Introduction
MicroRNAs (miRNAs or miRs) play important roles in
gene regulation; more than half of human genes are known to be
regulated by miRNAs. miRNAs are also involved in numerous
pathological pathways/process, such as the inflammatory response
(1,2) and cancer development (3,4), as
well as in the formation/development of ischemic stroke (IS)
(5-9).
Gene polymorphisms, including the Leu33Pro
polymorphism (5), the
apolipoprotein E gene polymorphism (among the Chinese population)
(6,7), the Gly82Ser polymorphism (8) and the breast susceptibility gene 2
rs9534275 polymorphism (9), have
been reported to be associated with an increased risk of IS.
Several studies have assessed the association between the miR-499
(rs3746444) polymorphism and the risk of IS; however, the varying
conclusions reached by the different studies are contradictory.
Jeon et al (10) reported
no difference in the allele frequencies of the miR-499 polymorphism
between patients with IS and the controls; a previous meta-analysis
(11) based on 3 studies reported
a similar finding. On the contrary, Darabi et al (12) reported that the miR-499 A/G
polymorphism was significantly associated with an increased risk of
IS.
Thus, whether the miR-499 polymorphism increases the
risk of IS remains controversial. The present study aimed to assess
this association through a systematic review and meta-analysis.
Odds ratios (ORs) and 95% confidence intervals (CIs) were used to
quantitatively estimate the association in 5 models, including the
allelic model (G allele vs. A allele), the dominant model (GG + AG
vs. AA), the recessive model (GG vs. AG + AA), the heterozygote
model (AG vs. AA) and the homozygote model (GG vs. AA).
Materials and methods
Inclusion and exclusion criteria
Studies were selected under the following
considerations: i) Studies should use a case-control design; ii)
the subjects should include both patients with IS and healthy
controls; iii) there must be extractable data on allele frequencies
and genotype frequencies; iv) the subjects/models studied should be
human, instead of animals or cells.
Studies were excluded if any of the following
conditions were met: i) Duplication studies; ii) case reports,
letters, editorials, or reviews; iii) studies not written in the
English language; iv) studies without extractable data on allele
frequencies or genotype frequencies.
Databases and search strategy
A comprehensive literature search was performed
using the PubMed, Embase, Science Direct and google scholar
databases (from inception to June 15, 2020). The following terms
were used to perform the literature search: rs3746444, miRNA-499,
microRNA-499, miR-499; with a combination of stroke, ischemic,
ischemia, ischemic stroke, cerebrovascular and cerebrovascular
disease.
Data extraction
The data were extracted independently by 2
researchers using a predesigned data extraction form; the following
information was extracted: i) Name of the first author; ii) year of
publication; iii) country where the study was conducted; iv) the
study design; v) sample size; vi) gene sequencing method; vii)
P-values of Hardy-Weinberg equilibrium (HWE) of the control group.
Most importantly, the frequencies of genotypes of the patients with
IS and the controls were extracted.
Assessment of methodological
quality
Two researchers used the Newcastle-Ottawa scale
(13) to assess the quality of the
included studies independently. For the Newcastle-Ottawa scale, a
full score is 9; a score of 5-9 indicates a high methodological
quality, whereas a score of 0-4 indicates poor quality. Any
disagreement would be resolved by other authors following a
comprehensive reassessment. Low-quality studies were excluded from
the current meta-analysis.
Data synthesis and statistical
analysis
Prior to the meta-analysis, the heterogeneity across
studies was assessed by the Q test and the I2 statistic.
I2 describes the percentage of variability caused by
heterogeneity rather than by chance. An I2 <50%
indicates a small heterogeneity; consequently a fixed-effects model
would be used to perform the meta-analysis, otherwise a
random-effects model would be used, and a sensitivity analysis
(13) would be performed in the
case of a large heterogeneity. Odds ratios (ORs) with 95%
confidence intervals (CIs) were used to estimate the association
between the miR-499 (rs3746444) polymorphism and the risk of IS.
Egger's test (14) was used to
determine whether there was a publication bias. The significance
level was set as 0.05, two-tailed. All the analyses were performed
using the open source R program (version 4.0.1).
