Introduction
Metastasis accounts for up to 90% of cancer-related
deaths, driven by complex interplay between malignant cells and the
tumor microenvironment (1–4). It is a progressive, multistep process
in which cancer cells disseminate from the primary tumor, invade
surrounding tissue, survive in circulation and colonize distant
organs (5,6). A deeper understanding of how cancer
cells deviate from normal programs of growth, motility and
stromal-immune crosstalk is essential for identifying potential
targets and optimal time windows for detection, prevention and
treatment of the metastatic diseases.
Lung cancer is the most common and most fatal cancer
worldwide, with 2.5 million new cases (12.4% of all global cancers)
and 1.8 million deaths (18.7% of global cancer deaths) in 2022
(7); it remains the leading cause
of cancer mortality largely due to the high incidence of metastasis
(8,9). Non-small cell lung cancer (NSCLC)
constitutes 80–85% of all lung cancer cases (10). While surgical resection benefits
early-stage NSCLC, a large proportion of patients present with
advanced disease, rendering surgery unfeasible. These patients
often experience poor survival and diminished quality of life, with
limited survival gains from conventional treatments such as
chemotherapy or radiotherapy. Prolonged exposure to radiotherapy or
chemotherapy frequently induces resistance (11). In clinical practice, multiple
therapeutic strategies have been developed to overcome drug
resistance and for patients with lung cancer with targetable driver
gene mutations, such as EGFR or ALK, the treatment paradigm has
evolved. While starting with first-generation inhibitors (such as
gefitinib and crizotinib) was practiced previously, current
first-line standards often involve newer agents (such as
osimertinib and alectinib). Switching to next-generation inhibitors
upon resistance remains the standard strategy (12). For instance, osimertinib effectively
treats EGFR T790M-mediated resistance after first-generation
EGFR-tyrosine kinase inhibitors (13,14).
Combination approaches that leverage non-overlapping mechanisms,
such as pairing targeted therapy with chemotherapy or
anti-angiogenic agents, or using dual-target inhibitors, which aim
to suppress parallel pathways, thereby delaying the emergence of
resistant clones (15). For
patients progressing after targeted therapy or chemotherapy, immune
checkpoint inhibitors [such as programmed cell death protein 1
(PD-1)/programmed death-ligand (PD-L1) antibodies] offer an
orthogonal strategy independent of traditional cytotoxic pathways
(16,17). These agents act by reinvigorating
anti-tumor T cells, with enhanced efficacy in subsets characterized
by high tumor mutation burden (TMB) or PD-L1 expression (18). These precision treatments have
improved outcomes, yet benefit remains heterogeneous and often
transient.
Given these advances, a focus of current clinical
research is identifying patients most likely to benefit from these
therapies early in the treatment course. However, relying solely on
biomarkers such as PD-L1 and TMB has notable limitations (19). Among advanced NSCLC with PD-L1
expression ≥50%, response rates to single-agent immunotherapy are
~44% (20). Similarly, for TMB ≥10
mutations per megabase, response rates remain <30% (21). While combining immunotherapy with
chemotherapy increases objective response rates to 50–70%,
high-dose chemotherapy can significantly damage key effector
subsets, including CD8+ T cells, potentially shortening
the durability of response relative to immunotherapy alone
(22,23). These gaps underscore the need for
functional, patient-specific models that more faithfully
recapitulate tumor-immune-stromal interactions for drug testing and
response prediction.
Accurate modeling of the tumor microenvironment is
crucial for effective drug screening, particularly for
immunotherapies. Traditional models, such as cell lines and
xenografts, have considerable constraints. Extended passaging of
cell lines (>10–15 passages) leads to loss of the original
intratumoral heterogeneity, and xenografts may not faithfully
recapitulate human stromal and immune contexts due to interspecies
differences (24,25). Most patient-derived organoid (PDO)
models similarly lack critical microenvironment elements, such as
vasculature and immune components (26). Even in vascularized organoids, the
absence of perfusion often results in vascular regression. To
address this, researchers have developed approaches combining
vascularized organoids with immune co-cultures (27). Reconstructing the tumor immune
microenvironment by co-culturing tumor-infiltrating lymphocytes
(TILs) with tumor cells shows promise for more accurately
predicting responses to immune checkpoint inhibitors.
Lung cancer organoid-based metastasis models utilize
three-dimensional (3D) culture systems that preserve genomic
diversity and phenotypic heterogeneity of patient tumors. These
models can incorporate key microenvironmental components, including
stromal and immune cells, which enables simulation of critical
steps of the metastatic cascade and provides a powerful platform
for investigating metastasis-driving mechanisms.
A key aspect of developing physiologically relevant
lung cancer organoids is the choice of supporting scaffold. Early
models predominantly relied on commercially available basement
membrane extracts (such as Matrigel) (28). While instrumental for initial
success, these matrices suffered from ill-defined composition,
batch-to-batch variability and non-physiological mechanical
properties. Consequently, there is a growing emphasis on defined
and tunable scaffolds, such as synthetic polyethylene glycol (PEG)
hydrogels (29) and decellularized
extracellular matrix (dECM) hydrogels (30), to provide more precise control over
biochemical cues and biophysical properties, improving
reproducibility and enabling hypothesis-driven testing of
matrix-regulated metastatic behaviors.
The present review summarized recent advances in
lung cancer organoid metastasis models, with emphasis on their use
in elucidating metastatic mechanisms, optimizing drug screening and
resolving tumor heterogeneity. Methodological considerations,
persistent technical challenges, barriers to clinical translation
and ethical considerations were discussed. The future directions
described includes interdisciplinary collaborations involving
multi-omics and spatial profiling, artificial intelligence
(AI)-assisted analytics, immune-competent and vascularized
platforms, and standardization effort. The present review aimed to
provide methodological guidance for basic research and a conceptual
foundation for developing precision treatment strategies.
Literature search methodology
To comprehensively review advances in this field, a
systematic literature search of the PubMed database (https://pubmed.ncbi.nlm.nih.gov/) was conducted
for publications from 2009 to 2025, prioritizing studies from the
past 5 years, using the terms ‘lung cancer organoids’, ‘metastasis
model’, ‘patient-derived organoids’, ‘tumor microenvironment’ and
‘drug screening’, alone and in combination, to identify original
research articles, high-quality reviews and key methodological
papers. Emerging lung cancer organoid-based metastasis models
recapitulate critical steps of the metastatic cascade, yielding
mechanistic insights and informing therapeutic discovery.
Definitions and stem cell origin of
organoids
Stem cells are central to life sciences research and
are defined by two core properties: Self-renewal and multipotent
differentiation. Self-renewal is the capacity to maintain a stable
stem cell pool through mitotic division, generating daughter cells
with the same genetic characteristics (31). Multipotent differentiation is the
ability to give rise to diverse cell types under specific
environmental conditions and signaling cues, thereby contributing
to tissue formation (32).
Organoids are 3D in vitro constructs derived from stem cells
that recapitulate the cellular composition, spatial organization
and selected functions of their native organs. Organoids provide
essential platforms for studying developmental biology and disease
pathogenesis (33).
Organoids derived from stem cells
Since the pioneering development of intestinal
organoids by Sato et al (34) in 2009, stem cell-derived organoid
technology has rapidly expanded across multiple tissues, including
the brain, liver and diverse tumor types, offering new
opportunities for drug screening and disease modeling. Brain
organoids serve as a prominent example, as they are constructed
through the systematic differentiation of neural stem cells and
precise cellular interactions, ultimately forming highly complex
neural networks. These networks not only exhibit functions similar
to those of the natural brain but also provide essential biological
markers for investigating brain development and neurodegenerative
diseases (35). Leveraging this
advantage, researchers have systematically studied the
pathogenesis, pathological evolution and potential therapeutic
targets of neurodegenerative diseases such as Alzheimer's disease
and Parkinson's disease using in vitro models that closely
mimic in vivo physiological conditions (36,37).
PDOs
In oncology research, tumor organoids derived from
patient samples often retain the phenotypic characteristics,
genomic heterogeneity and mutational landscape of the original
tumor. Tumor organoids derived from patient samples preserve the
architectural, molecular and genetic features of the primary tumor,
including structure, function, mutation spectrum and gene
expression profile. These organoids are effectively used for
disease characterization, mechanism exploration, high-throughput
drug screening and evaluation, discovery of innovative therapeutic
targets and potential compounds and the development of personalized
treatment strategies based on individual tumor characteristics
(38).
