1. Introduction
Neurodegenerative diseases (NDDs) are the second
leading cause of mortality worldwide, constituting an increasing
public health concern (1).
Alzheimer's disease (AD) and Parkinson's disease (PD) are the two
most common NDDs, affecting 35 million and 6 million individuals
worldwide, respectively (2).
Dementia currently affects ~50 million individuals, with
projections suggesting an increase to 130 million by the year 2050.
AD is the predominant NDD, including 60-70% of all cases of
dementia (3). According to the
Alzheimer's Association, an estimated 6.7 million Americans aged
≥65 years are currently living with Alzheimer's dementia, a number
projected to nearly double to 13.8 million by 2060 in the absence
of disease-modifying treatments. AD is the sixth-leading cause of
mortality in the USA overall, and the fifth-leading cause among
those aged ≥65 years. In 2019, 121,499 deaths were attributed to
AD, and between 2000 and 2019, deaths from AD increased by
>145%, in contrast to declines in stroke, heart disease and
HIV-related mortality (4). The
World Health Organization (WHO) reports that the prevalence of PD
has multiplied over the past 25 years, affecting >8.5 million
individuals worldwide. In 2019, PD was responsible for 329,000
deaths, more than double the number of deaths that occurred in
2000, and resulted in 5.8 million disability-adjusted life years,
which represents an 81% increase since the year 2000(5). Motor neuron disease, commonly known
as amyotrophic lateral sclerosis (ALS), affects individuals
globally, with an incidence rate of ~2 per 100,000 person-years, a
prevalence of 6 to 9 per 100,000 person-years, and a lifetime risk
estimated at 1 in 350(6). As the
population increases and society ages, a greater number of
individuals are attaining ages associated with a high prevalence of
neurological illnesses. The etiology of NDD is multifaceted and
intricate. Progress in genomic technology has revealed mutations
linked to disease (7). However, in
the case of NDDs, such as AD, PD and ALS, a considerable number of
sporadic and even familial cases have uncleared genetic origins.
Furthermore, not all identified mutations are fully penetrant or
result in disease. Instead, a combination of genetic risk factors
may affect the vulnerability of an individual to developing NDDs
(8). Single-nucleotide
polymorphism (SNP)-based heritability estimates range from ~16
to36% for PD, 8 to 61% for ALS, and 38 to 66% for AD. These
estimations nonetheless indicate that non-genetic variables have a
significant effect (9). As a
result, it is generally acknowledged that environmental exposures,
also known as the exposome, play a major role in the onset and
course of NDD (10).
Generally, NDDs are gradual, irreversible and linked
to functional loss. NDDs manifest physiologically as demyelination,
dendritic loss and neuronal death (11). A slow and cumulative loss of
cognitive abilities (dementia) and movement abilities (ataxia)
results from the degeneration of neural structures, which may lead
to mental impairment, functional loss and debilitation. Despite
being more common among the elderly, NDDs may affect individuals of
any age (12). The early
identification of NDDs is crucial for facilitating rapid therapies
and controlling these progressive disorders efficiently. There is
increasing interest in identifying early diagnostic tools and novel
treatment strategies for NDDs (13). Traditional biomarkers, including
protein biomarkers, exosomes and microRNAs (miRNAs), exhibit
promise in detecting neural dysfunction prior to the appearance of
clinical symptoms (14-16).
Researchers investigate these biomarkers, combined with other
laboratory and biochemical indicators, for their potential in early
diagnosis and evaluation of disease development (17).
The requirement for biological material and
inpatient treatment limits the use of analog biomarkers for
identifying NDDs (18). Although
these challenges exist, the progress in bioassays and the
identification of biological indicators in blood, urine, tissue,
plasma and serum indicates the potential for overcoming these
limitations. However, the complete verification of the therapeutic
efficacy of these biomarkers remains elusive (19). Further research is warranted to
standardize these findings and to evaluate their effectiveness in
identifying the early stages of the illness. A search for an
optimal biomarker for NDDs continues to guarantee a reliable and
accurate diagnosis in the earliest clinical phases. Conversely,
digital technologies that provide objective, high-frequency data
are being investigated to solve the existing subjective assessments
of NDDs (20).
In recent years, artificial intelligence (AI) has
emerged as a transformative tool in health care (21). Machine learning (ML), a subset of
AI, has been increasingly favored over other deep learning or
traditional statistical methods due to its ability to learn complex
patterns from high-dimensional data without extensive feature
engineering. ML has demonstrated significant potential in enhancing
the early diagnosis, disease monitoring and predictive models of
NDDs (22). ML algorithms can
analyze complex, high-dimensional biological datasets to identify
patterns associated with disease onset and progression. By
integrating neuroimaging, genetic, molecular, and behavioral data,
ML models also improve diagnostic accuracy and facilitate
personalized treatment approaches (23). Additionally, wearable sensors and
remote monitoring systems leverage ML to track disease symptoms in
real-time, provoiding a non-invasive and scalable approach to early
diagnosis (24).
