1. Introduction
Papillary thyroid carcinoma (PTC). Thyroid
cancer, originating from thyroid follicular epithelial cells, is
the most common type of cancer of the endocrine system, exhibiting
a rapid increase in mortality rate worldwide (1-3).
There are 4 subtypes of thyroid cancer, including papillary thyroid
cancer (PTC), follicular thyroid cancer (FTC), anaplastic thyroid
cancer (ATC) and medullary thyroid cancer (MTC) (4). PTC accounts for >80% of all
thyroid cancer cases (1,5,6).
Among all types of thyroid cancers, the incidence of PTC is much
higher and is often observed mainly among younger patients
(7). Genetic mutations and
environmental exposure are considered risk factors of PTC (8). The optimal treatment period for PTC
is the early stages of the disease; this period is often surpassed
by the time of diagnosis, due to the slow growth rate and atypical
symptoms (9). Despite the increase
in the 5-year survival rate up to 90% and favorable prognosis in
the majority of patients in the case of prompt treatment, including
thyroidectomy and adjuvant radioactive iodine therapy, certain
aggressive phenotypes of PTC, such as extra-thyroidal extension,
multifocal tumors, and lymph node and distant metastases lead to a
poor prognosis (10,11). Hence, it is of utmost importance to
identify novel diagnostic and therapeutic targets for PTC.
It has recently been demonstrated that genetic
factors affect thyroid cancer progression (12). Various molecular deregulations
direct the tumorigenicity of PTC. BRAFV600E is known as a highly
specific prognostic factor for patients with PTC. It functions by
activating mitogen-activated protein kinase (MAPK) that has
previously been proven to participate in various human cancers
(13-15).
Although the diagnostic strategies for thyroid cancer have greatly
improved, fine needle aspiration (FNA) cytology remains the gold
standard technique, which has a success rate of 70% in determining
the findings (16,17). Therefore, it is vital to identify
novel diagnostic, prognostic and therapeutic biomarkers in order to
diagnose thyroid cancer during the early stages.
Characteristics and roles of circular
RNAs (circRNAs)General characteristics
Non-coding RNAs (ncRNAs) form the majority of the
human transcriptome. A number of regulatory mechanisms and
pathophysiological pathways are controlled by these ncRNAs,
including microRNAs (miRNAs or miRs), long non-coding RNAs
(lncRNAs) and circRNAs (18). The
existence of circRNAs in human cells was first observed and
reported by Kos et al in 1986(19). Recently, more investigations have
been conducted on the function of circRNAs in various diseases and
circRNAs have become a hotspot in cancer research. circRNAs are a
product of a back-splicing mechanism containing a single-stranded
covalently closed-loop structure possessing neither 5'-3' polarity
nor a polyadenylated tail (18,20).
These characters render them more stable and resistant to RNase R
(21). circRNAs are mainly located
in the cytoplasm and are found in a wide range of living organisms
(22). Given their interesting
features, the present review article aimed to summarize their main
functions in cancer.
circRNAs as miRNA sponges. Predominantly,
circRNAs act as miRNA sponges and regulate the cell progression and
cell cycle by modulating gene expression or protein-generation in
transcriptional or post-transcriptional stages (23). Similar to other ncRNAs, circRNAs
have been found to play a pivotal role in the onset of diseases
(24,25). Recent studies have demonstrated
that a number of circRNAs function as tumor-promoters or
tumor-suppressors. For instance, the overexpression of circCDR1as
in osteosarcoma cells sponges miR-7 and suppresses the inhibitory
effects of miR-7 on osteosarcoma cell progression (26,27).
In addition, the upregulation of circRNA HIPK3 has been shown to
promote the development of gall bladder cancer by sponging miR-124
and acting as a tumor-promoter (28).
Role of circRNAs in cancer. miRNAs have
previously been used as molecular biomarkers in various types of
cancer (29). Currently,
researchers have found that the up- or downregulation of certain
circRNAs can affect tumor progression in some diseases (30-32).
However, the investigations on the functions of circRNA in
cancerous tissues are insufficient, and their reliability for being
used as diagnostic, prognostic or therapeutic biomarkers is
indeterminate. circRNAs are resistant to exonucleases and RNase R,
and these characters render them more stable than other types of
ncRNAs (21,33). Hence, circRNAs may be acknowledged
as ideal biomarkers for the diagnosis of cancer tissues.
2. Roles of circRNAs in PTC cell
progression.
