppresses cell proliferation (69). Similarly, SOX4 supports NSCLC cell
proliferation indirectly by promoting migration, invasion and EMT
(21). Analyses of patients with
NSCLC has revealed significantly elevated SOX4 expression in tumor
tissue, identifying it as an independent prognostic marker
(35). However, not all SOX family
members serve as oncogenes. For example, SOX17 is downregulated in
lung adenocarcinoma, and its upregulation suppresses NSCLC cell
proliferation, suggesting a potential tumor-suppressive function
(63).
The SOX family serves a key role in the regulation
of apoptosis in NSCLC cells, constituting an essential component of
its involvement in tumor pathophysiology (70). SOX6 not only inhibits the
proliferation of lung adenocarcinoma cells but also promotes
apoptosis by modulating the expression of key proteins such as p53,
cyclin-dependent kinase inhibitor 1A, cyclin D1 and β-catenin,
thereby impacting cell cycle control and apoptosis-associated
signaling pathways (71). In
studies of erlotinib resistance in NSCLC cells, OTU
domain-containing 1 (OTUD1) enhances cellular sensitivity to
erlotinib by inhibiting YAP1 nuclear translocation, accompanied by
the inactivation of the SOX9/secreted phosphoprotein 1 pathway
(72,73). Notably, overexpression of SOX9
reverses the sensitizing effects of OTUD1, indicating a role for
SOX9 in apoptosis and drug resistance mechanisms in NSCLC (72). In addition, SOX9, together with
STAT3, forms a differential regulatory network that may contribute
to erlotinib resistance by modulating cellular proliferation and
survival signaling pathways (73).
Furthermore, SOX-associated signaling pathways are also involved in
the regulation of apoptosis. For example, in endometrial cancer
cells, propofol inhibits proliferation, migration and invasion, and
promotes apoptosis by downregulating SOX4 expression, which is
associated with inactivation of the Wnt/β-catenin signaling
pathway, indicating that the Wnt/β-catenin-SOX4 axis may be involve
in the apoptosis of NSCLC cells (74). Furthermore, SOX4 affects the
proliferation and apoptosis of NSCLC cells by regulating cell
cycle-associated proteins (33).
For example, SOX4 may alter the expression of proteins such as
Cyclin D1, promoting the transition of cells from the G1 to the S
phase, thereby promoting cell proliferation. By regulating the
expression of apoptosis-associated proteins such as Bax and Bcl-2,
SOX4 affects the occurrence of apoptosis (33). circ_0089823 affects cell
proliferation and apoptosis by regulating SOX4. Knockdown of
circ_0089823 inhibits the proliferation of NSCLC cells, induces
cell cycle arrest and apoptosis (75). Overexpression of circ_0089823
promotes malignant behavior such as cell proliferation, and SOX4 is
positively regulated by circ_0089823. Silencing SOX4 can counteract
the effect of overexpression of circ_0089823 on NSCLC cells
(75).
In the invasion and metastasis of NSCLC, multiple
signaling pathways and molecular mechanisms are associated with the
SOX family (76). For example, in
LC, SOX1 expression is markedly downregulated, which contributes to
tumor initiation and progression. SOX1 suppresses the malignant
progression of NSCLC by inhibiting the hairy and enhancer of split
1 factor, thereby suppressing anchorage-independent growth,
invasion and metastatic behavior (77). Furthermore, SOX4 promotes tumor
invasion and metastasis by upregulating LEM domain containing 1,
which activates the PI3K/Akt signaling pathway in colorectal
cancer, implying that SOX4 may facilitate NSCLC progression through
a similar mechanism (78). In
addition, miR-363-3p can inhibit the migration, invasion and EMT of
NSCLC cells by targeting SOX4. The overexpression of miR-363-3p
inhibits cell migration and invasion, while knockdown of miR-363-3p
shows the opposite effect (79).
Further studies have found that miR-363-3p directly binds the
3′-untranslated region of SOX4 and negatively regulates its
expression, and neural precursor cell expressed developmentally
downregulated 9 or SOX4 knockdown can salvage the
translocation-promoting effect of antagomiR-363-3p (80–82).
In lung adenocarcinoma, SOX30 inhibits Wnt signaling by directly
suppressing the transcription of β-catenin, thereby blocking cell
migration and invasion. High SOX30 expression is associated with
better patient prognosis (76,83).
Additionally, interactions between SOX genes and other molecular
regulators influence NSCLC metastasis (76). For example, elevated expression of
lncRNA KCNQ1OT1 is notably associated with tumor size, TNM stage
and lymph node metastasis in NSCLC. This lncRNA promotes
proliferation, migration and invasion via the miR-129-5p/Jagged1
pathway. While SOX genes are not directly involved in this pathway,
the associated signaling networks suggest potential regulatory
interplay (84). Moreover, miR-548l
suppresses NSCLC cell migration and invasion by targeting the AKT1
signaling pathway, indicating SOX genes may participate in
coordinated regulation of invasion and metastasis through crosstalk
with such pathways (85).
The SOX family serves a notable role in the
development of drug resistance in NSCLC (18,90).
For example, SOX2 is associated with resistance to paclitaxel and
platinum-based chemotherapeutics. In studies using A549 NSCLC cell
lines (68,91), SOX2 enhances resistance to
paclitaxel by promoting the transcription of chloride voltage-gated
channel 3 (ClC-3). Knockdown of either SOX2 or ClC-3 significantly
decreases drug resistance (68).
SOX2 overexpression attenuates the Wnt/β-catenin signaling activity
in both lung adenocarcinoma A549 cells and cisplatin-resistant
counterpart A549/DDP cells through upregulation of GSK3β, a key
negative regulator of this pathway (91). Additionally, SOX2 modulates
resistance to cisplatin through the AP endonuclease 1 (APE1)
signaling pathway, and silencing SOX2 restores cisplatin
sensitivity (69). Other SOX family
members are also implicated in resistance mechanisms. For example,
upregulation of SOX4 is associated with chemoresistance in NSCLC
(92,93). miR-129-2 enhances chemosensitivity
by targeting SOX4 and inducing apoptosis, indicating modulation of
SOX4 levels influences drug response (92). In-depth exploration of the molecular
mechanisms underlying SOX-mediated resistance in NSCLC may provide
valuable insights for the development of novel therapeutic
strategies.
Given the critical role of the SOX family in NSCLC
development, individualized therapies targeting these genes hold
promise. Analyzing the expression profiles, mutation status and
associated signaling pathways of SOX genes in tumors enables
precise patient stratification, providing a basis for tailoring
individualized treatment strategies. For example, in patients with
high SOX gene expression associated with poor prognosis, targeted
interventions [such as small-molecule inhibitors, RNA interference
(RNAi) or immunotherapy-based combination strategies] can be
developed or selected to suppress SOX gene function. Integrating
these findings with clinical features and molecular markers may
further refine treatment regimens and enhance therapeutic efficacy
(17). Moreover, advancements in
drug development technologies have facilitated the creation of
SOX-specific targeted therapies. These drugs precisely target SOX
genes or their related signaling pathways, minimizing toxicity to
normal cells while improving treatment safety and effectiveness
(94,95). Additionally, integrating genetic
backgrounds and lifestyle factors contributes to more precise
individualized therapies, improving NSCLC prognosis.
