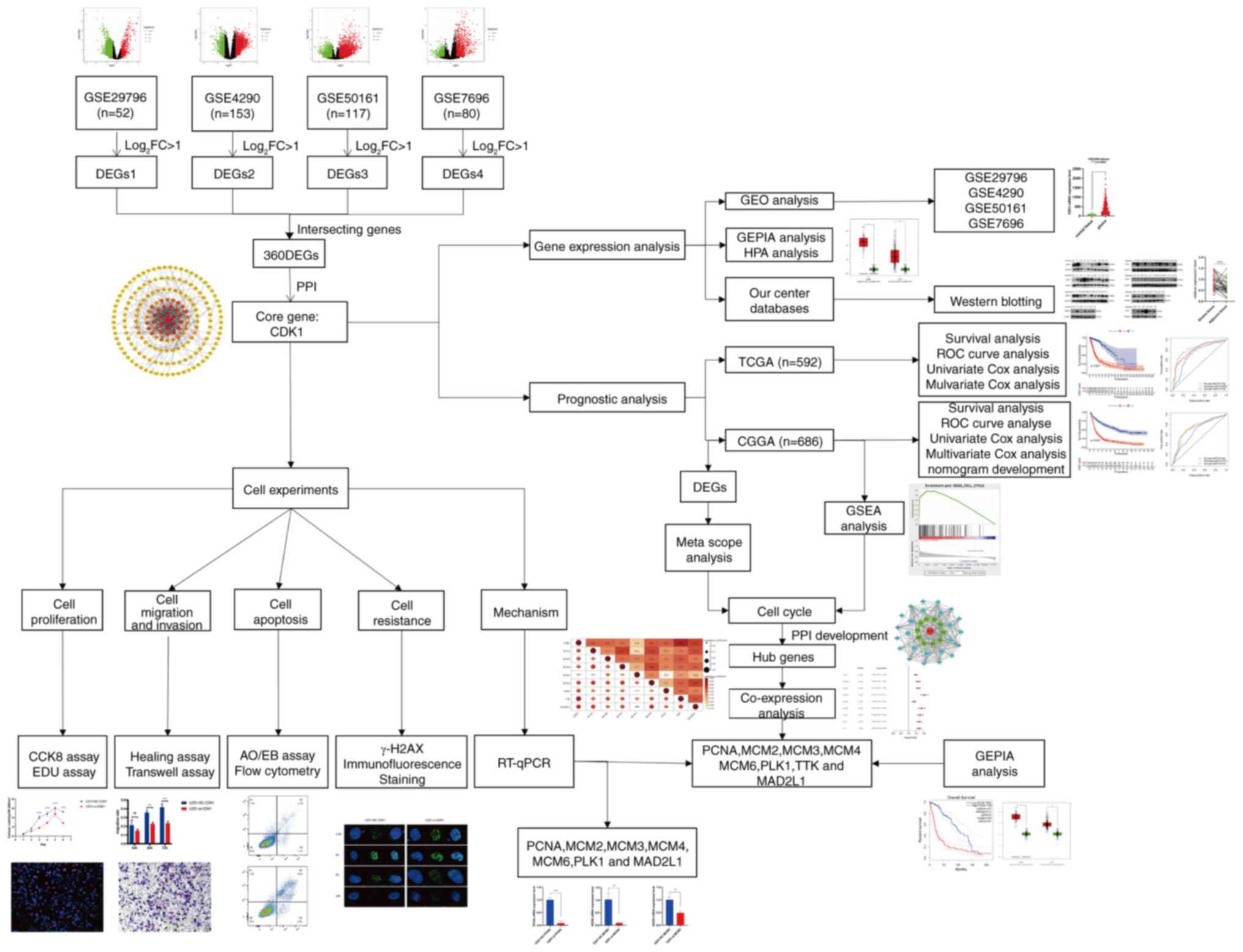
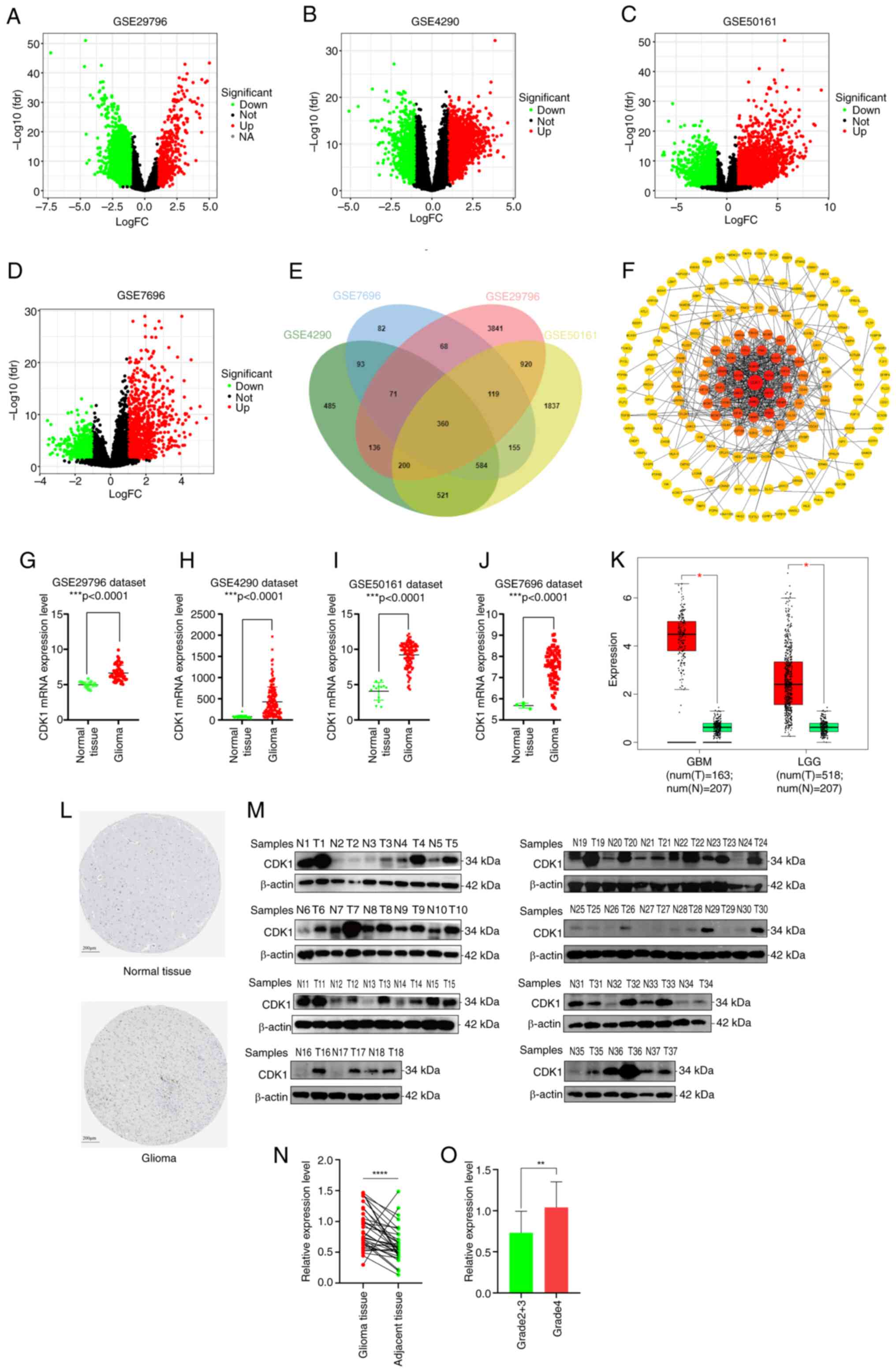
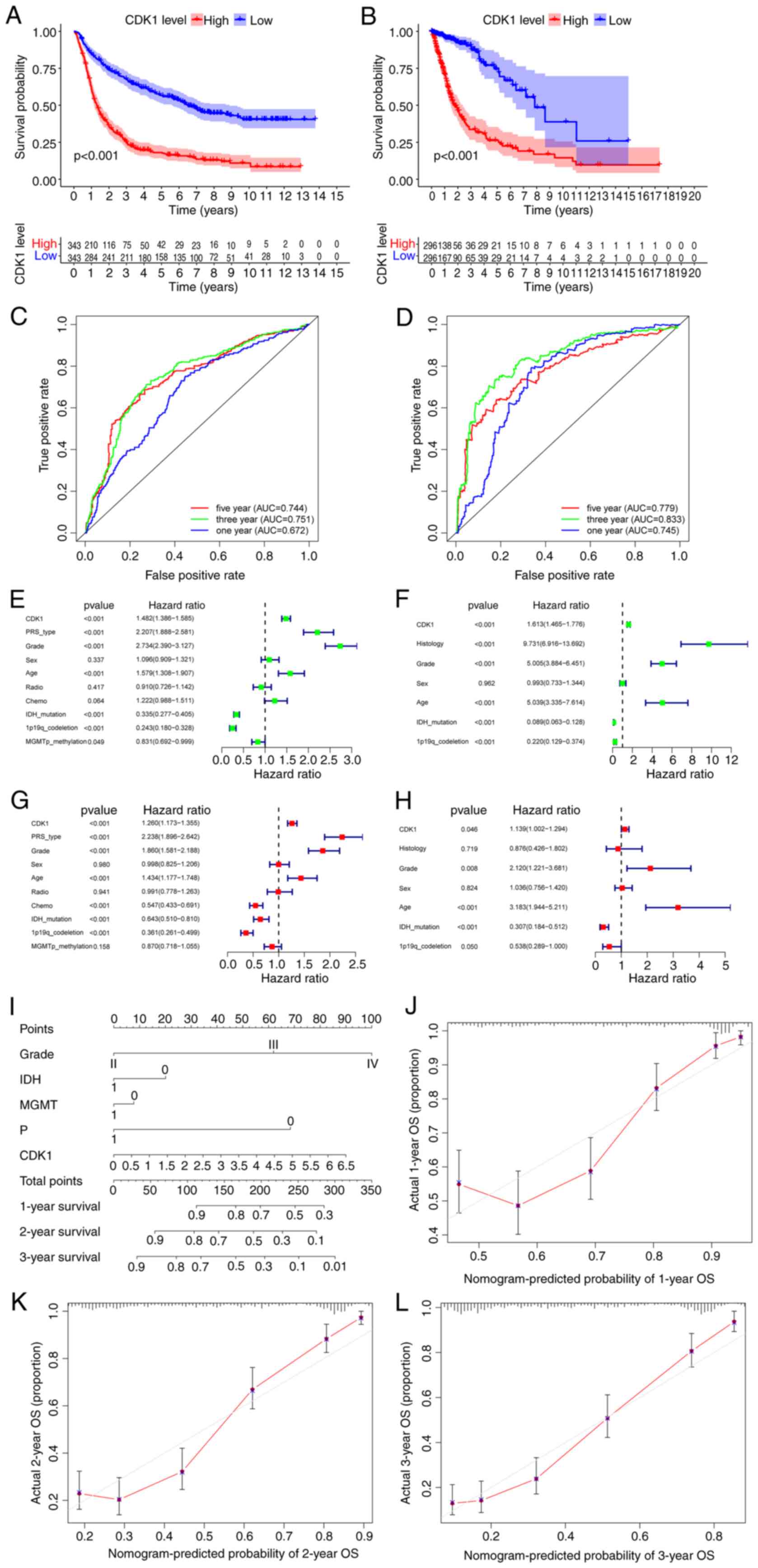
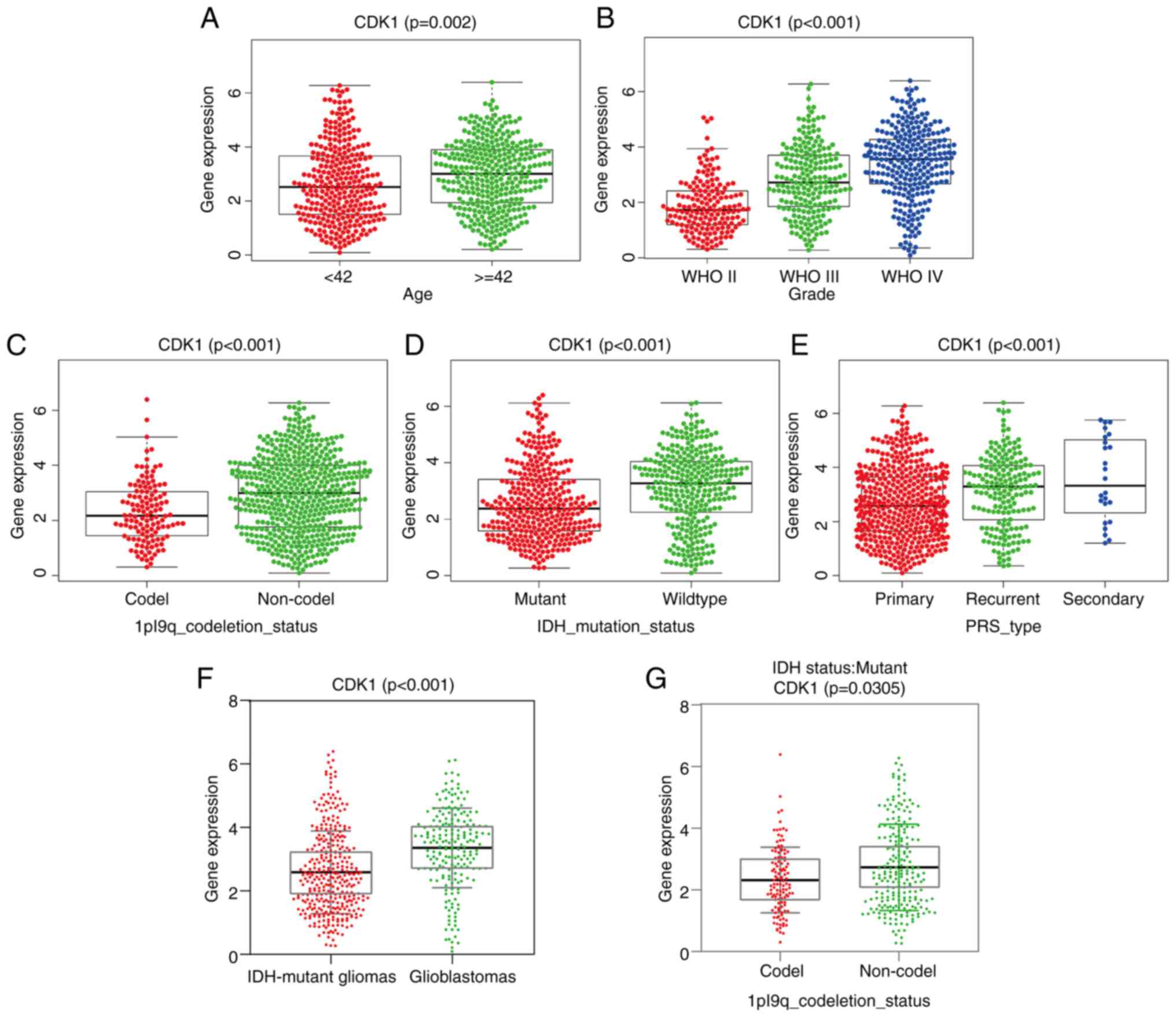
|
1
|
Ostrom QT, Gittleman H, Truitt G, Boscia
A, Kruchko C and Barnholtz-Sloan JS: CBTRUS statistical report:
Primary brain and other central nervous system tumors diagnosed in
the United States in 2011–2015. Neuro Oncol. 20:iv1–iv86. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rong L, Li N and Zhang Z: Emerging
therapies for glioblastoma: Current state and future directions. J
Exp Clin Cancer Res. 41:1422022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu MW and Quail DF: Immunotherapy for
glioblastoma: Current progress and challenges. Front Immunol.