The present review discusses the use of ML in the
early diagnosis of NDDs, emphasizing key areas, such as biomarker
discovery, genetic analysis, neuroimaging and cognitive assessment.
It also explains the advantages of ML over traditional methods in
capturing complex associations and improving predictive accuracy.
The essential ML techniques, feature selection strategies and data
preprocessing methods relevant to biomedical fields are emphasized.
Additionally, the improved diagnostic accuracy and the ability to
address challenges related to data consistency and privacy using
combined multimodal data sources are discussed. By reviewing the
latest advances in ML-based NDD research, the present review aimed
to provide insight into the role of AI in the early detection and
management of NDDs.
2. Pathophysiology of neurodegenerative
diseases: Overview
AD is marked by a slow and advancing
neurodegeneration due to the death of neuronal cells, significantly
affecting cognitive abilities. This neurodegenerative process
usually begins in the entorhinal cortex of the hippocampus, an area
vital for memory processing (25).
The formation of neurofibrillary tangles is composed of
phosphorylated tau protein, strongly associated with cognitive
impairment, compared to the amyloid plaques. Neurofibrillary
tangles and amyloid plaques are essential for the neuropathological
diagnosis of AD (11).
Neurofibrillary tangles first develop in the entorhinal cortex and
hippocampus before moving to the isocortex, which is how AD
proceeds stereotypically. This progression is divided into phases
that correspond to the clinical presentation of dementia and
indicate the growing severity of the disease (26).
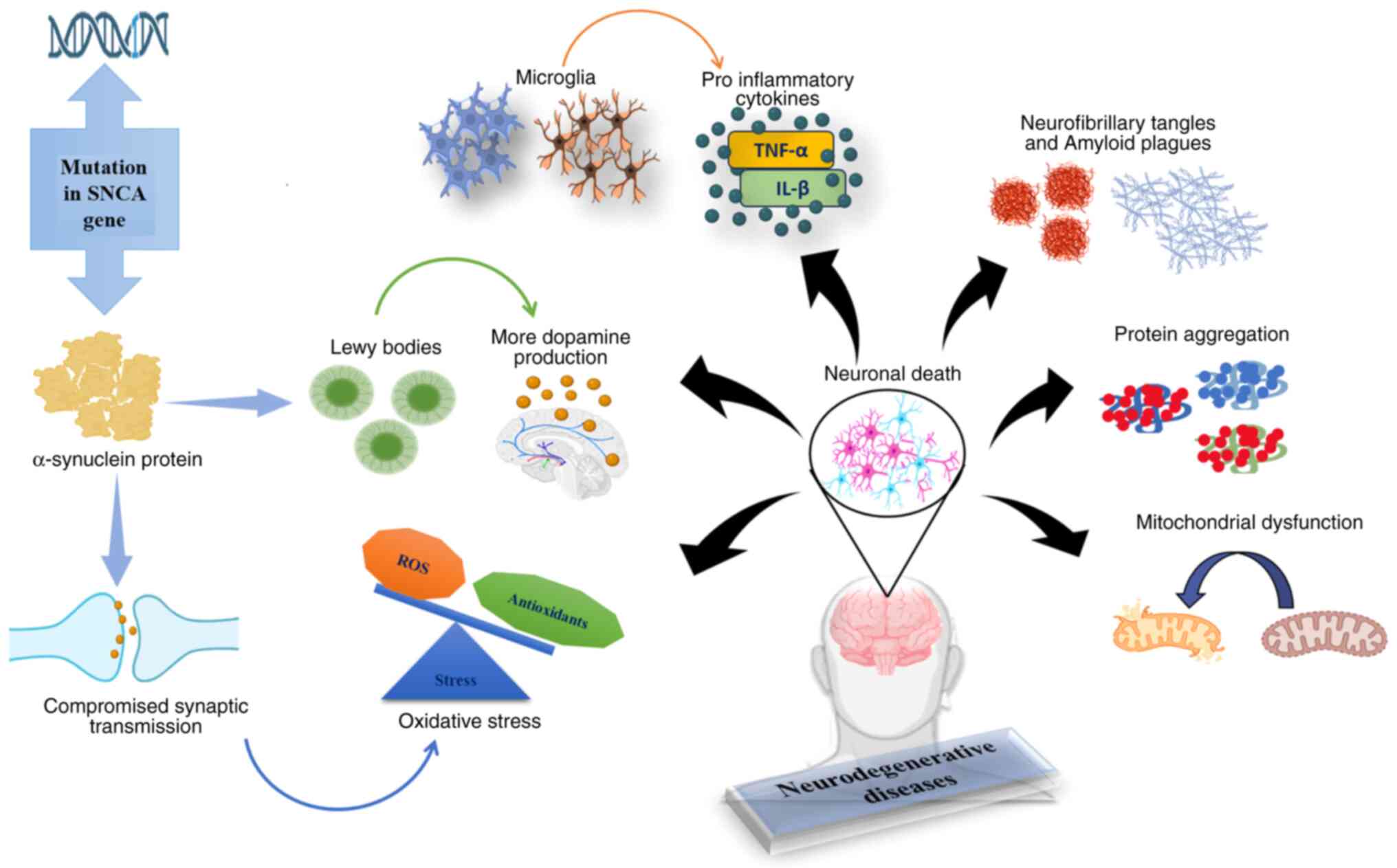
The degeneration of dopaminergic neurons in the
substantia nigra is the main characteristic of PD, a complex
neurodegenerative illness that causes motor symptoms, such as
bradykinesia, stiffness and tremors (27). The first known gene linked to PD is
the synuclein alpha (SNCA) gene, which codes for α-synuclein. PD
with autosomal-dominant inheritance patterns showed that an early
onset may be due to mutations in SNCA (28). The α-synuclein protein, a key
component of Lewy bodies in the brains of patients with PD,
destroys dopaminergic neurons. More dopamine may worsen
dopaminergic neuron degeneration. While α-synuclein is advantageous
for dopaminergic neurons, its overexpression may destroy them when
paired with dopamine (29). SNCA
aggregation interferes with cellular function, resulting in
compromised synaptic transmission and increased oxidative stress,
which exacerbates neuronal cell death (30). Mutations in genes, such as SNCA,
leucine-rich repeat kinase 2, PTEN-induced putative kinase 1,
Parkin RBR E3 ubiquitin protein ligase, protein deglycase and
glucosylceramidase beta 1 cause ~10-15% of cases of familial PD
(31). Neuroinflammation
significantly contributes to the progression of PD, with microglial
activation noted in post-mortem studies of affected individuals.
This dysfunctional immune response can worsen neuronal stress and
death, as microglia may release pro-inflammatory cytokines that aid
in neurodegeneration. Increased levels of inflammatory markers,
such as IL-1β and TNF-α have been linked to the severity and
progression of the disease (Fig.
1) (32).
Several pathways that interfere with the
pathogenesis of ALS, such as mitochondrial dysfunction,
neuroinflammation, oxidative stress, axonal damage, protein
aggregation and excitotoxicity, have been suggested to play a role
(33). TAR DNA-binding protein 43
(TDP-43) is the primary component of inclusions observed in >95%