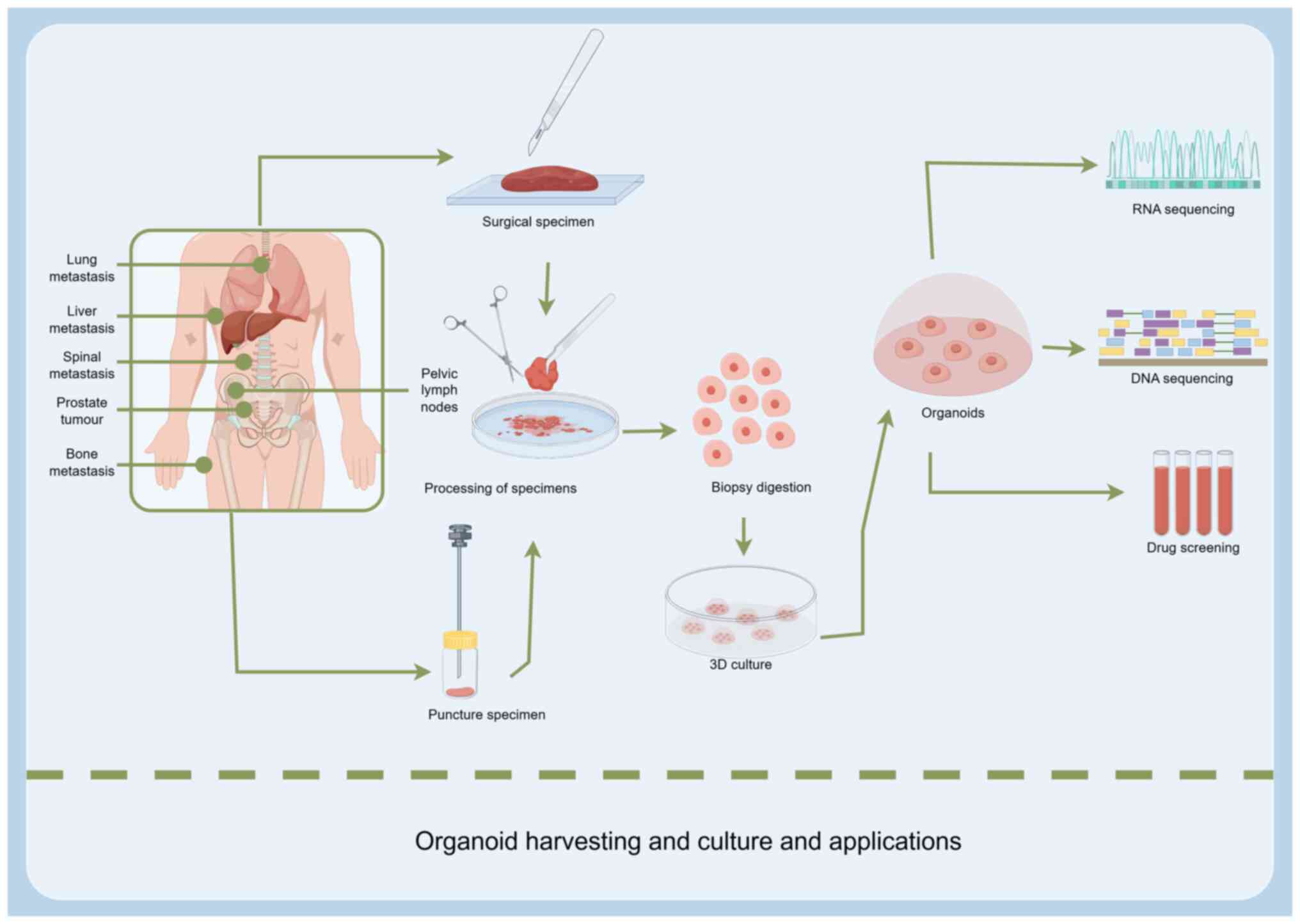
The construction of lung cancer organoid metastasis
models begins with the acquisition of suitable tissue samples.
Primary lung cancer tissues, obtained from surgical resections or
biopsies are commonly used (39,40).
To investigate metastatic mechanisms, tissues from metastatic
lesions are particularly valuable, including samples from brain,
bone or liver metastases, which directly reflect the
characteristics of tumor cells within site-specific metastatic
microenvironments. For example, a previous study successfully
established organoid models from surgical specimens of brain
metastases in patients with lung cancer, providing essential tools
for exploring the mechanisms of brain metastasis (41).
Malignant pleural effusions (MPEs), including
pleural effusion and ascitic fluids, represent an important source
for organoid construction. MPEs are rich in tumor cells, and their
collection is relatively noninvasive (42). Mazzocchi et al (43) successfully developed 3D lung cancer
organoids from pleural effusion. These organoids, composed of tumor
and stromal cells embedded in a hydrogel that mimics the ECM,
exhibited alveolar-like structures and in vivo-like drug
responses. The study confirmed that pleural effusion-derived
organoids are suitable for investigating lung adenocarcinoma (LUAD)
and that 3D cultures outperform two-dimensional (2D) cultures in
disease modeling and drug screening. In another study, Wang et
al (44) established lung
cancer organoids from both tumor tissue and MPE. It was found that
MPE significantly enhanced organoid formation, reduced culture time
by >50% and altered drug sensitivity profiles, resulting in
increased resistance to certain drugs. Mechanistically, although
MPE did not alter the genomic composition of the organoids, it
influenced stem cell distribution and cell architecture, including
expansion of the extracellular space. Gene expression analyses
revealed significant enrichment of pathways related to the ECM and
mitochondrial function.
Distinct challenges in developing lung
cancer organoids
The development of lung cancer organoids presents
unique challenges that are not typically encountered in other
organoid systems. A primary obstacle is the need for multiple,
subtype-specific culture protocols to account for the substantial
histological and genetic diversity between NSCLC and small cell
lung cancer (SCLC). Furthermore, the high clinical mortality
associated with metastasis necessitates modeling organ-specific
metastatic niches, such as those in the brain and bone. In
addition, given the focus on guiding precision therapies,
particularly immunotherapy, there is a pressing need to develop
sophisticated, immune-competent co-culture models. This combination
of factors makes lung cancer organoids a highly specialized and
complex platform.
Advantages of organoids as drug
screening platforms
Organoids provide physiologically relevant platforms
for drug screening and sensitivity testing by closely replicating
the in vivo architecture and functional characteristics of
human tissues. Furthermore, organoids retain tissue-specific
features and preserve critical cellular interactions, and capture
tumor heterogeneity more accurately. This makes them especially
valuable for personalized treatment strategies (45). Drug responses can be evaluated by
treating patient-derived tumor organoids (PDTOs) with a range of
therapeutic agents and measuring efficacy through multiple
analytical readouts. In oncology, organoid-based models are widely
used to assess cancer drug sensitivity and to predict therapeutic
outcomes, covering a variety of agents, including targeted
therapies, immune monotherapies, combination immunotherapies and
anti-angiogenic drugs (46,47). For example, clinical studies have
employed organoid drug sensitivity assays to anticipate
chemotherapy efficacy (48,49).
Modeling key steps of the metastatic cascade
in organoids
Lung cancer organoid models are powerful tools for
deconstructing the complex, multistep process of metastasis into
discrete, investigable mechanisms. By engineering specific culture
conditions, these models can replicate key biological events,
including ECM remodeling, epithelial-mesenchymal transition (EMT)
and tumor-immune interactions, which collectively drive metastatic
progression.
Simulating metastatic microenvironment
conditions
To mimic the in vivo tumor microenvironment,
lung cancer organoids are cultured in 3D matrices such as Matrigel,
which provide both structural support and biochemical cues
(50). Cultures are maintained in a
base medium (such as advanced DMEM/F12) supplemented with growth
factors and nutrients essential for organoid survival and expansion
(51,52). Furthermore, media are tailored to
histological subtypes. NSCLC organoids typically require the
addition of EGF, FGF, R-spondin1 and Noggin, whereas SCLC organoids
often need additional factors to maintain their neuroendocrine
characteristics (53) Further
optimization is necessary to model organ-specific metastasis more
accurately. For brain metastasis models, neural-supportive medium
components such as Neurobasal™ (Gibco; Thermo Fisher
Scientific, Inc.) are incorporated into the medium (54). Culture conditions, including oxygen
tension, temperature and humidity, are tightly controlled. Hypoxia,
in particular, can preserve cancer stem-like phenotypes and
upregulate metastasis-associated programs, providing a more
physiologically relevant context for studying tumor invasion,
spread and therapy response (55,56).
Modeling ECM remodeling
The 3D scaffold not only supports organoid growth
but also serves as a modifiable ECM surrogate. Tumor organoids
dynamically remodel their surrounding matrix to facilitate
invasion, which can be captured by live imaging using fluorescent
ECM components (57,58). Parallel biochemical analyses of
conditioned media reveals elevated levels of matrix
metalloproteinases and other ECM-modifying enzymes, providing
quantitative readouts of invasive potential (59).
Inducing EMT
EMT is a key transcriptional reprogramming event
that confers migratory and invasive capacities. In organoid
systems, EMT can be reproducibly elicited by microenvironmental
stimuli. Exposure to hypoxia or exogenous transforming growth
factor-β (TGF-β) drives molecular and morphological shifts,
including downregulation of epithelial markers (such as
E-cadherin), upregulation of mesenchymal markers (such as vimentin)
and morphological transitions from smooth, spherical structures to
irregular, invasive protrusions. This phenotypic change, often
mediated by effectors, directly enhances tumor cell migration and
invasion, thereby modeling early metastatic steps (60).
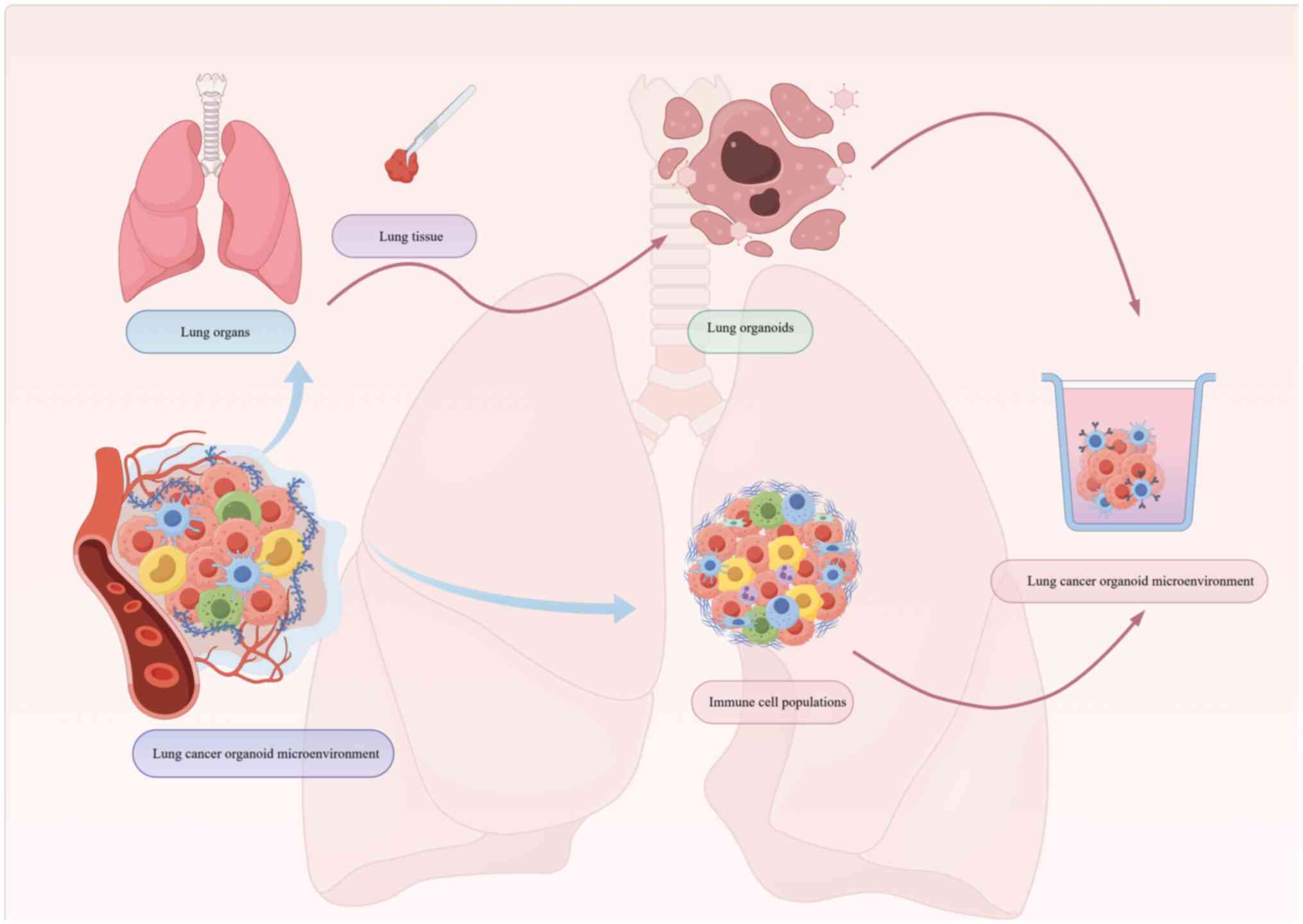
Reconstituting tumor-immune
interactions
Immune-organoid co-culture systems are crucial for
elucidating the dynamic interactions between tumor cells and the
immune microenvironment, particularly the mechanisms by which
cancer cells evade immune surveillance during metastasis. Lung
cancer organoids can be directly co-cultured with autologous immune
components, such as TILs or peripheral blood mononuclear cells
(Fig. 1) (61), to recapitulate patient-specific
immune responses. This setup enables real-time assessment of immune
cell cytotoxicity and tumor cell lysis, and it captures adaptive
immune evasion mechanisms, including the upregulation of checkpoint
molecules such as PD-L1 on organoid cells. These co-culture models
also provide a physiologically relevant platform for evaluating
immunotherapies, including immune checkpoint inhibitors such as
anti-PD-1/PD-L1 antibodies, by quantifying treatment-induced
organoid death (62).