Targeting SOX genes represents a promising
therapeutic strategy for NSCLC, aiming to suppress tumor growth by
modulating SOX gene expression or activity. RNAi has been employed
to silence SOX gene expression, altering the biological behavior of
NSCLC cells. For example, in cisplatin-resistant NSCLC cell lines,
SOX2 upregulation promotes resistance via APE1 signaling. Small
interfering (si)RNA-mediated SOX2 knockdown (siSOX2) inhibits
colony formation, decreases cell viability, enhances apoptosis and
restores cisplatin sensitivity. Combined siSOX2 and cisplatin
treatment inhibits tumor progression in vitro, with low SOX2
expression linked to better patient survival (69). Silencing SOX4 has been shown to
inhibit proliferation, migration and invasion of osteosarcoma
cells, while also inducing apoptosis (96). These findings suggest that targeting
SOX4 may similarly regulate progression of NSCLC (96). Additionally, several novel
small-molecule compounds have been developed to target SOX
proteins: Nobiletin, a small molecule, binds to the transcription
factor SOX5, exhibiting synergistic cytotoxic effects when combined
with doxorubicin in SOX5-overexpressing cells (97). This highlights a potential
combination therapy approach for NSCLC. Furthermore, the
development of targeted drugs based on signaling pathways involving
SOX genes has become a focal point of research (98–100).
In NSCLC, histone deacetylase 7 (HDAC7) promotes tumor
proliferation and metastasis by activating the β-catenin/FGF18
pathway, suggesting that targeting HDAC7 or its associated
signaling cascades may serve as a novel therapeutic strategy
(98). Moreover, CRISPR/Cas9
gene-editing technology offers a tool to modify SOX genes (101). Although this approach currently
faces technical challenges (off-target effects, delivery
challenges, editing efficiency) (102,103) for clinical translation, it holds
promise for personalized cancer therapies. To the best of our
knowledge, no clinical trials involving siRNA therapy targeting SOX
genes have been reported to date. The majority of research remains
at the preclinical stage (69,96,97),
encompassing in vitro studies, animal models and the
optimization of small molecule delivery systems. This delay in
clinical translation may be attributed to challenges associated
with siRNA delivery, including issues of stability, targeting
efficiency and off-target effects (104,105). Although clinical research on siRNA
therapies targeting SOX genes in LC remains in its early stages,
siRNA technology has demonstrated notable clinical progress across
various oncological indications, providing robust support for its
potential application in malignancy. For example, NBF-006, a lipid
nanoparticle-formulated siRNA targeting the KRAS G12D mutation
(present in ~25% of patients with NSCLC), has been evaluated in a
Phase I clinical trial (106) for
KRAS-mutant NSCLC and pancreatic and colorectal cancer. This trial
has demonstrated sustained KRAS silencing within tumors and
improved safety and tolerability compared with small-molecule
inhibitors (106). In glioblastoma
multiforme (GBM), NU-0129, a gold nanoparticle-conjugated siRNA
targeting bcl-2-like protein 12, an anti-apoptotic factor
upregulated in GBM, achieves localized tumor delivery via
convection-enhanced delivery. In patients with recurrent GBM, it
induces tumor cell apoptosis without evidence of neurotoxicity,
progressing to a Phase Ib expansion study (107). Additionally, CALAA-01, a
cyclodextrin-based nanoparticle delivery system to target the M2
subunit of ribonucleotide reductase, was investigated in a Phase I
trial (108) across various solid
tumors, including ovarian and peritoneal cancer. Tumor biopsy
confirmed activation of the RNAi mechanism, with clinical outcomes
indicating partial responses in 11% of patients and disease
stabilization in 48% (108,109).
Collectively, these early-phase clinical studies validate the
feasibility and therapeutic potential of siRNA-based approaches in
multiple types of cancer. They also provide a key foundation for
the development of novel RNAi strategies targeting the SOX pathway,
offering promise for advancing precision therapeutics in NSCLC and
optimization and innovation in its treatment paradigm.
Immunotherapy harnesses the host immune system to
target tumor cells, and SOX genes may play a pivotal regulatory
role within the tumor immune microenvironment. The integration of
SOX family-targeted therapies with immunotherapy offers new
possibilities for the treatment of NSCLC. Expression of certain SOX
genes is notably associated with the quantity and type of
tumor-infiltrating immune cells, suggesting their potential
influence on immunotherapy efficacy (39,110).
For example, expression of SOXF family genes is positively
associated with CD4+ T cell infiltration, a
characteristic that may modulate tumor response to immunotherapy
(60). In NSCLC, SOX2 upregulates
IL6 via FOS-like antigen 2, promoting inflammation and metastasis
while suppressing CD8+ T cell infiltration via cyclic
GMP-AMP synthase/STING degradation (111). SOX2/SOX9 enable natural killer
cell evasion by downregulating major histocompatibility complex
class I markers in cancer cells. SOX expression is associated with
checkpoints and TMB, predicting immunotherapy response. Targeting
SOX may enhance PD-1 blockade and radioimmunotherapy (112). Furthermore, targeting SOX genes
can enhance the effectiveness of immunotherapy in NSCLC. For
example, the use of proteolysis-targeting chimeras (PROTACs) to
degrade EGFR L858R has been shown to downregulate PD-L1 and
indoleamine 2,3-dioxygenase 1 protein levels, thereby amplifying
anti-tumor immune responses and providing an approach to NSCLC
immunotherapy (113).
Additionally, combining immune checkpoint inhibitors with
modulators targeting SOX gene-associated signaling pathways in
NSCLC immunotherapy has also shown enhanced efficacy (114). For instance, tumor-intrinsic SOX2
signaling in NSCLC promotes the recruitment of regulatory T cells
(Tregs) by upregulating CCL2, thereby mediating resistance to ICIs.
Depletion of Tregs or inhibition of the SOX2 pathway restores T
cell infiltration and markedly suppresses tumor growth, suggesting
that targeting the SOX2 pathway may synergistically enhance the
therapeutic efficacy of ICIs (110). This strategy may optimize the
tumor immune microenvironment and improve therapeutic outcomes,
warranting further exploration and validation.
NSCLC, the predominant subtype of LC, poses notable
clinical challenges due to its molecular heterogeneity, drug
resistance and low survival rate (115,116). The present review summarized the
key role of the SOX family of transcription factors in the
initiation, progression and treatment of NSCLC. Key members such as
SOX2, SOX4 and SOX9 notably influence the malignant progression of
NSCLC by regulating processes including tumor stemness, EMT, cell
proliferation, apoptosis, invasion, metastasis and drug resistance
(35,89,117).
