Introduction
The world has faced an unexpected public health
crisis since 2019 due to the severe acute respiratory syndrome
coronavirus type 2 (SARS-CoV-2) pandemic, known as COVID-19. The
COVID-19 epidemic has caused an astounding number of fatalities and
poses a distinct threat to the global food and public health
systems. The devastating economic and social effects of the
epidemic raise the prospect that many individualscould be pushed
into extreme poverty (1-4).
According to the World Health Organization (WHO) (https://covid19.who.int) (5), symptoms of COVID-19 can range from
fever and coughing to respiratory illness, bronchitis, renal
disease, and, in extreme cases, death (6,7). WHO
has documented 659,108,952 confirmed COVID-19 cases, including
6,684,756 fatalities. Up to December 21, 2022, 13,073,712,554 doses
of vaccine had been administered (https://covid19.who.int) (5).
Understanding the life cycle of SARS-CoV-2 and its
interactions with host cells is crucial for developing effective
strategies to fight the virus. The ~30-kb genome of SARS-CoV-2
shares significant similarities with other coronaviruses, such as
SAR-CoV and MERS-CoV, and contains two open reading frames (ORF)s1a
and ORF1b, capable of translating numerous structural and
non-structural proteins. The viral genome enters host cell
membranes with the aid of the spike protein, facilitating
attachment and membrane fusion via subunits S1 and S2. The rough
endoplasmic reticulum synthesizes the 1,273-amino acid polyprotein
precursor for the SARS-CoV-2 S glycoprotein, which is subsequently
processed into two subunits, S1 and S2. These subunits play key
roles in viral attachment to host cells and membrane fusion
(8,9).
The predominant type I transmembrane S glycoprotein
of the SARS-CoV-2 envelope incorporates with the cognate receptor
of the host cell through membrane fusion. The S-surface
glycoprotein specifies its target for host immune responses and
serves as a prime target for antibody neutralization. Moreover,
S-glycoprotein can promote the fusion of infected and uninfected
cells to produce multinucleated giant cells by entering the plasma
membrane through the secretory route (10,11).
This could enable direct virus transmission across cells and
possibly change the pathogen city of SARS-CoV-2. Spike proteins
function as antiviral medications and vaccines due to their
essential parts in viral infection and their ability to induce
protective innate immune and cell-mediated adaptive immunity in
susceptible hosts (12).
The four non-structural proteins identified in the
virus are RNA polymerase, helicase, papain-like (PLpro) and
3-chymotrypsin-like (3Clpro) proteases (13). The viral replication and
transcription are performed by proteases, Plpro and 3Clpro. Among
these, the 3Clpro plays a critical role in the ability of the
virusto reproduce (14). The key
protease, also known as Mpro, is 3Clpro, and it is crucial for
viral replication. One of the main targets for developing
anti-SARS-CoVdrugs is the protease 3Clpro, which generates 11 of
the 16 non-structural proteins (NSPs) that are produced when Plpro
and 3Clpro cleave the PP chain into NSPs (15,16).
According to apreviousstudy, the primary protease Mpro of COVID-19
shares 96% of its sequence with SARS-CoV (17). Despite being in development,
effective therapeutic and preventive methods, such as medications
and vaccines, are still lacking. Herein, it is necessary to
identify novel therapeutic candidates that specifically target
certain SARS-CoV-2 proteins to treat COVID-19. A computational
method for identifying possible therapeutic candidates counter to
the treatment of emerging infectious diseases such as COVID-19 is
to create drugs based on protein structures (18,19).
The majority of anti-infective drugs are derived
from secondary metabolites found in nature, sourced from microbial,
oceanic, or plant origins (20,21).
The pharmacological qualities of ~20% of all known plant species
have been investigated; phytochemicalanalyseshave improved
healthcare by aiding in the treatment of diseases such as cancer.
Plants are able to generate a wide range of bioactive substances;
fruits and vegetables, in particular, are able to store large
amounts of phytochemicals that guard against harm from free
radicals (22). The bioactive
compounds have been classified as alkaloids, saponins, diterpenes
and flavonoids. These bioactive compounds play a vital role in the
discovery of new drugs from natural plants.
1-Methylene-2b-hydroxymethyl-3,3-dimethyl-4b-(3-methylbut-2-enyl)-cyclohexane
(SL-MHDC) is an important secondary metabolite and is isolated from
Spinifex littoreus (S. littoreus) which belongs to
the family Poaceae (23). This
coastal grass thrives on dunes, characterized by extremely hard,
in-rolled and curved leaves, with rough margins and spiky tips. The
inflorescence of this plant is a raceme with an imbricate spikelet.
This species forms substantial colonies and stabilizes dunes making
it an effective sand binder (24).
The ethnomedical history of S. littoreus indicates that the
root extract of S. littoreus has been traditionally
administered orally for the treatment of digestive disorders in the
Purba Medinipur District of West Bengal, India (25). Root decoction is also used to treat
joint and muscle pain (26).
Furthermore, the therapeutic potential of this plant has been
studied in order to elucidate its antimicrobial, antioxidant,
anti-inflammatory and analgesic properties (27). This plant is also used
traditionally as a diuretic medicine. The methanol and aquatic
extract of this herb have been demonstrated in numerous
pharmacological tests to have analgesic, anti-inflammatory and
antibacterial effects (28).
However, no therapeutic evidence is available for the antiviral
property of the plant S. littoreus, as well as for the
phytocompound MHDC. Dampalla et al (29) conducted a structure-guided design
of conformationally constrained cyclohexane inhibitors of
SARS-CoV-2 3CL protease. Considering this, the present study
performed molecular docking analysis of the phytocompound MHDC
identified from the plant S. littoreus against the 3CL main
protease and spike glycoprotein of SARS-CoV-2(30).
The present study aimed to investigate the
phytochemical diversity of S. littoreus (Burm.f.) Merr.
through preliminary phytochemical screening techniques and to
validate the findings using gas chromatography-mass spectrometry
(GC-MS) analysis. Furthermore, the present study sought to identify
the inhibitory effects of the bioactive compound SL-MHDCon the key
proteins of SARS-CoV-2, i.e., the spike protein. By leveraging the
phytochemical diversity of this coastal grass, the authors aimed to
contribute to the ongoing efforts to mitigate the devastating
impact of COVID-19 on global health.
Materials and methods
Plant collection
S. littoreus was collected from Kunthukal
Beach, Pamban, Tamil Nadu, India (latitude: 9.25323; longitude:
79.21829). The collected plants were authenticated, and the
prepared herbarium sheets of the plantwas submitted to the Rapinat
Herbarium and Centre for Molecular Systematics, St. Joseph's
College, Tiruchirappalli, Tamil Nadu, India. (Voucher nos. J.V 001
and J.V 002).
Plant extraction
The young leaves of S. littoreus were
separated from the plant, cleaned and shade-dried. The dried S.
littoreus leaves were mechanically pulverized. The powdered
sample of S. littoreus was extracted with methanol and
chloroform through the Soxhlet apparatus (Borosil Scientific
Ltd.)for 16 h. To concentrate the collected extract, it was
subjected to a rotary evaporatorand weighed. The extraction process
was repeated until obtaining 5 g of powdered form of the extracts
(31).
Preliminary phytochemical screening.
Qualitative analysis
The qualitative analysis of carbohydrates, proteins,
amino acids, tannins, saponins, flavonoids, alkaloids, terpenoids,
steroids, phenols, glycosides, quinones, coumarins and
anthraquinones was performed in six different solvent-mediated
(aqueous, acetone, ethanol, methanol, chloroform and ethyl acetate)
extracts of S. littoreus. All the chemicals and glassware
used for the phytochemical screening were sourced from Royal
Scientific Suppliers.
Carbohydrates. A total of two drops of
Molisch reagent were added to 2 ml S. littoreus extract.
Subsequently, 2 ml concentrated H2SO4 were
added on the sides of the tube. The formation of purple colour was
an indicator of the presence of carbohydrates (32).
Proteins. A few drops reagent were combined
with 2 ml S. littoreus extract. The presence of proteinswas
indicated by the production of a white precipitate, which
subsequently turns brick red in colour (33).
Amino acids. In a boiling water bath, 1 ml
S. littoreus extract along with 1 drop of ninhydrin solution
was heated. The development of a purple colour indicated the
presence of amino acids (32).
Tannins. In a test tube, 0.5 g powdered S.
littoreus extract was heated in 20 ml distilled water, filtered
and treated with 0.1% iron III chloride (FeCl3). The
samples were then observed for the development of brownish-green or
blue-black coloration, indicating the presence of tannins (33).
Terpenoids. In a test tube, 5 ml S.
Littoreusextract were used along with 2 ml CHCl3.
Subsequently, 3 ml concentrated H2SO4 were
gently added to the mixture to form a layer. In the case that
terpenoids are present, an interface with a reddish-brown colour
will form (33).
Alkaloids (Mayer's test). A test tube was filled
with 1 ml S. littoreus extract and 1 ml potassium mercuric
iodide solution (Mayer's reagent). When this mixture was shaken
gently, aprecipitate will be formed, confirming the presence of
alkaloids (33).
Quinines. To 10 mg S. littoreus
extract in isopropyl alcohol, one drop of concentrated sulfuric
acid was added. The presence of quinones was indicated by the
formation of a red colour (33).
Steroids. A total of 5 ml S. littoreus
extract was mixed with 2 ml chloroform and concentrated
H2SO4. A red colour appeared in the lower
chloroform layer, indicating the presence of steroids (34).
Coumarin. A total of 2 ml S. littoreus
extract was mixed with 3 ml 10% NaOH. The presence of coumarin was
indicated by a yellow coloration (35).
Saponins. A total of 20 ml distilled water
and 2 g powdered S. littoreus extract were heated together
in a water bath and then filtered. Subsequently, 10 ml of the
filtered sample were mixed with 5 ml distilled water in a test tube
and vigorously shaken to form a stable, long-lasting froth. This
was followed by the addition of three drops of extra virginolive
oil to the froth to create an emulsion, indicating the presence of
saponins (34).
Phenols. A few drops of FeCl3 and
1 ml water were added to a test tube along with ~2 ml of the S.
littoreus extract. The presence of a blue, green, red, or
purple colour indicates the presence of phenols (35).
Anthraquinones (Borntrager's test). A total
of 1 ml S. littoreus extract was added to a mixture of
diluted ammonia, chloroform, or benzene. The presence of
anthraquinone derivatives causes the ammonical layer to change
colour from pink to bright red (35).
Flavonoids. S. littoreus extract was
added to a test tube along with a few drops of a 1% NH3
solution. Yellow colour concludes the presence of flavonoids
(36).