Results
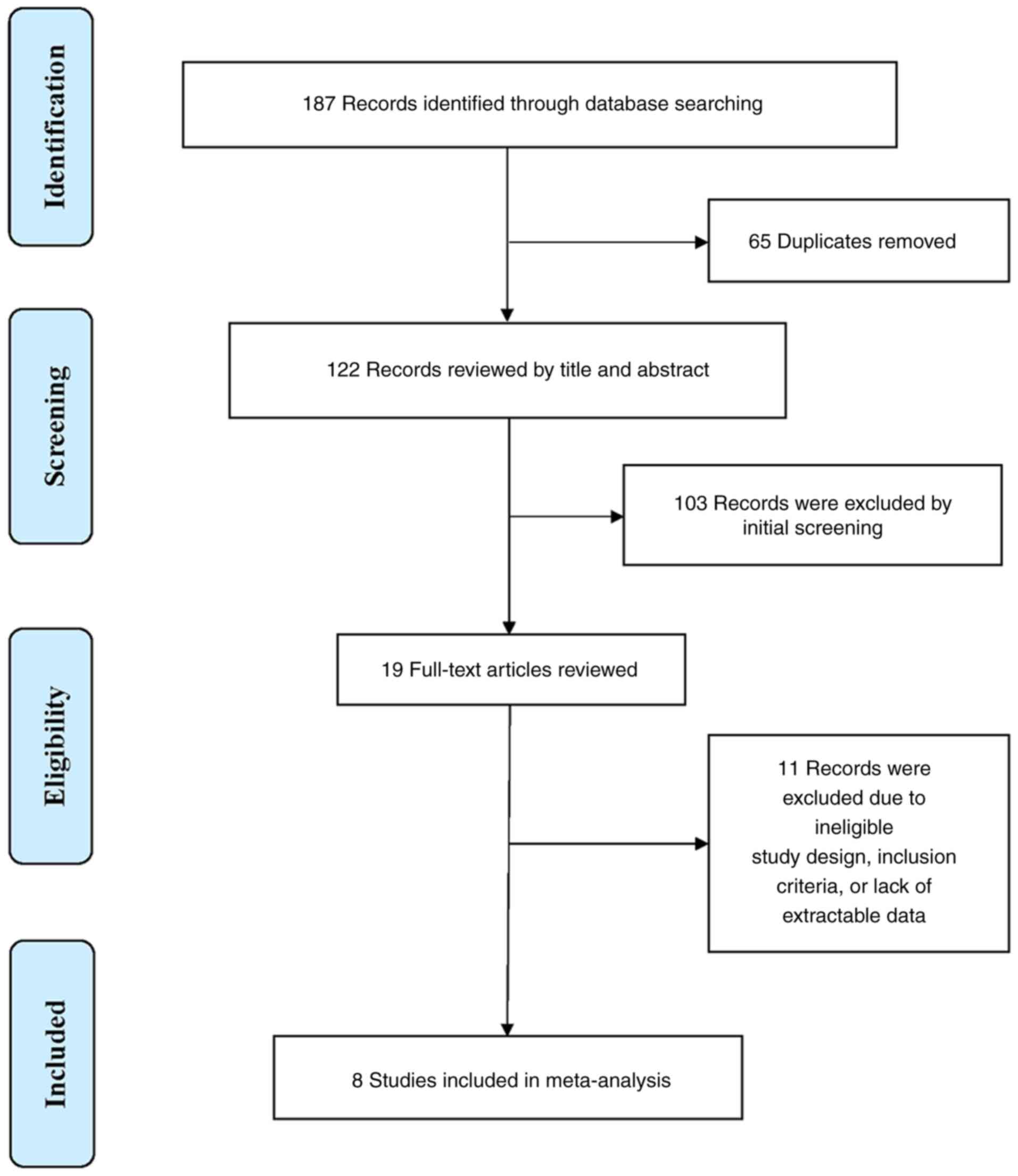
Study selection
A total of 187 studies were identified through
initial database searching. A total of 65 duplicates were removed,
and 122 records were reviewed for title and Abstract. In total, 103
irrelevant records and studies not written in the English language
were further excluded. A total of 19 articles were full-text
reviewed, and 11 records were further excluded due to ineligible
study design or the lack of extractable data. The details of the
study selection process are presented in Fig. 1.
Characteristics of included
studies
A total of 8 studies (10,12,15-20),
involving 3,400 patients with IS and 3,652 controls, met the
inclusion and exclusion criteria. The characteristics of the
included studies are presented in Table I. A total of 5 studies were carried
out in China, 2 in Korea and one in Iran. All studies were
hospital-based studies, and used a case-control study design.
