Cancer remains one of the most serious diseases that
threaten human health, with metastasis being the leading cause of
cancer-related deaths worldwide and posing a major challenge in
cancer treatment (1,2). Metastasis is a complex process
involving multiple steps (3),
including intravasation, extravasation, migration and regeneration.
During metastasis, tumor cells from the primary site invade distant
tissues through the bloodstream, where they form metastatic foci
(4). In recent years, the
prevailing view has been that metastatic disease is usually
extensive and incurable. However, the advent of immunotherapy has
led to significant breakthroughs in cancer treatment. When combined
with surgery, radiotherapy and chemotherapy, immunotherapy can
improve patient survival (5).
Tissue biopsy is among the clinical methods used to guide
immunotherapy (6). However,
traditional tissue biopsies are not 100% accurate and have several
limitations: They are invasive (7),
have limited sensitivity and specificity, provide only local tissue
samples, and fail to capture tumor heterogeneity (8). Circulating tumor cells (CTCs) are a
cornerstone of liquid biopsy and offer undeniable advantages that
can compensate for some of the limitations of traditional tissue
biopsy. They are non-invasive (9),
easy to administer, and more patient friendly. Additionally, they
address the issue of tumor heterogeneity, which enables more
effective monitoring of tumor progression through serial testing
and provides valuable insights to guide treatment decisions
(10–12).
CTCs are tumor cells that are shed from primary
tumors or metastatic sites and enter the peripheral bloodstream
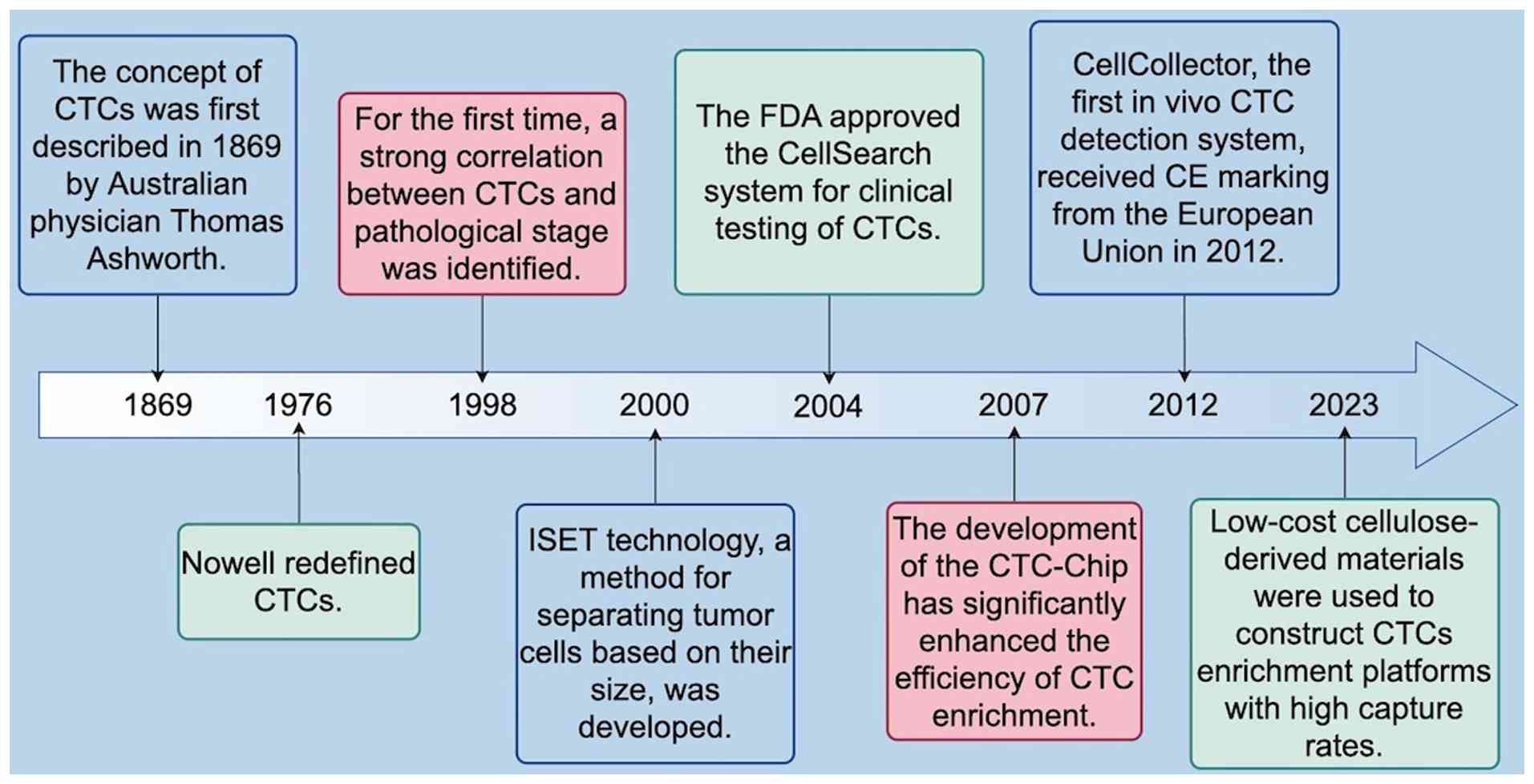
(13,14). The concept of CTCs was first
introduced in 1869 by the Australian physician Thomas Ashworth, who
observed cells in the blood of a patient with metastatic cancer
that were similar to the primary tumor cells (15,16).
However, given the technical limitations of that time period, CTCs
were not further examined (17). It
was not until 1976 that Nowell redefined CTCs as tumor cells
originating from primary or metastatic tumors that have acquired
the ability to detach from the basement membrane and invade the
vasculature through tissue stroma. In 1998, Peck et al
(18) explored the feasibility of
using CTC assays for the rapid assessment of chemotherapeutic
responsiveness. Their study revealed that serial assays of changes
in CTC counts correlated with tumor burden and therapeutic response
in patients. Furthermore, in 2004, a prospective multicenter study
demonstrated that the number of CTCs in peripheral blood prior to
treatment was an independent predictor of progression-free survival
(PFS) and overall survival (OS) in patients with metastatic breast
cancer (19,20). That same year, the U.S. Food and
Drug Administration (FDA) approved the CellSearch System for the
treatment of metastatic colorectal, breast and prostate cancers
(21). With continued technological
advancements, the CTC chip developed by Nagrath et al
(22) in 2007 markedly improved the
efficiency of CTC enrichment. This microfluidic platform enables
the efficient and selective isolation of viable CTCs from
peripheral whole blood under precisely controlled laminar-flow
conditions. Using this system, CTCs were successfully detected in
115 of 116 peripheral blood samples (99%) obtained from patients
with metastatic lung, prostate, pancreatic, breast, and colon
cancers. The detected CTC counts ranged from 5–1,281 cells/ml of
blood, with an approximate purity of 50% (22). However, most CTC enrichment
techniques during that period were performed in vitro.
Hartmann et al (23)
reported that this approach is inherently limited by the minimal
number of CTCs that can be captured from small-volume blood
samples. Moreover, preanalytical handling procedures, including
sample transport, preservation, centrifugation, and subsequent
in vitro enrichment and isolation, frequently introduce
artifactual alterations, such as cellular contamination, loss,
inactivation and morphological distortion (23). To overcome these challenges, the
first in vivo CTC detection system, known as Cell Collector,
received CE certification from the European Union in 2012. The
cells captured by this system require no pretreatment, such as
transfection or fluorescent labeling, and can be directly
identified within the immunomagnetic separation device. This
feature effectively minimizes the detrimental effects associated
with clinical sample collection, preprocessing, and CTC isolation
on cellular integrity and overall quality (24). Compared with the Cell Collector
system, the CTC enrichment platform developed in recent years by
Hazra et al (25), which
employs cellulose derivatives, may reduce costs and thus improve
affordability for a larger patient population; moreover, this
method achieves an overall capture efficiency of ~40%. Despite
these advancements, isolating CTCs from blood with high sensitivity
and specificity remains a significant challenge (26). Most patients with cancer have
extremely low concentrations of CTCs in their peripheral
circulating blood (from 1–100 cells/ml) (27), and factors such as blood shear
stress and immune system activity result in a short half-life for
CTCs, usually from 1–2.5 h (28,29).
Additionally, CTCs exhibit significant heterogeneity across
individuals and tumor types, even within the same patient. With the
rapid advancement of new digital technology, researchers have
established a new interdisciplinary field of medical, industrial
integration by combining biology, chemistry, physics, computer
science and medicine. This integration has led to the development
of innovative technologies for the capture, detection, and analysis
of CTCs. These advances have significantly facilitated the clinical
application of CTCs in cancer screening, treatment response
monitoring and prognosis (28).
A search of the PubMed database through December
2023 using ‘CTCs’ as a keyword retrieved more than 33,000 studies.
The number of publications per year has shown an upward trend,
reaching more than 2,000 by 2020. Although there has been a slight
decline since then, the annual number of relevant publications
still exceeds 1,500, confirming continued interest in advancing the
technology and developing new applications. These findings indicate
that research on CTCs is advancing rapidly, yielding significant
findings (Fig. 1). The present
study provides a systematic comparison of the advantages and
disadvantages of the current methods for collecting and identifying
CTCs and assessing their clinical applicability in
immunotherapy.