Glycosides. S. littoreus extract was
used along with 2 ml acetic acid and 2 ml chloroform. Once the
mixture was cooledat room temperature, concentrated
H2SO4 was added. The steroidal glycosides can
be identified by their green colour appearance (37).
Quantitative screening of
phytochemicals
The presence of alkaloids, terpenoids, phenolics,
flavonoids and tannins was quantitatively estimated with their
respective standard using the following methods:
Quantitative estimation of alkaloids. To
estimate the quantity of alkaloids in S. littoreus, the
extracts were treated with 5 ml bromocresol green and PBS. Atropine
was used as a standard solution. Alkaloids were measured in
milligrams of atropine equivalents per gram (mg AE/g) of chloroform
extract of sample (38).
Quantification of terpenoids. A total of 500
g S. littoreus leaf powder was soaked in ice-cold 95%
methanol and then centrifuged at 4,000 x g for 15 min at room
temperature. The supernatant was mixed with 1.5 ml chloroform and
100 ml concentrated sulfuric acid. Following incubation at room
temperature for 1.5-2 h in the dark, a brown precipitate appeared
at the bottom, which was dissolved in 1.5 ml 95% methanol, and its
absorbance was measured in the colorimeterat 538 nm (39).
Quantification of flavonoids. A colorimetric
test was used to determine the quantity of flavonoids in the S.
littoreus extract. To 1 ml of the extract, 0.3 ml of 10%
aluminum chloride, 5% sodium nitrite and 2 ml of NaOH were added.
The absorption spectrum of this mixture was measured using
UV-visible instrumentat 510 nm. Similarly, a set of quercetin
solutions was prepared and measured as standards. Flavonoid
concentration was expressed as mg of quercetin equivalents per gram
of extract (mg QE/g) (40).
Quantification of tannins. A total of 1 ml
S. littoreus extract was mixed with 0.5 ml Folin-Ciocalteu
reagent and 1 ml of 35% Na2CO3, followed by
the addition of 7.5 ml distilled water. This mixture was then
incubated at 30˚C for 30 min. Similarly, a set of tannic acid
solutions was prepared, and the optical density was measured at 700
nm with an UV/visible spectrophotometer. The amount of tannins in
the S. littoreus extract was expressed in milligrams of
tannic acid equivalent per gram (mg TAE/g) (41).
Determination of total phenolics. A total of
1 ml S. littoreus extract was dispersed in 9 ml of distilled
water along with 1 ml of Folin-Ciocalteu and 10 ml of 7%
Na2CO3 solution. This was incubated at 30˚C
for 2 h at room temperature. Gallic acid was used as standard.
Gallic acid equivalents (mg GAE/g) were used to represent mg of
total phenolic component per gram of extract (42).
GC-MS analysis
The phytoconstituents present in the methanolic
ethanol, ethyl acetate and chloroform extracts were screened using
GC-MS analysis. The experimental condition of the instrument was a
fused silica column filled with Elite-5MS (5% biphenyl, 95%
dimethylpolysiloxane, 30 m, 0.25 mm ID, 250 m df) utilized in the
analysis by the Clarus 680 GC, and the Helium carrier gas was used
to separate the components at a constant flow rate of 1 ml/min. The
injector temperature was set to 260˚C. The operating parameters of
the mass detector were 240˚C for the transfer line, 240˚C for the
ion source, 70 eV for the electron impact in the ionization mode,
0.2 sec for each scan, and 0.1 sec between scans. The component
spectra were compared to a database of component spectrums
maintained in the GC-MS NIST (2008) library (43,44).
No standard compounds were used for the spectral comparison.
Drug suitability and pharmacokinetic
profile
The physicochemical, pharmacokinetic and
drug-likeliness properties of the compound SL-MHDC were predicted
using the SwissADME server (http://www.swissadme.ch/) to optimize the compound
based on its bioavailability, safety and efficacy (45,46).
Molecular docking. Ligand
preparation
Based on the GC-MS results of S. littoreus,
the chemical structure of the phytocompound SL-MHDCwas retrieved
from the PubChem database (PubChem CID: 550196) as an optimized 3D
ligand molecule. The downloaded ligand was prepared using the
Autodock MGL application (https://autodock.scripps.edu/) by detecting its
torsion root and saved in pdbqt format. This was then used for
docking with AutodockVina (47,48).
Target protein preparation. The target
protein selected for the study was the structure of SARS-CoV-2
XBB.1. The structure was downloaded from the protein data bank (PDB
ID: 8IOV) in PDB format. The chain B spike glycoprotein (RBD) of
XBB.1 was used as a target receptor for the docking analyses. The
protein preparation was performed by removing the water molecules
and the heteroatom within the structure. The structure was then
examined for the missing residues and it was saved in pdbqt format.
Both the optimized protein and ligand were then subjected to
docking analysis using Autodock 4.2.6 software (49) to identify the affinity of the
GC-MS-derived compound towards the SARS-CoV-2 XBB.1. The optimal
pose with the least energy of binding was used for further
analyses.
Molecular dynamics simulations (MDS). MDS is
a computational paradigm that enables atomistic interrogation of
biomolecular systems, which serves as a cornerstone in
pharmaco-reconnaissance. Thus, MDS were executed using Desmond
module (Schrödinger Suite) (50)
(https://www.schrodinger.com/platform/products/desmond/).
The SARS-CoV-2 XBB.1 spike glycoprotein complexed with SL-MHDC was
solvated via system builder by employing TIP4P aqueous solvation
within an orthorhombic solvent box. Moreover, the cell size was
49.6x68.1x51.7 Å, as well as the solvent buffer extending 10 Å,
which is beyond the protein in all Cartesian directions. As a
corollary, OPLS_2005 force field was deployed to parameterize
molecular energetics. Judiciously, electrostatic neutrality was
achieved by the incorporation of counter ions (Na+ and
Cl-) that was followed by constrained energy
minimization with the exorbitant of 2,500 iterations, as well as
the convergence thresholds of 1 kcal/mol/Å. Furthermore, the
present study exploited NPT ensemble with the Nose-Hoover
thermostat algorithm which is lauded for thermal fluctuations set
at a reference temperature of 300 K. A >30 nsec trajectory was
generated and the post-simulation analyses encompassed with
Root-mean-square deviation (RMSD) and the interrogation of
SARS-CoV-2 XBB.1 integrated with SL-MHDC complex persistent was
designated against the binding locus.
Results
Screening of phytoconstituents
The preliminary phytochemical screening of S.
littoreus was carried out to identify the presence of secondary
metabolites, such as alkaloids, flavonoids, tannins, saponins,
glycosides, quinones, coumarins, steroids, phenols, terpenoids and
anthraquinones using six different solvents (aqueous, acetone,
ethanol, methanol, chloroform and ethyl acetate). Among these, the
chloroform extract revealed sevensecondary metabolites and one
primary metabolitein S. littoreus followed by the acetone,
methanol and ethanol extracts, which revealed sixsecondary
metabolites and one primary metabolite. The ethyl acetate extract
only revealed four compounds (Table
I).
 | Table IQualitative assessment of
phytochemicals. |
Table I
Qualitative assessment of
phytochemicals.
| | Solvents |
|---|
| Metabolites | Aqueous | Acetone | Ethanol | Methanol | Chloroform | Ethyl acetate |
|---|
| Primary
metabolites | | | | | | |
|
Carbohydrates | + | - | + | + | + | - |
|
Proteins | - | - | - | - | - | - |
|
Amino
acids | - | - | - | - | - | - |
| Secondary
metabolites | | | | | | |
|
Alkaloids | - | - | + | + | + | - |
|
Flavonoids | - | + | + | + | + | + |
|
Tannins | + | + | - | + | - | - |
|
Saponins | + | - | + | - | + | - |
|
Glycosides | + | + | - | - | - | - |
|
Quinones | - | + | + | + | + | + |
|
Coumarins | + | + | + | + | + | + |
|
Steroids | - | - | + | + | + | + |
|
Phenols | + | + | - | - | - | - |
|
Terpenoids | - | - | - | - | + | - |
|
Anthraquinones | - | - | - | - | - | - |
The total amount of terpenoids, phenols, tannins,
alkaloids and flavonoids present in the S. littoreus extract
was estimated using quantitative analysis with their respective
standards. The findings indicated the presence of all five examined
compounds in the S. littoreus plant, albeit in varying
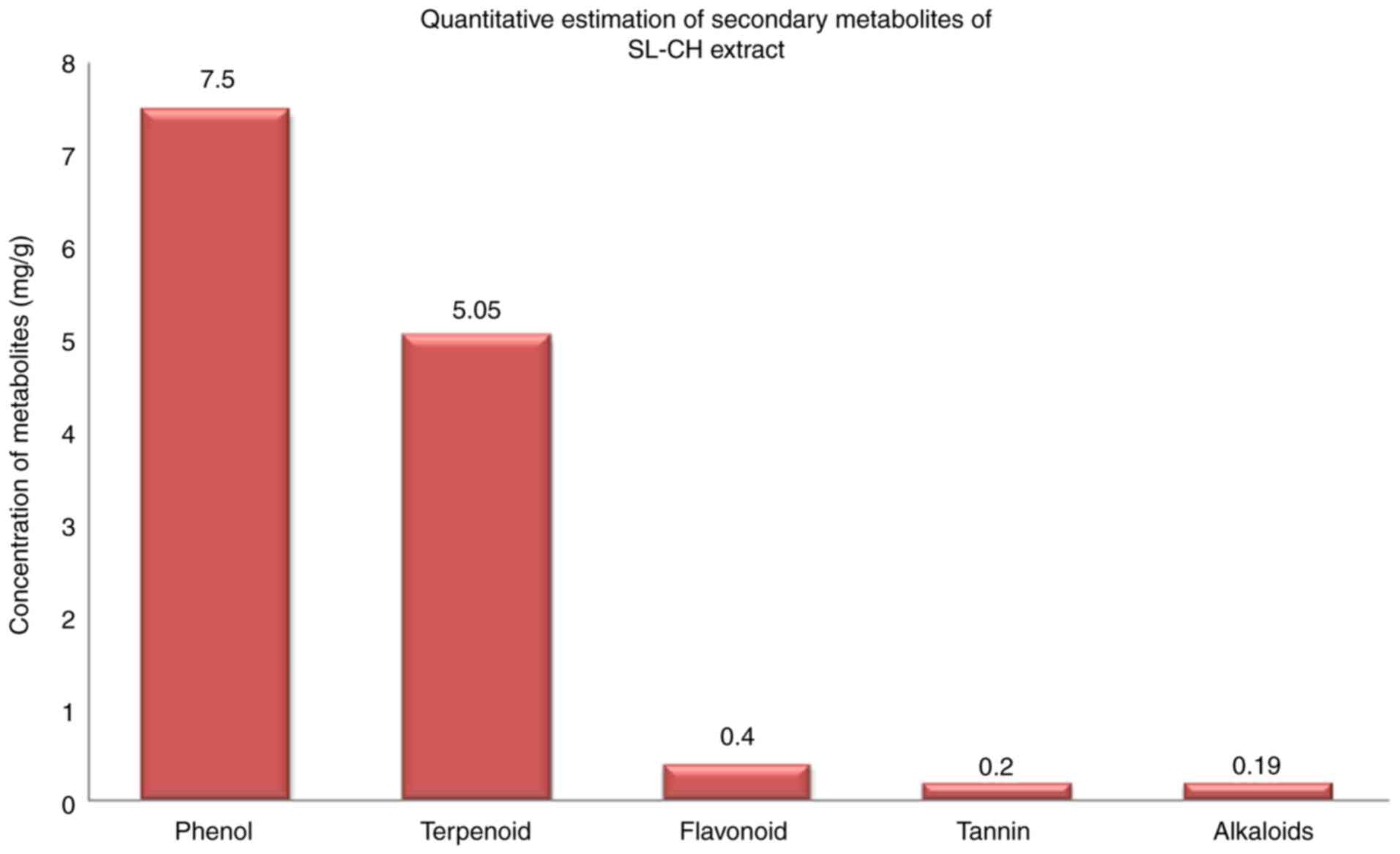
concentrations: Phenol (7.5 mg/g), terpenoids (5.05 mg/g),
flavonoids (0.4 mg/g), tannin (0.2 mg/g) and alkaloids (0.19 mg/g).