of patients with ALS. This RNA- and DNA-binding protein is critical
for key processes, including transcription, splicing and RNA
transport (34). TDP-43 mostly
exists in the nucleus; however, in ALS, it may be mislocalized to
the cytoplasm, resulting in nuclear depletion and protein
aggregation (35). Protein clumps
impair cellular protein homeostasis, eliciting stress. Molecular
chaperones facilitate the refolding of misfolded proteins, whereas
excess aggregates are eliminated by the ubiquitin-proteasome system
or lysosomal autophagy (36). The
buildup of misfolded superoxide dismutase 1 (mSOD1) in the
mitochondria adversely affects spinal motor neurons and skeletal
muscles, resulting in the release of aberrant ATP, elevated
reactive oxygen species production and apoptosis (37). A dominant missense mutation in the
SOD1 gene, which is a major cause of ALS, results in the creation
of insoluble, ubiquitin-positive inclusion bodies in motor neurons.
While chaperones play a role in protein folding, SOD1 aggregates
capture heat shock proteins, leading to endoplasmic reticulum
stress and the accumulation of toxic substances (38). Autophagy mitigates mutant SOD1
toxicity, yet it often proves inadequate, resulting in the
accumulation of aggregates and higher cell mortality rates
(39). Genetic mutations are key
factors in the pathophysiology of ALS. Of note, >20 genes have
been shown to be associated with ALS, with the most prevalent
mutations identified in the C9 or f72, TDP-43, ubiquitin-2, VCP,
TANK-binding kinase 1, SOD1, TARDBP and FUS genes (40).
3. Diagnostic challenges and limitations of
neurodegenerative disease
The diagnosis of NDDs is difficult since symptoms
often develop gradually. Numerous NDDs have overlapping symptoms,
potentially resulting in misdiagnosis (41). Furthermore, the dependence on
clinical criteria implies that a number of pathological alterations
may remain undetected until significant brain damage has occurred.
This delay in diagnosis may lead to lost possibilities for early
intervention (42). Discrimination
and misinformation about cognitive decline could prevent
individuals from seeking therapy, delaying early identification
(43). Current diagnostic methods
depend on clinical assessments and standard neuropsychological
testing, which may be inadequate for detecting early underlying
pathologies in NDDs. Blood biomarkers, such as neurofilament light
chain, phosphorylated tau, amyloid-β and total tau, have been
proposed to assist in diagnosis (44). A notable issue is the fluctuation
in biomarker levels, which are affected by variables, such as age,
sex and comorbidities, potentially confusing interpretations. While
several biomarkers have impressive sensitivity, their specificity
for NDDs is often inadequate, resulting in possible false positives
(45). Cerebrospinal fluid (CSF)
biomarkers serve as direct indicators of the central nervous
system, offering insights into pathological processes (46). Lumbar puncture for CSF collection
is invasive and may be poorly tolerated. Not all healthcare
environments provide it, and it may be costly. The conditions of
sample processing and analysis may also influence diagnostic
accuracy (47). Imaging
biomarkers, such as diffusion imaging, magnetic resonance imaging
(MRI) and positron emission tomography (PET), allow for the
visualization of brain changes. These approaches detect
neurodegenerative processes before symptoms appear. Magnetic
resonance elastography examines tissue properties to enhance early
diagnosis (48). Advanced imaging
techniques, such as PET scans, may be costly and less accessible.
