1. Introduction
The ARID1A gene
The AT-rich interactive domain-containing protein 1A
(ARID1A) gene is located on chromosome 1p36.11, a genetic site
often found to be deleted in a wide range of human cancers
(1). It contains 20 exons and
encodes two functionally identical 2285 and 2086 amino acid
isoforms of the ARID1A protein, also known as Brahma-related
associated factor 250a (BAF 250a), SWI/SNF-related
matrix-associated actin-dependent regulators of chromatin factor 1
(SMARCF1) or p270 (2,3). ARID1A is a large nucleocytoplasmic
protein of 250 kDa, expressed in almost all tissues, whose
stability depends on its cellular location (4-6).
Nuclear ARID1A is significantly more unstable than its cytoplasmic
counterpart, as it degrades rapidly, dependent on the
ubiquitin-proteasome system of the nucleus, demonstrating
significant fluctuations of expression during the cell cycle
(4). Normally, the accumulation
of the produced protein is detected during the G0-G1 phase of the
cell cycle, while a strong reduction occurs during the G2-S phase
(7).
The first significant evidence showing correlation
between tumourigenicity and the reduced expression of ARID1A
protein emerged in the middle of the past decade as the result of a
cancer profiling array containing complementary cDNA from various
tumour tissue cells and simultaneous northern, Southern and western
blot analyses of several human cancer cell lines (6). In 2010, two next generation
sequencing-based studies of highly aggressive ovarian cancers
revealed a high frequency of inactivating ARID1A mutations
(8,9), thus initiating further investigation
and confirmation of its reliable (bona fide) tumour suppressor role
in a wide range of malignancies (10-13).
Nowadays, it is considered that inactivating mutations of the gene
affect the biological behaviour of tumours; hence, recent clinical
studies examine their prognostic impact on the clinical outcome of
cancer (14).
ARID1A protein and SWI/SNF
complexes
ARID1A is a member of a large family of proteins
that contain a characteristic ARID domain of approximately 100
amino acids, which binds DNA fragments rich in adenine-thymine (AT)
and was originally discovered in 1995 in the Drosophila dead
ringer protein and in the murine B cell-specific transcription
factor Bright (5,15-17).
Later, researchers demonstrated the presence of at least fourteen
human homologous proteins, without sequential specificity
requirement during DNA binding, including the ARID1A and the
encoded by the paralog gene ARID1B protein, that are mutually
exclusive, found in cells at a 3.5:1 ratio, basic subunits of the
BAF subfamily of human switching mate/sucrose non-fermenting
(SWI/SNF), ATP-dependent chromatin remodeling complexes (6,17-19).
Phylogenetically conserved SWI/SNF protein
complexes, whose name derives from the type of yeast
Saccharomyces cerevisiae in which they were discovered
(20,21), are employed by transcriptional
activators of the genes in order to reconstruct chromatin and break
its structural constraints that prevent transcriptional proteins
from accessing the genome (22-25).
Using the energy released from the hydrolysis of ATP, the SWI/SNF
mobilize histone octamers and interrupt their interactions with
nucleosomal DNA, releasing the transcribed part of the helix
(26-29),
while altering the sensitivity of the restructured nucleoprotein to
the digestive activity of nucleases (30) and enhancing the affinities of the
gene promoters with the TATA-box-binding-protein (TBP) and the
basic translation machinery (31). The role of SWI/SNF is considered
to be critical for the regulation of gene expression, cellular
proliferation, apoptosis, differentiation and the repair of genetic
material (32-34).
The majority of the one hundred members of the
hSWI/SNF family, alias BAF or SMARC, depending on the type of cell
in which they are contained, may consist of combinations of 8-15
protein subunits and their isoforms, encoded in total by 26 genes,
whose mutations were found to be involved in 20% of all human
cancers (19,23,32,35-38).
The main structural feature and catalytic trunk of the complexes is
the ATPase, belonging to the superfamily II of helicases that
converts the chemical energy of one hydrolysed ATP molecule into
mechanical motion of 1 bp step along the DNA double helix (2,24,39). Based on the type of helicase, BRM
(SMARCA2) or BRG-1 (SMARCA4), by 74% identical to each other,
hSWI/SNF are divided into two corresponding subfamilies: hSWI/SNF-A
(BAF) and hSWI/SNF-B (PBAF) (2,40-42).
ARID1A is the largest, non-catalytic BAF subunit,
with the main property of conferring target specificity on the
complex and directing the ATPase activity, as the ARID domain binds
across to at least 50 bp of specific nucleosomal DNA constituting
the origin of chromatin remodeling (21,40,43,44). Its guiding effect, according to
the prevailing theory, is attributed on the one hand to the
interactions of the protein with the transcription factors, the
hormone nuclear receptors and the p53, p21 (CDKN1A), SMAD3
proteins, via the C-terminal peptide-rich binding loci
(LXXLL-leucin rich motifs), while on the other hand to the ARID
domain-mediated high affinity between chromatin and SWI/SNF
(4,36,40,45,46). Recently, ARID1A has also been
found to be indirectly involved in the modification of histones, by
binding to histone H2B as E3 ligase of ubiquitin (23).
Epigenetic regulation of ARID1A
expression in physiological cell processes
Effective cellular homeostasis, normal development
and tissue-specific differentiation in multicellular organisms
require the translation of the same genotype into different
phenotypes and depend on the epigenetic regulation of gene
expression, that determines the cell identity and establishes
heritable, not associated with the DNA sequence expression patterns
through a complex system of mechanisms, including DNA methylation,
histone modifications and miRNA inhibition (47-49).
Although the cell-specific molecular mechanisms mediating
transcriptional responses to environmental and developmental
signals remain poorly understood, ARID1A as a cell cycle regulator
that affects cell growth and differentiation, undergoes spatial and
temporal epigenetic modifications, depending on the cell type and
the developmental process (50,51).
Sun et al previously reported that ARID1A
protein was absent in neonatal mouse liver until the tenth day of
life, thus allowing rapid cell proliferation, while its expression
was physiologically downregulated following surgically- and
chemically-induced injuries in mouse models, promoting liver
regeneration and ear hole wound healing (52). Similar to tissue regeneration,
embryonic development requires unique gene expression patterns that
facilitate the reorganization of tissue structural design (53). In fact, the presence of ARID1A
protein in the nucleus of mouse embryonic stem cells has been
proven to be essential for their differentiation, pluripotency and
early germ-layer formation, by coordinating the expression of key
developmental and pluripotent genes (54,55). Similarly, Han et al
demonstrated that the universal expression of ARID1A across
different lineages of mouse hematopoietic stem cells was
determinant for their frequency and function, regulating the
production of mature blood cells, while the gene expression was
relatively lower in mature myeloid cells (56). As regards mouse cardiogenesis and
cardiac progenitor cell differentiation, distinct gene expression
patterns have been observed for BAF complex subunits (57). Specifically, ARID1A has been shown
to be expressed to a great extent in the early developing heart, in
order to selectively control the differentiation of second heart
field cardiac progenitor cells into beating cardiomyocytes,
although it is downregulated during the development and initiation
of cardiac trabeculation (58).
Furthermore, the epigenetic regulation of ARID1A in response to DNA
damage seems to play a key role in DNA repair and genome integrity
maintenance. As a matter of fact, ARID1A appears accumulated in DNA
double-strand breaks sites, recruited through its interactions with
the ataxia telangiectasia and RAD3-related protein (ATR), in order
to support and diffuse damage signals within mammalian cells, and
enable the access of the non-homologous end joining (NHEJ)
pathway-related repair proteins to the break sites (59,60).
Abnormal epigenetic regulation of
ARID1A expression
Wiegand et al detected the loss of ARID1A
expression, not attributable to gene mutations in 11% of ovarian
clear cell carcinomas and 9% of endometrioid carcinomas, raising a
matter of epigenetic silencing (9), which also leads to deviated
chromatin remodeling, and subsequently to the deregulated
expression of 99 target genes involved in carcinogenesis (61). Considering that almost 40% of the
human gene-promoters incorporate regions of several kb, rich in
cytosines preceding guanines commonly called CpG islands, whose
methylation carried out by DNA methyltransferases (DNMTs) represses
transcription (62,63), it is not surprising that among the
known epigenetic mechanisms, the altered patterns of DNA
methylation silencing established tumour suppressor genes have been
recognised as a consistent molecular characteristic of human
tumours since 1991, and are currently regarded as the most
important epigenetic mark, critically involved in tumourigenesis
(64-67).
Aiming to investigate the underlying reasons for the
ARID1A low mRNA expression in invasive breast cancers, Zhang et
al demonstrated that it was not associated with genetic
alterations and reported that 86.45% of low expression patients
exhibited a >2-fold aberrant increase in ARID1A promoter
methylation, often accompanied by repressive histone modifications,
while in 81.8% of patients with a high expression, the promoter
methylation was found to be decreased 2-fold (68). The key role of DNA
hypermethylation in ARID1A protein loss during gastric cancer
progression was demonstrated by the fact that the de-methylation of
gastric cancer cell lines restored the expression of ARID1A
(69), while a recent study,
focusing on decreased ARID1A expression during the pathogenesis of
endometriosis, revealed that oxidative stress stimulates the
expression of DNMT1 and causes ARID1A downregulation due to
promoter hypermethylation (70).
In the opposite direction, ARID1A promoter was found to be
unmethylated during the investigation of its methylation status in
The Cancer Genome Atlas (TCGA) Illumina Infinium dataset from 50
representative clear cell renal cell carcinomas (ccRCCs), drawing
the attention towards other mechanisms of epigenetic silencing
(71).
Concerning the suppressed ARID1A expression in
ccRCC, a recent study identified ARID1A as a direct downstream
target of microRNA (miRNA or miR)-144-3p, whose upregulation
provoked a significant decrease in ARID1A mRNA and protein levels
(72). miRNAs were identified for
the first time in 1993, as non-coding RNA molecules that regulated
larval transition and neuronal development in Caenorhabditis
elegans (73). Since then,
these small RNAs consisting of approximately 21-25-nucleotides have
been known to repress gene expression at the post-transcriptional
level through direct interactions with specific target mRNAs, to
regulate various developmental and physiological cellular processes
via different expression patterns (74) and when abnormally expressed, to
function as oncogenes or tumour suppressors, depending on the
cellular circumstance and the function of their target genes
(75).
Further studies have highlighted the implications of
overexpressed miRNAs targeting ARID1A by binding to its 3'
untranslated region (3'UTR). The investigation of the strong
association between Helicobacter pylori (H. Pylori)
infection and gastric carcinogenesis indicated that the H.
Pylori virulence factor CagA triggers the nuclear factor
(NF)-κΒ pathway and stimulates the expression of miR-223-3p, which
in turn functions as an oncomiRNA by downregulating ARID1A
(76). Two studies have reported
the ARID1A-associated oncogenic action of miR-31. The first one
demonstrated that the early upregulation of miR-31 due to EGFR
activation in head and neck squamous cell carcinoma (HNSCC) caused
enhanced oncogenicity and stemness by directly targeting ARID1A and
inhibiting its expression (77),
while the second identified miR-31 high levels as the cause of
ARID1A silencing in cervical cancer cell lines and tissues
(78). Yang et al also
studied cervical cancer tissues and detected the presence of
significantly increased miR-221 and miR-222 that simultaneously
bind to the ARID1A 3'UTR and inhibit its expression, thus inducing
cancer cell proliferation and invasion (79). Of note, the findings of Li et
al described an inverse regulatory axis of events, suggesting
that the loss of the expression of ARID1A in pancreatic cancer
cells upregulates miR-503, which in turn inhibits cell senescence
and promotes mutant KRASG12D induced tumourigenesis by targeting
another cell cycle regulator, the cyclin dependent kinase inhibitor
2A (CDKN2-A) (80).
ARID1A: Mutational profile and tumour
suppressing role
A total of 97% of inactivating ARID1A somatic
mutations that lead to the reduction or complete loss of protein
expression are nonsense, point and insertion or deletion frameshift
mutations, distinctive of tumour suppressor genes, that have been
found to be distributed throughout its length (3,9,12,13). The consequential abnormal mRNA
often carries premature stop codons and is translated into a
truncated protein, functionally degraded, either due to misfolding
or as it is partially incomplete, resulting in the disturbance of
the normal levels of nuclear ARID1A and the destabilization of
SWI/SNF complexes (46,81-83).