For example, SOX2 sustains tumor stemness to promote chemotherapy
resistance, SOX4 modulates EMT and angiogenesis and SOX9
collaborates with EGFR/KRAS signaling pathways to drive tumor
invasion. Conversely, other SOX family members, such as SOX17,
exhibit tumor-suppressive potential, underscoring the dual
functionality within the SOX family (63).
Molecular mechanistic studies have revealed that SOX
genes participate in NSCLC pathogenesis through intricate signaling
networks (such as Wnt/β-catenin and PI3K/AKT) and epigenetic
regulation (118,119). Clinically, dysregulated SOX
expression is associated with patient outcomes, highlighting the
potential of SOX family members as diagnostic biomarkers and
therapeutic targets (76,120). In parallel, certain emerging
technologies (such as single-cell sequencing and CRISPR/Cas9)
provide novel tools to determine the roles of the SOX family, while
the development of SOX-targeted therapy (including RNAi and
PROTACs) and their integration with immunotherapy offer new avenues
for precision NSCLC therapy. However, clinical translatability
remains a challenge. siRNA-based RNAi, for example, excels in
sequence-specific SOX silencing, with preclinical NSCLC models
showing decreased tumor growth and stemness following SOX2 or SOX9
knockdown (69,121,122). However, its limitations include
poor in vivo stability (rapid nuclease degradation and renal
clearance, often limiting half-life to <24 h), inefficient cell
uptake due to negative charge, endosomal entrapment and potential
off-target effects or immune activation in the fibrotic NSCLC tumor
microenvironment (123–125). PROTACs induce sustained
ubiquitin-mediated degradation of SOX proteins, targeting
undruggable surfaces without relying on enzymatic pockets,
potentially overcoming compensatory upregulation seen in SOX family
members. In NSCLC, PROTACs have shown promise against transcription
factors such as STAT3 analogs, but face hurdles in E3 ligase
selectivity (risking off-target toxicity) and dependency on
endogenous ubiquitin machinery (126,127). CRISPR/Cas9 offers permanent
genomic editing of SOX loci, bypassing delivery instability by
enabling knockout or base editing in preclinical patient-derived
tumor xenograft models (128–130), where SOX9 ablation halts
metastasis more durably than siRNA. However, its application is
constrained by vector-associated immunogenicity, potential
off-target genomic alterations and ethical and regulatory barriers
(17,131,132).
Taken together, the present findings highlight the
promise and the limitations of SOX-targeted strategies in NSCLC.
Future studies should focus on optimizing delivery platforms,
enhancing therapeutic specificity and integrating SOX-based
interventions with existing targeted therapies and immunotherapies.
Addressing these translational barriers may unlock the clinical
potential of SOX-targeted approaches to develop more personalized
and effective treatment strategies for NSCLC.
Not applicable.
The present study was supported by the National Natural Science
Foundation of China (grant nos. U24A20765 and T2321005), China
Postdoctoral Science Foundation (grant no. 2024M762310), Jiangsu
Provincial Science and Technology Plan Special Fund (grant no.
BM2023003), Jiangsu Provincial Medical Key Discipline (grant no.
ZDXK202247) and the Priority Academic Program Development of the
Jiangsu Higher Education Institutes.
Not applicable.
LYM conceived the study and edited the manuscript.
KWW, YQL, ZNG, LS, XLD, LSL, TH, YCB, and CRH performed the
literature review and prepared the figures.. KWW, YQL and ZNG wrote
the manuscript. Data authentication is not applicable. All authors
have read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Tung CH, Huang MF, Liang CH, Wu YY, Wu JE,
Hsu CL, Chen YL and Hong TM: α-Catulin promotes cancer stemness by
antagonizing WWP1-mediated KLF5 degradation in lung cancer.
Theranostics. 12:1173–1186. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He Q, Yang L, Gao K, Ding P, Chen Q, Xiong
J, Yang W, Song Y, Wang L, Wang Y, et al: FTSJ1 regulates tRNA
2′-O-methyladenosine modification and suppresses the malignancy of
NSCLC via inhibiting DRAM1 expression. Cell Death Dis. 11:3482020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jia Z, Wang K, Duan Y, Hu K, Zhang Y, Wang
M, Xiao K, Liu S, Pan Z and Ding X: Claudin1 decrease induced by
1,25-dihydroxy-vitamin D3 potentiates gefitinib resistance therapy
through inhibiting AKT activation-mediated cancer stem-like
properties in NSCLC cells. Cell Death Discov. 8:1222022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou Y, Dang J, Chang KY, Yau E, Aza-Blanc
P, Moscat J and Rana TM: miR-1298 inhibits Mutant KRAS-driven tumor
growth by repressing FAK and LAMB3. Cancer Res. 76:5777–5787. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Skoulidis F and Heymach JV: Co-occurring
genomic alterations in non-small-cell lung cancer biology and
therapy. Nat Rev Cancer. 19:495–509. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang P, Yorke E, Mageras G, Rimner A,
Sonke JJ and Deasy JO: Validating a predictive atlas of tumor
shrinkage for adaptive radiotherapy of locally advanced lung
cancer. Int J Radiat Oncol Biol Phys. 102:978–986. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lahiri A, Maji A, Potdar PD, Singh N,
Parikh P, Bisht B, Mukherjee A and Paul MK: Lung cancer
immunotherapy: Progress, pitfalls, and promises. Mol Cancer.
22:402023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sarkar A and Hochedlinger K: The sox
family of transcription factors: Versatile regulators of stem and
progenitor cell fate. Cell Stem Cell. 12:15–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saloni, Sachan M, Rahul Verma RS and Patel
GK: SOXs: Master architects of development and versatile emulators
of oncogenesis. Biochim Biophys Acta Rev Cancer. 1880:1892952025.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tang Q, Chen J, Di Z, Yuan W, Zhou Z, Liu
Z, Han S, Liu Y, Ying G, Shu X and Di M: TM4SF1 promotes EMT and
cancer stemness via the Wnt/β-catenin/SOX2 pathway in colorectal
cancer. J Exp Clin Cancer Res. 39:2322020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bagati A, Kumar S, Jiang P, Pyrdol J, Zou
AE, Godicelj A, Mathewson ND, Cartwright ANR, Cejas P, Brown M, et
al: Integrin αvβ6-TGFβ-SOX4 pathway drives immune evasion in
triple-negative breast cancer. Cancer Cell. 39:54–67.e9. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma XL, Hu B, Tang WG, Xie SH, Ren N, Guo L
and Lu RQ: CD73 sustained cancer-stem-cell traits by promoting SOX9
expression and stability in hepatocellular carcinoma. J Hematol
Oncol. 13:112020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shang J, Zheng Y, Mo J, Wang W, Luo Z, Li
Y, Chen X, Zhang Q, Wu K, Liu W and Wu J: Sox4 represses host
innate immunity to facilitate pathogen infection by hijacking the
TLR signaling networks. Virulence. 12:704–722. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zu F, Chen C, Geng Q, Li H, Chan B, Luo G,
Wu M, Ilmer M, Renz BW, Bentum-Ennin L, et al: Smad2 cooperating
with TGIF2 contributes to EMT and cancer stem cells properties in
pancreatic cancer via co-targeting SOX2. Int J Biol Sci.