Construction strategies and key technologies
for lung cancer organoid metastasis models
Leveraging organoids for lung cancer metastasis
research requires a multifaceted approach. Key elements include
developing subtype-tailored culture protocols, creating customized
models through gene editing, recapitulating metastatic steps via
in vivo transplantation, and directly capturing
disseminating cells using circulating tumor cell (CTC)-derived
organoids (Fig. 2; Table I).
 | Table I.Comparative analysis of primary
strategies for establishing lung cancer organoid metastasis
modelsa. |
Table I.
Comparative analysis of primary
strategies for establishing lung cancer organoid metastasis
modelsa.
| Strategy | Core
advantages | Limitations | Relative cost | Translational
readiness | Primary application
scenarios |
|---|
| PDOs | Preserves genomic
heterogeneity, phenotype, and drug response of he toriginal tumor.
Short establishment time (weeks), suitable for high-throughput
screening. Provides a platform for personalized medicine. | Suboptimal overall
culture success rate (particularly for SCLC and rare subtypes).
Difficult to fully recapitulate the complete tumor microenvironment
(for example, vasculature and immune components). | Medium-high | Intermediate (used
in retrospective studies guiding clinical therapy, requires
prospective trial validation) | Disease modeling,
drug sensitivity testing, personalized therapy exploration, tumor
heterogeneity research |
|
|
| Lack of
standardized protocols. |
|
|
|
| Gene-edited
organoids | Enables precise
study of causal roles of specific genes in metastasis. Allows
creation of customized models driven by specific driver
mutations. | Ethical and safety
considerations. Technically complex, requires specialized
platform. | High | Low (primarily a
powerful basic research tool; direct clinical translation path is
long) | Mechanism studies,
target validation, gene function research, overcoming drug
resistance |
|
| Useful for gene
knockout, knock in, and correcting resistance mutations | Single-gene editing
may not mimic the complex polygenic background of human
cancer. |
|
|
|
| In vivo
organoid transplantation | Provides the most
physiologically relevant in vivo microenvironment; models
the entire metastatic cascade. The ‘gold standard’ for validating
the tumorigenicity and metastatic potential of organoids. | Time-consuming
(months). Immunodeficient mouse models lack a human immune system,
limiting immunotherapy research. | High | Intermediate (core
component of preclinical research but species differences are a
translational bottleneck) | Validation of in
vitro findings, studying the metastatic cascade, preclinical
drug efficacy evaluation. |
|
| Evaluates drug
efficacy within a whole body physiological context. |
|
|
|
|
| CTC-derived
organoids (liquid biopsy). | Minimally invasive
sample acquisition Directly derived from metastatic cell
populations, representing real time tumor status. Potential to
capture cell subpopulations with high metastatic potential. | Extreme rarity of
CTCs in blood, high technical challenge. Very low culture success
rate (for example, ~58% in prostate cancer) Requires tumor-specific
culture conditions | Very high | Low (the technology
itself is immature, but has great future translational potential as
an extension of liquid biopsy) | Metastasis
mechanism research, dynamic therapy monitoring, personalized
therapy for metastatic disease |
Specific culture requirements for
primary lung cancer subtypes
The successful establishment of lung cancer
organoids critically depends on tailoring culture conditions to
each subtype, reflecting their distinct cellular origins and
oncogenic drivers.
i) LUAD organoids
LUAD often originates from alveolar type II (AT2)
cells and thus requires culture conditions that sustain AT2
stemness and proliferative capacity. The base medium typically
includes essential mediators such as EGF, FGF-7/10, Noggin and
R-spondin 1 to promote proliferation and maintain an AT2-like
state. This protocol was further optimized by inhibiting the BMP
signaling pathway and supplementing with ligands such as
heregulin-β1 to better mimic the alveolar niche, as demonstrated in
feeder-free systems that enable long-term expansion and modeling of
LUAD (63,64).
ii) Lung squamous cell carcinoma
(LUSC) organoids
LUSC, which arises from bronchial basal cells, often
relies more on ECM-derived physical and biochemical cues than
exogenous growth factors. A robust LUSC model was established using
a base medium without the addition of growth factors such as
R-spondin, EGF or BMP-4. The success of this system hinged on a
tailored ECM composed of Matrigel and collagen I, as well as the
strategic initiation of cultures from pre-formed cell spheroids.
This approach reproducibly generated organoids that self-organize
into a characteristic hierarchical structure with a p63-positive
basal cell layer and inward differentiation, providing a
distinctive model for studying LUSC biology (65).
iii) SCLC organoids and metastatic
modeling
The high-grade neuroendocrine phenotype of SCLC
necessitates specialized culture conditions. A base media typically
includes B27 and N2 supplements, key mitogens (bFGF, EGF and
insulin), hydrocortisone and ROCK inhibitors to prevent anoikis and
support suspension growth. To model metastasis, particularly brain
metastasis, advanced platforms co-culture SCLC cells with human
cerebral organoids in a Matrigel-based 3D environment. When
combined with modulation of key pathways such as Notch and
longitudinal monitoring using live-cell imaging, this platform
enables real-time assessment of invasion, colonization and subtype
plasticity within the brain microenvironment (66,67).
Application of gene editing
technologies
Patient-derived lung cancer organoids faithfully
replicate the pathological and genomic characteristics of their
source tumors, preserving key driver alterations, while maintaining
malignant cell phenotypes (68).
Using gene editing technologies such as the CRISPR/Cas9 system
(69), lung cancer organoids can be
genetically modified to create metastasis models driven by specific
gene mutations. By knocking out or inserting key genes associated
with lung cancer metastasis, researchers can study gene function
and its impact on the metastatic process.
For example, the CRISPR/Cas9 system has been used to
knock out the SOX9 gene in human embryonic stem cells prior
to directed differentiation into lung organoids. While SOX9
deletion did not affect epithelial cell differentiation, it
impaired proliferation and increased apoptosis. Upon
transplantation under the renal capsule of immunodeficient mice or
orthotopically into bleomycin-injured mouse lungs, SOX9
inactivation was found to affect the expression of specific cell
markers and cell maturation. Techniques such as immunofluorescence
and histological analysis revealed these changes, which provided
direct evidence for the role of SOX9 in human lung
epithelial development (70).
Application of functionalized hydrogel
scaffolds in organoid culture
Gmeiner et al (71) (2020) successfully established small
cell lung cancer PDX-derived organoid models using HyStem-HP
hydrogel scaffolds and demonstrated that the novel drug CF10 could
overcome tumor chemoresistance. Functionalized hydrogels are
defined scaffolds with well-defined compositions and precisely
tunable properties. As a result, they are increasingly supplanting
animal-derived matrices such as Matrigel, whose undefined
composition and batch variability limit reproducibility and
mechanistic control. Current advancements are primarily focused on
two main directions. The first involves synthetic hydrogels, such
as PEG and polyisocyanopeptides, which provide a highly
controllable microenvironment for organoids through the grafting of
cell-adhesive peptides (such as RGD and GFOGER), and through
fine-tuning of mechanical properties (such as stiffness and
viscoelasticity). This approach enables precise investigation of
the role of mechanical signals in organoid growth, EMT and invasion
(72). The other direction focuses
on engineered natural polymers, such as organ-specific dECM,
peptide-functionalized nanocellulose and alginate (73). These materials retain
tissue-specific biological signals and enhance functionality
through chemical modifications, effectively promoting structural
maturity and functional complexity of organoids (such as
vascularization). The establishment of these functionalized
hydrogel scaffold systems has significantly improved the
reliability and utility of organoids in standardized modeling,
disease mechanism research, drug screening and clinical translation
(74–76).
In vivo transplantation and
comparative analysis of preclinical models
The metastatic cascade unfolds within a complex
physiological milieu that is difficult to fully recapitulate in
vitro. Consequently, the in vivo transplantation of
organoids serves as a critical intermediary, validating their
tumorigenic and metastatic capacities in a more physiologically
relevant framework for investigation. For example, orthotopic
transplantation of lung cancer organoids into the lungs of
immunodeficient mice can reproduce the native tumor
microenvironment, enabling comprehensive observation of the
metastatic continuum from local invasion to distant colonization
(77).