Mutations of tumour suppressor genes usually include alterations in
both alleles. However, in the case of ARID1A, one allele mutation
is sufficient to cause the loss of ARID1A expression in the
majority of heterozygous tumours, thus indicating genetic
haplodeficiency (3,8,10).
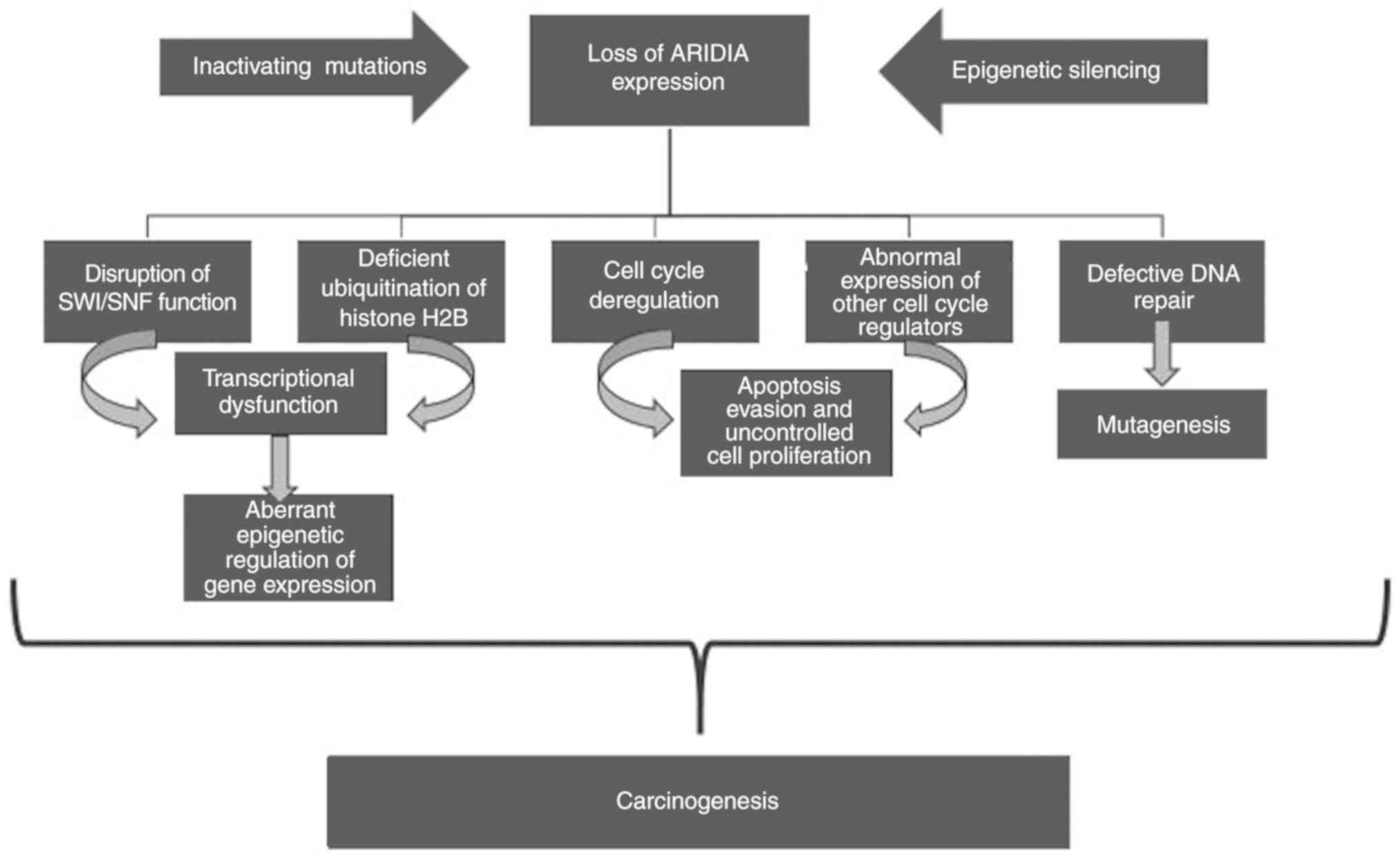
Fig. 1 illustrates
the effects of the loss of ARID1A expression and its impact on
carcinogenesis, although the precise mechanisms that triggers
cancer development have not yet been fully elucidated. ARID1A
expression disorders disrupt the function of SWI/SNF and the
chromatin remodeling mechanism, causing epigenetic abnormalities in
gene expression with severe consequent effects in the cell identity
and possible carcinogenicity (41,84-86).
In addition, functional studies of ARID1A have demonstrated that
its tumour-suppressive action lies both in the control of cellular
proliferation and in maintaining the integrity of the genetic
material, which is why both roles of guardian and caretaker of the
genome are attributed to it (3,13).
As regards the regulation of cell proliferation,
experimental studies of a wide range of ovarian, endometrial,
breast, stomach, liver and other cancer cell lines have
demonstrated that the loss of ARID1A expression, either due to a
mutation and epigenetic mechanism, or due to the in vitro
silencing of the gene promotes tumour expansion, while the
experimental restoration of the normal levels of the protein
suppresses cancer cell proliferation (11,13,87-90),
confirming that cell cycle inhibition and differentiation requires
a high concentration of ARID1A at the G0-G1
checkpoint (44). According to
recent research findings, mutant ARID1A diverts cell proliferation
by triggering the phosphatidylinositol 3-kinase
PI3K/serine-threonine kinase AKT/mammalian target of rapamycin mTOR
pathway (91-93),
affects the expression of other cell cycle regulators, such as the
c-MYC gene and promotes carcinogenicity in synergy with concurrent
mutations of other tumour suppressors such as TP53, PTEN and
SMARCB1 (44,46,94) or oncogenes, such as PIK3CA
(95,96).
As a genome caretaker, ARID1A contributes to the
prevention of chromosome and gene structural abnormalities through
its direct interactions as a SWI/SNF subunit with topoisomerase
IIα, which ensures the effective decatenation of sister chromatids
during meiotic anaphase, thus preventing potential aneuploidies,
polyploidies and sequence mutations (97). The contribution of the gene to DNA
repair is considered to be equally important, as ARID1A is involved
in the SWI/SNF recruitment of repair proteins, such as BRCA1 at the
lesion sites (98), while the
loss of its expression is significantly associated with
microsatellite instability (MSI) in endometrial, gastric and
colorectal cancers (89,91,99,100).
Aims of this bibliographic review
Although the tumour-suppressive role of ARID1A is
now considered to be unquestionable, the investigation of the
mutant gene's diagnostic significance and prognostic role in the
outcome of various malignancies, has yielded controversial results.
It is indicative that the loss of protein expression occurs in 6.5%
of patients with squamous cervical cancer and is associated with
significantly reduced overall survival (101,102), while it has no effect on the
prognosis of patients with ovarian clear cell carcinoma, in which
the gene mutation frequency reaches 55% (103,104). However, three recent
meta-analyses of published studies on gastrointestinal (81,105), gynaecological and urological
cancers (11) present a general
tendency to increased cancer-associated mortality in ARID1A
protein-negative patients, in comparison to those who are positive,
expressing the protein at normal levels. In particular, Luchini
et al associated the loss of ARID1A with increased
cancer-specific mortality following the meta-analysis of pooling
data from three studies, one on gynaecological and two on
urological cancers, while the gathered data from seven studies,
five on gynaecological and two on urological cancers revealed no
significant difference in cancer recurrence between ARID1A negative
and positive patients (11).
Given that abnormalities in ARID1A expression affect
both tumourigenesis and disease prognosis differently, this study
has been based on the review of relevant research articles
published in peer-reviewed journals, regardless of geographical
origins. The aim was thus to record the incidence of inactivated
ARID1A in human malignant tumours, to examine its diagnostic
significance and to determine its prognostic value in the outcome
of cancer, following a brief description of the most typical
methods applied to detect protein expression abnormalities and gene
mutations.
2. Summary of the materials and methods used
for analysis in previous studies
Analytical samples
The samples mainly analysed by researchers were
derived from contemporaneous or archived primary tumour resections,
originating from cancer patients regardless of sex, nationality and
age, whereas adjacent healthy tissue samples were used as negative
controls. Basic prerequisites for inclusion were the identification
of the histopathological type, the grading and the staging of the
tumours, according to the World Health Organization classification
(WHO), the TNM Staging System of the American Joint Committee on
Cancer (AJCC) and the International Federation of Gynecology and
Obstetrics (FIGO) guidelines in cases of gynaecological cancers,
while the pre-excision subjection to chemotherapy or preoperative
radiotherapy was the most frequently used exclusion criterion
(106-113).
Depending on the method selected for the detection
of ARID1A mutations and for the examination of protein expression
in the cell nuclei, the majority of the samples were fresh-frozen
tissues and formalin-fixed, paraffin-embedded histological
preparations (88,111,114,115), while in some cases as subject of
the study were selected cancer cell lines of various tissues
(41,46,90,116). Exceptionally, the ARID1A
expression levels in glioma patients were determined in blood serum
due to the intracranial tumour (117).
Survival data
In order to investigate the prognostic role of the
gene, the survival data of respective patients in previous studies
were collected after clinical follow-up for an average of a
five-year period (102,107,117). Among the time parameters
analysed were overall survival (OS), progression-free survival
(PFS) and cancer recurrence, corresponding to the time interval
from the date of diagnosis or ablation until up to documented
death, to the noticeable worsening of the disease and to recurrent
cancer diagnosis (46,111,112,115,118).
Immunohistochemistry (IHC)
The most commonly used methodology for controlling
nuclear ARID1A expression in cancer tissues was found to be IHC,
which was carried out by the deparaffinisation and rehydration of
whole tissue sections or specific microarrays, 4-5-µm-thick
(82,88,107,109,110,115,119). Usually, the preparations were
initially immersed in antigen retrieval solution to amplify the
imminent immune complex and after the endogenous peroxidase and the
non-specific background were blocked, the tissues were incubated
with primary, monoclonal mouse anti-ARID1A antibody or polyclonal
rabbit anti-ARID1A, followed by secondary antibody labelled with
horseradish peroxidase (46,82,108,110-112,119).
The immunoreactive signal was most frequently amplified with
diaminobenzidine followed by competitive nuclear haematoxylin
staining, while the evaluation and yield of the tissue
immunoreactivity score were based on the percentage extent of the
immunohistochemical expression and the intensity of staining of the
ARID1A-positive nuclei (41,108,110,115).
ARID1A reverse
transcription-quantitative PCR (RT-qPCR) and western blot
analysis
Along with the immunohistochemical method, the
expression levels of ARID1A messenger RNA (mRNA) were also examined
by RT-qPCR, following the isolation of total RNA from fresh
tissues, most commonly using ΤRIzol reagent and following
purification with the RNeasy mini kit (82,88,111,118). RNA was processed with the TaqMan
Reverse Transcription Reagents kit and RT-qPCR of the complementary
DNA (cDNA) was performed on a 7900H Fast Real Time PCR System with
a forward 5'→3' primer (CCCCTCAATGACCTCCAGTA) and a reverse 3'→5'
primer (ATCCCTGATGTGCTCACTCC) (88,107).
Equally common, for the validation of the IHC and
RT-qPCR analyses, western blot analysis has been used to detect
potential aberrations in ARID1A expression levels in cells,
following extraction and measurement of cellular proteins by using
the T-PER reagent and the BCA Assay kit, respectively (46,88,107,116,118). Subsequently, in previous
studies, polyacrylamide gel electrophoresis was performed in order
to separate the proteins and transfer them to a nitrocellulose
membrane where the immunoblotting was carried out using a primary
anti-ARID1A antibody and a secondary horseradish
peroxidase-labelled antibody (46,118).
Sporadically and due to specific research
requirements, additional laboratory methods have been used, such as
high-performance liquid chromatography (HPLC) (117), cDNA microarrays (41), northern and Southern blot analyses
(6).
Next-generation sequencing: The most
sophisticated and specific approach
The genetic analyser Illumina Genome Analyzer II has
been widely used in order to detect and accurately determine the
profile of ARID1A mutations in cancer genomes by applying various
NGS methods. Such methods are RNA sequencing (9), whole exome sequencing (WES)
(120-122)
and exome sequencing of specific genes (targeted NGS) (8,106,107,114).