The concentrations of phenol, terpenoids, flavonoids, tannin and
alkaloids in the S. Littoreus leaflet extract are
illustrated in Fig. 1. Among these
compounds, phenol and terpenoids accumulated at higher levels in
the S. Littoreus leaflet. Additionally, the leaflet extract
of S. Littoreus also contained trace levels of alkaloids,
tannins and flavonoids.
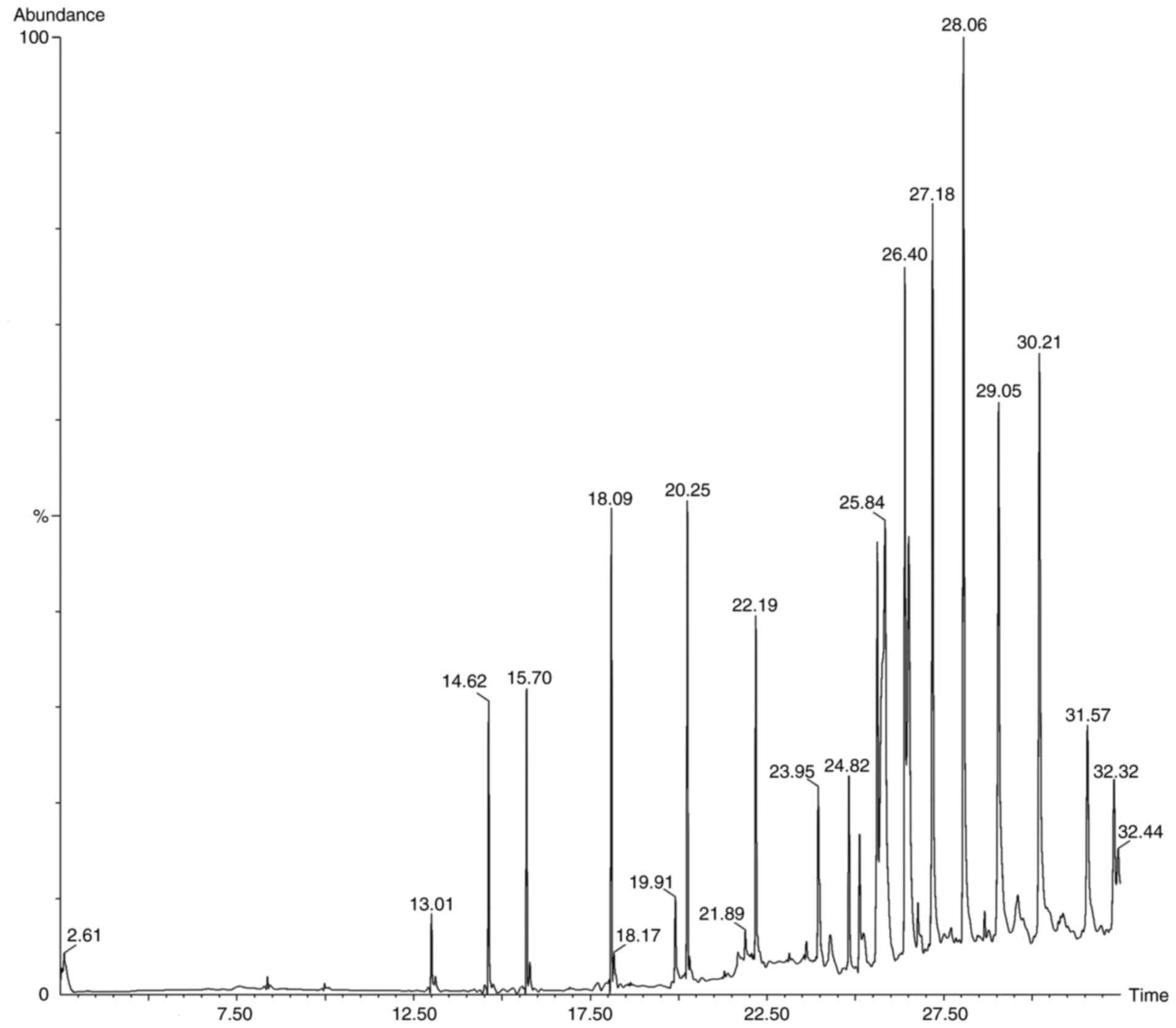
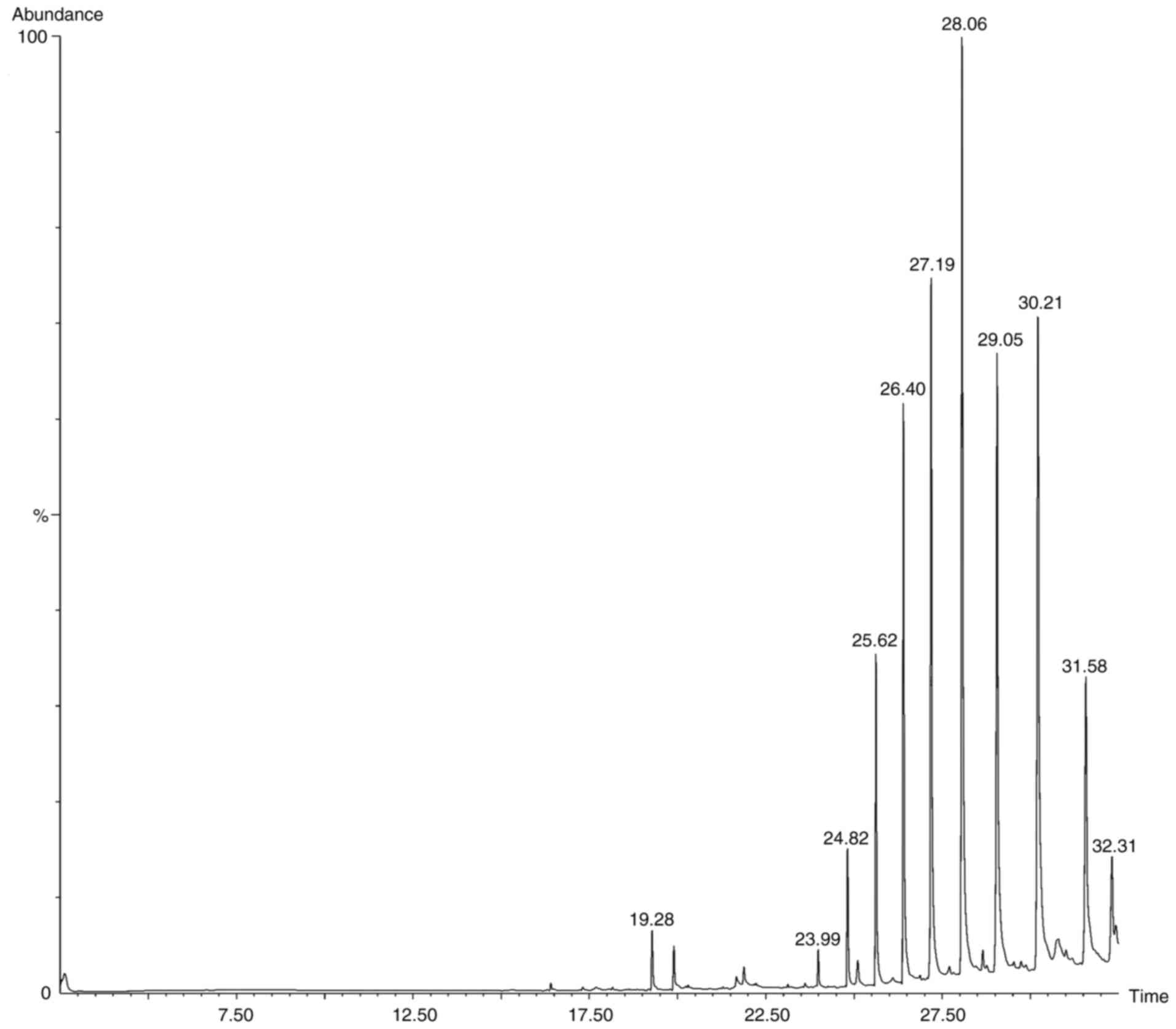
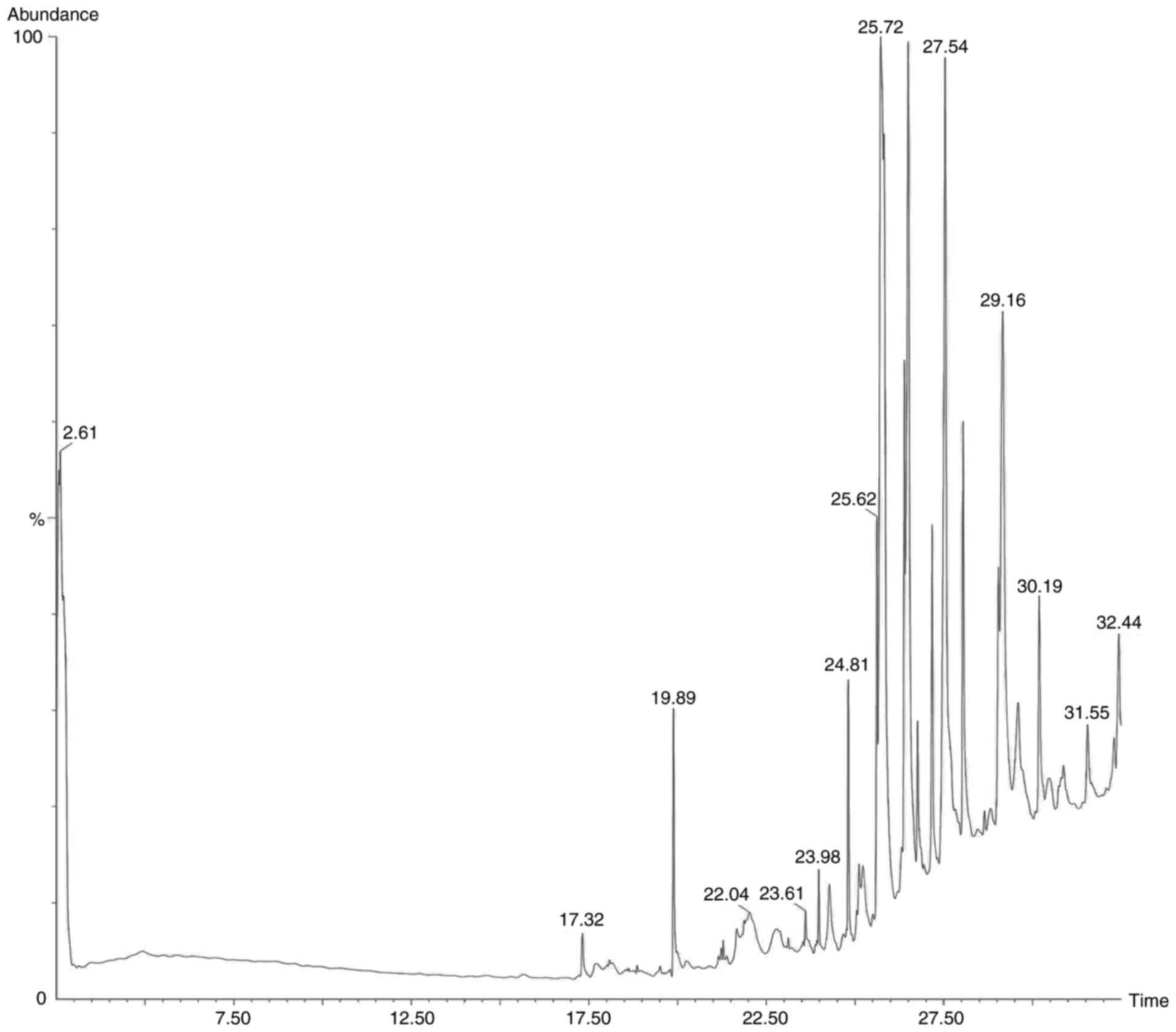
Following the preliminary screening, GC-MS analysis
of the ethanol, methanol, chloroform and ethyl acetate extracts of
S. Littoreus leaves was performed; this revealed the
presence of various active phytocompounds. A total of 33 different
compounds were eluted from this plant. Among these 33 compounds,