Organoid transplantation for
metastasis research
The utility of organoid transplantation has been
extensively demonstrated across multiple cancer types. In
colorectal cancer, organoid libraries retain stable pathological
and genetic characteristics. GFP-labeled organoids derived from
patient-derived xenografts (PDXs), when transplanted into
immunodeficient mice, facilitate visualization and quantification
of micrometastatic lesions (such as hepatic metastases following
splenic injection) (78).
Comparable strategies have been employed in models of lung, breast
(79), pancreatic (80) and brain (81) cancer, effectively recapitulating
tumor dynamics and supporting preclinical evaluation of therapeutic
efficacy.
By constructing humanized lung cancer PDX models
through engraftment of human hematopoietic stem cells into
immunodeficient mice to reconstitute a human immune system,
researchers can effectively evaluate the efficacy of PD-1/PD-L1
inhibitors such as pembrolizumab and nivolumab (82).
A comparative perspective on
preclinical models
While organoid transplantation is a versatile tool
in metastasis research, it is one of several available preclinical
models. A comparative evaluation of these mainstream platforms
underscores their distinct advantages and optimal applications,
with PDOs occupying a unique and increasingly important niche in
translational studies.
i) PDOs vs. PDXs
Comparative studies have highlighted the distinct
advantages of PDOs over PDXs in several key aspects. PDOs can often
be established within weeks, which is significantly faster than the
months typically required for PDXs. PDOs are more cost-effective
and offer greater scalability, making them suitable for
high-throughput in vitro studies (83). A critical limitation of conventional
PDX models, which are grown in immunodeficient mice, is the absence
of a functional human immune system, restricting their application
in immunotherapy research. Humanized PDX models address this issue
to some extent, but still face challenges such as incomplete immune
reconstitution. Additionally, murine stromal cells gradually
replace the human tumor microenvironment in PDXs, potentially
altering key aspects of tumor biology. Although PDOs also face
challenges in fully replicating the tumor microenvironment, they
offer greater flexibility for incorporating human immune components
through co-culture systems (84).
ii) Characteristics and limitations of
spheroids
Spheroids are simple 3D cell aggregates that are
valuable for high-throughput drug screening and for studying basic
nutrient gradients. However, they generally lack the complex and
architecturally accurate tissue morphology, cellular diversity
(including stromal components) and long-term functional
differentiation that characterize PDOs derived from stem or
progenitor cells. These shortcomings limit their ability to mimic
the complexity of native tissues (85,86).
iii) Comparative analysis of PDOs and
lung-on-a-chip models
Organ-on-a-chip platforms are highly effective at
engineering precise physiological microenvironments, incorporating
fluid flow, mechanical forces and multi-cellular interactions to
model dynamic processes such as vascular perfusion and immune cell
trafficking. Their limitation often stems from the use of
established cell lines, which may not fully represent the
patient-specific genomic heterogeneity. PDOs complement these
systems by offering a genetically accurate, patient-derived tissue
source, which can be integrated into chip-based devices to create
more personalized and representative models (87).
iv) Comparative analysis of PDOs and
genetically engineered mouse models (GEMMs)
GEMMs are an optimal model for studying
tumorigenesis and metastasis within an intact, immunocompetent
organism and in the context of developing tissues. However, their
primary limitations include long timelines, high costs and inherent
species differences, which can hinder the direct translation of
findings to human patients. PDOs offer a more rapid, human-based
platform for personalized screening and genetic manipulation,
providing a valuable complement to GEMMs (88).
v) Integrating preclinical models:
Synergistic applications
These models are not mutually exclusive, and they
are highly complementary. Insights gained from high-throughput drug
screening in PDOs can be validated within the more physiologically
intact contexts of PDXs or GEMMs. Conversely, PDOs can be
integrated into organ-on-a-chip devices to create more complex,
human-relevant organoid-on-a-chip systems. The choice of model
depends on the specific research question, with PDOs offering a
balanced platform that preserves human tumor heterogeneity while
enabling scalable in vitro experimentation.
vi) Advances in patient-derived models
for studying metabolism-immunity interplay in lung cancer
PDOs and patient-derived organotypic tissue cultures
(PD-OTCs) have recently demonstrated notable value in
recapitulating the lung cancer tumor microenvironment and
elucidating its functional dynamics, particularly the interactions
between metabolism and immunity. Using stable isotope-resolved
metabolomics, Fan et al (89) found that PD-OTCs more accurately
reproduce complex metabolic reprogramming features observed in
patients, including enhanced glycolysis, nucleotide synthesis and
anaplerotic reactions, compared with PDX models of NSCLC. Building
on this platform, the study directly uncovered key regulatory
mechanisms along the metabolism-immunity axis. It demonstrated that
immunomodulators (such as β-glucan) and checkpoint inhibitors can
reprogram the metabolic state of immune cells, effectively
reversing immunosuppression by shifting TAMs from the M2
anti-inflammatory phenotype to the M1 pro-inflammatory phenotype.
These experimental findings provide empirical support for the
theoretical framework proposed in the study by Huang et al
(90), which suggested that
metabolic networks within the tumor microenvironment coordinate
immune responses and drive immune evasion. In summary, PDOs and
PD-OTCs have emerged as key bridges connecting the theory of
metabolic reprogramming with the practice of immunotherapy,
establishing a methodological foundation for developing
individualized combination strategies that target the
metabolism-immunity axis.
CTC-derived organoids
By contrast, CTC-derived organoids are established
from tumor cells shed into the bloodstream from primary or
metastatic sites, thereby capturing dynamic and comprehensive
tumor-related information. Culturing organoids from sources such as
intestinal tissues is relatively straightforward, as these models
can be stably cultured and passaged in defined media and matrix
gels using well-established protocols. However, culturing CTC
organoids poses greater technical challenges. CTCs are extremely
rare in peripheral blood, typically on the order of 1 to 10
cells/ml, making their isolation and enrichment particularly
demanding. CTCs derived from different tumor types require
tumor-specific culture conditions, further contributing to low
success rates (91,92). In prostate cancer, for example, the
reported success rate of culturing CTCs is ~5.8% (93).
Regarding practical applications, general organoids
are widely used to study organ development, disease modeling and
drug testing. CTC organoids are particularly valuable for
investigating tumor metastasis, personalized cancer therapy and
drug resistance mechanisms. In terms of cellular characteristics,
organoids derived from normal tissue stem cells typically exhibit
stable morphology and function, while those derived tumor tissues
retain some malignant features of the original tumor cells. CTC
organoids display a high degree of heterogeneity that reflects the
diversity of tumor cells during the metastatic process, and they
may exhibit stronger metastatic potential (94).
CTC organoids derived from blood samples of patients
with SCLC can simulate the tumor microenvironment and support drug
response testing. By preserving tumor cell heterogeneity, CTC
organoids can improve prediction of drug efficacy relative to
traditional preclinical models. This enables clinicians to identify
effective therapeutic agents tailored to individual patients,
prioritize drugs with higher sensitivity, avoid those with
potential resistance and reduce adverse effects associated with
ineffective treatments. Ultimately, this approach can improve
clinical outcomes and enhance patient life quality. Beyond their
utility in guiding personalized therapy, CTC organoids serve as a
powerful tool for investigating the biological features of SCLC,
including the mechanisms of tumor initiation, progression and
metastasis (94,95). They also offer insights into the
development of drug resistance and inform strategies to overcome
it. From a translational perspective, molecular analysis of CTC
organoids can facilitate the discovery of novel therapeutic
targets, assist in evaluating the efficacy and safety of emerging
drug candidates and support early-stage clinical trials due to
their operational simplicity, cost-effectiveness and relatively
short culture time (96).
Application of lung cancer organoid
metastasis models in research
Mechanistic studies of metastasis
Lung cancer organoid-based metastasis models provide
an effective platform for in-depth exploration of metastatic
mechanisms. Research on metastasis-related genes and signaling
pathways has revealed that numerous genes serve critical roles in
lung cancer metastasis. For example, deletion of the histone
methyltransferase gene KMT2C promotes distant metastasis in
SCLC through DNMT3A-mediated epigenetic reprogramming. In
lung cancer organoid models, KMT2C knockout markedly
increases organoid invasiveness and metastatic potential. Further
analyses have revealed associated alterations in histone
modification and DNA methylation patterns, which regulate
downstream genes involved in metastatic progression (97).
These models are also instrumental for studying the
impact of the tumor microenvironment on metastasis. Cellular
components such as tumor-associated macrophages (TAMs) and
fibroblasts interact with tumor cells and modulate their metastatic
behavior. Co-culture experiments involving TAMs and lung cancer
organoids have demonstrated that macrophages secrete cytokines that
enhance organoid invasion and migration. This pro-metastatic effect
has been further validated in vivo, where the presence of
TAMs significantly promoted metastatic spread following
transplantation (98).
Drug screening and efficacy
evaluation
Lung cancer organoid metastasis models are valuable
for drug screening and efficacy evaluation. They support
high-throughput screening of anti-metastatic drugs by evaluating
treatment-induced changes in cancer cell migration, invasion and
expression of metastasis-associated markers. Lung cancer organoid
metastasis models are also well suited for assessing the
synergistic effects of combination therapies, thereby informing
optimization of clinical treatment strategies. In addition,
correlating genetic characteristics and molecular marker expression
in patient-derived lung cancer organoids with clinical metastasis
data may facilitate the development of predictive models for
metastatic risk. For example, elevated expression of specific genes
or proteins could indicate increased metastatic risk and poor
prognosis, aiding early intervention and improved outcomes
(99).
Metastasis is a major determinant of prognosis in
lung cancer. A previous study established lung cancer organoids
from malignant serous effusions that retained key tumor
characteristics and showed high genomic concordance with the
original tumors. The Lung Cancer Organoids-Drug Sensitivity Test
(LCO-DST) demonstrated strong predictive performance, with a
sensitivity of 84.0%, specificity of 82.8% and accuracy of 83.3%
for distinguishing drug-sensitive from drug-resistant patients.
This test effectively predicted responses to targeted therapies and
chemotherapy. Notably, lung cancer organoids exhibit both stability
and inter-patient heterogeneity. Organoids derived at different
time points from the same patient maintained consistent genomic
features, while organoids from different patients showed distinct
morphologies and drug sensitivities. These findings highlight the
potential of LCO-DST for predicting the effectiveness of
combination therapies in advanced lung cancer (100).
