Introduction
It is estimated that one in four people will develop
cancer at some point in their lives. Cancer is the first or second
leading cause of premature death in many countries, with deaths
occurring before the age of 70 years. In the female population, the
incidence and mortality from breast cancer are increasing rapidly
worldwide, with one in five women being diagnosed with breast
cancer during their lifetime (1).
An estimated 43,170 deaths from breast cancer are expected
worldwide in 2023 (2).
Understanding tumor biology is essential to
elucidate the reasons behind the high incidence of cancer
worldwide, especially breast cancer. Detailed analysis of tumor
pathophysiology allows us to identify the cellular and molecular
processes that contribute to the development and progression of
cancer (3–5). Furthermore, this knowledge is crucial
for identifying biological markers that may influence cancer
progression, enabling the formulation of more effective diagnostic
and therapeutic strategies (6–9).
microRNA (miRNA or miR), a post-transcriptional gene modulating
non-coding small molecule, can target multiple genes, and is key to
tumorigenic progression and immune response (10–12).
In patients with breast cancer, altered miRNAs are
linked to different tumor hallmark pathways including the presence
of Tumor infiltrating lymphocytes (13,14).
Among the different types of biomarkers the microRNAs are a strong
candidate to be incorporated into the clinical workflow, due to
their stability in the body fluids used for diagnostic analysis,
making them attractive biological entities (15,16).
Moreover, by investigating the miRNAs that trigger and sustain
tumor growth, may facilitate development of targeted interventions
that aim to improve clinical outcomes and increase patient survival
(17). miR-155 has been linked to
tumor biology in breast cancer and immune response such as the
presence or absence of TILs, demonstrating the strong interplay in
tumour biology (18–20).
In the present review, we appraised the current
understanding of the miRNAs 155 and TILs in the breast cancer
context, more specifically the Triple-Negative subtype. TNBC,
presents, gene expression profiling with intrinsic subtypes of
female breast cancer, which may correspond to distinct etiological
pathways and hold significant therapeutic implications, and impact
mortality risk (21).
BC biology and classification
Tumor biology and pathological
characteristics
The mammary parenchyma comprises ductal epithelium
organized in a bilayer arrangement: The luminal epithelial layer
comprises cuboidal or columnar cells and the basal layer contains
contractile myoepithelial cells. The stromal component is composed
of connective tissue, blood vessels and neural elements (22). While tumors of non-epithelial
origin, such as sarcomas or skin tumors, can arise in the breast,
these cases are rare and not classified as BC despite their
anatomical location (23).
On macroscopic pathological examination, lesions
manifest as firm, grayish white, gritty masses that randomly invade
the surrounding tissue, resulting in an irregular formation, termed
stellate configuration. Microscopically, the lesions demonstrate
cords and nests of tumor cells with cytological features ranging
from indolent to highly malignant. As the malignant cells
infiltrate the mammary stroma and adipose tissue, they induce a
fibrotic response, frequently producing a clinically palpable mass
with radiological density and solid ultrasonographic
characteristics typical of invasive carcinoma (24,25).
Invasive ductal carcinoma represents the most common
type of BC, accounting for 40–70% of cases. Other histological
subtypes include invasive lobular carcinoma (5–15%), apocrine
carcinoma (4%), mucinous carcinoma (2%), tubular carcinoma (1.6%),
micropapillary carcinoma (0.9–2.0%), metaplastic carcinoma
(0.2–1.0%) and cribriform carcinoma (0.4%). Invasive ductal
carcinoma is also referred to as invasive carcinoma of no special
type or invasive carcinoma not otherwise specified (4).
Certain invasive lobular carcinomas exhibit
macroscopic features similar to those of invasive ductal carcinoma.
Consequently, immunohistochemical analysis is necessary to evaluate
the expression of E-cadherin protein, which mediates epithelial
cell adhesion. This protein demonstrates positive expression in
ductal tumors while being characteristically absent in lobular
neoplasms (26).
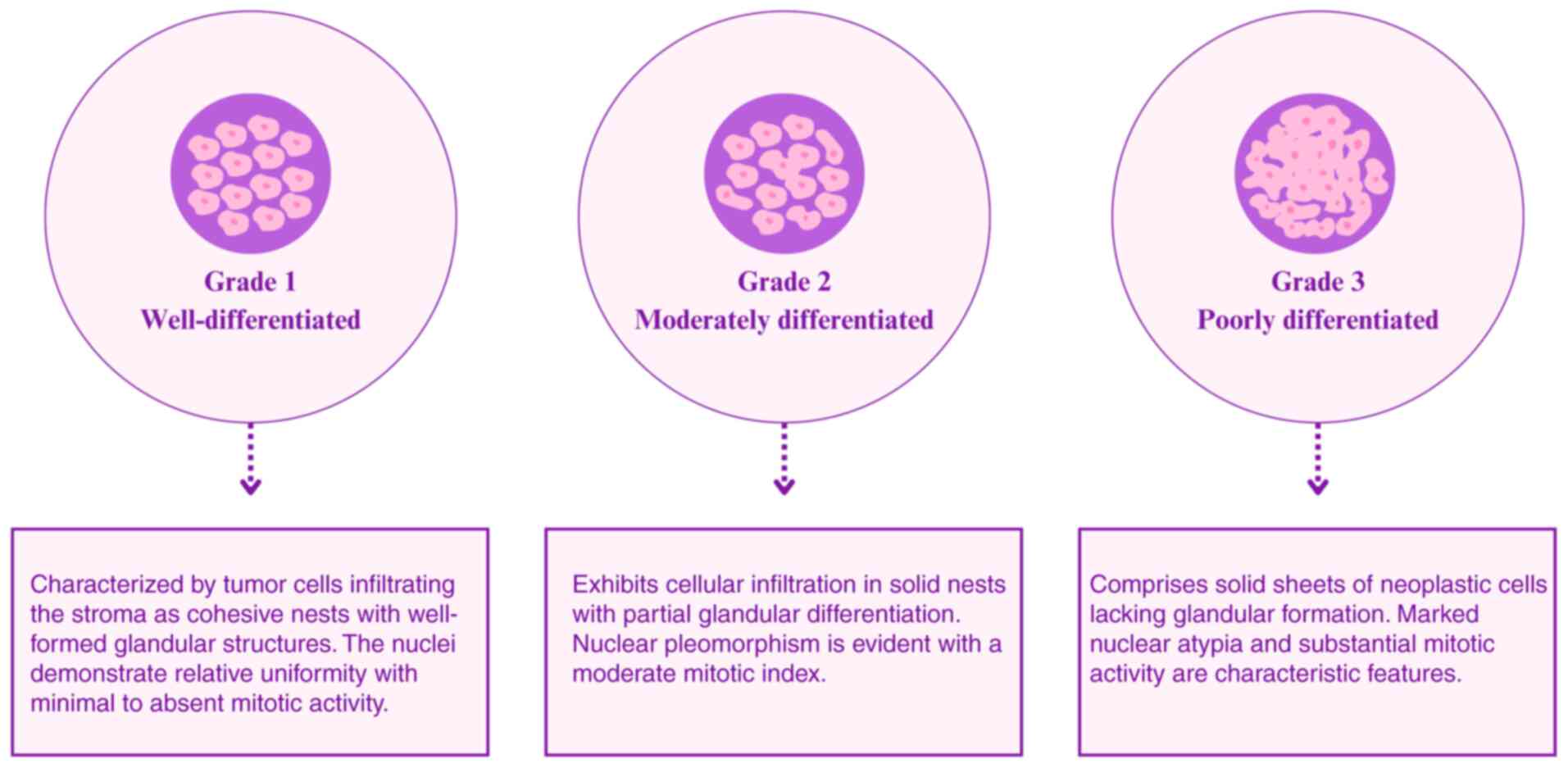
Invasive carcinomas are classified into three
distinct grades based on a comprehensive evaluation of
architectural and cytological features. This classification is
performed using a standardized scoring system that reflects the
degree of cell differentiation (Fig.
1) (27). Histological analysis
at the molecular level is key to grading invasive carcinoma.
Histopathological classification
The histological classification of BC is key for
tumor characterization (Table I).
Moreover, molecular markers serve a pivotal role in guiding
appropriate therapeutic strategies. The molecular classification
system developed by Perou et al (28) remains widely implemented in
contemporary clinical practice.
 | Table I.Histological subtypes of breast
cancer. |
Table I.
Histological subtypes of breast
cancer.
| Characteristic | Invasive breast
carcinoma of no special type | Invasive lobular
carcinoma | Invasive mucinous
carcinoma | Rare entities |
|---|
|
Characterization | Heterogeneous group
that cannot be categorized into any other group | Tumor cells that
are discohesive and typically arranged in a single file or
Indian-file pattern, often distributed across a desmoplastic
stroma. | Cluster of low- to
moderate-grade tumor cells floating in a pool of extracellular
mucin. | Tubular carcinoma
Cribriform carcinoma Mucinous carcinoma Mucinous cystadenocarcinoma
Carcinoma with apocrine differentiation Metaplastic carcinoma |
| Frequency of
cases | 80% | 5–15% | 2% | 1% |
| Biomarkers | ER and HER2 | ER+,
HER2− and aberrant E-cadherin | ER+,
PR+ and HER2− | Varies according to
cancer subtype |
The diverse spectrum of BC encompasses distinct
molecular subtypes, exhibiting specific characteristics that guide
therapeutic strategies. Consequently, all newly diagnosed BC must
undergo immunohistochemistry (IHC) to identify the expression of
estrogen receptor (ER), progesterone receptor (PR), and human
epidermal growth factor receptor 2 (HER2). This molecular
characterization provides essential information for both prognostic
assessment and therapeutic decision-making (Table II) (29).
 | Table II.Molecular subtypes of breast cancer
based on receptor status (ER, PR, HER2), proliferation index
(Ki-67), histological grade and clinical parameters. |
Table II.
Molecular subtypes of breast cancer
based on receptor status (ER, PR, HER2), proliferation index
(Ki-67), histological grade and clinical parameters.
|
|
| Luminal B |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Luminal A | HER2- | HER2+ | HER2+ | TN |
|---|
| Biomarkers | ER+ | ER+ | ER+ | ER- | ER- |
|
| PR+ | PR- | PR-/+ | PR- | PR- |
|
| HER2- | HER2- | HER2+ | HER2+ | HER2- |
|
| Ki-67 low | Ki-67 high | Ki-67 low/high | Ki-67 high | Ki-67 high |
| Frequency of
cases | 40–50% | 20–30% |
| 10–20% | 15–20% |
| Target Therapy | Tamoxifen | Tamoxifen |
| Herceptin | - |
| Response to
Therapies | Endocrine | Endocrine
Chemotherapy | Chemotherapy | Chemotherapy |
|
|
|
|
|
| PARP
Inhibitors |
| Prognosis | Good | Intermediate |
| Poor | Poor |
| Observations | Controlled cell
growth | Fast cancer cell
growth | Overexpression of
HER2; Faster growth than luminals subtypes | Aggressive subtype;
occurs more often in younger women; highest association with BRCA1
mutations |
A tumor is considered positive for ER and PR when
these receptors are present in >1% of tumor cells, representing
~80% of BC cases (30). HER2
upregulation occurs in 15–20% of patients and is characterized by
intense membrane staining in >10% of invasive tumor cells (IHC
3+) or by HER2 gene amplification, determined through fluorescence
in situ hybridization (FISH), with a HER2/chromosome 17 centromeric
probe ratio ≥2.0 and ≥4 HER2 copy signals/cell. Tumors that do not
express any of these factors are classified as triple-negative
(TN)BC, corresponding to 10–15% of cases (31).
Hormone receptor-positive
(HR+) tumors
Tumors that test positive for the hormone receptors
estrogen (ER) or progesterone (PR) are classified as hormone
receptor-positive (HR+) and are typically categorized as luminal
subtypes, according to Perou et al (28). The receptors are located in target
cells and serve as ligand-dependent transcription factors. When
estrogen or an estrogen analog bind ER in the cell nucleus, a
conformational change occurs in the binding domain, enabling
interactions with coactivators if the ligand is an agonist and
blocking these interactions if it is an antagonist, thereby
affecting the transcription rates of estrogen-responsive genes
(32).
The updated American Society of Clinical
Oncology/College of American Pathologists guidelines established
that cancers with ER-positive staining in 1–10% of cells should be
classified as ER low positive (33). Limited data currently supports
endocrine therapy in this context, as these tumors typically
exhibit behavioral patterns more closely resembling TN rather than
luminal BC (34).
Superexpressing HER2 tumors
The HER2 oncogene belongs to the epidermal growth
factor receptor family. These receptors play a fundamental role in
activating signal transduction pathways responsible for epithelial
cell proliferation and differentiation, as well as angiogenesis
(35). High levels of HER2
expression indicate that patients may benefit from
receptor-targeted therapies.
HER2 is primarily identified through western
blotting, ELISA or IHC. In cases of indeterminate results,
alternative techniques may be employed, including ISH, FISH,
chromogenic or silver-enhanced ISH or PCR (36). HER2 expression is classified as
negative (IHC 0/1+), indeterminate (IHC 2+), or positive (IHC 3+)
based on staining intensity and tumor cell percentage, with
indeterminate and positive results requiring confirmatory testing
(30).
TNBC
TNBC refers to BC that does not express ER and PR,
or with IHC staining levels <1%. Additionally, the HER2 status
is 0 or 1+, with negative hybridization (FISH) for HER2+
expression (37).
Triple-negative breast cancer (TNBC) is considered
more aggressive than other breast cancer subtypes. It lacks
specific targeted therapies, such as hormone therapy used for
luminal tumors or anti-HER2 agents indicated for HER2 IHC 3+ cases.
TNBC accounts for approximately 15% of breast cancer diagnoses
worldwide and is more frequently observed in female patients under
the age of 40 years (21).
TNBC is characterized by distinct risk factors
supported by epidemiological evidence (38). BRCA mutations, particularly in
BRCA1, are found in up to 20% of patients with TNBC, indicating a
notable genetic predisposition. Also, African-American patients
demonstrate higher susceptibility compared with Caucasian patients,
as documented in population-based studies (39,40).
Premenopausal status is also a relevant risk factor (21). The absence of targeted therapies for
TNBC has driven extensive research to identify predictive
biomarkers for treatment response (41–43).
Investigations into immune system interactions with TNBC have
yielded notable insights into treatment responsiveness (44–46).
Tumor-infiltrating lymphocytes (TILs) in
BC
Role of TILs in tumor progression and
clinical value
Tumor biological behavior is influenced by its
intrinsic characteristics and the tumor microenvironment, which
interacts with the cancerous cells. This environment is formed by
various structures and substances, such as vessels, fibroblasts,
myofibroblasts and inflammatory cells (47).
Genetic alterations leading to malignancy are common
in somatic cells, and the immune system serves a crucial role in
the elimination or inactivation of abnormal cells. Studies in mice
have demonstrated an increased incidence of malignant neoplasms in
immunodeficient individuals, highlighting the importance of
immunity in cancer prevention (48–50).
TIL-mediated immunoediting:
Elimination and escape mechanism
TILs are used for tumor classification in the
clinical setting. These lymphocytes dynamically engage with other
immune system and cancer cells, termed cancer immunoediting cell
neoplasia, serving roles that can be either favorable or
detrimental to the tumor (51).
This process comprises three phases: Elimination, equilibrium and
escape (52). The elimination phase
occurs when tumor antigens are presented to CD8+
cytotoxic lymphocytes by cells, in combination with human leukocyte
antigen molecules for recognition by the T cell receptor (53). Most TILs in cancer are of T cell
phenotype, including CD4+ lymphocytes (helper cells) and
CD8+ (cytotoxic cells). CD4+ T lymphocytes
are essential for priming tumor-specific CD8+ TILs, in
addition to supporting their expansion and memory (54). Tumor elimination also involves the
action of natural killer (NK) cells and macrophages in antigen
identification. NK cells target tumor cells that have managed to
escape from cytotoxic lymphocyte control (55).
During the equilibrium phase, the FOXP3 protein
plays a critical role in the generation of CD4+
regulatory T cells (Tregs), which promote immunosuppression,
resulting in immunological tolerance for CD8+ cells.
Excessive FOXP3 expression is associated with Treg proliferation
and severe immunodeficiency, while its absence leads to immune
system activation. FOXP3 is also involved in immune escape
mechanisms, with impacts on low survival in breast cancer, as it
acts by decreasing the immune response in the tumor (56). In the equilibrium phase, the tumor
microenvironment presents a high proportion of cytotoxic cells and
a low proportion of Tregs due to decreased FOXP3 protein, allowing
progression to the next phase (57).
The third stage of the process is immune system
evasion, which can occur through decreased immunological
recognition due to antigen loss in neoplastic cells, increased cell
resistance or survival, reducing apoptosis through STAT3 or BCL-2
activation and development of an immunosuppressive
microenvironment. The latter involves the expression of cytokines
(VEGF, IL-10, TGF-β) and immunoregulatory molecules, including the
B7 family and CD3 expression (52).
The B7 family comprise the programmed cell death protein 1 (PD-1),
also known as CD279, programmed death-ligand 1 (PD-L1), cytotoxic
T-Lymphocyte associated protein 4 CTLA-4, V-domain Ig suppressor of
T cell Activation (VISTA), B7 homolog 4 and B and T Lymphocyte
Attenuator (BTLA).
Prospective use of TILs in precision
oncology
Characterization of immune evasion mechanisms
expands prognostic and therapeutic frameworks in oncology.