Polymerase chain reaction-restriction fragment length polymorphism
(PCR-RFLP) was the most popular method for genotype sequencing. The
age and sex of the patients in the selected studies were matched
between the patients with IS and the controls in each study. Only
in 1 study, the P-value of HWE was <0.05.
 | Table ICharacteristics of the included
studies. |
Table I
Characteristics of the included
studies.
| | Sample size | |
|---|
| Author | Year | Country | Study design | Cases | Controls | Genotyping
methods | P-value (HWE) | (Refs.) |
|---|
| Jeon et
al | 2013 | Korea | Hospital-based | 678 | 553 | PCR-RFLP | 0.740 | (10) |
| Liu et al | 2014 | China | Hospital-based | 296 | 391 | PCR-RFLP | 0.394 | (16) |
| Huang et
al | 2015 | China | Hospital-based | 531 | 531 | TaqMan | 0.002 | (15) |
| Luo et al | 2017 | China | Hospital-based | 298 | 303 | RT-PCR | 0.447 | (17) |
| Darabi et
al | 2019 | Iran | Hospital-based | 470 | 489 | PCR-RFLP | 0.061 | (12) |
| Zhu et
al | 2018 | China | Hospital-based | 296 | 378 | PCR-RFLP | 0.512 | (18) |
| Hong et
al | 2019 | Korea | Hospital-based | 264 | 455 | PCR | 0.999 | (19) |
| Zhu et
al | 2020 | China | Hospital-based | 567 | 552 | PCR-LDR | 0.630 | (20) |
Study quality assessment
The mean Newcastle-Ottawa score of the included
studies was 7 (Table II); thus,
the overall quality of the included studies was high.
 | Table IIMethodological quality of the studies
by the Newcastle-Ottawa scale. |
Table II
Methodological quality of the studies
by the Newcastle-Ottawa scale.
| | Included studies,
author, year (Refs.) |
|---|
| Score | Items | Jeon et al,
2013(10) | Liu et al,
2014(16) | Huang et al,
2015(15) | Luo et al,
2017(17) | Darabi et
al, 2019(12) | Zhu et al,
2018(18) | Hong et al,
2019(19) | Zhu et al,
2020(20) |
| Selection. | Adequate definition
of patient cases | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| | Representativeness
of patient cases | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| | Selection of
controls | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| | Definition of
controls | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Comparability | Control for
important/additional factor | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 |
| Exposure. | Ascertainment of
exposure (blinding) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| | Same ascertainment
method for participants | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| | Non-response
ratea | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Total
scoreb | | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
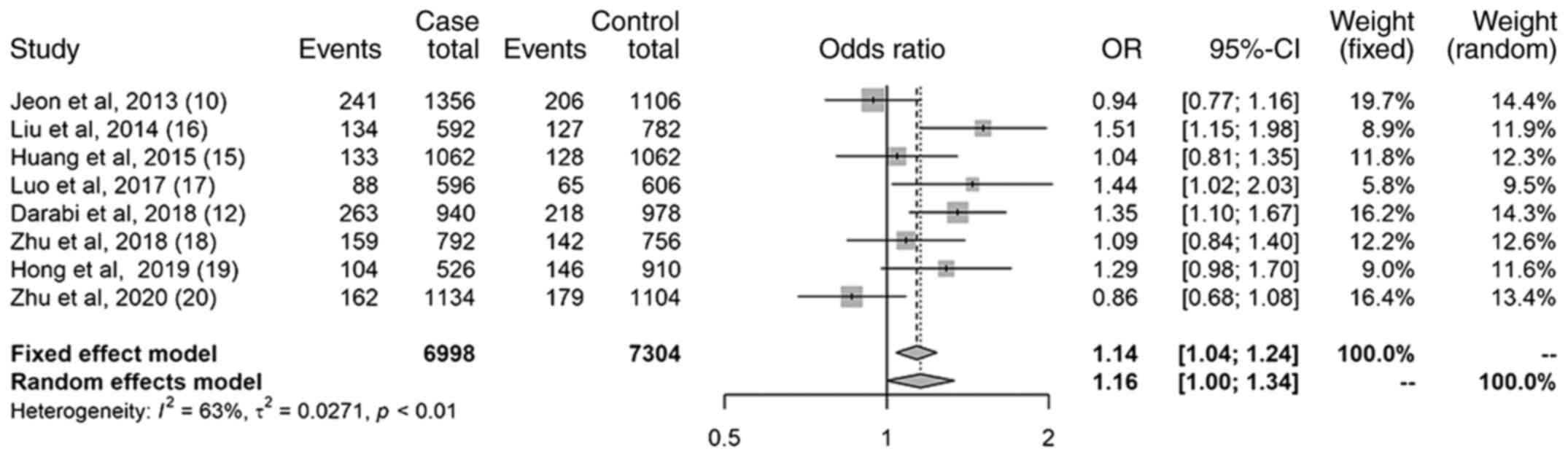
Results of the allelic model
The results of the allelic model revealed a
statistically significant association between the miR-499
(rs3746444) polymorphism and an increased risk of IS (Fig. 2); the OR was 1.16 (95% CI,
1.00-1.34). However, the I2 was 63%, and there was a
large heterogeneity across the included studies. A sensitivity
analysis was further performed. It was found that after omitting
the study of Zhu et al, 2020(20), the I2 decreased to 51.3%
(data not shown). Moreover, the conclusion remained unaltered; the
association between the miR-499 (rs3746444) polymorphism and the
increased risk of IS was still significant; the OR was 1.19 (95%
CI, 1.08-1.31; P<0.05) (data not shown).