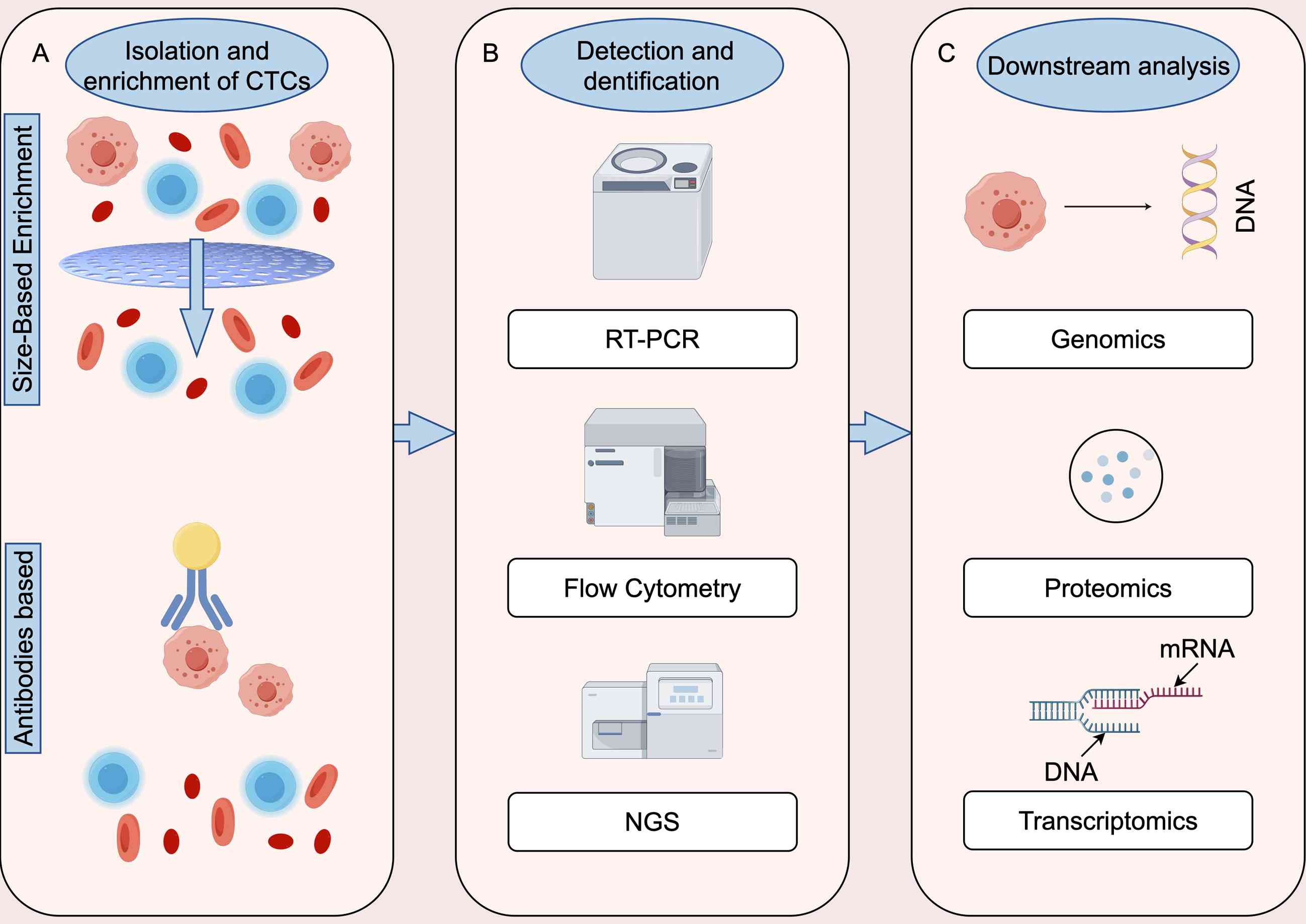
In general, technologies for identifying CTCs
involve three critical steps: Capture and enrichment, detection and
identification, and release for further analysis (30). As noted by Wang et al
(31), capture and enrichment
methods rely primarily on the interaction between specific
materials and certain physical, chemical or biological properties
of CTCs. After collection, various identification methods can be
used, such as flow cytometry, fluorescence microscopy and DNA
sequencing. Released CTCs are typically utilized for downstream
analyses, such as genomics, transcriptomics, proteomics, and
immunofluorescence assays of cultured cells (31). Enrichment methods for CTCs are based
on certain biological and physical properties (32,33).
Since most tumors originate from epithelial tissues, epithelial
cell adhesion molecules (EpCAMs) are the most commonly used
biomarkers for CTCs. Physically, CTCs can be differentiated by
properties such as size, density, charge, and deformability
(34–37). In recent years, detection techniques
that integrate both the physical and biological properties of CTCs
have emerged, offering enhanced efficiency and sensitivity
(38,39) (Fig.
2).
Studies have shown that CTCs are typically larger
and firmer than hematopoietic cells are, and numerous CTC
enrichment techniques have been developed on the basis of these
physical properties (40). For
example, the epithelial tumor cell size separation (ISET) method
primarily uses a two-dimensional membrane microfilter and is the
first assay to allow the direct filtration of peripheral blood. In
this system, blood samples are diluted 1:10 and passed through a
polycarbonate track-etch-type membrane with 8-µm pores (41,42).
CTCs, which are generally larger than peripheral blood leukocytes,
are retained, while the other cells pass through. However, this
method is not very efficient. A similar system, the ScreenCell,
enriches CTCs by processing blood samples through a microporous
membrane filter with 8 µm pore size. Unlike ISET, ScreenCell system
can consistently process large numbers of blood samples with high
throughput (43). In addition, the
ScreenCell system offers other advantages, including the use of
cellular and molecular technologies for identifying CTCs and
analyzing their underlying genetic abnormalities, which facilitate
the easy analysis of tumor cells extracted onto the filter
(44). Furthermore, the cells
isolated from blood maintain a high level of activity and
integrity, allowing for in vitro tissue culture. To minimize
cell damage and preserve cell viability, a flexible miniature
spring array device ensures the harvesting of CTCs with greater
viability (45). This device uses a
pressure-regulated system to capture CTCs with an efficiency of up
to 90%. However, experiments have shown that the device tends to
yield variable CTC counts across different samples, making it less
reliable (46). There are also 3D
membrane microfilters, such as the Parsortix system (47), Resettable Cell Trap (RCT) (48) and FaCTChecker and Cluster-Chip,
which allow for the separation of individual CTCs and/or CTC
clusters, depending on the size and/or variability of the CTCs
(38). Unlike the FaCTChecker
microfilter, the Parsortix system employs a horizontal
configuration rather than a vertical one. This microdevice features
a stepped architecture with channel widths progressively narrowing
to ≤10 µm. CTCs larger than the channel width become trapped in the
gaps, while smaller cells pass through. After CTC capture, reverse
flow releases the captured CTCs for collection and subsequent
molecular characterization. The RCT employs a strategy similar to
that of Parsortix; however, rather than reducing the channel
height, the device uses pneumatically controlled microvalves to
modulate the channel aperture and capture CTCs within pockets that
are taller than the main channel. When the microvalves are relaxed,
the aperture widens, enabling the release of the captured CTCs
(44). Both 2D and 3D membrane
microfilters have distinct characteristics in the separation and
enrichment of CTCs, with the choice depending primarily on the
enrichment principle of the specific method. With respect to
microfiltration techniques, the cross-sectional area of 2D surfaces
is more critical, whereas volumetric measurements are more relevant
for 3D shapes in flow separation. However, since the size of CTCs
may vary in size during aggregation, apoptosis, and different
stages of the cell cycle and can partially overlap with the size of
leukocytes, false-positive results may occur during the enrichment
process (38). Although size-based
enrichment of CTCs is a simple and label-free technique for
high-throughput assays, the capture rates and specificity are
influenced by the size heterogeneity of CTCs, necessitating
combination with other assays to improve accuracy.
In addition to size-based enrichment methods,
density-based enrichment is a workable strategy with distinct
advantages. One commonly used density gradient centrifugation
technique involves the use of a sucrose polymer with a high
synthetic molecular weight (Ficoll-Paque; GE Healthcare) as the
primary density gradient medium (49). The high centrifugal force applied in
this method creates a density gradient and the size and density of
the cells determines their sedimentation rate. The cell loading and
harvesting procedures must be determined empirically as they are
significantly influenced by the Ficoll-Paque, the rotor, and the
type of centrifuge tube used. Excessive cell loss and low
separation efficiency for CTCs isolated by Ficoll-based density
gradient centrifugation have been attributed to density-related
toxicity (38,50). The OncoQuick gradient system
addresses these problems by minimizing cell loss and contamination,
thereby achieving higher CTC enrichment rates than the traditional
Ficoll-Paque technology does (38,51).
The dielectrophoretic array (DEPArray) system,
developed in 2013, is a semiautomated platform capable of selecting
and isolating individual CTCs and small populations of cells
(52). The primary advantage of the
DEPArray system lies in its ability to identify multiparameter
immunofluorescence staining characteristics of individual cells
before isolation. Additionally, it can be integrated into a
continuous, semiautomated workflow platform, such as with the
CellSearch System. However, the system's advanced capabilities for
imaging and isolating individual CTCs have significant drawbacks,
including high costs and an average cell loss of ~40% during sample
preparation and separation (52).
To address these challenges and achieve higher separation
resolution, researchers developed an integrated dielectrophoretic
(DEP) and magnetophoretic (MAP) microfluidic chip in 2022. This
integrated MAP-DEP approach enables the separation of various types
of CTCs on the basis of their size and dielectric properties
(53).
In 2019, an investigator introduced a novel
multiflow microfluidic (MFM) system designed for the high-purity
separation of CTCs. By utilizing the inherent differences in the
inertial migration of cells flowing within microchannels, the MFM
system demonstrated a sensitivity that achieved over 87% purity in
label-free separations. However, the system has serious
limitations. Its sensitivity and recovery rates are compromised
when CTCs exhibit significant size overlap with other cell types.
Additionally, compared with other label-free separation systems,
the device operates at a relatively low flow rate. To address these
limitations, a new class of collectors based on similar principles,
known as inertial microfluidic labyrinth devices, was developed
(54). These devices incorporate 12
distinct arrays of microfilters and combine inertial focusing with
Dean flow dynamics. This approach achieves size-based cell
separation by continuously focusing all cells while selectively
isolating CTCs from smaller blood cells at the device's outlet.
Current CTC and CTC cluster separation techniques often rely on
biomarker-dependent antibody-based capture. However, compared with
individual CTCs, CTC clusters pose unique challenges: They have a
lower surface-area-to-volume ratio (55) and often exhibit mixed
epithelial-mesenchymal phenotypes, resulting in reduced antibody
binding regions (56).
Consequently, these methods may result in lower enrichment rates
for CTCs and CTC clusters with stem-like or mesenchymal
characteristics. By contrast, physical property-based separation
methods, such as inertial microfluidic labyrinth devices, avoid the
biases inherent in molecular marker-dependent systems. These
advanced devices offer a more effective solution for separating
CTCs and CTC clusters, including those with diverse phenotypes,
because they rely on size-based rather than molecular-based
characteristics (54).
In summary, CTC enrichment methods based on physical
properties are label-free and relatively simple to perform.
However, they often suffer from limited enrichment purity and a
risk of cell loss. As a result, they are best suited for
preliminary enrichment, and their combination with immunoaffinity
methods may offer a more effective strategy to increase detection
sensitivity and specificity.
CTCs are typically enriched through positive
selection using antibodies targeting tumor-associated antigens.
These antigens include epithelial cell adhesion molecule (EpCAM),
mesenchymal markers, cytokeratins (CKs), mucin-1, human epidermal
growth factor receptor 2 (HER2) and epithelial growth factor
receptor (EGFR), among others (57,58).
Alternatively, CTCs can be enriched through negative selection by
removing unwanted leukocytes using antibodies against markers such
as CD45; however, the EpCAM-dependent positive selection technique
is the most widely used (59,60).