PD-1/PD-L1 checkpoint inhibition has demonstrated substantial
efficacy across multiple malignancies, like cutaneous squamous cell
carcinoma, with notable response rates in TNBC (58). This approach exemplifies successful
translation of immunobiological mechanisms into effective clinical
intervention (59). Similarly,
CTLA-4 blockade improves survival outcomes in patients with
melanoma (60). Recent
investigations have identified microRNA (miRNA or miR)-155 as a key
immunomodulatory factor influencing effector T cell function and
potentially predicting therapeutic response (18,61–63).
Integration of multidimensional biomarkers, including PD-L1
expression, tumor mutational burden, microsatellite instability,
miR-155 profile and TIL quantification, provides essential
stratification parameters, enabling precision in patient selection
and treatment optimization protocols (18).
T cell activation requires interaction between
complementary costimulatory molecules (B7 on antigen-presenting
cells and CD28 on T cells), providing essential secondary
signaling. Optimal T cell activation depends on the synchronous
delivery of both antigenic peptide recognition and costimulatory
signaling (64). In the absence of
co-stimulation, antigenic peptide presentation fails to induce
complete T cell activation, instead promoting immunological
tolerance (65). The degree of T
cell activation is determined by the balance between co-stimulation
and co-suppression. Clinical trials have demonstrated that PD-1
pathway blockade with anti-PD-1 or anti-PD-L1 therapy enhances T
cell-mediated anticancer responses without causing serious adverse
events (66,67).
Beyond intrinsic T cell regulation, their activation
is influenced by extrinsic factors. Cytokines such as IL-2,
released by CD4+ T helper (Th) cells (Th1 and Th17),
serve a direct role in promoting the expansion of cancer-specific T
cells (68). However, these
regulatory cells inhibit specific T cell function, inducing
immunosuppression and decreasing immunotherapy efficacy (69).
TILs Working Group consider breast carcinomas rich
in inflammatory infiltrates when they exhibit >50% lymphocytic
presence in the tumor stroma (70).
A previous study showed that, in this context, TNBC is associated
with improved progression-free survival (PFS) and overall survival
(OS) (42). A meta-analysis
including >22,000 patients demonstrated the presence of
CD8+ lymphocytes is associated with favorable prognosis,
while FOXP3 expression is associated with lower OS and PFS rates
(71).
In tumors treated with neoadjuvant chemotherapy,
high TIL levels favor the achievement of pathological complete
response (pCR) (72,73). A study involving 3,771 patients with
different molecular subtypes of BC undergoing neoadjuvant
chemotherapy classified TIL expression into three categories
(74): Low (0–10%), intermediate
(11–59%) and high (60–100%). In patients with luminal cancer, pCR
occurred in 45 (6%) of 759 patients with low TILs, 48 (11%) of 435
with intermediate TILs and 49 (28%) of 172 with high TILs. In
HER2-positive subtype, pCR was observed in 194 (32%) of 605
patients with low TILs, 198 (39%) of 512 with intermediate TILs and
127 (48%) of 262 with high TIL levels. In patients with TNBC, pCR
was achieved in 80 (31%) of 260 patients with low, 117 (31%) of 373
with intermediate and 136 (50%) of 273 with high TIL levels. In
univariate analysis, a 10% increase in TILs demonstrated a hazard
ratio of 0.93 for disease-free survival (DFS) and 0.92 for OS in
TNBC, showing negative results in other tumor subtypes (74).
A study of 1,966 patients with TNBC revealed a 94%
recurrence-free survival (RFS) and 95% OS rate in stage I patients
with TIL levels ≥50% with TNBC (75). In patients with TIL levels <30%,
RFS and OS rates were 78 and 82%, respectively, in patients who did
not undergo neoadjuvant therapy. These results confirm TIL
abundance in tissue as a notable prognostic factor for patients
with TNBC (75).
The clinical evidence regarding the prognostic and
predictive value of TILs in BC, particularly in TNBC, affirms their
potential use as a clinically actionable biomarker (76). However, the implementation of TILs
assessment in routine clinical practice necessitates standardized
methodological approaches and clearly defined threshold values for
optimal patient stratification. Given the continuous nature of TIL
measurements and their varying importance across molecular
subtypes, establishing clinically relevant cut-off values is a key
step toward integrating this immunological parameter into treatment
decision protocols.
Clinically relevant cut-off values for TILs
in pre-neoadjuvant biopsy samples
Proportion of TILs in BC
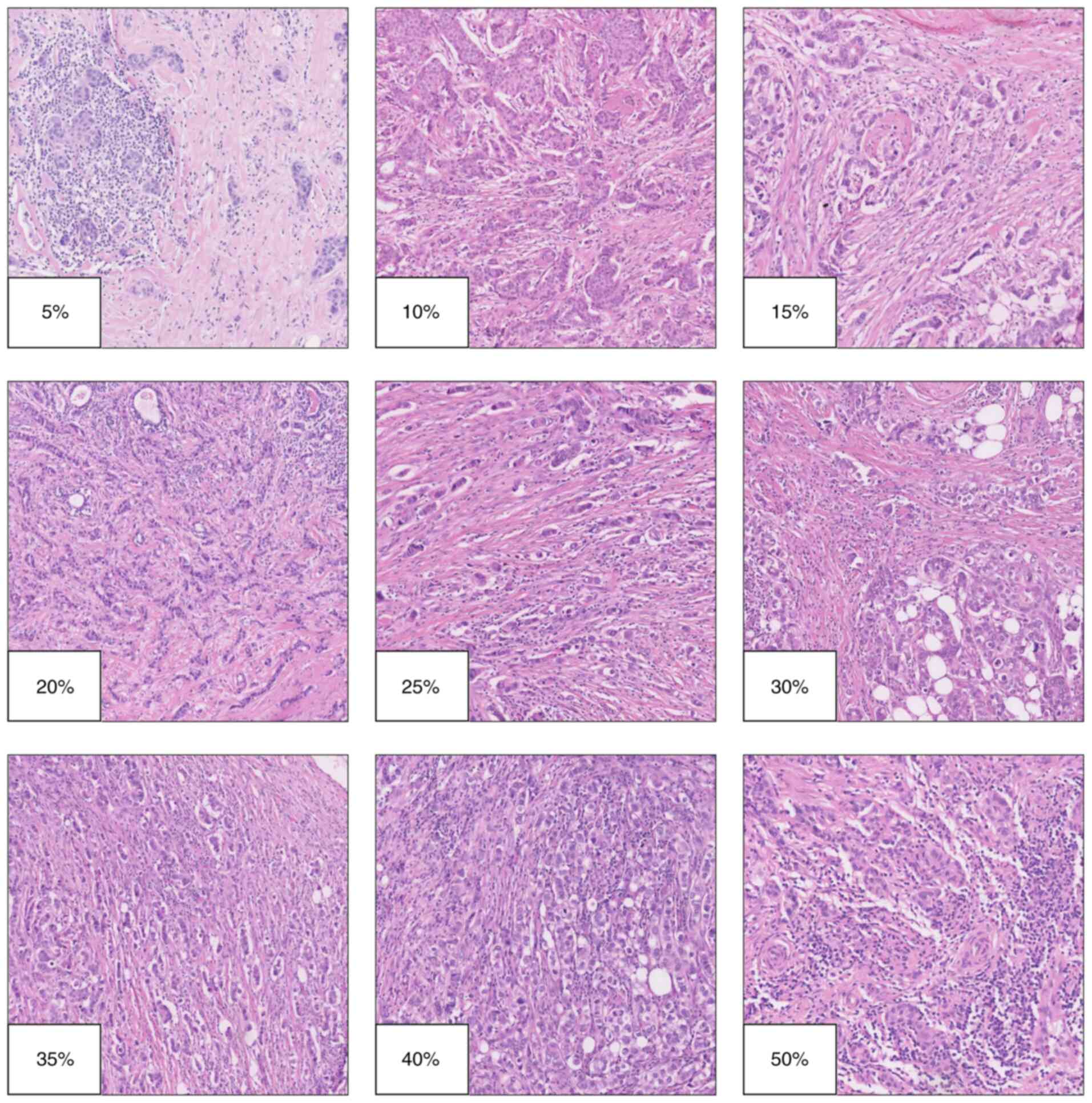
Numerous critical aspects must be considered when
analyzing TILs. First, the pathologist-reported value refers to the
percentage of TILs. The proportion of TILs represents the area of
tumor stroma infiltrated total lymphocytes relative to the total
area of stroma examined by the pathologist and should be measured
using the hematoxylin-eosin method (Fig. 2). Adherence to recommendations by
the TILs Working Group is essential for accuracy and
reproducibility (70,77).
Higher levels of lymphocytic infiltration serve as
predictors of pCR (78). However, a
formal recommendation of a clinically relevant cut-off value
categorizing breast tumors as having high lymphocytic infiltration
is lacking, complicating the establishment of a universal standard
for this variable as a therapeutic response biomarker.
Meta-analyses reveal cut-off values ranging from 10 to 60%
(79,80).
The relevance of high TILs as biomarkers of good
therapeutic response has more clinical value for TN tumors compared
with other BC subtypes (81). Ochi
et al (82) categorized
tumors into three groups: Low TILs (0–9%), intermediate (10–49%)
and high TILs (≥50%), the latter being known as
lymphocyte-predominant (LP)BC. The pCR rates in TN tumors with low
TILs are 4%, compared with 43.6% in those with intermediate or high
TILs. In HER2-positive tumors, pCR rates are 26 vs. 51.9%,
respectively, and significant in both cases (82). Loi et al (83) observed higher TILs in TNBC (n=134)
and HER2+ (n=209) compared with luminal BC subtypes
(n=591) (83). The second quartile
median of TILs expression was 25.0, 15.0 and 7.5% for these tumor
types, respectively. Despite substantial variability in
infiltration percentages for TNBC, HER2+ and luminal
groups, the upper quartile showed 40.0, 30.0 and 12.5%
infiltration, respectively. This aligns with findings of Stanton
et al (78,84), which demonstrated TNBC and
HER2+ as most frequent subtype of LPBC (78,84)
Denkert et al (74)
reinforced these findings, reporting high TIL percentages (>60%)
in 30% of TNBC tumors, 19% in HER2+ and 13% in
luminal-HER2-negative tumors (74).
Loi et al (83) identified
TILs (LPBC with a cut-off ≥50%) as predictors of good prognosis for
distant disease-free survival in TNBC but not in HER2+
or luminal BC (83). Additionally,
Russo et al (85) supported
these findings by using TILs ≥30% for LPBC classification, a
cut-off derived from Liu et al (86). In an analysis of 41 TNBC samples,
Russo et al (85) found that
34% had TILs >30%, with a pCR rate of 78.6% (11/14) compared
with 14.8% (4/27) in those with TILs <30%. Furthermore, a
significant pCR rate of 71.4% was noted in HER2+
individuals with TILs >30%.
Thresholds for LPBCs: Issues and
sensitivity limitations
A fundamental aspect of using TILs for prognosis is
determining the appropriate percentage of lymphocytic infiltration
that characterizes LPBCs. A widely accepted cut-off value is 50%
for high lymphocytic infiltration, as recommended by the TILs
Working Group (70). Establishing
this threshold is key for accurately identifying LPBCs, which can
impact treatment decisions and prognostic evaluation.
Salgado et al (70) proposed LPBC classification for
tumors with ‘more lymphocytes than tumor cells’, meaning they
exhibit 50–60% lymphocytic infiltration (70). Denkert et al (87) reported that 40% of patients with
LPBC (>50% TILs) achieved pCR, compared with 7% of patients
without lymphocytic infiltration (87). Cut-off values of 50 or 60% predict
both short-term pCR responses and long-term OS and DFS (Table III) (82,83,88–91)
.
 | Table III.TIL thresholds and study
characteristics. |
Table III.
TIL thresholds and study
characteristics.
| First author(s),
year | Cut-off value,
% | Breast
subtypes | Lymphocyte
type | Number of
patients | Duration of
follow-up, months | Evaluation
indicator | Country | Outcome | (Refs.) |
|---|
| Dieci et al,
2014 | 60 | TNBC | All | 278 | 76 | OS, DFS | France | High-TIL (n=27):
91% 5-year OS (95% CI, 68–97%). Low-TIL: 55% 5-year OS (95% CI
48–61%) | (88) |
| Loi et al,
2019 | 50 | HER2+,
HR+, TNBC | All | 1,010 | 62 | OS, DFS | Australia | TNBC (n=134): 10%
increase in TILs associated with decreased distant recurrence (HR
0.77, 95% CI 0.61–0.98, P=0.02). | (95) |
|
|
|
|
|
|
|
|
| HER2+ BC
(n=209): 10% increase in lymphocytic infiltration associated with
decreased distant recurrence in trastuzumab-treated patients (P=
0.025). |
|
| Ochi et al,
2019 | 50 | HER2+,
TNBC | All | 209 | 120 | DFS, pCR | Japan | Low pre-NAT TILs
associated with lower pCR rate: TNBC (4.0 vs. 43.6%);
HER2+ BC (26.0 vs. 51.9%). | (82) |
|
|
|
|
|
|
|
|
| In TNBC with RD:
Low pre-NAT TILs associated with shorter RFS (HR 3.844, P=0.024).
Low post-NAT TILs demonstrate a non-significant association with
shorter RFS (HR 2.836, P=0.061). No association between TIL change
and RFS in TNBC or HER2+ BC |
|
| Cerbelli et
al, 2017 | 50 | TNBC | All | 54 | - | pCR | Italy | pCR achieved in 35%
of cases. Univariate analysis: PD-L1 expression in ≥25% of
neoplastic cells associated with pCR (P=0.024). ≥50% sTILs
associated with higher pCR frequency (P<0.001). Multivariate
analysis: PD-L1 expression on tumor cells significantly associated
with pCR (OR 1.13, 95% CI 1.01–1.27). | (91) |
| Van Bockstal et
al, 2020 | 40 | HER2+,
TNBC | All | 35 | 8 | pCR | Belgium | High sTILs (≥40%)
significantly associated with increased pCR rate | (92) |
| Loi et al,
2014 | 30 | TNBC | All |
2,148a | >78 | DFS, OS | Australia, France,
Italy, Finland, Belgium, Germany and USA | 736 iDFS and 548
D-DFS events; 533 deaths. sTILs are a significant prognostic
biomarker for: iDFS (HR 0.87, 95% CI 0.83–0.91, P<0.001), D-DFS
(HR 0.83, 95% CI 0.79–0.88, P<0.001), OS (HR 0.84, 95% CI
0.79–0.89, P<0.001) Node-negative patients with sTILs ≥30%,
showed the following outcomes at 3 years outcomes: iDFS, 92% (95%
CI 89–98%), D-DFS, 97% (95% CI 95–99%), OS, 99% (95% CI
97–100%) | (83) |
| Park et al,
2019 | 30 | TNBC | All |
476b | 96 | DFS, OS | France, Italy, | A total of 107
deaths, 173 iDFS and 118 D-DFS events. sTILs have independent
prognostic value for iDFS | (96) |
|
|
|
|
|
|
|
| South Korea | (HR 0.90, 95% CI
0.82–0.97, P<0.01), D-DFS (HR 0.86, 95% CI 0.77–0.95,
P<0.01), OS (HR 0.88, 95% CI 0.79–0.98, P=0.015). |
|
|
|
|
|
|
|
|
|
| Patients with stage
I tumors and sTILs ≥30% (n=74) had the following 5-year outcomes:
iDFS, 91% (95% CI 84–96%), D-DFS, 97% (95% CI 93–100%), OS, 98%
(95% CI 95–100%). |
|
| Russo et al,
2019 | 30 | HER2+,
TNBC | All | 187 | 62.5 | OS, pCR | Venezuela | pCR rate: TILs ≥30%
= 58.5% (odds ratio, 8.85); TILs <30% = 11% (P<0.001) | (85) |
|
|
|
|
|
|
|
|
| Association in
HER2+ and TN subtypes. No association between TILs and
OS (P=0.834) or DFS (P=0.937) |
|
| Floris et
al, 2021 | 30 | TNBC | All | 445 | 91.56 | pCR | Belgium | High sTIL
associated with pCR in lean (OR 4.24; 95% CI 2.10–8.56, P<0.001)
but not heavier patients (OR 1.48, 95% CI 0.75–2.91, P=0.26) | (93) |
|
|
|
|
|
|
|
|
| High sTIL
associated with increased event-free survival in lean patients (HR
0.22, 95% CI 0.08–0.62, P=0.004) but not heavier patients (HR 0.53,
95% CI 0.26–1.08, P=0.08) Similar results for OS |
|
| Dieci et al,
2020 | 30 | TNBC | All | 244 | 81.6 | pCR | Italy | TILs confirmed as
independent prognostic factor. PD-L1 is a prognostic biomarker: LR
χ2 4.60, P=0.032 (model with classical factors,
including age, stage, histological grade and TIL 10% increments).