Most commonly, following homogenisation and the
lysis of fresh tissue, genomic gDNA has been isolated using the
Qiagen Blood and Cell Culture Mini kit and following qualitative
examination by electrophoresis, it has been quantitated
photometrically (9,114,118). Using the DNA Sample Prep Reagent
Set 1 kit, the enzymatic fragmentation of gDNA, multiplex PCR with
T4 or Taq DNA polymerase and ligation of initiating (adapters) and
sample-identifiability (barcodes) sequences at the ends of
single-stranded fragments have been typically conducted by thermal
cycling, in order to finalize the library according to the Illumina
protocol and capture the coding sequences with the Human SureSelect
All Exon kit (8,107,114,122-124).
The programs ExonPrimer and Primer3Plus have been utilised for the
design of primers, whereas the PCR products have been purified with
the ExoSAP-IT PCR Purification kit (114).
To complete the sequencing of the DNA libraries, new
synthesis cycles have been performed by the addition of four
reverse endings nucleotides (A, T, G and C) labelled with different
fluorescent dyes, whose detection from the optical system of
Illumina Genome Analyzer II gave imaging data in the form of
chromatograms, subsequently processed by algorithms programs and
compared to reference genomes (Genome Browsers) (107,114,118,120,123,124).
Statistical analyses
Differences in the levels of ARID1A protein and mRNA
expression between cancer tissues and normal controls have been
analysed by paired-samples Students' t-tests, whereas for the
association between ARIDIA expression and the clinicopathological
characteristics of tumours, depending on the type of findings,
various methods have been applied, such as the χ2 test,
Fisher's exact test, and non-parametric McNemar, Wilcoxon and
Kruskal-Wallis techniques (82,88,111,112,117). The associations between
continuous variables have been evaluated by a Spearman's
correlation coefficient (82).
Statistically significant differences were considered data
presenting a value of P<0.05 (88,112,115,119). As regards the findings of the
NGS methods, the Benjamini-Hochberg multiple test correction method
was used to estimate the false discovery rate adaptive P-values
(106,120,122).
Survival data analyses have been performed using
Kaplan-Meier survival curves, evaluated against the log-rank test,
while the Cox proportional hazard model was used for the
correlation between expression, survival and clinicopathological
characteristics (102,107,110,115,118,119).
3. Summary of the findings of previous
studies regarding ARID1A in cancer
Ovarian cancers
Genomic sequencing and the investigation of ARID1A
immunoreactivity in the most common ovarian tumour subtype, which
is ovarian clear cell carcinoma (OCCC), revealed ARID1A mutations
and loss of protein expression that ranged between 46-57 and
41-62%, respectively with the exception of sporadic deviations
(8,9,96,103,104,113,119,125-133).
Mutations in the specific genomic area of interest have also been
identified in 30 and 21% of endometriosis-associated ovarian
cancers (EnAOCs) (9,113), while protein loss ranges between
31-55% (9,103,126,127,131,134). Zero percentages of mutations and
expression loss have been detected in high-grade serum
adenocarcinoma (HG-SAC) and low-grade serum adenocarcinoma (LG-SAC)
(9,87,132), whereas in the rare cases of
adenosquamous carcinoma (ASQ), adenosarcoma (ASA) and ovarian
atypical proliferative seromucinous tumours (OAPSMT), high
frequencies of ARID1A mutations have been attributed to the limited
number of samples (126,135). As regards mucinous
adenocarcinoma (MAC), two immunoreactivity studies have revealed
the loss of ARID1A expression in 0 and 27% of samples respectively,
while the sequence analysis detected 0% mutations (87,131). The analytical findings obtained
from the studies of all ovarian cancer subtypes are presented in
Table I.
 | Table ILoss of ARID1A protein expression and
gene mutation frequencies in histological subtypes of invasive
epithelial ovarian cancers including OCCC, EnAOC, SAC, HG-SAC,
LG-SAC, MAC, ASQ, ASA and OAPSMT. |
Table I
Loss of ARID1A protein expression and
gene mutation frequencies in histological subtypes of invasive
epithelial ovarian cancers including OCCC, EnAOC, SAC, HG-SAC,
LG-SAC, MAC, ASQ, ASA and OAPSMT.
| Author/(Refs.) | Cancer subtype | Protein loss
(%) | Mutation (%) |
|---|
| Wiegand et
al (9) | OCCC | 55/132(42) | 55/119(46) |
| | EnAOC | 39/125(31) | 10/33(30) |
| | HG-SAC | 12/198(6) | 0/76 (0) |
| Jones et al
(8) | OCCC | N/A | 24/42(57) |
| Maeda et al
(128) | OCCC | 88/149(59) | 9/12(75) |
| Guan et al
(87) | HG-SAC | 0/221 (0) | 0/32 (0) |
| | LG-SAC | 0/15 (0) | 0/19 (0) |
| | MAC | 0/36 (0) | 0/5 (0) |
| Ayhan et al
(103) | OCCC | 18/24(75) | N/A |
| | EnAOC | 11/20(55) | N/A |
| Katagiri et
al (132) | OCCC | 9/60(15) | N/A |
| | HG-SAC | 0/17 (0) | N/A |
| Lowery et al
(127) | OCCC | 34/82(41) | N/A |
| | EnAOC | 62/130(48) | N/A |
| Samartzis et
al (133) | OCCC | 5/23(22) | N/A |
| | EnAOC | 13/28(46) | N/A |
| | SAC | 7/63(11) | N/A |
| | MAC | 4/15(27) | N/A |
| Wu et al
(135) | OAPSMT | 8/24(33) | N/A |
| Xiao et al
(96) | OCCC | 15/26(58) | N/A |
| Yamamoto et
al (130) | OCCC | 40/90(44) | N/A |
| Yamamoto et
al (104) | OCCC | 23/42(55) | N/A |
| Lai et al
(126) | OCCC | 20/40(50) | N/A |
| | EnAOC | 13/33(39) | N/A |
| | SAC | 2/4(50) | N/A |
| | ASQ | 1/1(100) | N/A |
| | ASA | 1/1(100) | N/A |
| Huang et al
(125) | OCCC | 35/68(51) | N/A |
| McConechy et
al (134) | LG-EnAOC | N/A | 9/30(30) |
| | HG-EnAOC | N/A | 0/3 (0) |
| Wiegand et
al (113) | OCCC | N/A | 17/31(55) |
| | EnAOC | N/A | 5/24(21) |
| | SAC | N/A | 0/35 (0) |
| Wu et al
(129) | OCCC | 115/191(50) | N/A |
| Itamochi et
al (119) | OCCC | 44/112(39) | N/A |
| | HG-SAC | 8/108(7) | N/A |
| Murakami et
al (131) | OCCC | 23/39(56) | 24/39(62) |
Four studies on the effects of the mutant gene on
the prognosis of ovarian cancer found no significant differences in
tumour progression, clinical status and OS between ARID1A
protein-positive and -negative patients (127,128,130,136). By contrast, Ayhan et al
associated the loss of ARID1A expression with cancer stages I and
II and Itamochi et al with a significant reduction of OS of
patients at these specific cancer stages (103,119). Parallel studies have reported a
simultaneous overstimulation of the PI3K/AKT/mTOR signalling
pathway, strong resistance to chemotherapy, reduced PFS, as well as
an unaffected OS of the ARID1A protein-negative patients at III and
IV cancer stages (104,125,132,137).
Endometrial and cervical cancers
Three studies reported ARID1A mutation frequencies
of 40-55 and 10% in the endometrial endometrioid carcinoma (EEC)
and endometrial serous carcinoma (ESC) subtypes of endometrial
cancer, respectively (87,138,139),
whereas the loss of protein expression ranged between 20-26% in
endometrial clear cell carcinoma (ECCC) (140-143),
19-34% in EEC (87,142-144),
0-18% in ESC (87,142,143) and 14% in endometrial
carcinosarcoma (ECS) (143). Two
immunohistochemical studies on cervical cancer subtypes
demonstrated loss of ARID1A protein in 24-31 and 6.5-16% of
cervical adenocarcinoma (CAC) and cervical squamous cell carcinoma
(CSQC), respectively (101,102). The analytical findings obtained
from the studies of endometrial and cervical cancer subtypes are
presented in Table II.
 | Table IILoss of ARID1A protein expression and
gene mutation frequencies in endometrial cancer subtypes including
EEC, ESC, ECCC and ECS, and in cervical cancer subtypes including
the most common CSQC and the rare CAC. |
Table II
Loss of ARID1A protein expression and
gene mutation frequencies in endometrial cancer subtypes including
EEC, ESC, ECCC and ECS, and in cervical cancer subtypes including
the most common CSQC and the rare CAC.
|
Authors/(Refs.) | Cancer subtype | Protein loss
(%) | Mutation (%) |
|---|
| Guan et al
(87) | EEC | 15/58(26) | 10/25(40) |
| | ESC | 0/17 (0) | N/A |
| Wiegand et
al (143) | EEC | 73/214(34) | N/A |
| | ESC | 17/95(18) | N/A |
| | ECCC | 6/23(26) | N/A |
| | ECS | 18/127(14) | N/A |
| Fadare et al
(140) | ECCC | 5/22(23) | N/A |
| Katagiri et
al (102) | CAC | 14/45(31) | N/A |
| | CSQC | 3/46 (6.5) | N/A |
| Liang et al
(139) | EEC | N/A | 82/186(44) |
| Cho et al
(101) | CAC | 6/25(24) | N/A |
| | CSQC | 19/116(16) | N/A |
| Fadare et al
(141) | ECCC | 10/50(20) | N/A |
| Kandoth et
al (138) | EEC | N/A | 73/186(55) |
| | ESC | N/A | 4/42(10) |
| Rahman et al
(144) | EEC | 27/111(24) | N/A |
| Werner et al
(142) | EEC | 84/436(19) | N/A |
| | ESC | 1/44(3) | N/A |
| | ECCC | 4/19(21) | N/A |
Two studies have reported the loss of ARID1A
immunoreactivity exclusively in stages III and IV of EEC without an
impact on OS or PFS survival (140,141). By contrast, two parallel studies
revealed reduced PFS due to resistance to chemotherapy and high
metastasis of EEC ARID1A protein-negative tumours at the early
stages (102,142). As for the investigation of the
prognostic value of the gene in the outcome of cervical cancer,
only one of the two relevant studies revealed a significant
reduction in the OS of ARID1A-deficient patients (101,102).
Breast cancers
Studies on ARID1A mutations frequency and the loss
of protein expression in unspecified breast cancer subtypes have
yielded widely variable results ranging between 4-37% (10,90,145) and 1-65% (41,87,90,108,143,146-148)
(Table III), while the findings
of three survival analyses converged with each other, pointing out
the significantly reduced OS and PFS survival of ARID1A
protein-negative patients (41,108,147).
 | Table IIILoss of ARID1A protein expression and
gene mutation frequencies in breast cancer. |
Table III
Loss of ARID1A protein expression and
gene mutation frequencies in breast cancer.
|
Authors/(Refs.) | Protein loss
(%) | Mutation (%) |
|---|
| Guan et al
(87) | 1/91(1) | N/A |
| Wiegand et
al (143) | 11/315(3) | N/A |
| Cornen et al
(145) | N/A | 95/256(37) |
| Jones et al
(10) | N/A | 4/114(4) |
| Mamo et al
(90) | 151/236(64) | 11/82(13) |
| Zhang et al
(146) | 63/112(56) | N/A |
| Zhao et al
(147) | 324/496(65) | N/A |
| Cho et al
(108) | 150/476 (31.5) | N/A |
| Takao et al
(41) | 63/127(50) | N/A |
| Ünçel et al
(148) | 123/92(42) | N/A |
The investigation of the correlation between ARID1A
expression and breast cancer clinicopathologic parameters have led
to different conclusions as well. Cornen et al associated
the low ARID1A expression with an advanced clinical stage and
high-grade invasive tumours, ER and PR negativity, HER2 positivity
and poor-prognosis molecular subtypes (145). Ünçel et al confirmed that
the loss of ARID1A was strongly associated with ER/PR negativity
and tumour aggressiveness, but also reported that no significant
association was found between ARID1A expression and molecular
subtypes of breast cancer (148). Another study linked the reduced
expression of ARID1A with ER/PR/HER2 triple-negative tumours, TP53
mutation and a higher Ki-67 labelling index, resulting in tumours
of a larger size and higher stage (146), while the contradictory findings
of Cho et al associated a low expression of ARID1A with
lymph node metastasis and an advanced pathological stage, but also
with a low histological grade, a low Ki-67 labelling index and a
negative p53 expression, features broadly recognized as indicators
of auspicious prognosis (108).