Currently, several methods are popular for the
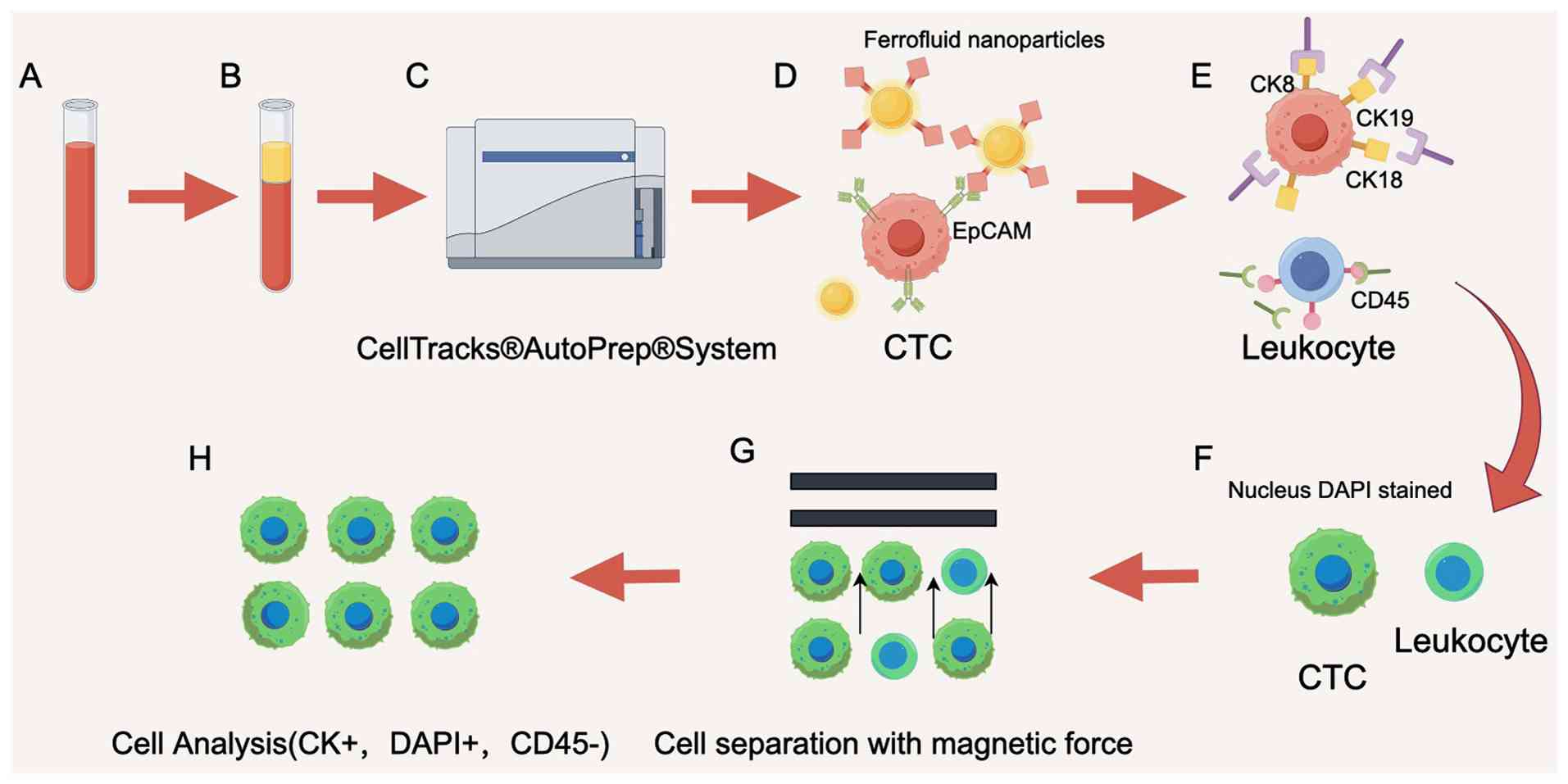
positive enrichment of CTCs. The CellSearch system, which uses an
EpCAM-based immunoassay, is the first and only FDA-approved CTC
assay (61) for colorectal, breast,
prostate and other tumors (62).
The CellSearch system's principle involves the use of ferromagnetic
fluid nanoparticle-labeled antibodies that specifically bind to
EpCAM, which is highly expressed on the surface of a wide range of
tumor cells. CTCs are then separated from other blood cells by
means of a magnet. The detailed collection process is illustrated
in Fig. 3. However, EpCAM-antibody
interactions can cause CTCs to adhere to the device's surface,
making their release difficult. To address the shortcomings of the
CellSearch system, researchers developed MagSweeper, an
immunomagnetic cell separator that enriches CTCs while eliminating
nonmagnetic particle-bound cells (63).
Advances in microfluidic chip technology and
nanomaterials have further improved the sensitivity, specificity,
and cost efficiency of CTC detection (32). A key feature of microfluidic chips
is their flexible combination and large-scale integration of
multiple functional units on a miniature platform. These chips
represent an interdisciplinary achievement combining physics,
chemistry, material science, medical science, mechanics, optics,
mechanical engineering and bioengineering. Microfluidic devices
utilizing antigen-antibody interactions can be categorized into two
types: Microcolumns (requiring chemical modification of the
monoclonal antibody coating) and devices using antibody-coated
surfaces (38). The CTC microarray
isolates target cells by enabling the interaction of CTCs with an
array of 78,000 microcolumns coated with EpCAM antibodies under
precisely controlled laminar flow conditions. This method does not
require pretreatment of the samples and preserves cellular activity
(22).
The size-determined immunocapture chip, a
deterministic lateral displacement (DLD) microarray, captures CTCs
with high sensitivity, efficiency and purity. This chip features
microcolumns that are surface modified with EpCAM antibodies.
Relatively large CTCs are deflected laterally as they collide with
microposts, prolonging their flow time, while smaller blood cells
advance with the laminar flow (64)
and selectively increase the probability of interaction with the
microposts. This method achieves capture efficiencies exceeding
92%, with a purity of 82% (65,66).
Another approach employs microfluidic encapsulated microarrays with
a staggered herringbone design on channel ceilings, increasing
cell-surface contact points. This design significantly enhances
capture efficiency while preserving cellular activity, enabling
downstream analyses such as cell culture and genomic studies.
However, CTCs undergoing epithelial-mesenchymal transition (EMT)
exhibit increased invasive and metastatic potential (67). During the EMT, the expression of
tumor markers on the cell surface is downregulated (68,69),
posing significant challenges for devices that rely exclusively on
anti-EpCAM antibodies for CTC capture (70). A synergistic, dual-antibody chip
based on the DLD principle partially addresses this challenge. In
this approach, anti-ASGPR and anti-EpCAM antibodies are
simultaneously modified in parallel on a microfluidic synergistic
chip, with a capture efficiency of more than 87% per channel
(71).
Over time, advancements in research have
transitioned CTC capture technology from first-generation chips,
such as the CTC-Chip, to second-generation chips, such as the
HB-Chip, GO-Chip and GEM-Chip. Currently, a third-generation
microfluidic chip, called the CTC-iChip, has emerged as a
recognized innovation in the field (72–74).
Compared with first-generation CTC microarrays, second-generation
microarray technology, specifically surface capture devices, is
simpler to fabricate and more efficient at capturing CTCs. However,
these devices have serious limitations, as the captured CTCs are
immobilized on the device surface, making their recovery for
downstream genomic analyses challenging. While trypsin digestion
can be used to release the captured cells, this process may remove
some receptor proteins from the surface of the CTCs and interfere
with subsequent analyses.
Third-generation microfluidic chip technology, such
as the inertial focusing-enhanced microfluidic system (CTC-iChip),
enables the efficient negative depletion of anuclear RBCs and
platelets. This is achieved through the hydrodynamic separation of
nucleated leukocytes and CTCs in plasma, followed by inertial
focusing into a single stream. Magnetic bead-labeled leukocytes are
then efficiently removed by magnetophoresis, resulting in a
solution enriched with CTCs with both epithelial and mesenchymal
features (78). However, this
system has notable drawbacks, including high initial cost a long
setup time, limited ability to analyze single-cell molecules,
reliance on multiple manually attached chips, and challenges in
clinical implementation (32).
To reduce the cost of capturing CTCs, a recent study
proposed the use of EpCAM-resistant nanofiber cellulose scaffolds
with immobilized Fe3O4 NPs for harvesting
CTCs (25). Cellulose
nanostructures can be categorized into two main types: Cellulose
nanocrystals and cellulose nanofibers (CNFs). These nanomaterials
offer several advantages, including relatively high aspect ratios,
large specific surface areas, biodegradability, environmental
sustainability, biocompatibility and non-toxicity (25). Compared with fibrous scaffolds,
rigid nanocrystal scaffolds interact more efficiently with CTCs
(25); however, CNFs have a higher
aspect ratio, which increases the probability of intermolecular
interactions. Kumar et al (79) developed a CNF-based microfluidic
chip with a capture efficiency of 79–98%. However, its average flow
rate of 5 µl/min poses a significant limitation for processing
large volumes of blood, as it reduces the point of contact
necessary for effective CTC capture (79). By contrast, a microchip developed by
Cui et al (80) and
integrated with ZnO nanowires achieved a high CTC capture
efficiency of 91.1±5.5% while maintaining excellent cell viability
(96%). Notably, the platform also enabled damage-free release of
captured cells. This advancement in microarray technology
facilitates single-cell analysis and holds considerable potential
for advancing personalized medicine, particularly in the
development of therapeutics targeting cancer stem cells (80). Researchers have also recently
developed a nanoplatform called NICHE for determining the
relationships between host genes and the microenvironment in living
cells (81). This innovative
platform combines microfluidics and nanofluidics to efficiently
capture CTCs from blood samples. The platform then integrates
stable multichannel fluorescent probes into live CTCs, enabling
rapid in situ quantification of target gene expression. The
NICHE microfluidic device is unique in that it allows simultaneous
gene expression and phenotypic analyses on individual cells in
situ. It also has the potential to generate predictive indices
for screening patients who are likely to benefit from ICIs.
The decreased expression of EpCAM on the surface of
cells undergoing EMT results in a reduced capture rate when an
EpCAM-dependent positive enrichment method is used (38). To overcome these shortcomings,
label-free and negative enrichment methods have been developed.
Similar to the previously mentioned CTC-iChip, a 3D microfluidic
device has been developed and fabricated for the negative
enrichment of CTCs from whole blood samples (82). The device captures unlabeled
leukocytes on its inner surface using antibodies targeting
leukocyte-specific membrane antigens and separates red blood cells
and platelets from CTCs according to their small size. This method
is compatible with subsequent molecular, immunocytochemical and
functional analyses.
In conclusion, positive enrichment methods offer
superior specificity and purity, making them effective for
enriching CTCs with known markers. However, their reliance on
specific markers may cause them to miss certain CTC subtypes.
