Introduction
The National Cancer Institute of the United States
estimated that approximately 175,000 new prostate cancer cases will
be identified in 2020, while there will be a projected 32,000
cancer-related deaths among American males. Approximately 14% of
men will acquire the disease throughout their lifetimes (1). To date, radiation therapy,
chemotherapy and hormone therapy are the most common treatments for
PC. Radiation therapy is not ideal as it destroys surrounding
healthy cells and tissues, leading to a number of side-effects.
Hormone treatment implies clinical castration and the usage of
anti-androgens, that can have adverse effects and can affect the
lifestyle of the recipient (2).
Moreover, PC treatments involve the use of chemotherapeutic agents,
such as docetaxel. Docetaxel only increases the survival time of
the patient by approximately 100 days, primarily due to the
emergence of resistance (3).
In previous studies by the authors, protein kinase
C-ι (PKC-ι) was recognized as a major driving factor of prostate
and melanoma carcinogenesis, and was therefore proposed as a
potential and novel therapeutic target (4-10).
PKC is a part of the protein kinase enzyme class that
post-translationally modifies certain other proteins and
participates in a variety of cellular signaling cascades. A total
of 15 PKC isoforms are recognized in humans; these are further
grouped as classical, novel and atypical PKCs (aPKCs). aPKCs
comprise two structurally and functionally distinctive isoforms;
PKC-ζ and PKC-ι (11-13).
Other than PC and melanoma, PKC-ι has been found to function as an
oncogene in several other cancer types, such as neuroblastoma,
ovarian cancer and glioma, where the upregulation of PKC-ι
expression has been shown to be associated with a low survival rate
(10,11,14).
In previous studies by the authors, it was also reported that
higher levels of PKC-ι were observed in DU-145 and PC-3 cells
compared to undetectable levels in normal tissues and the normal
prostate epithelial cell line, RWPE-1 (7,10).
In addition, it was demonstrated that PKC-ι phosphorylates to
activate IκB kinase (IKKα/β). This p-IKKα/β activation triggers the
dissociation of nuclear factor of κ light polypeptide gene enhancer
in B-cells inhibitor (IκB) from the nuclear factor NF-κB complex,
which leads to the ubiquitination of IκB. IκB releasing from NF-κB
activates the translocation of NF-κB to the nucleus. Previous
studies by the authors suggested that PKC-ι specific leads to the
suppression of NF-κB nuclei translocation, thereby causing a
downregulation of NF-κB activity (9,10).
These findings indicated that the inhibition of PKC-ι not only
impaired pathways regulated by PKC-ι, but also downgraded its
protein expression (7-10).
There are data to suggest that PKC-ι maintains a self-propagative
mechanism, as observed in certain other cancer-related cycles, such
as the transformation of the growth factor (TGF)-β and
CD147(15). Since transcription
factors play a pivotal role in gene expression, the aim of the
present study was to determine which transcription factors were key
PKC-ι regulators, as well as which pathways were integral
components for these transcription factors.
In the present study, the outcomes of the knockdown
of the expression of c-Jun, FOXO1, PKC-ι and NF-κB are
demonstrated, giving emphasis to pathways associated with PKC-ι.
The findings suggested that c-Jun is a crucial transcriptional
activator, while FOXO1 functions as a transcriptional suppressor of
PRKCI expression. The roles performed by these transcription
factors were determined in an inflammatory process that promotes
PKC-ι and is dependent on PKC-ι for the continuation of the
process. Moreover, the pathway through which cytokines stimulate
PKC-ι expression to release activated NF-κB is demonstrated, which
leads to the production of additional cytokines; this process is
used in certain types of cancer as part of a loop through which to
grow and propagate. In addition, IL-8 promotes c-Jun and NF-κB
signaling to enhance PKC-ι production. On the other hand, PKC-ι
inhibition induces the production of ICAM-1, which promotes FOXO1
to reduce PKC-ι production. In general, these findings indicate
that with a dynamic and closely controlled expression profile,
PKC-ι plays a central role in the development of prostate cancer.
The specific inhibition of PKC-ι may interact with its own
regulatory process, contributing to a distortion of its oncogenic
function in prostate cancer.
Materials and methods
Materials and reagents
[4-(5-Amino-4-carbamoylimidazol-1-yl)-2,3-dihydroxycyclopentyl]
methyl dihydrogen phosphate (ICA-1T) was purchased by Therachem and
the NF-κB-specific inhibitor,
4-methyl-N1-(3-phenylpropyl)-1,2-benzenediamine (JSH-23, J4455),
was purchased from Sigma-Aldrich; Merck KGaA. Sterile distilled
water was used as the solvent for the inhibitors. The following
materials were acquired: Antibodies against PKC-ι (610175, BD
Biosciences); NF-κB p65 (sc-372-G, Santa Cruz Biotechnology, Inc.);
p-PKC-ι (T555; ab5813, Abcam); FOXO1 (2880S), p-FOXO1 (T24; 9464S),
c-Jun (9165S), p-c-Jun (S73; 3270S), mammalian target of rapamycin
(mTOR; 2972S), p-AKT (S473; 4059S), signal transducer and activator
of transcription (STAT)3 (9139S) and STAT5 (25656S) (all from Cell
Signaling Technology, Inc.); and β-actin (A3854, Sigma-Aldrich;
Merck KGaA); enhanced chemiluminescence solution (34080, Pierce;
Thermo Fisher Scientific, Inc.); human small interfering RNA
(siRNA) for PKC-ι (SR303741), c-Jun (SR302499), FOXO1 (SR301618),
early growth response 1 (EGR1; SR301358), paired box gene 3 (PAX3;
SR303360), interferon regulatory factor 9 (IRF9; SR307030), NF-κB
p65 (SR321602) (all from Origene Technologies, Inc.); DPBS without
magnesium and calcium ions (Dulbecco's phosphate-buffered saline,
D8537), Trypsin-ethylenediaminetetraacetic acid (EDTA; T4049,
Sigma-Aldrich; Merck KGaA); recombinant protein tumor necrosis
factor (TNF)-α (human, 10602HNAE25, Thermo Fisher Scientific,
Inc.).
Cells and cell culture
DU-145 (ATCC® HTB81™) and PC-3
(ATCC® CRL-1435™) cells were obtained from the American
Type Tissue Culture Collection (ATCC). All cell lines were
authenticated by ATCC using karyotyping, morphology and PCR-based
approaches. Early passages of cells were cryo-preserved in liquid
nitrogen and cells of early passages were resuscitated from liquid
nitrogen for experiments. A temperature of 37˚C and 5%
CO2 were maintained as the cell culture conditions. EMEM
(ATCC 30-2003) and RPMI-1640 media (ATCC 30-2001) were used with
fetal bovine serum (FBS, 10% v/v) and penicillin (5 µg/ml) for the
DU-145 and PC-3 cells, respectively.
Identification of c-Jun and FOXO1 as
probable transcription factors to bind to the PRKCI promoter
region
The gene sequence of PRKCI which is located on
chromosome 3 between bp170222365-170305981 (3q26.2) was acquired
from ensemble.org (ENSG00000163558) (16,17).
The promoter sequence was identified by comparing the forward
strand sequence with the Eukaryotic Promoter Database (EPD,
https://epd.vital-it.ch/index.php). The
sequence between bp170220768-170225128 was selected and which
contained the promoter, promoter flank, enhancer and a motif
feature. Transcription factors that exhibited a probability to bind
with an accuracy >90% were selected using PROMO; a virtual
laboratory for reviewing transcription factor binding sites in DNA
selected sequences (http://alggen.lsi.upc.es/). The PROMO results were
also compared with the Genomatix Matinspector results to confirm
the accuracy.
Knockdown of c-Jun, FOXO1, PKC-ι and
NF-κB gene expression by siRNA
The PC-3 and DU-145 (1x105) cells were
seeded in T25 flasks and at 24 h post-seeding, siRNA (30 nM)
transfections were conducted against scrambled siRNA for 2 days
using ‘siTran’ siRNA transfection reagent (TT300002) from Origene
Technologies, Inc. according to the manufacturer's recommended
ratios. The cell pellets were collected at the end of the 48-h
incubation period and cell lysates were prepared using cell lysis
buffer (C7027, Thermo Fisher Scientific, Inc.). Western blot and
densitometric analyses were executed as previously described by
Ratnayake et al (9,18).
Prostate cancer cellular cytokine
expression analysis
The cytokine array kit (ARY005B, R&D Systems)
contained with an enzyme-linked immunosorbent assay (ELISA) was
used for the experiment. Approximately 1x105 cells were
cultured in T25 flasks (PC-3 and DU-145) and at 24 h post-plating,
the cells were treated with a ICA-1T (2.5 µM) for 2 consecutive
days at 24-h intervals. The cells were then collected and cell
lysates were prepared and administered according to the
manufacturer's instructions. The experiment was repeated and TNF-α
(250 ng/ml) was added to the flasks 30 min prior to the harvesting
point. Total protein (150 µg) was used from each sample and
introduced to the immunoblots provided and chemiluminescence images
were acquired using ‘ECL Western Blotting Substrate’ (Thermo
Fischer Scientific, PI32106). These images were then analyzed as
instructed in the cytokine array kit manual (ARY005B).