Microarray analysis has indicated that certain
circRNAs are differently expressed between PTC and adjacent normal
tissues; however, their function and downstream mechanisms remain
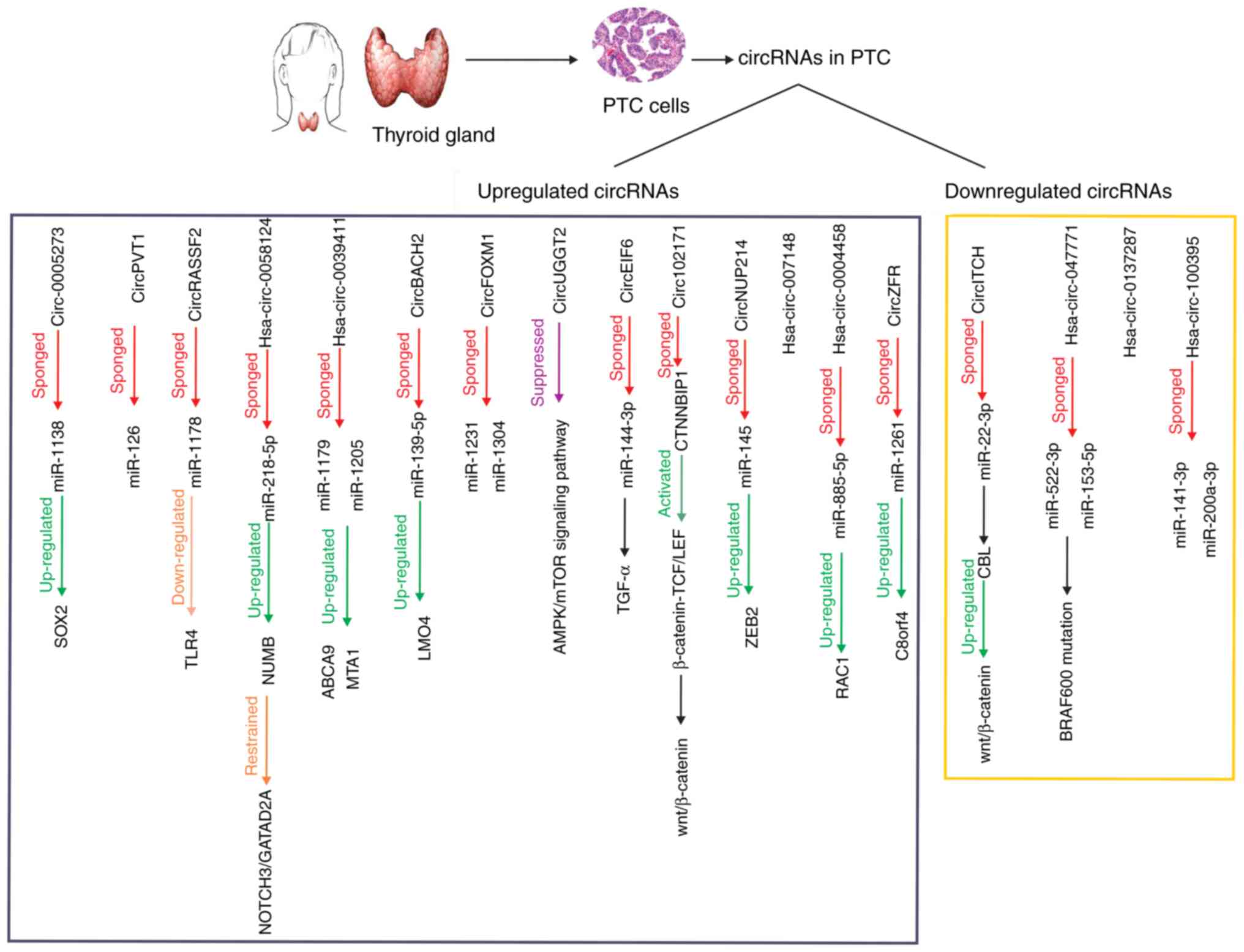
largely unknown (34). A graphical
abstract of the identified circRNAs that are dysregulated in PTC
cells and their downstream signaling pathways is depicted in
Fig. 1. Conducted studies on the
role of circRNAs in PTC are limited; however, circRNAs have a more
stable structure, they are considered as ideal biomarkers for
diagnosis (33). In the present
review, the deregulated circRNAs in PTC are discussed, with
particular focus on their regulatory mechanisms and functions.
Upregulated circRNAs in PTC
hsa-circ-0058124
Conducting RT-qPCR, hsa-circ-0058124, located on
chromosome 2 and generated from the fibronectin1 gene (35,36),
has been demonstrated to be upregulated in PTC tissues and cell
lines. Cell fraction assay and FISH have exhibited that this
circRNA is mainly available in the nucleus and sometimes in the
cell cytoplasm (37). The
overexpression of hsa-circ-0058124 has been shown to result in the
poor prognosis of patients with PTC; in addition, a larger tumor
size, advanced stage, extra-thyroidal extension, lymph node
metastasis and distant metastasis are more often observed in
patients with a higher expression of hsa-circ-0058124. Western blot
analysis and dual-luciferase assay have revealed that
hsa-circ-0058124 upregulates NUMB expression by sponging miR-218-5p
to suppress the NOTCH3 signaling pathway. hsa-circ-0058124
functions by restraining the NOTCH3/GATAD2A cascade (37). These findings suggest that
hsa-circ-0058124 regulates PTC progression and functions as an
oncogene; thus, it can be used as an ideal prognostic biomarker for
patients with PTC.
hsa-circ-0039411. The overexpression of
hsa-circ-0039411, located on chromosome 16, has been proven by
conducting RT-qPCR in PTC tissues and cell lines (38). Gain- and loss-of-function and CCK-8
assays have demonstrated that hsa-circ-0039411 is positively
associated with cell growth. Flow cytometric analysis have also
shown that the downregulation of hsa-circ-0039411 leads to PTC cell
apoptosis. Transwell assay has also confirmed that PTC cell
migration and invasion are positively controlled by this biomarker
(39). RT-qPCR, along with
dual-luciferase assay, have revealed that hsa-circ-0039411 sponges
miR-1179 and miR-1205, and two potential binding sites for miR-1205
have been recognized. hsa-circ-0039411 upregulates the expression
of ABCA9 and MTA1 by sponging miR-1179 and miR-1205, respectively,
and thus performs an oncogenic role in PTC cell progression
(39). It can thus be concluded
that hsa-circ-0039411 may be used as a therapeutic target in
patients with PTC.
circBACH2. Cai et al have reported
that circBACH2, also known as hsa-circ-0001627 and derived from
exon 2 of chromosome 6, is upregulated in PTC tissues and cell
lines. Previously, ROC curve analysis determined that circBACH2
expression level was upregulated in PTC tissues in comparison with
adjacent non-tumor tissues; a longer survival lifetime was
recognized among patients with higher expression levels of
circBACH2 by conducting Kaplan-Meier analysis (40). Previously, luciferase reporter
assay also proved that miR-139-5p had some potential binding sites
on circBACH2, and this circRNA functioned by sponging miR-139-5p.
RT-qPCR, western blot analysis and CCK-8 assay revealed that
circBACH2 sponged miR-139-5p and suppressed its inhibitory effect
on LIM domain only 4 (LMO4) (40).
These results indicate that circBACH2 mediates PTC cell progression
and invasion by regulating the miR-139-5p/LMO4 axis.
circFOXM1. circFOXM1, also known as
circ-0025033, is a product of the FOXM1 gene and is mapped on
chromosome 12. In a previous study, by performing circRNA
microarray analysis and RT-qPCR, it was found that the expression
level of circFOXM1 was upregulated in PTC tissues and cell lines.
RT-qPCR, CCK-8 assay and colony formation assay also revealed that
circFOXM1 affected cell proliferation. Cell apoptosis and
metastasis were also found to be controlled by circFOXM1, and the
overexpression of this biomarker promoted PTC cell migration and
invasion (38). In previous
studies, bioinformatics analysis also demonstrated that circFOXM1
functioned as a sponge for both miR-1231 and miR-1304. These two
miRNAs were reported to function as tumor-suppressors (41-43).