LR χ2 6.50, P=0.011 (model with classical factors and
TIL >30 vs. <30%). | (94) |
|
|
|
|
|
|
|
|
| In patients treated
with neoadjuvant chemotherapy, FOXP3 is a prognostic biomarker in
addition to classical factors, including age, stage, histologic
grade -, TILs, and pCR (LR χ2 5.01, P=0.025). |
|
|
|
|
|
|
|
|
|
| In patients without
pCR, CD8 and PD-L1 expression significantly increase from baseline
to residual disease |
|
| Jimenez et
al, 2022 | 20 | TNBC | All | 80 | - | pCR | USA | 38/145 extracted
MRIRF significantly associated with pCR; 5 non-redundant imaging
features. | (215) |
|
|
|
|
|
|
|
|
| MRIRF model
accuracy (P=0.001, 72.7% PPV, 72.0% NPV). TIL model accuracy
(P=0.038; 65.5% PPV, 72.6% NPV). Combined MRIRF and TIL model has
improved prognostic accuracy (P<0.001, 90.9% PPV; 81.4%
NPV)-AUC: 0.632 (TIL); 0.712 (MRIRF); 0.752 (TIL + MRIRF). |
|
| Asano et al,
2018 | 10 | HER2+,
TNBC | All | 177 | 40 | OS, DFS, pCR,
HR | Japan | High-(n=96) vs.
low-TIL (n=81): TNBC and HER2+ BC more frequent (P<0.001 and
P=0.040, respectively); higher pCR rate (P=0.003). | (216) |
|
|
|
|
|
|
|
|
| In TNBC and HER2+
BC, pCR rate is higher in high-TIL group (P=0.013 and P=0.014,
respectively). |
|
|
|
|
|
|
|
|
|
| Multivariable
analysis: High-TIL status independently predicts favorable
prognosis (HR=0.24, P=0.023 and HR=0.13, P=0.036). Biopsy specimens
from local recurrence following NAT show decreased TIL. |
|
| Song et al,
2017 | 10 | TNBC | All, T, B | 180 | 34.9 | DFS, pCR | Korea | Mean number of HEVs
in pre-NAT biopsy, 12 (range, 0–72). TILs, TLSs, HEV density and
CXCL13 expression are associated with each other | (217) |
|
|
|
|
|
|
|
|
| Higher
CD8+ cell density and CXCL13 expression are
significantly associated with better DFS rate |
|
| Khoury et
al, 2018 | 46 | HER2+,
TNBC luminal | All | 331 | - | pCR | Canadian | pCR rate: TBNC
30.9% (29/95); HER2+ 32.5% (25/77); Luminal 5.6%
(9/159) | (218) |
|
|
|
|
|
|
|
|
| Independent
predictors of pCR: Luminal subtype iTIL (OR=1.44, 95% CI 1.08–1.9,
P=0.013), TNBC subtype sTIL (OR=1.68, 95% CI 1.29–2.18, P=0.001)
and iTIL (OR=1.31, 95% CI 1.05–1.63, P=0.017). |
|
|
|
|
|
|
|
|
|
| HER2+
subtype: sTIL and iTIL do not predict pCR. |
|
| Ruan et al,
2018 | 20 or 10 | TNBC | All | 166 | - | pCR | China | Univariate logistic
regression analysis: sTILs (P=0.0001) and iTILs (P=0.001) are
associated with pCR. Multivariate logistic regression analysis:
sTILs (P=0.006) and iTILs (P=0.04) independent predictors of pCR
ROC curve analysis of TNBCs with >20% sTILs (P=0.001) or >10%
iTILs (P=0.003) associated with higher pCR rate. Multivariate
analysis: 20% sTILs (P=0.005) independently predicts pCR | (219) |
Using a >50% TIL cut-off as a significant
infiltration marker has limitations regarding sensitivity, as
relatively few breast tumors reach this cut-off. Stanton et
al (78,84) found that ~20% of TN and
HER2+ tumors exhibit LPBC with >50% TILs (78,84).
Denkert et al (47) assessed
3,771 samples, reporting pCR in 50% of high-TIL TNBC tumors and 48%
of high-TIL HER2+ tumors (>60% TILs) (74). pCR rates for TNBC and
HER2+ with intermediate infiltration (11–59%) were 31
and 39%, respectively. Hence, a cut-off between 11 (low
infiltration) and 50% (high infiltration) could enhance sensitivity
for identifying individuals with favorable prognosis without
compromising the discriminative accuracy.
Comparison of accuracy and sensitivity
in LPBCs using lymphocyte infiltration thresholds between 30 and
50%
Studies have used cut-offs between 30 and 50% for
defining LPBC (Table III). An
analysis from Belgium employed a 40% cut-off (92), while other studies in Venezuela,
Belgium and Italy used a 30% cut-off (85,93,94).
The prognostic significance of TILs in early-stage TNBC was
demonstrated by two pivotal multicenter studies (95) (96).
Firstly, a retrospective individual patient data (IPD)
meta-analysis was conducted using data from 2,148 early-stage TNBC
patients across 9 international studies from Australia, Europe
(France, Italy, Finland, Belgium, Germany), and the United States.
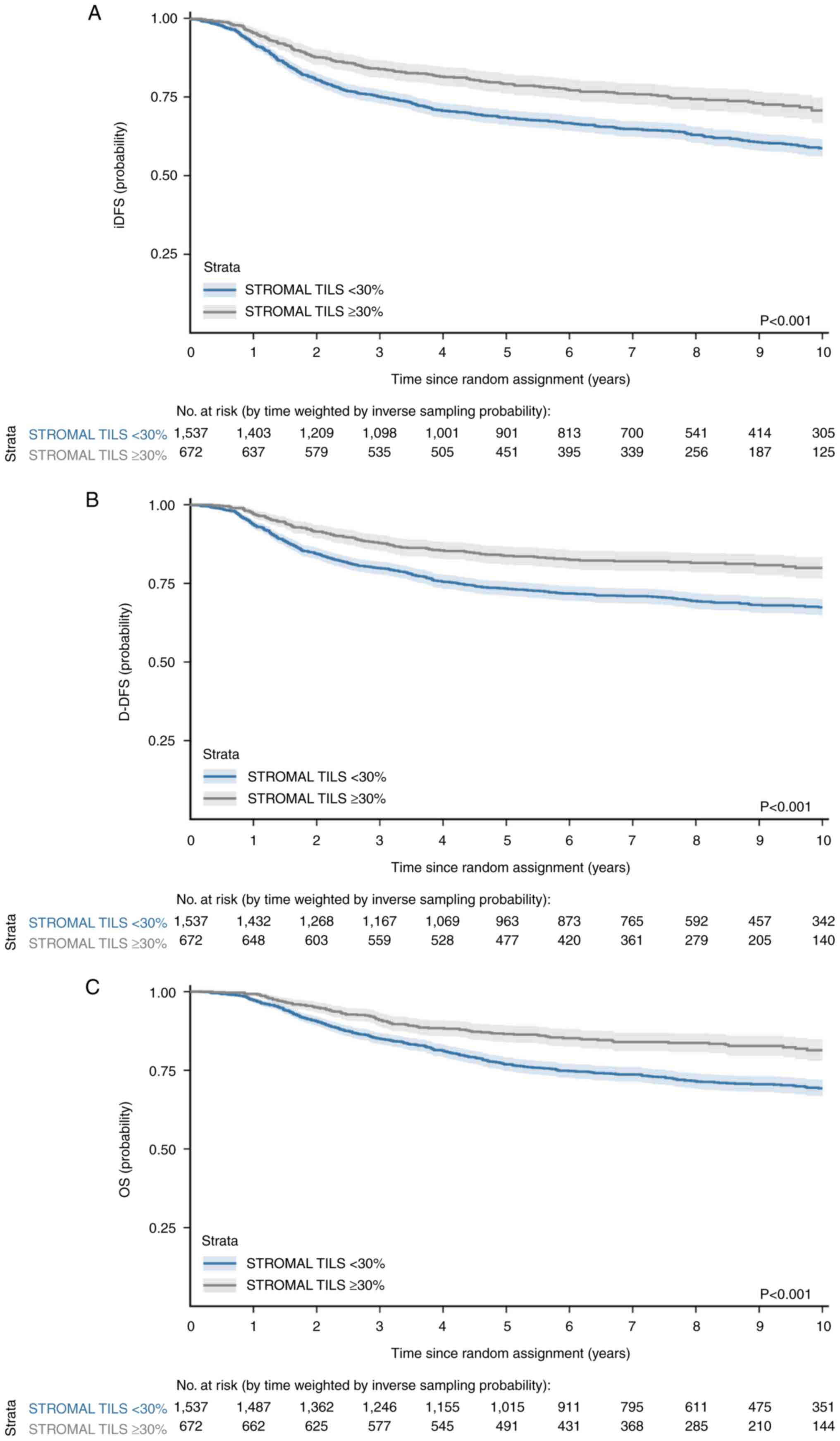
The analysis demonstrated that a stromal TIL cut-off of ≥30% was
associated with significantly better prognosis in early-stage TNBC
patients (95). The second study
was a retrospective pooled analysis of 476 early-stage TNBC
patients from 4 centers who did not receive adjuvant chemotherapy,
showing that Stage I patients with sTILs ≥30% had excellent 5-year
survival (91–98%) without any systemic treatment (96). The former study evaluated invasive
(i)DFS (primary endpoint), distant (D-)DFS and OS, treating TILs as
a continuous variable. Treatments included either an anthracycline
or a combination of anthracycline and taxane. Patients with
node-negative TILs ≥30% showed three-year iDFS of 92% (95% CI,
89–98%), D-DFS of 97% (95% CI, 95–99%) and OS of 99% (95% CI,
97–100%; Fig. 3). The
aforementioned study recommend integrating TILs into
clinical-pathological diagnostic models (97). In 2,148 individuals, TIL exhibited
an interquartile range from 10 to 30%, with a median of 15%.
Notably, about one-third of patients showed ≥30% TILs, broadening
the pool of individuals benefiting from the biomarker compared with
studies using 50% TILs, which was present in one-fifth of patients
(78,84). The 30% cut-off aligns with Q3
indicating 30% lymphocytic infiltration as the threshold parameter
(95).
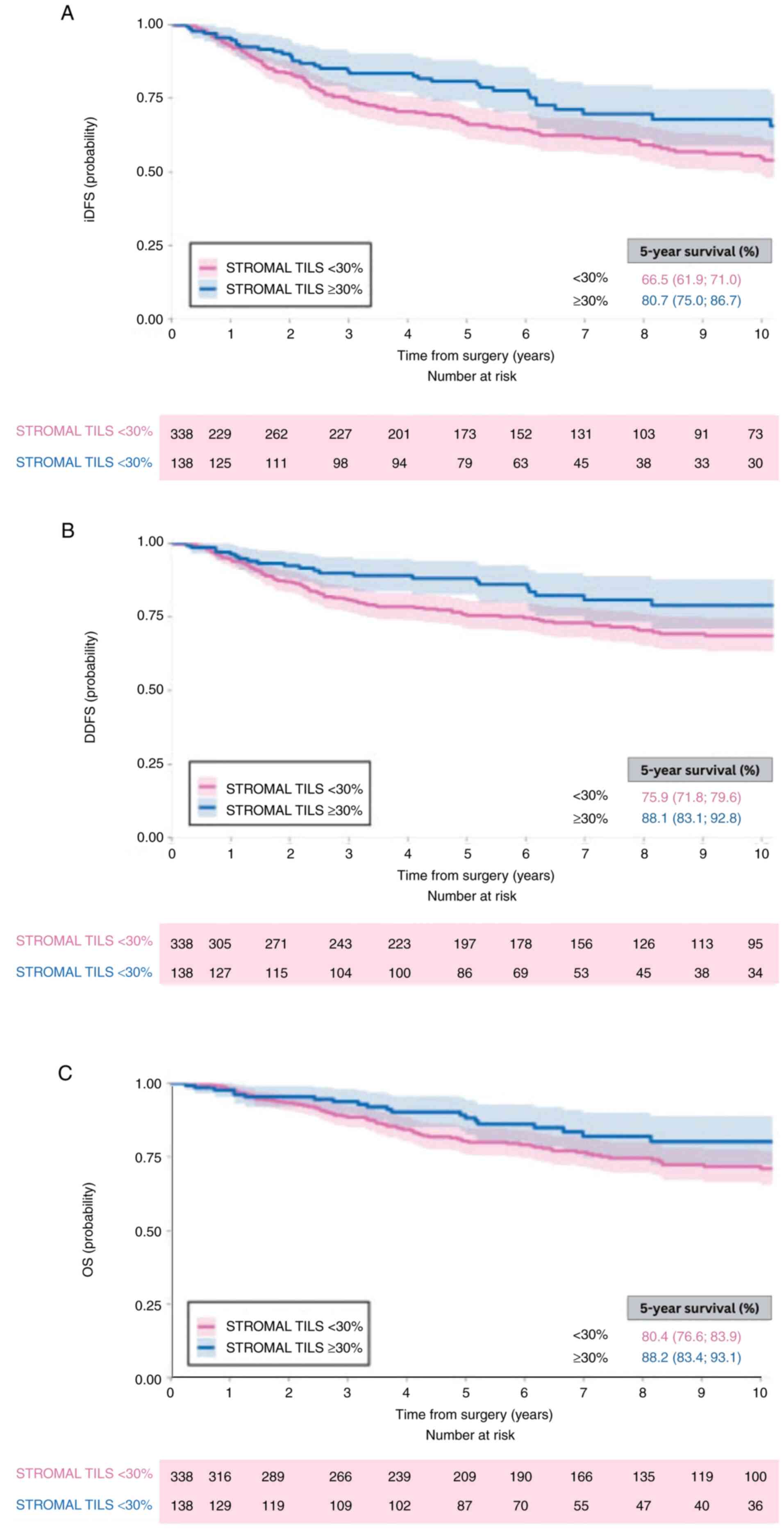
Park et al (96) investigated whether TILs ≥30%
identify those who might not require adjuvant chemotherapy
(96). Patients with stage I TNBC
(n=74) presented a 5-year iDFS of 91% (95% CI 84–96%), D-DFS of 97%
(95% CI 93–100%) and OS of 98% (95% CI 95–100%; Fig. 4). The aforementioned study used the
Q3-based cut-off of 30%, consistent with Loi et al (95). Compared with the aforementioned
study, the median was lower (10 vs. 15%), while the third quartile
remained at 30%, confirming its consistency. Stromal TILs ≥30%
could signify a subgroup of patients with stage I TNBC with
excellent prognosis without adjuvant therapy. This suggests
vulnerable groups, such as the elderly or those with comorbidities,
may be spared from the toxicity and costs of adjuvant chemotherapy
without jeopardizing survival rates. Moreover, TIL evaluation at
30% may yield good reproducibility among pathologists, as at this
level, visual differences are easier to be detected. Nonetheless,
the use of biomarkers necessitates caution concerning established
clinical diagnostic criteria, such as tumor size and lymph node
status, to refine pathological analysis and therapeutic insights
for each clinical case (98).
Russo et al (85), Floris et al (93) and Dieci et al (94) demonstrated good discriminative
capacity with a TIL cut-off of 30% (85,93,94).
Russo et al (85) reported a
five-fold greater incidence of pCR for patients with TILs ≥30%
compared with those with TILs <30% (58.5 vs. 11.0%) (85).
TIL thresholds of 30 or 50% not only serve as
promising biomarkers for guiding chemotherapy de-escalation in
early-stage TNBC, but also inform therapeutic decisions regarding
immune checkpoint inhibitors. Measuring TIL and PD-L1 expression
aids in identifying immune-enriched tumors, thereby enhancing
selection of patients with advanced TNBC or HER2+ BC who
are likely to respond to PD-1/PD-L1 inhibitors (99).
Comparative analyses between 30 and
50% thresholds for LPBCs
The recommendation to adopt 30 or 50% as clinical
cut-off values for LPBCs is based on their predictive value for pCR
and favorable prognostic outcomes in BC. TILs ≥50% are associated
with both short-term pCR responses and long-term OS as well as DFS
(82,83,88–91).
This threshold was initially proposed by the TILs Working Group
(70).
Further studies have indicated that cut-off values
<50% can also predict favorable prognosis, with the majority
using 30% as the cut-off value (85,93,94).
Specifically, two studies employed TILs ≥30%: The first identified
patients who would benefit from adjuvant therapy, while the second
identified individuals with a favorable prognosis who may not
require adjuvant chemotherapy (95,96).
In conclusion, both cut-off values for LPBCs are
effective in identifying patients who are likely to benefit from
therapeutic intervention. The 30% threshold is particularly
inclusive, enabling the identification of a broader patient
population compared with the stricter 50%, which is less frequently
achieved in clinical practice (78,84).
miR-155 and TIL activity
The prognostic value of TILs in TNBC is mediated
through complex molecular regulatory networks. Post-transcriptional
gene regulation via miRNAs is a critical mechanism in the
modulation of the tumor inflammatory landscape, with miR-155
emerging as a key mediator. miRNAs are a class of small,
non-coding, single-stranded RNAs, typically 18–25 nucleotides in
length, that play a crucial role in the post-transcriptional
regulation of gene expression. By directly binding to the 3′
untranslated regions (UTRs) of target messenger RNAs (mRNAs),
miRNAs promote mRNA degradation or inhibit translation (100,101). This control of protein synthesis
allows miRNAs to function as modulators of gene expression rather
than as complete silencers (100,101).