Certain procedures expose patients to radiation, raising safety
concerns. Furthermore, outcomes may differ based on patient
attributes and circumstances, resulting in possible
misinterpretations (49). Genetic
biomarkers, such as mutations, SNPs and miRNAs, provide insight
into disease causes and susceptibility. Identifying effective
biomarkers for NDDs is challenging due to the intricate connections
between hereditary and environmental variables (50). Furthermore, genetic markers may
vary across populations, affecting their effectiveness and
therapeutic significance. Initial genetic testing prompts ethical
issues, including privacy, potential discrimination, and the
psychological impact on those at risk (51).
4. Fundamentals of machine learning models
and techniques
Recent research highlights the potential of emerging
technologies to enhance diagnostics. There is growing interest in
the use of ML to analyze diagnostic data effectively (52). ML will be crucial in developing
learning healthcare systems that integrate various data sources
with complex algorithms. This will provide continuous,
data-informed insight to enhance biomedical research, public health
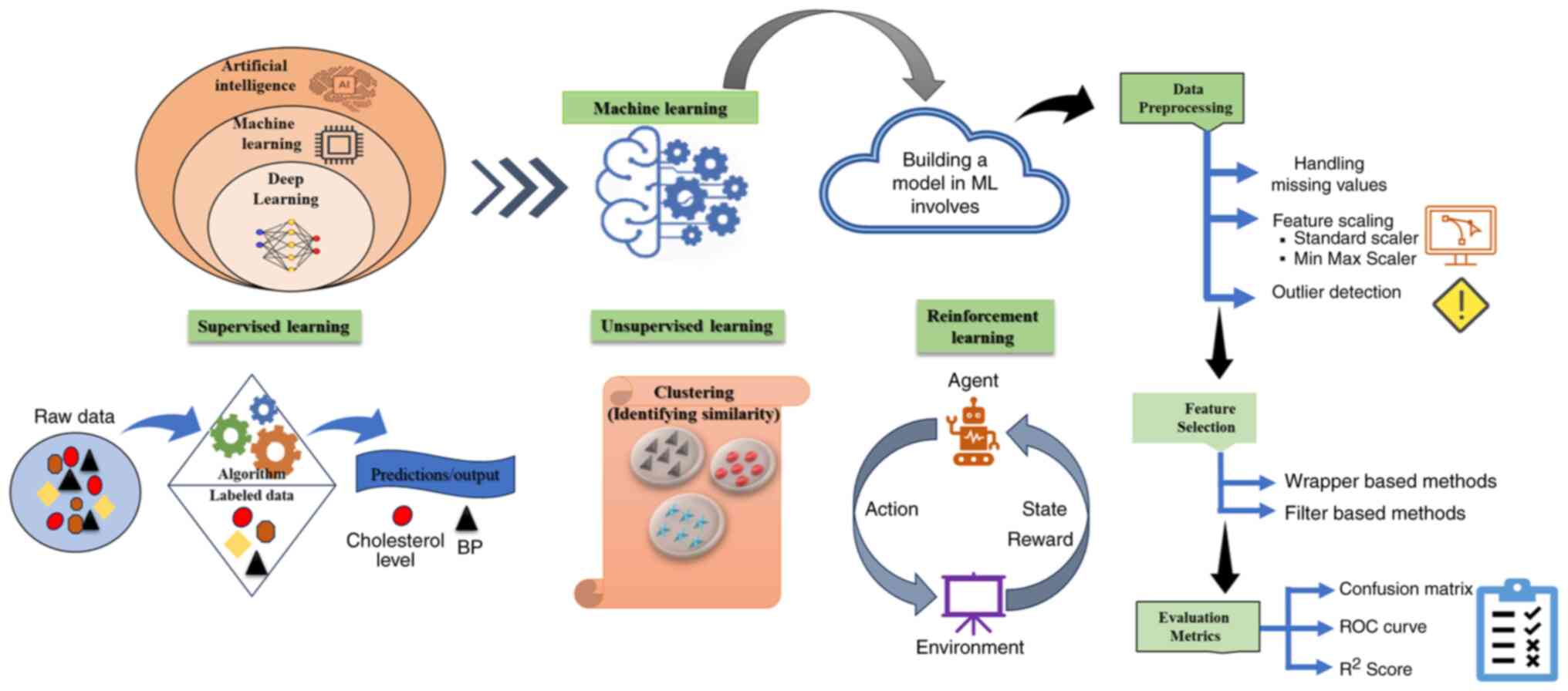
and the quality of healthcare (53). The majority of ML methods can be
grouped into three categories, with supervised ML being the first.