21:524–543. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Balgkouranidou I, Chimonidou M, Milaki G,
Tsaroucha E, Kakolyris S, Georgoulias V and Lianidou E: SOX17
promoter methylation in plasma circulating tumor DNA of patients
with non-small cell lung cancer. Clin Chem Lab Med. 54:1385–1393.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cao YH, Ding J, Tang QH, Zhang J, Huang
ZY, Tang XM, Liu JB, Ma YS and Fu D: Deciphering cell-cell
interactions and communication in the tumor microenvironment and
unraveling intratumoral genetic heterogeneity via single-cell
genomic sequencing. Bioengineered. 13:14974–14986. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dehshahri A, Biagioni A, Bayat H, Lee EHC,
Hashemabadi M, Fekri HS, Zarrabi A, Mohammadinejad R and Kumar AP:
Editing SOX genes by CRISPR-Cas: Current insights and future
perspectives. Int J Mol Sci. 22:113212021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tripathi SK and Biswal BK: SOX9 promotes
epidermal growth factor receptor-tyrosine kinase inhibitor
resistance via targeting β-catenin and epithelial to mesenchymal
transition in lung cancer. Life Sci. 277:1196082021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hushmandi K, Saadat SH, Mirilavasani S,
Daneshi S, Aref AR, Nabavi N, Raesi R, Taheriazam A and Hashemi M:
The multifaceted role of SOX2 in breast and lung cancer dynamics.
Pathol Res Pract. 260:1553862024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen S, Xu Y, Chen Y, Li X, Mou W, Wang L,
Liu Y, Reisfeld RA, Xiang R, Lv D and Li N: SOX2 gene regulates the
transcriptional network of oncogenes and affects tumorigenesis of
human lung cancer cells. PLoS One. 7:e363262012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, Chen P, Zu L, Liu B, Wang M and Zhou
Q: MicroRNA-338-3p suppresses metastasis of lung cancer cells by
targeting the EMT regulator Sox4. Am J Cancer Res. 6:127–140.
2016.PubMed/NCBI
|
|
22
|
Liang Z, Xu J and Gu C: Novel role of the
SRY-related high-mobility-group box D gene in cancer. Semin Cancer
Biol. 67:83–90. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jones K, Ramirez-Perez S, Niu S,
Gangishetti U, Drissi H and Bhattaram P: SOX4 and RELA function as
transcriptional partners to regulate the expression of
TNF-responsive genes in fibroblast-like synoviocytes. Front
Immunol. 13:7893492022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xia X, Huo W, Wan R, Zhang L, Xia X and
Chang Z: Molecular cloning and expression analysis of Sox3 during
gonad and embryonic development in Misgurnus
anguillicaudatus. Int J Dev Biol. 61:565–570. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Baroti T, Schillinger A, Wegner M and
Stolt CC: Sox13 functionally complements the related Sox5 and Sox6
as important developmental modulators in mouse spinal cord
oligodendrocytes. J Neurochem. 136:316–328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wan H, Liao J, Zhang Z, Zeng X, Liang K
and Wang Y: Molecular cloning, characterization, and expression
analysis of a sex-biased transcriptional factor sox9 gene of mud
crab Scylla paramamosain. Gene. 774:1454232021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Olbromski M, Podhorska-Okołów M and
Dzięgiel P: Role of SOX protein groups F and H in lung cancer
progression. Cancers (Basel). 12:32352020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Toschi L, Finocchiaro G, Nguyen TT, Skokan
MC, Giordano L, Gianoncelli L, Perrino M, Siracusano L, Di Tommaso
L, Infante M, et al: Increased SOX2 gene copy number is associated
with FGFR1 and PIK3CA gene gain in non-small cell lung cancer and
predicts improved survival in early stage disease. PLoS One.
9:e953032014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pradhan S, Guddattu V and Solomon MC:
Association of the co-expression of SOX2 and Podoplanin in the
progression of oral squamous cell carcinomas-an immunohistochemical
study. J Appl Oral Sci. 27:e201803482019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Choi CM, Jang SJ, Park SY, Choi YB, Jeong
JH, Kim DS, Kim HK, Park KS, Nam BH, Kim HR, et al:
Transglutaminase 2 as an independent prognostic marker for survival
of patients with non-adenocarcinoma subtype of non-small cell lung
cancer. Mol Cancer. 10:1192011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li C, He B, Huang C, Yang H, Cao L, Huang
J and Hu C: Sex-determining region Y-box 2 promotes growth of lung
squamous cell carcinoma and directly targets cyclin D1. DNA Cell
Biol. 36:264–272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu B, Liu Y, Zou J, Zou M and Cheng Z:
Smoking is associated with lung adenocarcinoma and lung squamous
cell carcinoma progression through inducing distinguishing lncRNA
alterations in different genders. Anticancer Agents Med Chem.
22:1541–1550. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou Y, Wang X, Huang Y, Chen Y, Zhao G,
Yao Q, Jin C, Huang Y, Liu X and Li G: Down-regulated SOX4
expression suppresses cell proliferation, metastasis and induces
apoptosis in Xuanwei female lung cancer patients. J Cell Biochem.
116:1007–1018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang D, Hao T, Pan Y, Qian X and Zhou D:
Increased expression of SOX4 is a biomarker for malignant status
and poor prognosis in patients with non-small cell lung cancer. Mol
Cell Biochem. 402:75–82. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wen T, Zhang X, Gao Y, Tian H, Fan L and
Yang P: SOX4-BMI1 axis promotes non-small cell lung cancer
progression and facilitates angiogenesis by suppressing ZNF24. Cell
Death Dis. 15:6982024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu X, Wang Y, Zhou G, Zhou J, Tian Z and
Xu J: circGRAMD1B contributes to migration, invasion and
epithelial-mesenchymal transition of lung adenocarcinoma cells via
modulating the expression of SOX4. Funct Integr Genomics.
23:752023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sasaki A, Abe H, Mochizuki S, Shimoda M
and Okada Y: SOX4, an epithelial-mesenchymal transition inducer,
transactivates ADAM28 gene expression and co-localizes with ADAM28
at the invasive front of human breast and lung carcinomas. Pathol
Int. June 7–2018.(Epub ahead of print). View Article : Google Scholar
|
|
38
|
Sun J, Xiong Y, Jiang K, Xin B, Jiang T,
Wei R, Zou Y, Tan H, Jiang T, Yang A, et al: Hypoxia-sensitive long
noncoding RNA CASC15 promotes lung tumorigenesis by regulating the
SOX4/β-catenin axis. J Exp Clin Cancer Res. 40:122021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu J, Zhu MX, Li KS, Peng L and Zhang PF:
Circular RNA drives resistance to anti-PD-1 immunotherapy by
regulating the miR-30a-5p/SOX4 axis in non-small cell lung cancer.