Using lung cancer brain metastasis organoid models
as an example, researchers have evaluated the impact of various
therapeutic agents on the growth and invasion of brain metastases.
Treating lung cancer brain metastases (LCBM) remains a significant
clinical challenge, underscoring the importance of drug screening.
Traditional cancer cell lines and PDX models have notable
limitations; cell lines often lack in vivo tumor
heterogeneity and PDX models face interspecies incompatibilities
and low establishment efficiency (101). By contrast, LCBM organoids
recapitulate the molecular and phenotypic features of metastatic
tumors. They can be used to screen targeted therapies based on
specific genetic alterations such as EGFR or ALK
mutations, to identify potential radiosensitizers and evaluate the
combined radio-chemotherapy efficacy. With access to large-scale
drug libraries, organoid models enable high-throughput drug
screening, providing strong support for LCBM drug development and
clinical treatment selection (101).
Research on tumor heterogeneity
Tumor heterogeneity is a critical factor in the
development, therapeutic resistance and metastasis of lung cancer.
The dynamic evolution of tumor heterogeneity in space (between
primary and metastatic sites and within different tumor regions)
and in time (before and after treatment or at different disease
stages), poses significant challenges for constructing effective
metastasis models (102–104). Compared with traditional cell
lines and GEMMs, lung cancer organoids more accurately preserve
genomic diversity, phenotypic variations and key components of the
tumor microenvironment, including co-culture systems that
incorporate stromal and immune cells from the original tumors
(105).
Single-cell sequencing studies (106–109) have demonstrated that lung cancer
organoids retain subclonal structures and transcriptomic
heterogeneity comparable to those of the primary tumors. These
organoids are also enriched for invasive front cell populations,
particularly cells undergoing EMT, which serve crucial roles in
metastasis (110). However,
replicating the dynamic evolution of heterogeneity within
organoid-based metastasis models remains technically challenging.
As the metastatic cascade involves selection of heterogeneous CTCs,
current organoid systems are limited in their ability to mimic
circulatory dynamics, including fluid shear stress and
trans-endothelial migration. To address these limitations,
integration with organ-on-a-chip technologies will be necessary to
achieve more physiologically relevant spatiotemporal reconstruction
(111–113).
LCBMs are a common and severe complication in
advanced diseases. Brain metastases exhibit higher median absolute
deviation and higher mutation allele tumor heterogeneity scores
than primary tumors, indicating greater genomic diversity (114). In addition, single-cell RNA
sequencing has revealed marked differences in gene expression
between tumor cells from brain metastases and those from primary
sites, highlighting cell-level heterogeneity that may influence
disease progression and therapeutic response (115). In the future, the integration of
spatial transcriptomics, live-cell imaging and AI-driven algorithms
for quantifying heterogeneity will enable lung cancer organoid
models to capture dynamic tumor evolution at single-cell
resolution. These advances will support early prediction and
precise intervention for metastatic risk.
Challenges and future directions in lung
cancer organoid metastasis models
Clinical translation of lung cancer organoid
metastasis models faces several intertwined technical,
translational and ethical challenges. Overcoming these obstacles is
essential for advancing the field, and future development will
depend on integrating innovative technologies and establishing
standardized practices. Addressing these challenges will enable
more effective and personalized approaches to lung cancer research
and treatment.
Current challenges. i) Technical
hurdles in model construction
The success rate of organoid culture remains
suboptimal for certain lung cancer subtypes, such SCLC and
sarcomatoid carcinoma, due to their limited stemness and strong
dependence on the ECM (116). A
major bottleneck is the widespread use of ill-defined matrices such
as Matrigel, which are affected by batch-to-batch variability,
undefined composition and non-physiological mechanical properties.
Although dECM provides a more native biochemical environment,
donor-dependent variability complicates standardization (117–119).
Reconstructing the tumor microenvironment also
presents significant challenges. Co-culture systems that integrate
immune cells are hindered by poor immune cell infiltration into
dense hydrogel domes and by the difficulty of sourcing authentic
TILs. These limitations impede accurate modeling of complex tumor
microenvironment interactions that are crucial for studying
tumor-immune dynamics and metastasis (120). In addition, current models are
limited in ability to replicate full cellular diversity and complex
stromal interactions, including functional vasculature, which
limits the ability to model tumor growth, metastatic dissemination
and dynamic interactions between tumor cells and their
microenvironment.
ii) Barriers to clinical translation
and ethical considerations
From a clinical perspective, the absence of
standardized protocols for model construction, culture and
assessment severely limits reproducibility and hinders large-scale
application. High costs associated with tissue acquisition,
organoid culture and molecular analyses present significant
economic barriers. Additionally, there is a lack of large-scale,
multi-center clinical validation to confirm the predictive accuracy
of these models for drug response and personalized therapy, which
impedes the broader clinical adoption (121).
Ethically, the collection and use of patient-derived
tissues requires rigorous informed consent processes. The
application of advanced techniques such as gene editing introduces
additional ethical complexities, requiring clear regulatory
frameworks for safety assessment and intellectual property
management.
Integrated future directions
To address these challenges, the field is moving
towards a multi-faceted strategy that leverages cutting-edge
technologies and promotes standardization.
i) Advancing model fidelity through
technology integration
Integrating multi-omics technologies, including
genomics, transcriptomics, proteomics and metabolomics, with
organoid models will provide a comprehensive molecular view of
metastasis. This approach may reveal novel drivers, elucidate
dysregulated metabolic pathways and uncover complex molecular
interactions within the tumor microenvironment. Combining these
high-throughput techniques with organoid models will deepen
mechanistic insights and accelerate the development of more
targeted and effective therapies (122).
A key goal in cancer research is the development of
sophisticated immune-organoid models. By co-culturing lung cancer
organoids with autologous immune cells, including T cells and
natural killer cells, or by utilizing humanized mouse models,
researchers can better replicate crucial tumor-immune interactions.
These models provide a powerful platform for evaluating
immunotherapy efficacy and for studying mechanisms of immune
evasion. Understanding how tumors interact with immune cells in a
more physiologically relevant context can accelerate the discovery
of novel therapeutic strategies and improve the precision of cancer
immunotherapy (123–128).
The synergy between organoid models and liquid
biopsy is also highly promising. Analysis of circulating tumor DNA
(ctDNA) from patient plasma or from organoid culture supernatants,
provides a non-invasive method to track tumor evolution in
real-time. This approach can guide drug selection in organoid
models, enabling personalized and adaptive treatment strategies. By
incorporating ctDNA analysis, researchers can continuously monitor
treatment response and identify emerging resistance mechanisms,
creating a closed-loop feedback system that enhances the precision
and flexibility of oncology therapies. This combination has the
potential to transform personalized cancer care by aligning
treatments with the evolving molecular landscape of each tumor
(91,129–131). Single-cell RNA sequencing is
another pivotal tool; it can resolve cellular heterogeneity within
organoids and their tumor microenvironment, validate model fidelity
and identify rare cell populations critical to metastasis research
(106).
ii) Leveraging AI and
standardization
The application of AI and machine learning is poised
to transform the field. AI algorithms can analyze high-content
imaging data from organoids to automate quantification of
metastatic phenotypes, including invasion and migration, which
streamlines the screening process (132–134). Furthermore, AI can integrate
multi-omics and drug response data to build predictive models for
metastatic risk and therapy optimization, thereby guiding
personalized treatment decisions. These advances must be supported
by rigorous standardization. Harmonized protocols for tissue
processing, culture conditions and quality control are essential to
ensure reproducibility, enable multi-center collaborations and
facilitate clinical adoption. In parallel, optimizing workflows and
reducing costs are important for improving accessibility.
Conclusion
Lung cancer organoid metastasis models have emerged
as pivotal platforms for deciphering metastatic mechanisms and
advancing precision therapy. Despite persistent challenges in
standardization and tumor microenvironment recapitulation, future
investigations should prioritize the following directions:
Integrating multi-omics with spatial transcriptomics to
systematically resolve spatiotemporal dynamics of metastasis;
developing immunocompetent organoid co-culture systems to deepen
understanding of tumor-immune interactions; employing chemically
defined functionalized hydrogels for precise modulation of the
tumor microenvironment; and combining artificial intelligence with
liquid biopsy to establish dynamic drug response prediction models.
Through interdisciplinary integration and standardized framework
development, these organoid models will accelerate anti-metastatic
drug discovery and ultimately facilitate the clinical translation
of precision medicine for lung cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Xi'an Science and
Technology Program (grant no. 2024JH-ZCLGG-0040), the Wu Jieping
Medical Foundation Clinical Research Special Grant Fund (grant no.
320.6750.2023-17-23), the Technology Program (grant no.
2024YXYJ0133), The Xi'an Science and Technology Program (grant no.
24YXYJ0179), Fundamental Research Funds for the Central
Universities (‘Free Exploration’ Teacher Project) of Xi'an Jiaotong
University (grant no. xzy012025151), the Beijing KC Medical
Development Foundation Research Project (grant no.
KC2023-JX-0288-PM92) and the China Health and Medical Development
Foundation (grant no. chmdf2024-xrzx09-20).
Availability of data and materials
Not applicable.
Authors' contributions
JJ conceived the study. JJ, GMD, QG, ZYZ, XYL, FSH,
SNL, JQM, JB, HW and ZZ wrote and reviewed the manuscript. All
authors have read and approved the final manuscript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
El-Kenawi A, Hänggi K and Ruffell B: The
immune microenvironment and cancer metastasis. Cold Spring Harb
Perspect Med. 10:a0374242020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Seyfried TN and Huysentruyt LC: On the
origin of cancer metastasis. Crit Rev Oncog. 18:43–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peitzsch C, Tyutyunnykova A, Pantel K and
Dubrovska A: Cancer stem cells: The root of tumor recurrence and
metastases. Semin Cancer Biol. 44:10–24. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sulekha Suresh D and Guruvayoorappan C:
Molecular principles of tissue invasion and metastasis. Am J
Physiol Cell Physiol. 324:C971–C991. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jin K, Li T, van Dam H, Zhou F and Zhang
L: Molecular insights into tumour metastasis: Tracing the dominant
events. J Pathol. 241:567–577. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kalita B and Coumar MS: Deciphering
molecular mechanisms of metastasis: Novel insights into targets and
therapeutics. Cell Oncol (Dordr). 44:751–775. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smolarz B, Łukasiewicz H, Samulak D,
Piekarska E, Kołaciński R and Romanowicz H: Lung
cancer-epidemiology, pathogenesis, treatment and molecular aspect
(review of literature). Int J Mol Sci. 26:20492025. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Orooji N, Babaei S, Fadaee M,
Abbasi-Kenarsari H, Eslami M, Kazemi T and Yousefi B: Novel
therapeutic approaches for non-small cell lung cancer: An updated
view. J Drug Target. 33:1306–1321. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Q, Abdo R, Iosef C, Kaneko T,
Cecchini M, Han VK and Li SS: The spatial transcriptomic landscape
of non-small cell lung cancer brain metastasis. Nat Commun.