Immunopaired antibody detection
assay
Approximately 1x105 cells (PC-3 and
DU-145) were cultured in T25 flasks and ICA-1T (2.5 µM) treatments
were conducted as descirbed above. Cells were then collected and
cell lysates was prepared to contain the final total protein
concentration >2 µg/ml. Samples were sent to ActivSignal, LLC
for analysis. The ActivSignal IPAD assay is a multiplex ELISA-based
proprietary tool for evaluating multiple signaling cascades
considering both upstream and downstream targets. In total, >20
signaling pathways were examined at once in a single well by
assessing the expression or protein phosphorylation of 70 human
proteins targets.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was performed on RNA isolated from PC-3 and
DU-145 cell lysates collected following ICA-1T (with or without
TNF-α) or siRNA treatments (as described in above) against their
respective controls. The detailed procedure was previously
described by Ratnayake et al (18). Total RNA was isolated from the cell
pellets using RNA lysis buffer (RNeasy mini kit, 74104) from
Qiagen, Inc. RNA was reverse transcribed into cDNA with You-Prime
First Strand Beads (27-9264-01) form GE Healthcare. qPCR was
performed on cDNA using the QuantStudio3 Real-Time PCR system
(Thermo Fisher Scientific, Inc.). The following primers were used:
PKC-ι forward, TTGCAATGAGGTTCGAGACA and reverse,
CTGAGATGATACTGTACACGGG; c-Jun forward, GTGCCGAAAAAGGAAGCTGG and
reverse, CTGCGTTAGCATGAGTTGGC; FOXO1 forward,
ATGGCTTGGTGTCTTTCTTTTCT and reverse, TGTGGCTGACAAGACTTAACTCAA; IL-8
forward, CAGAGACAGCAGAGCACAC and reverse, ATCAGGAAGGCTGCCAAGAG;
ICAM-1 forward, GGGAACAACCGGAAGGTGTA and reverse,
CAGTTCCACCCGTTCTGGAG; and β-actin forward, AGAGCTACGAGCTGCCTGAC and
reverse, AGCACTGTGTTGGCGTACAG which was used as an internal
control. PCR reactions conditions were used as explained by Livak
and Schmittgen (19). PCR
reactions used SYBR-Green PCR Mix (Applied Biosystems). cDNA was
denatured at 95˚C for 10 min, followed by 40 cycles of denaturing
at 95˚C for 20 sec and an annealing stage of 65˚C for 40 sec.
QuantStudio Software 2.0 was used to quantify gene expression by
the 2-ΔΔCq method (Thermo Fisher Scientific, Inc.).
Statistical analysis
All data are presented as the means ± SD.
Statistical analysis was carried out using one or two-way ANOVA
followed by Tukey's HSD test as multiple comparisons tests using
the statistical research online tool ‘VassarStats’. P-values ≤0.05
or ≤0.01 were considered to indicate statistically significant or
highly statistically significant differences, respectively.
Results
The unique PRKCI sequence, carefully selected to
contain the promoter, promoter flank, enhancer and a motif element,
was 4,360 bp in length (chr3; bp170220768-170225128). The promoter
allowed TFs to bind and start transcription, while the enhancer
ensured a regulating area on the flank that promoted transcription
factor binding. By having only transcription factors, which bind
within a dissimilarity range of approximately 10%, potential hits
were narrowed down to achieve a high specificity. After analyzing
the results, approximately 70 transcription factor hits to the
target were obtained. c-Jun, ISGF3, PAX3, EGR1 and FOXO1 were
identified as the top 5 transcription factors with the greatest
likelihood of binding to the PRKCI gene sequence.
c-Jun and FOXO1 are the two key TFs of
PKC-ι expression in PC-3 and DU-145 cells
As presented in Fig.
1, the results of western blot analysis revealed that each
siRNA transfection targeting FOXO1 and c-Jun markedly diminished
the expression levels of those targets. siRNA against FOXO1
significantly knocked down FOXO1 by 64% (P≤0.05) and 27% (P≤0.05),
while p-FOXO1 (T24) by 19% (P≤0.05) and 79% (P≤0.05) in the PC-3
and DU-145 cells, respectively. siRNA against c-Jun knocked down
c-Jun by 48% (P≤0.05) and 73% (P≤0.05), while p-c-Jun (S73) by 26%
(P≤0.05) and 43% (P≤0.05) in the PC-3 and DU-145 cells,
respectively. These findings indicated that transfection with siRNA
knocked down the respective target expression. Only the knockdown
of c-Jun and FOXO1 in both cell lines was shown to have an affect
on PKC-ι levels. The diminution of FOXO1 by siRNA increased total
PKC-ι expression by 33% (P≤0.05) and 9% (P≤0.05) in the PC-3 and
DU-145 cells, respectively. The diminution of c-Jun by siRNA
diminished PKC-ι expression by 42% (P≤0.05) and 24% (P≤0.05) in the
PC-3 and DU-145 cells, respectively. Similarly, lower levels of
FOXO1 due to transfection with siRNA augmented p-PKC-ι (T555)
expression by 18% (P≤0.05) and 22% (P≤0.05) in the PC-3 and DU-145
cells, respectively. Of note, c-Jun diminution decreased PKC-ι
(T555) expression by 36% (P≤0.05) and 13% (P≤0.05) in the PC-3 and
DU-145 cells, respectively. The knockdown of the expression of
ISGF3, EGR1 and PAX3 did not exert notable effect on PKC-ι
expression or on its phosphorylated protein levels; thus, these
data were not included in this manuscript. Hence, FOXO1 and c-Jun
were selected for use in the following experiments.
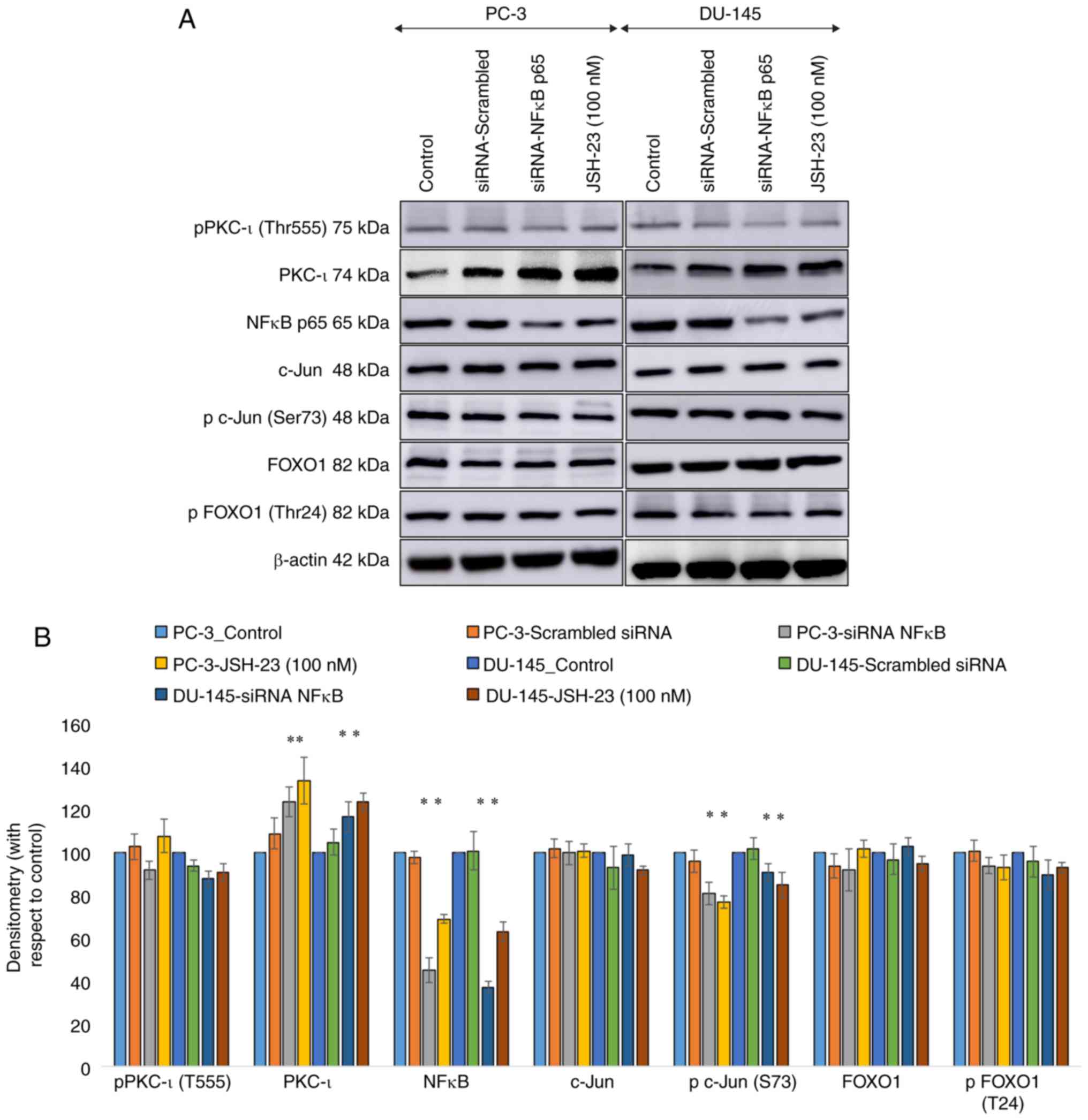
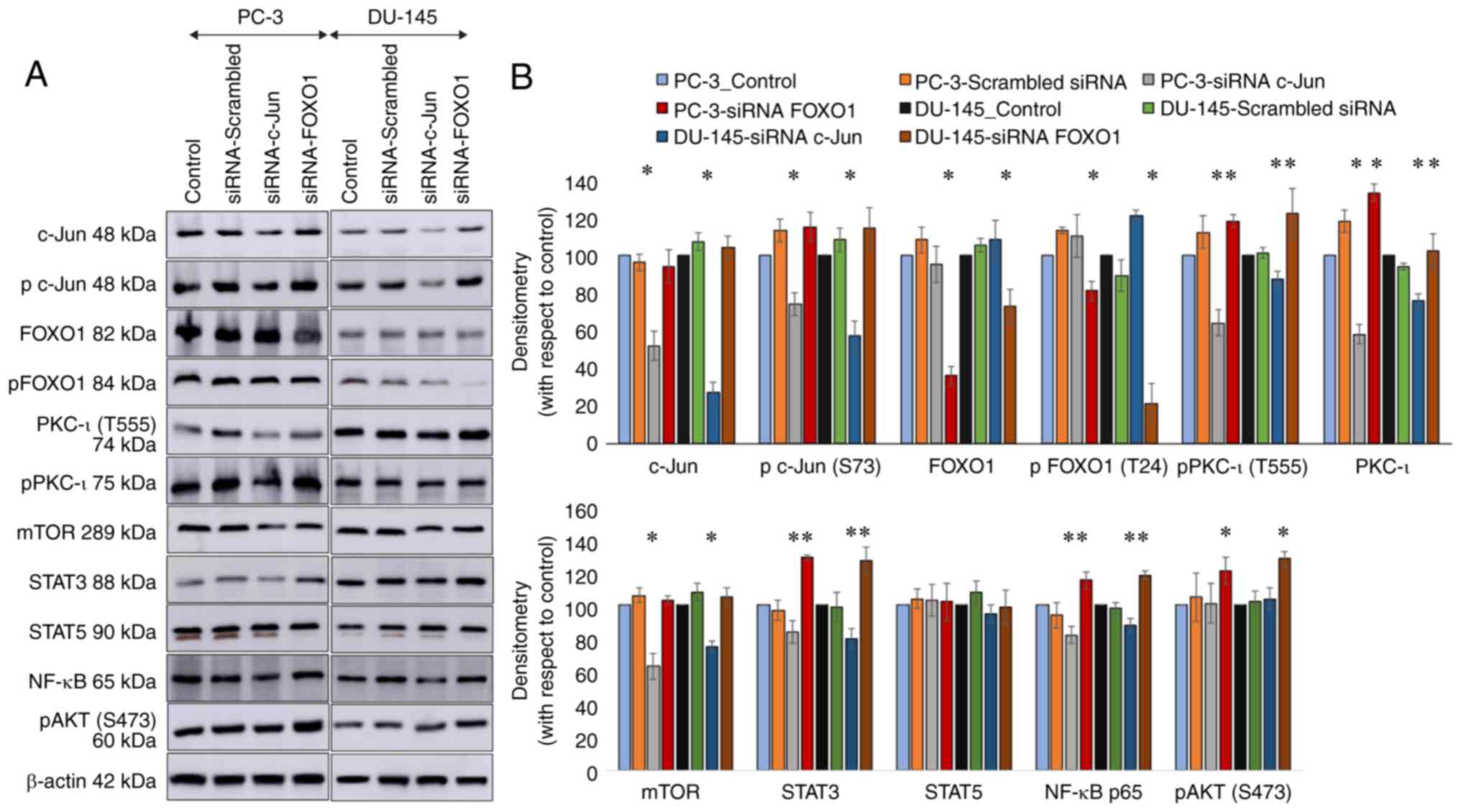 | Figure 1Effect of RNA interference (siRNA) of
the transcription factors, c-Jun and FOXO1, in two prostate cancer
cell lines (PC-3 and DU-145). (A) Expression of the protein levels
of phosphor-PKC-ι (T555), total PKC-ι, c-Jun, phosphor-c-Jun (S73),
FOXO1, phosphor-FOXO1 (T24), mTOR, STAT3, STAT5, NF-κB p65 and
phosphor-AKT (S473) following the siRNA knockdown of the expression
FOXO1 and c-Jun for PC-3 and DU-145 cell lines. Total protein (80
µg) was loaded into each well and β-actin was used as the internal
control in each western blot. (B) Representative densitometry
values for the western blots shown in (A) Experiments (n=3) were
performed in each trial and representative bands are shown.