Cancer Drug Resist. 5:261–270. 2022.PubMed/NCBI
|
|
40
|
Fois SS, Paliogiannis P, Zinellu A, Fois
AG, Cossu A and Palmieri G: Molecular epidemiology of the main
druggable genetic alterations in non-small cell lung cancer. Int J
Mol Sci. 22:6122021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Capaccione KM, Hong X, Morgan KM, Liu W,
Bishop JM, Liu L, Markert E, Deen M, Minerowicz C, Bertino JR, et
al: Sox9 mediates Notch1-induced mesenchymal features in lung
adenocarcinoma. Oncotarget. 5:3636–3650. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhong H, Lu W, Tang Y, Wiel C, Wei Y, Cao
J, Riedlinger G, Papagiannakopoulos T, Guo JY, Bergo MO, et al:
SOX9 drives KRAS-induced lung adenocarcinoma progression and
suppresses anti-tumor immunity. Oncogene. 42:2183–2194. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen NM, Singh G, Koenig A, Liou GY, Storz
P, Zhang JS, Regul L, Nagarajan S, Kühnemuth B, Johnsen SA, et al:
NFATc1 links EGFR signaling to induction of Sox9 transcription and
acinar-ductal transdifferentiation in the pancreas.
Gastroenterology. 148:1024–1034.e9. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ling S, Chang X, Schultz L, Lee TK, Chaux
A, Marchionni L, Netto GJ, Sidransky D and Berman DM: An
EGFR-ERK-SOX9 signaling cascade links urothelial development and
regeneration to cancer. Cancer Res. 71:3812–3821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xue VW, Ng SSM, Tsang HF, Wong HT, Leung
WW, Wong YN, Wong YKE, Yu ACS, Yim AKY, Cho WCS, et al: The
non-invasive diagnosis of colorectal cancer via a SOX9-based gene
panel. Clin Exp Med. 23:2421–2432. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ramakrishnan AB, Burby PE, Adiga K and
Cadigan KM: SOX9 and TCF transcription factors associate to mediate
Wnt/β-catenin target gene activation in colorectal cancer. J Biol
Chem. 299:1027352023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou H, Qin Y, Ji S, Ling J, Fu J, Zhuang
Z, Fan X, Song L, Yu X and Chiao PJ: SOX9 activity is induced by
oncogenic Kras to affect MDC1 and MCMs expression in pancreatic
cancer. Oncogene. 37:912–923. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kopp JL, von Figura G, Mayes E, Liu FF,
Dubois CL, Morris JP IV, Pan FC, Akiyama H, Wright CV, Jensen K, et
al: Identification of Sox9-dependent acinar-to-ductal reprogramming
as the principal mechanism for initiation of pancreatic ductal
adenocarcinoma. Cancer Cell. 22:737–750. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zou H, Wang S, Wang S, Wu H, Yu J, Chen Q,
Cui W, Yuan Y, Wen X, He J, et al: SOX5 interacts with YAP1 to
drive malignant potential of non-small cell lung cancer cells. Am J
Cancer Res. 8:866–878. 2018.PubMed/NCBI
|
|
50
|
Chen D, Wang R, Yu C, Cao F, Zhang X, Yan
F, Chen L, Zhu H, Yu Z and Feng J: FOX-A1 contributes to
acquisition of chemoresistance in human lung adenocarcinoma via
transactivation of SOX5. EBioMedicine. 44:150–161. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen R, Zhang C, Cheng Y, Wang S, Lin H
and Zhang H: LncRNA UCC promotes epithelial-mesenchymal transition
via the miR-143-3p/SOX5 axis in non-small-cell lung cancer. Lab
Invest. 101:1153–1165. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen X, Fu Y, Xu H, Teng P, Xie Q, Zhang
Y, Yan C, Xu Y, Li C, Zhou J, et al: SOX5 predicts poor prognosis
in lung adenocarcinoma and promotes tumor metastasis through
epithelial-mesenchymal transition. Oncotarget. 9:10891–10904. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xue JD, Xiang WF, Cai MQ and Lv XY:
Biological functions and therapeutic potential of SRY related high
mobility group box 5 in human cancer. Front Oncol. 14:13321482024.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jethon A, Pula B, Olbromski M, Werynska B,
Muszczynska-Bernhard B, Witkiewicz W, Dziegiel P and
Podhorska-Okolow M: Prognostic significance of SOX18 expression in
non-small cell lung cancer. Int J Oncol. 46:123–132. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Olbromski M, Grzegrzolka J,
Jankowska-Konsur A, Witkiewicz W, Podhorska-Okolow M and Dziegiel
P: MicroRNAs modulate the expression of the SOX18 transcript in
lung squamous cell carcinoma. Oncol Rep. 36:2884–2892. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
de Souza Silva FH, Underwood A, Almeida
CP, Ribeiro TS, Souza-Fagundes EM, Martins AS, Eliezeck M,
Guatimosim S, Andrade LO, Rezende L, et al: Transcription factor
SOX3 upregulated pro-apoptotic genes expression in human breast
cancer. Med Oncol. 39:2122022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Guo Y, Yin J, Tang M and Yu X:
Downregulation of SOX3 leads to the inhibition of the
proliferation, migration and invasion of osteosarcoma cells. Int J
Oncol. 52:1277–1284. 2018.PubMed/NCBI
|
|
58
|
Del Puerto HL, Miranda APGS, Qutob D,
Ferreira E, Silva FHS, Lima BM, Carvalho BA, Roque-Souza B, Gutseit
E, Castro DC, et al: Clinical correlation of transcription factor
SOX3 in cancer: Unveiling its role in tumorigenesis. Genes (Basel).
15:7772024. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Qiu M, Chen D, Shen C, Shen J, Zhao H and
He Y: Sex-determining region Y-box protein 3 induces
epithelial-mesenchymal transition in osteosarcoma cells via
transcriptional activation of Snail1. J Exp Clin Cancer Res.
36:462017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wu D, Jiang C, Zheng JJ, Luo DS, Ma J, Que
HF, Li C, Ma C, Wang HY, Wang W and Xu HT: Bioinformatics analysis
of SOXF family genes reveals potential regulatory mechanism and
diagnostic value in cancers. Ann Transl Med. 10:7012022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cook M, Qorri B, Baskar A, Ziauddin J,
Pani L, Yenkanchi S and Geraci J: Small patient datasets reveal
genetic drivers of non-small cell lung cancer subtypes using
machine learning for hypothesis generation. Exp Med. 4:428–440.