This method trains a model on input characteristics with known
results. In medicine, it may link height, weight and smoking status
to the 5-year diabetes risk. After training, the system will
predict fresh data outcomes with discrete or continuous scores
(54). Unlike supervised learning,
unsupervised learning operates without a predetermined outcome.
This strategy involves algorithms independently detecting patterns
without human involvement. Consequently, unsupervised algorithms
are investigative and intended to reveal hidden patterns or
clusters within datasets (55).
Reinforcement learning entails a system engaging with its
environment, promoting favorable actions and discouraging
unfavorable actions. These approaches are used in a number of
medical operations, including disease diagnosis (56). Deep learning (DL), a branch of ML,
is characterized by the use of several layers, each signifying
different levels of abstraction. In this framework, each layer
evaluates the information obtained from the previous layer and
transmits the results to the subsequent layer (57).
Selection of features, data
preprocessing and assessment of matrices for biomedical
applications
Feature selection is a common method in ML to reduce
dimensionality by identifying a subset of relevant features based
on established criteria (58).
Reducing noise and removing non-informative features are essential
to tackle the ‘curse of dimensionality’, which arises when the
number of features exceeds the number of observations (59). Feature selection allows for the
identification of high-risk genes associated with cancer. As
microarray gene expression data are high-dimensional, it is
essential to perform critical feature extraction techniques,
including the t-test, Wilcoxon sign rank sum test test, random
forest, Boruta and LASSO, among others (60). Feature selection techniques may be
classified as filters, embedding methods and wrappers according to
their association with the learning algorithm (61). Data preprocessing entails the
preparation of raw data to render it appropriate for ML analysis.
This phase is essential in biological applications where data may
often be noisy or partial. Data preparation techniques include
normalization, management of missing values and outlier
identification (62). Data
preprocessing includes data cleansing and feature engineering. Data
cleaning removes duplicate, incorrect, irrelevant and missing data.
This requires a detailed knowledge of the data, its collection
context, and the application of the model in the environment.
Clinicians and data scientists from different fields need to
collaborate to clean data (63).
Feature engineering employs a range of statistical methods to
transform data into a format that ML algorithms can use more
effectively. Typical procedures in feature engineering comprise
transformation, dimensionality reduction, data type conversion,
data normalization and feature selection, all aimed at fulfilling
the requirements of ML algorithms (64). ML performance measures are
essential for assessing diagnostic models in healthcare. Standard
metrics include classification and regression measures, which need
to be analyzed in light of class imbalance, prevalence and
cost-benefit trade-offs (65).
Effective validation methods, including cross-validation and
distinct test sets, are crucial to prevent data leakage and provide
impartial estimates. In binary classification tasks, measurements
such as sensitivity, specificity, and the area under the ROC curve