Densitometry values are reported as the means ± SD. Statistical
significance is indicated by an asterisk (*P≤0.05). |
In addition to the total and p-PKC-ι levels, the
levels of mTOR, STAT3, STAT5, NF-κB p65 and p-AKT (S473) were
determined following transfection with c-Jun and FOXO1 siRNA.
Transfection with FOXO1 siRNA increased the expression of STAT3 by
29% (P≤0.05) and 27% (P≤0.05) in the PC-3 and DU-145 cells,
respectively. On the other hand, transfection with c-Jun siRNA
reduced STAT3 expression by 16% (P≤0.05) and 20% (P≤0.05) in PC-3
and DU-145 cells, respectively. Of note, transfection with FOXO1
and c-Jun siRNA did not exert a significant effect on STAT5
expression. The knockdown of c-Jun by siRNA deceased mTOR
expression by 37% (P≤0.05) and 25% (P≤0.05) in the PC-3 and DU-145
cells, respectively. FOXO1 diminution did not alter the protein
levels of mTOR. Transfection with FOXO1 siRNA increased the
expression of p-AKT (S473) by 21% (P≤0.05) and 28% (P≤0.05) in the
PC-3 and DU-145 cells, respectively. Transfection with c-Jun siRNA
did not exert a significant effect on the levels of p-AKT (S473).
Transfection with FOXO1 siRNA increased the expression of NF-κB p65
by 15% (P≤0.05) and 18% (P≤0.05) in the PC-3 and DU-145 cells,
respectively. On the other hand, the knockdown of c-Jun
significantly deceased NF-κB p65 expression by 18% (P≤0.05) and 12%
(P≤0.05) in the PC-3 and DU-145 cells, respectively (Fig. 1).
c-Jun and FOXO1 regulate atypical
PKC-ι expression through NF-κB and STAT3 signaling in prostate
cancer cells
As presented in Fig.
2, the findings of western blot analysis revealed that the
knockdown of NF-κB expression by siRNA substantially increased the
overall PKC-ι levels by 24 % (P≤0.05) and 17% (P≤0.05) in the PC-3
and DU-145 cells, respectively. The expression of p-PKC-ι (T555)
was not significantly altered. Of note, the FOXO1 and p-FOXO1
levels were not affected as a result of NF-κB depletion. Notably,
the p-c-Jun (S73) level was significantly decreased upon NF-κB
depletion. The total c-Jun levels were not affected as a result of
NF-κB depletion. Similar outcomes were acquired with JSH-23 (100
nM) treatments. JSH-23 is an established NF-κB specific inhibition
available on the market.
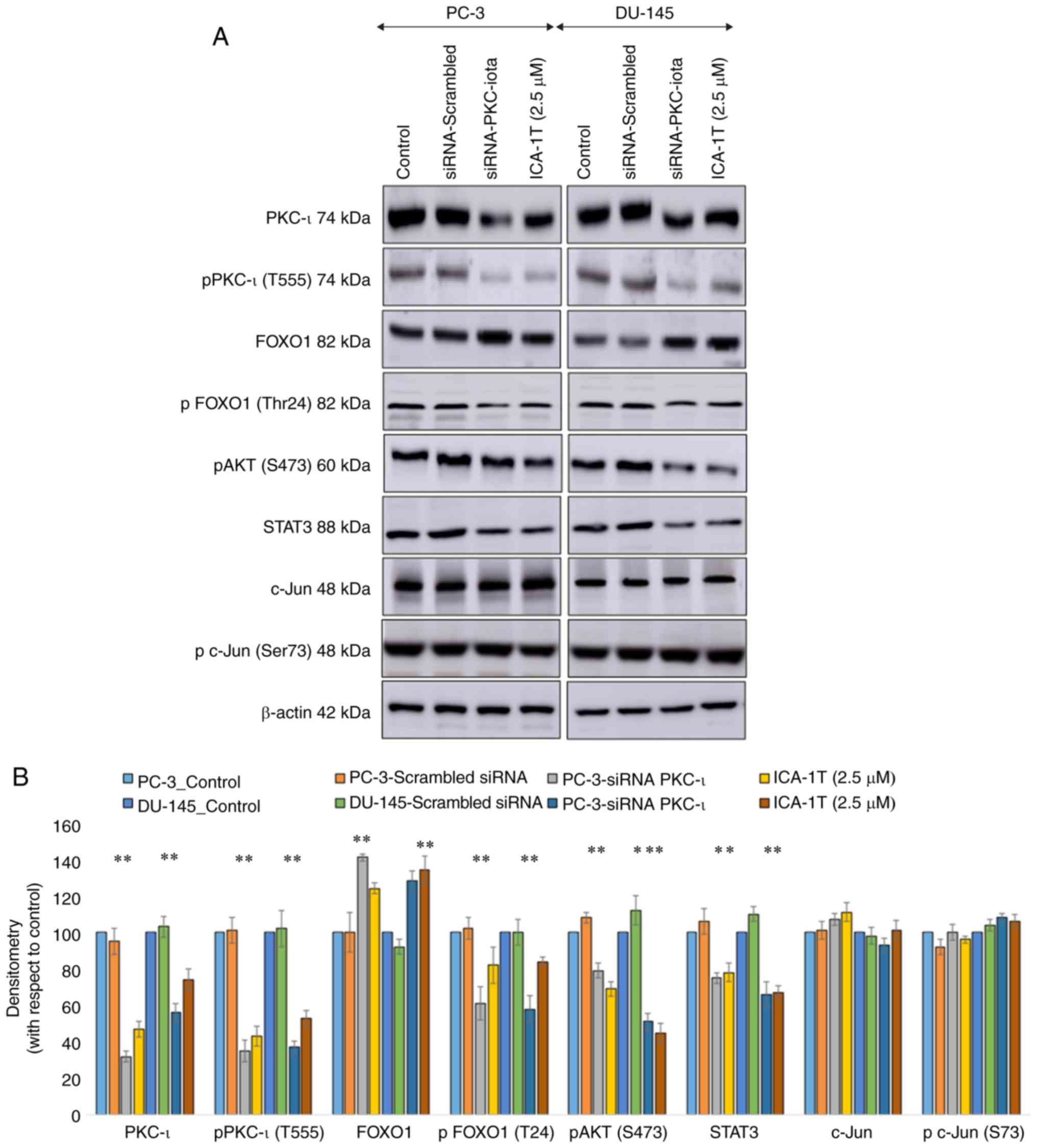
In addition, Fig. 3
demonstrates the effects of PKC-ι knockdown or specific inhibition
using ICA-1T on total PKC-ι, p-PKC-ι (T555), FOXO1, p-FOXO1, p-AKT
(S473), STAT3, c-Jun and p-c-Jun (S73) expression. Transfection
siRNA against PKC-ι and ICA-1T treatment yielded similar results.
The total PKC-ι and p-PKC-ι levels decreased significantly (P≤0.05)
with PKC-ι knockdown or inhibition. Of note, upon the depletion of
PKC-ι, the total FOXO1 levels increased (P≤0.05), while the levels
of p-FOXO1 decreased, indicating an upregulation of FOXO1 activity.
Similarly, both the p-AKT (S473) and STAT3 levels significantly
(P≤0.05) decreased owing to the decrease in PKC-ι expression. On
the other hand, the levels of c-Jun or p-c-Jun were not
substantially altered following the depletion of PKC-ι.