1-methylene-2β-hydroxymethyl-3,3-dimethyl-4β-(3-methylbut-2-enyl)-cyclohexane
(28.834%), hexadecane (19.962%), tetratetracontane (18.416%),
octacosane (16.136%), octadecanal (15.833%), heptacosane (14.574%),
2-methyl-3-(3-methyl-but-2-enyl)-2-(4-methyl-pent-3-enyl)-oxetane
(14.434%) had the high percentage area. The names, molecular
formulas and area percentages of the screened phytoconstituents are
presented in Table II, and the
biological activities with the 2D structures of the compounds are
presented in Table SI. The
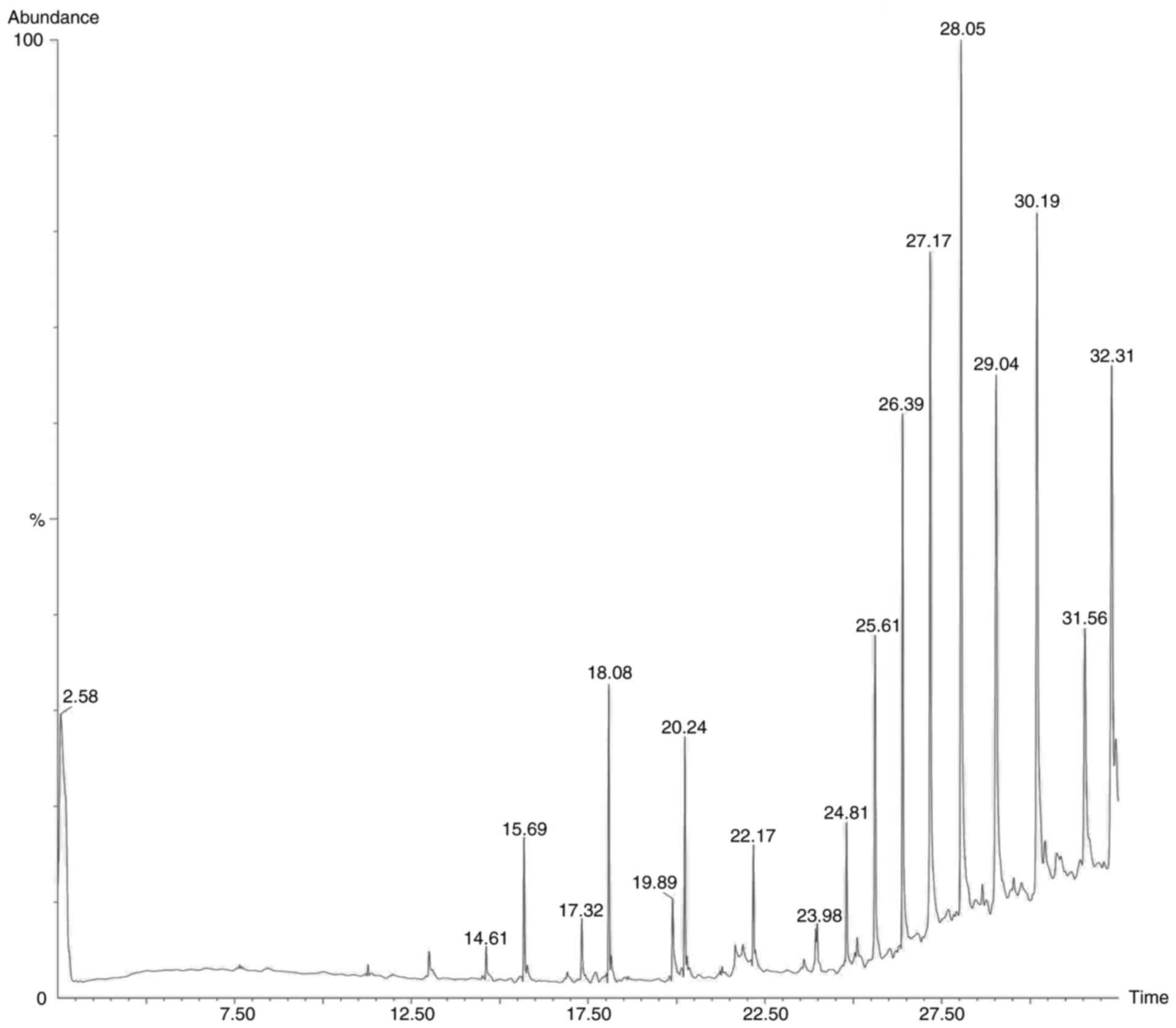
chromatogram images of the tested samples are illustrated in
Fig. 2, Fig. 3, Fig.
4 and Fig. 5.
 | Table IIPhytoconstituents present in the
extract of Spinifex littoreus (Burm.f.) Merr. |
Table II
Phytoconstituents present in the
extract of Spinifex littoreus (Burm.f.) Merr.