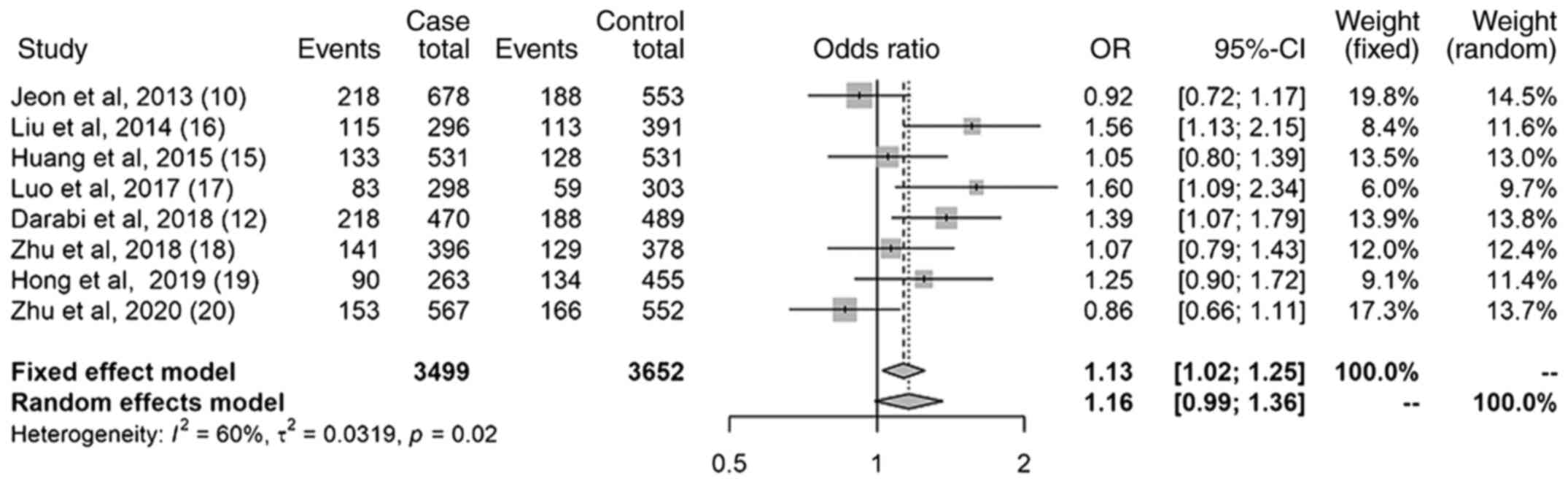
Results of the dominant model
The results of the dominant model revealed a
non-significant association between the miR-499 (rs3746444)
polymorphism and the risk of IS (Fig.
3); the OR was 1.16 (95% CI, 0.99-1.36). The I2 was
60%; a sensitivity analysis was subsequently performed. After
omitting the study of Zhu et al, 2020(20), the I2 dropped to 50.9%
(data not shown). Notably, the conclusion changed, and there was a
statistically significant association between the miR-499
(rs3746444) polymorphism and the risk of IS; the OR was 1.19 (95%
CI, 1.07-1.33) (data not shown).