Conversely, negative enrichment methods can capture a more diverse
range of CTCs but generally result in lower enrichment purity and a
risk of cell loss. In simple terms, positive enrichment methods are
best suited for applications requiring high sensitivity and
specificity, whereas negative enrichment methods are more
appropriate for capturing a heterogeneous CTC population,
particularly when markers are unknown or inconsistently expressed.
A combined approach may provide a more thorough CTC detection
strategy, despite being more complex to implement. In addition to
the above-described platforms, numerous potential CTC enrichment
methods have been developed in recent years (Table SI).
Although there are numerous complex methods for
enriching CTCs, the detection procedure is still relatively
straightforward. The current identification methods include
immunofluorescence, reverse transcription-polymerase chain reaction
(RT-PCR) and next-generation sequencing (NGS).
This technique involves labeling enriched cells
after fixation using fluorescent markers such as anti-human
pancytokeratin (CK), anti-mouse CD45, DAPI, and a cocktail of
antibodies targeting cell surface proteins such as EpCAM, EGFR and
HER2. CTCs are identified as
DAPI+/CK+/CD45− cells (83). The primary advantage of this method
is its convenience in observing the phenotype and morphology of
cells under a fluorescence microscope (84). However, the expression of protein
markers often lacks stability and cannot be used to distinguish
between viable and non-viable cells (83,85).
RT-PCR can be used to detect target cells by reverse
transcription, in which the enzyme reverse transcriptase converts
RNA into complementary DNA using a specific primer (86). While this method allows for the
genetic identification of target cells, it cannot distinguish
non-viable cells from viable cells, limiting its application in
analyses requiring live cells, such as assessments of cellular
deformability and drug responses.
This method enables the detection of specific
proteins secreted by CTCs. It involves culturing enriched CTCs and
tagging secreted proteins with fluorescently labeled antibodies.
Each immunofluorescent spot represents a living cell, offering high
specificity and stability (87).
However, this technique requires sufficient accumulation of
secreted proteins to form immune spots, which can be
time-consuming.
FISH can be used to detect CTCs by labeling abnormal
chromosomal numbers or structures specific to tumor cells. This
genetic method offers high sensitivity and specificity for
identifying CTCs but it can suffer interference from abnormal
chromosomes in the patient's blood and the limited scope of
chromosomal probes, which can detect only a limited number of
specific markers (54,88).
NGS provides insights into genetic structures and
expression states at the single-cell level, allowing for the
genotyping of CTCs and the detection of tumor-specific mutations
(89). While this method offers
unparalleled precision, highly effective upstream enrichment
techniques and stringent QC are needed to ensure the purity and
abundance of CTCs (90) (Table SII).
Immunotherapy involves activating immune cells for
the recognition and destruction of cancer cells (91). Common immune checkpoints include
cytotoxic T-lymphocyte-associated antigen 4, programmed cell death
protein 1 (PD-1), programmed cell death ligand 1 (PD-L1),
lymphocyte activation gene 3, B7-H3 and T-cell immunoglobulin and
mucin-domain containing-3. Among these, PD-1 and PD-L1 are key
molecules in the immune system, and play crucial roles in the
immune escape of tumor cells (92).
PD-1 is an immunosuppressive molecule that is primarily expressed
on activated T and B cells, whereas PD-L1 is predominantly
upregulated in various tumor cells (93). When PD-L1 binds to PD-1 on T cells,
it promotes tumor immune escape. PD-1 inhibitors work by blocking
the interaction between PD-1 and PD-L1 (94), thereby restoring the antitumor
activity of immune cells, enhancing the immune response, reducing
tumor cell proliferation and metastasis, and increasing patient
survival rates (95,96). Numerous studies have demonstrated
the important clinical value of CTCs in immunotherapy.
Numerous studies have shown the usefulness of CTCs
in clinical prognosis; however, their role in predicting the
response to tumor immunotherapy has been relatively unexplored.
Research has demonstrated that the presence of CTCs serves as an
independent prognostic indicator of shorter overall survival (OS)
in patients with various cancers, including non-small cell lung
cancer (NSCLC) (97). Lin et
al (98) reported a significant
reduction in the number of CTCs in the peripheral blood of patients
with stage IV NSCLC who were treated with NK cell immunotherapy on
Days 7 and 30 of treatment. These findings suggest that CTCs may
serve as valuable biomarkers for evaluating treatment efficacy
(98). A different study revealed
that the presence of CTCs was a predictor of poor durable response
rates following immune checkpoint inhibitor (ICI) therapy. A
durable response is defined as stable disease or a partial or
complete response without disease progression at six months
(99). The prognostic and
predictive value of CTCs was further demonstrated in a phase 1b
study using lysosomal virus immunotherapy, where a reduction in CTC
counts was observed in baseline-positive patients who responded
effectively to treatment. Moreover, a study by Castello et
al (100) revealed that the
mean CTC count was significantly lower in patients with NSCLC
treated with ICIs than in untreated patients. The results of the
aforementioned study also revealed that combining the mean CTC
count with the median metabolic tumor volume was associated with
PFS and OS after 8 weeks of ICI treatment (100). Importantly, CTC counts were
identified as independent predictors of both PFS and OS (101,102). Gu et al (103) developed a risk model to predict
prognosis on the basis of aberrantly methylated DNA in CTCs from
patients with lung adenocarcinoma (LUAD). In the aforementioned
study, patients were categorized into high-risk and low-risk groups
on the basis of their median risk score. High-risk patients were
associated with poor prognosis, whereas low-risk patients
demonstrated hyperimmunocompetence, which correlated with favorable
prognosis. Univariate and multivariate Cox regression analyses
confirmed that the risk score was an independent prognostic factor
for survival in patients with LUAD (103).
Antitumor immunotherapeutic strategies have emerged
as promising approaches for the treatment of various cancers. The
expression level of PD-L1 is a crucial indicator of the immune
status of patients with cancer and a key marker for predicting the
efficacy of immunotherapy (104).
Studies have suggested that PD-L1 expression serves as a predictive
biomarker for immunotherapy response in certain solid tumors,
including NSCLC, melanoma and renal cell carcinoma (95,105,106). For example, in a study of
immunotherapy for NSCLC, patients with PD-L1-negative CTCs at
baseline, as well as at 3 and 6 months of nivolumab treatment,
achieved favorable clinical benefits at the 6-month mark. By
contrast, tumor progression occurred in patients with
PD-L1-positive CTCs. These findings suggest that PD-L1 expression
on CTCs could serve as a predictive marker for the immunotherapy
response (107). Dall' Olio et
al (108) reported that
pretreatment with PD-L1-positive CTCs was associated with improved
survival, whereas posttreatment with PD-L1-positive CTCs was
correlated with worse survival in patients with advanced NSCLC
receiving PD-L1/PD-1 inhibitors as second- or third-line therapy
(108). In a phase 1 trial of the
PD-1 inhibitor IBI308 for the treatment of gastrointestinal tumors,
the presence of CTCs with high PD-L1 expression was associated with
significantly better disease control rates. The abundance of
PD-L1-positive CTCs at baseline was also found to predict PFS,
suggesting that monitoring CTC dynamics could provide an early
indication of therapeutic response.
The authors' hypothesis that CTCs can serve as
predictive biomarkers of tumor immunotherapy efficacy was also
supported by a study on metastatic genitourinary tumors.
PD-L1-positive CTCs, elevated CTC counts, and specific CTC
morphological subtypes were associated with shorter OS from
immunotherapy for metastatic genitourinary cancers (109). Combining an assessment of CTC
cluster abundance with single CTC counts significantly improved
prognostic accuracy (110). Thus,
CTCs not only are useful for assessing the prognosis of patients
with cancer, but they also serve as important predictive biomarkers
for monitoring the efficacy of immunotherapy and developing robust
personalized treatment strategies. Most studies have focused on
NSCLC, colorectal cancer, breast cancer, prostate cancer and
melanoma; the predictive value of CTCs in other cancer types
requires further exploration.
Tumor metastasis and recurrence remain the primary
causes of cancer-related death and present difficult treatment
challenges. The development and successful application of ICIs
represent major breakthroughs in tumor therapy in recent years.
While it is not yet clear whether CTCs are more susceptible to
interventions targeting PD-L1 than cells in solid tumors are,
activating the immune system by blocking PD-L1 expression on CTCs
and reducing their numbers in peripheral circulation could decrease
the likelihood of malignant tumor recurrence and metastasis,
representing a promising new therapeutic approach (8,111).
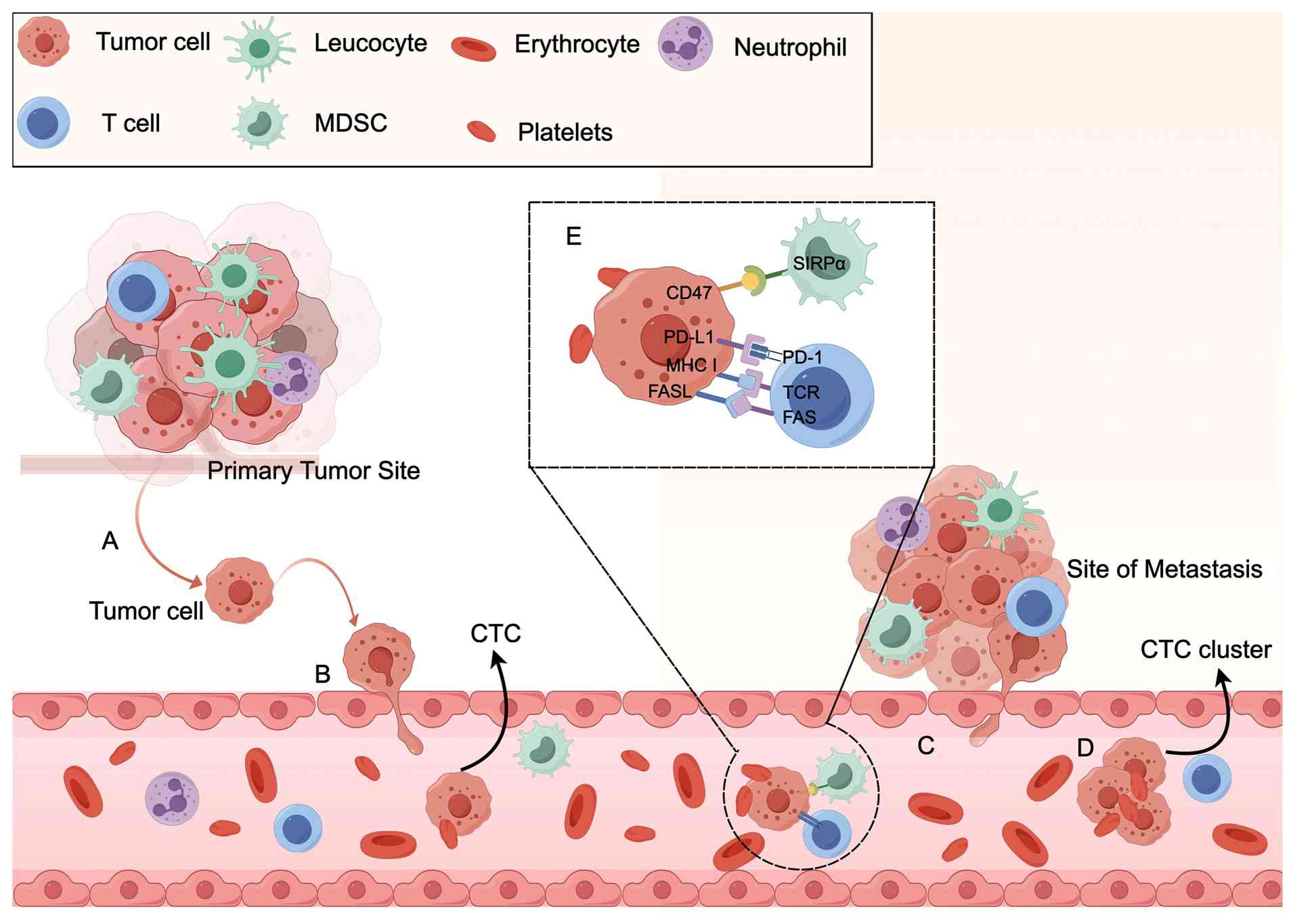
In addition to PD-L1, CD47 is recognized as a crucial immune
checkpoint that is highly expressed on tumor cells. CD47 interacts
with signal-regulated protein alpha on macrophages, transmitting
inhibitory signals that suppress phagocytosis (112). Thus, PD-L1 acts as the key ‘don't
find me’ signal for the adaptive immune system, whereas CD47
functions as the ‘don't eat me’ signal for the innate immune system
and regulates the adaptive immune response (113,114). Targeting both PD-L1 and CD47
simultaneously with specific antibodies has been shown to be more
effective at suppressing lung metastases than targeting PD-L1 or
CD47 alone (114) (Fig. 4).