In a previous study by Pan et al, CCK-8 and Transwell assay
conducted demonstrated that circFOXM1 negatively regulated the
expression of miR-1231 and miR-1304, and played an oncogenic role
in patients with PTC (38). These
data indicate that circFOXM1 promotes PTC progression by sponging
miR-1231 and miR-1304.
circUGGT2. Zhou et al described
circUGGT2, also known as hsa-circ-0008274 and located on chromosome
11, as an upregulated biomarker in PTC tissues and cell lines
(44). These data were obtained
using bioinformatics analysis and RT-qPCR. In addition, in that
same study, it was found out that the high expression of circUGGT2
was associated with TNM stage and lymph node metastasis, but not
with age, sex, extra-thyroidal extension, primary tumor and tumor
size. Furthermore, MTT assay was conducted and it was shown that
circUGGT2 regulated PTC cell proliferation. In the same study,
further investigations revealed that the invasion of PTC cells was
contributed to circUGGT2. Western blot analysis was also performed
to explore the downstream mechanisms. The results revealed that the
overexpression of circUGGT2 led to the suppression of the
5'AMP-activated protein kinase (AMPK)/mammalian target of rapamycin
(mTOR) signaling pathway (44).
The AMPK/mTOR signaling pathway has been shown to inhibit autophagy
and increase cell proliferation in both normal and tumor tissues
(45). Taken together, it can be
concluded that circUGGT2 mediates PTC cell progression and invasion
by regulating the AMPK/mTOR signaling pathway.
circEIF6. In a previous study, bioinformatics
and RT-qPCR confirmed that circEIF6 (hsa-circ-0060060) was
upregulated in PTC tissues and cell lines. Conducting sequence
analysis, two potential binding sites were assessed between
circEIF6 and miR-144-3p. RT-qPCR also revealed that the level of
miR-144-3p was decreased in PTC tissues and cell lines.
Dual-luciferase reporter assay proved that circEIF6 sponged
miR-144-3p and regulated the expression of TGF-α as a result in
cisplatin-treated cells (46).
Previous studies have reported miR-144-3p can function as either
tumor-promoter or tumor-suppressor (47,48).
Previously, the results of MTT assay and flow cytometry
demonstrated that the overexpression of circEIF6 enhanced cell
proliferation and autophagy, and suppressed cell apoptosis
(46). These data suggest that
circEIF6 promotes PTC progression by activating the
miR-144-3p/TGF-α signaling pathway.
circ-102171. circ-102171 has been shown to be
significantly upregulated in PTC tissues and cell lines. In a
previous study, functional assays, including CCK-8 and colony
formation assays, demonstrated that circ-102171 regulated the
proliferation and apoptosis of PTC cells, and affected their cell
cycle. Performing Transwell assay indicated that the knockdown of
circ-102171 suppressed the migration and invasion of PTC cells.
Using silver staining, MS identification, biotin-labeled probes and
RNA-EMSA assay, it was also found that circ-102171 interacted with
CTNNBIP1 protein in PTC cells (49). CTNNBIP1 has been shown to be
negatively associated with the Wnt/β-catenin pathway by regulating
the β-catenin-TCF/LEF cascade. As a result, the overexpression of
circ-102171 leads to the increased activation of the Wnt/β-catenin
signaling pathway (50). In a
previous study, RT-qPCR and functional experiments revealed that
CTNNBIP1 was downregulated in PTC cells, and this protein
functioned as a tumor-suppressor (49). To sum up, these data confirm that
circ-102171 controls PTC cell progression and invasion by
activating the Wnt/ β-catenin pathway by regulating CTNNBIP1.
circNUP214. circNUP214, also known as
hsa-circ-0089153, is located on chromosome 9. In a previous study,
bioinformatics and RT-qPCR revealed that this circRNA was
significantly upregulated in PTC tissues and cell lines. Localized
by fluorescence in situ hybridization, circNUP214 was found
to be mainly located in the cytoplasm. CCK-8 and colony formation
assays revealed that the expression of circNUP214 was directly
associated with the proliferation and apoptosis of PTC cells.
Transwell assays also demonstrated that the circNUP214 expression
level was associated with PTC cell migration and invasion. By
conducting the dual-luciferase assay, it was also confirmed that
circNUP214 enhanced zinc finger E-box-binding homeobox 2 (ZEB2)
expression by sponging miR-145(51). ZEB2 has previously been reported to
function as an oncogene in various tumor tissues (52,53).
These results suggest that the overexpression of circNUP214
promotes PTC cell proliferation and invasion by sponging miR-145 in
a ZEB2-dependent manner.
hsa-circ-007148. In a previous study,
bioinformatics and RT-qPCR validated that hsa-circ-007148
expression was enhanced in PTC tissues and cell lines. It was also
found that the overexpression of hsa-circ-007148 was significantly
associated with lymph node metastasis; however, no other
clinicopathological association was detected. ROC curve analysis
also indicated that hsa-circ-007148 may be used as a diagnostic
biomarker for distinguishing PTC tissues (12). These data suggest that the
upregulation of hsa-circ-007148 plays an oncogenic role in patients
with PTC.
hsa-circ-0004458. hsa-circ-0004458, located
on chromosome 8: 18656804-18662408 with 448 nucleotides in length,
was first detected in gastric cancer and PTC. The upregulation of
this biomarker was previously confirmed among PTC tissues and cell
lines by performing RT-qPCR. It was also proven that
hsa-circ-0004458 expression was associated with tumor size,
invasion, lymphatic metastasis, distant metastasis and TNM stage.
Following the knockdown of hsa-circ-0004458, a reduction in tumor
growth was also observed, as well as cell cycle arrest.
Dual-luciferase assay confirmed that hsa-circ-0004458 modulated
RAC1 by sponging miR-885-5p (54).
RAC1 is a critical protein in underlying mechanisms responsible for
cell growth, migration and the activation of various protein
kinases (55,56). These findings indicate that the
hsa-circ-0004458/miR-885-5p/RAC1 signaling pathway regulates PTC
cell progression and invasion.
circZFR. circZFR, also known as
hsa-circ-0072088 and mapped on chromosome 5p13.3, plays a role in
various tumor-regulating mechanisms (57,58).