13:59832022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xie S, Wu Z, Qi Y, Wu B and Zhu X: The
metastasizing mechanisms of lung cancer: Recent advances and
therapeutic challenges. Biomed Pharmacother. 138:1114502021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim SY, Park HS and Chiang AC: Small cell
lung cancer: A review. JAMA. 333:1906–1917. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Malapelle U, Ricciuti B, Baglivo S, Pepe
F, Pisapia P, Anastasi P, Tazza M, Sidoni A, Liberati AM, Bellezza
G, et al: Osimertinib. Recent Results Cancer Res. 211:257–276.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang JC, Ahn MJ, Kim DW, Ramalingam SS,
Sequist LV, Su WC, Kim SW, Kim JH, Planchard D, Felip E, et al:
Osimertinib in pretreated T790M-positive advanced non-small-cell
lung cancer: AURA study phase II extension component. J Clin Oncol.
35:1288–1296. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Patil BR, Bhadane KV, Ahmad I, Agrawal YJ,
Shimpi AA, Dhangar MS and Patel HM: Exploring the structural
activity relationship of the Osimertinib: A covalent inhibitor of
double mutant EGFRL858R/T790M tyrosine kinase for the
treatment of non-small cell lung cancer (NSCLC). Bioorg Med Chem.
109:1177962024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhan Z, Chen B, Xu S, Lin R, Chen H, Ma X,
Lin X, Huang W, Zhuo C, Chen Y and Guo Z: Neoadjuvant chemotherapy
combined with antiangiogenic therapy and immune checkpoint
inhibitors for the treatment of locally advanced gastric cancer: A
real-world retrospective cohort study. Front Immunol.
16:15182172025. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Y, Chen Z, Chen R, Fang C, Zhang C,
Ji M and Yang X: Immunotherapy-based combination strategies for
treatment of EGFR-TKI-resistant non-small-cell lung cancer. Future
Oncol. 18:1757–1775. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li J, Yao X, Qiu L, Zhang R and Wang G:
Research progress in immune checkpoint inhibitors combination
therapy applied to non-small cell lung cancer after EGFR
mutation-targeted therapy resistance. Zhongguo Fei Ai Za Zhi.
26:392–399. 2023.(In Chinese). PubMed/NCBI
|
|
18
|
Gotwals P, Cameron S, Cipolletta D,
Cremasco V, Crystal A, Hewes B, Mueller B, Quaratino S,
Sabatos-Peyton C, Petruzzelli L, et al: Prospects for combining
targeted and conventional cancer therapy with immunotherapy. Nat
Rev Cancer. 17:286–301. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rizzo A, Ricci AD and Brandi G: PD-L1,
TMB, MSI, and other predictors of response to immune checkpoint
inhibitors in biliary tract cancer. Cancers (Basel). 13:5582021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hektoen HH, Tsuruda KM, Fjellbirkeland L,
Nilssen Y, Brustugun OT and Andreassen BK: Real-world evidence for
pembrolizumab in non-small cell lung cancer: A nationwide cohort
study. Br J Cancer. 132:93–102. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Griffiths AD, Young RO, Yuan Y, Chaudhary
MA, Lee A, Gordon J and McEwan P: Cost-effectiveness of nivolumab
plus ipilimumab versus chemotherapy for previously untreated
metastatic NSCLC using mixture-cure survival analysis based on
CheckMate 227 5-year data. Pharmacoecon Open. 9:247–257. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du Z, Ge X, Li Y, Qin Y, Fan H, Lv Y, Du X
and Liu Z: Clinical characteristics and survival outcomes of
long-term responders for advanced nonsmall cell lung cancer
patients with first-line PD-1/PD-L1 inhibitors: A multicenter
retrospective study. Postgrad Med J. 101:1072–1080. 2025.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ul Bassar W, Ogedegbe OJ, Qammar A, Sumia
F, Ul Islam M, Chaudhari SS, Ntukidem OL and Khan A: Efficacy of
tislelizumab in lung cancer treatment: A systematic review and
meta-analysis of randomized controlled trials. Cureus.
17:e806092025.PubMed/NCBI
|
|
24
|
Prasad CP, Tripathi SC, Kumar M and
Mohapatra P: Passage number of cancer cell lines: Importance,
intricacies, and way-forward. Biotechnol Bioeng. 120:2049–2055.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mestas J and Hughes CCW: Of mice and not
men: Differences between mouse and human immunology. J Immunol.
172:2731–2738. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guan X, Wu D, Zhu H, Zhu B, Wang Z, Xing
H, Zhang X, Yan J, Guo Y and Lu Y: 3D pancreatic ductal
adenocarcinoma desmoplastic model: Glycolysis facilitating stemness
via ITGAV-PI3K-AKT-YAP1. Biomater Adv. 170:2142152025. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Smabers LP, Wensink E, Verissimo CS,
Koedoot E, Pitsa KC, Huismans MA, Higuera Barón C, Doorn M,
Valkenburg-van Iersel LB, Cirkel GA, et al: Organoids as a
biomarker for personalized treatment in metastatic colorectal
cancer: Drug screen optimization and correlation with patient
response. J Exp Clin Cancer Res. 43:612024. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao KY, Du YX, Cao HM, Su LY, Su XL and
Li X: The biological macromolecules constructed Matrigel for
cultured organoids in biomedical and tissue engineering. Colloids
Surf B Biointerfaces. 247:1144352025. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fu S, Zhu X, Huang F and Chen X: Anti-PEG
antibodies and their biological impact on PEGylated drugs:
Challenges and strategies for optimization. Pharmaceutics.
17:10742025. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu L, Yuhan J, Yu H, Zhang B, Huang K and
Zhu L: Decellularized extracellular matrix for remodeling
bioengineering organoid's microenvironment. Small. 19:e22077522023.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He S, Nakada D and Morrison SJ: Mechanisms
of stem cell self-renewal. Annu Rev Cell Dev Biol. 25:377–406.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dalton S: Linking the cell cycle to cell
fate decisions. Trends Cell Biol. 25:592–600. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Corrò C, Novellasdemunt L and Li VSW: A
brief history of organoids. Am J Physiol Cell Physiol.
319:C151–C165. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sato T, Vries RG, Snippert HJ, van de
Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters
PJ and Clevers H: Single Lgr5 stem cells build crypt-villus
structures in vitro without a mesenchymal niche. Nature.
459:262–265. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou Y, Song H and Ming GL: Genetics of
human brain development. Nat Rev Genet. 25:26–45. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Zeng PM, Wu J and Luo ZG: Advances
and applications of brain organoids. Neurosci Bull. 39:1703–1716.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kanupriya K, Pal Verma S, Sharma V, Mishra
I and Mishra R: Advances in human brain organoids: Methodological
innovations and future directions for drug discovery. Curr Drug Res
Rev. 17:360–374. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu H, Jiao D, Liu A and Wu K: Tumor
organoids: Applications in cancer modeling and potentials in
precision medicine. J Hematol Oncol. 15:582022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li Z, Yu L, Chen D, Meng Z, Chen W and
Huang W: Protocol for generation of lung adenocarcinoma organoids
from clinical samples. STAR Protoc. 2:1002392020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Phifer CJ, Bergdorf KN, Bechard ME,
Vilgelm A, Baregamian N, McDonald OG, Lee E and Weiss VL: Obtaining
patient-derived cancer organoid cultures via fine-needle
aspiration. STAR Protoc. 2:1002202020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim SY, Kim SM, Lim S, Lee JY, Choi SJ,
Yang SD, Yun MR, Kim CG, Gu SR, Park C, et al: Modeling clinical
responses to targeted therapies by patient-derived organoids of
advanced lung adenocarcinoma. Clin Cancer Res. 27:4397–4409. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mazzocchi A, Devarasetty M, Herberg S,
Petty WJ, Marini F, Miller L, Kucera G, Dukes DK, Ruiz J, Skardal A
and Soker S: Pleural effusion aspirate for use in 3D lung cancer
modeling and chemotherapy screening. ACS Biomater Sci Eng.
5:1937–1943. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mazzocchi A, Dominijanni A and Soker S:
Pleural effusion aspirate for use in 3D lung cancer modeling and
chemotherapy screening. Methods Mol Biol. 2394:471–483. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang L, Yu Y, Fang Y, Li Y, Yu W, Wang Z,
Lv J, Wang R and Liang S: Malignant pleural effusion facilitates
the establishment and maintenance of tumor organoid biobank with
multiple patient-derived lung tumor cell sources. Exp Hematol
Oncol. 13:1152024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Driehuis E, Kretzschmar K and Clevers H:
Establishment of patient-derived cancer organoids for
drug-screening applications. Nat Protoc. 15:3380–3409. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Taverna JA, Hung CN, Williams M, Williams
R, Chen M, Kamali S, Sambandam V, Hsiang-Ling Chiu C, Osmulski PA,
Gaczynska ME, et al: Ex vivo drug testing of patient-derived lung
organoids to predict treatment responses for personalized medicine.