CTCs were first discovered more than a century ago,
and extensive studies have since demonstrated their importance in
clinical applications. CTCs offer the potential to detect relevant
target lesions at an early stage, even before imaging can reveal a
tumor. They can also be used in combination with other tumor
markers to guide therapy, monitor patients' postoperative
treatment, and predict prognosis. Despite the promising results
from studies of these novel applications, significant challenges
remain before CTCs can be fully integrated into clinical
practice.
The rarity and heterogeneity of CTCs present major
obstacles. While recent advances in CTC enrichment and detection
technology have been achieved, differences in equipment and
detection methods can lead to differences in the interpretation of
results. Currently, standardized, robust CTC detection procedures
are lacking. Moreover, most existing CTC enrichment and detection
methods are expensive, limiting their widespread use in clinical
settings. To address this issue, numerous researchers are now
focusing on utilizing nanoscale materials to develop more
cost-effective platforms for CTC separation and enrichment. While
laboratory testing of these approaches has shown excellent results,
large-scale experiments are needed to validate their feasibility
for clinical applications.
In summary, CTCs not only are useful for assessing
the prognosis of patients with cancer in clinical applications, but
they also serve as important predictive biomarkers for monitoring
the effectiveness of immunotherapy and personalized treatment
strategies. However, current research on CTCs has focused primarily
on a limited number of cancer types, and their predictive value in
other cancers requires further investigation. Immunotherapy is a
prominent research focus for the treatment of malignant tumors, but
no large studies have been conducted that definitively show the
efficacy of using CTCs as targets for tumor immunotherapy. Using
immunotherapy to target CTCs has the potential to reduce the risk
of metastasis and recurrence in malignant tumors, particularly in
postoperative patients, offering a greater hope for permanent
remission.
In conclusion, advancements in CTC isolation and
detection technologies, along with their gradual integration into
clinical applications, hold great potential as key components of
precision individualized medicine in the future.
Not applicable.
The present study was sponsored by the Technological Innovation
R & D Project of Chengdu Science and Technology Program (grant
no. 2022-YF05-01950-SN) and the CSCO-Merck Oncology Research Fund -
Key Project (grant no. Y-MSD2020-0354).
Not applicable.
PZ and QY conducted data mining, writing, draft, and
figure preparation. YH and YF conceptualized, designed and
supervised the study, and revised the manuscript. LZ and KX
reviewed and edited the manuscript. All authors read and approved
the final version of the manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Feng Z, Wu J, Lu Y, Chan YT, Zhang C, Wang
D, Luo D, Huang Y, Feng Y and Wang N: Circulating tumor cells in
the early detection of human cancers. Int J Biol Sci. 18:3251–3265.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang W, Xu F, Yao J, Mao C, Zhu M, Qian
M, Hu J, Zhong H, Zhou J, Shi X and Chen Y: Single-cell metabolic
fingerprints discover a cluster of circulating tumor cells with
distinct metastatic potential. Nat Commun. 14:24852023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Kenawi A, Hänggi K and Ruffell B: The
immune microenvironment and cancer metastasis. Cold Spring Harb
Perspect Med. 10:a0374242020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pereira-Veiga T, Schneegans S, Pantel K
and Wikman H: Circulating tumor cell-blood cell crosstalk: Biology
and clinical relevance. Cell Rep. 40:1112982022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schmidt C: The benefits of immunotherapy
combinations. Nature. 552:S67–S69. 2017. View Article : Google Scholar
|
|
6
|
Brozos-Vázquez EM, Díaz-Peña R,
García-González J, León-Mateos L, Mondelo-Macía P, Peña-Chilet M
and López-López R: Immunotherapy in nonsmall-cell lung cancer:
current status and future prospects for liquid biopsy. Cancer
Immunol Immunother. 70:1177–1188. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu H, Zuo Y, Gao S, Liu Y, Liu T, He S,
Wang M, Hu L, Li C and Yu Y: Circulating tumor cell phenotype
detection and epithelial-mesenchymal transition tracking based on
dual biomarker co-recognition in an integrated PDMS chip. Small.
20:e23103602024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhong X, Zhang H, Zhu Y, Liang Y, Yuan Z,
Li J, Li J, Li X, Jia Y, He T, et al: Circulating tumor cells in
cancer patients: Developments and clinical applications for
immunotherapy. Mol Cancer. 19:152020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh B, Arora S, D'Souza A, Kale N, Aland
G, Bharde A, Quadir M, Calderón M, Chaturvedi P and Khandare J:
Chemo-specific designs for the enumeration of circulating tumor
cells: Advances in liquid biopsy. J Mater Chem B. 9:2946–2978.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ye Q, Ling S, Zheng S and Xu X: Liquid
biopsy in hepatocellular carcinoma: Circulating tumor cells and
circulating tumor DNA. Mol Cancer. 18:1142019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li M, Li L, Zheng J, Li Z, Li S, Wang K
and Chen X: Liquid biopsy at the frontier in renal cell carcinoma:
recent analysis of techniques and clinical application. Mol Cancer.
22:372023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shuai Y, Ma Z, Ju J, Wei T, Gao S, Kang Y,
Yang Z, Wang X, Yue J and Yuan P: Liquid-based biomarkers in breast
cancer: Looking beyond the blood. J Transl Med. 21:8092023.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jou HJ, Ho HC, Huang KY, Chen CY, Chen SW,
Lo PH, Huang PW, Huang CE and Chen M: Isolation of TTF-1 positive
circulating tumor cells for single-cell sequencing by using an
automatic platform based on microfluidic devices. Int J Mol Sci.
23:151392022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu CM, Tang M, Feng J, Xia HF, Wu LL, Pang
DW, Chen G and Zhang ZL: A liquid biopsy-guided drug release system
for cancer theranostics: Integrating rapid circulating tumor cell
detection and precision tumor therapy. Lab Chip. 20:1418–1425.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ashworth T: A case of cancer in which
cells similar to those in the tumours were seen in the blood after
death. Aust Med J. 14:146–147. 1869.
|
|
16
|
Nasr MM and Lynch CC: How circulating
tumor cluster biology contributes to the metastatic cascade: From
invasion to dissemination and dormancy. Cancer Metastasis Rev.
42:1133–1146. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smit DJ, Pantel K and Jücker M:
Circulating tumor cells as a promising target for individualized
drug susceptibility tests in cancer therapy. Biochem Pharmacol.
188:1145892021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peck K, Sher YP, Shih JY, Roffler SR, Wu
CW and Yang PC: Detection and quantitation of circulating cancer
cells in the peripheral blood of lung cancer patients. Cancer Res.
58:2761–2765. 1998.PubMed/NCBI
|
|
19
|
Cristofanilli M, Budd GT, Ellis MJ,
Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ,
Terstappen LW and Hayes DF: Circulating tumor cells, disease
progression, and survival in metastatic breast cancer. N Engl J
Med. 351:781–791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Magri V, Marino L, Nicolazzo C, Gradilone
A, De Renzi G, De Meo M, Gandini O, Sabatini A, Santini D, Cortesi
E and Gazzaniga P: Prognostic role of circulating tumor cell
trajectories in metastatic colorectal cancer. Cells. 12:11722023.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Allard WJ, Matera J, Miller MC, Repollet
M, Connelly MC, Rao C, Tibbe AG, Uhr JW and Terstappen LW: Tumor
cells circulate in the peripheral blood of all major carcinomas but
not in healthy subjects or patients with nonmalignant diseases.
Clin Cancer Res. 10:6897–6904. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nagrath S, Sequist LV, Maheswaran S, Bell
DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky
A, et al: Isolation of rare circulating tumour cells in cancer
patients by microchip technology. Nature. 450:1235–1239. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hartmann C, Patil R, Lin CP and Niedre M:
Fluorescence detection, enumeration and characterization of single
circulating cells in vivo: Technology, applications and future
prospects. Phys Med Biol. 63:01TR012017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang M, Xia HF, Xu CM, Feng J, Ren JG,
Miao F, Wu M, Wu LL, Pang DW, Chen G and Zhang ZL: Magnetic chip
based extracorporeal circulation: A new tool for circulating tumor
cell in vivo detection. Anal Chem. 91:15260–15266. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hazra RS, Kale N, Boyle C, Molina KB,
D'Souza A, Aland G, Jiang L, Chaturvedi P, Ghosh S, Mallik S, et
al: Magnetically-activated, nanostructured cellulose for efficient
capture of circulating tumor cells from the blood sample of head
and neck cancer patients. Carbohydr Polym. 323:1214182024.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Q, Zhang X, Xie P and Zhang W:
Liquid biopsy: An arsenal for tumour screening and early diagnosis.