| Chloroform | Methanol | Ethanol | Ethyl acetate |
|---|
| Name of the
compound | Molecular
formula | RT | Area % | Name of the
compound | Molecular
formula | RT | Area % | Name of the
compound | Molecular
formula | RT | Area % | Name of the
compound | Molecular
formula | RT | Area % |
|---|
| Phenol, 2,4-bis
(1,1-dimethyl) |
C14H22O | 14.618 | 2.537 |
1,2-benzenedicarboxylic acid, butyl octyl
ester |
C20H30O4 | 19.885 | 2.118 | Methanediamine,
n,n,n',n'-tetraethyl |
C9H22N2 | 19.275 | 1.090 | 3-tetradecane,
(z) |
C14H28 | 15.689 | 1.921 |
| 3-tetradecene,
(Z) |
C14H28 | 15.699 | 2.498 |
Alpha-bisabolol |
C15H26O | 24.287 | 1.229 | N-hexadecanoic
acid |
C16H32O | 19.896 | 0.918 |
1,6;3,4-dianhydro-2-deoxybeta-d-lyxo-hexopyranose |
C6H8O3 | 17.324 | 1.094 |
| 3-octadecene,
(E) |
C18H36 | 18.095 | 3.790 |
Hexatriacontane |
C36H74 | 24.813 | 2.076 |
1-iodo-2-methylundecane |
C12H25I | 23.987 | 0.683 | 3-tetradecane,
(z) |
C14H28 | 18.085 | 3.427 |
| N-Hexadecanoic
acid |
C16H32O2 | 19.911 | 0.939 |
1-iodo-2-methylundecane |
C12H25I | 25.618 | 2.854 | Hexadecane |
C16H34 | 24.818 | 2.361 | N-hexadecanoic
acid |
C16H32O | 19.891 | 1.593 |
| N-tetracosanol |
C24H50O | 20.246 | 3.852 |
1-methylene-2b-hydroxymethyl-3,3-dimethyl-4b-(3-methylbut-2-enyl)-cyclohexane |
C15H26O | 25.728 | 28.834 | Hexadecane |
C16H34 | 25.623 | 5.859 |
N-tetracosanol-1 |
C24H50O | 20.236 | 2.714 |
| 1-hexacosanol |
C26H54O | 22.187 | 3.300 |
1-iodo-2-methylundecane |
C12H25I | 26.388 | 3.628 | Sulfurus acid,
butyl decyl ester |
C14H30O3S | 26.398 | 10.570 | Oleic acid |
C18H34O2 | 21.661 | 1.135 |
| 1-hexacosanol |
C26H54O | 23.947 | 1.541 |
2-methyl-3-(3-methyl-but-2-enyl)-2-(4-methyl-pent-3-enyl)-oxetane |
C15H26O | 26.503 | 14.434 | Hexadecane |
C16H34 | 27.189 | 13.820 | 1-docosene |
C22H44 | 22.171 | 1.852 |
|
1-octonal,2-butyl |
C12H26O | 23.987 | 0.807 |
5-ethyl-1-nonane |
C11H22 | 26.768 | 1.199 | Hexadecane |
C16H34 | 28.064 | 19.962 |
1-iodo-2-methylundecane |
C12H25I | 24.813 | 2.223 |
| Hexadecane |
C16H34 | 24.818 | 2.139 |
1-iodo-2-methylundecane |
C12H25I | 27.173 | 3.313 | Octacosane |
C28H58 | 29.054 | 14.993 | Nonadecane |
C19H40 | 25.613 | 5.161 |
| DI-N-Octyl
phthalate |
C24H38O4 | 25.118 | 1.250 | Octadecanal |
C18H36O | 27.539 | 15.833 |
Tetratetracontane |
C44H90 | 30.220 | 18.416 | Hexadecane |
C16H34 | 26.393 | 7.837 |
| Nonadecane |
C19H40 | 25.623 | 3.660 |
Hexatriacontane |
C36H74 | 28.049 | 4.191 | Nonadecane |
C19H40 | 31.575 | 8.984 | Hexadecane |
C16H34 | 27.173 | 11.655 |
|
1-Methylene-2b-hydroxymethyl-3,3-dimethyl-4b-(3-methylbut-2-enyl)-cyclohexane |
C15H26O | 25.838 | 12.558 | Octadecylpentaflu
oropropionate |
C21H37F5O | 29.039 | 2.659 | Octadecanol |
C18H36O | 32.316 | 2.345 | Heptacosane |
C27H56 | 28.049 | 14.574 |
| Nonadecane |
C19H40 | 26.403 | 7.461 | 1-hexacosanol |
C26H54O | 29.169 | 11.664 | | | | |
Tetratetracontane |
C44H90 | 29.039 | 11.422 |
|
2-methyl-3-(3-methyl-but-2-enyl)-2-(4-methyl-pent-3-enyl)-oxetane |
C15H26O | 26.513 | 9.114 | Hexadecanal |
C16H32O | 29.594 | 2.114 | | | | | Octacosane |
C28H58 | 30.200 | 16.136 |
| Hexadecane |
C16H34 | 27.184 | 9.437 | 1-decanol,
2-ethyl |
C12H26O | 30.195 | 2.677 | - | - | - | - |
Cis-9,10-epoxyoctadecane-1-ol |
C18H36O2 | 30.425 | 1.004 |
| Hexadecane |
C16H34 | 28.064 | 11.368 | Undecanal |
C11H22O | 31.545 | 1.176 | - | | - | - |
Hexatriacontane |
C36H74 | 31.555 | 6.475 |
| Tetracontane, 3,5,
24-trimethyl |
C43H88 | 29.059 | 9.041 | - | - | - | | - | - | - | - | Pentadecanal |
C15H30O | 32.306 | 9.777 |
| Nonadecane |
C19H40 | 30.210 | 8.803 | - | - | - | - | - | - | - | - | - | - | - | - |
|
1-octanol,2-butyl |
C12H26O | 31.571 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Pentadecanal |
C15H30O | 32.316 | - | - | - | - | - | - | - | - | - | - | - | - | - |
Assessment of drug-likeliness and
pharmacokinetic profiles
The compound selected from the GC-MS analysis was
subjected to in silico drug-likeliness and pharmacokinetic
prediction to investigate its absorption, distribution, metabolism
and toxicity profiles, and to evaluate whether there are any
violations in Lipinski's rule of 5. According to Lipinski's rule of
5, the molecular weight should be <500 g/mol, not >10 g/mol,
with >5 hydrogen bond acceptors and donors, and a log P-value
>5. It is important to consider any bioactive compounds as a
lead molecule in the process of drug discovery. From the results of
ADME analysis, it was identified that there were 0 violations in
Lipinski's' rule of 5, the molecular weight of the compound was
observed to be 222.37 g/mol, there was 1 hydrogen bond donor and
acceptor, the MLogP of the compound was 3.56 (Table III). Additionally, the
pharmacokinetic properties of the compounds were evaluated,
including the calculation of inhibition, which includes the
subfamilies of cytochrome P450, such as CYP2C19, CYP2C9, CYP1A2,
CYP2D6 and CYP3A4, and BBB permeant. All these results, including
the drug likeliness of the compounds are presented in Tables IV and V. This suggests that the compound has
drug-like properties, as it meets the Lipinski's and Ghose's rules,
which evaluate the properties for drug-likeliness, indicating that
it falls within the range of properties commonly found in known
drugs. The compound meets Veber's criteria suggesting its good oral
availability potential. In addition, the compound meets the Egan
rule, indicating it is less likely to be a P-glycoprotein
substrate.
 | Table IIIPhysicochemical properties of the
compound selected through gas chromatography-mass spectrometry
analysis. |
Table III
Physicochemical properties of the
compound selected through gas chromatography-mass spectrometry
analysis.
| Compound name | Molecular weight
g/mol (<500) | H-bond acceptors
(<10) | H-bond donors
(<5) | MlogP
(<4.15) | Lipinski
violations |
|---|
|
1-Methylene-2b-hydroxymethyl-3,3-dimethyl-4b-(3-methyl
but-2-enyl)-cyclohexane | 222.37 | 1 | 1 | 3.56 | 0 |
 | Table IVPharmacokinetic properties of the
compound selected from chromatography-mass spectrometry
analysis. |
Table IV
Pharmacokinetic properties of the
compound selected from chromatography-mass spectrometry
analysis.
| Compound name | Gi absorption | BbbPermeant | P-Gp substrate | Cyp1a2
inhibitor | Cyp2c19
inhibitor | Cyp2c9
inhibitor | Cyp2d6
inhibitor | Cyp3a4
inhibitor |
|---|
|
1-Methylene-2b-hydroxymethyl-3,3-dimethyl-4b-(3-methylbut-2-enyl)-cyclohexane | High | Yes | No | No | No | Yes | No | No |
 | Table VDrug-likeness of the compound
selected through chromatography-mass spectrometry analysis computed
using SWISS-ADME. |
Table V
Drug-likeness of the compound
selected through chromatography-mass spectrometry analysis computed
using SWISS-ADME.
| Compound name | Lipinski | Ghose | Veber | Egan | Muegge | Bioavailability
score |
|---|
|
1-Methylene-2b-hydroxymethyl-3,3-dimethyl-4b-(3-methylbut-2-enyl)-cyclohexane | Yes; 0
violation | Yes | Yes | Yes | No, 1 violation:
heteroatoms <2 | 0.55 |
Docking analyses results
To evaluate the activity of the compound SL-MHDC
against the SARS-CoV-2 XBB.1 spike RBD (PDB ID: 8IOV), both the
target protein and the selected ligand were subjected to docking
analyses. The computational docking was performed using the
AutoDock tool, which generated binding modes in the lowest energy
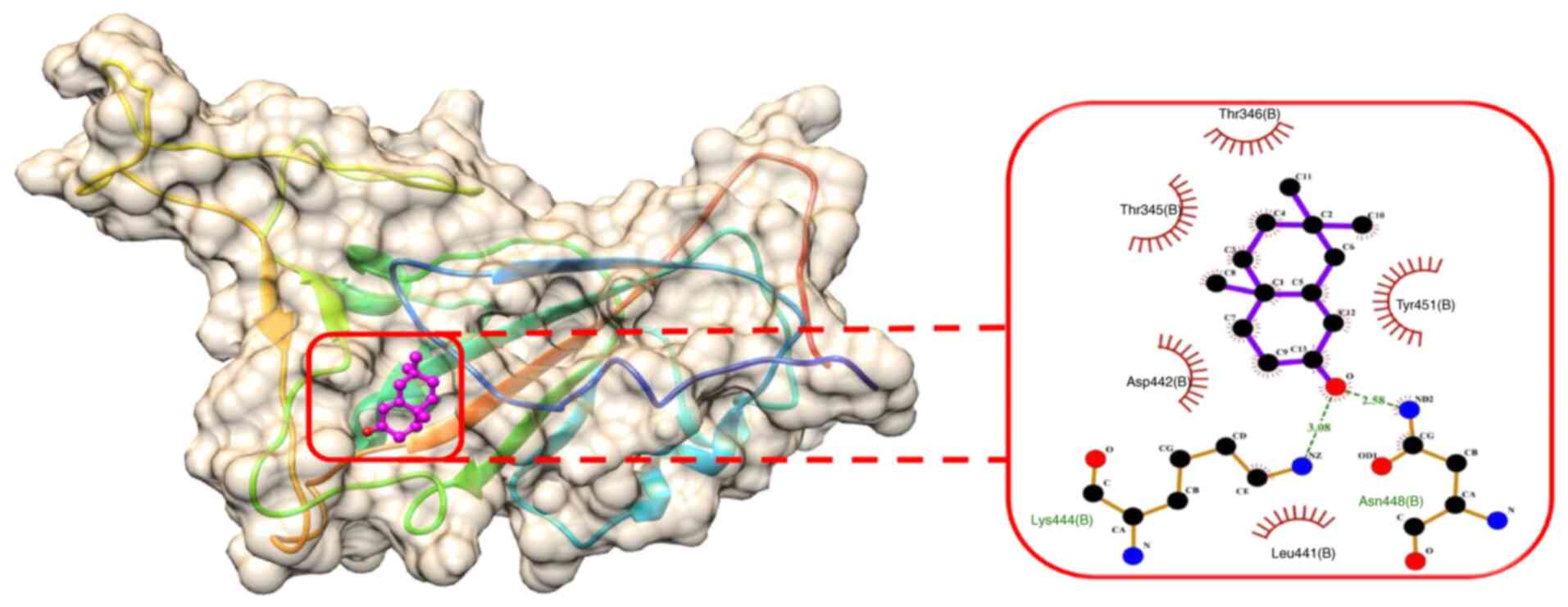
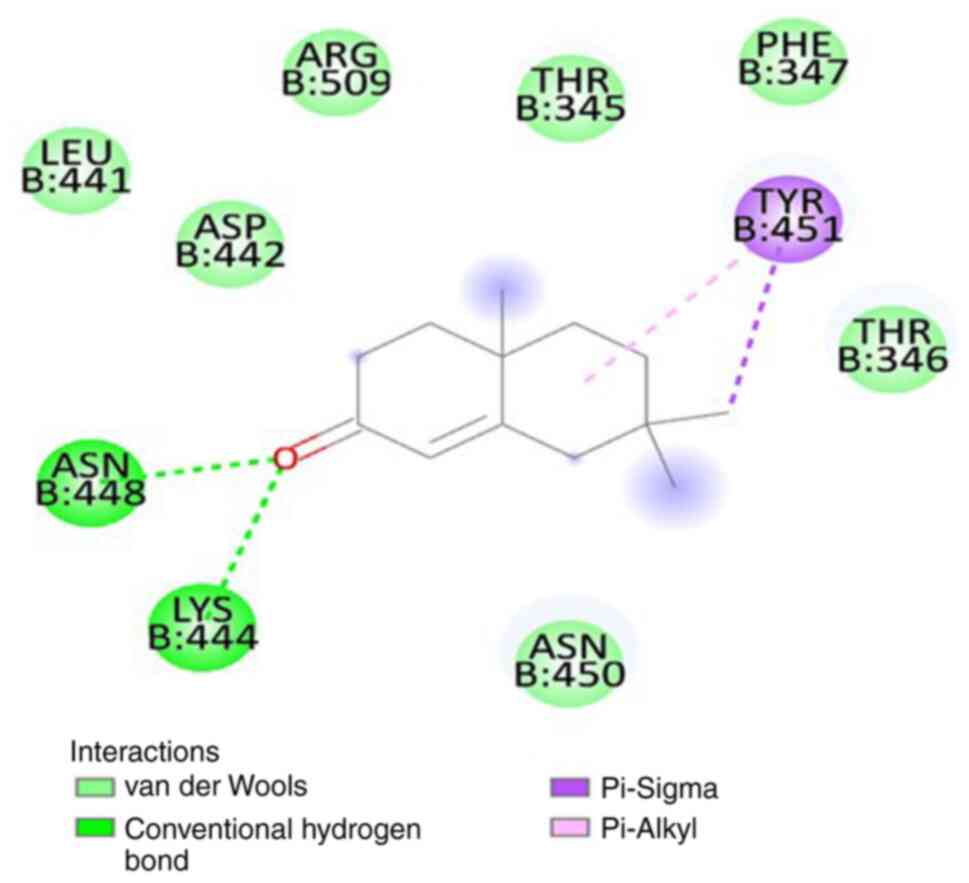
state. The binding energy of SL-MHDC with the XBB.1 was observed to
be -4.86 kcal/mol. This interaction formed two conventional
hydrogen bonds with the amino acid residues LYS444 (B) and ASN 448
(B) (Fig. 6) in the spike
glycoprotein. The bond lengths were measured as 3.08 and 2.58 Å,
respectively (Table VI). The
other non-bonded interactions include the Vander der Waals
interaction formed by the residues, such as LEU 441 (B), ASP 442
(B), ARG 509 (B), THR 345 (B), PHE 347 (B), THR 346 (B), ASN 450
(B), the Pi-sigma and Pi-Alkyl interaction was formed by TYR 451
(B) (Fig. 7). Hence, from the
molecular docking it could be inferred that the compound SL-MHDC
S. littoreus can be used as a putative inhibitor potent drug
against the spike glycoprotein of XBB.1.