2023. View Article : Google Scholar
|
|
62
|
Hayano T, Garg M, Yin D, Sudo M, Kawamata
N, Shi S, Chien W, Ding LW, Leong G, Mori S, et al: SOX7 is
down-regulated in lung cancer. J Exp Clin Cancer Res. 32:172013.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wu X, Liu H, Zhang M, Ma J, Qi S, Tan Q,
Jiang Y, Hong Y and Yan L: miR-200a-3p promoted cell proliferation
and metastasis by downregulating SOX17 in non-small cell lung
cancer cells. J Biochem Mol Toxicol. 36:e230372022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zang K, Yu ZH, Wang M, Huang Y, Zhu XX and
Yao B: SOX2 como posible biomarcador pronóstico y diana molecular
en el cáncer de pulmón: Metaanálisis. Rev Clin Esp (Barc).
222:584–592. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang S, Li Z, Li P, Li L, Liu Y, Feng Y,
Li R and Xia S: SOX2 promotes radioresistance in non-small cell
lung cancer by regulating tumor cells dedifferentiation. Int J Med
Sci. 20:781–796. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yun HS, Baek JH, Yim JH, Um HD, Park JK,
Song JY, Park IC, Kim JS, Lee SJ, Lee CW and Hwang SG: Radiotherapy
diagnostic biomarkers in radioresistant human H460 lung cancer
stem-like cells. Cancer Biol Ther. 17:208–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Gomez-Casal R, Bhattacharya C, Ganesh N,
Bailey L, Basse P, Gibson M, Epperly M and Levina V: Non-small cell
lung cancer cells survived ionizing radiation treatment display
cancer stem cell and epithelial-mesenchymal transition phenotypes.
Mol Cancer. 12:942013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Huang Y, Wang X, Hu R, Pan G and Lin X:
SOX2 regulates paclitaxel resistance of A549 non-small cell lung
cancer cells via promoting transcription of ClC-3. Oncol Rep.
48:1812022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Chen TY, Zhou J, Li PC, Tang CH, Xu K, Li
T and Ren T: SOX2 knockdown with siRNA reverses cisplatin
resistance in NSCLC by regulating APE1 signaling. Med Oncol.
39:362022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Han F, Liu W, Jiang X, Shi X, Yin L, Ao L,
Cui Z, Li Y, Huang C, Cao J and Liu J: SOX30, a novel epigenetic
silenced tumor suppressor, promotes tumor cell apoptosis by
transcriptional activating p53 in lung cancer. Oncogene.
34:4391–4402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lv L, Zhou M, Zhang J, Liu F, Qi L, Zhang
S, Bi Y and Yu Y: SOX6 suppresses the development of lung
adenocarcinoma by regulating expression of p53, p21CIPI,
cyclin D1 and β-catenin. FEBS Open Bio. 10:135–146. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu H, Zhong L, Lu Y, Liu X, Wei J, Ding
Y, Huang H, Nie Q and Liao X: Deubiquitylase OTUD1 confers
Erlotinib sensitivity in non-small cell lung cancer through
inhibition of nuclear translocation of YAP1. Cell Death Discov.
8:4032022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yu Y, Luo Y, Zheng Y, Zheng X, Li W, Yang
L and Jiang J: Exploring the mechanism of non-small-cell lung
cancer cell lines resistant to epidermal growth factor receptor
tyrosine kinase inhibitor. J Cancer Res Ther. 12:121–125. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Du Q, Liu J, Zhang X, Zhang X, Zhu H, Wei
M and Wang S: Propofol inhibits proliferation, migration, and
invasion but promotes apoptosis by regulation of Sox4 in
endometrial cancer cells. Braz J Med Biol Res. 51:e68032018.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li J, Zhu Z, Li S, Han Z, Meng F and Wei
L: Circ_0089823 reinforces malignant behaviors of non-small cell
lung cancer by acting as a sponge for microRNAs targeting SOX4.
Neoplasia. 23:887–897. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Han F, Zhang MQ, Liu WB, Sun L, Hao XL,
Yin L, Jiang X, Cao J and Liu JY: SOX30 specially prevents
Wnt-signaling to suppress metastasis and improve prognosis of lung
adenocarcinoma patients. Respir Res. 19:2412018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chang SY, Wu TH, Shih YL, Chen YC, Su HY,
Chian CF and Lin YW: SOX1 functions as a tumor suppressor by
repressing HES1 in lung cancer. Cancers (Basel). 15:22072023.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Li D, Wang D, Liu H and Jiang X: LEM
domain containing 1 (LEMD1) transcriptionally activated by
SRY-related high-mobility-group box 4 (SOX4) accelerates the
progression of colon cancer by upregulating phosphatidylinositol
3-kinase (PI3K)/protein kinase B (Akt) signaling pathway.
Bioengineered. 13:8087–8100. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Jin L, Chen C, Huang L, Sun Q and Bu L:
Long noncoding RNA NR2F1-AS1 stimulates the tumorigenic behavior of
non-small cell lung cancer cells by sponging miR-363-3p to increase
SOX4. Open Med (Wars). 17:87–95. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Chang J, Gao F, Chu H, Lou L, Wang H and
Chen Y: miR-363-3p inhibits migration, invasion, and
epithelial-mesenchymal transition by targeting NEDD9 and SOX4 in
non-small-cell lung cancer. J Cell Physiol. 235:1808–1820. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ye Q, Raese R, Luo D, Cao S, Wan YW, Qian
Y and Guo NL: MicroRNA, mRNA, and proteomics biomarkers and
therapeutic targets for improving lung cancer treatment outcomes.
Cancers (Basel). 15:22942023. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Martínez-Espinosa I, Serrato JA,
Cabello-Gutiérrez C, Carlos-Reyes Á and Ortiz-Quintero B:
Mechanisms of microRNA regulation of the epithelial-mesenchymal
transition (EMT) in lung cancer. Life (Basel).
14:14312024.PubMed/NCBI
|
|
83
|
Han F, Liu WB, Shi XY, Yang JT, Zhang X,
Li ZM, Jiang X, Yin L, Li JJ, Huang CS, et al: SOX30 inhibits tumor
metastasis through attenuating Wnt-signaling via transcriptional
and posttranslational regulation of β-catenin in lung cancer.
EBioMedicine. 31:253–266. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Wang Y, Zhang L, Yang J and Sun R: LncRNA
KCNQ1OT1 promotes cell proliferation, migration and invasion via
regulating miR-129-5p/JAG1 axis in non-small cell lung cancer.