Cancer Treat Rev. 129:1027742024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ghassemi P, Ren X, Foster BM, Kerr BA and
Agah M: Post-enrichment circulating tumor cell detection and
enumeration via deformability impedance cytometry. Biosens
Bioelectron. 150:1118682020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nikanjam M, Kato S and Kurzrock R: Liquid
biopsy: Current technology and clinical applications. J Hematol
Oncol. 15:1312022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kowalik A, Kowalewska M and Góźdź S:
Current approaches for avoiding the limitations of circulating
tumor cells detection methods-implications for diagnosis and
treatment of patients with solid tumors. Transl Res. 185:58–84.e15.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
You Q, Peng J, Chang Z, Ge M, Mei Q and
Dong WF: Specific recognition and photothermal release of
circulating tumor cells using near-infrared light-responsive 2D
MXene simplenanosheets@hydrogel
membranes. Talanta. 235:1227702021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang K, Wang X, Pan Q and Zhao B: Liquid
biopsy techniques and pancreatic cancer: diagnosis, monitoring, and
evaluation. Mol Cancer. 22:1672023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lin D, Shen L, Luo M, Zhang K, Li J, Yang
Q, Zhu F, Zhou D, Zheng S, Chen Y and Zhou J: Circulating tumor
cells: Biology and clinical significance. Signal Transduct Target
Ther. 6:4042021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu Y, Zhang Y, Chen Y, Wang X, Kang K, Zhu
N, Wu Y and Yi Q: Floating immunomagnetic microspheres for highly
efficient circulating tumor cell isolation under facile magnetic
manipulation. ACS Sens. 8:1858–1866. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yeo D, Kao S, Gupta R, Wahlroos S, Bastian
A, Strauss H, Klemm V, Shrestha P, Ramirez AB, Costandy L, et al:
Accurate isolation and detection of circulating tumor cells using
enrichment-free multiparametric high resolution imaging. Front
Oncol. 13:11412282023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Labib M and Kelley SO: Circulating tumor
cell profiling for precision oncology. Mol Oncol. 15:1622–1646.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen J, Li D, Zhou C, Zhu Y, Lin C, Guo L,
Le W, Gu Z and Chen B: Principle superiority and clinical
extensibility of 2D and 3D charged nanoprobe detection platform
based on electrophysiological characteristics of circulating tumor
cells. Cells. 12:3052023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Khan T, Becker TM, Po JW, Chua W and Ma Y:
Single-circulating tumor cell whole genome amplification to unravel
cancer heterogeneity and actionable biomarkers. Int J Mol Sci.
23:83862022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dianat-Moghadam H, Azizi M, Eslami S Z,
Cortés-Hernández LE, Heidarifard M, Nouri M and Alix-Panabières C:
The role of circulating tumor cells in the metastatic cascade:
Biology, technical challenges, and clinical relevance. Cancers
(Basel). 12:8672020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hyun KA and Jung HI: Advances and critical
concerns with the microfluidic enrichments of circulating tumor
cells. Lab Chip. 14:45–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shimmyo N, Furuhata M, Yamada M, Utoh R
and Seki M: Process simplification and structure design of
parallelized microslit isolator for physical property-based capture
of tumor cells. Analyst. 147:1622–1630. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang Q, Wang FB, Yuan CH, He Z, Rao L,
Cai B, Chen B, Jiang S, Li Z, Chen J, et al: Gelatin
nanoparticle-coated silicon beads for density-selective capture and
release of heterogeneous circulating tumor cells with high purity.
Theranostics. 8:1624–1635. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tamminga M, Andree KC, Hiltermann TJN,
Jayat M, Schuuring E, van den Bos H, Spierings DCJ, Lansdorp PM,
Timens W, Terstappen LWMM and Groen HJM: Detection of circulating
tumor cells in the diagnostic leukapheresis product of
non-small-cell lung cancer patients comparing
cellsearch® and ISET. Cancers (Basel). 12:8962020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Barr J, Chudasama D, Rice A, Karteris E
and Anikin V: Lack of association between Screencell-detected
circulating tumour cells and long-term survival of patients
undergoing surgery for non-small cell lung cancer: A pilot clinical
study. Mol Clin Oncol. 12:191–195. 2020.PubMed/NCBI
|
|
44
|
Ferreira MM, Ramani VC and Jeffrey SS:
Circulating tumor cell technologies. Mol Oncol. 10:374–394. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Y, Wang B, Cai J, Yang Y, Tang C,
Zheng X, Li H and Xu F: Enrichment and separation technology for
evaluation of circulating tumor cells. Talanta. 282:1270252025.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Harouaka RA, Zhou MD, Yeh YT, Khan WJ, Das
A, Liu X, Christ CC, Dicker DT, Baney TS, Kaifi JT, et al: Flexible
micro spring array device for high-throughput enrichment of viable
circulating tumor cells. Clin Chem. 60:323–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gkountela S, Castro-Giner F, Szczerba BM,
Vetter M, Landin J, Scherrer R, Krol I, Scheidmann MC, Beisel C,
Stirnimann CU, et al: Circulating tumor cell clustering shapes DNA
methylation to enable metastasis seeding. Cell. 176:98–112.e14.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kurma K, Eslami-S Z, Alix-Panabières C and
Cayrefourcq L: Liquid biopsy: Paving a new avenue for cancer
research. Cell Adh Migr. 18:1–26. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jia Y, Xu H, Li Y, Wei C, Guo R, Wang F,
Wu Y, Liu J, Jia J, Yan J, et al: A Modified ficoll-paque gradient
method for isolating mononuclear cells from the peripheral and
umbilical cord blood of humans for biobanks and clinical
laboratories. Biopreserv Biobank. 16:82–91. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
He S, Wei J, Ding L, Yang X and Wu Y:
State-of-the-arts techniques and current evolving approaches in the
separation and detection of circulating tumor cell. Talanta.
239:1230242022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang Y, Li Y and Tan Z: A review of
enrichment methods for circulating tumor cells: From single
modality to hybrid modality. Analyst. 146:7048–7069. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Peeters DJ, De Laere B, Van den Eynden GG,
Van Laere SJ, Rothé F, Ignatiadis M, Sieuwerts AM, Lambrechts D,
Rutten A, van Dam PA, et al: Semiautomated isolation and molecular
characterisation of single or highly purified tumour cells from
CellSearch enriched blood samples using dielectrophoretic cell
sorting. Br J Cancer. 108:1358–1367. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhao K, Zhao P, Dong J, Wei Y, Chen B,
Wang Y, Pan X and Wang J: Implementation of an integrated
dielectrophoretic and magnetophoretic microfluidic chip for CTC
isolation. Biosensors (Basel). 12:7572022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zeinali M, Lee M, Nadhan A, Mathur A,
Hedman C, Lin E, Harouaka R, Wicha MS, Zhao L, Palanisamy N, et al:
High-throughput label-free isolation of heterogeneous circulating
tumor cells and CTC clusters from non-small-cell lung cancer
patients. Cancers (Basel). 12:1272020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Boya M, Ozkaya-Ahmadov T, Swain BE, Chu
CH, Asmare N, Civelekoglu O, Liu R, Lee D, Tobia S, Biliya S, et
al: High throughput, label-free isolation of circulating tumor cell
clusters in meshed microwells. Nat Commun. 13:33852022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Fabisiewicz A and Grzybowska E: CTC
clusters in cancer progression and metastasis. Med Oncol.
34:122017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Niu Z, Kozminsky M, Day KC, Broses LJ,
Henderson ML, Patsalis C, Tagett R, Qin Z, Blumberg S, Reichert ZR,
et al: Characterization of circulating tumor cells in patients with
metastatic bladder cancer utilizing functionalized microfluidics.
Neoplasia. 57:1010362024. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Vidlarova M, Rehulkova A, Stejskal P,
Prokopova A, Slavik H, Hajduch M and Srovnal J: Recent advances in
methods for circulating tumor cell detection. Int J Mol Sci.
24:39022023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Sajay BN, Chang CP, Ahmad H, Khuntontong
P, Wong CC, Wang Z, Puiu PD, Soo R and Rahman AR: Microfluidic
platform for negative enrichment of circulating tumor cells. Biomed
Microdevices. 16:537–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ma S, Zhou M, Xu Y, Gu X, Zou M,
Abudushalamu G, Yao Y, Fan X and Wu G: Clinical application and
detection techniques of liquid biopsy in gastric cancer. Mol
Cancer. 22:72023. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lopes C, Piairo P, Chícharo A, Abalde-Cela
S, Pires LR, Corredeira P, Alves P, Muinelo-Romay L, Costa L and
Diéguez L: HER2 expression in circulating tumour cells isolated
from metastatic breast cancer patients using a size-based
microfluidic device. Cancers (Basel). 13:44462021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chowdhury T, Cressiot B, Parisi C,
Smolyakov G, Thiébot B, Trichet L, Fernandes FM, Pelta J and
Manivet P: Circulating tumor cells in cancer diagnostics and
prognostics by single-molecule and single-cell characterization.
ACS Sens. 8:406–426. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Talasaz AH, Powell AA, Huber DE, Berbee
JG, Roh KH, Yu W, Xiao W, Davis MM, Pease RF, Mindrinos MN, et al:
Isolating highly enriched populations of circulating epithelial
cells and other rare cells from blood using a magnetic sweeper
device. Proc Natl Acad Sci USA. 106:3970–3975. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shi J, Zhao C, Shen M, Chen Z, Liu J,
Zhang S and Zhang Z: Combination of microfluidic chips and
biosensing for the enrichment of circulating tumor cells. Biosens
Bioelectron. 202:1140252022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ahmed MG, Abate MF, Song Y, Zhu Z, Yan F,
Xu Y, Wang X, Li Q and Yang C: Isolation, detection, and
antigen-based profiling of circulating tumor cells using a
size-dictated immunocapture chip. Angew Chem Int Ed Engl.
56:10681–10685. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu HY, Koch C, Haller A, Joosse SA, Kumar
R, Vellekoop MJ, Horst LJ, Keller L, Babayan A, Failla AV, et al:
Evaluation of microfluidic ceiling designs for the capture of
circulating tumor cells on a microarray platform. Adv Biosyst.
4:e19001622020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kulus M, Farzaneh M, Bryja A, Zehtabi M,
Azizidoost S, Abouali Gale Dari M, Golcar-Narenji A, Ziemak H,
Chwarzyński M, Piotrowska-Kempisty H, et al: Phenotypic transitions
the processes involved in regulation of growth and proangiogenic
properties of stem cells, cancer stem cells and circulating tumor
cells. Stem Cell Rev Rep. 20:967–979. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Gwak H, Park S, Kim J, Lee JD, Kim IS, Kim
SI, Hyun KA and Jung HI: Microfluidic chip for rapid and selective
isolation of tumor-derived extracellular vesicles for early
diagnosis and metastatic risk evaluation of breast cancer. Biosens
Bioelectron. 192:1134952021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Cortés-Hernández LE, Eslami-S Z,
Costa-Silva B and Alix-Panabières C: Current applications and
discoveries related to the membrane components of circulating tumor
cells and extracellular vesicles. Cells. 10:22212021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Shahhosseini R, Pakmehr S, Elhami A,
Shakir MN, Alzahrani AA, Al-Hamdani MM, Abosoda M, Alsalamy A,
Mohammadi-Dehcheshmeh M, Maleki TE, et al: Current biological
implications and clinical relevance of metastatic circulating tumor
cells. Clin Exp Med. 25:72024. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhu L, Lin H, Wan S, Chen X, Wu L, Zhu Z,
Song Y, Hu B and Yang C: Efficient isolation and phenotypic
profiling of circulating hepatocellular carcinoma cells via a
combinatorial-antibody-functionalized microfluidic synergetic-chip.