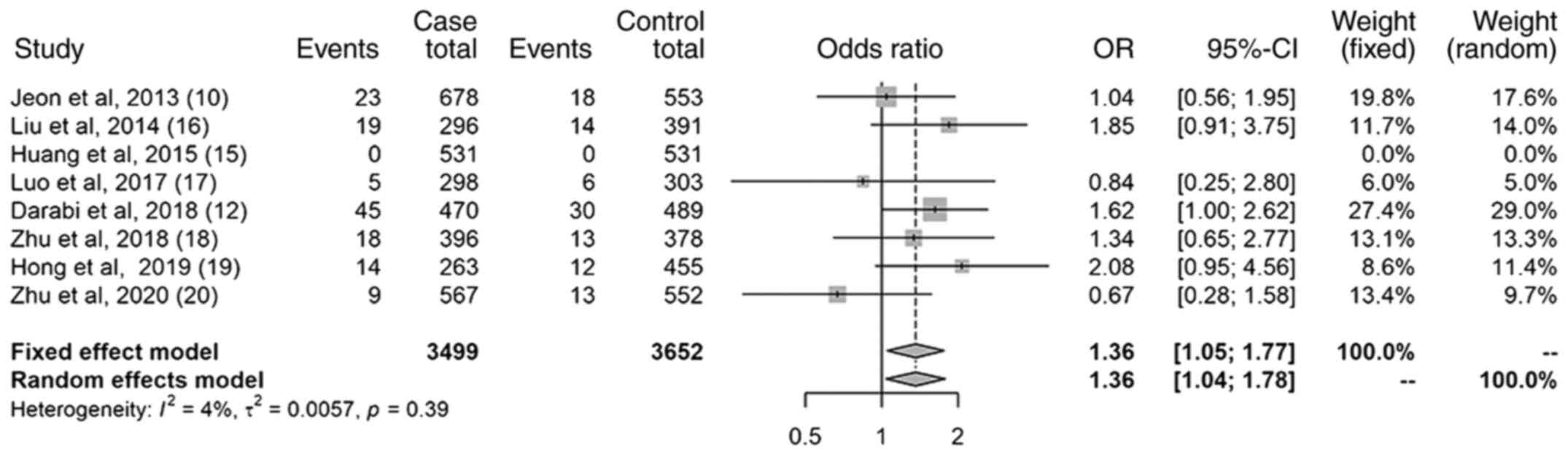
Results of the recessive model
The results of the recessive model (Fig. 4) revealed a statistically
significant association between the miR-499 (rs3746444)
polymorphism and the increased risk of IS; the OR was 1.36 (95% CI,
1.05-1.77). The heterogeneity was small (I2=4%).
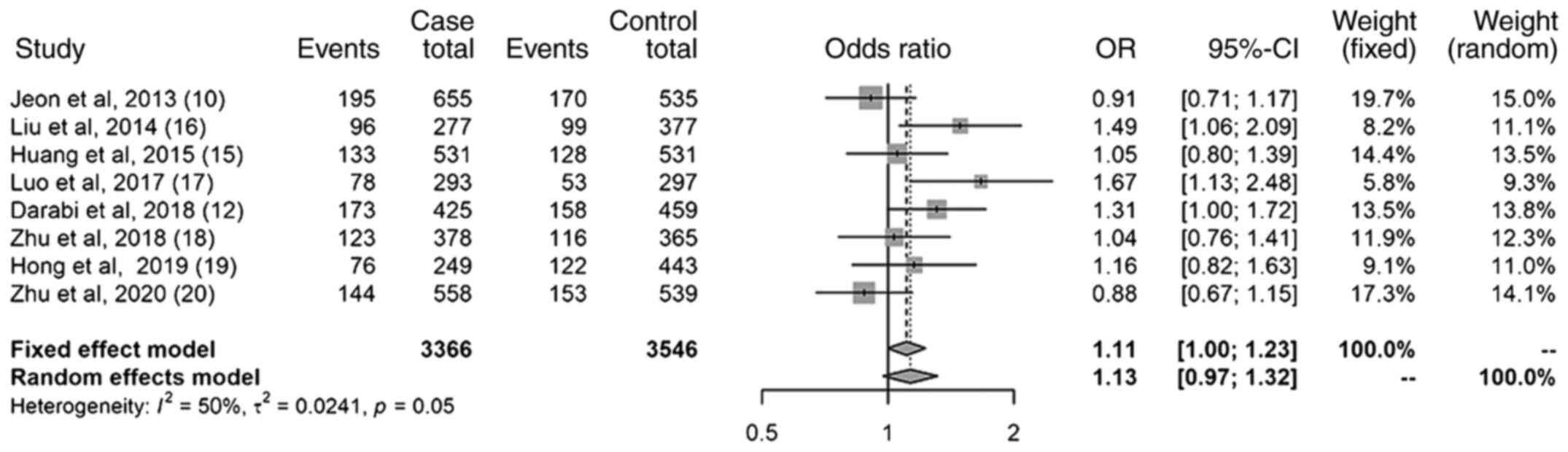
Results of the heterozygote model
The results of the heterozygote model (Fig. 5) also revealed a statistically
significant association between the miR-499 (rs3746444)
polymorphism and the increased risk of IS; the OR was 1.11 (95% CI,
1.00-1.23). The I2 was 50%; the heterogeneity was not
large.