12:6763012021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Louis DN, Perry A, Wesseling P, Brat DJ,
Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM,
Reifenberger G, et al: The 2021 WHO classification of tumors of the
central nervous system: A summary. Neuro Oncol. 23:1231–1251. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bruford EA, Braschi B, Denny P, Jones TEM,
Seal RL and Tweedie S: Guidelines for human gene nomenclature. Nat
Genet. 52:754–758. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Śledzińska P, Bebyn MG, Furtak J,
Kowalewski J and Lewandowska MA: Prognostic and predictive
biomarkers in gliomas. Int J Mol Sci. 22:103732021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qu S, Huang C, Zhu T, Wang K, Zhang H,
Wang L, Xu R, Zheng H, Yuan X, Liu G, et al: OLFML3, as a potential
predictor of prognosis and therapeutic target for glioma, is
closely related to immune cell infiltration. VIEW. 4:202200522023.
View Article : Google Scholar
|
|
9
|
Alleman K, Knecht E, Huang J, Zhang L, Lam
S and DeCuypere M: Multimodal deep learning-based prognostication
in glioma patients: A systematic review. Cancers (Basel).
15:5452023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yuan Y, Zhang X, Wang Y, Li H, Qi Z, Du Z,
Chu YH, Feng D, Hu J, Xie Q, et al: Multimodal data integration
using deep learning predicts overall survival of patients with
glioma. VIEW. 5:202400012024. View Article : Google Scholar
|
|
11
|
Xu S, Tang L, Li X, Fan F and Liu Z:
Immunotherapy for glioma: Current management and future
application. Cancer Lett. 476:1–12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang K, Wu Z, Zhang H, Zhang N, Wu W, Wang
Z, Dai Z, Zhang X, Zhang L, Peng Y, et al: Glioma targeted therapy:
Insight into future of molecular approaches. Mol Cancer. 21:392022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Le Rhun E, Preusser M, Roth P, Reardon DA,
van den Bent M, Wen P, Reifenberger G and Weller M: Molecular
targeted therapy of glioblastoma. Cancer Treat Rev. 80:1018962019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Malumbres M: Cyclin-dependent kinases.
Genome Biol. 15:1222014. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han Z, Jia Q, Zhang J, Chen M, Wang L,
Tong K, He W, Zhang Y, Zhu W, Qin J, et al: Deubiquitylase YOD1
regulates CDK1 stability and drives triple-negative breast cancer
tumorigenesis. J Exp Clin Cancer Res. 42:2282023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeng K, Li W, Wang Y, Zhang Z, Zhang L,
Zhang W, Xing Y and Zhou C: Inhibition of CDK1 overcomes
oxaliplatin resistance by regulating ACSL4-mediated ferroptosis in
colorectal cancer. Adv Sci (Weinh). 10:e23010882023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen J, Gong X, Tan S, Zhang Y, Xia R, Xu
S, Wang S, Zhou H, Jiang Y, Zhao T, et al: CDK1 acts as a
prognostic biomarker associated with immune infiltration in
pan-cancer, especially in gastrointestinal tumors. Curr Med Chem.
32:4836–4857. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xue J, Song Y, Xu W and Zhu Y: The
CDK1-related lncRNA and CXCL8 mediated immune resistance in lung
adenocarcinoma. Cells. 11:26882022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sung WW, Lin YM, Wu PR, Yen HH, Lai HW, Su
TC, Huang RH, Wen CK, Chen CY, Chen CJ and Yeh KT: High
nuclear/cytoplasmic ratio of Cdk1 expression predicts poor
prognosis in colorectal cancer patients. BMC Cancer. 14:9512014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xi Q, Huang M, Wang Y, Zhong J, Liu R, Xu
G, Jiang L, Wang J, Fang Z and Yang S: The expression of CDK1 is
associated with proliferation and can be a prognostic factor in
epithelial ovarian cancer. Tumour Biol. 36:4939–4948. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang C, Elkahloun AG, Robertson M, Gills
JJ, Tsurutani J, Shih JH, Fukuoka J, Hollander MC, Harris CC,
Travis WD, et al: Loss of cytoplasmic CDK1 predicts poor survival
in human lung cancer and confers chemotherapeutic resistance. PLoS
One. 6:e238492011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
van Nes JG, Smit VT, Putter H, Kuppen PJ,
Kim SJ, Daito M, Ding J, Shibayama M, Numada S, Gohda K, et al:
Validation study of the prognostic value of cyclin-dependent kinase
(CDK)-based risk in Caucasian breast cancer patients. Br J Cancer.