Anal Chem. 92:15229–15235. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Stott SL, Hsu CH, Tsukrov DI, Yu M,
Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK,
et al: Isolation of circulating tumor cells using a
microvortex-generating herringbone-chip. Proc Natl Acad Sci USA.
107:18392–18397. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham
TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF, et al:
Sensitive capture of circulating tumour cells by functionalized
graphene oxide nanosheets. Nat Nanotechnol. 8:735–741. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sheng W, Ogunwobi OO, Chen T, Zhang J,
George TJ, Liu C and Fan ZH: Capture, release and culture of
circulating tumor cells from pancreatic cancer patients using an
enhanced mixing chip. Lab Chip. 14:89–98. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Rostami P, Kashaninejad N, Moshksayan K,
Saidi MS, Firoozabadi B and Nguyen NT: Novel approaches in cancer
management with circulating tumor cell clusters. Journal of
Science: Advanced Materials and Devices. 4:1–18. 2019.
|
|
76
|
Sun N, Zhang C, Wang J, Yue X, Kim HY,
Zhang RY, Liu H, Widjaja J, Tang H, Zhang TX, et al: Hierarchical
integration of DNA nanostructures and NanoGold onto a microchip
facilitates covalent chemistry-mediated purification of circulating
tumor cells in head and neck squamous cell carcinoma. Nano Today.
49:1017862023. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kolb HC, Finn MG and Sharpless KB: Click
chemistry: Diverse chemical function from a few good reactions.
Angew Chem Int Ed Engl. 40:2004–2021. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Fachin F, Spuhler P, Martel-Foley JM, Edd
JF, Barber TA, Walsh J, Karabacak M, Pai V, Yu M, Smith K, et al:
Monolithic chip for high-throughput blood cell depletion to sort
rare circulating tumor cells. Sci Rep. 7:109362017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kumar T, Soares RRG, Ali Dholey L,
Ramachandraiah H, Aval NA, Aljadi Z, Pettersson T and Russom A:
Multi-layer assembly of cellulose nanofibrils in a microfluidic
device for the selective capture and release of viable tumor cells
from whole blood. Nanoscale. 12:21788–21797. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Cui H, Liu Q, Li R, Wei X, Sun Y, Wang Z,
Zhang L, Zhao XZ, Hua B and Guo SS: ZnO nanowire-integrated
bio-microchips for specific capture and non-destructive release of
circulating tumor cells. Nanoscale. 12:1455–1463. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Dong Z, Wang Y, Xu G, Liu B, Wang Y,
Reboud J, Jajesniak P, Yan S, Ma P, Liu F, et al: Genetic and
phenotypic profiling of single living circulating tumor cells from
patients with microfluidics. Proc Natl Acad Sci USA.
121:e23151681212024. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Chu CH, Liu R, Ozkaya-Ahmadov T, Swain BE,
Boya M, El-Rayes B, Akce M, Bilen MA, Kucuk O and Sarioglu AF:
Negative enrichment of circulating tumor cells from unmanipulated
whole blood with a 3D printed device. Sci Rep. 11:205832021.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ramirez AB, Bhat R, Sahay D, De Angelis C,
Thangavel H, Hedayatpour S, Dobrolecki LE, Nardone A, Giuliano M,
Nagi C, et al: Circulating tumor cell investigation in breast
cancer patient-derived xenograft models by automated
immunofluorescence staining, image acquisition, and single cell
retrieval and analysis. BMC Cancer. 19:2202019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ren F, Fei Q, Qiu K, Zhang Y, Zhang H and
Sun L: Liquid biopsy techniques and lung cancer: diagnosis,
monitoring and evaluation. J Exp Clin Cancer Res. 43:962024.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Koinis F, Zafeiriou Z, Messaritakis I,
Katsaounis P, Koumarianou A, Kontopodis E, Chantzara E, Aidarinis
C, Lazarou A, Christodoulopoulos G, et al: Prognostic role of
circulating tumor cells in patients with metastatic
castration-resistant prostate cancer receiving cabazitaxel: A
prospective biomarker study. Cancers (Basel). 15:45112023.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Eftekhari A, Alipour M, Chodari L, Maleki
Dizaj S, Ardalan M, Samiei M, Sharifi S, Zununi Vahed S, Huseynova
I, Khalilov R, et al: A comprehensive review of detection methods
for SARS-CoV-2. Microorganisms. 9:2322021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Cieślikowski WA, Budna-Tukan J,
Świerczewska M, Ida A, Hrab M, Jankowiak A, Mazel M, Nowicki M,
Milecki P, Pantel K, et al: Circulating tumor cells as a marker of
disseminated disease in patients with newly diagnosed high-risk
prostate cancer. Cancers (Basel). 12:1602020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Yang D, Yang X, Li Y, Zhao P, Fu R, Ren T,
Hu P, Wu Y, Yang H and Guo N: Clinical significance of circulating
tumor cells and metabolic signatures in lung cancer after surgical
removal. J Transl Med. 18:2432020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Lei Y, Tang R, Xu J, Wang W, Zhang B, Liu
J, Yu X and Shi S: Applications of single-cell sequencing in cancer
research: progress and perspectives. J Hematol Oncol. 14:912021.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Kojima M, Harada T, Fukazawa T, Kurihara
S, Touge R, Saeki I, Takahashi S and Hiyama E: Single-cell
next-generation sequencing of circulating tumor cells in patients
with neuroblastoma. Cancer Sci. 114:1616–1624. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Rzhevskiy A, Kapitannikova A, Malinina P,
Volovetsky A, Aboulkheyr Es H, Kulasinghe A, Thiery JP,
Maslennikova A, Zvyagin AV and Ebrahimi Warkiani M: Emerging role
of circulating tumor cells in immunotherapy. Theranostics.
11:8057–8075. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhang Y and Zheng J: Functions of immune
checkpoint molecules beyond immune evasion. Adv Exp Med Biol.
1248:201–226. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Kloten V, Lampignano R, Krahn T and
Schlange T: Circulating Tumor Cell PD-L1 expression as biomarker
for therapeutic efficacy of immune checkpoint inhibition in NSCLC.
Cells. 8:8092019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Kennedy LC, Lu J, Kuehn S, Ramirez AB, Lo
E, Sun Y, U'Ren L, Chow LQM, Chen Z, Grivas P, et al: Liquid biopsy
assessment of circulating tumor Cell PD-L1 and IRF-1 expression in
patients with advanced solid tumors receiving immune checkpoint
inhibitor. Target Oncol. 17:329–341. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Hou W, Zhou X, Yi C and Zhu H: Immune
check point inhibitors and immune-related adverse events in small
cell lung cancer. Front Oncol. 11:6042272021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Ouyang Y, Liu W, Zhang N, Yang X, Li J and
Long S: Prognostic significance of programmed cell death-ligand 1
expression on circulating tumor cells in various cancers: A
systematic review and meta-analysis. Cancer Med. 10:7021–7039.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Papadaki MA, Messaritakis I, Fiste O,
Souglakos J, Politaki E, Kotsakis A, Georgoulias V, Mavroudis D and
Agelaki S: Assessment of the efficacy and clinical utility of
different circulating tumor cell (CTC) detection assays in patients
with chemotherapy-naïve advanced or metastatic non-small cell lung
cancer (NSCLC). Int J Mol Sci. 22:9252021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Lin M, Liang SZ, Shi J, Niu LZ, Chen JB,
Zhang MJ and Xu KC: Circulating tumor cell as a biomarker for
evaluating allogenic NK cell immunotherapy on stage IV non-small
cell lung cancer. Immunol Lett. 191:10–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Tamminga M, de Wit S, Hiltermann TJN,
Timens W, Schuuring E, Terstappen LWMM and Groen HJM: Circulating
tumor cells in advanced non-small cell lung cancer patients are
associated with worse tumor response to checkpoint inhibitors. J
Immunother Cancer. 7:1732019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Castello A, Carbone FG, Rossi S, Monterisi
S, Federico D, Toschi L and Lopci E: Circulating tumor cells and
metabolic parameters in NSCLC patients treated with checkpoint
inhibitors. Cancers (Basel). 12:4872020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Banys-Paluchowski M, Fehm TN, Grimm-Glang
D, Rody A and Krawczyk N: Liquid biopsy in metastatic breast
cancer: Current role of circulating tumor cells and circulating
tumor DNA. Oncol Res Treat. 45:4–11. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Heidrich I, Ačkar L, Mossahebi Mohammadi P
and Pantel K: Liquid biopsies: Potential and challenges. Int J
Cancer. 148:528–545. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Gu X, Huang X, Zhang X and Wang C:
Development and validation of a DNA methylation-related classifier
of circulating tumour cells to predict prognosis and to provide a
therapeutic strategy in lung adenocarcinoma. Int J Biol Sci.
18:4984–5000. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Peng QH, Wang CH, Chen HM, Zhang RX, Pan
ZZ, Lu ZH, Wang GY, Yue X, Huang W and Liu RY: CMTM6 and PD-L1
coexpression is associated with an active immune microenvironment
and a favorable prognosis in colorectal cancer. J Immunother
Cancer. 9:e0016382021. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wang X, Teng F, Kong L and Yu J: PD-L1
expression in human cancers and its association with clinical
outcomes. Onco Targets Ther. 9:5023–5039. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Yao S, Han Y, Yang M, Jin K and Lan H:
Integration of liquid biopsy and immunotherapy: Opening a new era
in colorectal cancer treatment. Front Immunol. 14:12928612023.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Ju S, Chen C, Zhang J, Xu L, Zhang X, Li
Z, Chen Y, Zhou J, Ji F and Wang L: Detection of circulating tumor
cells: Opportunities and challenges. Biomark Res. 10:582022.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Dall'Olio FG, Gelsomino F, Conci N,
Marcolin L, De Giglio A, Grilli G, Sperandi F, Fontana F,
Terracciano M, Fragomeno B, et al: PD-L1 Expression in circulating
tumor cells as a promising prognostic biomarker in advanced
non-small-cell lung cancer treated with immune checkpoint
inhibitors. Clin Lung Cancer. 22:423–431. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Chalfin HJ, Pramparo T, Mortazavi A,
Niglio SA, Schonhoft JD, Jendrisak A, Chu YL, Richardson R, Krupa
R, Anderson AKL, et al: Circulating tumor cell subtypes and T-cell
populations as prognostic biomarkers to combination immunotherapy
in patients with metastatic genitourinary cancer. Clin Cancer Res.