ELISA suggests the involvement of
multiple pathways; JNK/c-Jun, NF-κB/AKT/FOXO1 and STAT3 for the
regulation PKC-ι of expression
The specific inhibitor, ICA-1T, was to inhibit
PKC-ι, permitting us to gain a clearer view of the mechanisms
through which multiple cellular signaling pathways may affect PC-3
and DU-145 cells in vitro owing to PKC-ι regulation. The
IPAD assay is an ELISA series, which enables several proteins to be
identified simultaneously. Fig. 4
demonstrates the changes in the expression of CD44, E-cadherin,
caspase-3, H2AX, IκB and Myc, and the degree pf phosphorylation of
p-4E-BP1 (T37/46), p-AKT (S473), p-β-catenin (S33/37), p-HER3
(Y1289), pIKKαβ (S176/180), p-JNK (T183), p-mTOR (S2448), p-NF-κB
p65 (S536), p-Met1, p-STAT3 (Y705), p-STAT5 (Y694) and p-ZAP70
(Y493), as a result of ICA-1T inhibition against the respective
control samples for both PC-3 and DU-145 cell lines. The present
study observed that the levels of p-JNK (T183) p-mTOR (S2448) and
p-ZAP70 (Y493) significantly increased in DU-145 cells following
PKC-ι inhibition, while those of p-AKT (S473) and p-β-catenin
(S33/37) significantly decreased in DU-145 and PC-3 cells,
respectively.
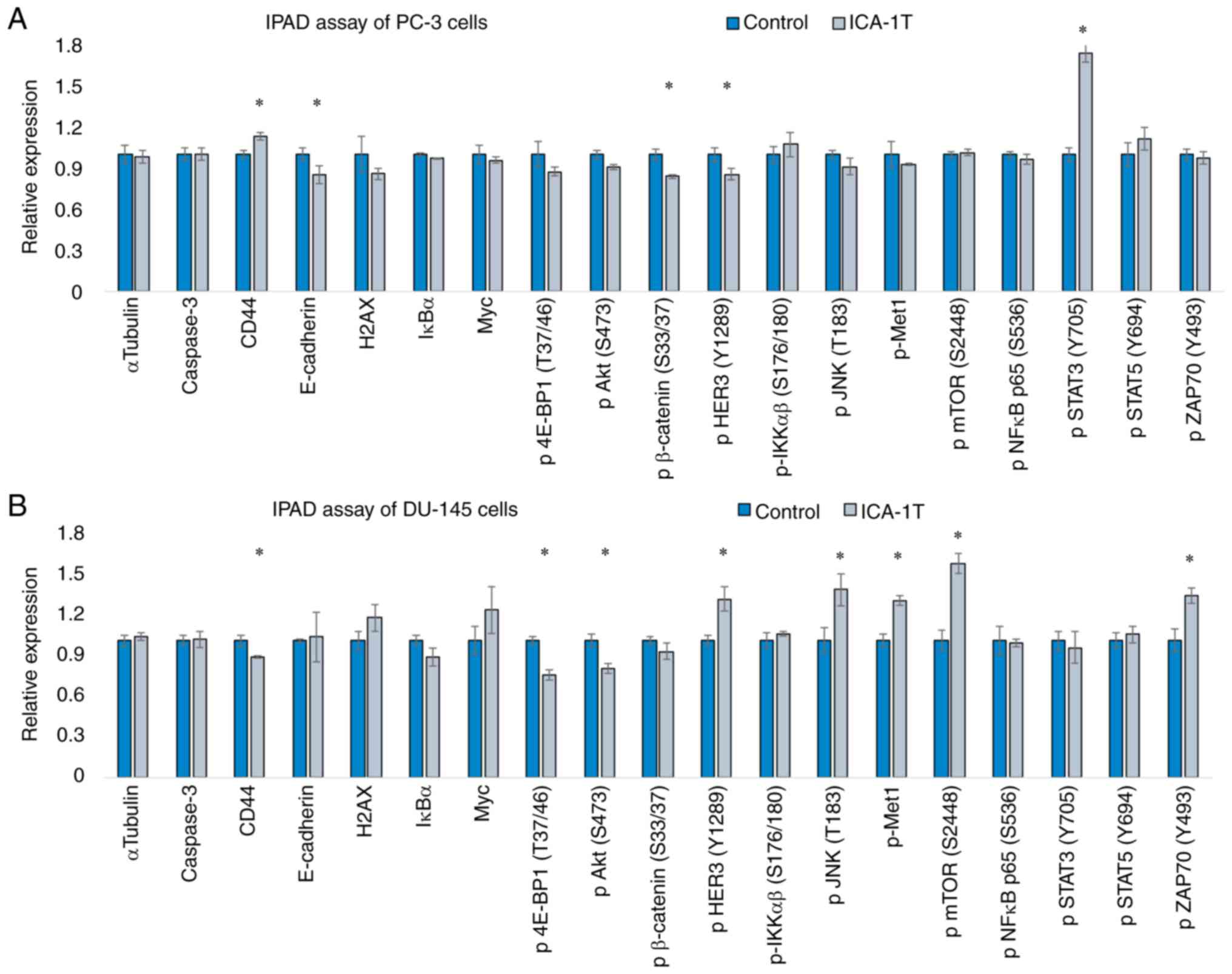 | Figure 4Immunopaired antibody detection assay
(IPAD) for PC-3 and DU-145 cells. (A and B) expression of IPAD
assay targets for PC-3 and DU-145 cell lines, respectively.
Approximately 1x105 cells were cultured in T75 flasks and 24 h
post-plating, fresh medium was supplied and the cells were treated
with either volume of sterile water (control) or the IC50
concentration of ICA-1T (2.5 µM). Additional concentrations were
supplied every 24 h during a 3-day incubation period. The cells
were then lysed and prepared lysates with the final total protein
concentration to be >2 µg/ml and then sent to ActivSignal, LLC
facility to conduct the IPAD assay. IPAD platform is a proprietary
multiplexed ELISA technology for analyzing the activity of multiple
signaling pathways in one reaction. Activities of multiple
signaling pathways were monitored simultaneously in a single well
through assessing the expression or protein phosphorylation of 25
target human proteins, such as caspase-3, CD44, CHOP, E-cadherin,
IκBα, Myc, NOTCH, p-4E-BP1, p-AKT (S473), p-β-catenin, p-HER3,
p-IRS-1, p-JNK, p-MEK1, p-mTOR, p-NF-κB, p-NUMB, p-SMAD1, p-SMAD2,
p-STAT3, p-STAT5, p-YAP1, p-ZAP70, p21 and PARP. α-tubulin and
β-tubulin were used as internal controls in each trial. Experiments
(n=3) were performed in each cell lines and the means ± SD are
plotted. Statistical significance is indicated by an asterisk
(*P≤0.05). |
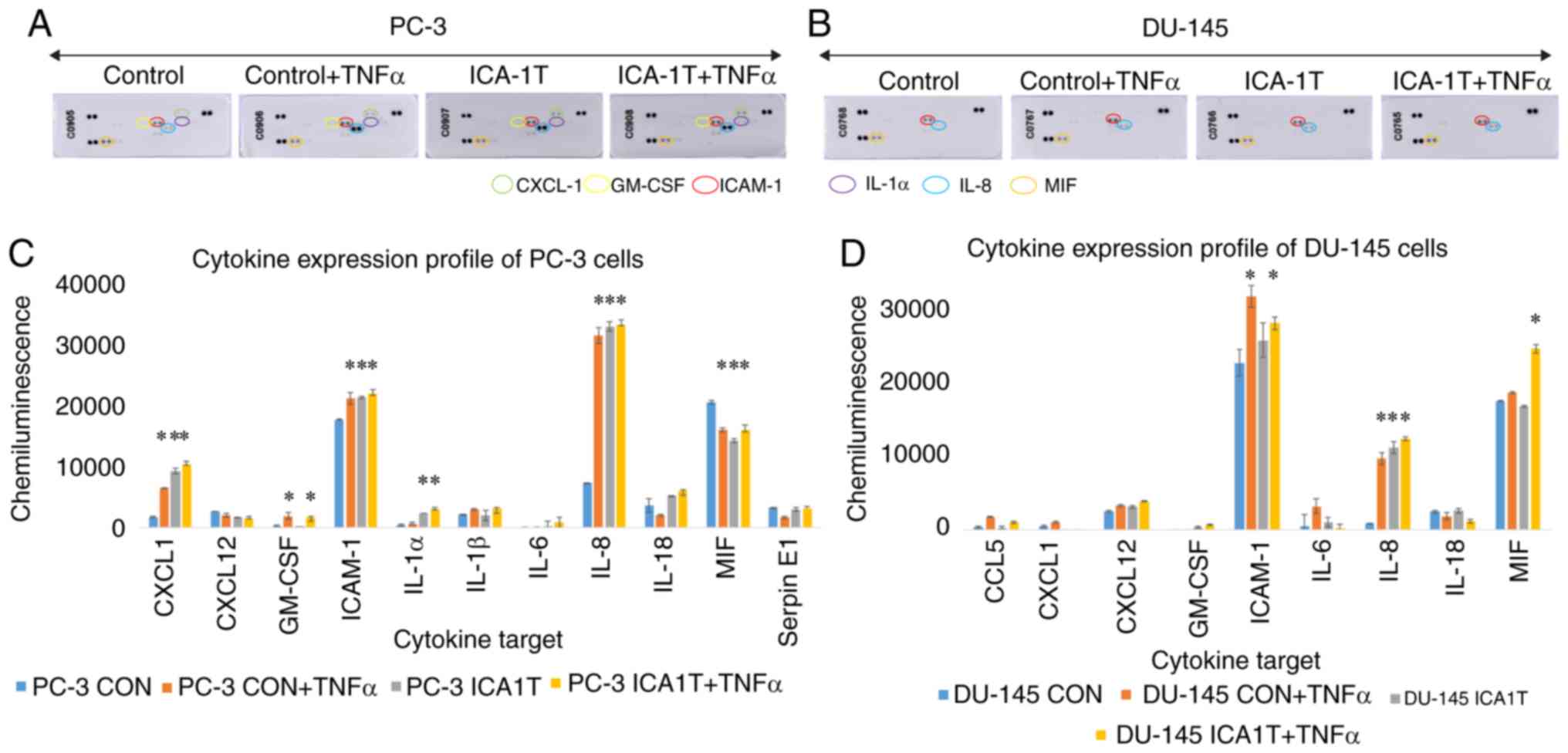
IL-8/c-Jun and ICAM-1/FOXO1 affect
PKC-ι regulation positively and negatively
As revealed in Fig.