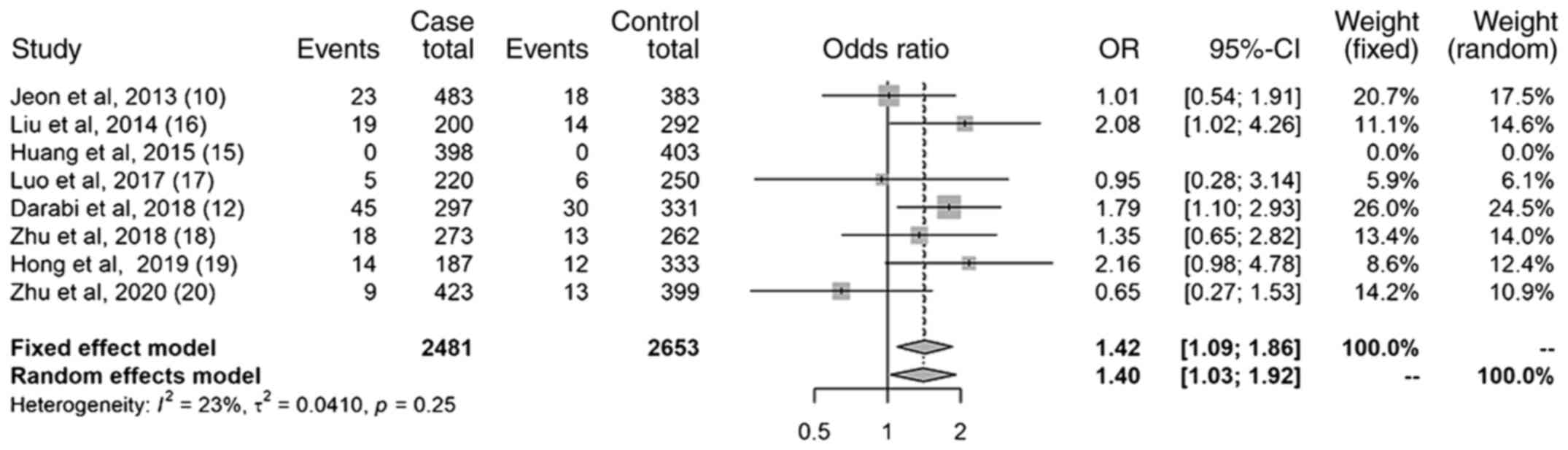
Results of the homozygote model
The results of the homozygote model (Fig. 6) revealed a statistically
significant association between the miR-499 (rs3746444)
polymorphism and the increased risk of IS; the OR was 1.42 (95% CI,
1.09-1.86). The heterogeneity was small (I2=23%).
Publication bias
There was no publication bias for all the 5 models
according to results of Eggers' test (Table III).
 | Table IIIEgger's test results. |
Table III
Egger's test results.
| Model | t value | P-value |
|---|
| Allele | 0.818 | 0.440 |
| Dominant | 2.223 | 0.067 |
| Recessive | -0.969 | 0.377 |
| Heterozygous | 2.124 | 0.053 |
| Homozygous | -0.898 | 0.410 |
Discussion
The present meta-analysis aimed to determine whether
the miR-499 (rs3746444) polymorphism had an influence on the risk
of IS based on case-control studies. The main finding was that the
miR-499 (rs3746444) polymorphism was associated with an increased
risk of IS; the association was statistically significant in the
allelic model, the recessive model, the heterozygote model and the
homozygote model. Furthermore, by sensitivity analysis, the
dominant model was found to be significant.
In a previous review article, Zou et al
(21) concluded that the miR-499
(rs3746444) polymorphism may not be associated with a risk of IS in
Asian populations. However, in the present study, neither the
allelic model, dominant model, recessive model, homozygote model,
or the heterozygote model were found to be significant in the study
by Zou et al. However, as 3 Chinese articles were included
in the review article by Zou et al (21), it is difficult for researchers who
do not speak Chinese to repeat the results.
The present study attempted to explain why findings
by the previous review article were contrary to those of the
present study. Considering that the previous review article focused
on a Chinese population, the present study performed a subgroup
analysis in the meta-analysis, and studies were divided by
ethnicity into subgroup 1 (studies based on Chinese population) and
subgroup 2 (studies based on non-Chinese population). Subgroup
analysis revealed that the homozygote model and the recessive model
of subgroup 1 was indeed non-significant; however, the allelic
model, the dominant model, and the heterozygote model still
exhibited a significant association between the miR-499 (rs3746444)
polymorphisms and the increased risk of IS. Notably, there was a
tendency that the ORs of subgroup 2 were larger than those of
subgroup 1. The results of subgroup analysis indicated that the
association between the miR-499 (rs3746444) polymorphism and the
risk of IS may vary for different ethnicities (data not shown). The
relatively small sample size of the study by Zou et al
(21) (there were only 4 studies
if the 3 articles written in Chinese were not counted) may be
another reason why non-significant results were found, and that is
why an updated meta-analysis is of utmost importance.