Biogenesis and function
In animals, miRNAs are transcribed by RNA polymerase
II (Pol II) as primary transcripts known as primary miRNAs
(pri-miRNAs), which adopt a characteristic hairpin structure
(Fig. 5). These pri-miRNAs are
processed in the nucleus by the enzymes Drosha and DiGeorge
syndrome critical region 8 DGCR8 (DGCR8) is a protein that serves a
crucial role in the maturation of microRNAs to generate precursor
miRNAs (pre-miRNAs), which are transported to the cytoplasm by the
protein exportin-5. In the cytoplasm, a second processing step is
performed by the enzyme Dicer, in association with the
double-stranded RNA-binding proteins trans-activation response
RNA-binding protein. It's a cellular protein that primarily binds
to the trans-activation response RNA-binding protein (TRBP) or
protein activator of PKR (Protein Kinase R), that are
double-stranded RNA-binding proteins resulting in a miRNA duplex
18–25 nucleotides in length. Subsequently, the passenger (sense)
strand is degraded, while the guide (antisense) strand,
representing the mature miRNA, is incorporated into the RNA-induced
silencing complex (RISC). Within RISC, argonaute proteins serve a
central role in directing the complex to target mRNAs containing
partially complementary sequences within their 3′ UTRs. This
interaction leads to gene silencing through either translational
suppression or mRNA destabilization (102–105).
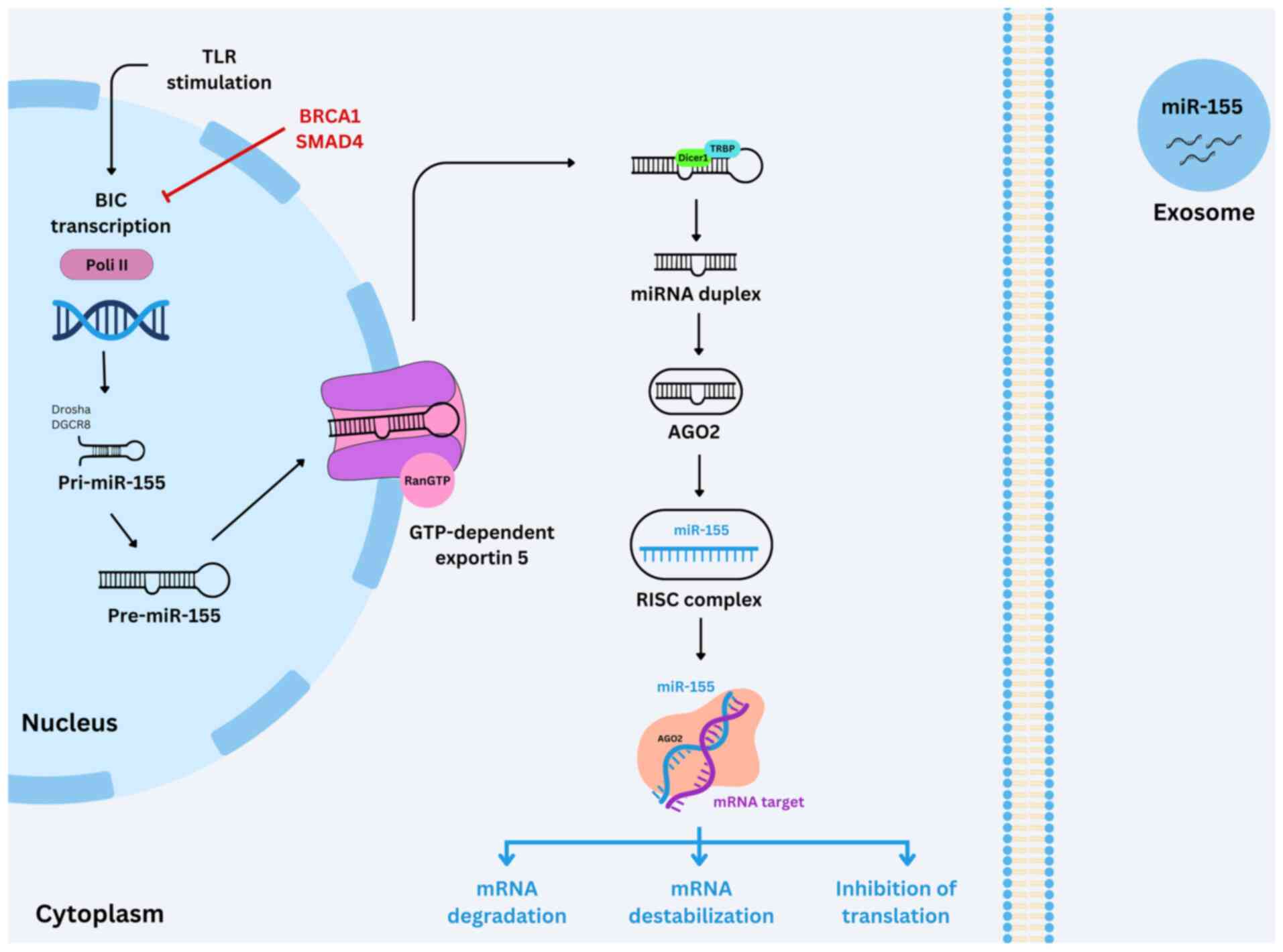 | Figure 5.Biogenesis and function of miR-155.
TLR stimulation initiates BIC transcription by Pol II in the
nucleus, producing pri-miR-155 (negatively regulated by
BRCA1/SMAD4). Drosha/DGCR8 processing generates pre-miR-155, which
is exported to the cytoplasm via exportin 5. Cytoplasmic
Dicer1/TRBP cleaves pre-miR-155 into a duplex that incorporates
into AGO2, forming the RISC complex. Mature miR-155 mediates mRNA
degradation, destabilization or translational inhibition. miR-155
can be packaged into exosomes for intercellular signaling. miR,
microRNA; TLR, toll-like receptor; pri-miR, primary microRNA;
DGCR8, DiGeorge syndrome critical region 8 protein; TRBP,
transactivation response RNA binding protein; AGO, argonaut
proteins; RISC, RNA-induced silencing; BIC, B cell integration
cluster; Pol, polymerase. |
Evolutionarily conserved across a range of
organisms, miRNAs comprise ~1% of the human genome. Despite their
small size, these regulatory RNAs exert post-transcriptional
control over more than one-third of all protein-coding genes,
underscoring their key role in numerous cellular processes and
disease pathogenesis (106,107)
. miRNA dysregulation is implicated in various pathological
conditions, most notably cancer, where altered expression patterns
contribute to tumor initiation, progression, and metastasis
(20,105,108). Therefore, understanding the role
of miRNAs in oncogenesis is key for the development of novel
diagnostic and therapeutic strategies (20,105,108)
The first miRNA sequence database, established in
2002, contained 506 entries across six organisms (109,110). By 2010, the number of annotated
human miRNAs had grown to 1,424 (111). However, miRNAs may be reclassified
over time due to initial misannotations, often stemming from high
sequence similarity between precursor miRNAs and minor sequence
variants (112).
miRNAs in BC
The search for predictive biomarkers to improve
early cancer diagnosis and therapeutic outcomes increasingly
emphasizes the analysis of molecular signatures in normal tissue
prior to the clinical manifestation of disease (112–114). In BC, miRNAs have been
consistently identified as dysregulated in both tissue and
serum/plasma samples (115–117).
These alterations contribute to early disease development and
progression by modulating the post-transcriptional expression of
proto-oncogenes and tumor suppressor genes. Such findings
underscore the potential of miRNAs as valuable biomarkers for early
detection and targeted intervention in BC (118,119).
The prognostic value of miRNAs in BC connects
directly to critical genomic alterations. Genomic instability, a
key hallmark of malignancy, affects and is influenced by miRNA
expression patterns. This enables tumor cells to acquire multiple
growth advantages such as autonomous growth signaling, resistance
to inhibitory stimuli, apoptosis evasion, sustained angiogenesis
and enhanced metastatic capacity (120). This instability also affects miRNA
expression, disrupting key regulatory pathways involved in tumor
initiation and progression. miRNAs have emerged as key regulators
in the interplay between cancer cells and the immune
microenvironment (121). Moreover,
miRNAs promote genomic instability by influencing DNA double-strand
break repair, mismatch repair mechanisms and DNA methylation
patterns (100)
miRNAs are key regulators of the interplay between
cancer cells and the immune system, functioning as either oncogenes
or tumor suppressors depending on the cellular context. They
influence key biological processes including tumorigenesis, cell
proliferation, apoptosis and metastasis. Notably, miR-21 and
miR-155 are frequently upregulated across numerous types of cancer,
like lung cancer or prostate and hematologic tumor, highlighting
their prominent roles in tumor initiation and progression (122).
miRNAs exhibit context-dependent functions. For
example, miR-10b, miR-103/107 and miR-30d have been characterized
as oncogenic, whereas miR-31, miR-29 and members of the miR-200
family act as tumor suppressors (123). Additionally, miR-7 is a negative
regulator of epidermal growth factor receptor expression in BC
cells, further illustrating the diverse mechanisms by which miRNAs
modulate oncogenic signaling pathways (124).
In BC, miRNAs serve influential roles across all
molecular subtypes, with specific subsets contributing to key
regulatory pathways. For example, miR-21, miR-18a/b, miR-193,
miR-302, miR-92, let-7, miR-22, miR-221/222, miR-449a/b and the
miR-17-92 cluster modulate ER signaling, a pathway central to
HR-positive BC. Furthermore, HER2 protein, a major oncogenic driver
in HER2+ BC, is regulated by miR-125a/b, highlighting
the diverse roles of miRNAs in shaping the molecular landscape of
BC (125).
Beyond their regulatory functions, miRNAs serve as
promising diagnostic and prognostic biomarkers in BC research
(116). Their dysregulation in
tumor development and progression supports their potential use in
detection, diagnosis, subtype classification and targeted therapy
(117). Distinct miRNA expression
profiles reliably differentiate BC tissue from normal counterparts
(121). For example, upregulation
of miR-21, miR-106a and miR-155, along with reduced expression of
miR-126, miR-199a and miR-335, is observed in BC compared with
normal tissues (126).
The first comprehensive study evaluating miRNA
expression in BC tissue analyzed 76 tumor samples and identified 29
miRNAs with differential expression compared with normal breast
tissue; among these, miR-10b, miR-125b, miR-145, miR-21 and miR-155
were key candidates, with miR-155 demonstrating the most pronounced
dysregulation (127).
miR-21 is one of the most extensively studied
miRNAs in BC and is associated with aggressive tumor
characteristics, including advanced clinical stage, high
histological grade and HR-negative status (128–130). Its expression is positively
regulated by TGF-β, and its upregulation is observed in BC tissue
(131).
Further research has revealed subtype-specific
miRNA expression patterns (132,133). For example, miR-342 expression is
elevated in ER+ and HER2+ tumors but
decreased in TNBC (134).
Conversely, miR-520 is upregulated in HR− tumors,
suggesting distinct functional roles across BC subtypes (135). Analysis examining 309 miRNAs in 93
breast tumor samples identified 31 miRNAs associated with favorable
prognostic features, including low histological grade and
ER+ status (136).
Notably, miR-155 is upregulated in BC tissues compared with normal
controls and elevated in ER− compared with
ER+ tumors (136).
Disruption of miRNA expression patterns can lead to
widespread cell dysfunction, including aberrant cell cycle
regulation and uncontrolled tumor proliferation (16).
miR-155
miR-155 is an evolutionarily conserved molecule
encoded by the host gene MIR155HG, located on chromosome 21 at the
21q21.3 locus (137). This gene
spans ~13 kilobases and comprises three exons, with the third exon
containing a 1,500-base-pair primary transcript, pri-miR-155, which
undergoes sequential processing to produce the mature miR-155
(138). Initially identified as
the B cell integration cluster, MIR155HG was first recognized for
its role as a retroviral integration site in B cell lymphoma in
both human and animal models, marking its role in oncogenesis
(139). Mature miR-155 exists in
two isoforms: miR-155-5p and miR-155-3p, each exerting distinct
regulatory effects. miR-155-3p enhances the production of IFN-α/β
by promoting degradation of interleukin-1 receptor-associated
kinase 3, whereas miR-155-5p suppresses IFN-α/β production by
targeting Transforming growth factor-β (TGF-β)-activated kinase 1,
also known as MAP3K7 binding protein 2 (138,140,141). miR-155-5p also modulates immune
responses and contributes to drug resistance, while miR-155-3p is
implicated in multiple types of cancer, like lung cancer and
glioblastoma (141). These diverse
functions underscore the multifaceted role of miR-155 in cancer
progression and immune regulation (20,138,140–143).
miR-155 has emerged as a promising therapeutic
target due to its involvement in immunosuppressive mechanisms and
its potential to enhance antitumor immune responses. This miRNA
serves a key role in augmenting T cell-mediated immunity by
improving T cell functionality, memory formation, cytotoxic
activity and IFN-γ production (12). However, while enhancing miRNA
expression can potentiate T cell responses against tumors, IFN-γ
signaling induces the expression of PD-L1, which may paradoxically
promote a pro-tumorigenic environment (12).
miR-155 is one of the most extensively
characterized miRNAs in BC (19,20,142,144–148). Initially classified as an oncomiR,
it has been implicated in promoting tumorigenesis and disease
progression. Elevated expression of miR-155 is associated with
tumor development, poor prognosis and resistance to therapy in both
solid tumors and hematological malignancies (138,141). Functioning as an oncogene, miR-155
targets genes involved in immune regulation, DNA repair, hypoxia
responses and inflammation, thereby influencing both tumor cell
behavior and the tumor microenvironment (149). Studies have reported that high
miR-155 expression is associated with improved OS in various
cancers, including BC (19). This
may be attributed to its dual role in both the innate and adaptive
immune systems, as it is expressed in immune as well as in tumor
cells (150). Consequently, the
functional impact of miR-155 in BC remains context-dependent, and
whether its expression is pro- or antitumorigenic is under
investigation (12,18).
In patients with BC, circulating levels of miR-155
are significantly elevated compared with healthy individuals.
Moreover, miR-155 levels tend to decrease following adjuvant cancer
treatment, such as surgery and chemotherapy and are highest in
tumor with diameter >5 cm, suggesting its potential utility as a
biomarker for therapeutic monitoring. Elevated circulating miR-155
is also linked to early-stage tumor recurrence, further supporting
its relevance in disease surveillance (151,152).
Upregulation of miR-155 is associated with
treatment resistance, notably in trastuzumab-resistant BC. In
early-stage patients receiving trastuzumab-based therapy, elevated
levels of circulating exosomal miR-155 are an independent predictor
of poor event-free survival. Similarly, in metastatic patients
undergoing trastuzumab-containing regimens, high circulating
miR-155 levels are associated with decreased PFS. These findings
underscore the prognostic value of miR-155 in predicting adverse
clinical outcomes in both early-stage and metastatic BC (153).
In another study (154), patients with early-stage and
metastatic BC were stratified into high and low miR-155 expression
groups. Among early-stage patients, decreased miR-155 expression
was associated with shorter DFS compared with those with higher
expression. Furthermore, miR-155 expression was significantly lower
in patients who experienced relapse than in those who remained
relapse-free, further supporting its potential prognostic value in
BC (154).
A systematic review (147) of 28 studies found that miR-155
overexpression is associated with higher tumor grade and advanced
staging. However, inconsistencies were noted across studies
regarding the association between miR-155 levels and HR status, and
no definitive association was observed between miR-155 expression
and other key patient prognostic factors (147).
The diagnostic utility of circulating miR-155 also
varies across studies (148,151). Notably, serum-derived miR-155 has
greater diagnostic accuracy compared with plasma-derived miR-155
(155). This discrepancy may stem
from the coagulation process, which influences the extracellular
miRNA profile in blood. Despite these findings, the limited number
of studies examining plasma miR-155 underscores the need for
further investigation to establish its diagnostic potential in BC
(148).
In metastasis, exosomes (nanometer-sized vesicles
capable of transporting various biomolecules, including miRNAs) are
hypothesized to mediate the transfer of malignant traits to distant
cells. miR-155 is enriched in exosomes derived from metastatic BC,
suggesting a role in promoting metastatic behavior (156). This is consistent with studies
linking miR-155 to chemoresistance, as it has also been detected in
exosomes isolated from cancer stem and drug-resistant tumor cells
(157,158). Furthermore, a panel of circulating
miRNAs, including miR-155, is elevated in the plasma of patients
with non-metastatic BC prior to treatment, with levels declining
following therapy, supporting its potential use as a biomarker for
therapeutic monitoring (159).
miR-155 is a promising therapeutic target due to
its involvement in immunoregulatory pathways and its capacity to
enhance antitumor immune responses. It serves a critical role in
augmenting T cell-mediated immunity by enhancing T cell
functionality, memory formation, cytotoxic activity and IFN-γ
production. However, while modulation of miR-155 expression
strengthens antitumor T cell responses, the resulting increase in
IFN-γ signaling may induce upregulation of PD-L1, thereby
contributing to an immunosuppressive tumor microenvironment
(160).
The search for biomarkers in TNBC has prompted
extensive investigation into miRNAs as potential therapeutic
targets, providing deeper insight into tumor biology (161,162). miR-155 has emerged as a key
regulator of cell proliferation, in part due to its role in
maintaining thiamine metabolism in TNBC, as well as its involvement
in promoting TGF-β-induced epithelial-mesenchymal transition
(12). Although miR-155 is
typically characterized as an oncogenic miRNA that promotes tumor
growth, angiogenesis and aggressiveness, emerging evidence suggests
a context-dependent, protective role in TNBC (163). In this subtype, miR-155
upregulation is associated with improved survival outcomes. A
systematic review and meta-analysis demonstrated that low miR-155
expression is predictive of poor OS in patients with TNBC,
potentially due to its influence on molecular pathways involved in
DNA damage response and repair (163).
miR-155 modulates cell pathways
involved in TIL activity
Although BC has not traditionally been considered
an immunogenic tumor, the presence of TILs has been consistently
documented and is associated with favorable clinical outcomes,
particularly in patients with TNBC subtype (75,83,164).