are often used (Fig. 2) (66). Researchers and clinicians must
comprehend these parameters to evaluate ML studies objectively and
determine how they could affect patient treatment (67). When assessing ML models, it is
crucial to consider the sample size as well as the issues of
overfitting and underfitting. Researchers have created tools to
compute and visualize many performance indicators, thereby aiding
in the comparison and understanding of ML models (68).
5. Machine learning in biomarker discovery
and analysis
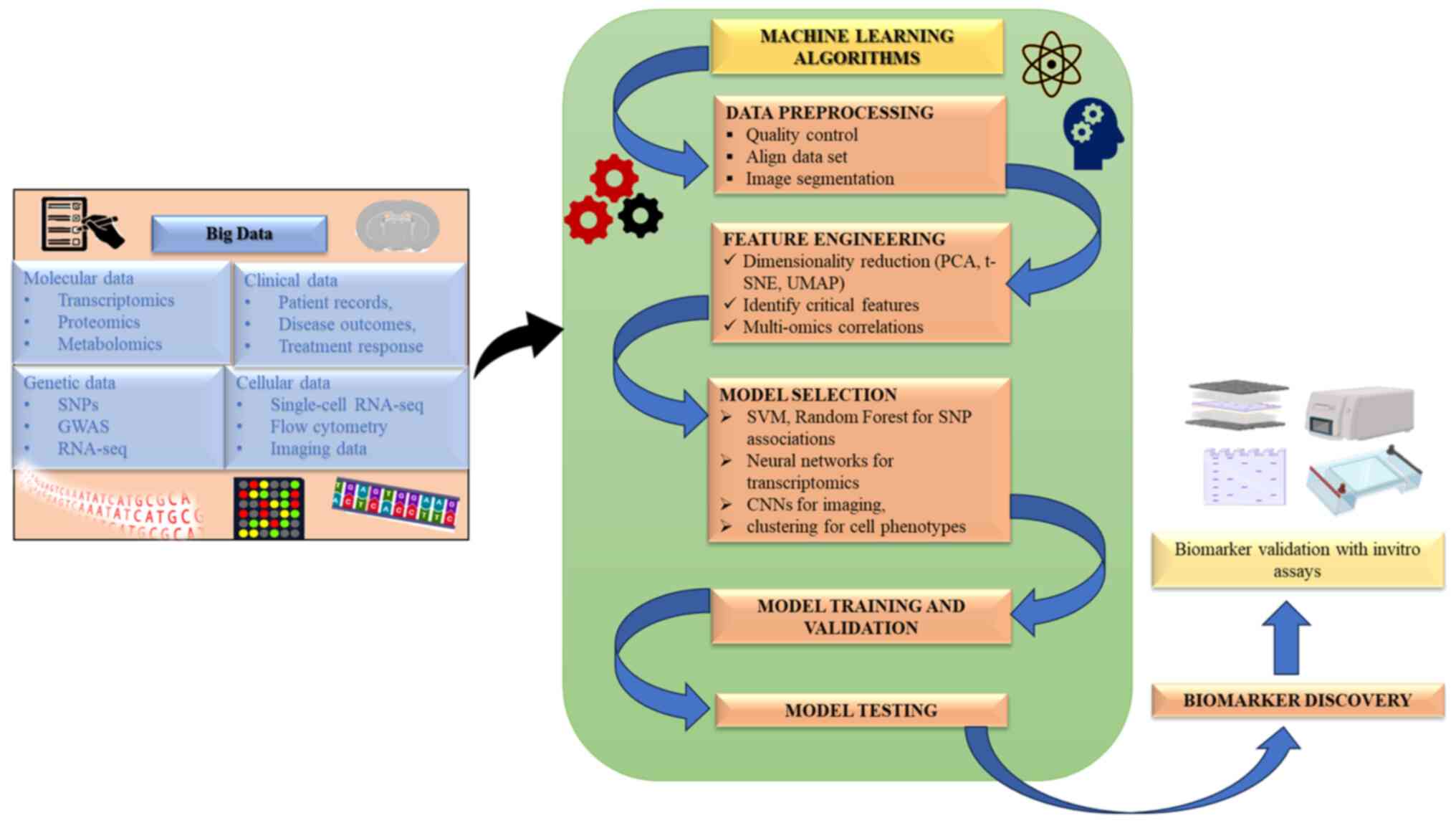
ML is an effective tool for the diagnosis of various
diseases and analyzing data. ML approaches, such as DL and support
vector machines (SVMs), examine intricate data from genomics,
proteomics and imaging to identify molecular signatures and
biomarkers (Fig. 3) (69). These approaches provide advantages
over traditional statistical techniques in handling large,
high-dimensional datasets. However, challenges such as data privacy
and overfitting persist. Explainable ML models could mitigate these
issues by providing mechanistic insights into predictions, thus
improving the robustness and reliability of biomarker
discovery.
ML used in genetic biomarkers
Recent research has investigated the use of ML to
detect genetic biomarkers for the diagnosis of NDDs. Broadly, these
studies fall into three categories, such as: i) Large-scale
genomic/transcriptomic analyses; ii) miRNA and blood transcript
investigations; and iii) DNA methylation or SNP-based
approaches.
In a large-scale genomic study, Lam et al
analyzed clinical and genetic data from the UK Biobank to create
models that predict motor neuron disease, AD, PD and myasthenia
gravis, achieving 88.3% accuracy. They discovered common genetic
risk loci shared across NDDs, although reliance on a single biobank
may limit generalizability (70).
Similarly, transcriptomic and clinical/laboratory data integrating
with ML has been used to detect early comorbidities and cognitive
impairment with improved accuracy (71). For miRNA and blood transcript-based
biomarkers, Li et al (72)
applied a feature that differentiates normal and neurodegenerative
disease subgroups using computational analysis. Boruta's feature
selection removed irrelevant features, although mRMR and MCFS
prioritized the remaining ones. The appropriate miRNA biomarker set
was established, and the correlation between candidate features and
NDDs was confirmed. Other studies using random forest classifiers
on blood transcript data have reported high sensitivity and
specificity in distinguishing AD, PD and ALS from controls,
although small sample sizes raise concerns about model robustness
(73). In the area of epigenetic
and SNP-based approaches, Ren et al (74) applied random forest feature
selection and ROC diagnostic analysis of genes exhibiting varied
methylation patterns to identify optimal gene biomarkers for AD.
Differential methylation was identified in eight genes: STAMBPL1,
ANKRD34B, FAM82A1, CDKN1C, NOG, CORO2 B and TXNIP. MYNN was the
optimal biomarker for AD (74).
Although promising, such findings require replication in
independent cohorts. Furthermore, ADNI-1 and WGS datasets have been
leveraged to evaluate millions of SNPs, with ML algorithms (SMO,
NB, TAN and K2) achieving exceptionally high accuracies (98-99.75%)
using 500 SNPs. However, these near-perfect results raise the
possibility of overfitting, emphasizing the need for validation on
external datasets (75). Deep
learning approaches have also demonstrated considerable potential.