The association between the miR-499 polymorphism and
the increased risk of IS was understandable: It has been reported
that hsa-miR-499 plays a role in cell apoptosis under ischemic
conditions (22); the mechanisms
involved may be that miR-499 inhibits cell apoptosis by suppressing
the dephosphorylation of dynamin-related protein-1 (Drp1) that is
mediated by calcineurin. miR-499 affects the regulation of
C-reactive protein (CRP) (23),
and C-reactive protein has been reported to be a cause of cerebral
ischemia (24). Similarly, TNF-α
(25), another general cause of
cerebral ischemia, has been reported to be regulated by miR-499;
both CRP and TNF-α are activated under conditions of stress
(26). Furthermore, it has been
reported that the thrombosis and inflammation pathways of the.
In conclusion, miR-circulation system can be
influenced by the miR-499 (rs3746444) polymorphism; an A:U pair to
a G:U mismatch of hsa-miR-499 precursor caused by miR-499
(rs3746444) (27,28) polymorphism can alter the
function/expression of mature hsa-miR-499, and the regulation of
its' target mRNAs. The miR-499 polymorphism is associated with a
risk of IS, and the G allele increases the risk of IS.
There are two limitations to the current
meta-analysis. On the one hand, all the included studies used a
case-control design; there may thus be a selection bias, which
could enhance or weaken the true association between miR-499
polymorphism and the risk of IS. On the other hand, subgroup
analysis based on 8 studies was not sufficient; thus, whether the
association between the miR-499 polymorphism and risk of IS is
dependent on ethnicity remains a matter of debate.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used and/or analyzed during the current
study is available from the corresponding author on reasonable
request.
Authors' contributions
KC designed the study. XHL and MLL performed the
literature search and manuscript writing. RL extracted the data for
the analysis. YPX performed the quality control of the extracted
data. MZ and QRL performed the data analysis. WZ and TLZ
interpreted the data and revised the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Olivieri F, Capri M, Bonafe M, Morsiani C,
Jung HJ, Spazzafumo L, Vina J and Suh Y: Circulating miRNAs and
miRNA shuttles as biomarkers: Perspective trajectories of healthy
and unhealthy aging. Mech Ageing Dev. 165:162–170. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Xu XM and Zhang HJ: MiRNAs as new
molecular insights into inflammatory bowel disease: Crucial
regulators in autoimmunity and inflammation. World J Gastroenterol.
22:2206–2218. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Qadir MI and Faheem A: MiRNA: A diagnostic
and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene
Expr. 27:197–204. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang
L, Zhang H, Wang W, Zhu J, Cheng W, et al: Six serum-based miRNAs
as potential diagnostic biomarkers for gastric cancer. Cancer
Epidemiol Biomarkers Prev. 26:188–196. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kekomaki S, Hamalainen L,
Kauppinen-Makelin R, Palomaki H, Kaste M and Kontula K: Genetic
polymorphism of platelet glycoprotein IIIa in patients with acute
myocardial infarction and acute ischaemic stroke. J Cardiovasc
Risk. 6:13–17. 1999.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gu L, Su L, Chen Q, Liang B, Qin Y, Xie J,
Wu G, Yan Y, Long J, Wu H, et al: Association between the
apolipoprotein E gene polymorphism and ischemic stroke in Chinese
populations: New data and meta-analysis. Exp Ther Med. 5:853–859.
2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ariyaratnam R, Casas JP, Whittaker J,
Smeeth L, Hingorani AD and Sharma P: Genetics of ischaemic stroke
among persons of non-European descent: A meta-analysis of eight
genes involving approximately 32,500 individuals. PLoS Med.
4(e131)2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ma WQ, Qu QR, Zhao Y and Liu NF:
Association of RAGE gene Gly82Ser polymorphism with coronary artery
disease and ischemic stroke: A systematic review and meta-analysis.
Medicine (Baltimore). 95(e5593)2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Miao L, Yin RX, Yang S, Huang F, Chen WX
and Cao XL: Association between single nucleotide polymorphism
rs9534275 and the risk of coronary artery disease and ischemic
stroke. Lipids Health Dis. 16(193)2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jeon YJ, Kim OJ, Kim SY, Oh SH, Oh D, Kim
OJ, Shin BS and Kim NK: Association of the miR-146a, miR-149,
miR-196a2, and miR-499 polymorphisms with ischemic stroke and
silent brain infarction risk. Arterioscler Thromb Vasc Biol.