100:494–500. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Piao J, Zhu L, Sun J, Li N, Dong B, Yang Y
and Chen L: High expression of CDK1 and BUB1 predicts poor
prognosis of pancreatic ductal adenocarcinoma. Gene. 701:15–22.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jones MJ and Jones MC: Cell cycle control
by cell-matrix interactions. Curr Opin Cell Biol. 86:1022882024.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lemmens B, Hegarat N, Akopyan K,
Sala-Gaston J, Bartek J, Hochegger H and Lindqvist A: DNA
replication determines timing of mitosis by restricting CDK1 and
PLK1 activation. Mol Cell. 71:117–128.e113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ci M, Zhao G, Li C, Liu R, Hu X, Pan J,
Shen Y, Zhang G, Li Y, Zhang L, et al: OTUD4 promotes the
progression of glioblastoma by deubiquitinating CDK1 and activating
MAPK signaling pathway. Cell Death Dis. 15:1792024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi YX, Zhu T, Zou T, Zhuo W, Chen YX,
Huang MS, Zheng W, Wang CJ, Li X, Mao XY, et al: Prognostic and
predictive values of CDK1 and MAD2L1 in lung adenocarcinoma.
Oncotarget. 7:85235–85243. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Faienza F, Polverino F, Rajendraprasad G,
Milletti G, Hu Z, Colella B, Gargano D, Strappazzon F, Rizza S,
Vistesen MV, et al: AMBRA1 phosphorylation by CDK1 and PLK1
regulates mitotic spindle orientation. Cell Mol Life Sci.
80:2512023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Auvergne RM, Sim FJ, Wang S,
Chandler-Militello D, Burch J, Al Fanek Y, Davis D, Benraiss A,
Walter K, Achanta P, et al: Transcriptional differences between
normal and glioma-derived glial progenitor cells identify a core
set of dysregulated genes. Cell Rep. 3:2127–2141. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun L, Hui AM, Su Q, Vortmeyer A,
Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey
R, et al: Neuronal and glioma-derived stem cell factor induces
angiogenesis within the brain. Cancer Cell. 9:287–300. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Griesinger AM, Birks DK, Donson AM, Amani
V, Hoffman LM, Waziri A, Wang M, Handler MH and Foreman NK:
Characterization of distinct immunophenotypes across pediatric
brain tumor types. J Immunol. 191:4880–4888. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lambiv WL, Vassallo I, Delorenzi M, Shay
T, Diserens AC, Misra A, Feuerstein B, Murat A, Migliavacca E,
Hamou MF, et al: The Wnt inhibitory factor 1 (WIF1) is targeted in
glioblastoma and has a tumor suppressing function potentially by
induction of senescence. Neuro Oncol. 13:736–747. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao Z, Zhang KN, Wang Q, Li G, Zeng F,
Zhang Y, Wu F, Chai R, Wang Z, Zhang C, et al: Chinese glioma
genome atlas (CGGA): A comprehensive resource with functional
genomic data from Chinese glioma patients. Genomics Proteomics
Bioinformatics. 19:1–12. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cancer Genome Atlas Research Network, .
Comprehensive genomic characterization defines human glioblastoma
genes and core pathways. Nature. 455:1061–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brat DJ, Verhaak RG, Aldape KD, Yung WK,
Salama SR, Cooper LAD, Rheinbay E, Miller CR, Vitucci M, Morozova
O, et al: Comprehensive, integrative genomic analysis of diffuse
lower-grade gliomas. N Engl J Med. 372:2481–2498. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou Y, Zhou B, Pache L, Chang M,
Khodabakhshi AH, Tanaseichuk O, Benner C and Chanda SK: Metascape
provides a biologist-oriented resource for the analysis of
systems-level datasets. Nat Commun. 10:15232019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu L, Chai R, Zhao Z, Wang Q and Jiang T:
Role of the tumor microenvironment in shaping IDH-wildtype glioma
plasticity, and potential therapeutic strategies. Cancer Biol Med.