In a previous study, bioinformatics and RT-qPCR revealed the
upregulated expression of this biomarker in PTC tissues and cell
lines. Moreover, the overexpression of circZFR in patients with PTC
exhibited a positive association with clinical severity, including
TNM stage and distant metastasis. Kaplan-Meier analysis also
indicated that a higher expression of circZFR was associated with a
poor prognosis. The effects of circZFR on cell proliferation and
invasion were also confirmed by performing CCK-8, colony formation
and Transwell assays, respectively. To explore the underlying
mechanisms, a dual-luciferase reporter assay was conducted and it
was validated that circZFRmodulated C8orf4 expression by sponging
miR-1261(59). C8orf4 is known as
thyroid cancer 1 (TC1) and has been previously shown to play an
oncogenic role in other cancerous tissues (60,61).
In conclusion, these data illustrate that circZFR regulates PTC
cell proliferation and invasion via the miR-1261/C8orf4 axis.
circRNA plasmacytoma variant translocation gene 1
(circPVT1). circPVT1, located on chromosome 8q24, has been
reported to promote PTC progression through different pathways.
Exploring the potential role of circPVT1 in PTC, it was found that
circPVT1 was upregulated in PTC tissues. Further investigations
revealed that the T stage, lymph node metastasis and survival
status were associated with a higher expression of circPVT1. Still,
no association was found with age, sex and the ATA risk of
patients. In addition, the overexpression of circPVT1 promoted
apoptosis and inhibited the migration and invasion of PTC cells.
Luciferase reporter assay demonstrated that circPVT1 had putative
binding sites for miR-126. RIP assay and RT-qPCR indicated that
circPVT1 sponged miR-126 and a reduction in miR-126 level occurred
by circPVT1 overexpression (62).
It has previously been demonstrated that miR-126 functions as a
tumor-suppressor (63). In
summary, the upregulation of circPVT1 promotes the progression of
PTC cells by sponging miR-126.
circRASSF2. hsa-circ-0059354, also known as
circRASSF2 and derived from the RASSF2 gene on chromosome 20:
4760668-4766974, was previously found to be upregulated in PTC
tissues and cell lines. Microarray analysis indicated that
circRASSF2 was overexpressed in PTC tumor tissues with a
>10-fold change. Further investigations confirmed that the
higher expression of circRASSF2 was associated with tumor stage and
lymph node metastasis. Colony formation and Transwell assays
demonstrated that the overexpression of circRASSF2 promoted cell
proliferation, and enhanced the cell migratory and invasive
capabilities. Dual-luciferase reporter assay confirmed that
circRASSF2 functioned as a sponge for miR-1178. Functional
experiments also revealed that Toll-like receptor (TLR)4 was a
direct target of miR-1178 in PTC tissues (64). Thus, these data suggest that
circRASSF2 sponges miR-1178 and regulates PTC cell proliferation
and invasion by targeting TLR4.
circ-0005273. By conducting RT-qPCR, a
previous study found that circ-0005273 was upregulated in PTC
tissues and cell lines. circ-0005273 was found to be mainly located
in the cell cytoplasm and the presence of this biomarker indicated
a poor prognosis of patients with PTC. Functional experiments
revealed that circ-0005273 promoted PTC tumor growth and
progression. Further investigations indicated that there were
potential binding sites on circ-0005273 for miR-1138. CCK-8, colony
formation and Transwell assays demonstrated that circ-0005273
sponged miR-1138 and suppressed its inhibitory effect on
sex-determining region Y (SRY)-box 2 (SOX2) (65). Thus, circ-0005273 plays an
oncogenic role in PTC cells by regulating the
circ-0005273/miR-1138/ SOX2 axis.
Downregulated circRNAs in PTC tissues
hsa-circ-100395
In a previous study, microarray analysis revealed
that hsa-circ-100395 was downregulated in PTC tissues and cell
lines. The lower expression of this biomarker was validated by
conducting RT-qPCR and bioinformatics analysis. Functional
experiments indicated that hsa-circ-100395 was associated with
miR-141-3p and miR-200a-3p in PTC tissues. These two miRNAs were
overexpressed in PTC tissues as a result of a downregulation that
occurred in hsa-circ-100395 expression. Downstream cancer-related
genes were sponged by miR-141-3p and miR-200a-3p and this led to
PTC cell progression. Accordingly, hsa-circ-100395 regulated PTC
cell progression by modulating the
hsa-circ-100395/miR-141-3p/miR-200a-3p axis (34).
circITCH
circITCH plays a vital role in a variety of
downstream mechanisms involved in tumorigenesis (66). Wang et al reported a
downregulation in the expression of this biomarker in PTC tissues
and cell lines, validated by RT-qPCR. CircITCH expression was also
shown to be positively associated with clinical stage and the lymph
node metastasis of patients with PTC. CCK-8 and Transwell assays
demonstrated that the higher expression of circITCH led to the
inhibition of the proliferation and invasion of PTC cells.
Luciferase reporter assays indicated that circITCH functioned by
sponging miR-22-3p. Moreover, functional experiments revealed that
miR-22-3p had 3 potential binding sites for CBL (67). As has been previously demonstrated,
CBL regulates PTC progression by regulating the Wnt/β-catenin
pathway (68). To sum up, circITCH
functions as a tumor-suppressor by regulating the
circITCH/miR-22-3p/CBL/β-catenin pathway, and regulating the
proliferation and invasion of PTC cells.
hsa-circ-047771. In a previous study
microarray and RT-qPCR analysis revealed that hsa-circ-047771 was
downregulated in PTC tissues and cell lines (12). Further investigations illustrated
that there was a significant association between hsa-circ-047771
expression and BRAFV600 mutation, lymph node metastasis and TNM
stage, whereas no association was observed with other
clinicopathological features (12). BRAFV600 mutation has been used as a
poor prognostic marker in patients with PTC (69,70).
In a previous study, ROC curve analysis demonstrated that
hsa-circ-047771 was a diagnostic biomarker for the differentiation
of PTC tissues from adjacent normal tissues. Functional experiments
also reported that hsa-circ-047771 targeted miR-522-3p/miR-153-5p,
and that these miRNAs were upregulated in PTC tissues (12). It can thus be concluded from these
data that the downregulation of hsa-circ-047771 results in PTC cell
progression in a miR-522-3p/miR-153-5p-dependent manner.
hsa-circ-0137287. In a previous study RT-qPCR
demonstrated that hsa-circ-0137287, located on chromosome 8:
92301363-92307931, was downregulated in PTC tissues and cell lines.