In TNBC, an evaluation of plasma miR-155 expression revealed that
lower levels are significantly associated with shorter median DFS
and OS. Notably, in multivariate analysis, miR-155 emerged as the
only independent predictor of decreased DFS (154). Given the key immunoregulatory
functions of miR-155, including its roles in B lymphocyte and
CD4+ T cell differentiation, as well as Treg activation,
these findings suggest that elevated plasma miR-155 levels may
reflect a more robust antitumor immune response (154).
miR-155 upregulation enhances TIL
activity in tumor immunoediting
Studies using T cell-specific Dicer knockout mice
have demonstrated impaired T cell development, aberrant Th cell
differentiation and altered cytokine production, underscoring the
key role of miRNAs in T cell maturation and function (165,
166).
miR-155 has been identified as a key regulator of
Treg and Th17 cell differentiation through its direct targeting of
suppressor of cytokine signaling 1 (SOCS1), a negative regulator of
the JAK/STAT signaling pathway. By suppressing SOCS1, miR-155
enhances phosphorylation of STAT5 and STAT3, potentially by
alleviating SOCS1-mediated inhibition. Furthermore, miR-155
promotes Th17 cell differentiation and effector function via
distinct mechanisms. It facilitates activation of the IL-6/STAT3
signaling pathway, which is key for Th17 lineage commitment and
IL-17A production (167).
Additionally, miR-155 counteracts the inhibitory effects of IL-10
and TGF-β1 on IL-17A expression, thereby augmenting the
pro-inflammatory potential of Th17 cells (167).
Wang et al (18) identified a positive association
between the expression levels of miR-155 in breast tumors and the
presence of antitumoral immune cells, such as CD8+ T
cells and M1 macrophages. Conversely, they noted a negative
association with protumoral cell types, including Tregs and M2
macrophages. The forced overexpression of miR-155 enhances the
production of chemokines CXCL9, CXCL10 and CXCL11, which is driven
by SOCS1 inhibition and an increased ratio of phosphorylated
STAT1/STAT3. Furthermore, a convolutional neural network analysis
confirmed that elevated levels of miR-155 are associated with an
increased proportion of TILs, emphasizing its role in enhancing
both innate and adaptive immunity (18).
In a mouse model with miR-155-overexpressing
tumors, there was an increase in immune checkpoint molecules such
as PD-L1 and CTLA4, which are known to inhibit antitumor activity
(18). This upregulation was also
observed in human BC tissue miR-155 overexpression results in
increased PD-L1 expression in both human and murine BC cell lines,
suggesting a potential sensitivity to immune checkpoint blockade
therapy (18,168).
miR-155 inhibits SOCS1, which affects
the activity of tumor-infiltrating lymphocytes (TILs)
The downregulation of SOCS1 by direct miR-155
targeting serving a pivotal role in enhancing TILs activity in
TNBC. SOCS1, a member of the STAT-induced STAT inhibitor family,
negatively modulates pro-inflammatory cytokines via two mechanisms.
Firstly, SOCS1 catalyzes the ubiquitination of signaling
intermediates recruited by SOCS1. Secondly, it directly inhibits
the JAK/STAT pathway (169),
thereby preventing excessive levels of pro-inflammatory cytokines,
such as IFN (170) and ILs
(171).
Increased IFN-γ is associated with more effective
anti-tumor response and with an enhancement of T cell response
(138,141). Therefore, by targeting SOCS1,
miR-155 indirectly promotes IFN-γ production, resulting in more
active TILs, which, in TNBC, is associated with a positive
prognosis. However, in other types of cancer, such as colorectal
cancer (172) and pancreatic
cancer (173), SOCS1 is considered
a tumor suppressor, and downregulation of SOCS1 can be associated
with tumor progression (174).
PI3K/AKT is modulated by
miR-155/SOCS1, leading to enhanced TIL activity in the tumor
microenvironment
SOCS1 suppresses the PI3K/AKT pathway, which is
involved in cell proliferation, meaning miR-155 also promotes cell
proliferation. SH-2 containing inositol 5′ polyphosphatase 1
(SHIP1), an inositol phosphatase, together with PTEN is a key
negative regulator of PI3K/AKT and miR-155 directly targets its
expression; when miR-155 is upregulated, SHIP1 expression is
reduced, and cell proliferation is promoted together with increased
pro-inflammatory cytokine levels. AKT promotes the production of
ILs such as IL-6 and IL-12. High TIL levels influence the migration
and infiltration of TILs into the tumor site. By increasing
chemokine production and cell motility, this pathway may promote
the recruitment of TILs into the tumor microenvironment, improving
the immune response (175).
miR-155 can also enhance the cytotoxic activity of CD8+
T cells, promoting tumor cell death (175).
miR-155 triggers a suppressive cascade
that reduces Treg function
SOCS1 maintains FOXP3 expression and Treg cell
stability under inflammatory conditions. Treg cells serve an
immunosuppressive role in the tumor microenvironment. The
transcription factor FOXP3 is key for the normal development of
Tregs; decreased FOXP3 is associated with higher proportions of
cytotoxic cells, which can lead to a better prognosis. High
expression of FOXP3 is associated with immune tolerance of cancer
cells. During the equilibrium phase of the immune response in the
tumor microenvironment, there is a low proportion of Tregs due to
decreased FOXP3 expression, and a high proportion of cytotoxic
cells (56,57,176,177).
miR-155 suppresses expression of
PD-L1, enhancing TIL activity in TNBC
PD-L1 is a cell surface protein that serves a
critical role in suppressing the immune response; its expression on
tumor cells is a mechanism for evading immunosurveillance. The
interplay between miR-155 and PD-L1 has notable implications for
TIL activity in TNBC. When miR-155 downregulates PD-L1, T cell
inhibition is reduced, thus enhancing the anti-tumor immune
response mediated by TILs (178).
Studies have confirmed that miR-155 regulates TILs by targeting
specific regions of PD-L1 mRNA (179,180).
However, the increase in pro-inflammatory cytokines
such as TNF-α and IFN-γ can indirectly increase PD-L1 expression;
the net result of this interaction will dictate the immune evasion
in TNBC (181).miR-155 inhibits
the IL-6/STAT3 pathway. IL-6 and miR-155 form a positive feedback
loop: IL-6 stimulates the expression of miR-155, which inhibits
SOCS. By inhibiting SOCS, miR-155 allows increased signaling of
cytokines, such as IL-6, further promoting the activation of the
IL-6/STAT3 pathway. When SOCS1 is inhibited by miR-155, the
IL-6/STAT3 pathway becomes more active, leading to increased
production of IL-6 and other pro-inflammatory cytokines.
IL-6 is a pleiotropic cytokine that serves an
important role in physiological processes, including cell
proliferation, immune surveillance, acute inflammation, metabolism
and bone remodeling. IL-6 binds to the IL-6 receptor, which
subsequently binds to the glycoprotein 130 receptor creating a
signal transducing hexameric receptor complex. JAKs are recruited
and activated; activated JAK phosphorylates STAT3 for activation,
leading to gene regulation. Constitutively active IL-6/JAK/STAT3
signaling promotes cancer cell proliferation and invasiveness while
suppressing apoptosis, and STAT3 enhances IL-6 signaling to promote
an inflammatory feedback loop (182).
The IL-6/STAT3 pathway is a key signaling pathway
in cancer. IL-6 activates STAT3, which regulates the expression of
genes involved in cell proliferation, survival, and inflammation.
Targeting this IL-6 has a dual effect on BC which may either be
tumorigenic or antitumorigenic. The tumorigenic effect is caused by
inhibiting apoptosis, triggering the survival of tumor cells, and
allowing metastasis. IL-6 stimulates miR-155 expression, which
targets SOCS, hence promoting the progression of BC (183). Blocking IL-6 pathway can prevent
this progression via the inhibition of tumor migration and invasion
(184).
Upregulation of miR-155 in dendritic
cells (DCs) induces T cell proliferation and IFN-γ and IL-2
secretion
miR-155 promotes DC maturation and increases the
expression of MHC Class II (MHCII), CD86, CD40 and CD83, which are
key for T cell activation and the generation of effective TILs
(185). This enhances the ability
of DCs to present antigens to CD4+ T cells, a key step
in activating the adaptive immune response and generating effective
TILs (55). By increasing CD86
expression, miR-155 enhances the costimulatory signal needed for T
cell activation, promoting a more robust immune response and
enhancing TIL function. CD40 signaling is vital for effective
antigen-presenting cells (APC) activation, which is a key step in
generating potent TIL responses. As a marker of mature DCs, the
presence of CD83+ DCs in the tumor microenvironment
indicates a more active T cell response, enhancing anti-tumor
immunity through TILs. miR-155 promotes the production of IL12p70,
a cytokine that promotes the differentiation of CD4+ T
into Th1 cells, enhances CD8+ cytotoxic T cell activity
and stimulates IFN-γ production. These factors that contribute to
an enhanced anti-tumor immune response and are necessary for
effective TIL function (53,184,185).
miR-155 exerts an inhibitory effect on
ecto-5′-nucleotidase (NT5E) expression
NT5E is an enzyme that produces immunosuppressive
adenosine in the tumor microenvironment by hydrolyzing AMP. This
adenosine helps cancer cells evade destruction by cytotoxic T
lymphocytes. Inhibition of NT5E could enhance the ability of the
immune system to target and kill tumor cells. NT5E exists in two
isoforms, with the shorter isoform able to bind to the longer one
and cause its degradation. Several studies have shown that
miR-155-5p can directly inhibit NT5E expression by binding its 3′
UTR (186,187).
Dual functionality of miR-155 in
BC
miR-155 exemplifies the complex, context-dependent
functionality characteristic of miRNAs in BC pathophysiology
(138,141). miRNA expression profiles vary
across the five molecular subtypes of BC, creating unique molecular
environments that modulate miRNA function (136). Alteration of the level of
expression and activity of the enzymes involved in the biogenesis
of the miRNA may drive the unique profiles observed in BC subtypes
(138,141). Robust inverse associations have
been established between miR-155 expression and ER positivity,
demonstrating a direct association with TNBC phenotypes (136).
Interaction with other non-coding RNA regulatory
mechanisms and target availability determined by competing
endogenous RNAs (ceRNAs) may explain the positive prognostic
association observed with miR-155 expression in TNBC (188). Genomic alterations characteristic
of TNBC, including mutations and copy number variations, may modify
the expression of transcripts and UTRs, thereby altering the
abundance of miRNA response elements. These molecular perturbations
disrupt ceRNA network homeostasis, potentially influencing miRNA
sequestration dynamics, and target transcript regulation, including
the positive prognostic association of miR-155 expression in TNBC
genomic contexts-which is characterized by high mutational burden,
genomic instability and heterogeneity (189).
The unique TNBC microenvironment affects miR-155
functionality through cell-type specific regulatory networks and
signaling cascades (138,190). Furthermore, miR-155 demonstrates
notable stage-specific effects; elevated expression may promote
recurrence in early-stage disease but is associated with improved
outcomes in established TNBC. This temporal heterogeneity is
complemented by spatial expression gradients within the tumor
architecture, emphasizing the contextual nature of miR-155 function
in BC progression (190).
The complex regulatory mechanisms of miR-155 within
the TNBC microenvironment illustrate the key intersection between
basic molecular oncology and translational medicine. The
context-dependent functions of miR-155 presents both challenges and
opportunities for clinical application, and barriers remain in
translating these molecular insights into practical clinical
tools.
Translating scientific findings into
clinical practice and limitations
Strategies for modifying miRNA content
in tumor cells
Wang et al (18) demonstrated that miRNAs notably
enhance the inflammatory state of breast tumors (18) . This finding is important because
tumors characterized by a high percentage of TILs, specifically
those classified as LPBC (TILs ≥30 or 50%), are associated with
improved outcomes for anticancer therapy (70,95,96).
The relationship between TILs and tumor response offers a
compelling rationale for exploring strategies to enhance lymphocyte
infiltration into tumors, thereby generating tumors with an
enhanced immune response dubbed as ‘hot tumors’. This are
characterized by proinflammatory cytokines and T cell
infiltration.
Approaches to modulate the tumor microenvironment
using checkpoint inhibitor have been investigated to convert ‘cold’
into ‘hot’ tumors, which are expected to respond more effectively
to immune-modulatory agents (191,192). Wang et al (18) demonstrated that increased expression
of miR-155 enhances the recruitment of antitumor immune cells to
the tumor microenvironment, effectively transforming ‘cold’ into
‘hot’ tumors (18).
Modifying miRNA levels in cells is accomplished
through two primary methods: Increasing their concentration with
target mimics known as miRNA mimics or decreasing their levels
using antisense sequences that inhibit specific targets, referred
to as antimiRs. Both strategies can be implemented using
oligonucleotides or viral vectors. Advancements in biotechnology
have improved the stability of synthetic RNA nucleotides,
facilitating their use alongside nanoparticles that exhibit high
transfection efficiency (193).
This progress in nanotechnology, in conjunction with enhancements
in oligonucleotide chemistry, has accelerated the development of
RNA interference therapeutics (194,195). Challenges in optimizing the use of
short RNA nucleotides include improving targeted delivery,
enhancing exosomal delivery, using both viral and non-viral vectors
and minimizing toxicity and costs (196,197).
The biotechnological development of a miRNA-based
strategy aimed at producing an approved drug necessitates an
organized workflow. This process begins with a preclinical stage
focused on establishing the proof-of-concept, which involves
demonstrating that the alteration of specific miRNAs yields a
notable anticancer effect. This is followed by preclinical studies
using animal models, ultimately leading to clinical trials designed
to secure approval for the treatment as an anticancer therapy
(198–200).
miRNAs as blood-based biomarkers for
BC
Wang et al (18) demonstrated that miR-155 levels in
blood samples from patients are an indicator of the tumor
inflammatory state (18).
Specifically, higher levels of miR-155 in serum are associated with
increased expression of this miRNA in tumor samples, as well as an
enhanced immune status.
The biotechnological development of blood-based
biomarkers necessitates a carefully designed strategy to ensure
safe translation to clinical practice. Standardized protocols and
testing targets through multicenter studies are crucial for
effectively translating miRNA-based signatures to the clinic. One
of the challenges of standardization is identifying suitable
reference genes to normalize quantitative PCR results.
Additionally, following analytical guidelines for accurate data
quantification is key. Multicenter studies involving large,
independent cohorts of patients from various countries,
representing diverse ethnic groups, are required to validate the
proposed miRNA-based biomarkers (201,202).
A typical study involves at least two phases, each
with distinct patient cohorts, as illustrated by Zou et al
(203), which developed a panel of
serum miRNAs for BC screening. In the initial phase, known as the
discovery cohort, researchers assessed a broad range of targets
(324 miRNAs) to identify the most promising candidates whose
expression levels significantly differ between patients and healthy
controls. The targets selected from this phase, often referred to
as the ‘miRNA signature’, are then evaluated in a validation cohort
(203). This systematic approach
ensures that the identified miRNA signatures possess clinical
relevance across different populations, enhancing their potential
application in clinical settings. The next step involves conducting
clinical trials to evaluate the accuracy of the selected miRNAs in
diagnosing the intended disease. These trials are key for
establishing the clinical use of biomarkers. Previous reviews have
highlighted ongoing trials focused on validating miRNA-based
biomarkers (147,148,151,155). In cancer such lymphoma, leukemia
and most solid cancers it is consider an oncogenic miRNA.
Differently, in TNBC it is linked with a better prognosis due to
the regulation of TILs (204–207).
Limitations
Mounting evidence supports the consensus that the
proportion of TILs is one of the most reliable biomarkers for
prognosticating outcomes in TNBC (78,79).
However, determining this proportion can be subjective, heavily
relying on the pathologist microscopic evaluation. Previous studies
have identified reproducibility issues among different pathologists
(77,208). To address this limitation,
multicenter initiatives have been launched, particularly by the
TILs Working Group, to provide standardized guidelines and training
(70,78). While TILs are valuable biomarkers,
standardization is crucial for developing laboratory assessments
that effectively guide treatment decisions and prognosis in
clinical settings (209,210).
The analysis of miRNAs in tumor biology has been
advanced by large datasets, such as The Cancer Genome Atlas
(211), which provide clinical
information alongside mRNA and miRNA expression patterns. This
highlights the complexity of the BC transcriptome, characterized by
clusters of expressed genes (212). Each miRNA can regulate hundreds of
genes, and the miRNome of tumor cells often exhibits multiple
dysregulated miRNAs. Thus, it is key to investigate the role of
specific miRNAs to ascertain their involvement in cancer biology
and the inflammatory status of tumors (213,214).
Research has elucidated the roles of specific
miRNAs in TNBC and their potential as biomarkers for TIL
infiltration and tumor outcomes, particularly miR-155. Wang et
al (18) revealed a significant
association between miR-155 levels in blood and tumor samples, as
well as with cytokines CCL5 and CXCL9/10/11. The sample size for
biomarker validation is key, highlighting the need for multicenter
studies. Developing multi-target signatures can enhance the
accuracy of miRNA biomarkers, making it essential to evaluate
metrics such as area under the curve, accuracy, sensitivity and
specificity, typically defined using receiver operating
characteristic curves (201–203).