 | Table VIDocking results of
1-methylene-2b-hydroxymethyl-3,3-dimethyl-4b-(3-methylbut-2-enyl)-cyclohexane
with SARS-CoV-2 XBB.1. |
Table VI
Docking results of
1-methylene-2b-hydroxymethyl-3,3-dimethyl-4b-(3-methylbut-2-enyl)-cyclohexane
with SARS-CoV-2 XBB.1.
| Compound name | Protein ID | Binding energy
(kcal/mol) | No. of H-bonds | H-Bond forming
residues | Bond length
(Å) |
|---|
|
1-Methylene-2b-hydroxymethyl- | 8IOV | -4.86 | 2 | LYS444 | 3.08 2.58 |
|
3,3-dimethyl-4b-(3-methylbut-2-enyl)-cyclohexane | | | | ASN448 | |
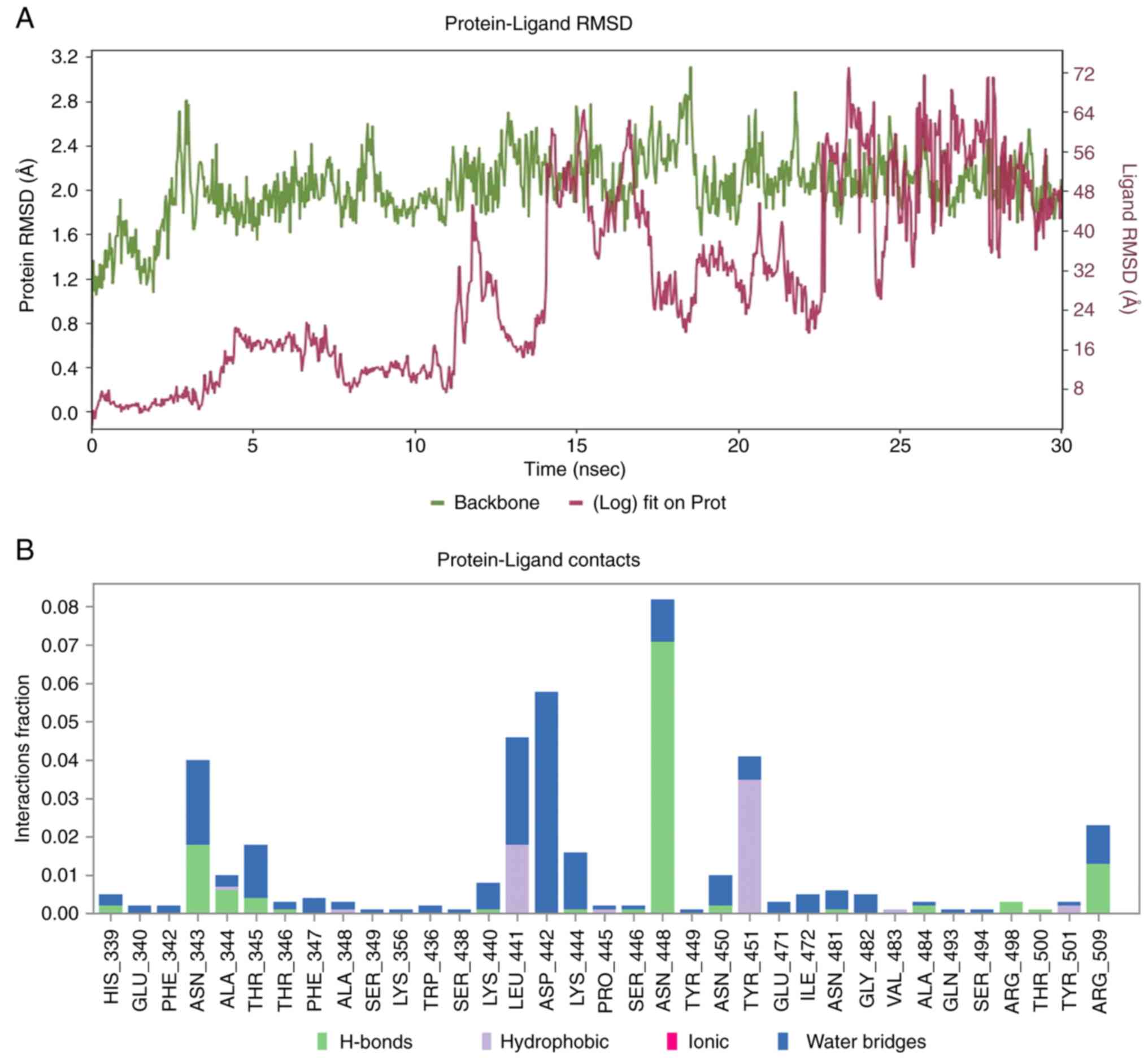
Molecular dynamics evaluation of the
XBB.1 - SL-MHDC complex
In order to assess conformational stability and
interaction fidelity of the SARS-CoV-2 XBB.1 spike glycoprotein
complexed with the phyto-ligand SL-MHDC, explicit MDS were
effectuated for a 30 nsec. The simulation encompassed with the
canonical analyses including RMSD profiling and protein-ligand
contact mapping (Fig. 8).
Discussion
Phytochemicals are non-nutritive plant compounds
considered to be beneficial for human health due to their potential
therapeutic properties (51).
Worldwide, due to their perceived effectiveness, scientists are
investigating the potential use of pharmacologically active
compounds derived from therapeutic plants. Of note, 80% of
individuals globally utilise herbal medications due to their
effectiveness, affordability, non-narcotic nature and lack of
adverse effects (52). The present
study aimed to identify a potent anti-coronaviral compound from a
coastal grass S. Littoreus by screening its
phytoconstituents. The preliminary phytochemical screening
of S. Littoreus leaflet revealed the presence of major
constituents, such as alkaloids, flavonoids, tannins, saponins,
glycosides, quinones, coumarins, steroids, phenols and terpenoids.
Chandran et al (53) in
2014 also supported the presence of phytocompounds of alkaloids,
flavonoids, saponins and phenol in S. littoreus. These
compounds are considered therapeutically significant metabolites
due to their pharmacological activities, such as anti-malarial,
anticancer, anti-microbial, anti-ulcer, anti-inflammatory and
diuretic actions (54-58).
Quantitative analysis revealed the marked abundance of phenolic
compounds and terpenoids. Phenolic compounds play a vital role in
the plant by protecting them from UV radiations, pathogenic
microorganisms and parasites (59). Additionally, they can serve as
sources of anti-carcinogenic and anti-mutagenic agents (60). Terpenoids play a crucial role in
plant defence against biotic and abiotic stresses and as signal
molecules for pollination insects (61). Along with this, they are also
effective in preventing and treating various diseases, including
cancer. They do have pharmacological importance as antimicrobial,
antifungal, antiparasitic, antiviral, anti-allergenic,
antispasmodic, antihyperglycemic, anti-inflammatory and
immunomodulatory agents (62). A
trace amount of flavonoids, tannin and alkaloids was also recorded,
which in plants serve as a defence system and possesses
antimicrobial and anti-inflammatory properties (63). While quantitative determination is
a key technique used to set the standard of a crude medication,
solvent efficiency in phytochemical extraction provides an early
indicator of medicinal grade (64).
One of the most effective methods for determining
the components of volatile matter, including alcohols, esters,
acids and long-chain hydrocarbons, is GC-MS. Considering the
molecular formula, peak area and retention time, it was established
that the phytochemical substances were what they claimed to be
(65). The GC-MS results in the
present study revealed the presence of pharmaceutically important
phytoconstituents. GC-MS in conjunction with methanolic and aqueous
extraction has been extensively utilized to discover phytochemicals
of clinical significance (66,67).
Tetradecanoic acid can be used as a lubricant,
hypercholesterolemic, anticancer, antioxidant and cosmetic.
Hexadecanoic acid functions as a pesticide, flavouring agent,
5-alpha-reductase inhibitor, lubricant, haemolytic,
antifibrinolytic, nematicide and anti-alopecic. Octadecanoic acid,
or stearic acid, is used in the cosmetics, flavour and perfumery
industries (68). Heptacosane,
heptadecane, octacosane and pentacosane have all been shown to have
antioxidant activity (69-71).