Convolutional neural network (CNN) models applied to blood-based
biomarkers for AD and PD have yielded strong predictive
performance, with 81% accuracy and ROC AUC values reaching up to
0.889 and 0.743, respectively (76). Research has demonstrated CNNs
applied to microarray data, attaining 95-96% accuracy following
dimensionality reduction (PCA and SVD) and data augmentation to
mitigate overfitting. While these findings are encouraging,
heterogeneity in datasets and limited real-world testing remain as
major challenges (77). Overall,
ML has proven to be highly effective in identifying diverse genetic
and molecular biomarkers for NDDs, providing strong predictive
accuracy and the potential to enhance early diagnosis. However,
numerous studies are constrained by small sample sizes, reliance on
single datasets and risks of overfitting. To enable clinical
translation, future research is warranted to emphasize validation
across larger, more diverse and independent cohorts. A summary of
ML approaches applied to genetic and epigenetic biomarkers in NDDs
is depicted in Table I (78-87).
 | Table IMachine learning approaches for
identifying genetic and epigenetic biomarkers in NDDs. |
Table I
Machine learning approaches for
identifying genetic and epigenetic biomarkers in NDDs.
| First author, year
of publication | Biomarker type | Dataset | ML Method | Accuracy | Key findings | (Refs.) |
|---|
| Wang, 2025 | Purine metabolism
genes (PMGs) | GSE6613,
GSE7621 | Lasso regression,
SVM-RFE | AUC=0.769 with a
low error rate of 0.231 | The diagnostic
capacity of these nine PMGs in distinguishing PD was shown to be
significant. | (78) |
| Huang, 2021 | Epigenetic
bio-markers (brain CpG methylation sites) | Six AD-related
brain datasets (cohorts) | EWASplus
(supervised machine learning) | ROC/AUC=
0.831/0.962 | Predicted hundreds
of novel brain CpGs linked to AD; some loci were tested in the lab;
found genes that are rich in kinases and interact with known AD
genes; EWAS coverage goes beyond array-based approaches. | (79) |
| Alamro, 2023 | Gene expression
biomarkers (hub genes, feature-selected genes, miRNAs, TF JUN) | datasets of brain
tissue in the Gene Expression Omnibus (GEO) database (GSE5281,
GSE48350, and GSE1297) | Machine learning
and deep learning (LASSO, Ridge; hub gene ranking: Degree, MNC,
MCC, BC, Closeness, Stress Centrality) | AUC=0.979 (for 5
genes from LASSO and Ridge) | Identified five
genes that accurately differentiate Alzheimer's disease from
healthy controls; 70% of the hub genes that are turned on are known
to be targets for AD; 6 miRNAs and TF JUN are connected to hub
genes; Overlapping hub genes limit the search for new AD
targets. | (80) |
| Madar, 2021 | Differentially
expressed genes (CNPY3, GPR84, HIST1H2AB, HIST1H2AE, IFNAR1, LMO3,
MYO18A, N4BP2L1, PML, SLC4A4, ST8SIA4, TLE1) | HG-U133_Plus_2
platform GDS2795 GDS4136 | SMO/SVM, Logit
Boost, other classifiers | Achieved 85 to 90%
accuracy | Identified 13
significant DEGs expressed in brain tissue; co-expression networks
validated; J48 emerged as the best classifier for distinguishing AD
vs. controls | (81) |
| Lin, 2022 | Blood-based gene
biomarkers (29 genes, 31 probes) | ADNI database | Random Forest with
feature selection | AUC=0.841
(cross-validation), 0.775 (test set); 97% concordance for
high-score patients | Found gene
biomarkers that may help predict stable MCI patients; a
low-invasive, cost-effective way to screen people; and a possible
first-tier diagnostic tool for precision medicine. | (82) |
| Sharma, 2021 | Genetic biomarkers
(CORO1C, SLC25A46, RAE1,ANKIB1, CRLF3, PDYN, and non-coding RNAs
AK057435, BC037880) | Microarray datasets
from four brain regions: Prefrontal cortex, Middle temporal gyrus,
Hippocampus, Entorhinal cortex | Ensemble of Random
Forest and LASSO (feature selection and classification) | 99% average
accuracy (5-fold cross-validation) | Identified unique
and clinically important genetic indicators for Alzheimer's disease
across several brain areas, using uncharacterized non-coding RNAs
as possible differentiators. | (83) |
| Augustine,
2022 | Blood-based gene
biomarkers (DEGs from microarray) | Three independent
PD microarray datasets (blood samples); independent test:
GSE72267 | Two-layer embedded
wrapper feature selection and classification with 9 ML models,
including SVM-R, DNN | AUC=0.821 (SVM-R),
0.82 (DNN) on the independent dataset | Found a strong
blood-based gene signature that can be used to detect early signs
of PD; verified its reliability by comparing it to existing
signatures and combining several datasets. better ability to
forecast. | (84) |
| Sekaran, 2023 | Gene expression
biomarkers (ORAI2, STIM1, TRPC3, TPI1 + other candidate genes) | GEO database
(Accession: GSE36980). AD blood samples from frontal, hippocampal,
and temporal regions vs. non-AD controls. | Supervised ML
classifiers (Naive Bayes with 5-fold cross-validation, plus other
ML algorithms; model interpretation with explainable AI) | 100% accuracy Naive
Bayes, 5-fold CV | Identified 34
(frontal), 60 (hippocampal), and 28 (temporal) genes as biomarkers.