An important consideration in using miRNAs to
assess the inflammatory status of breast tumors is whether the
miRNA is expressed by cancer cells or lymphocytes. This distinction
is key, as tumors exhibit varying levels of lymphocyte infiltration
(18). In tumors with a higher
proportion of TILs, miR-155 content primarily reflects
lymphocyte-derived miRNAs. Conversely, in tumors with minimal
lymphocyte infiltration, miR-155 expression is predominantly
derived from cancer cells. This limitation warrants caution in
interpreting miRNAs as biomarkers, despite the potential of miRNAs
as targets for developing prognostic assays in BC.
Conclusion
Considering the complexity and diversity of BC
subtypes, as well as the impact of this variation on diagnosis,
prognosis and therapeutic planning, the use of tumor biomarkers has
relevance in the field of breast oncology. miR-155 is a promising
biomarker for patient stratification in TNBC due to its
multifaceted enhancement of TIL activity, operating via key
molecular pathways including SOCS1 suppression, PI3K/AKT modulation
and IL-6/STAT3 signaling. The detection of miR-155 in both tumor
tissue and circulation, coupled with its role in augmenting DC
maturation and T cell responses, suggests its potential use as a
predictive marker for immunotherapy efficacy.
As elevated TIL presence is associated with
improved survival outcomes in TNBC, miR-155 expression profiling
may facilitate more precise therapeutic stratification, advancing
personalized treatment approaches in this aggressive BC subtype.
Properly designed preclinical studies assessing efficacy, toxicity
and pharmacokinetic parameters, followed by clinical trials, are
key in the development of miRNA-based therapies aimed at improving
treatment outcomes in TNBC.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Ministry of Education
(grant no. 9249 TED/MEC), the National Council for Scientific and
Technological Development, the Coordination for the Improvement of
Higher Education Personnel, the Federal District Research Support
Foundation (00193-00001738/2022-51) and Foundation for Scientific
and Technological Enterprises-Brasília (01/2024).
Availability of data and materials
Not applicable.
Authors' contributions
MMAV conceived the study and wrote and edited the
manuscript. FS performed the literature review and wrote and edited
the manuscript. SSARF constructed figures and edited the
manuscript. MEX and RTA performed the literature review and editing
the manuscript. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, AI tools were
used to improve the readability and language of the manuscript or
to generate images, and subsequently, the authors revised and
edited the content produced by the AI tools as necessary, taking
full responsibility for the ultimate content of the present
manuscript.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48.
2023.PubMed/NCBI
|
|
3
|
Blackadar CB: Historical review of the
causes of cancer. World J Clin Oncol. 7:54–86. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tan PH, Ellis I, Allison K, Brogi E, Fox
SB, Lakhani S, Lazar AJ, Morris EA, Sahin A, Salgado R, et al: The
2019 World Health Organization classification of tumours of the
breast. Histopathology. 77:181–185. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marra A, Trapani D, Viale G, Criscitiello
C and Curigliano G: Practical classification of triple-negative
breast cancer: Intratumoral heterogeneity, mechanisms of drug
resistance, and novel therapies. NPJ Breast Cancer. 6:542020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma Y and Gamagedara S: Biomarker analysis
for oncology. Biomark Med. 9:845–850. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aronson JK and Ferner RE: Biomarkers-A
general review. Curr Protoc Pharmacol. 76:9.23.1–9.23.17.
2017.PubMed/NCBI
|
|
8
|
Barzaman K, Karami J, Zarei Z,
Hosseinzadeh A, Kazemi MH, Moradi-Kalbolandi S, Safari E and
Farahmand L: Breast cancer: Biology, biomarkers, and treatments.
Int Immunopharmacol. 84:1065352020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tarighati E, Keivan H and Mahani H: A
review of prognostic and predictive biomarkers in breast cancer.
Clin Exp Med. 15:1–16. 2022.
|
|
10
|
Yi M, Xu L, Jiao Y, Luo S, Li A and Wu K:
The role of cancer-derived microRNAs in cancer immune escape. J
Hematol Oncol. 13:252020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mu D, Han B, Huang H, Zheng Y, Zhang J and
Shi Y: Unraveling the advances of non-coding RNAs on the tumor
microenvironment: Innovative strategies for cancer therapies. J
Transl Med. 23:6142025. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abdul Manap AS, Wisham AA, Wong FW, Najmi
HR, Ng ZF and Diba RS: Mapping the function of MicroRNAs as a
critical regulator of tumor-immune cell communication in breast
cancer and potential treatment strategies. Front Cell Dev Biol.
12:13907042024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Z, Yang X, Shen J, Xu J, Pan M, Liu J
and Han S: Gene expression patterns associated with
tumor-infiltrating CD4+ and CD8+ T cells in invasive breast
carcinomas. Hum Immunol. 82:279–287. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fu Y, Yang Q, Xu N and Zhang X: MiRNA
affects the advancement of breast cancer by modulating the immune
system's response. Biochim Biophys Acta Mol Basis Dis.
1871:1677592025. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Galvão-Lima LJ, Morais AHF, Valentim RAM
and Barreto EJSS: miRNAs as biomarkers for early cancer detection
and their application in the development of new diagnostic tools.
Biomed Eng Online. 20:212021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cuk K, Zucknick M, Heil J, Madhavan D,
Schott S, Turchinovich A, Arlt D, Rath M, Sohn C, Benner A, et al:
Circulating microRNAs in plasma as early detection markers for
breast cancer. Int J Cancer. 132:1602–1612. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bedard PL, Hyman DM, Davids MS and Siu LL:
Small molecules, big impact: 20 years of targeted therapy in
oncology. Lancet. 395:1078–1088. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Wang Q, Guan Y, Sun Y, Wang X,
Lively K, Wang Y, Luo M, Kim JA, Murphy EA, et al: Breast cancer
cell-derived microRNA-155 suppresses tumor progression via
enhancing immune cell recruitment and antitumor function. J Clin
Invest. 132:e1572482022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu X, Chang Q, Wang H, Qian H and Jiang
Y: Discovery and function exploration of microRNA-155 as a
molecular biomarker for early detection of breast cancer. Breast
Cancer. 28:806–821. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu Y, Hong Q, Lu F, Zhang Z, Li J, Nie Z
and He B: The diagnostic and prognostic value of miR-155 in
cancers: An updated meta-analysis. Mol Diagn Ther. 27:283–301.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Trivers KF, Lund MJ, Porter PL, Liff JM,
Flagg EW, Coates RJ and Eley JW: The epidemiology of
triple-negative breast cancer, including race. Cancer Causes
Control. 20:1071–1082. 2009.PubMed/NCBI
|
|
22
|
Lucena C, Paulinelli R and Pedrini JL:
Oncoplastia e reconstrução mamária. (1st ed.). Vol. 1. Rio de
Janeiro: MEDBOOK. 2017.3–16
|
|
23
|
Radu I, Scripcariu V, Panuța A, Rusu A,
Afrăsânie VA, Cojocaru E, Aniței MG, Alexa-Stratulat T, Terinte C,
Șerban CF and Gafton B: Breast sarcomas-how different are they from
breast carcinomas? Clinical, pathological, imaging and treatment
insights. Diagnostics (Basel). 13:13702023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Orvieto E, Maiorano E, Bottiglieri L,
Maisonneuve P, Rotmensz N, Galimberti V, Luini A, Brenelli F, Gatti
G and Viale G: Clinicopathologic characteristics of invasive
lobular carcinoma of the breast: Results of an analysis of 530
cases from a single institution. Cancer. 113:1511–1520. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Elston CW and Ellis IO: pathological
prognostic factors in breast cancer. I. The value of histological
grade in breast cancer: Experience from a large study with
long-term follow-up. Histopathology. 19:403–410. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moran MS, Yang Q and Haffty BG: The yale
university experience of early-stage invasive lobular carcinoma
(ILC) and invasive ductal carcinoma (IDC) treated with breast
conservation treatment (BCT): Analysis of clinical-pathologic
features, long-term outcomes, and molecular expression of COX-2,
Bcl-2, and p53 as a function of histology. Breast J. 15:571–578.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jenkins S, Kachur ME, Rechache K, Wells JM
and Lipkowitz S: Rare breast cancer subtypes. Curr Oncol Rep.
23:542021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Perou CM, Sùrlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wolff AC, Hammond MEH, Hicks DG, Dowsett
M, McShane LM, Allison KH, Allred DC, Bartlett JMS, Bilous M,
Fitzgibbons P, et al: Recommendations for human epidermal growth
factor receptor 2 testing in breast. J Clin Oncol. 31:118–145.
2013. View Article : Google Scholar
|
|
31
|
Parise CA, Bauer KR, Brown MM and Caggiano
V: Breast cancer subtypes as defined by the estrogen receptor (ER),
progesterone receptor (PR), and the human epidermal growth factor
receptor 2 (HER2) among women with invasive breast cancer in
California, 1999–2004. Breast J. 15:593–602. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang P, Chandra V and Rastinejad F:
Structural overview of the nuclear receptor superfamily: Insights
into physiology and therapeutics. Annu Rev Physiol. 72:247–272.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim MC, Park MH, Choi JE, Kang SH and Bae
YK: Characteristics and prognosis of estrogen receptor low-positive
breast cancer. J Breast Cancer. 25:318–326. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Allison KH, Hammond MEH, Dowsett M,
Mckernin SE, Carey LA, Fitzgibbons PL, Hayes DF, Lakhani SR,
Chavez-MacGregor M, Perlmutter J, et al: Estrogen and progesterone
receptor testing in breast cancer: ASCO/CAP guideline update. J
Clin Oncol. 38:1346–1366. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Giatromanolaki A, Koukourakis MI,
Simopoulos C, Polychronidis A, Gatter KC, Harris AL and Sivridis E:
c-erbB-2 related aggressiveness in breast cancer is hypoxia
inducible factor-1 dependent. Clin Cancer Res. 10:7972–7977. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wolff AC, Hammond ME, Allison KH, Harvey
BE, Mangu PB, Bartlett JMS, Bilous M, Ellis IO, Fitzgibbons P,
Hanna W, et al: Human epidermal growth factor receptor 2 testing in
breast cancer: American Society of Clinical Oncology/College of
American Pathologists clinical practice guideline focused update. J
Clin Oncol. 36:2105–2122. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hammond MEH, Hayes DF, Dowsett M, Allred
DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS,
Hayes M, et al: American society of clinical oncology/college of
american pathologists guideline recommendations for
immunohistochemical testing of estrogen and progesterone receptors
in breast cancer. J Clin Oncol. 28:2784–2795. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Akshata Desai KA: Triple Negative Breast
Cancer-An Overview. Hereditary Genetics. 2012. View Article : Google Scholar
|
|
39
|
Millikan RC, Newman B, Tse CK, Moorman PG,
Conway K, Dressler LG, Smith LV, Labbok MH, Geradts J, Bensen JT,
et al: Epidemiology of basal-like breast cancer. Breast Cancer Res
Treat. 109:123–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gonzalez-Angulo AM, Timms KM, Liu S, Chen
H, Litton JK, Potter J, Lanchbury JS, Stemke-Hale K, Hennessy BT,
Arun BK, et al: Incidence and outcome of BRCA mutations in
unselected patients with triple receptor-negative breast cancer.
Clin Cancer Res. 17:1082–1089. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou Y, Tao L, Qiu J, Xu J, Yang X, Zhang
Y, Tian X, Guan X, Cen X and Zhao Y: Tumor biomarkers for
diagnosis, prognosis and targeted therapy. Signal Transduct Target
Ther. 9:1322024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
da Silva JL, Nunes NC, Izetti P, de
Mesquita GG and de Melo AC: Triple negative breast cancer: A
thorough review of biomarkers. Crit Rev Oncol Hematol.
145:1028552020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yadav BS, Chanana P and Jhamb S:
Biomarkers in triple negative breast cancer: A review. World J Clin
Oncol. 6:252–263. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bai X, Ni J, Beretov J, Graham P and Li Y:
Immunotherapy for triple-negative breast cancer: A molecular
insight into the microenvironment, treatment, and resistance. J
Natl Cancer Cent. 1:75–87. 2021.PubMed/NCBI
|
|
45
|
Anayyat U, Ahad F, Muluh TA, Zaidi SAA,
Usmani F, Yang H, Li M, Hassan HA and Wang X: Immunotherapy:
Constructive approach for breast cancer treatment. Breast Cancer
(Dove Med Press). 15:925–951. 2023.PubMed/NCBI
|
|
46
|
Dvir K, Giordano S and Leone JP:
Immunotherapy in breast cancer. Int J Mol Sci. 25:75172024.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
da Cunha BR, Domingos C, Stefanini AC,
Henrique T, Polachini GM, Castelo-Branco P and Tajara EH: Cellular
interactions in the tumor microenvironment: The role of secretome.
J Cancer. 10:4574–4587. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
O'Sullivan T, Saddawi-Konefka R, Vermi W,
Koebel WV, Arthur C, White JM, Uppaluri R, Andrews DM, Ngiow SF,
Teng MW, et al: Cancer immunoediting by the innate immune system in
the absence of adaptive immunity. J Exp Med. 209:1869–1882. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chiu PPL, Ivakine E, Mortin-Toth S and
Danska JS: Susceptibility to lymphoid neoplasia in immunodeficient
strains of nonobese diabetic mice. Cancer Res. 62:5828–5834.
2002.PubMed/NCBI
|
|
50
|
Dunn GP, Old LJ and Schreiber RD: The
immunobiology of cancer immunosurveillance and immunoediting.
Immunity. 21:137–148. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Muenst S, Läubli H, Soysal SD, Zippelius
A, Tzankov A and Hoeller S: The immune system and cancer evasion
strategies: Therapeutic concepts. J Intern Med. 279:541–562. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mittal D, Gubin MM, Schreiber RD and Smyth
MJ: New insights into cancer immunoediting and its three component
phases-elimination, equilibrium and escape. Curr Opin Immunol.
27:16–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Palucka K and Banchereau J: Cancer
immunotherapy via dendritic cells. Nat Rev Cancer. 12:265–277.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Janssen EM, Lemmens EE, Wolfe T, Christen
U, von Herrath MG and Schoenberger SP: CD4+ T cells are required
for secondary expansion and memory in CD8+ T lymphocytes. Nature.
421:852–856. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Catalán E, Charni S, Jaime P, Aguiló JI,
Enríquez JA, Naval J, Pardo J, Villalba M and Anel A: MHC-I
modulation due to changes in tumor cell metabolism regulates tumor
sensitivity to CTL and NK cells. Oncoimmunology. 4:e9859242015.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hori S, Nomura T and Sakaguchi S: Control
of regulatory T cell development by the transcription factor Foxp3.
Science. 299:1057–1061. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sakaguchi S, Miyara M, Costantino CM and
Hafler DA: FOXP3+ regulatory T cells in the human immune system.
Nat Rev Immunol. 10:490–500. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Mc Neil V and Lee SW: Advancing cancer
treatment: A review of immune checkpoint inhibitors and combination
strategies. Cancers (Basel). 17:14082025. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cortes J, Cescon DW, Rugo HS, Nowecki Z,
Im SA, Yusof MM, Gallardo C, Lipatov O, Barrios CH, Holgado E, et
al: Pembrolizumab plus chemotherapy versus placebo plus
chemotherapy for previously untreated locally recurrent inoperable
or metastatic triple-negative breast cancer (KEYNOTE-355): A
randomised, placebo-controlled, double-blind, phase 3 clinical
trial. Lancet. 396:1817–1828. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hodi FS, O'Day SJ, McDermott DF, Weber RW,
Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel
JC, et al: Improved survival with ipilimumab in patients with
metastatic melanoma. N Engl J Med. 363:711–723. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang Z, Huang Q, Yu L, Zhu D, Li Y, Xue
Z, Hua Z, Luo X, Song Z, Lu C, et al: The role of miRNA in tumor
immune escape and mirna-based therapeutic strategies. Front
Immunol. 12:8078952022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tang WW, Battistone B, Bauer KM, Weis AM,
Barba C, Fadlullah MZH, Ghazaryan A, Tran VB, Lee SH, Agir ZB, et
al: A microRNA-regulated transcriptional state defines intratumoral
CD8+ T cells that respond to immunotherapy. Cell Rep.
44:1153012025. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sharma S, Opyrchal M and Lu X: Harnessing
tumorous flaws for immune supremacy: Is miRNA-155 the weak link in
breast cancer progression? J Clin Invest. 132:e1630102022.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang M, Yin B, Wang HY and Wang RF:
Current advances in T-cell-based cancer immunotherapy.
Immunotherapy. 6:1265–1278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen L and Flies DB: Molecular mechanisms
of T cell co-stimulation and co-inhibition. Na Rev Immunol.
13:227–242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Naidoo J, Page DB, Li BT, Connell LC,
Schindler K, Lacouture ME, Postow MA and Wolchok JD: Toxicities of
the anti-PD-1 and anti-PD-L1 immune checkpoint antibodies. Ann
Oncol. 26:2375–2391. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Brahmer JR, Tykodi SS, Chow LQM, Hwu WJ,
Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al:
Safety and activity of anti-PD-L1 antibody in patients with
advanced cancer. N Engl J Med. 366:2455–2465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Shevach EM: Application of IL-2 therapy to
target T regulatory cell function. Trends Immunol. 33:626–632.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang HY and Wang RF: Regulatory T cells
and cancer. Curr Opin Immunol. 19:217–223. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Salgado R, Denkert C, Demaria S, Sirtaine
N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL,
Penault-Llorca F, et al: The evaluation of tumor-infiltrating
lymphocytes (TILS) in breast cancer: Recommendations by an
international TILS working group 2014. Ann Oncol. 26:259–271. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Mao Y, Qu Q, Chen X, Huang O, Wu J and
Shen K: The prognostic value of tumor-infiltrating lymphocytes in
breast cancer: A systematic review and meta-analysis. PLoS One.