5, immunoblot analysis of cytokines in the PC-3 and DU-145 cell
lines demonstrated a significant increase in the levels of IL-8 and
ICAM-1 in the cells treated with ICA-1T. IL-1α, IL-1β, IL-18,
CXCL-1, CXCL-12, GM-SCF, MIF and Serpin E1 were also found at
detectable levels, although the levels of these cytokines were not
altered substantially owing to PKC- ι inhibition, apart from
CXCL-1, which exhibited a significant (P≤0.05) change in PC-3
cells. These results of PKC-ι inhibition by ICA-1T were compared to
the samples treated with TNF-α prior to extraction. TNF-α, a
cytokine known to upregulate NF-κB signaling, did lead to a
significant change in the expression profiles with ICA-1T
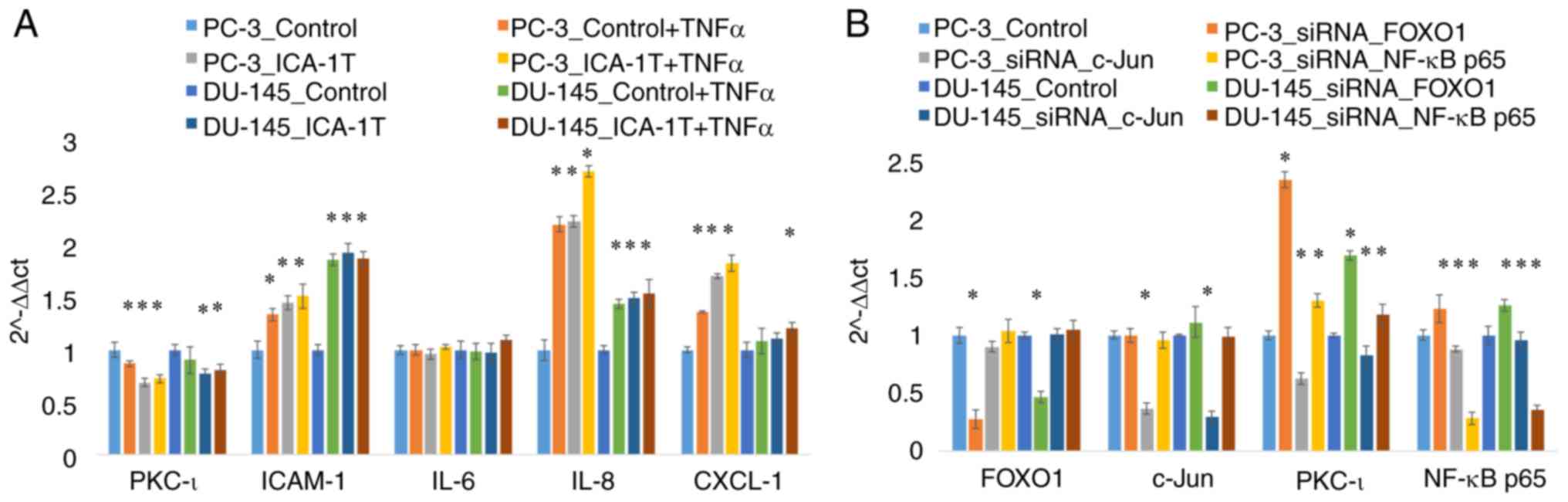
treatments. As shown in Fig. 6A,
RT-qPCR analyses were also conducted for these samples for which
the western blot data was presented in Fig. 5. As shown in Fig. 6A, the PKC-ι mRNA levels
significantly decreased by 32% (P≤0.05) and 23% (P≤0.05) in the
PC-3 and DU-145 cells treated with ICA-1T, respectively. Along with
PKC-ι depletion, ICAM-1 expression increased significantly by 45%
(P≤0.05) and 93% (P≤0.05) in the PC-3 and DU-145 cells,
respectively. Additionally, the IL-8 levels also increased
significantly by 123% (P≤0.05) and 50% (P≤0.05) in the PC-3 and
DU-145 cells, respectively.
Fig. 6B
demonstrates the mRNA levels of PKC-ι, c-Jun, FOXO1 and NF-κB in
the cells subjected to the knockdown of FOXO1, c-Jun and NF-κB p65
by siRNA for both cell lines with respect to the controls. Fig. 6B demonstrates the results of mRNA
expression analysis following transfection of the cells with siRNA
against FOXO1, c-Jun and NF-κB in which the western blot analysis
data are presented in Figs.
1-3. The mRNA analysis for these siRNA transfections confirmed
the western blot analysis observations presented in Figs.
1-3. The diminution of FOXO1 led to an increase in PKC-ι
expression by 134 and 68% (P≤0.05) in the PC-3 and DU-145 cells,
respectively. Additionally, the diminution of c-Jun expression
decreased PKC-ι expression by 38 and 18% (P≤0.05) in the PC-3 and
DU-145 cells, respectively. These outcomes confirmed that FOXO1
functions as a transcriptional deactivator for expressing the PRKCI
gene, while c-Jun functions as a transcriptional activator.
Fig. 7 presents a
graphical overview of PKC-ι expression modulation in prostate
cancer cells based on the present study current and on previous
evidence (7,10,18).
This illustration reveals the connections between multiple pathways
of JNK, NF-κB, AKT/FOXO1 and STAT3 in relation to PKC-ι regulation.
It indicates that PKC-ι plays a vital role in controlling its
expression via the c-Jun and FOXO1 transcriptional
activation/deactivation. Owing to c-Jun transcriptional function,
PKC-ι is overexpressed with the aid of pro-survival, oncogenic
STAT3, NF-κB/PI3K/AKT and signaling cascades. PKC-ι inhibition
using ICA-1T pledges an interruption to PKC-ι expression cycles
through the downregulation of the NF-κB pathway by limiting IKKα/β
due to the limitation of activated p-PKC-ι. It caused a suppression
of NF-κB transcriptional activity and IL-8. Due to the lack of
NF-κB stimulation, IL-8 accumulates in the cytosol and does not
perform its intended paracrine further upregulation of PI3K/AKT
signaling. AKT signaling decreases due to the lack of cytokine
activation, such as IL-8, which ultimately contributes to FOXO1
upregulation. FOXO1 adversely governs the expression of PKC-ι and
also decreases the function of JNKs to postpone its activation of
c-Jun which upregulates the expression of PKC-ι. Moreover, FOXO1
downregulates STAT3 and NF-κB signaling. The cycle persists and
contributes to the further downregulation of NF-κB and c-Jun, and
the upregulation of FOXO1, decreasing PKC-ι expression. The whole
process began upon the inhibition of PKC-ι. As a result of this
signaling alteration, the total PKC-ι level decreases in the tested
prostate cancer cells. The results of the present study closely
reinforce the findings of our previous study, wherein precise
inhibition utilizing PKC-ι inhibitors decreased overall PKC
quantities (10).
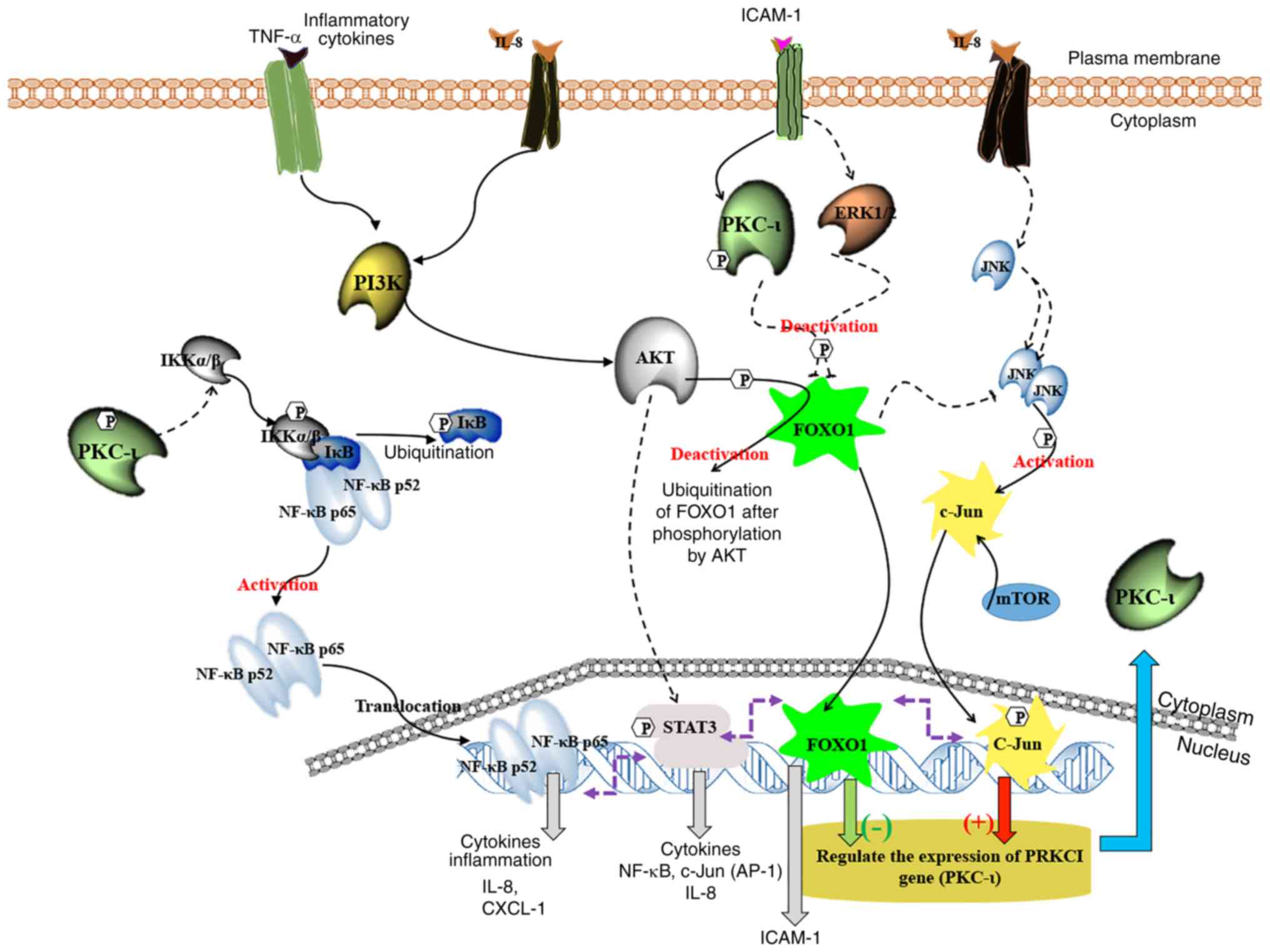 | Figure 7A schematic summary of the regulation
of the expression of PKC-ι in PC-3 and DU-145 cell lines. This
model depicts how the crosstalk occurs between the NF-κB,
PI3K/AKT/FOXO1, JNK/c-Jun and STAT3/5 signaling pathways during the
PKC-ι regulation. It is shown that PKC-ι plays a very important
role in the regulation of its expression in a complex signaling
network through the transcriptional activation/deactivation of
c-Jun and FOXO1. The PKC-ι-specific inhibition by ICA-1T,
downregulates the NF-κB, STAT3 and IL-8 activities. As a result,
the activity of AKT decreases, which leads to the upregulation of
FOXO1, which turns out to be the most important transcription
factor regulating PKC-ι expression upon receiving stimulation from
ICAM-1. FOXO1 downregulates the expression of PKC-ι, suppressing
JNK activity to attenuate the activation of c-Jun. This reduces
c-Jun expression. This whole process continues and leads to the
further downregulation of NF-κB and c-Jun, while upregulating
FOXO1, which leads to the continuation of the depletion of PKC-ι
expression in the cell lines. PKC-ι inhibition leads to a decrease
in its own production while enhancing multiple
antitumor/pro-apoptotic signaling. |
Discussion
In our previous study, the selective binding of
ICA-1T to an allosteric site located in the C-lobe of PKC-ι kinase
domain was recognized. This binding led to the inhibition of PKC-ι
activity (9). This consequentially
leads to a reduction in cellular differentiation, proliferation,
migration and invasion, whilst simultaneously driving the apoptosis
of prostate cancer cells via the diminution of the NF-κB pathway
in vitro. Subsequently, PKC-ι was established as a key
factor in the induction of cell growth, differentiation and
survival (8,9,11).