Undecanal and N-tetracosanol have potential antiviral properties
(72). The compound
1-methylene-2b-hydroxymethyl-3,3-dimethyl-4b-(3-methylbut-2-enyl)-cyclohexane
wasthe first hit in the methanolic and chloroform extract of S.
littoreus.
As there were no violations in Lipinski's rule of 5,
the compound was taken as a lead compound for further analysis.
Additionally, the binding affinity between the selected target and
the compound indicated the lowest energy binding of -4.86 kcal/mol,
with two hydrogen bonds formed between residues LYS444 and ASN448,
respectively. Furthermore, the stability of the complex was
evaluated by the RMSD (Fig. 8A);
it was inferred that complex became equilibrated around 13 nsec and
it fluctuates around its average value. According to the
protein-ligand contact analysis (Fig.
8B), residues interactions with the SL-MHDC were meticulously
monitored throughout the simulation. Protein-ligand contacts are
stratified into hydrogen bonding, hydrophobic interactions, ionic
linkages and water-mediated bridges. A total of 35 residues were
engaged with SL-MHDC among which His339, Asn343, Ala344, Thr345,
Asn448, Asn450, Ala484, Arg498, and Arg509 were identified as
critical constituents of the XBB.1 variant. Notably, Ala348,
Leu441, Tyr451 and Tyr501 manifested with hydrophobic contacts,
while His339, Asn343, Ala344, Thr345, Thr346, Lys440, Lys444,
Ser446, Asn448, Asn450, Ala484, Arg498, Thr500 and Arg509 exhibited
hydrogen bonding. Particularly, Asn448 demonstrated persistent
hydrogen bonding throughout the simulation which also corroborating
interactional role during molecular docking (Figs. 6 and 7). Furthermore, Asn343, Ala344, Thr345,
Asn448, Arg498 and Arg509 were implicated in hydrogen bonding with
water bridge formation, which underscores the multifaceted
capabilities of ligand anchorage. This suggests that the compound
could effectively inhibit the Spike glycoprotein. Furthermore, the
presence of all these essential components in S. Littoreus
enhances their potential therapeutic value in combating
life-threatening diseases such as COVID-19.
In conclusion, in the present study, an innovative
in silico molecular docking approach was utilized to
pinpoint a putative compound for combating severe acute respiratory
syndrome caused by coronavirus. The analysis identified the
bioactive
compound1-methylene-2b-hydroxymethyl-3,3-dimethyl-4b-(3-methylbut-2-enyl)-cyclohexane,
from the leaf extracts of coastal grass as a potential inhibitor
against the SARS-CoV-2 XBB.1 spike glycoprotein (PDB ID: 8IOV).
Furthermore, in order to validate the docking results and to shed
light on to the stability of protein-ligand interaction, MDS were
conducted. Through this process, the binding potential of the
complex under dynamic physiological conditions and its structural
stability were demonstrated. These findings indicate a possible
interaction that necessitates further analysis to determine the
therapeutic potential of the compound. Additionally, the present
study focused on the test compounds rather than known inhibitors,
such as remdesivir and nirmatrelvir; thus, such comparisons need to
be included in future studies. Moreover, further investigations of
the influence of the compound on the spike-angiotensin-converting
enzyme 2 interface could provide a more detailed understanding of
its antiviral potential. Nevertheless, further research on its
in vitro and in vivo efficacy against SARS.CoV-2 is
imperative for possible clinical translation and patient
well-being.
Supplementary Material
2D structure and biological
applications of phytocompounds of Spinifex littoreus.
Acknowledgements
The authors are very much grateful to the Rapinat
Herbarium and Centre for Molecular Systematics, St. Joseph's
College (Autonomous), Tiruchirappalli, Tamil Nadu, India for their
support in authenticating the experimental plant.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JV, INPA and SS conceived and designed the study.
JV, INPA, JSM, SKRN, VCS and SS surveyed the scientific literature.
JV, INPA, JSM and SS analysed data and wrote the draft manuscript.
JV, INPA, SKRN, VCS and SS interpreted the data and reviewed the
manuscript. INPA and SS revised the manuscript. All authors have
read and approved the final manuscript. INPA and SS confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Morens DM and Fauci AS: Emerging pandemic
diseases: How we got to COVID-19. Cell. 182:1077–1092.
2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang W, Xu Y, Gao R, Lu R, Han K, Wu G and
Tan W: Detection of SARS-CoV-2 in different types of clinical
specimens. JAMA. 323:1843–1844. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Zaki AM, van Boheemen S, Bestebroer TM,
Osterhaus AD and Fouchier RA: Isolation of a novel coronavirus from
a man with pneumonia in Saudi Arabia. N Engl J Med. 367:1814–1820.
2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zhu N, Zhang D, Wang W, Li X, Yang B, Song
J, Zhao X, Huang B, Shi W, Lu R, et al: A novel coronavirus from
patients with Pneumonia in China, 2019. N Engl J Med. 382:727–733.
2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Pilia E, Belletti A, Fresilli S, Lee TC,
Zangrillo A, Finco G and Landoni G: full anticoagulation. The
effect of heparin full-dose anticoagulation on survival of
hospitalized, non-critically Ill COVID-19 patients: A meta-analysis
of high quality studies. Lung. 201:135–147. 2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mehta OP, Bhandari P, Raut A, Kacimi SEO
and Huy NT: Coronavirus disease (COVID-19): Comprehensive review of
clinical presentation. Front Public Health.
8(582932)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Mustafa M, Abbas K, Ahmad R, Ahmad W,
Tantry IQ, Islam S, Moinuddin Alam M, Hassan M, Usmani N and Habib
S: Unmasking vulnerabilities in the age of COVID-19 (Review). World
Acad Sci J. 7(2)2025.
|
|
8
|
Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu
G, Lee PW and Tang Y: admet SAR: A comprehensive source and free
tool for assessment of chemical ADMET properties. J Chem Inf Model.
52:3099–3105. 2012.
|
|
9
|
Low KO, Md Iqbal N, Ahmad A and Johari N:
Detection of SARS-CoV-2 Delta, Omicron and XBB variant using
colorimetric reverse-transcription loop-mediated isothermal
amplification and specific primers. World Acad Sci J.
7(50)2025.
|
|
10
|
Hoffmann M, Kleine-Weber H, Schroeder S,
Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH,
Nitsche A, et al: SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2
and is blocked by a clinically proven protease inhibitor. Cell.
181:271–280.e8. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Selvavinayagam ST, Karishma SJ, Hemashree
K, Yong YK, Suvaithenamudhan S, Rajeshkumar M, Aswathy B, Kalaivani
V, Priyanka J, Kumaresan A, et al: Clinical characteristics and
novel mutations of omicron subvariant XBB in Tamil Nadu, India - a
cohort study. Lancet Reg Health Southeast Asia.
19(100272)2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Malik YS, Sircar S, Bhat S, Sharun K,
Dhama K, Dadar M, Tiwari R and Chaicumpa W: Emerging novel
coronavirus (2019-nCoV)-current scenario, evolutionary perspective
based on genome analysis and recent developments. Vet Q. 40:68–76.
2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zumla A, Chan JF, Azhar EI, Hui DS and
Yuen KY: Coronaviruses-drug discovery and therapeutic options. Nat
Rev Drug Discov. 15:327–347. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
De Wit E, van Doremalen N, Falzarano D and
Munster VJ: SARS and MERS: Recent insights into emerging
coronaviruses. Nat Rev Microbiol. 14:523–534. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Anand K, Ziebuhr J, Wadhwani P, Mesters JR
and Hilgenfeld R: Coronavirus main proteinase (3CLpro) structure:
Basis for design of anti-SARS drugs. Science. 300:1763–1767.
2003.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Needle D, Lountos GT and Waugh DS:
Structures of the Middle East respiratory syndrome coronavirus
3C-like protease reveal insights into substrate specificity. Acta
Crystallogr D Biol Crystallogr. 71:1102–1111. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Xu J, Zhao S, Teng T, Abdalla AE, Zhu W,
Xie L, Wang Y and Guo X: Systematic comparison of two
animal-to-human transmitted human coronaviruses: SARS-CoV-2 and
SARS-CoV. Viruses. 12(244)2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Talluri S: Computational protein design of
bacteriocins based on structural scaffold of aureocin A53. Int J.
Bioinformatics Research and Applications. 15:129–143. 2019.
|
|
19
|
Talluri S: Molecular docking and virtual
screening based prediction of drugs for COVID-19. Comb Chem High
Throughput Screen. 24:716–728. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Jalota K, Sharma V, Agarwal C and Jindal
S: Eco-friendly approaches to phytochemical production: Elicitation
and beyond. Nat Prod Bioprospect. 14(5)2024.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang YM, Ran XK, Riaz M, Yu M, Cai Q, Dou
DQ, Metwaly AM, Kang TG and Cai DC: Chemical constituents of stems
and leaves of Tagetespatula L. and its fingerprint. Molecules.
24(3911)2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Altemimi A, Lakhssassi N, Baharlouei A,
Watson DG and Lightfoot DA: Phytochemicals: Extraction, isolation,
and identification of bioactive compounds from plant extracts.
Plants (Basel). 6(42)2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Vedhamani J, Ajithkumar IP and Jemima EA:
Exploring the in vitro antimicrobial, antioxidant, and anticancer
potentials of Spinifex littoreus Burm f. Merr. against human
cervical cancer. Plant Sci Today. 11:242–251. 2024.
|
|
24
|
Connor HE: Breeding systems in
Indomalesian Spinifex (Paniceae: Gramineae). BLUMEA. 41:445–454.
1996.
|
|
25
|
Sen UK and Bhakat RK: Ethnobotanical study
on sand-dune based medicinal plants and traditional therapies in
coastal Purba Medinipur District, West Bengal, India. Eur J Med
Plants. 26:1–19. 2019.
|
|
26
|
Neamsuvan O, Singdam P, Yingcharoen K and
Sengnon N: A survey of medicinal plants in mangrove and beach
forests from sating Phra Peninsula, Songkhla Province, Thailand. J
Med Plants Res. 6:2421–2437. 2012.
|
|
27
|
Shakila K: Bioactive Flavonids of Spinifex
littoreus (Burm. f.) merr-investigation by elisa method. J Stress
Physiol Biochem. 16:107–112. 2020.
|
|
28
|
Thirunavukkarasu P, Ramanathan T, Ramkumar
L and Balasubramanian T: Anti microbial effect of a coastal sand
dune plant of Spinifex littoreus (Burm. f.) Merr. Curr Res J Biol
Sci. 2:283–285. 2010.
|
|
29
|
Dampalla CS, Kim Y, Bickmeier N,
Rathnayake AD, Nguyen HN, Zheng J, Kashipathy MM, Baird MA,
Battaile KP, Lovell S, et al: Structure-guided design of
conformationally constrained cyclohexane inhibitors of severe acute
respiratory syndrome coronavirus-2 3CL protease. J Med Chem.