Takao et al reported that the partial loss of ARID1A
expression was associated with a poor prognosis and a worse PFS of
patients with invasive ductal carcinoma, whilst the severe protein
loss did not affect the prognosis (41). Notably, the following
comprehensive gene expression analysis of cultured cancer breast
cells revealed that the downregulation of ARID1A mRNA by 20% caused
an increased expression of the breast cancer-promoting gene,
RAB11FIP1, while the >50% deficiency led to decreased RAB11FIP1
protein levels (41). It is worth
mentioning that although 5-10% of breast cancer worldwide is
attributed to pathogenic variants of the breast cancer driver
genes, BRCA1/2 (149,150), to date, no association has been
reported between ARID1A expression and the BRCA status.
Gastric cancers
The genomic analyses of gastric cancers have
reported ARID1A mutations ranging between 8-29% (10,100,151), while the loss of protein
expression has been found in 11-51% of tumours, often associated
with MSI and Epstein-Barr infection (46,69,87,89,100,111,115,143,152-155)
(Table IV). Two studies have
reported that the loss of expression of ARID1A was unrelated to
gastric cancer clinical characteristics (115,154). On the contrary, Yan et al
associated the reduced ARID1A expression with CDH1 silencing and
subsequent decreased E-cadherin levels that enhance gastric cancer
migration and invasion, leading to local lymph node metastasis and
tumour infiltration (46).
Another study associated the loss of ARID1A expression with higher
T stage infiltration, but not with distant or lymph node metastasis
(111), while the findings of
Kim et al associated the loss of ARID1A with poorly
differentiated subtypes located in the upper third of the stomach,
showing frequent vascular invasion (89).
 | Table IVLoss of ARID1A protein expression and
gene mutation frequencies in gastric cancer. |
Table IV
Loss of ARID1A protein expression and
gene mutation frequencies in gastric cancer.
|
Authors/(Refs.) | Protein loss
(%) | Mutation (%) |
|---|
| Guan et al
(87) | 5/45(11) | N/A |
| Wang et al
(100) | 38/109(35) | 32/109(29) |
| Wiegand et
al (143) | 26/180(14) | N/A |
| Abe et al
(152) | 95/857(11) | N/A |
| Jones et al
(10) | N/A | 10/100(10) |
| Wang et al
(111) | 115/224(51) | N/A |
| Zang et al
(151) | N/A | 9/110(8) |
| Wiegand et
al (115) | 39/173 (22.5) | N/A |
| 2 cohorts | 16/80(20) | N/A |
| Yan et al
(46) | 44/183(24) | N/A |
| Ibarrola-Villava
et al (154) | 14/33(42) | N/A |
| Kim et al
(89) | 62/191 (32.5) | N/A |
| Han et al
(153) | 88/417(21) | N/A |
| Kim et al
(155) | 52/350(15) | N/A |
| Aso et al
(69) | 103/516(20) | N/A |
The strong positive association of ARID1A
deficiency with EBV positivity, high MSI and the loss of mismatch
repair (MMR) protein expression has been consistently reported
(69,89,100,115,151-153).
In particular, Wang et al detected inactivating ARID1A
mutations and protein loss in 83% of gastric cancers with MSI and
in 73% of those carrying EBV infection (100), while two studies reported that
ARID1A deficiency was significantly more frequent in EBV-positive
and MLH1-negative gastric carcinomas, suggesting that the
EBV-associated promoter hypermethylation downregulates the
expression of both genome guardians (69,152). Of note, among the two MMR genes,
Kim et al confirmed a positive correlation between ARID1A
and MLH1 decreased levels in gastric tumours, but found no
association with MLH2 expression (89). Zang et al detected ARID1A
mutations in 8% of tumours characterised by concurrent MSI and
PIK3CA mutations (151), while
no significant association was reported between the loss of ARID1A
expression and HER2 amplification (115). The negative association between
ARID1A and TP53 mutations has been reported in four studies,
highlighting the mutual exclusivity of the two tumour suppressors
(69,100,115,153).
Two studies associated the decreased expression of
ARID1A and genetic alterations with a significantly improved OS and
prognosis (100,154), contrary to the findings of five
studies that reported a high tumour differentiation and a
significantly reduced PFS of ARID1A protein-negative patients
(46,111,152,153,155).
Liver, pancreatic, gallbladder,
intestinal, oesophageal, thyroid and lung cancers
Table V lists the
results of genomic and immunohistochemical analyses of liver,
pancreatic, gallbladder, intestinal, oesophageal, thyroid and lung
cancer tissues. Among all, the highest frequency of ARID1A
mutations of 39% was found in colorectal tumours (114) and the lowest of 8% in cancers of
the pancreas, the duodenum and the lung (10,118,156). Although protein loss was
detected mostly at a low rate 0-16% of malignancies (87,109,116,143,157-161),
two studies reported the loss of ARID1A immunoreactivity in 64% of
liver tumours (88) and in 25.8%
of colorectal cancers (112).
 | Table VLoss of ARID1A protein expression and
gene mutation frequencies in liver, pancreatic, gallbladder,
intestinal, oesophageal, thyroid and lung cancers. |
Table V
Loss of ARID1A protein expression and
gene mutation frequencies in liver, pancreatic, gallbladder,
intestinal, oesophageal, thyroid and lung cancers.
| Cancer type |
Authors/(Refs.) | Protein loss
(%) | Mutation (%) |
|---|
| Liver | Guan et al
(87) | 0/41 (0) | N/A |
| | Fujimoto et
al (157) | N/A | 15/147(10) |
| | Guichard et
al (158) | 20/125(16) | 20/125(16) |
| | He et al
(88) | 41/64(64) | N/A |
| Pancreas | Guan et al
(87) | 4/48(8) | N/A |
| | Wiegand et
al (143) | 5/85(6) | N/A |
| | Jones et al
(10) | N/A | 10/119(8) |
| | Zhang et al
(116) | 10/73(7) | N/A |
| Gallbladder | Guan et al
(87) | 2/27(7) | N/A |
| | Jiao et al
(121) | N/A | 9/64(14) |
| | Ahn et al
(106) | N/A | 25/183(14) |
| Colorectal | Guan et al
(87) | 2/49(4) | N/A |
| | Wiegand et
al (143) | 2/250(1) | N/A |
| | Jones et al
(10) | N/A | 12/119(10) |
| | Cajuso et al
(114) | N/A | 18/46(39) |
| | Wei et al
(112) | 54/209 (25.8) | N/A |
| | Sen et al
(161) | 24/164 (14.6) | N/A |
| | Lee et al
(160) | 12/196(6) | N/A |
| Ampulla of
vater | Nastase et
al (118) | N/A | 4/49 (8.2) |
| Duodenum | Nastase et
al (118) | N/A | 2/6(33) |
| Oesophagus | Streppel et
al (159) | 12/98(12) | 3/20(15) |
| | Drage et al
(109) | 12/120(10) | N/A |
| Thyroid | Wiegand et
al (143) | 5/35(14) | N/A |
| Lung | Imielinski et
al (156) | N/A | 15/183(8) |
As regards colorectal cancer, Wei et al
reported that the loss of ARID1A expression was significantly
associated with a late TNM stage, distant metastasis and poor
pathological differentiation, but did not seem to affect the tumour
T stage, size or location (112), in partial accordance with the
findings of Lee et al that associated the loss of ARID1A
expression with expanding tumour borders, but negative lymphatic
invasion (160). In parallel,
two studies reported the positive association between ARID1A
mutations and MSI colorectal cancers (10,114), while Cajuso et al, based
on a limited number of study samples, questioned the relevance of
mutual exclusiveness between ARID1A and TP53 mutations in
colorectal cancer (114). Of
note, the molecular analyses of two KRAS wild-type and two
KRASG13D colorectal cancer cell lines, subjected to
CRISPR/Cas9-mediated ARID1A deletion, led Sen et al to the
conclusion that KRAS mutated colorectal cancer cells are
particularly dependent on ARID1A presence, as their proliferation
proved to be severely impaired by its absence, due to the decreased
activity of specific enhancers bound by ARID1A and the AP1
transcription factors, that subsequently caused the down regulation
of 48 genes (161).
Survival analyses have not detected a statistically
significant difference in OS between ARID1A protein-positive and
-negative patients with oesophageal, pancreatic and colorectal
cancers (109,112,116). On the contrary, in the case of
hepatocellular carcinoma, the loss of expression has been shown to
be associated with a poor prognosis and high metastaticity of the
tumour (88), whereas in the case
of cancer of the ampulla of Vater, it was found that the mutation
of the gene is associated with an increased overall survival
(118).
Bladder, renal and prostate
cancers
Three sequencing analyses of bladder cancer genomes
have revealed a mean ARID1A mutation frequency of 13.5% (107,120,162), while IHC analyses have detected
the loss of protein expression in 0, 2 and 67% of renal
malignancies (82,87,143), in 13% of bladder tumours
(110) and in 8% of prostate
cancers (10) (Table VI). Two survival studies have
found that ARID1A mutation is associated with the reduced OS and
PFS of bladder and kidney cancer patients (82,107), while Faraj et al reported
that ARID1A protein loss was associated with the first stage of
bladder cancer and positively affected prognosis (110).
 | Table VILoss of ARID1A protein expression and
gene mutation frequencies in bladder, renal and prostate
cancers. |
Table VI
Loss of ARID1A protein expression and
gene mutation frequencies in bladder, renal and prostate
cancers.
| Cancer type |
Authors/(Refs.) | Protein loss
(%) | Mutation (%) |
|---|
| Bladder | Gui et al
(120) | N/A | 13/97(13) |
| | Balbás-Martínez
et al (107) | N/A | 6/52(12) |
| | Guo et al
(162) | N/A | 15/99(15) |
| | Faraj et al
(110) | 16/122(13) | N/A |
| Renal | Guan et al
(87) | 0/73 (0) | N/A |
| | Wiegand et
al (143) | 1/58(2) | N/A |
| | Lichner et
al (82) | 53/79(67) | N/A |
| Prostate | Jones et al
(10) | 2/23(8) | N/A |
Nervous and lymphatic system
cancers
ARID1A mutations detected in myeloblastoma,
neuroblastoma, Burkitt lymphoma and Waldenstrӧm macroglobulinemia
genomes ranged between 2-17% (10,122,163,164), while the loss of protein
expression was found in 75% of the glioma serum samples (117) (Table VII). The parallel investigation
of the effect of the inactivated gene on the prognosis of patients
with neuroblastoma and glioma revealed a significant reduction in
the OS of ARID1A protein-negative patients (117,122).
 | Table VIILoss of ARID1A protein expression and
gene mutation frequencies in nervous (myeloblastoma, neuroblastoma,
glioma) and lymphatic (macroglobulinemia Waldenstrӧm, Burkitt
lymphoma) system cancers. |
Table VII
Loss of ARID1A protein expression and
gene mutation frequencies in nervous (myeloblastoma, neuroblastoma,
glioma) and lymphatic (macroglobulinemia Waldenstrӧm, Burkitt
lymphoma) system cancers.
| Cancer type |
Authors/(Refs.) | Protein loss
(%) | Mutation (%) |
|---|
| Myeloblastoma | Jones et al
(10) | N/A | 3/125(2) |
| Neuroblastoma | Sausen et al
(122) | N/A | 4/71(6) |
| Glioma | Tan et al
(117) | 62/83(75) | N/A |
| Macroglobulinemia
Waldenstrӧm | Treon et al
(163) | N/A | 5/30(17) |
| Burkitt
lymphoma | Giulino-Roth et
al (164) | N/A | 5/29(17) |
4. Discussion
The dynamic remodeling of chromatin is a key
mechanism for proper cellular function, as it enables the
transcription, replication and repair of genetic material,
regulates gene expression, while at the same time prevents
chromosome breakage and supports the exact DNA distribution during
cell divisions, thus ensuring the preservation of the cellular
phenotype across generations (165). In order to respond to
environmental stimuli and developmental signals that require the
activation or suppression of particular genes, chromatin's
structure is appropriately remodeled by the SWI/SNF protein
complexes, which bind to the onset loci of the upcoming
transcriptional activity under the guidance of the ARID1A subunit
and rearrange the array of nucleosomes along the double stranded
helix length (32,37). The mutation of the ARID1A gene
diverts the remodeling mechanism and besides its epigenetic
implications, results involved in carcinogenesis in many other ways
that have not been fully elucidated yet (4).