19:1423–1427. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Weller M and Le Rhun E: How did lomustine
become standard of care in recurrent glioblastoma? Cancer Treat
Rev. 87:1020292020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nabors LB, Portnow J, Ahluwalia M,
Baehring J, Brem H, Brem S, Butowski N, Campian JL, Clark SW,
Fabiano AJ, et al: Central nervous system cancers, Version 3.2020,
NCCN Clinical practice guidelines in oncology. J Natl Compr Canc
Netw. 18:1537–1570. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Humphrey PA, Moch H, Cubilla AL, Ulbright
TM and Reuter VE: The 2016 WHO classification of tumours of the
urinary system and male genital organs-part B: Prostate and bladder
tumours. Eur Urol. 70:106–119. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li K, Wang R, Gu Z, Weng W, Liu W, Huang
Y, Wu J, Zhang Z, Yang S, Su J, et al: Serum metabolic profiling
enables diagnosis, prognosis, and monitoring for brainstem gliomas.
Nat Commun. 16:61082025. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen R, Smith-Cohn M, Cohen AL and Colman
H: Glioma subclassifications and their clinical significance.
Neurotherapeutics. 14:284–297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xie B, Wang S, Jiang N and Li JJ: Cyclin
B1/CDK1-regulated mitochondrial bioenergetics in cell cycle
progression and tumor resistance. Cancer Lett. 443:56–66. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Guan T, Li M, Song Y, Chen J, Tang J,
Zhang C, Wen Y, Yang X, Huang L, Zhu Y, et al: Phosphorylation of
USP29 by CDK1 governs TWIST1 stability and oncogenic functions. Adv
Sci (Weinh). 10:e22058732023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
López-Otín C, Blasco MA, Partridge L,
Serrano M and Kroemer G: Hallmarks of aging: An expanding universe.
Cell. 186:243–278. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chatsirisupachai K, Lesluyes T, Paraoan L,
Van Loo P and de Magalhães JP: An integrative analysis of the
age-associated multi-omic landscape across cancers. Nat Commun.
12:23452021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang HY, Tang K, Liang TY, Zhang WZ, Li
JY, Wang W, Hu HM, Li MY, Wang HQ, He XZ, et al: The comparison of
clinical and biological characteristics between IDH1 and IDH2
mutations in gliomas. J Exp Clin Cancer Res. 35:862016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nambirajan A, Suri V, Kedia S, Goyal K,
Malgulwar PB, Khanna G, Panda PK, Gulati S, Garg A and Sharma MC:
Paediatric diffuse leptomeningeal tumor with glial and neuronal
differentiation harbouring chromosome 1p/19q co-deletion and H3.3
K27M mutation: Unusual molecular profile and its therapeutic
implications. Brain Tumor Pathol. 35:186–191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang Q, Bode AM and Zhang T: Targeting
CDK1 in cancer: Mechanisms and implications. NPJ Precis Oncol.
7:582023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu Y, Zhou F, Ali H, Lathia JD and Chen
P: Immunotherapy for glioblastoma: Current state, challenges, and
future perspectives. Cell Mol Immunol. 21:1354–1375. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Pećina-Šlaus N, Zottel A, Škripek Ž,
Puljko B, Dumančić F, Bukovac A, Jovčevska I and Kafka A: In silico
analysis reveals distinct changes in markers of epithelial to
mesenchymal transition in glioma subtypes. Biomol Biomed.