Cancer Cell Int. 20:1442020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Liu C, Yang H, Xu Z, Li D, Zhou M, Xiao K,
Shi Z, Zhu L, Yang L and Zhou R: microRNA-548l is involved in the
migration and invasion of non-small cell lung cancer by targeting
the AKT1 signaling pathway. J Cancer Res Clin Oncol. 141:431–441.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wang X, Chen Y, Wang X, Tian H, Wang Y,
Jin J, Shan Z, Liu Y, Cai Z, Tong X, et al: Stem cell factor SOX2
confers ferroptosis resistance in lung cancer via upregulation of
SLC7A11. Cancer Res. 81:5217–5229. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Loh JJ and Ma S: Hallmarks of cancer
stemness. Cell Stem Cell. 31:617–639. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wan X, Ma D, Song G, Tang L, Jiang X, Tian
Y, Yi Z, Jiang C, Jin Y, Hu A and Bai Y: The SOX2/PDIA6 axis
mediates aerobic glycolysis to promote stemness in non-small cell
lung cancer cells. J Bioenerg Biomembr. 56:323–332. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yan F, Teng Y, Li X, Zhong Y, Li C, Yan F
and He X: Hypoxia promotes non-small cell lung cancer cell
stemness, migration, and invasion via promoting glycolysis by
lactylation of SOX9. Cancer Biol Ther. 25:23041612024. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Jia Z, Zhang Y, Yan A, Wang M, Han Q, Wang
K, Wang J, Qiao C, Pan Z, Chen C, et al: 1,25-Dihydroxyvitamin D3
signaling-induced decreases in IRX4 inhibits NANOG-mediated cancer
stem-like properties and gefitinib resistance in NSCLC cells. Cell
Death Dis. 11:6702020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
He J, Shi J, Zhang K, Xue J, Li J, Yang J,
Chen J, Wei J, Ren H and Liu X: Sox2 inhibits Wnt-β-catenin
signaling and metastatic potency of cisplatin-resistant lung
adenocarcinoma cells. Mol Med Rep. 15:1693–1701. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhou W, Cai C, Lu J and Fan Q: miR-129-2
upregulation induces apoptosis and promotes NSCLC chemosensitivity
by targeting SOX4. Thorac Cancer. 13:956–964. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Huang Q, Xing S, Peng A and Yu Z: NORAD
accelerates chemo-resistance of non-small-cell lung cancer via
targeting at miR-129-1-3p/SOX4 axis. Biosci Rep.
40:BSR201934892020. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Xia Y, Tang G, Chen Y, Wang C, Guo M, Xu
T, Zhao M and Zhou Y: Tumor-targeted delivery of siRNA to silence
Sox2 gene expression enhances therapeutic response in
hepatocellular carcinoma. Bioact Mater. 6:1330–1340.
2020.PubMed/NCBI
|
|
95
|
Masarwy R, Breier D, Stotsky-Oterin L,
Ad-El N, Qassem S, Naidu GS, Aitha A, Ezra A, Goldsmith M,
Hazan-Halevy I and Peer D: Targeted CRISPR/Cas9 lipid nanoparticles
elicits therapeutic genome editing in head and neck cancer. Adv Sci
(Weinh). 12:e24110322025. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chen D, Hu C, Wen G, Yang Q, Zhang C and
Yang H: DownRegulated SOX4 expression suppresses cell
proliferation, migration, and induces apoptosis in osteosarcoma in
vitro and in vivo. Calcif Tissue Int. 102:117–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Adham AN, Abdelfatah S, Naqishbandi A,
Sugimoto Y, Fleischer E and Efferth T: Transcriptomics, molecular
docking, and cross-resistance profiling of nobiletin in cancer
cells and synergistic interaction with doxorubicin upon SOX5
transfection. Phytomedicine. 100:1540642022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Guo K, Ma Z, Zhang Y, Han L, Shao C, Feng
Y, Gao F, Di S, Zhang Z, Zhang J, et al: HDAC7 promotes NSCLC
proliferation and metastasis via stabilization by deubiquitinase
USP10 and activation of β-catenin-FGF18 pathway. J Exp Clin Cancer
Res. 41:912022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Taniguchi J, Pandian GN, Hidaka T, Hashiya
K, Bando T, Kim KK and Sugiyama H: A synthetic DNA-binding
inhibitor of SOX2 guides human induced pluripotent stem cells to
differentiate into mesoderm. Nucleic Acids Res. 45:9219–9228. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Panda M, Tripathi SK and Biswal BK: SOX9:
An emerging driving factor from cancer progression to drug
resistance. Biochim Biophys Acta Rev Cancer. 1875:1885172021.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Liu Z, Liao Z, Chen Y, Zhou L, Huangting W
and Xiao H: Research on CRISPR/system in major cancers and its
potential in cancer treatments. Clin Transl Oncol. 23:425–433.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Fu Y, Foden JA, Khayter C, Maeder ML,
Reyon D, Joung JK and Sander JD: High-frequency off-target
mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat
Biotechnol. 31:822–826. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Komor AC, Kim YB, Packer MS, Zuris JA and
Liu DR: Programmable editing of a target base in genomic DNA
without double-stranded DNA cleavage. Nature. 533:420–424. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Hu B, Zhong L, Weng Y, Peng L, Huang Y,
Zhao Y and Liang XJ: Therapeutic siRNA: State of the art. Signal
Transduct Target Ther. 5:1012020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Bobbin ML and Rossi JJ: RNA interference
(RNAi)-based therapeutics: Delivering on the promise? Annu Rev
Pharmacol Toxicol. 56:103–122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Cina C, Majeti B, O'Brien Z, Wang L,
Clamme JP, Adami R, Tsang KY, Harborth J, Ying W and Zabludoff S: A
novel lipid nanoparticle NBF-006 encapsulating glutathione
S-transferase P siRNA for the treatment of KRAS-driven non-small
cell lung cancer. Mol Cancer Ther. 24:7–17. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Kumthekar P, Ko CH, Paunesku T, Dixit K,
Sonabend AM, Bloch O, Tate M, Schwartz M, Zuckerman L, Lezon R, et
al: A first-in-human phase 0 clinical study of RNA
interference-based spherical nucleic acids in patients with
recurrent glioblastoma. Sci Transl Med. 13:eabb39452021. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zuckerman JE, Gritli I, Tolcher A, Heidel
JD, Lim D, Morgan R, Chmielowski B, Ribas A, Davis ME and Yen Y:
Correlating animal and human phase Ia/Ib clinical data with
CALAA-01, a targeted, polymer-based nanoparticle containing siRNA.
Proc Natl Acad Sci USA. 111:11449–11454. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Molinar C, Tannous M, Meloni D, Cavalli R
and Scomparin A: Current status and trends in nucleic acids for
cancer therapy: A focus on polysaccharide-based nanomedicines.
Macromol Biosci. 23:e23001022023. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Torres-Mejia E, Weng S, Whittaker CA,
Nguyen KB, Duong E, Yim L and Spranger S: Lung cancer-intrinsic
SOX2 expression mediates resistance to checkpoint blockade therapy
by inducing treg-dependent CD8+ T-cell exclusion. Cancer Immunol
Res. 13:496–516. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Njouendou AJ, Szarvas T, Tiofack AAZ,
Kenfack RN, Tonouo PD, Ananga SN, Bell EHMD, Simo G, Hoheisel JD,
Siveke JT and Lueong SS: SOX2 dosage sustains tumor-promoting
inflammation to drive disease aggressiveness by modulating the
FOSL2/IL6 axis. Mol Cancer. 22:522023. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Jiang J, Wang Y, Sun M, Luo X, Zhang Z,
Wang Y, Li S, Hu D, Zhang J, Wu Z, et al: SOX on tumors, a comfort
or a constraint? Cell Death Discov. 10:672024. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Wang K and Zhou H: Proteolysis targeting
chimera (PROTAC) for epidermal growth factor receptor enhances
anti-tumor immunity in non-small cell lung cancer. Drug Dev Res.