Laboratory studies investigating the frequency of
ARID1A mutations and the loss of homologous protein expression have
been conducted worldwide over the past decade and have spread to
almost the entire spectrum of human cancers. Most of these have
focused on the association between oncogenesis and gene
inactivation in gynaecological and gastrointestinal cancers,
revealing a substantive association, while concrete-positive
indications have been reported by the majority of malignant
neoplasms studies consolidating its tumour-suppressive role.
However, the contemporary scientific community has not precisely
defined yet the diagnostic significance and the clinical
implications of the mutant ARID1A, as the findings of genomic and
immunohistochemical analyses show a high degree of heterogeneity
among the various cancer tissues, whereas wide ranges of variations
are observed even between cancers of the same type and subtype
(13).
Highest mutation frequencies, often >50%, have
been recorded in ovarian, endometrial and breast cancer tissues,
attributable to the hormone-dependent nature of these specific
malignancies and the interactions of the gene with nuclear hormone
receptors during transcriptional regulation (12). Mutation frequencies ranging
between 8-39% have been detected in gastrointestinal cancers
(10,100,114,118,151), while <10% rates have been
detected in lung, prostate, pancreatic and intracranial tumours
(10,122,156). The findings of three
immunohistochemistry-based studies of the same ovarian tumour
subtype OCCC, reporting the loss of ARID1A expression in 75, 22 and
15% of the samples (103,131,132),
are indicative of the wide heterogeneity between the results of
various studies, although the reliability of the IHC method is
unquestionable. In fact, a study based on the concurrent
comparative analyses of ARID1A mutational status and
immunoreactivity reported the concordance of results in 91% of the
OCCC ovarian cancers examined, demonstrating 100% sensitivity and
66% specificity of the IHC method (128).
The findings of the relatively limited number of
survival analyses conducted to investigate the prognostic value of
the ARID1A gene in the outcome of cancer treatment appear to be
controversial as well. Actually, relevant studies have reported
adverse, beneficial or absolutely no effect of protein loss on the
biological behaviour and metastaticity of tumours, the outcome of
chemotherapy, the recurrence of cancer, the PFS and the OS of
cancer patients, thus clarifying that the abnormal expression of
ARID1A affects the prognosis in different ways, mainly depending on
the type, stage and grade of the tumour (107,110,117,136).
Among the factors that have been found to influence
the prognosis of specific malignancies, the existence of
concomitant PIK3CA, TP53, EZH2 and KRAS mutations has been proven
to be of great significance and has led to the deterioration of
ARID1A-deficient gynaecological cancers, while in gastric
malignancies the synergy of ARID1A protein loss with the expression
of E-cadherin, MSI and the simultaneous presence of Epstein-Barr
virus was detected in highly aggressive tumours (46,89,100,152,153). In addition to the
clinicopathological characteristics of the tumours, the
heterogeneity of the research findings is attributed to other
factors as well, such as the occasionally limited number of
samples, the use of different antibodies in the immunoassays
conducted, the general characteristics, the clinical condition and
the racial origin of the patients (12,81).
5. Conclusion and future therapeutic
perspectives
The enlightenment of the diagnostic significance
and the prognostic role of the ARID1A gene in cancer entails the
exclusion of the specific parameters, which embroil the research
findings, hence requires new highly specialized research approaches
to tumours and patients with common clinicopathological and
anthropological characteristics (11). The identification of the
association between the mutational status of the gene, the stages
and the grading of malignant tumours can lead on the one hand to
the development of an early diagnosis methodology and on the other
hand to the identification and distinction between low and high
risk patients in order to be subjected to a more personalized and
targeted therapeutic intervention, thus avoiding the effects of
over-treatment (11,12,113).
Undoubtedly, the full decoding of the ARID1A tumour
suppressor mechanism and the development of targeted gene therapy
are part of the future field of investigation. Moreover, in
exploiting the proven interaction of the gene with the
PI3K/AKT/mTOR intracellular signalling pathway, whose diversion
disrupts cell proliferation and leads to carcinogenesis,
contemporary experimental studies have attempted to develop novel
chemotherapeutic regimens for the treatment of ARID1A
protein-negative tumours, including inhibitors of the kinases PI3K
and AKT, among which buparlisib and the combination of
MK-2206/perifosine were the most effective (92,93), in contrast to cytostatic cisplatin
which proved to be ineffective in suppressing ARID1A negative,
ovarian cancer cell lines (14).
Parallel therapeutic approaches are currently
focused on the epigenetic aberrations and the deficient DNA damage
responses caused by the loss of ARID1A expression, aiming to
identify a potential ARID1A-synthetic lethality target. Recently,
Bitler et al reported that the pharmacological inhibition of
the histone deacetylase 6 (HDAC6) in mouse models of ARID1A-mutated
ovarian tumours suspended the tumour expansion and improved the
survival of the treated mice (166), while another study reported the
synthetic lethality between EZH2 methyltransferase inhibition and
ARID1A-mutated ovarian cancer cell lines (167). According to the in vitro
and in vivo findings of Shen et al, PARP inhibitors,
already known to be selectively lethal to cells carrying BRCA1 or
BRCA2 mutations, two proteins involved in the DNA damage signalling
pathway similarly to ARID1A, represent a potential therapeutic
strategy for ARID1A mutant tumours as well (60).
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
VB conceived and ENP designed this review article.
ENP and VB collected and evaluated the research articles included
in this review. VB supervised the project. ENP wrote the manuscript
and designed the figure. VB revised the manuscript critically for
important intellectual content. All authors agreed to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of any part of the work are
appropriately investigated and resolved. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bagchi A and Mills AA: The quest for the
1p36 tumor suppressor. Cancer Res. 68:2551–2556. 2008.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Reisman D, Glaros S and Thompson EA: The
SWI/SNF complex and cancer. Oncogene. 28:1653–1668. 2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wu JN and Roberts CW: ARID1A mutations in
cancer: Another epigenetic tumor suppressor? Cancer Discov.
3:35–43. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Guan B, Gao M, Wu CH, Wang TL and Shih IM:
Functional analysis of in-frame indel ARID1A mutations reveals new
regulatory mechanisms of its tumor suppressor functions. Neoplasia.
14:986–993. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Samartzis EP, Noske A, Dedes KJ, Fink D
and Imesch P: ARID1A mutations and PI3K/AKT pathway alterations in
endometriosis and endometriosis-associated ovarian carcinomas. Int
J Mol Sci. 14:18824–18849. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang X, Nagl NG Jr, Flowers S, Zweitzig D,
Dallas PB and Moran E: Expression of p270 (ARID1A), a component of
human SWI/SNF complexes, in human tumors. Int J Cancer.
112(636)2004.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Flores-Alcantar A, Gonzales-Sandoval A,
Escalante-Alcalde D and Lomeli H: Dynamics of expression of ARID1A
and ARID1B subunits in mouse embryos and in cells during the cell
cycle. Cell Tissue Res. 345:137–148. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Jones S, Wang TL, Shih IeM, Mao TL,
Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr, Vogelstein B, et
al: Frequent mutations of chromatin remodeling gene ARID1A in
ovarian clear cell carcinoma. Science. 330:228–231. 2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wiegand KC, Shah SP, Al-Agha OM, Zhao Y,
Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et
al: ARID1A mutations in endometriosis-associated ovarian
carcinomas. N Engl J Med. 363:1532–1543. 2010.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jones S, Meng L, Parsons DW, Zhang X,
Wesseling J, Kristel P, Schmidt MK, Markowitz S, Yan H, Bigner D,
et al: Somatic mutations in the chromatin remodeling gene ARID1A
occur in several tumor types. Hum Mutat. 33:100–103.
2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Luchini C, Veronese N, Solmi M, Cho H, Kim
JH, Chou A, Gill AJ, Faraj SF, Chaux A, Netto GJ, et al: Prognostic
role and implications of mutation status of tumor suppressor gene
ARID1A in cancer: A systematic review and meta-analysis.
Oncotarget. 6:39088–39097. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mao TL and Shih IeM: The roles of ARID1A
in gynecologic cancer. J Gynecol Oncol. 24:376–381. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wu RC, Wang TL and Shih IeM: The emerging
roles of ARID1A in tumor suppression. Cancer Biol Ther. 15:655–664.
2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lyu C, Zhang Y, Zhou X and Lang J: ARID1A
gene silencing reduces the sensitivity of ovarian clear cell
carcinoma to cisplastin. Exp Ther Med. 12:4067–4071.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gregory SL, Kortschak DR, Kallionis B and
Saint R: Characterization of the dead ringer gene identifies a
novel, highly conserved family of sequence-specific DNA-binding
proteins. Mol Cell Biol. 16:792–799. 1996.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Herrscher RF, Kaplan MH, Lelsz DL, Das C,
Scheuermann R and Tucker PW: The immunoglobulin heavy-chain
matrix-associating regions are bound by Bright: A B cell-specific
trans-activator that describes a new DNA-binding protein family.
Genes Dev. 9:3067–3082. 1995.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Patsialou A, Wilsker D and Moran E:
DNA-binding properties of ARID family proteins. Nucleic Acids Res.
33:66–80. 2005.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Dallas PB, Pacchione S, Wilsker D, Bowrin
V, Kobayashi R and Moran E: The human SWI-SNF complex protein p270
is an ARID family member with non-sequence-specific DNA binding
activity. Mol Cell Biol. 20:3137–3146. 2000.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang X, Haswell JR and Roberts CW:
Molecular pathways: SWI/SNF (BAF) complexes are frequently mutated
in cancer-mechanisms and potential therapeutic insights. Clin
Cancer Res. 20:21–27. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Carlson M, Osmond BC and Botstein D:
Mutants of yeast defective in sucrose utilization. Genetics.
98:25–40. 1981.PubMed/NCBI
|
|
21
|
Wilsker D, Patsialou A, Zumbrun SD, Kim S,
Chen Y, Dallas PB and Moran E: The DNA-binding properties of the
ARID-containing subunits of yeast and mammalian SWI/SNF complexes.
Nucleic Acids Res. 32:1345–1353. 2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kwon H, Imbalzano AN, Khavari PA, Kingston
RE and Green MR: Nucleosome disruption and enhancement of activator
binding by a human SWI/SNF complex. Nature. 370:477–481.
1994.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li XS, Trojer P, Matsumura T, Treisman JE
and Tanese N: Mammalian SWI/SNF-a subunit BAF250/ARID1 is an E3
ubiquitin ligase that targets histone H2B. Mol Cell Biol.
30:1673–1688. 2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Narlicar GJ, Sundaramoorthy R and
Owen-Hughes T: Mechanisms and functions of ATP-dependent
chromatin-remodeling enzymes. Cell. 154:490–503. 2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Smith CL and Peterson CL: A conserved
Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin
remodeling. Mol Cell Biol. 25:5880–5892. 2005.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Becker PB: Nucleosome sliding: Facts and
fiction. EMBO J. 21:4749–4753. 2002.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cairns BR: Chromatin remodeling: Insights
and intrigue from single-molecule studies. Nat Struct Mol Biol.
14:989–996. 2007.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hargreaves DC and Crabtree GR:
ATP-dependent chromatin remodeling: Genetics, genomics and
mechanisms. Cell Res. 21:396–420. 2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Vignali M, Hassan AH, Neely KE and Workman
JL: ATP-dependent chromatin-remodeling complexes. Mol Cell Biol.
20:1899–1910. 2000.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Schnitzler G, Sif S and Kingston RE: Human
SWI/SNF interconverts a nucleosome between its base state and a
stable remodeled state. Cell. 94:17–27. 1998.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Imbalzano AN, Kwon H, Green MR and
Kingston RE: Facilitated binding of TATA-binding protein to
nucleosomal DNA. Nature. 370:481–485. 1994.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bartolomew B: Regulating the chromatin
landscape: Structural and mechanistic perspectives. Annu Rev
Biochem. 83:671–696. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jeong KW, Lee YH and Stallcup MR:
Recruitment of the SWI/SNF chromatin remodeling complex to steroid
hormone-regulated promoters by nuclear receptor coactivator
flightless-I. J Biol Chem. 284:29298–29309. 2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ronan JL, Wu W and Crabtree GR: From
neural development to cognition: Unexpected roles for chromatin.
Nat Rev Genet. 14:347–359. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Decristofaro MF, Betz BL, Rorie CJ,
Reisman DN, Wang W and Weissman BE: Characterization of SWI/SNF
protein expression in human breast cancer cell lines and other
malignancies. J Cell Physiol. 186:136–145. 2001.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Inoue H, Furukawa T, Giannakopoulos S,
Zhou S, King DS and Tanese N: Largest subunits of the human SWI/SNF
chromatin-remodeling complex promote transcriptional activation by
steroid hormone receptors. J Biol Chem. 277:41674–41685.