Clinicopathological characteristics, including extra-thyroidal
extension, T stage, lymph node metastasis, microcarcinoma and tumor
size among patients with PTC, were primarily associated with
hsa-circ-0137287 expression. ROC curve analysis also determined
that hsa-circ-0137287 could be used as a diagnostic biomarker
(71). In conclusion, these pieces
of information indicate that hsa-circ-0137287 is downregulated in
PTC; however, further studies are required to explore the
downstream mechanisms.
3. Conclusion
Thyroid cancer is the most prevalent disorder of the
endocrine system. PTC is the most common type of thyroid cancer,
exhibiting an increase in incidence worldwide (72). It has recently been demonstrated
that the dysregulation of certain circRNAs results in PTC cell
progression or suppression (34).
These circRNAs may function as tumor-promoters or tumor-suppressors
and usually function by sponging different miRNAs. It has
previously been demonstrated that circRNAs possess an annular
structure and are thus known as a stable class of RNA molecules.
Based on their unique characteristics, circRNAs are promising
diagnostic and/or prognostic biomarkers in various types of cancer
(73). circRNAs contain multiple
miRNA binding sites, enabling them to sponge miRNAs and modulate
downstream mechanisms by controlling miRNA expression (74).
In PTC cells, researchers have discovered that
certain defined circRNAs target miRNAs and regulate cell
proliferation, invasion and apoptosis. However, there are some
circRNAs, including hsa-circ-007148(12) and hsa-circ-0137287(71), in PTC cells, for which the
downstream mechanisms have yet to be distinguished. The
overexpression of circRNAs in PTC [e.g., circBACH2(40) and circFOXM1(38)] function as tumor-promoters and
downregulated ones [e.g., circITCH (67) and hsa-circ-100395(34)] function as tumor-suppressors. These
circRNAs, their identified target genes/proteins, and their
biological functions are presented in Table I. Taken together, circRNAs with
their distinctive features, are more stable in cells and can even
cause drug resistance, thus suggesting that they may be used for
distinguishing and treating malignancies.
 | Table IExpression, gene locus, target
genes/proteins and biological function of circRNAs. |
Table I
Expression, gene locus, target
genes/proteins and biological function of circRNAs.
| No. | circRNA | Expression | Gene locus | Target genes/
proteins | Biological
function | Clinical value | (Refs.) |
|---|
| 1. |
hsa-circ-0058124 | Upregulation | Chr 2 | miR-218-5p NUMB
NOTCH3 GATAD2A | Cell viability,
colony formation, migration and invasion, cell cycle and
apoptosis | Prognostic and
therapeutic | (37) |
| 2. |
hsa-circ-0039411 | Upregulation | Chr 16 | miR-1179 miR-1205
ABCA9 MTA1 | PTC cell growth,
cell apoptosis, migration and invasion | Therapeutic | (39) |
| 3. | circBACH2
(Hsa-circ-0001627) | Upregulation | Chr 6:
90959407-90981660 | miR-139-5p
LMO4 | Cell proliferation,
migration and invasion | Diagnostic and
prognostic and therapeutic | (40) |
| 4. | circFOXM1
(Circ-0025033) | Upregulation | Chr 12:
2966846-2983691 | miR-1231
miR-1304 | Cell proliferation,
clonogenic ability, cell apoptosis and metastasis | Therapeutic | (38) |
| 5. | circUGGT2
(Hsa-circ-0008274) | Upregulation | Chr 11:
96485180-96489456 | AMPK/mTOR signaling
pathway | Cell proliferation
and invasion | Therapeutic | (44) |
| 6. | circEIF6
(Hsa-circ-0060060) | Upregulation | - | miR-144-3p
TGFα |
Cisplatin-resistance, cell proliferation
and autophagy regulation | Therapeutic | (46) |
| 7. | circ-102171 | Upregulation | - | CTNNBIP1
Wnt/β-catenin pathway TCF/LEF | Cell proliferation,
migration, invasion and apoptosis | Therapeutic | (49) |
| 8. | circNUP214
(Hsa-circ-0089153) | Upregulation | Chr 9 | miR-145 ZEB2 | Cell proliferation,
migration and invasion | Therapeutic | (51) |
| 9. | circ-ITCH | Downregulation | - | miR-22-3p CBL
Wnt/β-catenin | Cell proliferation,
invasion and apoptosis | Therapeutic | (67) |
| 10. |
hsa-circ-047771 | Downregulation | - | miR-522-3p
miR-153-3p | - | Diagnostic and
prognostic and therapeutic | (12) |
| 11. |
hsa-circ-007148 | Upregulation | - | - | - | Diagnostic and
prognostic and therapeutic | (12) |
| 12. |
hsa-circ-0004458 | Upregulation | Chr 8:
18656804-18662408 | miR-885-5p | Cell proliferation,
migration and invasion | Therapeutic | (54) |
| 13. | circZFR
(hsa-circ-0072088) | Upregulation | Chr 5p13.3 | miR-1261
C8orf4 | Cell proliferation
and invasion | Therapeutic | (59) |
| 14. |
hsa-circ-0137287 | Downregulation | Chr 8:
92301363-92307931 | - | Cell proliferation
and invasion | Diagnostic and
therapeutic | (71) |
| 15. |
hsa-circ-100395 | Downregulation | - | miR-141-3p
miR-200a-3p | - | - | (34) |
| 16. | circPVT1 | Upregulation | Chr 8q24 | miR-126 | Cell proliferation,
invasion and apoptosis | Prognostic and
therapeutic | (62) |
| 17. | circRASSF2
(hsa-circ-0059354) | Upregulation | Chr 20:
4760668-4766974 | miR-1178 TLR4 | Cell proliferation,
invasion and apoptosis | Diagnostic and
therapeutic | (64) |
| 18. | circ-0005273 | Upregulation | - | miR-1138 SOX2 | PTC tumor growth
and progression | Prognostic and
therapeutic | (65) |
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
DQ conceived and designed the study. NS and TU
contributed to the writing of the manuscript, and were involved in
the literature search. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cancer Genome Atlas Research Network.