11:e01525002016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
van den Ende NS, Nguyen AH, Jager A, Kok
M, Debets R and van Deurzen CHM: Triple-Negative breast cancer and
predictive markers of response to neoadjuvant chemotherapy: A
systematic review. Int J Mol Sci. 24:29692023. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Leon-Ferre RA, Polley MY, Liu H, Gilbert
JA, Cafourek V, Hillman DW, Elkhanany A, Akinhanmi M, Lilyquist J,
Thomas A, et al: Impact of histopathology, tumor-infiltrating
lymphocytes, and adjuvant chemotherapy on prognosis of
triple-negative breast cancer. Breast Cancer Res Treat. 167:89–99.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Denkert C, von Minckwitz G, Darb-Esfahani
S, Lederer B, Heppner BI, Weber KE, Budczies J, Huober J, Klauschen
F, Furlanetto J, et al: Tumour-infiltrating lymphocytes and
prognosis in different subtypes of breast cancer: A pooled analysis
of 3,771 patients treated with neoadjuvant therapy. Lancet Oncol.
19:40–50. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Leon-Ferre RA, Jonas SF, Salgado R, Loi S,
de Jong V, Carter JM, Nielsen TO, Leung S, Riaz N, Chia S, et al:
Tumor-Infiltrating lymphocytes in triple-negative breast cancer.
JAMA. 331:1135–1144. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Tan Q, Yin S, Zhou D, Chi Y, Man X and Li
H: Potential predictive and prognostic value of biomarkers related
to immune checkpoint inhibitor therapy of triple-negative breast
cancer. Front Oncol. 12:7797862022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
O'Loughlin M, Andreu X, Bianchi S,
Chemielik E, Cordoba A, Cserni G, Figueiredo P, Floris G, Foschini
MP, Heikkilä P, et al: Reproducibility and predictive value of
scoring stromal tumour infiltrating lymphocytes in triple-negative
breast cancer: A multi-institutional study. Breast Cancer Res
Treat. 171:1–9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Stanton SE and Disis ML: Clinical
significance of tumor-infiltrating lymphocytes in breast cancer. J
Immunother Cancer. 4:592016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Li S, Zhang Y, Zhang P, Xue S, Chen Y, Sun
L and Yang R: Predictive and prognostic values of tumor
infiltrating lymphocytes in breast cancers treated with neoadjuvant
chemotherapy: A meta-analysis. Breast. 66:97–109. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Gao G, Wang Z, Qu X and Zhang Z:
Prognostic value of tumor-infiltrating lymphocytes in patients with
triple-negative breast cancer: A systematic review and
meta-analysis. BMC Cancer. 20:1792020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Valenza C, Salimbeni BT, Santoro C,
Trapani D, Antonarelli G and Curigliano G: Tumor infiltrating
lymphocytes across breast cancer subtypes: Current issues for
biomarker assessment. Cancers (Basel). 15:7672023. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ochi T, Bianchini G, Ando M, Nozaki F,
Kobayashi D, Criscitiello C, Curigliano G, Iwamoto T, Niikura N,
Takei H, et al: Predictive and prognostic value of stromal
tumour-infiltrating lymphocytes before and after neoadjuvant
therapy in triple negative and HER2-positive breast cancer. Eur J
Cancer. 118:41–48. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Loi S, Michiels S, Salgado R, Sirtaine N,
Jose V, Fumagalli D, Kellokumpu-Lehtinen PL, Bono P, Kataja V,
Desmedt C, et al: Tumor infiltrating lymphocytes are prognostic in
triple negative breast cancer and predictive for trastuzumab
benefit in early breast cancer: Results from the FinHER trial. Ann
Oncol. 25:1544–1550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Stanton SE, Adams S and Disis ML:
Variation in the incidence and magnitude of tumor-infiltrating
lymphocytes in breast cancer subtypes: A systematic review. JAMA
Oncol. 2:1354–1360. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Russo L, Maltese A, Betancourt L, Romero
G, Cialoni D, De la Fuente L, Gutierrez M, Ruiz A, Agüero E and
Hernández S: Locally advanced breast cancer: Tumor-infiltrating
lymphocytes as a predictive factor of response to neoadjuvant
chemotherapy. Eur J Surg Oncol. 45:963–968. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Liu S, Duan X, Xu L, Xin L, Cheng Y, Liu
Q, Ye J, Zhang S, Zhang H, Zhu S, et al: Optimal threshold for
stromal tumor-infiltrating lymphocytes: Its predictive and
prognostic value in HER2-positive breast cancer treated with
trastuzumab-based neoadjuvant chemotherapy. Breast Cancer Res
Treat. 154:239–249. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Denkert C, Loibl S, Noske A, Roller M,
Müller BM, Komor M, Budczies J, Darb-Esfahani S, Kronenwett R,
Hanusch C, et al: Tumor-associated lymphocytes as an independent
predictor of response to neoadjuvant chemotherapy in breast cancer.
J Clin Oncol. 28:105–113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Dieci MV, Criscitiello C, Goubar A, Viale
G, Conte P, Guarneri V, Ficarra G, Mathieu MC, Delaloge S,
Curigliano G and Andre F: Prognostic value of tumor-infiltrating
lymphocytes on residual disease after primary chemotherapy for
triple-negative breast cancer: A retrospective multicenter study.
Ann Oncol. 25:611–618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yuan Y, Lee JS, Yost SE, Li SM, Frankel
PH, Ruel C, Schmolze D, Robinson K, Tang A, Martinez N, et al:
Phase II trial of neoadjuvant carboplatin and nab-paclitaxel in
patients with triple-negative breast cancer. Oncologist.
26:e382–e393. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Schmidt M, Weyer-Elberich V, Hengstler JG,
Heimes AS, Almstedt K, Gerhold-Ay A, Lebrecht A, Battista MJ,
Hasenburg A, Sahin U, et al: Prognostic impact of CD4-positive T
cell subsets in early breast cancer: A study based on the FinHer
trial patient population. Breast Cancer Res. 20:152018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Cerbelli B, Pernazza A, Botticelli A,
Fortunato L, Monti M, Sciattella P, Campagna D, Mazzuca F, Mauri M,
Naso G, et al: PD-L1 expression in TNBC: A predictive biomarker of
response to neoadjuvant chemotherapy? Biomed Res Int.
2017:17509252017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Van Bockstal MR, Noel F, Guiot Y, Duhoux
FP, Mazzeo F, Van Marcke C, Fellah L, Ledoux B, Berlière M and
Galant C: Predictive markers for pathological complete response
after neo-adjuvant chemotherapy in triple-negative breast cancer.
Ann Diagn Pathol. 49:1516342020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Floris G, Richard F, Hamy AS, Jongen L,
Wildiers H, Ardui J, Punie K, Smeets A, Berteloot P, Vergote I, et
al: Body mass index and tumor-infiltrating lymphocytes in
triple-negative breast cancer. J Natl Cancer Inst. 113:146–153.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Dieci MV, Tsvetkova V, Griguolo G,
Miglietta F, Tasca G, Giorgi CA, Cumerlato E, Massa D, Lo Mele M,
Orvieto E, et al: Integration of tumour infiltrating lymphocytes,
programmed cell-death ligand-1, CD8 and FOXP3 in prognostic models
for triple-negative breast cancer: Analysis of 244 stage I–III
patients treated with standard therapy. Eur J Cancer. 136:7–15.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Loi S, Drubay D, Adams S, Pruneri G,
Francis PA, Lacroix-Triki M, Joensuu H, Dieci MV, Badve S, Demaria
S, et al: Tumor-infiltrating lymphocytes and prognosis: A pooled
individual patient analysis of early-stage triple-negative breast
cancers. J Clin Oncol. 37:559–569. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Park JH, Jonas SF, Bataillon G,
Criscitiello C, Salgado R, Loi S, Viale G, Lee HJ, Dieci MV, Kim
SB, et al: Prognostic value of tumor-infiltrating lymphocytes in
patients with early-stage triple-negative breast cancers (TNBC) who
did not receive adjuvant chemotherapy. Ann Oncol. 30:1941–1949.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
TILs in Breast Cancer. International
Immuno-Oncology Biomarker Working Group on Breast Cancer
[Internet], . [cited 2025 Jul 7]. Available from. https://www.tilsinbreastcancer.org
|
|
98
|
Laenkholm AV, Callagy G, Balancin M,
Bartlett JMS, Sotiriou C, Marchio C, Kok M, Dos Anjos CH and
Salgado R: Incorporation of TILs in daily breast cancer care: How
much evidence can we bear? Virchows Arch. 480:147–162. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Loi S, Michiels S, Adams S, Loibl S,
Budczies J, Denkert C and Salgado R: The journey of
tumor-infiltrating lymphocytes as a biomarker in breast cancer:
Clinical utility in an era of checkpoint inhibition. Ann Oncol.
32:1236–1244. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Lovat F, Valeri N and Croce CM: MicroRNAs
in the pathogenesis of cancer. Semin Oncol. 38:724–733. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Titze-de-Almeida R and Titze-de-Almeida
SS: miR-7 replacement therapy in Parkinson's disease. Curr Gene
Ther. 18:143–153. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Fareh M, Yeom KH, Haagsma AC, Chauhan S,
Heo I and Joo C: TRBP ensures efficient Dicer processing of
precursor microRNA in RNA-crowded environments. Nat Commun.
7:136942016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Mashima R: Physiological roles of miR-155.
Immunology. 145:323–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function review. Cell. 116:281–297.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Shang R, Lee S, Senavirathne G and Lai EC:
microRNAs in action: Biogenesis, function and regulation. Nat Rev
Genet. 24:816–833. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Roberts JT and Borchert GM: Computational
prediction of microRNA target genes, target prediction databases,
and web resources. Methods Mol Biol. 1617:109–122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Kariuki D, Asam K, Aouizerat BE, Lewis KA,
Florez JC and Flowers E: Review of databases for experimentally
validated human microRNA-mRNA interactions. Database (Oxford).
25:baad0142023. View Article : Google Scholar
|
|
111
|
Griffiths-Jones S: The microRNA registry.
Nucleic Acids Res. 32:D109–D111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Nair VS, Maeda LS and Ioannidis JPA:
Clinical outcome prediction by MicroRNAs in human cancer: A
systematic review. J Natl Cancer Inst. 104:528–540. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Verret B, Bottosso M, Hervais S and
Pistilli B: The molecular predictive and prognostic biomarkers in
metastatic breast cancer: The contribution of molecular profiling.
Cancers (Basel). 14:42032022. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Koncina E, Haan S, Rauh S and Letellier E:
Prognostic and predictive molecular biomarkers for colorectal
cancer: Updates and challenges. Cancers (Basel). 12:3192020.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Chang JTH, Wang F, Chapin W and Huang RS:
Identification of MicroRNAs as breast cancer prognosis markers
through the cancer genome atlas. PLoS One. 11:e01682842016.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
van Schooneveld E, Wouters MCA, Van der
Auwera I, Peeters DJ, Wildiers H, Van Dam PA, Vergote I, Vermeulen
PB, Dirix LY and Van Laere SJ: Expression profiling of cancerous
and normal breast tissues identifies microRNAs that are
differentially expressed in serum from patients with (metastatic)
breast cancer and healthy volunteers. Breast Cancer Res.
14:R342012. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Feliciano A, González L, Garcia-Mayea Y,
Mir C, Artola M, Barragán N, Martín R, Altés A, Castellvi J,
Benavente S, et al: Five microRNAs in serum are able to
differentiate breast cancer patients from healthy individuals.
Front Oncol. 10:5862682020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Mihai AM, Ianculescu LM and Suciu N:
MiRNAs as potential biomarkers in early breast cancer detection: A
systematic review. J Med Life. 17:549–554. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Fu SW, Lee W, Coffey C, Lean A, Wu X, Tan
X, Man YG and Brem RF: miRNAs as potential biomarkers in early
breast cancer detection following mammography. Cell Biosci.
6:62016. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Medarova Z, Pantazopoulos P and Yoo B:
Screening of potential miRNA therapeutics for the prevention of
multi-drug resistance in cancer cells. Sci Rep. 10:19702020.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Navarro A, Marrades RM, Viñolas N, Quera
A, Agustí C, Huerta A, Ramirez J, Torres A and Monzo M: MicroRNAs
expressed during lung cancer development are expressed in human
pseudoglandular lung embryogenesis. Oncology. 76:162–169. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Singh R, Ha SE, Yu TY and Ro S: Dual roles
of miR-10a-5p and miR-10b-5p as tumor suppressors and oncogenes in
diverse cancers. Int J Mol Sci. 26:4152025. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Webster RJ, Giles KM, Price KJ, Zhang PM,
Mattick JS and Leedman PJ: Regulation of epidermal growth factor
receptor signaling in human cancer cells by MicroRNA-7. J Biol
Chem. 284:5731–5741. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Castañeda CA, Agullo-Ortuño MT, Vara JA,
Cortes-Funes H, Gomez HL and Ciruelos E: Implication of miRNA in
the diagnosis and treatment of breast cancer. Expert Rev Anticancer
Ther. 11:1265–1275. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Baylie T, Kasaw M, Getinet M, Getie G,
Jemal M, Nigatu A, Ahmed H and Bogale M: The role of miRNAs as
biomarkers in breast cancer. Front Oncol. 14:13748212024.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Yan LX, Huang XF, Shao Q, Huang MY, Deng
L, Wu QL, Zeng YX and Shao JY: MicroRNA miR-21 overexpression in
human breast cancer is associated with advanced clinical stage,
lymph node metastasis and patient poor prognosis. RNA.
14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Li S, Yang X, Yang J, Zhen J and Zhang D:
Serum microRNA-21 as a potential diagnostic biomarker for breast
cancer: A systematic review and meta-analysis. Clin Exp Med.
16:29–35.. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Syed RU, Banu H, Alshammrani A, Alshammari
MD, G SK, Kadimpati KK, Khalifa AAS, Aboshouk NAM, Almarir AM,
Hussain A and Alahmed FK: MicroRNA-21 (miR-21) in breast cancer:
From apoptosis dysregulation to therapeutic opportunities. Pathol
Res Pract. 262:1555722024. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Qian B, Katsaros D, Lu L, Preti M, Durando
A, Arisio R, Mu L and Yu H: High miR-21 expression in breast cancer
associated with poor disease-free survival in early stage disease
and high TGF-β1. Breast Cancer Res Treat. 117:131–140. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Wang Y, Zhang Y, Pan C, Ma F and Zhang S:
Prediction of poor prognosis in breast cancer patients based on
MicroRNA-21 expression: A meta-analysis. PLoS One. 10:e01186472015.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Bertoli G, Cava C and Castiglioni I:
Micrornas: New biomarkers for diagnosis, prognosis, therapy
prediction and therapeutic tools for breast cancer. Theranostics.
5:1122–1143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Romero-Cordoba S, Rodriguez-Cuevas S,
Rebollar-Vega R, Bautista-Pina V, Maffuz-Aziz A, Tagliabue E, Iorio
M, D'Ippolito E, Baroni S, Plantamura I and Hidalgo-Miranda A: A
microRNA signature identifies subtypes of triple-negative breast
cancer and reveals miR-342-3p as regulator of a lactate metabolic
pathway through silencing monocarboxylate transporter 1. Cancer
Res. 76:A472016. View Article : Google Scholar
|
|
135
|
Tariq M, Richard V and Kerin MJ: MicroRNAs
as molecular biomarkers for the characterization of basal-like
breast tumor subtype. Biomedicines. 11:30072023. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Blenkiron C, Goldstein LD, Thorne NP,
Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE,
Green AR, Ellis IO, et al: MicroRNA expression profiling of human
breast cancer identifies new markers of tumor subtype. Genome Biol.
8:R2142007. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Kalkusova K, Taborska P, Stakheev D and
Smrz D: The role of miR-155 in antitumor immunity. Cancers (Basel).
14:54142022. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Mahesh G and Biswas R: MicroRNA-155: A
master regulator of inflammation. J Interferon Cytokine Res.
39:321–330. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Cui B, Chen L, Zhang S, Mraz M, Fecteau
JF, Yu J, Ghia EM, Zhang L, Bao L, Rassenti LZ, et al: Micro
RNA-155 influences B-cell receptor signaling and associates with
aggressive disease in chronic lymphocytic leukemia. Blood.