82:422–429. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Jie C, Li R, Cheng Y, Wang Z, Wu Q and Xie
C: Prospects and feasibility of synergistic therapy with
radiotherapy, immunotherapy, and DNA methyltransferase inhibitors
in non-small cell lung cancer. Front Immunol. 14:11223522023.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Memon D, Schoenfeld AJ, Ye D, Fromm G,
Rizvi H, Zhang X, Keddar MR, Mathew D, Yoo KJ, Qiu J, et al:
Clinical and molecular features of acquired resistance to
immunotherapy in non-small cell lung cancer. Cancer Cell.
42:209–224.e9. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Rotow J and Bivona TG: Understanding and
targeting resistance mechanisms in NSCLC. Nat Rev Cancer.
17:637–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Justilien V, Walsh MP, Ali SA, Thompson
EA, Murray NR and Fields AP: The PRKCI and SOX2 oncogenes are
coamplified and cooperate to activate Hedgehog signaling in lung
squamous cell carcinoma. Cancer Cell. 25:139–151. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Wu JL, Xu CF, Yang XH and Wang MS:
Fibronectin promotes tumor progression through integrin
αvβ3/PI3K/AKT/SOX2 signaling in non-small cell lung cancer.
Heliyon. 9:e201852023. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Huang JQ, Wei FK, Xu XL, Ye SX, Song JW,
Ding PK, Zhu J, Li HF, Luo XP, Gong H, et al: SOX9 drives the
epithelial-mesenchymal transition in non-small-cell lung cancer
through the Wnt/β-catenin pathway. J Transl Med. 17:1432019.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Singh S, Trevino J, Bora-Singhal N,
Coppola D, Haura E, Altiok S and Chellappan SP: EGFR/Src/Akt
signaling modulates Sox2 expression and self-renewal of stem-like
side-population cells in non-small cell lung cancer. Mol Cancer.
11:732012. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Voronkova MA, Rojanasakul LW,
Kiratipaiboon C and Rojanasakul Y: The SOX9-aldehyde dehydrogenase
axis determines resistance to chemotherapy in non-small-cell lung
cancer. Mol Cell Biol. 40:e00307–19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Weina K and Utikal J: SOX2 and cancer:
Current research and its implications in the clinic. Clin Transl
Med. 3:192014. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ciccone G, Ibba ML, Coppola G, Catuogno S
and Esposito CL: The small RNA landscape in NSCLC: Current
therapeutic applications and progresses. Int J Mol Sci.
24:61212023. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Paul A, Muralidharan A, Biswas A, Kamath
BV, Joseph A and Alex AT: siRNA therapeutics and its challenges:
Recent advances in effective delivery for cancer therapy. OpenNano.
7:1000632022. View Article : Google Scholar
|
|
125
|
Khan P, Siddiqui JA, Lakshmanan I, Ganti
AK, Salgia R, Jain M, Batra SK and Nasser MW: RNA-based therapies:
A cog in the wheel of lung cancer defense. Mol Cancer. 20:542021.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Li JW, Zheng G, Kaye FJ and Wu L: PROTAC
therapy as a new targeted therapy for lung cancer. Mol Ther.
31:647–656. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Li S, Wang X, Huang J, Cao X, Liu Y, Bai
S, Zeng T, Chen Q, Li C, Lu C and Yang H: Decoy-PROTAC for specific
degradation of ‘undruggable’ STAT3 transcription factor. Cell Death
Dis. 16:1972025. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Ravindran Menon D, Luo Y, Arcaroli JJ, Liu
S, KrishnanKutty LN, Osborne DG, Li Y, Samson JM, Bagby S, Tan AC,
et al: CDK1 interacts with SOX2 and promotes tumor initiation in
human melanoma. Cancer Res. 78:6561–6574. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Maurizi G, Verma N, Gadi A, Mansukhani A
and Basilico C: SOX2 is required for tumor development and cancer
cell proliferation in osteosarcoma. Oncogene. 37:4626–4632. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Domenici G, Aurrekoetxea-Rodríguez I,
Simões BM, Rábano M, Lee SY, Millán JS, Comaills V, Oliemuller E,
López-Ruiz JA, Zabalza I, et al: A Sox2-Sox9 signalling axis
maintains human breast luminal progenitor and breast cancer stem
cells. Oncogene. 38:3151–3169. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wang Q, Li J, Zhu J, Mao J, Duan C, Liang
X, Zhu L, Zhu M, Zhang Z, Lin F and Guo R: Genome-wide CRISPR/Cas9
screening for therapeutic targets in NSCLC carrying wild-type TP53
and receptor tyrosine kinase genes. Clin Transl Med. 12:e8822022.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Liu B, Wang Z, Gu M, Wang J and Tan J:
Research into overcoming drug resistance in lung cancer treatment
using CRISPR-Cas9 technology: A narrative review. Transl Lung
Cancer Res. 13:2067–2081. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Li N and Li S: Epigenetic inactivation of
SOX1 promotes cell migration in lung cancer. Tumour Biol.
36:4603–4610. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Ai C, Huang Z, Rong T, Shen W, Yang F, Li
Q, Bi L and Li W: The impact of SOX4-activated CTHRC1
transcriptional activity regulating DNA damage repair on cisplatin
resistance in lung adenocarcinoma. Electrophoresis. 45:1408–1417.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Chen X, Zheng Q, Li W, Lu Y, Ni Y, Ma L
and Fu Y: SOX5 induces lung adenocarcinoma angiogenesis by inducing
the expression of VEGF through STAT3 signaling. Onco Targets Ther.
11:5733–5741. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhou Y, Zheng X, Chen LJ, Xu B and Jiang
JT: microRNA-181b suppresses the metastasis of lung cancer cells by
targeting sex determining region Y-related high mobility group-box
6 (Sox6). Pathol Res Pract. 215:335–342. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Sun QY, Ding LW, Johnson K, Zhou S, Tyner
JW, Yang H, Doan NB, Said JW, Xiao JF, Loh XY, et al: SOX7
regulates MAPK/ERK-BIM mediated apoptosis in cancer cells.
Oncogene. 38:6196–6210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Hao X, Han F, Ma B, Zhang N, Chen H, Jiang
X, Yin L, Liu W, Ao L, Cao J and Liu J: SOX30 is a key regulator of
desmosomal gene suppressing tumor growth and metastasis in lung
adenocarcinoma. J Exp Clin Cancer Res. 37:1112018. View Article : Google Scholar : PubMed/NCBI
|