Integrated genomic characterization of papillary thyroid carcinoma.
Cell. 159:676–690. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
La Vecchia C, Malvezzi M, Bosetti C,
Garavello W, Bertuccio P, Levi F and Negri E: Thyroid cancer
mortality and incidence: A global overview. Int J Cancer.
136:2187–2195. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Xing M: BRAF mutation in papillary thyroid
cancer: Pathogenic role, molecular bases, and clinical
implications. Endocr Rev. 28:742–762. 2007.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sheu SY, Grabellus F, Schwertheim S, Worm
K, Broecker-Preuss M and Schmid KW: Differential miRNA expression
profiles in variants of papillary thyroid carcinoma and
encapsulated follicular thyroid tumours. Br J Cancer. 102:376–382.
2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Xu B, Shao Q, Xie K, Zhang Y, Dong T, Xia
Y and Tang W: The long non-coding RNA ENST00000537266 and
ENST00000426615 influence papillary thyroid cancer cell
proliferation and motility. Cell Physiol Biochem. 38:368–378.
2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Saporito D, Brock P, Hampel H, Sipos J,
Fernandez S, Liyanarachchi S, de la Chapelle A and Nagy R:
Penetrance of a rare familial mutation predisposing to papillary
thyroid cancer. Fam Cancer. 17:431–434. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Clarke CA, Reynolds P, Oakley-Girvan I,
Lee E, Lu Y, Yang J, Moy LM, Bernstein L and Horn-Ross PL:
Indicators of microbial-rich environments and the development of
papillary thyroid cancer in the California teachers study. Cancer
Epidemiol. 39:548–553. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Viola D, Materazzi G, Valerio L, Molinaro
E, Agate L, Faviana P, Seccia V, Sensi E, Romei C, Piaggi P, et al:
Prophylactic central compartment lymph node dissection in papillary
thyroid carcinoma: Clinical implications derived from the first
prospective randomized controlled single institution study. J Clin
Endocrinol Metab. 100:1316–1324. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Haugen BR, Alexander EK, Bible KC, Doherty
GM, Mandel SJ, Nikiforov YE, Pacini F, Randolph GW, Sawka AM,
Schlumberger M, et al: 2015 American Thyroid Association management
guidelines for adult patients with thyroid nodules and
differentiated thyroid cancer: The American Thyroid Association
guidelines task force on thyroid nodules and differentiated thyroid
cancer. Thyroid. 26:1–133. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yin Y, Hong S, Yu S, Huang Y, Chen S, Liu
Y, Zhang Q, Li Y and Xiao H: miR-195 inhibits tumor growth and
metastasis in papillary thyroid carcinoma cell lines by targeting
CCND1 and FGF2. Int J Endocrinol. 2017(6180425)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ren H, Liu Z, Liu S, Zhou X, Wang H, Xu J,
Wang D and Yuan G: Profile and clinical implication of circular
RNAs in human papillary thyroid carcinoma. PeerJ.
6(e5363)2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xing M, Tufano RP, Tufaro AP, Basaria S,
Ewertz M, Rosenbaum E, Byrne PJ, Wang J, Sidransky D and Ladenson
PW: Detection of BRAF mutation on fine needle aspiration biopsy
specimens: A new diagnostic tool for papillary thyroid cancer. J
Clin Endocrinol Metab. 89:2867–2872. 2004.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kimura ET, Nikiforova MN, Zhu Z, Knauf JA,
Nikiforov YE and Fagin JA: High prevalence of BRAF mutations in
thyroid cancer: Genetic evidence for constitutive activation of the
RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma.
Cancer Res. 63:1454–1457. 2003.PubMed/NCBI
|
|
15
|
Kwak JY, Kim EK, Chung WY, Moon HJ, Kim MJ
and Choi JR: Association of BRAFV600E mutation with poor clinical
prognostic factors and US features in Korean patients with
papillary thyroid microcarcinoma. Radiology. 253:854–860.
2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Haugen BR: 2015 American Thyroid
Association management guidelines for adult patients with thyroid
nodules and differentiated thyroid cancer: What is new and what has
changed? Cancer. 123:372–381. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Vriens MR, Weng J, Suh I, Huynh N,
Guerrero MA, Shen WT, Duh QY, Clark OH and Kebebew E: MicroRNA
expression profiling is a potential diagnostic tool for thyroid
cancer. Cancer. 118:3426–3432. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kos A, Dijkema R, Arnberg AC, van der
Meide PH and Schellekens H: The hepatitis delta (delta) virus
possesses a circular RNA. Nature. 323:558–560. 1986.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Conn VM, Hugouvieux V, Nayak A, Conos SA,
Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta
C and Conn SJ: A circRNA from SEPALLATA3 regulates splicing of its
cognate mRNA through R-loop formation. Nat Plants.
3(17053)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chen Y, Li C, Tan C and Liu X: Circular
RNAs: A new frontier in the study of human diseases. J Med Genet.
53:359–365. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Qian L, Yu S, Chen Z, Meng Z, Huang S and
Wang P: The emerging role of circRNAs and their clinical
significance in human cancers. Biochim Biophys Acta Rev Cancer.
1870:247–260. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li Y, Hu J, Li L, Cai S, Zhang H, Zhu X,
Guan G and Dong X: Upregulated circular RNA circ_0016760 indicates
unfavorable prognosis in NSCLC and promotes cell progression
through miR-1287/GAGE1 axis. Biochem Biophys Res Commun.
503:2089–2094. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013.
|
|
26
|
Soghli N, Qujeq D, Yousefi T and Soghli N:
The regulatory functions of circular RNAs in osteosarcoma.
Genomics. S0888-7543:31052–31053. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Xu B, Yang T, Wang Z, Zhang Y, Liu S and
Shen M: CircRNA CDR1as/miR-7 signals promote tumor growth of
osteosarcoma with a potential therapeutic and diagnostic value.
Cancer Manag Res. 10:4871–4880. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kai D, Yannian L, Yitian C, Dinghao G, Xin
Z and Wu J: Circular RNA HIPK3 promotes gallbladder cancer cell
growth by sponging microRNA-124. Biochem Biophys Res Commun.