Lung Cancer. 190:1075332024. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xie C, Gu A, Khan M, Yao X, Chen L, He J,
Yuan F, Wang P, Yang Y, Wei Y, et al: Opportunities and challenges
of hepatocellular carcinoma organoids for targeted drugs
sensitivity screening. Front Oncol. 12:11054542023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Demyan L, Habowski AN, Plenker D, King DA,
Standring OJ, Tsang C, St Surin L, Rishi A, Crawford JM, Boyd J, et
al: Pancreatic cancer patient-derived organoids can predict
response to neoadjuvant chemotherapy. Ann Surg. 276:450–462. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Merrill NM, Kaffenberger SD, Bao L,
Vandecan N, Goo L, Apfel A, Cheng X, Qin Z, Liu CJ, Bankhead A, et
al: Integrative drug screening and multiomic characterization of
patient-derived bladder cancer organoids reveal novel molecular
correlates of gemcitabine response. Eur Urol. 86:434–444. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hughes CS, Postovit LM and Lajoie GA:
Matrigel: A complex protein mixture required for optimal growth of
cell culture. Proteomics. 10:1886–1890. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Seiji Y, Ito T, Nakamura Y,
Nakaishi-Fukuchi Y, Matsuo A, Sato N and Nogawa H: Alveolus-like
organoid from isolated tip epithelium of embryonic mouse lung. Hum
Cell. 32:103–113. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bhattacharya S, Calar K and de la Puente
P: Mimicking tumor hypoxia and tumor-immune interactions employing
three-dimensional in vitro models. J Exp Clin Cancer Res.
39:752020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Poggio F, Brofiga M, Callegari F, Tedesco
M and Massobrio P: Developmental conditions and culture medium
influence the neuromodulated response of in vitro cortical
networks. Annu Int Conf IEEE Eng Med Biol Soc. 2023:1–4.
2023.PubMed/NCBI
|
|
54
|
Valian N, Heravi M, Ahmadiani A and
Dargahi L: Comparison of rat primary midbrain neurons cultured in
DMEM/F12 and neurobasal mediums. Basic Clin Neurosci. 12:205–212.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ziółkowska-Suchanek I: Mimicking tumor
hypoxia in non-small cell lung cancer employing three-dimensional
in vitro models. Cells. 10:1412021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li Y, Zou J, Fang Y, Zuo J, Wang R and
Liang S: Lung tumor organoids migrate as cell clusters containing
cancer stem cells under hypoxic condition. Biol Cell.
117:e24000812025. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liu C, Pei M, Li Q and Zhang Y:
Decellularized extracellular matrix mediates tissue construction
and regeneration. Front Med. 16:56–82. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pasupuleti V, Vora L, Prasad R, Nandakumar
DN and Khatri DK: Glioblastoma preclinical models: Strengths and
weaknesses. Biochim Biophys Acta Rev Cancer. 1879:1890592024.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cui X, Liu H, Liu Y, Yu Z, Wang D, Wei W
and Wang S: Tissue-specific decellularized extracellular matrix
rich in collagen, glycoproteins, and proteoglycans and its
applications in advanced organoid engineering: A review. Int J Biol
Macromol. 315:1444692025. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Low RRJ, Fung KY, Gao H, Preaudet A,
Dagley LF, Yousef J, Lee B, Emery-Corbin SJ, Nguyen PM, Larsen RH,
et al: S100 family proteins are linked to organoid morphology and
EMT in pancreatic cancer. Cell Death Differ. 30:1155–1165. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Jeong SR and Kang M: Exploring
tumor-immune interactions in co-culture models of T cells and tumor
organoids derived from patients. Int J Mol Sci. 24:146092023.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yuan J, Li X and Yu S: Cancer organoid
co-culture model system: Novel approach to guide precision
medicine. Front Immunol. 13:10613882023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu Y, Chudgar N, Mastrogiacomo B, He D,
Lankadasari MB, Bapat S, Jones GD, Sanchez-Vega F, Tan KS, Schultz
N, et al: A germline SNP in BRMS1 predisposes patients with lung
adenocarcinoma to metastasis and can be ameliorated by targeting
c-fos. Sci Transl Med. 14:eabo10502022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Naranjo S, Cabana CM, LaFave LM, Romero R,
Shanahan SL, Bhutkar A, Westcott PMK, Schenkel JM, Ghosh A, Liao
LZ, et al: Modeling diverse genetic subtypes of lung adenocarcinoma
with a next-generation alveolar type 2 organoid platform. Genes
Dev. 36:936–949. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kawai S, Nakano K, Tamai K, Fujii E,
Yamada M, Komoda H, Sakumoto H, Natori O and Suzuki M: Generation
of a lung squamous cell carcinoma three-dimensional culture model
with keratinizing structures. Sci Rep. 11:243052021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Qu F, Brough SC, Michno W, Madubata CJ,
Hartmann GG, Puno A, Drainas AP, Bhattacharya D, Tomasich E, Lee
MC, et al: Crosstalk between small-cell lung cancer cells and
astrocytes mimics brain development to promote brain metastasis.
Nat Cell Biol. 25:1506–1519. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Quaranta V and Linkous A: Organoids as a
systems platform for SCLC brain metastasis. Front Oncol.
12:8819892022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Meng Y, Shu X, Yang J, Liang Y, Zhu M,
Wang X, Li Y and Kong F: Lung cancer organoids: A new strategy for
precision medicine research. Transl Lung Cancer Res. 14:575–590.
2025. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Butterfield GL, Reisman SJ, Iglesias N and
Gersbach CA: Gene regulation technologies for gene and cell
therapy. Mol Ther. 33:2104–2122. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Li L, Feng J, Zhao S, Rong Z and Lin Y:
SOX9 inactivation affects the proliferation and differentiation of
human lung organoids. Stem Cell Res Ther. 12:3432021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Gmeiner WH, Miller LD, Chou JW,
Dominijanni A, Mutkus L, Marini F, Ruiz J, Dotson T, Thomas KW,
Parks G and Bellinger CR: Dysregulated pyrimidine biosynthesis
contributes to 5-FU resistance in SCLC patient-derived organoids
but response to a novel polymeric fluoropyrimidine, CF10. Cancers
(Basel). 12:7882020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang Q and Zhang M: Recent advances in
lung cancer organoid (tumoroid) research (review). Exp Ther Med.
28:3832024. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Curvello R and Garnier G: Cationic
cross-linked nanocellulose-based matrices for the growth and
recovery of intestinal organoids. Biomacromolecules. 22:701–709.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Feng Y, He D and An X: Hydrogel
innovations for 3D organoid culture. Biomed Mater. 20:2025.
View Article : Google Scholar
|
|
75
|
Luo L, Liu L, Ding Y, Dong Y and Ma M:
Advances in biomimetic hydrogels for organoid culture. Chem Commun
(Camb). 59:9675–9686. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Li C, An N, Song Q, Hu Y, Yin W, Wang Q,
Le Y, Pan W, Yan X, Wang Y and Liu J: Enhancing organoid culture:
Harnessing the potential of decellularized extracellular matrix
hydrogels for mimicking microenvironments. J Biomed Sci. 31:962024.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Ishigamori R, Naruse M, Hirata A, Maru Y,
Hippo Y and Imai T: The potential of organoids in toxicologic
pathology: Histopathological and immunohistochemical evaluation of
a mouse normal tissue-derived organoid-based carcinogenesis model.
J Toxicol Pathol. 35:211–223. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yoshida GJ: Applications of
patient-derived tumor xenograft models and tumor organoids. J
Hematol Oncol. 13:42020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kabraji S, Ni J, Sammons S, Li T, Van
Swearingen AED, Wang Y, Pereslete A, Hsu L, DiPiro PJ, Lascola C,
et al: Preclinical and clinical efficacy of trastuzumab deruxtecan
in breast cancer brain metastases. Clin Cancer Res. 29:174–182.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Sereti E, Karagianellou T, Kotsoni I,
Magouliotis D, Kamposioras K, Ulukaya E, Sakellaridis N,
Zacharoulis D and Dimas K: Patient Derived Xenografts (PDX) for
personalized treatment of pancreatic cancer: Emerging allies in the
war on a devastating cancer? J Proteomics. 188:107–118. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Jeong H, Moon HE, Yun S, Cho SW, Park HR,
Park SH, Myung K, Kwon T and Paek SH: Enrichment of deleterious
mutated genes involved in ciliary function and histone modification
in brain cancer patient-derived xenograft models. Biomedicines.
11:29342023. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Liu W, Cui Y, Zheng X, Yu K and Sun G:
Application status and future prospects of the PDX model in lung
cancer. Front Oncol. 13:10985812023. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wang E, Xiang K, Zhang Y and Wang XF:
Patient-derived organoids (PDOs) and PDO-derived xenografts
(PDOXs): New opportunities in establishing faithful pre-clinical
cancer models. J Natl Cancer Cent. 2:263–276. 2022.PubMed/NCBI
|
|
84
|
Gao J, Lan J, Liao H, Yang F, Qiu P, Jin
F, Wang S, Shen L, Chao T, Zhang C and Zhu Y: Promising preclinical
patient-derived organoid (PDO) and xenograft (PDX) models in upper
gastrointestinal cancers: Progress and challenges. BMC Cancer.
23:12052023. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhuang P, Chiang YH, Fernanda MS and He M:
Using spheroids as building blocks towards 3D bioprinting of tumor
microenvironment. Int J Bioprint. 7:4442021. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Senrung A, Lalwani S, Janjua D, Tripathi
T, Kaur J, Ghuratia N, Aggarwal N, Chhokar A, Yadav J, Chaudhary A,
et al: 3D tumor spheroids: Morphological alterations a yardstick to
anti-cancer drug response. In Vitro Model. 2:219–248. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Mandrycky CJ, Howard CC, Rayner SG, Shin
YJ and Zheng Y: Organ-on-a-chip systems for vascular biology. J Mol
Cell Cardiol. 159:1–13. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Politi K and Pao W: How genetically
engineered mouse tumor models provide insights into human cancers.
J Clin Oncol. 29:2273–2281. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Fan TWM, Higashi RM and Lane AN: Metabolic
reprogramming in human cancer patients and patient-derived models.