It was also recognized that PKC-ι undergoes self-regulation as a
consequence of its inhibition and a decrease in its expression in
the PC-3 and DU-145 cell lines. Thus, the aim of the present study
was the identification of the underlying processes of PKC-ι
regulation in the aforementioned cell lines in vitro.
In order to investigate PKC-ι regulation and
expression, the roles of transcription factors which interacted
with the PRKC1 promoter region were investigated. The gene which
codes of PKC-ι is the PRKCI gene, which is positioned on chromosome
3 (3q26.2), which is an amplicon known to undergo replication
events (20). In order to deduce
key TFs in PRKCI regulation, a sequence encompassing the PRKCI
promotor with a motif feature was selected, as well as a promoter
flank and an enhancer. This was selected as it provides the ideal
platform in which the TFs can bind to regulate transcription. Two
systems, PROMO and Genomatix Matinspector, were utilized to predict
probable transcription factor bindings. This led to the
identification of 5 TFs two of which were FOXO1 and c-Jun.
Subsequently, these TFs were silenced in order to analyze the
downstream effect they would have on PKC-ι expression.
c-Jun was the first transcription factor found to be
associated with numerous types of cancer, including metastatic
breast cancer and non-small lung cancer (21). It functions through the formation
of an early response complex containing AP-1 and c-Fos (22). The activation of c-Jun occurs via
phosphorylation events by c-Jun N-terminal kinases (JNKs) on S63
and S73, and is regulated via multiple extracellular stimuli, i.e.,
cytokines (23). Upon
phosphorylation at S63 and S73, not only is c-Jun activated, but it
also leads to an increase in the transcription of c-Jun-targeted
genes. Extracellular signal-regulated kinase (ERK) is also
upregulated by activated c-Jun (24-26).
c-Jun is also known to promote the oncogenic transformation of
‘ras’ and ‘fos’ in several cancer types (27,28).
FOXO1 is known to play a role in regulating various metabolic
pathways, such as gluconeogenesis, adipogenesis and insulin
signaling. Similar to c-Jun, phosphorylation plays a crucial role
in FOXO1 function (29,30). FOXO1 is deactivated by AKT through
phosphorylation on T24, leading to the induction of nuclear
exclusion, which leads to ubiquitylation (31,32).
Therefore, it is important to note that the phosphorylation of
FOXO1 indicates its inactivation and the downregulation of FOXO1
signaling. In relation to cancer, FOXO1 is a well-established tumor
suppressor (33-35).
As such, there is a known association between FOXO dysregulation
and cancer progression, as it is also plays a role in both
intrinsic and extrinsic pathways of apoptosis (36,37).
Experiments in vitro and in vivo have confirmed that
the overexpression of FOXO1 causes a reduction in cell migration,
proliferation and tumorigenesis in cancer cells (38). Furthermore, ERK1/2, PKC-ι and AKT
can downregulate FOXO1(35). Thus,
in the present study, it was demonstrated that through the specific
inhibition of PKC-ι, the expression of active PKC-ι decreases,
which renders it ineffective in its role to deactivate FOXO1
through phosphorylation events. This is a crucial indication of
PKC-ι involvement in the regulation of its own expression, as PKC-ι
inhibition leads to the continuous upregulation of FOXO1.
At the same time, previous data have demonstrated
that the inhibition of PKC-ι causes the significant downregulation
of the PI3K/AKT pathway and in particular, downregulates the
activation of AKT (10). In the
present study, as shown in Figs. 3
and 6, NF-κB downregulation led to
elevated levels of active c-Jun (phospho c-Jun), which upregulated
PKC-ι expression. These results validate our previous observation
that PKC-ι inhibition, by which the phosphorylation of IKKα/β is
reduced, inhibits NF-κB activation and translocation to the nucleus
(10). Subsequently, NF-κB
depletion induces an increase in c-Jun expression, which then
attempts to increase the production of PKC-ι, which then needs to
phosphorylate IKKα/β to restore NF-κB signaling. The tight
regulation of PKC-ι expression through c-Jun may explain these
results as it enhances PRKCI transcription. There was also no
significant alteration in the levels of FOXO1 and phosphorylated
FOXO1 resulting from NF-κB siRNA knockdown. This suggests that the
downregulation of NF-κB does not disrupt PKC-ι expression through
FOXO1, but rather that c-Jun provides cancer cells with resistance
to apoptosis through interplay with NF-κB upon cytokine stimulation
(21). In our previous study, it
was demonstrated that in melanoma, TNF-α upregulates NF-κB,
phosphor-AKT and PKC-ι expression (9). However, the results of the present
study demonstrate that c-Jun ‘switches on’ PKC-ι expression and
FOXO1 ‘switches off’.
Apart from identifying c-Jun and FOXO1 out of 5 TFs
which could bind to the PKC-ι gene promoter region, other key
molecular factors were also identified. Through the conduction of
ELISA using IPAD assay and a cytokine array, crosstalks between
multiple pathways were examined. The data indicated links between
PKC-ι expression with cytokines IL-8 and ICAM-1, along with some
other key cellular signaling points.
As shown in Fig. 4,
the IPAD ELISA data revealed that there was a significant increase
in the expression levels of p-STAT3 (Y705), p-JNK (T183) and
p-mTOR, whilst displaying a significant decrease in p-AKT (S473),
p-β-catenin and CD44 levels. Moreover, it has been demonstrated
that irregular STAT3/5 is associated with the progression of
various cancer types (39-44).
Cell survival in multiple cancers has been shown to be induced by
upregulated STAT signaling, which is often stimulated by the
cytokines, IL-6 and IL-8 (39,40,45).
STAT3 signaling enhances the production of c-Jun, thereby inducing
c-Jun-targeted transcription (39,46).
The IPAD data of the present study strongly suggested that STAT3
was upregulated due to PKC-ι inhibition, suggesting that the
deprivation of PKC-ι tries to accelerate the production of c-Jun
through the upregulation of STAT3, JNK and mTOR. Connections
between the JNK pathway and FOXO1 have been explored in few studies
(35,47,48).
Hornsveld et al summarizes the tumor-suppressing features of
FOXO1 resulting in a decreased JNK activity (47). Whilst JNK activates c-Jun, by
contrast, PKC-ι inhibition renders it ineffective at increasing
c-Jun or phospho-c-Jun levels, as can be seen in Fig. 3. Instead, the FOXO1 levels were
increased, while the phosphor-FOXO1 levels along with the levels of
phosphor-AKT and STAT3 were reduced in both cell lines. This
demonstrates that the activation of FOXO1 leads to a reduction in
c-Jun levels by blocking the activity of phosphor-JNK. Therefore,
it was deduced that FOXO1 plays a major role in c-Jun regulation
only upon PKC-ι inhibition. This process likely employs multiple
mechanisms, such as JNK signaling inhibition, causing the further
retardation of PKC-ι expression, which will eventually lead to cell
cycle arrest. This is further corroborated by FOXO1 being
established as being able to induce cell cycle arrest. It
accomplishes this through the promotion of the transcription of
cell cycle kinase inhibitors or cyclin-dependent kinase inhibitor
(CKI). p21 and p27 are two of the most well-known FOXO-induced
downstream CKIs (35,47). FOXO1 has also been shown to be
associated with the induction of anoikis (apoptosis that occurs
when cells detach from the extracellular matrix) (47). Once again, this displays another
downstream effect of PKC-ι involvement, as the inhibition of its
expression augments FOXO1 antitumor activity.
As shown in Fig. 7,
it is summarized that the expression of PRKCI is negatively
affected by FOXO1, whilst being positively affected by c-Jun. The
inhibition of PKC-ι leads to the following downstream effects. The
downregulation of NF-κB activity through the lack of
phosphor-IKKα/β, decreases the levels of phosphor-AKT (S473),
thereby diminishing AKT activity. Subsequently the low activity of
AKT, along with PKC-ι, lead to the decreased phosphorylation of
FOXO1, also leading to elevated levels of active unphosphorylated
FOXO1. These elevated levels of activated FOXO1 lead to the further
suppression of PRKCI gene expression. This acts as a ‘switch off’
effect on PRKCI expression. PKC-ι downregulation also leads to
decreased STAT3, mTOR and JNK signaling. As a consequence, this
reduces c-Jun activity, leading to the cancellation of the positive
effects of c-Jun towards PKC-ι expression. Furthermore, STAT3 and
STAT5 upregulate NF-κB transcription in addition to c-Jun (46,49).