503:863–869. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ling H, Fabbri M and Calin GA: MicroRNAs
and other non-coding RNAs as targets for anticancer drug
development. Nat Rev Drug Discov. 12:847–865. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
30
|
Huang H, Wei L, Qin T, Yang N, Li Z and Xu
Z: Circular RNA ciRS-7 triggers the migration and invasion of
esophageal squamous cell carcinoma via miR-7/KLF4 and NF-κB
signals. Cancer Biol Ther. 20:73–80. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Arnaiz E, Sole C, Manterola L,
Iparraguirre L, Otaegui D and Lawrie CH: CircRNAs and cancer:
Biomarkers and master regulators. Semin Cancer Biol. 58:90–99.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chen F, Feng Z, Zhu J, Liu P, Yang C,
Huang R and Deng Z: Emerging roles of circRNA_NEK6 targeting
miR-370-3p in the proliferation and invasion of thyroid cancer via
Wnt signaling pathway. Cancer Biol Ther. 19:1139–1152.
2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Peng N, Shi L, Zhang Q, Hu Y, Wang N and
Ye H: Microarray profiling of circular RNAs in human papillary
thyroid carcinoma. PLoS One. 12(e0170287)2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Teng H, Mao F, Liang J, Xue M, Wei W, Li
X, Zhang K, Feng D, Liu B and Sun Z: Transcriptomic signature
associated with carcinogenesis and aggressiveness of papillary
thyroid carcinoma. Theranostics. 8:4345–4358. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Griffith OL, Melck A, Jones SJ and Wiseman
SM: Meta-analysis and meta-review of thyroid cancer gene expression
profiling studies identifies important diagnostic biomarkers. J
Clin Oncol. 24:5043–5051. 2006.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yao Y, Chen X, Yang H, Chen W, Qian Y, Yan
Z, Liao T, Yao W, Wu W, Yu T, et al: Hsa_circ_0058124 promotes
papillary thyroid cancer tumorigenesis and invasiveness through the
NOTCH3/GATAD2A axis. J Exp Clin Cancer Res. 38(318)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Pan Y, Xu T, Liu Y, Li W and Zhang W:
Upregulated circular RNA circ_0025033 promotes papillary thyroid
cancer cell proliferation and invasion via sponging miR-1231 and
miR-1304. Biochem Biophys Res Commun. 510:334–338. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yang Y, Ding L, Li Y and Xuan C:
Hsa_circ_0039411 promotes tumorigenesis and progression of
papillary thyroid cancer by miR-1179/ABCA9 and miR-1205/MTA1
signaling pathways. J Cell Physiol. 235:1321–1329. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Cai X, Zhao Z, Dong J, Lv Q, Yun B, Liu J,
Shen Y, Kang J and Li J: Circular RNA circBACH2 plays a role in
papillary thyroid carcinoma by sponging miR-139-5p and regulating
LMO4 expression. Cell Death Dis. 10(184)2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhang J, Zhang J, Qiu W, Zhang J, Li Y,
Kong E, Lu A, Xu J and Lu X: MicroRNA-1231 exerts a tumor
suppressor role through regulating the EGFR/PI3K/AKT axis in
glioma. J Neurooncol. 139:547–562. 2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wang H, Wu J, Luo W and Hu J: Low
expression of miR-1231 in patients with glioma and its prognostic
significance. Eur Rev Med Pharmacol Sci. 22:8399–8405.
2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Li CG, Pu MF, Li CZ, Gao M, Liu MX, Yu CZ,
Yan H, Peng C, Zhao Y, Li Y, et al: MicroRNA-1304 suppresses human
non-small cell lung cancer cell growth in vitro by targeting heme
oxygenase-1. Acta Pharmacol Sin. 38:110–119. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhou GK, Zhang GY, Yuan ZN, Pei R and Liu
DM: Has_circ_0008274 promotes cell proliferation and invasion
involving AMPK/mTOR signaling pathway in papillary thyroid
carcinoma. Eur Rev Med Pharmacol Sci. 22:8772–8780. 2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Cargnello M, Tcherkezian J and Roux PP:
The expanding role of mTOR in cancer cell growth and proliferation.
Mutagenesis. 30:169–176. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Liu F, Zhang J, Qin L, Yang Z, Xiong J,
Zhang Y, Li R, Li S, Wang H, Yu B, et al: Circular RNA EIF6
(Hsa_circ_0060060) sponges miR-144-3p to promote the
cisplatin-resistance of human thyroid carcinoma cells by autophagy
regulation. Aging (Albany NY). 10:3806–3820. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Xiao W, Lou N, Ruan H, Bao L, Xiong Z,
Yuan C, Tong J, Xu G, Zhou Y, Qu Y, et al: Mir-144-3p promotes cell
proliferation, metastasis, sunitinib resistance in clear cell renal
cell carcinoma by downregulating ARID1A. Cellular Cell Physiol
Biochem. 43:2420–2433. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhao Y, Xie Z, Lin J and Liu P: MiR-144-3p
inhibits cell proliferation and induces apoptosis in multiple
myeloma by targeting c-Met. Am J Transl Res. 9:2437–2446.
2017.PubMed/NCBI
|
|
49
|
Bi W, Huang J, Nie C, Liu B, He G, Han J,
Pang R, Ding Z, Xu J and Zhang J: CircRNA circRNA_102171 promotes
papillary thyroid cancer progression through modulating
CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin
Cancer Res. 37(275)2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Fu X, Zhu X, Qin F, Zhang Y, Lin J, Ding
Y, Yang Z, Shang Y, Wang L, Zhang Q and Gao Q: Linc00210 drives
Wnt/β-catenin signaling activation and liver tumor progression
through CTNNBIP1-dependent manner. Mol Cancer.
17(73)2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Li X, Tian Y, Hu Y, Yang Z, Zhang L and
Luo J: CircNUP214 sponges miR-145 to promote the expression of ZEB2
in thyroid cancer cells. Biochem Biophys Res Commun. 507:168–172.