25:2712–2736. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen H, Hu K, Xie Y, Qi Y, Li W, He Y, Fan
S, Liu W and Li C: CDK1 promotes epithelial-mesenchymal transition
and migration of head and neck squamous carcinoma cells by
repressing ∆Np63α-mediated transcriptional regulation. Int J Mol
Sci. 23:73852022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ren L, Yang Y, Li W, Zheng X, Liu J, Li S,
Yang H, Zhang Y, Ge B, Zhang S, et al: CDK1 serves as a therapeutic
target of adrenocortical carcinoma via regulating
epithelial-mesenchymal transition, G2/M phase transition, and
PANoptosis. J Transl Med. 20:4442022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ferretti LP, Lafranchi L and Sartori AA:
Controlling DNA-end resection: A new task for CDKs. Front Genet.
4:992013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tozaki Y, Aoki H, Kato R, Toriuchi K,
Arame S, Inoue Y, Hayashi H, Kubota E, Kataoka H and Aoyama M: The
combination of ATM and Chk1 inhibitors induces synthetic lethality
in colorectal cancer cells. Cancers (Basel). 15:7352023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liao H, Ji F, Geng X, Xing M, Li W, Chen
Z, Shen H and Ying S: CDK1 promotes nascent DNA synthesis and
induces resistance of cancer cells to DNA-damaging therapeutic
agents. Oncotarget. 8:90662–90673. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Suski JM, Braun M, Strmiska V and Sicinski
P: Targeting cell-cycle machinery in cancer. Cancer Cell.
39:759–778. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Masai H, Taniyama C, Ogino K, Matsui E,
Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, et
al: Phosphorylation of MCM4 by Cdc7 kinase facilitates its
interaction with Cdc45 on the chromatin. J Biol Chem.
281:39249–39261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
de Cárcer G, Venkateswaran SV, Salgueiro
L, El Bakkali A, Somogyi K, Rowald K, Montañés P, Sanclemente M,
Escobar B, de Martino A, et al: Plk1 overexpression induces
chromosomal instability and suppresses tumor development. Nat
Commun. 9:30122018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Pierantoni GM, Conte A, Rinaldo C,
Tornincasa M, Gerlini R, Valente D, Izzo A and Fusco A: Hmga1 null
mouse embryonic fibroblasts display downregulation of spindle
assembly checkpoint gene expression associated to nuclear and
karyotypic abnormalities. Cell Cycle. 15:812–818. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yang S, Yuan Y, Ren W, Wang H, Zhao Z,
Zhao H, Zhao Q, Chen X, Jiang X and Zhang L: MCM4 is a novel
prognostic biomarker and promotes cancer cell growth in glioma.
Front Oncol. 12:10043242022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li X, Tao Z, Wang H, Deng Z, Zhou Y and Du
Z: Dual inhibition of Src and PLK1 regulate stemness and induce
apoptosis through Notch1-SOX2 signaling in EGFRvIII positive glioma
stem cells (GSCs). Exp Cell Res. 396:1122612020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Deng S, Lu X, Wang X, Liang B, Xu H, Yang
D, Cui G, Yonemura A, Paine H, Zhou Y, et al: Overexpression of
TBX3 suppresses tumorigenesis in experimental and human
cholangiocarcinoma. Cell Death Dis. 15:4412024. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Reda M, Ngamcherdtrakul W, Nelson MA,
Siriwon N, Wang R, Zaidan HY, Bejan DS, Reda S, Hoang NH, Crumrine
NA, et al: Development of a nanoparticle-based immunotherapy
targeting PD-L1 and PLK1 for lung cancer treatment. Nat Commun.
13:42612022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Roskoski R Jr: Cyclin-dependent protein
serine/threonine kinase inhibitors as anticancer drugs. Pharmacol
Res. 139:471–488. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Gao JJ, Cheng J, Bloomquist E, Sanchez J,
Wedam SB, Singh H, Amiri-Kordestani L, Ibrahim A, Sridhara R,
Goldberg KB, et al: CDK4/6 inhibitor treatment for patients with
hormone receptor-positive, HER2-negative, advanced or metastatic
breast cancer: A US food and drug administration pooled analysis.
Lancet Oncol. 21:250–260. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Curigliano G and Loibl S: CDK4/6
inhibitors in breast cancer: One more step towards reduced
mortality. Lancet Oncol. 21:191–192. 2020. View Article : Google Scholar : PubMed/NCBI
|