2002.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kadoch C, Hargreaves DC, Hodges C, Elias
L, Ho L, Ranish J and Crabtree GR: Proteomic and bioinformatic
analysis of mammalian SWI/SNF complexes identifies extensive roles
in human malignancy. Nat Genet. 45:592–601. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Keenen B, Qi H, Saladi SV, Yeung M and de
la Serna IL: Heterogeneous SWI/SNF chromatin remodeling complexes
promote expression of microphthalmia-associated transcription
factor target genes in melanoma. Oncogene. 29:81–92.
2010.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tang L, Nogales E and Ciferri C: Structure
and function of SWI/SNF chromatin remodeling complexes and
mechanistic implications for transcription. Prog Biophys Mol Biol.
102:122–128. 2010.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ,
Archer TK and Wang W: A specificity and targeting subunit of a
human SWI/SNF family-related chromatin-remodeling complex. Mol Cell
Biol. 20:8879–8888. 2000.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Takao C, Morikawa A, Ohkubo H, Kito Y,
Saigo K, Sakuratami T, Futamura M, Takeuchi T and Yoshida K:
Downregulation of ARID1A, a component of the SWI/SNF chromatin
remodeling complex, in breast cancer. J Cancer. 8:1–8.
2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Inoue H, Giannakopoulos S, Parkhurst CN,
Matsumura T, Kono EA, Furukawa T and Tanese N: Target genes of the
largest human SWI/SNF complex subunit control cell growth. Biochem
J. 434:83–92. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Dechassa ML, Zhang B, Horowitz-Scherer R,
Persinger J, Woodcock CL, Peterson CL and Bartholomew B:
Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol.
28:6010–6021. 2008.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Nagl NG Jr, Wang X, Patsialou A, Van Scoy
M and Moran E: Distinct mammalian SWI/SNF chromatin remodeling
complexes with opposing roles in cell-cycle control. EMBO J.
26:752–763. 2007.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang X, Nagl NG, Wilsker D, Van Scoy M,
Pacchione S, Yaciuk P, Dallas PB and Moran E: Two related ARID
family proteins are alternative subunits of human SWI/SNF
complexes. Biochem J. 383:319–325. 2004.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yan HB, Wang XF, Zhang Q, Tang ZQ, Jiang
YH, Fan HZ, Sun Y, Yang PY and Liu F: Reduced expression of the
chromatin remodeling gene ARID1A enhances gastric cancer cell
migration and invasion via downregulation of E-cadherin
transcription. Carcinogenesis. 35:867–876. 2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Saghafinia S, Mina M, Riggi N, Hanahan D
and Ciriello G: Pan-cancer landscape of aberrant DNA methylation
across human tumors. Cell Rep. 25:1066–1080.e8. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Kamińska K, Nalejska E, Kubiak M,
Wojtysiak J, Żołna Ł, Kowalewski J and Lewandowska MA: Prognostic
and predictive epigenetic biomarkers in oncology. Mol Diagn Ther.
23:83–95. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Tsai HC and Baylin SB: Cancer epigenetics:
Linking basic biology to clinical medicine. Cell Res. 21:502–517.
2011.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Heinz S, Romanoski CE, Benner C and Glass
CK: The selection and function of cell type-specific enhancers. Nat
Rev Mol Cell Biol. 16:144–154. 2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Tillo D, Kaplan N, Moore IK,
Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal
E and Hughes TR: High nucleosome occupancy is encoded at human
regulatory sequences. PLoS One. 5(e9129)2010.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Sun X, Chuang JC, Kanchwala M, Wu L, Celen
C, Li L, Liang H, Zhang S, Maples T, Nguyen LH, et al: Suppression
of the SWI/SNF component Arid1a promotes mammalian regeneration.
Cell Stem Cell. 18:456–466. 2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wu S, Zhang R and Bitler BG: Arid1a
controls tissue regeneration. Stem Cell Investig.
3(35)2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lei I, West J, Yan Z, Gao X, Fang P,
Dennis JH, Gnatovskiy L, Wang W, Kingston RE and Wang Z: BAF250a
protein regulates nucleosome occupancy and histone modifications in
priming embryonic stem cell differentiation. J Biol Chem.
290:19343–19352. 2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Gao X, Tate P, Hu P, Tjian R, Skarnes WC
and Wang Z: ES cell pluripotency and germ-layer formation require
the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad
Sci USA. 105:6656–6661. 2008.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Han L, Madan V, Mayakonda A, Dakle P, Woon
TW, Shyamsunder P, Nordin HBM, Cao Z, Sundaresan J, Lei I, et al:
Chromatin remodeling mediated by ARID1A is indispensable for normal
hematopoiesis in mice. Leukemia. 33:2291–2305. 2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Hota SK, Johnson JR, Verschueren E, Thomas
R, Blotnick AM, Zhu Y, Sun X, Pennacchio LA, Krogan NJ and Bruneau
BG: Dynamic BAF chromatin remodeling complex subunit inclusion
promotes temporally distinct gene expression programs in
cardiogenesis. Development. 146(pii: dev174086)2019.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Lei I, Gao X, Sham MH and Wang Z: SWI/SNF
protein component BAF250a regulates cardiac progenitor cell
differentiation by modulating chromatin accessibility during second
heart field development. J Biol Chem. 287:24255–24262.
2012.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Watanabe R, Ui A, Kanno S, Ogiwara H,
Nagase T, Kohno T and Yasui A: SWI/SNF factors required for
cellular resistance to DNA damage include ARID1A and ARID1B and
show interdependent protein stability. Cancer Res. 74:2465–2475.
2014.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Shen J, Peng Y, Wei L, Zhang W, Yang L,
Lan L, Kapoor P, Ju Z, Mo Q, Shih IM, et al: ARID1A deficiency
impairs the DNA damage checkpoint and sensitizes cells to PARP
inhibitors. Cancer Discov. 5:752–767. 2015.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Lakshminarasimhan R, Andreu-Vieyra C,
Lawrenson K, Duymich CE, Gayther SA, Liang G and Jones PA:
Down-regulation of ARID1A is sufficient to initiate neoplastic
transformation along with epigenetic reprogramming in
non-tumorigenic endometriotic cells. Cancer Lett. 401:11–19.
2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Miranda TB and Jones PA: DNA methylation:
The nuts and bolts of repression. J Cell Physiol. 213:384–390.
2007.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Jang HS, Shin WJ, Lee JE and Do JT: CpG
and Non-CpG methylation in epigenetic gene regulation and brain
function. Genes (Basel). 8(pii: E148)2017.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Herman JG, Latif F, Weng Y, Lerman MI,
Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM, et al:
Silencing of the VHL tumor-suppressor gene by DNA methylation in
renal carcinoma. Proc Natl Acad Sci USA. 91:9700–9704.
1994.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Baylin SB, Makos M, Wu JJ, Yen RW, De
Bustros A, Vertino P and Nelkin BD: Abnormal patterns of DNA
methylation in human neoplasia: Potential consequences for tumor
progression. Cancer Cells. 3:383–390. 1991.PubMed/NCBI
|
|
66
|
Makos M, Nelkin BD, Lerman MI, Latif F,
Zbar B and Baylin SB: Distinct hypermethylation patterns occur at
altered chromosome loci in human lung and colon cancer. Proc Natl
Acad Sci USA. 89:1929–1933. 1992.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Qu Y, Dang S and Hou P: Gene methylation
in gastric cancer. Clin Chim Acta. 424:53–65. 2013.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Zhang X, Sun Q, Shan M, Niu M, Liu T, Xia
B, Liang X, Wei W, Sun S, Zhang Y, et al: Promoter hypermethylation
of ARID1A gene is responsible for its low mRNA expression in many
invasive breast cancers. PLoS One. 8(e53931)2013.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Aso T, Uozaki H, Morita S, Kumagai A and
Watanabe M: Loss of ARID1A, ARID1B, and ARID2 expression during
progression of gastric cancer. Anticancer Res. 35:6819–6827.
2015.PubMed/NCBI
|
|
70
|
Xie H, Chen P, Huang HW, Liu LP and Zhao
F: Reactive oxygen species downregulate ARID1A expression via its
promoter methylation during the pathogenesis of endometriosis. Eur
Rev Med Pharmacol Sci. 21:4509–4515. 2017.PubMed/NCBI
|
|
71
|
Ibragimova I, Maradeo ME, Dulaimi E and
Cairns P: Aberrant promoter hypermethylation of PBRM1, BAP1, SETD2,
KDM6A and other chromatin-modifying genes is absent or rare in
clear cell RCC. Epigenetics. 8:486–493. 2013.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Xiao W, Lou N, Ruan H, Bao L, Xiong Z,
Yuan C, Tong J, Xu G, Zhou Y, Qu Y, et al: Mir-144-3p promotes cell
proliferation, metastasis, sunitinib resistance in clear cell renal
cell carcinoma by downregulating ARID1A. Cell Physiol Biochem.
43:2420–2433. 2017.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854.
1993.PubMed/NCBI View Article : Google Scholar
|
|
74
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531.
2004.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Yang F, Xu Y, Liu C, Ma C, Zou S, Xu X,
Jia J and Liu Z: NF-κB/miR-223-3p/ARID1A axis is involved in
Helicobacter pylori CagA-induced gastric carcinogenesis and
progression. Cell Death Dis. 9(12)2018.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Lu WC, Liu CJ, Tu HF, Chung YT, Yang CC,
Kao SY, Chang KW and Lin SC: miR-31 targets ARID1A and enhances the
oncogenicity and stemness of head and neck squamous cell carcinoma.
Oncotarget. 7:57254–57267. 2016.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Wang N, Zhou Y, Zheng L and Li H: MiR-31
is an independent prognostic factor and functions as an oncomir in
cervical cancer via targeting ARID1A. Gynecol Oncol. 134:129–137.
2014.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Yang Y, Zhao X and Li HX: MiR-221 and
miR-222 simultaneously target ARID1A and enhance proliferation and
invasion of cervical cancer cells. Eur Rev Med Pharmacol Sci.
20:1509–1515. 2016.PubMed/NCBI
|
|
80
|
Li ZY, Zhu SS, Chen XJ, Zhu J, Chen Q,
Zhang YQ, Zhang CL, Guo TT and Zhang LM: ARID1A suppresses
malignant transformation of human pancreatic cells via mediating
senescence-associated miR-503/CDKN2A regulatory axis. Biochem
Biophys Res Commun. 493:1018–1025. 2017.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Kim YS, Jeong H, Choi JW, Oh HE and Lee
JH: Unique characteristics of ARID1A mutation and protein level in
gastric and colorectal cancer: A meta-analysis. Saudi J
Gastroenterol. 23:268–274. 2017.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Lichner Z, Scorilas A, White NM, Girgis
AH, Rotstein L, Wiegand KC, Latif A, Chow C, Huntsman D and Yousef
GM: The chromatin remodeling gene ARID1A is a new prognostic marker
in clear cell carcinoma. Am J Pathol. 182:1163–1170.
2013.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Takeda T, Banno K, Okawa R, Yanokura M,
Iijima M, Irie-Kunitomi H, Nakamura K, Iida M, Adachi M, Umene K,
et al: ARID1A gene mutation in ovarian and endometrial cancers
(Review). Oncol Rep. 35:607–613. 2016.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27.
2012.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Okawa R, Banno K, Iida M, Yanokura M,
Takeda T, Iijima M, Kunitomi-Irie H, Nakamura K, Adachi M, Umene K,
et al: Aberrant chromatin remodeling in gynecological cancer. Oncol
Lett. 14:5107–5113. 2017.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Wang Y, Wysocka J, Perlin JR, Leonelli L,
Allis CD and Coonrod SA: Linking covalent histone modifications to
epigenetics: The rigidity and plasticity of the marks. Cold Spring
Harb Symp Quant Biol. 69:161–169. 2004.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Guan B, Mao TL, Panuganti PK, Kuhn E,
Kurman RJ, Maeda D, Chen E, Jeng YM, Wang TL and Shih IeM: Mutation
and loss of expression of ARID1A in uterine low-grade endometrioid
carcinoma. Am J Surg Pathol. 35:625–632. 2011.PubMed/NCBI View Article : Google Scholar
|
|
88
|
He F, Li J, Xu JF, Zhang S, Xu Y, Zhao W,
Yin Z and Wang X: Decreased expression of ARID1A associates with
poor prognosis and promotes metastases of hepatocellular carcinoma.