Due to this, it was deduced that PKC-ι inhibition causes the
downregulation of NF-κB and STAT3, leading to a decrease in both
the transcription and activation of c-Jun. Therefore, these data
suggest that the PKC-ι levels were decreased when c-Jun expression
was silenced by siRNA (Figs. 1 and
6B).
In the present study, further in vitro
experiments (Figs. 5 and 6A) demonstrated the deviations in
cytokine expression (IL-8 and ICAM-1) in the PC-3 and DU-145 cells
upon PKC-ι knockdown. In both cell lines, the protein levels of
IL-8 and ICAM-1 (as well as their mRNA levels) were shown to
undergo a significant intensification following PKC-ι knockdown by
siRNA, as proven by western blot and RT-qPCR analyses. These data
suggest that PKC-ι self-regulation is involved in autocrine
signaling. Tumor cellular environments, with prostate cancer in
particular, are constantly exposed to a variety of immune cells and
inflammatory factors. The effects of which function to either
promote chronic inflammation or engage in antitumor activity
(50). Examples of these
inflammatory factors are cytokines; they play a crucial role in
controlling the tumor microenvironments (51). To achieve their functions,
cytokines utilize multiple signaling pathways. They can either act
to promote or downregulate tumor progression and metastasis.
Examples of tumor promoting cytokines are; as CXCL-1, CXCL-12,
IL-18, CXCL-10, IL-6 and IL-8. CXCL1, also known as melanoma
growth-stimulatory activity/growth-regulated protein α, functions
in processes of wound healing, angiogenesis and inflammation after
being secreted by cancer cells. It has also been linked to tumor
formation (52). Metastatic
regulation has also been linked to high levels of CXCL10/CXCR3,
with CXCL10 playing an important role in the promotion of tumor
growth and metastasis (52).
Metastatic regulation has also been linked to high levels of
CXCL10/CXCR3, with CXCL10 playing an important role in the
promotion of tumor growth and metastasis (52,53).
CXCL12 (stromal-derived factor-1) utilizes the receptors CXCR4 and
CXCR7 and it has been linked to playing a role in the regulation of
tumor metastasis. However, CXCL-10, CXCL-12 and IL-18 were not
observed as being significantly altered as result of PKC-ι
inhibition.
IL-6 has been linked to the stimulation of the
degradation of IκB-α, which in turn results in the upregulation of
NF-κB translocation. As previously demonstrated, PKC-ι stimulates
NF-κB translocation through IκB-α degradation (9). Upon translocation to the nucleus,
NF-κB induces cell survival via the transcription of multiple
survival factors and cytokines (39,45,53),
with IL-8 being one such cytokine. It plays a key role in the
regulation of polymorphonuclear neutrophil mobilization. It is also
associated with the extravasation in the steps of cancer
metastasis. IL-8 has been shown through studies to be regulated by
NF-κB in prostate cancer cells. As such, an increased IL-8
expression has been connected to the promotion of a favorable
microenvironment for metastasis (54,55).
Notably, the findings of the present study demonstrated that upon
transfection with PKC-ι siRNA, the IL-8 expression levels
increased. This may be a result of a backup mechanism in order to
upregulate IL-8. IL-8 also plays an essential role in upregulating
c-Jun through JNKs. As the inhibition of PKC-ι downregulates c-Jun,
the cells may be attempting to reinstate these downregulated
pathways by a higher IL-8 production. As shown in Fig. 7, the expression of IL-8 is
regulated through both NF-κB and STATs. These results indicate that
IL-8 is important in upregulating PKC-ι expression, activating
c-Jun, while deactivating FOXO1. Though it would appear that due to
the high activity of FOXO1, the effect of IL-8 are canceled
out.
Through utilizing an immune response, some cytokines
promote antitumor activity. One such cytokine is ICAM-1, which
plays a role in the immune response, including antigen recognition
and lymphocyte activation (56,57).
As such, ICAM-1 has beeb linked to the inhibition of tumor
progression via the inhibition of the PI3K/AKT pathway. In its role
in inhibiting this pathway, ICAM-1 exposes tumor cells to attack
and death through cytotoxic T-lymphocytes (57). ICAM-1 expression inhibition has
also been shown in clinical research to be associated with an
increased risk of metastasis within the first 5 years of ovarian
cancer diagnosis (57). Of note,
the results of the present study demonstrated that upon the
silencing of PKC-ι by siRNA, the ICAM-1 levels increased. This
confirmed that upon the knockdown of oncogenic PKC-ι,
antitumor/pro-apoptotic signaling was upregulated through an
autocrine manner via ICAM-1. Furthermore, these results demonstrate
that ICAM-1 plays an important downregulatory role in the
regulation of PKC-ι expression along with FOXO1, opposite to c-Jun
and IL-8.
To conclude, the results of the present study
illustrate that PKC-ι plays an imperative role in its own
expression via an intricate signaling grid that involves the
transcriptional activation/deactivation of c-Jun and FOXO1. The
inhibition of PKC-ι activity, based on its specific inhibition,
downregulates the NF-κB pathway along with the transcriptional
activity of STAT3 and IL-8. The results in a decrease in AKT
activity that leads to FOXO1 upregulation. FOXO1 was identified to
be the most important transcription factor when it comes to
regulating PKC-ι, along with ICAM-1 stimulation. FOXO1 negatively
regulates PKC-ι expression, diminishing JNK activity and further
suppressing the activation of c-Jun. The consequence of this
process is that it leads to the downregulation of NF-κB and c-Jun,
and further upregulates FOXO1. This continues to deplete the PKC-ι
expression, subsequently leading to a decrease in the total PKC-ι
levels in prostate cancer cells. The regulation of PKC-ι is
complex, and PKC-ι itself plays a key role in that process. As
such, when inhibited, it leads to a decrease in PKC-ι production,
prompting multiple antitumor/pro-apoptotic signaling. PKC-ι is
therefore a key factor to target when attempting to treat prostate
cancer in vitro. Finally, the results of the present study
demonstrate that PKC-ι is not only a novel biomarker to target for
personal therapeutics for prostate cancer, but also that ICA-1T
shows promise as one such therapy in relation to the proposed
mechanism.
Acknowledgements
The authors wish to acknowledge Mr. Andre
Apostolatos for his valuable ideas/suggestions for the
conceptualization of the study and experimental design of cytokine
analysis.
Funding
The authors acknowledge the financial contributions
of Mr. Gene Pranzo, the Leo and Anne Albert Charitable Trust, the
David Tanner Foundation, the Kyrias Foundation, the Frederick H.
Leonhardt Foundation, the Baker Hughes Foundation, the Brotman
Foundation of California, the Creag Foundation and the Irving S.
Cooper Family Foundation.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WSR and MAD were involved in the conceptualization
of the study. WSR, CAA and MAD were involved in the data analysis.
MAD and WSR were involved in the investigation. WSR and SB were
involved in cell culturing. WSR was involved in western blot
analysis. WSR and CAA were involved in the cytokine analysis, IPAD
assay and RT-qPCR. WSR and SB were involved in the writing of the
original draft. MAD was involved in reviewing the manuscript. WSR
and CAA edited the manuscript. MAD were involved in obtaining
resources and was also involved in the supervision of the study and
funding acquisition. All authors have read and approved the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Key Statistics for Prostate Cancer |
Prostate Cancer Facts.
|
|
2
|
Kume H, Kawai T, Nagata M, Azuma T,
Miyazaki H, Suzuki M, Fujimura T, Nakagawa T, Fukuhara H and Homma
Y: Intermittent docetaxel chemotherapy is feasible for
castration-resistant prostate cancer. Mol Clin Oncol. 3:303–307.
2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kharaziha P, Chioureas D, Rutishauser D,
Baltatzis G, Lennartsson L, Fonseca P, Azimi A, Hultenby K, Zubarev
R, Ullén A, et al: Molecular profiling of prostate cancer derived
exosomes may reveal a predictive signature for response to
docetaxel. Oncotarget. 6:21740–21754. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ratnayake WS and Acevedo-Duncan M:
Abstract 4569: Use of ACPD and ICA-1 as inhibitors of atypical
proteinkinase C-zeta (ζ) and iota (ι) in metastasized melanoma
cells. Cancer Res. 76:4569. 2016.
|
|
5
|
Ratnayake WS and Acevedo-Duncan M:
Abstract 862: Atypical protein kinase c inhibitors can repress
epithelial to mesenchymal transition (type III) in malignant
melanoma. Cancer Res. 77:862. 2017.
|
|
6
|
Ratnayake WS, Apostolatos CA and
Acevedo-Duncan M: Atypical protein kinase cs in melanoma
progression. Cutan Melanoma, 2019.
|
|
7
|
Apostolatos AH, Ratnayake WS, Smalley T,
Islam A and Acevedo-Duncan M: Abstract 2369: Transcription
activators that regulate PKC-iota expression and are downstream
targets of PKC-iota. Cancer Res. 77:2369. 2017.
|
|
8
|
Ratnayake WS, Apostolatos AH, Ostrov DA
and Acevedo-Duncan M: Two novel atypical PKC inhibitors; ACPD and
DNDA effectively mitigate cell proliferation and epithelial to
mesenchymal transition of metastatic melanoma while inducing
apoptosis. Int J Oncol. 51:1370–1382. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ratnayake WS, Apostolatos CA, Apostolatos
AH, Schutte RJ, Huynh MA, Ostrov DA and Acevedo-Duncan M: Oncogenic
PKC-ι activates Vimentin during epithelial-mesenchymal transition
in melanoma; a study based on PKC-ι and PKC-ζ specific inhibitors.
Cell Adhes Migr. 12:447–463. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Apostolatos AH, Ratnayake WS, Win-Piazza
H, Apostolatos CA, Smalley T, Kang L, Salup R, Hill R and
Acevedo-Duncan M: Inhibition of atypical protein kinase C-ι
effectively reduces the malignancy of prostate cancer cells by
downregulating the NF-κB signaling cascade. Int J Oncol.
53:1836–1846. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ratnayake W: Role of oncogenic protein
kinase C-iota in melanoma progression; A study based on atypical
protein kinase-C inhibitors (unpublished PhD thesis). University of
South Florida, 2019.
|
|
12
|
Manning G, Whyte DB, Martinez R, Hunter T
and Sudarsanam S: The protein kinase complement of the human
genome. Science. 298:1912–1934. 2002.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Regala RP, Weems C, Jamieson L, Khoor A,
Edell ES, Lohse CM and Fields AP: Atypical protein kinase C iota is
an oncogene in human non-small cell lung cancer. Cancer Res.