2018.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Liu Q, Chen J, Wang B, Zheng Y, Wan Y,
Wang Y, Zhou L, Liu S, Li G and Yan Y: miR-145 modulates
epithelial-mesenchymal transition and invasion by targeting ZEB2 in
non-small cell lung cancer cell lines. J Cell Biochem: Dec 7, 2018
(Epub ahead of print).
|
|
53
|
Brown CY, Dayan S, Wong SW, Kaczmarek A,
Hope CM, Pederson SM, Arnet V, Goodall GJ, Russell D, Sadlon TJ and
Barry SC: FOXP3 and miR-155 cooperate to control the invasive
potential of human breast cancer cells by down regulating ZEB2
independently of ZEB1. Oncotarget. 9:27708–27727. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Jin X, Wang Z, Pang W, Zhou J, Liang Y,
Yang J, Yang L and Zhang Q: Upregulated hsa_circ_0004458
contributes to progression of papillary thyroid carcinoma by
inhibition of miR-885-5p and activation of RAC1. Med Sci Monit.
24:5488–5500. 2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chen QY, Zheng Y, Jiao DM, Chen FY, Hu HZ,
Wu YQ, Song J, Yan J, Wu LJ and Lv GY: Curcumin inhibits lung
cancer cell migration and invasion through Rac1-dependent signaling
pathway. J Nutr Biochem. 25:177–185. 2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Becker MS, Müller PM, Bajorat J, Schroeder
A, Giaisi M, Amin E, Ahmadian MR, Rocks O, Köhler R, Krammer PH and
Li-Weber M: The anticancer phytochemical rocaglamide inhibits Rho
GTPase activity and cancer cell migration. Oncotarget.
7:51908–51921. 2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Ren S, Xin Z, Xu Y, Xu J and Wang G:
Construction and analysis of circular RNA molecular regulatory
networks in liver cancer. Cell Cycle. 16:2204–2211. 2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Liu T, Liu S, Xu Y, Shu R, Wang F, Chen C,
Zeng Y and Luo H: Circular RNA-ZFR inhibited cell proliferation and
promoted apoptosis in gastric cancer by sponging miR-130a/miR-107
and modulating PTEN. Cancer Res Treat. 50:1396–1417.
2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Wei H, Pan L, Tao D and Li R: Circular RNA
circZFR contributes to papillary thyroid cancer cell proliferation
and invasion by sponging miR-1261 and facilitating C8orf4
expression. Biochem Biophys Res Commun. 503:56–61. 2018.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Sunde M, McGrath KC, Young L, Matthews JM,
Chua EL, Mackay JP and Death AK: TC-1 is a novel tumorigenic and
natively disordered protein associated with thyroid cancer. Cancer
Res. 64:2766–2773. 2004.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Lei J, Li W, Yang Y, Lu Q, Zhang N, Bai G,
Zhong D, Su K, Liu B, Li X, et al: TC-1 overexpression promotes
cell proliferation in human non-small cell lung cancer that can be
inhibited by PD173074. PLoS One. 9(e100075)2014.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Tao L, Yang L, Tian P, Guo X and Chen Y:
Knockdown of circPVT1 inhibits progression of papillary thyroid
carcinoma by sponging miR-126. RSC Adv. 9:13316–13324. 2019.
|
|
63
|
Kitano M, Rahbari R, Patterson EE, Xiong
Y, Prasad NB, Wang Y, Zeiger MA and Kebebew E: Expression profiling
of difficult-to-diagnose thyroid histologic subtypes shows distinct
expression profiles and identify candidate diagnostic microRNAs.
Ann Surg Oncol. 18:3443–34452. 2011.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Wu G, Zhou W, Lin X, Sun Y, Li J, Xu H,
Shi P, Gao L and Tian X: CircRASSF2 Acts as ceRNA and promotes
papillary thyroid carcinoma progression through miR-1178/TLR4
signaling pathway. Mol Ther Nucleic Acids. 19:1153–1163.
2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Zhang W, Zhang H and Zhao X: Circ_0005273
promotes thyroid carcinoma progression by SOX2 expression. Endocr
Relat Cancer. 27:11–21. 2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Li F, Ma K, Sun M and Shi S:
Identification of the tumor-suppressive function of circular RNA
ITCH in glioma cells through sponging miR-214 and promoting linear
ITCH expression. Am J Transl Res. 10:1373–1386. 2018.PubMed/NCBI
|
|
67
|
Wang M, Chen B, Ru Z and Cong L: CircRNA
circ-ITCH suppresses papillary thyroid cancer progression through
miR-22-3p/CBL/β-catenin pathway. Biochem Biophys Res Commun.
504:283–288. 2018.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Shashar M, Siwak J, Tapan U, Lee SY, Meyer
RD, Parrack P, Tan J, Khatami F, Francis J, Zhao Q, et al: C-Cbl
mediates the degradation of tumorigenic nuclear β-catenin
contributing to the heterogeneity in Wnt activity in colorectal
tumors. Oncotarget. 7:71136–71150. 2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Lupi C, Giannini R, Ugolini C, Proietti A,
Berti P, Minuto M, Materazzi G, Elisei R, Santoro M, Miccoli P and
Basolo F: Association of BRAF V600E mutation with poor
clinicopathological outcomes in 500 consecutive cases of papillary
thyroid carcinoma. J Clin Endocrinol Metab. 92:4085–4090.
2007.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Elisei R, Ugolini C, Viola D, Lupi C,
Biagini A, Giannini R, Romei C, Miccoli P, Pinchera A and Basolo F:
BRAF(V600E) mutation and outcome of patients with papillary thyroid
carcinoma: A 15-year median follow-up study. J Clin Endocrinol
Metab. 93:3943–3949. 2008.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Lan X, Cao J, Xu J, Chen C, Zheng C, Wang
J, Zhu X, Zhu X and Ge M: Decreased expression of hsa_circ_0137287
predicts aggressive clinicopathologic characteristics in papillary
thyroid carcinoma. J Clin Lab Anal. 32(e22573)2018.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics 2018. CA Cancer J Clin. 68:7–30. 2018.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Lan X, Xu J, Chen C, Zheng C, Wang J, Cao
J, Zhu X and Ge M: The landscape of circular RNA expression
profiles in papillary thyroid carcinoma based on RNA sequencing.
Cell Physiol Biochem. 47:1122–1132. 2018.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Liu Q, Pan LZ, Hu M and Ma JY: Molecular
Network-Based identification of circular RNA-Associated ceRNA
network in papillary thyroid cancer. Pathol Oncol Res 2019 [Epub
ahead of print].
|