J Exp Clin Cancer Res. 34(47)2015.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Kim KJ, Jung HY, Oh MH, Cho H, Lee JH, Lee
HJ, Jang SH and Lee MS: Loss of ARID1A expression in gastric
cancer: Correlation with mismatch repair deficiency and
clinicopathologic features. J Gastric Cancer. 15:201–208.
2015.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Mamo A, Cavallone L, Tuzmen S, Chabot C,
Ferrario C, Hassan S, Edgren H, Kalliomeni O, Aleynikova O,
Przybytkowski E, et al: An integrated genomic approach identifies
ARID1A as a candidate tumor-suppressor gene in breast cancer.
Oncogene. 31:2090–2100. 2012.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Bosse T, ter Haar NT, Seeber LM, v Diest
PJ, Hes FJ, Vasen HF, Nout RA, Creutzberg CL, Morreau H and Smit
VT: Loss of ARID1A expression and its relationship with PI3K-Akt
pathway alterations, TP53 and microsatellite instability in
endometrial cancer. Mod Pathol. 26:1525–1535. 2013.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Lee D, Yu EJ, Ham IH, Hur H and Kim YS:
AKT-inhibition is an effective treatment strategy in
ARID1A-deficient gastric cancer cells. Onco Targets Ther.
10:4153–4159. 2017.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Samartzis EP, Gutsche K, Dedes KJ, Fink D,
Stucki M and Imesch P: Loss of ARID1A expression sensitizes cancer
cells to PI3K- and AKT-inhibition. Oncotarget. 5:5295–5303.
2014.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Guan B, Wang TL and Shih IeM: ARID1A, a
factor that promotes formation of SWI/SNF-mediated chromatin
remodeling, is a tumor suppressor in gynecologic cancers. Cancer
Res. 71:6718–6727. 2011.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Shain AH, Giacomini CP, Matsukuma K,
Karikari CA, Bashyam MD, Hidalgo M, Maitra A and Pollack JR:
Convergent structural alterations define SWItch/Sucrose
NonFermentable (SWI/SNF) chromatin remodeler as a central tumor
suppressive complex in pancreatic cancer. Proc Natl Acad Sci USA.
109:E252–E259. 2012.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Xiao W, Awadallah A and Xin W: Loss of
ARID1A/BAF250a expression in ovarian endometriosis and clear cell
carcinoma. Int J Clin Exp Pathol. 5:642–650. 2012.PubMed/NCBI
|
|
97
|
Dykhuizen EC, Hargreaves DC, Miller EL,
Cui K, Korshunov A, Kool M, Pfister S, Cho YJ, Zhao K and Crabtree
GR: BAF complexes facilitate decatenation of DNA by topoisomerase
IIα. Nature. 497:624–627. 2013.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Bochar DA, Wang L, Beniya H, Kinev A, Xue
Y, Lane WS, Wang W, Kashanchi F and Shiekhattar R: BRCA1 is
associated with a human SWI/SNF-related complex: Linking chromatin
remodeling to breast cancer. Cell. 102:257–265. 2000.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Putra J and Suriawinata AA: Clinical
significance of loss of ARID1A expression in colorectal and small
intestinal carcinoma. Clin Transl Gastroenterol.
6(e131)2015.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Wang K, Kan J, Yuen ST, Shi ST, Chu KM,
Law S, Chan TL, Kan Z, Chan AS, Tsui WY, et al: Exome sequencing
identifies frequent mutation of ARID1A in molecular subtypes of
gastric cancer. Nat Genet. 43:1219–1223. 2011.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Cho H, Kim JS, Chung H, Perry C, Lee H and
Kim JH: Loss of ARID1A/BAF250a expression is linked to tumor
progression and adverse prognosis in cervical cancer. Hum Pathol.
44:1365–1374. 2013.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Katagiri A, Nakayama K, Rahman MT, Rahman
M, Katagiri H, Ishikawa M, Ishibashi T, Iida K, Otsuki Y, Nakayama
S and Miyazaki K: Frequent loss of tumor suppressor ARID1A protein
expression in adenocarcinomas/adenosquamous carcinomas of the
uterine cervix. Int J Gynecol Cancer. 22:208–212. 2012.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Ayhan A, Mao TL, Seckin T, Wu CH, Guan B,
Ogawa H, Futagami M, Mizukami H, Yokoyama Y, Kurman RJ and Shih
IeM: Loss of ARID1A expression is an early molecular event in tumor
progression from ovarian endometriotic cyst to clear cell and
endometrioid carcinoma. Int J Gynecol Cancer. 22:1310–1315.
2012.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Yamamoto S, Tsuda H, Takano M, Tamai S and
Matsubara O: Loss of ARID1A protein expression occurs as an early
event in ovarian clear-cell carcinoma development and frequently
coexists with PIK3CA mutations. Mod Pathol. 25:615–624.
2012.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Yang L, Wei S, Zhao R, Wu Y, Qiu H and
Xiong H: Loss of ARID1A expression predicts poor survival prognosis
in gastric cancer: A systematic meta-analysis from 14 studies. Sci
Rep. 6(28919)2016.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Ahn DH, Javle M, Ahn CW, Jain A, Mikhail
S, Noonan AM, Ciombor K, Wu C, Shroff R, Chen JL and Bekaii-Saab T:
Next-generation sequencing survey of biliary tract cancer reveals
the association between tumor somatic variants and chemotherapy
resistance. Cancer. 122:3657–3666. 2016.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Balbás-Martinez C, Rodriguez-Pinilla M,
Casanova A, Dominguez O, Pisano DG, Gómez G, Lloreta J, Lorente JA,
Malats N and Real FX: ARID1A alterations are associated with
FGFR3-wild type, poor-prognosis, urothelial bladder tumors. PLoS
One. 8(e62483)2013.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Cho HD, Lee JE, Jung HY, Oh MH, Lee JH,
Jang SH, Kim KJ, Han SW, Kim SY, Kim HJ, et al: Loss of tumor
supressor ARID1A protein expression correlates with poor prognosis
in patients with primary breast cancer. J Breast Cancer.
18:339–346. 2015.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Drage MG, Tippayawong M, Agoston TA, Zheng
Y, Bueno R, Hornick JL, Odge RD and Srivastava A: Morphological
features and prognostic significance of ARID1A-deficient esophageal
adenocarcinomas. Arch Pathol Lab Med. 141:970–977. 2017.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Faraj SF, Chaux A, Gonzales-Roibon N,
Munari E, Ellis C, Driscoll T, Schoenberg MP, Bivalacqua TJ, Shih
IeM and Netto GJ: ARID1A immunohistochemistry improves outcome
prediction in invasive urothelial carcinoma of urinary bladder. Hum
Pathol. 45:2233–2239. 2014.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Wang DD, Chen YB, Pan K, Wang W, Chen SP,
Chen JG, Zhao JJ, Lv L, Pan QZ, Li YQ and Wang QJ: Decreased
expression of the ARID1A gene is associated with poor prognosis in
primary gastric cancer. PLoS One. 7(e40364)2012.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Wei XL, Wang DS, Xi SY, Wu WJ, Chen DL,
Zeng ZL, Wang RY, Huang YX, Jin Y, Wang F, et al: Clinicopathologic
and prognostic relevance of ARID1A protein loss in colorectal
cancer. World J Gastroenterol. 20:18404–18412. 2014.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Wiegand KC, Hennesy BT, Leung S, Wang Y,
Ju Z, McGahren M, Kalloger SE, Finlayson S, Stemke-Hale K, Lu Y, et
al: A functional proteogenomic analysis of endometrioid and clear
cell carcinomas using reverse phase protein array and mutation
analysis: Protein expression is histotype-specific and loss of
ARID1A/BAF250a is associated with AKT phosphorylation. BMC Cancer.
14(120)2014.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Cajuso T, Hӓnninen UA, Kondelin J, Gylfe
AE, Tanskanen T, Katainen R, Pitkӓnen E, Ristolainen H, Kaasinen E,
Taipale M, et al: Exome sequencing reveals frequent inactivating
mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite
unstable colorectal cancer. Int J Cancer. 135:611–623.
2014.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Wiegand KC, Sy K, Kalloger SE, Li-Chang H,
Woods R, Kumar A, Streutker CJ, Hafezi-Bakhtiari S, Zhou C, Lim HJ,
et al: ARID1A/BAF250a as a prognostic marker for gastric carcinoma:
A study of 2 cohorts. Hum Pathol. 45:1258–1268. 2014.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Zhang L, Wang C, Yu S, Jia C, Yan J, Lu Z
and Chen J: Loss of ARID1A expression correlates with tumor
differentiation and tumor progression stage in pancreatic ductal
adenocarcinoma. Technol Cancer Res Treat.
17(1533034618754475)2018.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Tan ZX, Liu HJ and Hou B: Decreased
expression of ARID1A is related to the poor prognosis of glioma
patients. Int J Clin Exp Pathol. 9:2009–2014. 2016.
|
|
118
|
Nastase A, Teo JY, Heng HL, Ng CC, Myint
SS, Rajaseragan V, Loh JL, Lee SY, Ooi LL, Chung AY, et al: Genomic
and proteomic characterization of ARID1A chromatin remodeller in
ampullary tumors. Am J Cancer Res. 7:484–502. 2017.PubMed/NCBI
|
|
119
|
Itamochi H, Oumi N, Oishi T, Shoji T,
Fujiwara J, Sugiyama T, Suzuki M, Kigawa J and Harada T: Loss of
ARID1A expression is associated with poor prognosis in patients
with stage I/II clear cell carcinoma of the ovary. Int J Clin
Oncol. 20:967–973. 2015.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao
S, Wu R, Chen C, Li X, Zhou L, et al: Frequent mutations of
chromatin remodeling genes in transitional cell carcinoma of the
bladder. Nat Genet. 43:875–878. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
121
|
Jiao Y, Pawlik TM, Anders RA, Selaru FM,
Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani
, et al: Exome sequencing identifies frequent inactivating
mutations in BAP1, ARID1A and PBRM1 in intrahepatic
cholangiocarcinomas. Nat Genet. 45:1470–1473. 2013.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Sausen M, Leary RJ, Jones S, Wu J,
Reynolds PC, Liu X, Blackford A, Parmigiani G, Diaz LA Jr,
Papadopoulos N, et al: Integrated genomic analyses identify ARID1A
and ARID1B alterations in the childhood cancer neuroblastoma. Nat
Genet. 45:12–17. 2013.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Illumina: An introduction to
Next-Generation Sequencing Technology. Illumina, Inc., 2017.
https://www.illumina.com/documents/products/illumina_sequencing_introduction.pdf.
|
|
124
|
Zhou X, Ren L, Meng Q, Li Y, Yu Y and Yu
J: The next-generation sequencing technology and application.
Protein Cell. 6:520–536. 2010.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Huang HN, Lin MC, Huang WC, Chiang YC and
Kuo KT: Loss of ARID1A expression and its relationship with
PI3K-Akt pathway alterations and ZNF217 amplification in ovarian
clear cell carcinoma. Mod Pathol. 27:983–990. 2014.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Lai CR, Hsu CY, Chen YJ, Yen MS, Chao KC
and Li AF: Ovarian cancers arising from endometriosis: A
microenvironmental biomarker study including ER, HNF1ß, p53, PTEN,
BAF250a, and COX-2. J Chin Med Assoc. 76:629–634. 2013.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Lowery WJ, Schildkraut JM, Akushevich L,
Bentley R, Marks JR, Huntsman D and Berchuck A: Loss of
ARID1A-associated protein expression is a frequent event in clear
cell and endometrioid ovarian cancers. Int J Gynecol Cancer.
22:9–14. 2012.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Maeda D, Mao TL, Fukayama M, Nakagawa S,
Yano T, Taketani Y and Shih leM: Clinicopathological significance
of loss of ARID1A immunoreactivity in ovarian clear cell carcinoma.