27:1391–1398. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Gu X, Wei S and Lv X: Circulating tumor
cells: From new biological insights to clinical practice. Signal
Transduct Target Ther. 9:2262024. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Pantel K and Alix-Panabières C: Crucial
roles of circulating tumor cells in the metastatic cascade and
tumor immune escape: Biology and clinical translation. J Immunother
Cancer. 10:e0056152022. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Jaiswal S, Jamieson CH, Pang WW, Park CY,
Chao MP, Majeti R, Traver D, van Rooijen N and Weissman IL: CD47 is
upregulated on circulating hematopoietic stem cells and leukemia
cells to avoid phagocytosis. Cell. 138:271–285. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Topalian SL, Drake CG and Pardoll DM:
Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor
immunity. Curr Opin Immunol. 24:207–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Lian S, Xie X, Lu Y and Jia L: Checkpoint
CD47 function on tumor metastasis and immune therapy. Onco Targets
Ther. 12:9105–9114. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Lian S, Xie R, Ye Y, Lu Y, Cheng Y, Xie X,
Li S and Jia L: Dual blockage of both PD-L1 and CD47 enhances
immunotherapy against circulating tumor cells. Sci Rep. 9:45322019.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Yu Y, Cheng Q, Ji X, Chen H, Zeng W, Zeng
X, Zhao Y and Mei L: Engineered drug-loaded cellular membrane
nanovesicles for efficient treatment of postsurgical cancer
recurrence and metastasis. Sci Adv. 8:eadd35992022. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Terme M, Ullrich E, Aymeric L, Meinhardt
K, Desbois M, Delahaye N, Viaud S, Ryffel B, Yagita H, Kaplanski G,
et al: IL-18 induces PD-1-dependent immunosuppression in cancer.
Cancer Res. 71:5393–5399. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Jacot W, Mazel M, Mollevi C, Pouderoux S,
D'Hondt V, Cayrefourcq L, Bourgier C, Boissiere-Michot F, Berrabah
F, Lopez-Crapez E, et al: Clinical correlations of programmed cell
death ligand 1 status in liquid and standard biopsies in breast
cancer. Clin Chem. 66:1093–1101. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Praharaj PP, Bhutia SK, Nagrath S, Bitting
RL and Deep G: Circulating tumor cell-derived organoids: Current
challenges and promises in medical research and precision medicine.
Biochim Biophys Acta Rev Cancer. 1869:117–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Riethdorf S, Fritsche H, Müller V, Rau T,
Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, et al:
Detection of circulating tumor cells in peripheral blood of
patients with metastatic breast cancer: A validation study of the
CellSearch system. Clin Cancer Res. 13:920–928. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Li F, Li J, Yuan Y, Zhang H, Zhang S, Bian
L, Wang T and Jiang Z: A real-world comparison of circulating tumor
cells in breast cancer from China: Novel device, CTC counts and its
overall survival. Heliyon. 10:e292172024. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Liu Z, Fusi A, Klopocki E, Schmittel A,
Tinhofer I, Nonnenmacher A and Keilholz U: Negative enrichment by
immunomagnetic nanobeads for unbiased characterization of
circulating tumor cells from peripheral blood of cancer patients. J
Transl Med. 9:702011. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Seyfoori A, Seyyed Ebrahimi SA, Samandari
M, Samiei E, Stefanek E, Garnis C and Akbari M:
Microfluidic-Assisted CTC isolation and in situ monitoring using
smart magnetic microgels. Small. 19:e22053202023. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Shen MJ, Olsthoorn RCL, Zeng Y, Bakkum T,
Kros A and Boyle AL: Magnetic-activated cell sorting using
coiled-coil peptides: An alternative strategy for isolating cells
with high efficiency and specificity. ACS Appl Mater Interfaces.
13:11621–11630. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zhang W, Duan X, Zhang Z, Yang Z, Zhao C,
Liang C, Liu Z, Cheng S and Zhang K: Combination of CT and
telomerase+ circulating tumor cells improves diagnosis of small
pulmonary nodules. JCI Insight. 6:e1481822021.PubMed/NCBI
|
|
126
|
Vasantharajan SS, Barnett E, Gray ES,
McCall JL, Rodger EJ, Eccles MR, Munro F, Pattison S and Chatterjee
A: Assessment of a size-based method for enriching circulating
tumour cells in colorectal cancer. Cancers (Basel). 14:34462022.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhang X, Xie P, Zhang K and Zhang W:
Circulating tumour cell isolation, analysis and clinical
application. Cell Oncol (Dordr). 46:533–544. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Lin E, Cao T, Nagrath S and King MR:
Circulating tumor cells: Diagnostic and therapeutic applications.
Annu Rev Biomed Eng. 20:329–352. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Pantel K and Alix-Panabières C: Liquid
biopsy and minimal residual disease-latest advances and
implications for cure. Nat Rev Clin Oncol. 16:409–424. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Kulasinghe A, Kenny L, Perry C, Thiery JP,
Jovanovic L, Vela I, Nelson C and Punyadeera C: Impact of
label-free technologies in head and neck cancer circulating tumour
cells. Oncotarget. 7:71223–71234. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Ye F, Wechsler J, Bouzidi A, Uzan G and
Naserian S: Fast and efficient isolation of murine circulating
tumor cells using screencell technology for pre-clinical analyzes.
Sci Rep. 14:150192024. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Philippron A, Depypere L, Oeyen S, De
Laere B, Vandeputte C, Nafteux P, De Preter K and Pattyn P:
Evaluation of a marker independent isolation method for circulating
tumor cells in esophageal adenocarcinoma. PLoS One.
16:e02510522021. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Andrikou K, Rossi T, Verlicchi A, Priano
I, Cravero P, Burgio MA, Crinò L, Bandini S, Ulivi P and Delmonte
A: Circulating tumour cells: Detection and application in advanced
non-small cell lung cancer. Int J Mol Sci. 24:160852023. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Çağlayan Arslan Z, Demircan Yalçın Y and
Külah H: Label-free enrichment of MCF7 breast cancer cells from
leukocytes using continuous flow dielectrophoresis.
Electrophoresis. 43:1531–1544. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Sarioglu AF, Aceto N, Kojic N, Donaldson
MC, Zeinali M, Hamza B, Engstrom A, Zhu H, Sundaresan TK, Miyamoto
DT, et al: A microfluidic device for label-free, physical capture
of circulating tumor cell clusters. Nat Methods. 12:685–691. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhou W, Zhu C, Shen P, Wang JF, Zhu G, Jia
Y, Wu Y, Wang S, Sun J, Yang F, et al: Hypoxia stimulates
CTC-platelet cluster formation to promote breast cancer metastasis.
IScience. 27:1095472024. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Boyer M, Cayrefourcq L, Garima F,
Foulongne V, Dereure O and Alix-Panabières C: Circulating tumor
cell detection and polyomavirus status in merkel cell carcinoma.
Sci Rep. 10:16122020. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Smit DJ and Pantel K: Circulating tumor
cells as liquid biopsy markers in cancer patients. Mol Aspects Med.
96:1012582024. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Karabacak NM, Spuhler PS, Fachin F, Lim
EJ, Pai V, Ozkumur E, Martel JM, Kojic N, Smith K, Chen PI, et al:
Microfluidic, marker-free isolation of circulating tumor cells from
blood samples. Nat Protoc. 9:694–710. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Sassi A and You L: Microfluidics-based
technologies for the assessment of castration-resistant prostate
cancer. Cells. 13:5752024. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Wei J, Deng W, Weng J, Li M, Lan G, Li X,
Ye L, Wang Y, Liu F, Ou H, et al: Epithelial-mesenchymal transition
classification of circulating tumor cells predicts clinical
outcomes in progressive nasopharyngeal carcinoma. Front Oncol.
12:9884582022. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Lopresti A, Malergue F, Bertucci F,
Liberatoscioli ML, Garnier S, DaCosta Q, Finetti P, Gilabert M,
Raoul JL, Birnbaum D, et al: Sensitive and easy screening for
circulating tumor cells by flow cytometry. JCI Insight.
5:e1281802019. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Kotsifaki A, Maroulaki S and Armakolas A:
Exploring the immunological profile in breast cancer: Recent
advances in diagnosis and prognosis through circulating tumor
cells. Int J Mol Sci. 25:48322024. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Mazard T, Cayrefourcq L, Perriard F,
Senellart H, Linot B, de la Fouchardière C, Terrebonne E, François
E, Obled S, Guimbaud R, et al: Clinical relevance of viable
circulating tumor cells in patients with metastatic colorectal
cancer: The COLOSPOT prospective study. Cancers (Basel).
13:29662021. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Soler A, Cayrefourcq L, Mazel M and
Alix-Panabières C: EpCAM-Independent enrichment and detection of
viable circulating tumor cells using the EPISPOT assay. Methods Mol
Biol. 1634:263–276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Xu C, Zhang Y, Fan X, Lan X, Ye X and Wu
T: An efficient fluorescence in situ hybridization (FISH)-based
circulating genetically abnormal cells (CACs) identification method
based on Multi-scale MobileNet-YOLO-V4. Quant Imaging Med Surg.
12:2961–2976. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Zhu Y and Lowe AC: Multiplexed
fluorescence in situ hybridization-based detection of circulating
tumor cells: A novel liquid-based technology to facilitate accurate
and early identification of non-small cell lung cancer patients.
Cancer Cytopathol. 128:518–519. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Kruse A, Abdel-Azim N, Kim HN, Ruan Y,
Phan V, Ogana H, Wang W, Lee R, Gang EJ, Khazal S and Kim YM:
Minimal residual disease detection in acute lymphoblastic leukemia.
Int J Mol Sci. 21:10542020. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zavridou M, Smilkou S, Tserpeli V, Sfika
A, Bournakis E, Strati A and Lianidou E: Development and analytical
validation of a 6-plex reverse transcription droplet digital PCR
assay for the absolute quantification of prostate cancer biomarkers
in circulating tumor cells of patients with metastatic
castration-resistant prostate cancer. Clin Chem. 68:1323–1335.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
De Luca G, Cardinali B, Del Mastro L,
Lastraioli S, Carli F, Ferrarini M, Calin GA, Garuti A, Mazzitelli
C, Zupo S and Dono M: Optimization of a WGA-free molecular
tagging-based NGS protocol for CTCs mutational profiling. Int J Mol
Sci. 21:43642020. View Article : Google Scholar : PubMed/NCBI
|