Cold Spring Harb Perspect Med. 15:a0415522025. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Huang K, Han Y, Chen Y, Shen H, Zeng S and
Cai C: Tumor metabolic regulators: Key drivers of metabolic
reprogramming and the promising targets in cancer therapy. Mol
Cancer. 24:72025. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhu Z, Hu E, Shen H, Tan J and Zeng S: The
functional and clinical roles of liquid biopsy in patient-derived
models. J Hematol Oncol. 16:362023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Lawrence R, Watters M, Davies CR, Pantel K
and Lu YJ: Circulating tumour cells for early detection of
clinically relevant cancer. Nat Rev Clin Oncol. 20:487–500. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Gao D, Vela I, Sboner A, Iaquinta PJ,
Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora
VK, et al: Organoid cultures derived from patients with advanced
prostate cancer. Cell. 159:176–187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Pan C, Wang X, Yang C, Fu K, Wang F and Fu
L: The culture and application of circulating tumor cell-derived
organoids. Trends Cell Biol. 35:364–380. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Huang L, Xu Y, Wang N, Yi K, Xi X, Si H,
Zhang Q, Xiang M, Rong Y, Yuan Y and Wang F: Next-generation
preclinical functional testing models in cancer precision medicine:
CTC-derived organoids. Small Methods. 8:e23010092024. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Pérez-Cabello JA, Artero-Castro A and
Molina-Pinelo S: Small cell lung cancer unveiled: Exploring the
untapped resource of circulating tumor cells-derived organoids.
Crit Rev Oncol Hematol. 207:1046222025. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Na F, Pan X, Chen J, Chen X, Wang M, Chi
P, You L, Zhang L, Zhong A, Zhao L, et al: KMT2C deficiency
promotes small cell lung cancer metastasis through DNMT3A-mediated
epigenetic reprogramming. Nat Cancer. 3:753–767. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Balážová K, Clevers H and Dost AFM: The
role of macrophages in non-small cell lung cancer and advancements
in 3D co-cultures. Elife. 12:e829982023. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lee D, Kim Y and Chung C: Scientific
validation and clinical application of lung cancer organoids.
Cells. 10:30122021. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Kim M, Mun H, Sung CO, Cho EJ, Jeon HJ,
Chun SM, Jung DJ, Shin TH, Jeong GS, Kim DK, et al: Patient-derived
lung cancer organoids as in vitro cancer models for therapeutic
screening. Nat Commun. 10:39912019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zheng M, Qu J, Xiang D and Xing L:
Organoids in lung cancer brain metastasis: Foundational research,
clinical translation, and prospective outlooks. Biochim Biophys
Acta Rev Cancer. 1880:1892352025. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Wu F, Fan J, He Y, Xiong A, Yu J, Li Y,
Zhang Y, Zhao W, Zhou F, Li W, et al: Single-cell profiling of
tumor heterogeneity and the microenvironment in advanced non-small
cell lung cancer. Nat Commun. 12:25402021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
de Sousa VML and Carvalho L: Heterogeneity
in lung cancer. Pathobiology. 85:96–107. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Goveia J, Rohlenova K, Taverna F, Treps L,
Conradi LC, Pircher A, Geldhof V, de Rooij LPMH, Kalucka J, Sokol
L, et al: An integrated gene expression landscape profiling
approach to identify lung tumor endothelial cell hetero-geneity and
angiogenic candidates. Cancer Cell. 37:21–36.e13. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Dijkstra KK, Cattaneo CM, Weeber F,
Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL,
Kaing S, Kelderman S, et al: Generation of tumor-reactive T cells
by co-culture of peripheral blood lymphocytes and tumor organoids.
Cell. 174:1586–1598.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Ren YF, Ma Q, Zeng X, Huang CX, Ren JL, Li
F, Tong JJ, He JW, Zhong Y, Tan SY, et al: Single-cell RNA
sequencing reveals immune microenvironment niche transitions during
the invasive and metastatic processes of ground-glass nodules and
part-solid nodules in lung adenocarcinoma. Mol Cancer. 23:2632024.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Raghavan S, Winter PS, Navia AW, Williams
HL, DenAdel A, Lowder KE, Galvez-Reyes J, Kalekar RL, Mulugeta N,
Kapner KS, et al: Microenvironment drives cell state, plasticity,
and drug response in pancreatic cancer. Cell. 184:6119–6137.e26.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Liu C, Li K, Sui X, Zhao T, Zhang T, Chen
Z, Wu H, Li C, Li H, Yang F, et al: Patient-derived tumor organoids
combined with function-associated ScRNA-Seq for dissecting the
local immune response of lung cancer. Adv Sci (Weinh).
11:e24001852024. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wu X, Li B, Wang Y, Xue J, Zhao H, Huang
Z, Zheng Z, Liang N and Wei Z: Microfluidic chip-based automatic
system for sequencing patient-derived organoids at the single-cell
level. Anal Chem. 96:17027–17036. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Landon-Brace N, Li NT and McGuigan AP:
Exploring new dimensions of tumor heterogeneity: The application of
single cell analysis to organoid-based 3D in vitro models. Adv
Healthc Mater. 12:e23009032023. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
R N, Aggarwal A, Sravani AB, Mallya P and
Lewis S: Organ-on-a-chip: An emerging research platform.
Organogenesis. 19:22782362023. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Shakeri A, Wang Y, Zhao Y, Landau S,
Perera K, Lee J and Radisic M: Engineering Organ-on-a-Chip systems
for vascular diseases. Arterioscler Thromb Vasc Biol. 43:2241–2255.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Nolan J, Pearce OMT, Screen HRC, Knight MM
and Verbruggen SW: Organ-on-a-chip and microfluidic platforms for
oncology in the UK. Cancers (Basel). 15:6352023. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Peng T, Ma X, Hua W, Wang C, Chu Y, Sun M,
Fermi V, Hamelmann S, Lindner K, Shao C, et al: Individualized
patient tumor organoids faithfully preserve human brain tumor
ecosystems and predict patient response to therapy. Cell Stem Cell.
32:652–669.e11. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Duan H, Ren J, Wei S, Yang Z, Li C, Wang
Z, Li M, Wei Z, Liu Y, Wang X, et al: Integrated analyses of
multi-omic data derived from paired primary lung cancer and brain
metastasis reveal the metabolic vulnerability as a novel
therapeutic target. Genome Med. 16:1382024. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Kozlowski MT, Crook CJ and Ku HT: Towards
organoid culture without Matrigel. Commun Biol. 4:13872021.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Kim S, Min S, Choi YS, Jo SH, Jung JH, Han
K, Kim J, An S, Ji YW, Kim YG and Cho SW: Tissue extracellular
matrix hydrogels as alternatives to Matrigel for culturing
gastrointestinal organoids. Nat Commun. 13:16922022. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Jamaluddin MFB, Ghosh A, Ingle A, Mohammed
R, Ali A, Bahrami M, Kaiko G, Gibb Z, Filipe EC, Cox TR, et al:
Bovine and human endometrium-derived hydrogels support organoid
culture from healthy and cancerous tissues. Proc Natl Acad Sci USA.
119:e22080401192022. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Isik M, Okesola BO, Eylem CC, Kocak E,
Nemutlu E, D'Este M, Mata A and Derkus B: Bioactive and chemically
defined hydrogels with tunable stiffness guide cerebral organoid
formation and modulate multi-omics plasticity in cerebral
organoids. Acta Biomater. 171:223–238. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Magré L, Verstegen MMA, Buschow S, van der
Laan LJW, Peppelenbosch M and Desai J: Emerging organoid-immune
co-culture models for cancer research: From oncoimmunology to
personalized immunotherapies. J Immunother Cancer. 11:e0062902023.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Thakar RG and Fenton KN: Bioethical
implications of organ-on-a-chip on modernizing drug development.
Artif Organs. 47:1553–1558. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
P T, S AK, U AN, S R, P S, M S and K S:
Artificial intelligence (AI) and liquid biopsy transforming early
detection of liver metastases in gastrointestinal cancers. Curr
Cancer Drug Targets. Jan 21–2025.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Polak R, Zhang ET and Kuo CJ: Cancer
organoids 2.0: Modelling the complexity of the tumour immune
microenvironment. Nat Rev Cancer. 24:523–539. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Oh B, Kim J, Kim N and Jeong Y: Lung
cancer organoid system to evaluate the cytotoxicity of natural
killer cells. Int J Stem Cells. 18:99–106. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Vella N, Fenech AG and Petroni Magri V: 3D
cell culture models in research: Applications to lung cancer
pharmacology. Front Pharmacol. 15:14380672024. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Gao M, Ding W, Wang Y, Li B, Huang Z,
Liang N and Wei Z: Quantitatively evaluating interactions between
patient-derived organoids and autologous immune cells by
microfluidic chip. Anal Chem. 96:13061–13069. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Evans K, Dabrowska C, Ng ME, Brainson CF
and Lee JH: Isolation, culture, and phenotypic analysis of murine
lung organoids. Methods Mol Biol. 2805:3–18. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Huang Z, Li B, Wang Y, Xue J, Wei Z, Liang
N and Li S: Application and research progress of lung cancer
organoid in precision medicine for lung cancer. Zhongguo Fei Ai Za
Zhi. 27:276–282. 2024.(In Chinese). PubMed/NCBI
|
|
129
|
Broutier L, Mastrogiovanni G, Verstegen
MM, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R,
Sidorova O, Gaspersz MP, et al: Human primary liver cancer-derived
organoid cultures for disease modeling and drug screening. Nat Med.
23:1424–1435. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Kato Y, Seishima R, Hattori K, Kato H,
Ishida H, Shigeta K, Okabayashi K, Sugihara E, Takimoto T, Nakamura
K, et al: Significance of homologous recombinant deficiency as a
biomarker for drug sensitivity in colorectal cancer. Br J Cancer.
132:533–542. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Lee MR, Woo SM, Kim MK, Han SS, Park SJ,
Lee WJ, Lee DE, Choi SI, Choi W, Yoon KA, et al: Application of
plasma circulating KRAS mutations as a predictive biomarker for
targeted treatment of pancreatic cancer. Cancer Sci. 115:1283–1295.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Yang J, Fischer NG and Ye Z:
Revolutionising oral organoids with artificial intelligence.
Biomater Transl. 5:372–389. 2024.PubMed/NCBI
|
|
133
|
Kerkar N and Hartjes K: Hepatitis C
virus-pediatric and adult perspectives in the current decade.
Pathogens. 14:112024. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Li Y, Du J, Deng S, Liu B, Jing X, Yan Y,
Liu Y, Wang J, Zhou X and She Q: The molecular mechanisms of
cardiac development and related diseases. Signal Transduct Target
Ther. 9:3682024. View Article : Google Scholar : PubMed/NCBI
|