64:10047–10058. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Nallaiyah VJ and Newton PAI: Antimicrobial
and larvicidal efficacy of the methanolic extract of Spinifex
littoreus (Burm F.) Merr. Biotech Res Asia. 21:1575–1582. 2024.
|
|
31
|
Redfern J, Kinninmonth M, Burdass D and
Verran J: Using soxhlet ethanol extraction to produce and test
plant material (essential oils) for their antimicrobial properties.
J Microbiol Biol Educ. 15:45–46. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Geetha TS and Geetha N: Phytochemical
screening, quantitative analysis of primary and secondary
metabolites of Cymbopogancitratus (DC) Stapf. leaves from
Kodaikanal hills, Tamilnadu. Int J Pharm Tech Res. 6:521–529.
2014.
|
|
33
|
Arya V, Thakur N and Kashyap CP:
Preliminary phytochemical analysis of the extracts of Psidium
leaves. Journal of Pharmacognosy and Phytochemistry. 1:1–5.
2012.
|
|
34
|
Mohammed J, Oba OA and Aydinlik NP:
Preliminary phytochemical screening, gc-ms, FT-IR analysis of
ethanolic extracts of Rosmarinus Officinalis, Coriandrum Sativum L.
and Mentha Spicata. Hacettepe J Biol Chem. 51:93–102. 2023.
|
|
35
|
Jose JK, Sudhakaran S, Sony J and Variyar
E: A comparative evaluation of anticancer activities of flavonoids
isolated from Mimosa pudica, Aloe vera and Phyllanthus niruri
against human breast carcinoma cell line (MCF-7) using MTT assay.
Int J Pharm Pharmaceut Sci. 6:319–322. 2014.
|
|
36
|
Ayoola GA, Coker HAB, Adesegun SA,
Adepoju-Bello AA, Obaweya K, Ezennia EC and Atangbayila TO:
Phytochemical screening and antioxidant activities of some selected
-3medicinal plants used for malaria therapy in Southwestern
Nigeria. Trop J Pharm Res. 7:1019–1024. 2008.
|
|
37
|
Yusuf AZ, Zakir A, Shemau Z, Abdullahi M
and Halima SA: Phytochemical analysis of the methanol leaves
extract of Paullinia pinnata linn. J. Pharmacognosy Phytother.
6:10–16. 2014.
|
|
38
|
Krishnaiah D, Sarbatly R and Bono A:
Phytochemical antioxidants for health and medicine: A move towards
nature. Biotechnol Mol Biol Rev. 1:097–104. 2007.
|
|
39
|
Sandosh TA, Peter MPJ and Raj JY:
Phytochemical analysis of Stylosanthesfruticosa using UV-VIS, FTIR
and GC-MS. Res J Chem Sci. 3:14–23. 2013.
|
|
40
|
Shamsa F, Monsef H, Ghamooshi R and
Verdian-Rizi M: Spectrophotometric determination of total alkaloids
in some Iranian medicinal plants. Thai J Pharm Sci. 32:17–20.
2008.
|
|
41
|
Ghorai N, Chakraborty S, Gucchait S, Saha
SK and Biswas S: Estimation of total Terpenoids concentration in
plant tissues using a monoterpene, Linalool as standard reagent.
Protocol Exchange. 1–6. 2012.
|
|
42
|
Marinova D, Ribarova F and Atanassova M:
Total phenolics and flavonoids in Bulgarian fruits and vegetables.
JU Chem Metal. 40:255–260. 2005.
|
|
43
|
Van Buren JP and Robinson WB: Formation of
complexes between protein and tannic acid. J Agric Food Chem.
17:772–777. 1969.
|
|
44
|
Taylor B: The International System of
Units (SI), 2008 Edition, Special Publication (NIST SP). National
Institute of Standards and Technology, Gaithersburg, MD, 2008.
Accessed on July 8, 2025. https://doi.org/10.6028/NIST.SP.330e2008.
|
|
45
|
Iordache A, Culea M, Gherman C and Cozar
O: Characterization of some plant extracts by GC-MS. Nucl Instrum
Methods Phys Res B. 267:338–342. 2009.
|
|
46
|
Daina A, Michielin O and Zoete V:
SwissADME: A free web tool to evaluate pharmacokinetics,
drug-likeness and medicinal chemistry friendliness of small
molecules. Sci Rep. 7(42717)2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Eberhardt J, Santos-Martins D, Tillack AF
and Forli S: AutoDock Vina 1.2.0: New docking methods, expanded
force field, and python bindings. J Chem Inf Model. 61:3891–3898.
2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Trott O and Olson AJ: AutoDock Vina:
Improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading. J Comput
Chem. 31:455–461. 2010.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Morris GM, Huey R, Lindstrom W, Sanner MF,
Belew RK, Goodsell DS and Olson AJ: AutoDock4 and AutoDockTools4:
Automated docking with selective receptor flexibility. J Comput
Chem. 30:2785–2791. 2009.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Bowers KJ, Chow E, Xu H, Dror RO, Eastwood
MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti
FD, et al: Scalable algorithms for molecular dynamics simulations
on commodity clusters'. Proceedings of the ACM/IEEE SC2006
Conference on High Performance Networking and Computing, New York,
NY, 11-17, 2006.
|
|
51
|
Ajuru MG, Williams LF and Ajuru G:
Qualitative and quantitative phytochemical screening of some plants
used in ethnomedicine in the Niger Delta region of Nigeria. J Food
Nutr Sci. 5:198–205. 2017.
|
|
52
|
Rechab SO, Ngugi CM, Maina EG, Madivoli
ES, Kareru PG, Mutembei JK, Kairigo PK, Cheruiyot K and Ruto MC:
Antioxidant activity and antimicrobial properties of
Entadaleptostachya and Prosopisjuliflora extracts. J
Med Plants Econ Dev. 2:1–8. 2018.
|
|
53
|
Chandran M, Vivek P and Kesavan D:
Phytochemical screening and anti-bacterial studies in salt marsh
plant extracts (Spinifex Littoreus (Burm. f) Merr. and
Heliotropium Curassavicum L.). Recent Trends Biotechnol Chem
Eng. 6:4307–11. 2014.
|
|
54
|
Koche D, Shirsat R and Kawale MA: An
overview of major classes of phytochemicals: their types and role
in disease prevention. Hislopia J. 9:1–11. 2016.
|
|
55
|
Shirsat R, Suradkar S and Koche D: Some
phenolic compounds from Salvia plebeia. Biosci Discov.
3:61–63. 2012.
|
|
56
|
Wink M, Schmeller T and Latz-Brüning B:
Modes of action of allelochemical alkaloids: Interaction with
neuroreceptors, DNA, and other molecular targets. J Chem Ecol.
24:1881–1937. 1998.
|
|
57
|
Langenheim JH: Higher plant terpenoids: A
phytocentric overview of their ecological roles. J Chem Ecol.
20:1223–1280. 1994.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Dudareva N, Pichersky E and Gershenzon J:
Biochemistry of plant volatiles. Plant Physiol. 135:1893–1902.
2004.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Dai J and Mumper RJ: Plant phenolics:
Extraction, analysis and their antioxidant and anticancer
properties. Molecules. 15:7313–7352. 2010.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Khatri S, Paramanya A and Ali A: Phenolic
acids and their health-promoting activity. Plant Human Health.
2:661–680. 2019.
|
|
61
|
Singh B and Sharma RA: Plant terpenes:
defense responses, phylogenetic analysis, regulation and clinical
applications. 3 Biotech. 5:129–151. 2015.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Thoppil RJ and Bishayee A: Terpenoids as
potential chemopreventive and therapeutic agents in liver cancer.
World J Hepatol. 3:228–249. 2011.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Subiono T, Sadarudin and Tavip MA:
Qualitative and quantitative phytochemicals of leaves, bark and
roots of Antiaris toxicaria Lesch, a promising natural
medicinal plant and source of pesticides. Plant Sci Today. 10:5–10.
2023.
|
|
64
|
Ouandaogo HS, Diallo S, Odari E and Kinyua
J: Phytochemical Screening and GC-MS Analysis of Methanolic and
Aqueous Extracts of Ocimum kilimandscharicum Leaves. ACS Omega.
8:47560–47572. 2023.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Konappa N, Udayashankar AC, Krishnamurthy
S, Pradeep CK, Chowdappa S and Jogaiah S: GC-MS analysis of
phytoconstituents from Amomum nilgiricum and molecular docking
interactions of bioactive serverogenin acetate with target
proteins. Sci Rep. 10(16438)2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Iikasha AMN, Bock R and Mumbengegwi DR:
Phytochemical screening and antibacterial activity of selected
medicinal plants against laboratory diarrheal bacteria strains. J
Pharmacogn Phytochem. 6:2337–2342. 2017.
|
|
67
|
Tiwari P, Kumar B, Kaur M, Kaur G and Kaur
H: Phytochemical screening and extraction: A review. Int Pharm Sci.
1:98–106. 2011.
|
|
68
|
Ponnamma SU and Manjunath K: GC-MS
Analysis of phytocomponents in the methanolic extract of
Justiciawynaadensis (nees) T. anders. Int J Pharm Bio Sci.
3:570–576. 2012.
|
|
69
|
Kim DH, Park MH, Choi YJ, Chung KW, Park
CH, Jang EJ, An HJ, Yu BP and Chung HY: Molecular study of dietary
heptadecane for the anti-inflammatory modulation of NF-kB in the
aged kidney. PLoS One. 8(e59316)2013.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Marrufo T, Nazzaro F, Mancini E, Fratianni
F, Coppola R, De Martino L, Agostinho AB and De Feo V: Chemical
composition and biological activity of the essential oil from
leaves of Moringa oleifera Lam. cultivated in Mozambique.
Molecules. 18:10989–11000. 2013.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Ma J, Xu RR, Lu Y, Ren DF and Lu J:
Composition, antiproliferative, and antioxidant activity of
cold-pressed seed oils from Paeonialactiflora and Paeoniaostii. J
Food Sci. 80:2465–2474. 2015.
|
|
72
|
Katz DH, Marcelletti JF, Khalil MH, Pope
LE and Katz LR: Antiviral activity of 1-docosanol, an inhibitor of
lipid-enveloped viruses including herpes simplex. Proc Natl Acad
Sci. 88:10825–10829. 1991.PubMed/NCBI View Article : Google Scholar
|