33:420–430. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Xiao Y, Bao MH, Luo HQ, Xiang J and Li JM:
A meta-analysis of the association between polymorphisms in
MicroRNAs and risk of ischemic stroke. Genes (Basel). 6:1283–1299.
2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Darabi H, Salmaninejad A, Jaripour ME,
Azarpazhooh MR, Mojarrad M and Sadr-Nabavi A: Association of the
genetic polymorphisms in immunoinflammatory microRNAs with risk of
ischemic stroke and subtypes in an Iranian population. J Cell
Physiol. 234:3874–3886. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wells GA, Shea B, O'Connell D, Peterson J,
Welch V, Losos M and Tugwell P: The Newcastle-Ottawa Scale (NOS)
for assessing the quality of nonrandomised studies in
meta-analyses. The Ottawa Hospital Research Institute. urihttp://www.ohri.ca/programs/clinical_epidemiology/oxford.aspsimplehttp://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
|
|
14
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Huang S, Zhou S, Zhang Y, Lv Z, Li S, Xie
C, Ke Y, Deng P, Geng Y, Zhang Q, et al: Association of the genetic
polymorphisms in pre-microRNAs with risk of ischemic stroke in a
Chinese population. PLoS One. 10(e117007)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liu Y, Ma Y, Zhang B, Wang SX, Wang XM and
Yu JM: Genetic polymorphisms in pre-microRNAs and risk of ischemic
stroke in a Chinese population. J Mol Neurosci. 52:473–480.
2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Luo HC, Luo QS, Wang CF, Lei M, Li BL and
Wei YS: Association of miR-146a, miR-149, miR-196a2, miR-499 gene
polymorphisms with ischemic stroke in a Chinese people. Oncotarget.
8:81295–81304. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhu X, Hou R, Ma A, Yang S and Pan X:
Associations of miR-146a, miR-149, miR-196a2, and miR-499
polymorphisms with ischemic stroke in the Northern Chinese Han
population. Med Sci Monit. 24:7366–7374. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Hong SJ, Kim SK, Yun DH, Chon J and Park
HJ: Association between MicroRNA-4669 polymorphism and ischemic
stroke in a Korean population. Dis Markers.
2019(7238319)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhu R, Wang QW, Zhao J, Liu X and He Z:
MiR-149 and miR-499 gene polymorphism and the incident of ischemic
stroke in the Asian population: From a case-control study to
meta-analysis. Clin Neurol Neurosurg. 193(105789)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zou D, Liu C, Zhang Q, Li X, Qin G, Huang
Q, Meng Y, Chen L and Wei J: Association between polymorphisms in
microRNAs and ischemic stroke in an Asian population: Evidence
based on 6,083 cases and 7,248 controls. Clin Interv Aging.
13:1709–1726. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wang JX, Jiao JQ, Li Q, Long B, Wang K,
Liu JP, Li YR and Li PF: MiR-499 regulates mitochondrial dynamics
by targeting calcineurin and dynamin-related protein-1. Nat Med.
17:71–78. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Yang B, Chen J, Li Y, Zhang J, Li D, Huang
Z, Cai B, Li L, Shi Y, Ying B and Wang L: Association of
polymorphisms in pre-miRNA with inflammatory biomarkers in
rheumatoid arthritis in the Chinese Han population. Hum Immunol.
73:101–106. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Cattin L, Da Col PG, Feruglio FS, Finazzo
L, Rimondi S, Descovich G, Manzato E, Zambon S, Crepaldi G, Siepi
D, et al: Efficacy of ciprofibrate in primary type II and IV
hyperlipidemia: The Italian multicenter study. Clin Ther.
12:482–488. 1990.PubMed/NCBI
|
|
25
|
El Gazzar M, Church A, Liu T and McCall
CE: MicroRNA-146a regulates both transcription silencing and
translation disruption of TNF-α during TLR4-induced gene
reprogramming. J Leukoc Biol. 90:509–519. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Calcagni E and Elenkov I: Stress system
activity, innate and T helper cytokines, and susceptibility to
immune-related diseases. Ann N Y Acad Sci. 1069:62–76.
2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Hu Z, Liang J, Wang Z, Tian T, Zhou X,
Chen J, Miao R, Wang Y, Wang X and Shen H: Common genetic variants
in pre-microRNAs were associated with increased risk of breast
cancer in Chinese women. Hum Mutat. 30:79–84. 2009.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Xiang Y, Fan S, Cao J, Huang S and Zhang
LP: Association of the microRNA-499 variants with susceptibility to
hepatocellular carcinoma in a Chinese population. Mol Biol Rep.
39:7019–7023. 2012.PubMed/NCBI View Article : Google Scholar
|