65:8905–8911. 2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Dey A, Patel R, Smalley T, Ratnayake WS,
Islam A and Acevedo-Duncan M: Abstract 244: Inhibition of atypical
PKC signaling enhances the sensitivity of glioblastoma cells
towards Temozolomide therapy. Cancer Res. 79:244. 2019.
|
|
15
|
Wu J, Lu M, Li Y, Shang YK, Wang SJ, Meng
Y, Wang Z, Li ZS, Chen H, Chen ZN and Bian H: Regulation of a
TGF-β1-CD147 self-sustaining network in the differentiation
plasticity of hepatocellular carcinoma cells. Oncogene.
35:5468–5479. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Venter JC, Adams MD, Myers EW, Li PW,
Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al:
The Sequence of the human genome. Science. 291:1304–1351.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fagerberg L, Hallström BM, Oksvold P,
Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S,
Danielsson A, Edlund K, et al: Analysis of the human
tissue-specific expression by genome-wide integration of
transcriptomics and antibody-based proteomics. Mol Cell Proteomics.
13:397–406. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ratnayake W, Apostolatos C, Breedy S,
Apostolatos A and Acevedo-Duncan M: FOXO1 regulates oncogenic PKC-ι
expression in melanoma inversely to c-Jun in an autocrine manner
via IL-17E and ICAM-1 activation. World Acad Sci J. 1:25–38.
2018.
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Butler AM, Buzhardt MLS, Erdogan E, Li S,
Inman KS, Fields AP and Murray NR: A small molecule inhibitor of
atypical protein kinase C signaling inhibits pancreatic cancer cell
transformed growth and invasion. Oncotarget. 6:15297–15310.
2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wisdom R, Johnson RS and Moore C: c-Jun
regulates cell cycle progression and apoptosis by distinct
mechanisms. EMBO J. 18:188–197. 1999.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Angel P, Hattori K, Smeal T and Karin M:
The jun proto-oncogene is positively autoregulated by its product,
Jun/AP-1. Cell. 55:875–885. 1988.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lopez-Bergami P, Huang C, Goydos JS, Yip
D, Bar-Eli M, Herlyn M, Smalley KS, Mahale A, Eroshkin A, Aaronson
S and Ronai Z: Rewired ERK-JNK signaling pathways in melanoma.
Cancer Cell. 11:447–460. 2007.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Vogt PK: Fortuitous convergences: The
beginnings of JUN. Nat Rev Cancer. 2:465–469. 2002.PubMed/NCBI View
Article : Google Scholar
|
|
25
|
Szabo E, Riffe ME, Steinberg SM, Birrer MJ
and Linnoila RI: Altered cJUN expression: An early event in human
lung carcinogenesis. Cancer Res. 56:305–315. 1996.PubMed/NCBI
|
|
26
|
Vleugel MM, Greijer AE, Bos R, van der
Wall E and van Diest PJ: c-Jun activation is associated with
proliferation and angiogenesis in invasive breast cancer. Hum
Pathol. 37:668–674. 2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Behrens A, Sibilia M and Wagner EF:
Amino-terminal phosphorylation of c-Jun regulates stress-induced
apoptosis and cellular proliferation. Nat Genet. 21:326–329.
1999.PubMed/NCBI View
Article : Google Scholar
|
|
28
|
Nateri AS, Spencer-Dene B and Behrens A:
Interaction of phosphorylated c-Jun with TCF4 regulates intestinal
cancer development. Nature. 437:281–285. 2005.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Rena G, Guo S, Cichy SC, Unterman TG and
Cohen P: Phosphorylation of the transcription factor forkhead
family member FKHR by protein kinase B. J Biol Chem.
274:17179–17183. 1999.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Nakae J, Kitamura T, Kitamura Y, Biggs WH,
Arden KC and Accili D: The forkhead transcription factor foxo1
regulates adipocyte differentiation. Dev Cell. 4:119–129.
2003.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Matsuzaki H, Daitoku H, Hatta M, Tanaka K
and Fukamizu A: Insulin-induced phosphorylation of FKHR (Foxo1)
targets to proteasomal degradation. Proc Natl Acad Sci USA.
100:11285–11290. 2003.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lu H and Huang H: FOXO1: A potential
target for human diseases. Curr Drug Targets. 12:1235–1244.
2011.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Borkhardt A, Repp R, Haas OA, Leis T,
Harbott J, Kreuder J, Hammermann J, Henn F, Lampert T, Harbott J,
et al: Cloning and characterization of AFX, the gene that fuses to
MLL in acute leukemias with a t(X;11)(q13;q23). Oncogene.
14:195–202. 1997.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Anderson MJ, Viars CS, Czekay S, Cavenee
WK and Arden KC: Cloning and characterization of three human
forkhead genes that comprise an FKHR-like gene subfamily. Genomics.
47:187–199. 1998.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang X, Tang N, Hadden TJ and Rishi AK:
Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta.
1813:1978–1986. 2011.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Farhan M, Wang H, Gaur U, Little PJ, Xu J
and Zheng W: FOXO signaling pathways as therapeutic targets in
cancer. Int J Biol Sci. 13:815–827. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fu Z and Tindall D: FOXOs, cancer and
regulation of apoptosis. Oncogene. 27:2312–2319. 2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang Y, Zhang L, Sun H, Lv Q, Qiu C, Che
X, Liu Z and Jiang J: Forkhead transcription factor 1 inhibits
endometrial cancer cell proliferation via sterol regulatory
element-binding protein 1. Oncol Lett. 13:731–737. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Hodge DR, Hurt EM and Farrar WL: The role
of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer.
41:2502–2512. 2005.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yue P and Turkson J: Targeting STAT3 in
cancer: How successful are we? Expert Opin Investig Drugs.
18:45–56. 2009.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Jing N and Tweardy DJ: Targeting Stat3 in
cancer therapy. Anticancer Drugs. 16:601–607. 2005.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Page BDG, Khoury H, Laister RC, Fletcher
S, Vellozo M, Manzoli A, Yue P, Turkson M, Minden MD and Gunning
PT: Small molecule STAT5-sh2 domain inhibitors exhibit potent
antileukemia activity. J Med Chem. 55:1047–1055. 2012.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Pardanani A, Lasho T, Smith G, Burns CJ,
Fantino E and Tefferi A: CYT387, a selective JAK1/JAK2 inhibitor:
In vitro assessment of kinase selectivity and preclinical studies
using cell lines and primary cells from polycythemia vera patients.
Leukemia. 23:1441–1445. 2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Rani A and Murphy JJ: STAT5 in cancer and
immunity. J Interferon Cytokine Res. 36:226–237. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Korneev KV, Atretkhany KSN, Drutskaya MS,
Grivennikov SI, Kuprash DV and Nedospasov SA: TLR-signaling and
proinflammatory cytokines as drivers of tumorigenesis. Cytokine.
89:127–135. 2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Zhang X, Wrzeszczynska MH, Horvath CM and
Darnell JE: Interacting regions in stat3 and c-jun that participate
in cooperative transcriptional activation. Mol Cell Biol.
19:7138–7146. 1999.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hornsveld M, Dansen TB, Derksen PW and
Burgering BMT: Re-evaluating the role of FOXOs in cancer. Semin
Cancer Biol. 50:90–100. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Sunters A, Madureira PA, Pomeranz KM,
Aubert M, Brosens JJ, Cook SJ, Burgering BMT, Coombes RC and Lam
EWF: Paclitaxel-induced nuclear translocation of FOXO3a in breast
cancer cells is mediated by c-Jun NH2-terminal kinase and Akt.
Cancer Res. 66:212–220. 2006.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Yuan ZL, Guan YJ, Wang LW, Wei W, Kane AB
and Chin YE: Central role of the threonine residue within the p+1
loop of receptor tyrosine kinase in STAT3 constitutive
phosphorylation in metastatic cancer cells. Mol Cell Biol.
24:9390–9400. 2004.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Antonicelli F, Lorin J, Kurdykowski S,
Gangloff SC, Naour RL, Sallenave JM, Hornebeck W, Grange F and
Bernard P: CXCL10 reduces melanoma proliferation and invasiveness
in vitro and in vivo. Br J Dermatol. 164:720–728. 2011.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zaynagetdinov R, Sherrill TP, Gleaves LA,
McLoed AG, Saxon JA, Habermann AC, Connelly L, Dulek D, Peebles RS
Jr, Fingleton B, et al: Interleukin-5 facilitates lung metastasis
by modulating the immune microenvironment. Cancer Res.
75:1624–1634. 2015.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Sun X, Cheng G, Hao M, Zheng J, Zhou X,
Zhang J, Taichman RS, Pienta KJ and Wang J: CXCL12/CXCR4/CXCR7
chemokine axis and cancer progression. Cancer Metastasis Rev.
29:709–722. 2010.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ishiguro H, Akimoto K, Nagashima Y, Kojima
Y, Sasaki T, Ishiguro-Imagawa Y, Nakaigawa N, Ohno S, Kubota Y and
Uemura H: aPKClamda/iota promotes growth of prostate cancer cells
in an autocrine manner through transcriptional activation of
interleukin-6. Proc Natl Acad Sci USA. 106:16369–16374.
2009.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Peng H, Chen P, Cai Y, Chen Y, Wu QH, Li
Y, Zhou R and Fang X: Endothelin-1 increases expression of
cyclooxygenase-2 and production of interlukin-8 in hunan pulmonary
epithelial cells. Peptides. 29:419–424. 2008.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Timani KA, Győrffy B, Liu Y, Mohammad KS
and He JJ: Tip110/SART3 regulates IL-8 expression and predicts the
clinical outcomes in melanoma. Mol Cancer. 17(124)2018.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Yang M, Liu J, Piao C, Shao J and Du J:
ICAM-1 suppresses tumor metastasis by inhibiting macrophage M2
polarization through blockade of efferocytosis. Cell Death Dis.
6(e1780)2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Groote ML, de Kazemier HG, Huisman C, Gun
BTF, van der Faas MM and Rots MG: Upregulation of endogenous ICAM-1
reduces ovarian cancer cell growth in the absence of immune cells.
Int J Cancer. 134:280–290. 2014.PubMed/NCBI View Article : Google Scholar
|