124:546–554. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Rai KR, Liao Y, Cai M, Qiu H, Wen F, Peng
M, Wang S, Liu S, Guo G, Chi X, et al: MIR155HG plays a bivalent
role in regulating innate antiviral immunity by encoding long
noncoding RNA-155 and microRNA-155-5p. mBio. 13:e02510222022.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Dawson O and Piccinini AM: miR-155-3p:
Processing by-product or rising star in immunity and cancer? Open
Biol. 12:2200702022. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Bayraktar R and Van Roosbroeck K: miR-155
in cancer drug resistance and as target for miRNA-based
therapeutics. Cancer Metastasis Rev. 37:33–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Zhou H, Huang X, Cui H, Luo X, Tang Y,
Chen S, Wu L and Shen N: miR-155 and its star-form partner miR-155*
cooperatively regulate type I interferon production by human
plasmacytoid dendritic cells. Blood. 116:5885–5894. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Pasculli B, Barbano R, Fontana A, Biagini
T, Di Viesti MP, Rendina M, Valori VM, Morritti M, Bravaccini S,
Ravaioli S, et al: Hsa-miR-155-5p up-regulation in breast cancer
and its relevance for treatment with poly[ADP-Ribose] polymerase 1
(PARP-1) inhibitors. Front Oncol. 10:14152020. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Kang Y, Cao X, Fan Y, Li Y, Xu T, Zhou Q
and He B: Exosome biomarkers in breast cancer: Systematic review
and meta-analysis. Clin Chim Acta. 574:1203422025. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Degheidy MS, Abou-Elalla AA, Kamel MM,
Abdel-Ghany S, Arneth B and Sabit H: Regulatory roles of
miR-155-5p, miR-21-5p, miR-93-5p, and miR-140-5p in breast cancer
progression. Curr Issues Mol Biol. 47:3772025. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Grimaldi AM, Nuzzo S, Condorelli G,
Salvatore M and Incoronato M: Prognostic and clinicopathological
significance of MiR-155 in breast cancer: A systematic review. Int
J Mol Sci. 21:58342020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Wang F, Wang J, Zhang H, Fu B, Zhang Y,
Jia Q and Wang Y: Diagnostic value of circulating miR-155 for
breast cancer: A meta-analysis. Front Oncol. 14:13746742024.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Tili E, Croce CM and Michaille JJ: miR-155
: On the crosstalk between inflammation and cancer. Int Rev
Immunol. 28:264–284. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Sun R, Kong X, Qiu X, Huang C and Wong PP:
the emerging roles of pericytes in modulating tumor
microenvironment. Front Cell Dev Biol. 9:6763422021. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Anwar SL, Tanjung DS, Fitria MS, Kartika
AI, Sari DNI, Rakhmina D, Wardana T, Astuti I, Haryana SM and
Aryandono T: Dynamic changes of circulating Mir-155 expression and
the potential application as a non-invasive biomarker in breast
cancer. Asian Pac J Cancer Prev. 21:491–497. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Bašová P, Pešta M, Sochor M and Stopka T:
Prediction potential of serum miR-155 and miR-24 for relapsing
early breast cancer. Int J Mol Sci. 18:21162017. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Zhang Z, Zhang L, Yu G, Sun Z, Wang T,
Tian X, Duan X and Zhang C: Exosomal miR-1246 and miR-155 as
predictive and prognostic biomarkers for trastuzumab-based therapy
resistance in HER2-positive breast cancer. Cancer Chemother
Pharmacol. 86:761–772. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Thomopoulou K, Papadaki C, Monastirioti A,
Koronakis G, Mala A, Kalapanida D, Mavroudis D and Agelaki S:
MicroRNAs regulating tumor immune response in the prediction of the
outcome in patients with breast cancer. Front Mol Biosci.
8:6685342021. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Schwarzenbach H, Hoon DSB and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
kia V, Paryan M, Mortazavi Y, Biglari A
and Mohammadi-Yeganeh S: Evaluation of exosomal miR-9 and miR-155
targeting PTEN and DUSP14 in highly metastatic breast cancer and
their effect on low metastatic cells. J Cell Biochem.
120:5666–5676. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Santos JC, da Silva Lima N, Sarian LO,
Matheu A, Ribeiro ML and Derchain SFM: Exosome-mediated breast
cancer chemoresistance via miR-155 transfer. Sci Rep. 8:8292018.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Liu F, Kong X, Lv L and Gao J: TGF-β1 acts
through miR-155 to down-regulate TP53INP1 in promoting
epithelial-mesenchymal transition and cancer stem cell phenotypes.
Cancer Lett. 359:288–298. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Khalighfard S, Alizadeh AM, Irani S and
Omranipour R: Plasma miR-21, miR-155, miR-10b, and Let-7a as the
potential biomarkers for the monitoring of breast cancer patients.
Sci Rep. 8:179812018. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Huffaker TB, Lee SH, Tang WW, Wallace JA,
Alexander M, Runtsch MC, Larsen DK, Thompson J, Ramstead AG, Voth
WP, et al: Antitumor immunity is defective in T cell-specific
microRNA-155- deficient mice and is rescued by immune checkpoint
blockade. J Biol Chem. 292:18530–18541. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Volovat SR, Volovat C, Hordila I, Hordila
DA, Mirestean CC, Miron OT, Lungulescu C, Scripcariu DV,
Stolniceanu CR, Konsoulova-Kirova AA, et al: MiRNA and LncRNA as
potential biomarkers in triple-negative breast cancer: A review.
Front Oncol. 10:5268502020. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Andrade F, Nakata A, Gotoh N and Fujita A:
Large miRNA survival analysis reveals a prognostic four-biomarker
signature for triple negative breast cancer. Genet Mol Biol.
43:e201802692020. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Santana TABDS, de Oliveira Passamai L, de
Miranda FS, Borin TF, Borges GF, Luiz WB and Campos LCG: The role
of miRNAs in the prognosis of triple-negative breast cancer: A
systematic review and meta-analysis. Diagnostics (Basel).
13:1272023. View Article : Google Scholar
|
|
164
|
Ibrahim E, Diab E, Hayek R, Hoyek K and
Kourie H: Triple-negative breast cancer: Tumor immunogenicity and
beyond. Int J Breast Cancer. 4:20979202024. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Muljo SA, Ansel KM, Kanellopoulou C,
Livingston DM, Rao A and Rajewsky K: Aberrant T cell
differentiation in the absence of Dicer. J Exp Med. 202:261–269.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Mi QS, Wang J, Liu Q, Wu X and Zhou L:
microRNA dynamic expression regulates invariant NKT cells. Cell Mol
Life Sci. 78:6003–6015. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X
and Liao YH: MicroRNA-155 modulates treg and Th17 cells
differentiation and Th17 cell function by targeting SOCS1. PLoS
One. 7:e460822012. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Touchaei AZ and Vahidi S: Unraveling the
interplay of CD8 + T cells and microRNA signaling in cancer:
Implications for immune dysfunction and therapeutic approaches. J
Transl Med. 22:11312024. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Liau NPD, Laktyushin A, Lucet IS, Murphy
JM, Yao S, Whitlock E, Callaghan K, Nicola NA, Kershaw NJ and Babon
JJ: The molecular basis of JAK/STAT inhibition by SOCS1. Nat
Commun. 9:15582018. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Alexander WS, Starr R, Fenner JE, Scott
CL, Handman E, Sprigg NS, Corbin JE, Cornish AL, Darwiche R,
Owczarek CM, et al: SOCS1 is a critical inhibitor of interferon γ
signaling and prevents the potentially fatal neonatal actions of
this cytokine. Cell. 98:597–608. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Davey GM, Starr R, Cornish AL, Burghardt
JT, Alexander WS, Carbone FR, Surh CD and Heath WR: SOCS-1
regulates IL-15-driven homeostatic proliferation of antigen-naive
CD8 T cells, limiting their autoimmune potential. J Exp Med.
202:1099–1108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Fujitake S, Hibi K, Okochi O, Kodera Y,
Ito K, Akiyama S and Nakao A: Aberrant methylation of SOCS-1 was
observed in younger colorectal cancer patients. J Gastroenterol.
39:120–124. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Fukushima N, Sato N, Sahin F, Su GH,
Hruban RH and Goggins M: Aberrant methylation of suppressor of
cytokine signalling-1 (SOCS-1) gene in pancreatic ductal neoplasms.
Br J Cancer. 89:338–343. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA and Kinzler KW: Cancer genome landscapes.
Science. 340:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Zonari E, Pucci F, Saini M, Mazzieri R,
Politi LS, Gentner B and Naldini L: A role for miR-155 in enabling
tumor-infiltrating innate immune cells to mount effective antitumor
responses in mice. Blood. 122:243–252. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Yu J, Mei J, Zuo D, Zhang M, Yu S, Li F,
Wang J, Bi D, Ma S, Wang J and Yin ZJ: Inflammatory factor-mediated
miR-155/SOCS1 signaling axis leads to Treg impairment in systemic
lupus erythematosus. Int Immunopharmacol. 141:1130132024.
View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Marson A, Kretschmer K, Frampton GM,
Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von
Boehmer H and Young RA: Foxp3 occupancy and regulation of key
target genes during T-cell stimulation. Nature. 445:931–935. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Piña-Sánchez P, Valdez-Salazar HA and
Ruiz-Tachiquín ME: Circulating microRNAs and their role in the
immune response in triple-negative breast cancer. Oncol Lett.
20:2242020. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Vaxevanis CK, Friedrich M, Tretbar SU,
Handke D, Wang Y, Blümke J, Dummer R, Massa C and Seliger B:
Identification and characterization of novel CD274 (PD-L1)
regulating microRNAs and their functional relevance in melanoma.
Clin Transl Med. 12:e9342022. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Tumeh PC, Harview CL, Yearley JH, Shintaku
IP, Taylor EJM, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu
V, et al: PD-1 blockade induces responses by inhibiting adaptive
immune resistance. Nature. 515:568–571. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Eichmüller SB, Osen W, Mandelboim O and
Seliger B: Immune modulatory microRNAs involved in tumor attack and
tumor immune escape. J Natl Cancer Inst. 109:doi:
10.1093/jnci/djx03. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Manore SG, Doheny DL, Wong GL and Lo HW:
IL-6/JAK/stat3 signaling in breast cancer metastasis: Biology and
treatment. Front Oncol. 12:8660142022. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Gyamfi J, Lee YH, Eom M and Choi J:
Interleukin-6/STAT3 signalling regulates adipocyte induced
epithelial-mesenchymal transition in breast cancer cells. Sci Rep.
8:88592018. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Taghikhani A, Hassan ZM, Ebrahimi M and
Moazzeni SM: microRNA modified tumor-derived exosomes as novel
tools for maturation of dendritic cells. J Cell Physiol.
234:9417–9427. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Yang P, Cao X, Cai H, Chen X, Zhu Y, Yang
Y, An W and Jie J: Upregulation of microRNA-155 enhanced migration
and function of dendritic cells in three-dimensional breast cancer
microenvironment. Immunol Invest. 50:1058–1071. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Kordaß T, Chao TY, Osen W and Eichmüller
SB: Novel microRNAs modulating ecto-5′-nucleotidase expression.
Front Immunol. 14:11993742023. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Hering C and Conover GM: Advancing
ischemic stroke prognosis: Key role of MIR-155 non-coding RNA. Int
J Mol Sci. 26:39472025. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Qattan A: Genomic alterations affecting
competitive endogenous RNAs (ceRNAs) and regulatory networks
(ceRNETs) with clinical implications in triple-negative breast
cancer (TNBC). Int J Mol Sci. 25:26242024. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Wilson TR, Udyavar AR, Chang CW, Spoerke
JM, Aimi J, Savage HM, Daemen A, O'Shaughnessy JA, Bourgon R and
Lackner MR: Genomic alterations associated with recurrence and TNBC
subtype in high-risk early breast cancers. Mol Cancer Res.
17:97–108. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Skourti E, Volpe A, Lang C, Johnson P,
Panagaki F and Fruhwirth GO: Spatiotemporal quantitative
microRNA-155 imaging reports immune-mediated changes in a
triple-negative breast cancer model. Front Immunol. 14:11802332023.
View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Duan Q, Zhang H, Zheng J and Zhang L:
Turning cold into hot: Firing up the tumor microenvironment. Trends
Cancer. 6:605–618. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Galon J and Bruni D: Approaches to treat
immune hot, altered and cold tumours with combination
immunotherapies. Nat Rev Drug Discov. 18:197–218. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Chen Y, Gao DY and Huang L: In vivo
delivery of miRNAs for cancer therapy: Challenges and strategies.
Adv Drug Deliv Rev. 81:128–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Haussecker D: Current issues of RNAi
therapeutics delivery and development. J Control Release.
195:49–54. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Whitehead KA, Langer R and Anderson DG:
Knocking down barriers: Advances in siRNA delivery. Nat Rev Drug
Discov. 8:129–138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Zhao J and Feng SS: Nanocarriers for
delivery of siRNA and co-delivery of siRNA and other therapeutic
agents. Nanomedicine (Lond). 10:2199–2228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Pagoni M, Cava C, Sideris DC, Avgeris M,
Zoumpourlis V, Michalopoulos I and Drakoulis N: miRNA-Based
technologies in cancer therapy. J Pers Med. 13:15862023. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
El Sayed SR, Cristante J, Guyon L, Denis
J, Chabre O and Cherradi N: Microrna therapeutics in cancer:
Current advances and challenges. Cancers (Basel). 29:26802021.
View Article : Google Scholar
|
|
199
|
Dasgupta I and Chatterjee A: Recent
advances in miRNA delivery systems. Methods Protoc. 4:102021.
View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Serpico D, Molino L and Di Cosimo S:
MicroRNAs in breast cancer development and treatment. Cancer Treat
Rev. 40:595–604. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
De Planell-Saguer M and Rodicio MC:
Analytical aspects of microRNA in diagnostics: A review. Anal Chim
Acta. 699:134–152. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Reid G, Kirschner MB and van Zandwijk N:
Circulating microRNAs: Association with disease and potential use
as biomarkers. Crit Rev Oncol Hematol. 80:193–208. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Zou R, Loke SY, Tang YC, Too HP, Zhou L,
Lee ASG and Hartman M: Development and validation of a circulating
microRNA panel for the early detection of breast cancer. Br J
Cancer. 126:472–481. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Ho PTB, Clark IM and Le LTT:
MicroRNA-based diagnosis and therapy. Int J Mol Sci. 23:71672022.
View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Kim T and Croce CM: MicroRNA: Trends in
clinical trials of cancer diagnosis and therapy strategies. Exp Mol
Med. 55:1314–132. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Pruneri G, Gray KP, Vingiani A, Viale G,
Curigliano G, Criscitiello C, Láng I, Ruhstaller T, Gianni L,
Goldhirsch A, et al: Tumor-infiltrating lymphocytes (TILs) are a
powerful prognostic marker in patients with triple-negative breast
cancer enrolled in the IBCSG phase III randomized clinical trial
22–00. Breast Cancer Res Treat. 158:323–331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Hendry S, Salgado R, Gevaert T, Russell
PA, John T, Thapa B, Christie M, van de Vijver K, Estrada MV,
Gonzalez-Ericsson PI, et al: Assessing tumor-infiltrating
lymphocytes in solid tumors: A practical review for pathologists
and proposal for a standardized method from the international
immunooncology biomarkers working group: Part 1: Assessing the host
immune response, TILs in invasive breast carcinoma and ductal
carcinoma in situ, metastatic tumor deposits and areas for further
research. Adv Anat Pathol. 24:235–251. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Ly A, Garcia V, Blenman KRM, Ehinger A,
Elfer K, Hanna MG, Li X, Peeters DJE, Birmingham R, Dudgeon S, et
al: Training pathologists to assess stromal tumour-infiltrating
lymphocytes in breast cancer synergises efforts in clinical care
and scientific research. Histopathology. 84:915–923. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Koboldt D, Fulton RS, Mclellan MD, Schmidt
H, Kalicki-Veizer J, McMichael JF, Fulton L, Dooling DJ, Ding L,
Mardis E, et al: Comprehensive molecular portraits of human breast
tumours. Nature. 490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Kalecky K, Modisette R, Pena S, Cho YR and
Taube J: Integrative analysis of breast cancer profiles in TCGA by
TNBC subgrouping reveals novel microRNA-specific clusters,
including miR-17-92a, distinguishing basal-like 1 and basal-like 2
TNBC subtypes. BMC Cancer. 20:1412020. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Chakraborty C, Sharma AR, Sharma G and Lee
SS: The Interplay among miRNAs, major cytokines, and cancer-related
inflammation. Mol Ther Nucleic Acids. 20:606–620. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Hill M and Tran N: miRNA interplay:
Mechanisms and consequences in cancer. Dis Model Mech.
14:dmm0476622021. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Jimenez JE, Abdelhafez A, Mittendorf EA,
Elshafeey N, Yung JP, Litton JK, Adrada BE, Candelaria RP, White J,
Thompson AM, et al: A model combining pretreatment MRI radiomic
features and tumor-infiltrating lymphocytes to predict response to
neoadjuvant systemic therapy in triple-negative breast cancer. Eur
J Radiol. 149:1102202022. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Asano Y, Kashiwagi S, Goto W, Takada K,
Takahashi K, Hatano T, Takashima T, Tomita S, Motomura H, Ohsawa M,
et al: Prediction of treatment response to neoadjuvant chemotherapy
in breast cancer by subtype using tumor-infiltrating lymphocytes.
Anticancer Res. 38:2311–2321. 2018.PubMed/NCBI
|
|
217
|
Song IH, Heo SH, Bang WS, Park HS, Park
IA, Kim YA, Park SY, Roh J, Gong G and Lee HJ: Predictive value of
tertiary lymphoid structures assessed by high endothelial venule
counts in the neoadjuvant setting of triple-negative breast cancer.
Cancer Res Treat. 49:399–407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Khoury T, Nagrale V, Opyrchal M, Peng X,
Wang D and Yao S: Prognostic significance of stromal versus
intratumoral infiltrating lymphocytes in different subtypes of
breast cancer treated with cytotoxic neoadjuvant chemotherapy. Appl
Immunohistochem Mol Morphol. 26:523–532. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Ruan M, Tian T, Rao J, Xu X, Yu B, Yang W
and Shui R: Predictive value of tumor-infiltrating lymphocytes to
pathological complete response in neoadjuvant treated
triple-negative breast cancers. Diagn Pathol. 13:662018. View Article : Google Scholar : PubMed/NCBI
|