Int J Mol Sci. 11:5120–5128. 2010.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Wu RC, Ayhan A, Maeda D, Kim KR, Clarke
BA, Shaw P, Chui MH, Rosen B, Shih leM and Wang TL: Frequent
somatic mutations of the telomerase reverse transcriptase promoter
in ovarian clear cell carcinoma but not in other major types of
gynecologic malignancies. J Pathol. 232:473–481. 2014.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Yamamoto S, Tsuda H, Takano M, Tamai S and
Matsubara O: PIK3CA mutations and loss of ARID1A protein expression
are early events in the development of cystic ovarian clear cell
adenocarcinoma. Virchows Arch. 460:77–87. 2012.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Murakami R, Matsumura N, Brown JB, Higasa
K, Tsutsumi T, Kamada M, Abou-Taleb H, Hosoe Y, Kitamura S,
Yamaguchi K, et al: Exome sequencing landscape analysis in ovarian
clear cell carcinoma shed light on key chromosomal regions and
mutation gene networks. Am J Pathol. 187:2246–2258. 2017.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Katagiri A, Nakayama K, Rahman MT, Rahman
M, Katagiri H, Nakayama N, Ishikawa M, Ishibashi T, Iida K,
Kobayashi H, et al: Loss of ARID1A expression is related to shorter
progression-free survival and chemoresistance in ovarian clear cell
carcinoma. Mod Pathol. 25:282–288. 2012.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Samartzis EP, Samartzis N, Noske A, Fedier
A, Caduff R, Dedes KJ, Fink D and Imesch P: Loss of
ARID1A/BAF250a-expression in endometriosis: A biomarker for risk of
carcinogenic transformation? Mod Pathol. 25:885–892.
2012.PubMed/NCBI View Article : Google Scholar
|
|
134
|
McConechy MK, Ding J, Senz J, Yang W,
Melnyk N, Tone AA, Prentice LM, Wiegand KC, McAlpine JN, Shah SP,
et al: Ovarian and endometrial endometrioid carcinomas have
distinct CTNNB1 and PTEN mutation profiles. Mod Pathol. 27:128–134.
2014.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Wu CH, Mao TL, Vang R, Ayhan A, Wang TL,
Kurman RJ and Shih leM: Endocervical-type mucinous borderline
tumors are related to endometrioid tumors based on mutation and
loss of expression of ARID1A. Int J Gynecol Pathol. 31:297–303.
2012.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Choi JY, Han HH, Kim YT, Lee JH, Kim BG,
Kang S and Cho NH: Ovarian clear cell carcinoma sub-typing by
ARID1A expression. Yonsei Med J. 58:59–66. 2017.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Yokoyama Y, Matsushita Y, Shigeto T,
Futagami M and Mizunuma H: Decreased ARID1A expression is
correlated with chemoresistance in epithelial ovarian cancer. J
Gynecol Oncol. 25:58–63. 2014.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Cancer Genome Atlas Research Network,
Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H,
Robertson AG, Pashtan I, Shen R, et al: Integrated genomic
characterization of endometrial carcinoma. Nature 497: 67-73,
2013.
|
|
139
|
Liang H, Cheung LW, Li J, Ju Z, Yu S,
Stemke-Hale K, Dogruluk T, Lu Y, Liu X, Gu C, et al: Whole-exome
sequencing combined with functional genomics reveals novel
candidate driver cancer genes in endometrial cancer. Genome Res.
22:2120–2129. 2012.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Fadare O, Renshaw IL and Liang SX: Does
the loss of ARID1A (BAF-250a) expression in endometrial clear cell
carcinomas have any clinicopathological significance? A pilot
assessment. J Cancer. 3:129–136. 2012.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Fadare O, Gwin K, Desouki MM, Crispens MA,
Jones HW III, Khabele D, Liang SX, Zheng W, Mohammed K, Hecht JL
and Parkash V: The clinicopathologic significance of p53 and
BAF-250a (ARID1A) expression in clear cell carcinoma of the
endometrium. Mod Pathol. 26:1101–1110. 2013.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Werner HM, Berg A, Wik E, Birkeland E,
Krakstad C, Kusonmano K, Petersen K, Kalland KH, Oyan AM, Akslen
LA, et al: ARID1A loss is prevalent in endometrial hyperplasia with
atypia and low-grade endometrioid carcinomas. Mod Pathol.
26:428–434. 2013.PubMed/NCBI View Article : Google Scholar
|
|
143
|
Wiegand KC, Lee AF, Al-Agha OM, Chow C,
Kalloger SE, Scott DW, Steidl C, Wiseman SM, Gascoyne RD, Gilks B
and Huntsman DG: Loss of BAF250a (ARID1A) is frequent in high-grade
endometrial carcinomas. J Pathol. 224:328–333. 2011.PubMed/NCBI View Article : Google Scholar
|
|
144
|
Rahman M, Nakayama K, Rahman MT, Katagiri
H, Katagiri A, Ishibashi T, Ishikawa M, Iida K and Miyazaki K:
Clinicopathologic analysis of loss of AT-Rich interactive domain 1A
expression in endometrial cancer. Hum Pathol. 44:103–109.
2013.PubMed/NCBI View Article : Google Scholar
|
|
145
|
Cornen S, Adelaide J, Bertucci F, Finetti
P, Guille A, Birnbaum DJ, Birnbaum D and Chaffanet M: Mutations and
deletions of ARID1A in breast tumors. Oncogene. 31:4255–4256.
2012.PubMed/NCBI View Article : Google Scholar
|
|
146
|
Zhang X, Zhang Y, Yang Y, Niu M, Sun S, Ji
H, Ma Y, Yao G, Jiang Y, Shan M, et al: Frequent low expression of
chromatin remodeling gene ARID1A in breast cancer and its clinical
significance. Cancer Epidemiol. 36:288–293. 2012.PubMed/NCBI View Article : Google Scholar
|
|
147
|
Zhao J, Liu C and Zhao Z: ARID1A: A
potential prognostic factor for breast cancer. Tumour Biol.
35:4813–4819. 2014.PubMed/NCBI View Article : Google Scholar
|
|
148
|
Ünçel M, Diniz G, Aköz G, Ekin ZY, Sayhan
S, Yardım S and Salimoğlu S: Loss of nuclear ARID-1A expressions is
associated with hormone receptor status in breast cancers. Eur J
Breast Health. 15:125–129. 2019.PubMed/NCBI View Article : Google Scholar
|
|
149
|
Momozawa Y, Iwasaki Y, Parsons MT,
Kamatani Y, Takahashi A, Tamura C, Katagiri T, Yoshida T, Nakamura
S, Sugano K, et al: Germline pathogenic variants of 11 breast
cancer genes in 7,051 Japanese patients and 11,241 controls. Nat
Commun. 9(4083)2018.PubMed/NCBI View Article : Google Scholar
|
|
150
|
Rajendran KB and Deng C: Characterization
of potential driver mutations involved in human breast cancer by
computational approaches. Oncotarget. 8:50252–50272.
2017.PubMed/NCBI View Article : Google Scholar
|
|
151
|
Zang ZJ, Cutcutache I, Poon SL, Zhang SL,
McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, et al:
Exome sequencing of gastric adenocarcinoma identifies recurrent
somatic mutations in cell adhesion and chromatin remodeling genes.
Nat Genet. 44:570–574. 2012.PubMed/NCBI View Article : Google Scholar
|
|
152
|
Abe H, Maeda D, Hino R, Otake Y, Isogai M,
Ushiku AS, Matsusaka K, Kunita A, Ushiku T, Uozaki H, et al: ARID1A
expression loss in gastric cancer: Pathway-dependent roles with and
without Epstein-Barr virus infection and microsatellite
instability. Virchows Arch. 461:367–377. 2012.PubMed/NCBI View Article : Google Scholar
|
|
153
|
Han N, Kim MA, Lee HS and Kim WH: Loss of
ARID1A expression is related to gastric cancer progression,
Epstein-Barr Virus infection, and mismatch repair deficiency. Appl
Immunohistochem Mol Morphol. 24:320–325. 2016.PubMed/NCBI View Article : Google Scholar
|
|
154
|
Ibarrola-Villava M, Llorca-Cardeñosa MJ,
Tarazona N, Mongort C, Fleitas T, Perez-Fidalgo JA, Roselló S,
Navarro S, Ribas G and Cervantes A: Deregulation of ARID1A, CDH1,
cMET and PIK3CA and target-related microRNA expression in gastric
cancer. Oncotarget. 6:26935–26945. 2015.PubMed/NCBI View Article : Google Scholar
|
|
155
|
Kim YB, Ham IH, Hur H and Lee D: Various
ARID1A expression patterns and their clinical significance in
gastric cancers. Hum Pathol. 49:61–70. 2016.PubMed/NCBI View Article : Google Scholar
|
|
156
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120.
2012.PubMed/NCBI View Article : Google Scholar
|
|
157
|
Fujimoto A, Totoki Y, Abe T, Boroevich KA,
Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya M, et al:
Whole-genome sequencing of liver cancers identifies etiological
influences on mutation patterns and recurrent mutations in
chromatin regulators. Nat Genet. 44:760–764. 2012.PubMed/NCBI View Article : Google Scholar
|
|
158
|
Guichard C, Amaddeo G, Imbeaud S, Ladeiro
Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M,
Degos F, et al: Integrated analysis of somatic mutations and focal
copy-number changes identifies key genes and pathways in
hepatocellular carcinoma. Nat Genet. 44:694–698. 2012.PubMed/NCBI View Article : Google Scholar
|
|
159
|
Streppel MM, Lata S, DelaBastide M,
Montgomery EA, Wang JS, Canto MI, Macgregor-Das AM, Pai S, Morsink
FH, Offerhaus GJ, et al: Next-generation sequencing of endoscopic
biopsies identifies ARID1A as a tumor-suppressor gene in Barrett's
esophagus. Oncogene. 33:347–357. 2014.PubMed/NCBI View Article : Google Scholar
|
|
160
|
Lee SY, Kim DW, Lee HS, Ihn MH, Oh HK,
Park DJ, Kim HH and Kang SB: Loss of AT-rich interactive domain 1A
expression in gastrointestinal malignancies. Oncology. 88:234–240.
2015.PubMed/NCBI View Article : Google Scholar
|
|
161
|
Sen M, Wang X, Hamdan FH, Rapp J, Eggert
J, Kosinsky RL, Wegwitz F, Kutschat AP, Younesi FS, Gaedcke J, et
al: ARID1A facilitates KRAS signaling-regulated enhancer activity
in an AP1-dependent manner in colorectal cancer cells. Clin
Epigenet. 11(92)2019.PubMed/NCBI View Article : Google Scholar
|
|
162
|
Guo G, Sun X, Chen C, Wu S, Huang P, Li Z,
Dean M, Huang Y, Jia W, Zhou Q, et al: Whole-genome and whole-exome
sequencing of bladder cancer identifies frequent alterations in
genes involved in sister chromatid cohesion and segregation. Nat
Genet. 45:1459–1463. 2013.PubMed/NCBI View Article : Google Scholar
|
|
163
|
Treon SP, Xu L, Yang G, Zhou Y, Liu X, Cao
Y, Sheehy P, Manning RJ, Patterson CJ, Tripsas C, et al: MYD88
L265P somatic mutation in Waldenström's macroglobulinemia. N Engl J
Med. 367:826–833. 2012.PubMed/NCBI View Article : Google Scholar
|
|
164
|
Giulino-Roth L, Wang K, MacDonald TY,
Mathew S, Tam Y, Cronin MT, Palmer G, Lucena-Silva N, Pedrosa F,
Pedrosa M, et al: Targeted genomic sequencing of pediatric Burkitt
lymphoma identifies recurrent alterations in antiapoptotic and
chromatin-remodeling genes. Blood. 120:5181–5184. 2012.PubMed/NCBI View Article : Google Scholar
|
|
165
|
Weaver IC, Korgan AC, Lee K, Wheeler RV,
Hundert AS and Goguen D: Stress and the emerging roles of chromatin
remodeling in signal integration and stable transmission of
reversible phenotypes. Front Behav Neurosci. 11(41)2017.PubMed/NCBI View Article : Google Scholar
|
|
166
|
Bitler BG, Wu S, Park PH, Hai Y, Aird KM,
Wang Y, Zhai Y, Kossenkov AV, Vara-Ailor A, Rauscher FJ III, et al:
ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell
Biol. 19:962–973. 2017.PubMed/NCBI View Article : Google Scholar
|
|
167
|
Bitler BG, Aird KM, Garipov A, Li H,
Amatangelo M, Kossenkov AV, Schultz DC, Liu Q, Shih IeM,
Conejo-Garcia JR, et al: Synthetic lethality by targeting EZH2
methyltransferase activity in ARID1A-mutated cancers. Nat Med.
21:231–238. 2015.PubMed/NCBI View Article : Google Scholar
|