ORAI2 is present in all areas. Pathway analysis connected ORAI2 to
STIM1 and TRPC3. Hub genes: TPI1, STIM1, TRPC3 → possible
involvement in the development of AD. ML and AI together may help
find medicinal targets. | (85) |
| Bhandari, 2023 | Blood-based gene
biomarkers | Gene Expression
Omnibus (GEO) database (GSE6613, GSE72267, GSE99039, GSE57475,
GSE18838) | Feature selection:
LASSO, Ridge regression; Classification: Logistic Regression, SVM;
Interpretation: SHAP (XAI) | All features were
achieved above 80% accuracy. | Important
blood-based gene biomarkers for PD found; some were also found in
other NDDs; XAI made it easier to understand for early
diagnosis. | (86) |
| Yu, 2024 | Genetic
biomarkers |
Electroencephalography (EEG) signals,
genotypes, and polygenic risk scores (PRSs) | Gradient Boosting
(XGB), Random Forest (RF), Support Vector Machine (SVM) | Accuracy: 0.920;
AUC: 0.916 (SVM) | The multimodal
integration of EEG and genetic data facilitated excellent diagnosis
accuracy, revealing substantial connections between EEG signals and
clinical variables, with SVM being the most effective in
differentiating AD from other disorders. | (87) |
Molecular and cellular biomarkers
identified through ML
ML has emerged as a powerful method for detecting
cellular and molecular biomarkers across the multiple diseases,
particularly in cancer research. By integrating high-throughput
omics data, such as transcriptomics, proteomics and genomics, ML
methods have achieved sensitivities as high as 95% in identifying
diagnostic and prognostic biomarkers (88,89).
These approaches are particularly valuable in interpreting complex
datasets generated from DNA/RNA sequencing, microarrays, and mass
spectrometry, enabling the discovery of biomarkers that were
previously difficult to detect (69). This highlights the strength of ML
in managing high-dimensional datasets where traditional statistical
approaches often fail.
Beyond classification accuracy, ML algorithms are
also applied to dynamic modelling of biological processes. For
example, they have been used to construct ordinary differential
equations (ODE) models of cancer signaling networks to find
biomarkers and therapeutic targets. Such ODE modelling and
tissue-level simulations may predict the necrosis, growth arrest,
cancer metastasis, and immune cell invasion (90). While innovative, these methods
require extensive validation AS they rely heavily on assumptions
about pathway interactions. Another key application of ML is
imaging-based biomarker discovery. Techniques, such as advanced
pattern analysis have revealed that imaging patterns can predict
the survival of patients with glioblastoma, with each subtype
exhibiting unique features. Factors, such as cell density,
infiltration, microvascularity and blood-brain barrier impairment
can be integrated to create predictive biomarkers that enhance
diagnosis and therapy (91). This
suggests that multimodal ML frameworks combining imaging with
molecular data could improve precision medicine in oncology.
Recent research has also demonstrated that ML may
identify new molecular markers in a broad spectrum of disorders.
Wang et al (92) used ML
techniques to examine RNA sequencing and microarray data collected
from the GEO database. They discovered essential immune cell types
and hub genes associated with unstable atherosclerotic plaques,
confirming indicators such as CD68, PAM and IGFBP6 by single-cell
RNA sequencing, demonstrating the strength of ML in integrating
bulk and single-cell data (92).
Similarly, Liang et al (93) applied SVM-RFE and LASSO regression
on GEO datasets and discovered APOLD1 and EPYC as pivotal
diagnostic genes for osteoarthritis. They further linked these
genes to immune cell activity through CIBERSORT analysis and
validated their findings with reverse transcription-polymerase
chain reaction and ROC assays, demonstrating the importance of
combining computational prediction with wet-lab validation
(93). In pancreatic cancer, ML
algorithms have discovered proteins, mRNAs, miRNAs and DNA
methylation patterns as potential subtype biomarkers. Integrative
profiling will improve treatment tactics by validating drug
sensitivity biomarkers using pattern recognition algorithms
(94). Likewise, in non-smoking
females with stage III non-small cell lung cancer, an analysis of
GDS3837 gene expression data using XGBoost achieved a robust AUC
score of 0.835, suggesting that these biomarkers may facilitate
early diagnosis and tailored treatment (95). The integration of ML with molecular
profiling methodologies can guide customized cancer therapies,
especially in the field of radiation (96). However, challenges remain, such as
small or heterogeneous sample sizes, risk of overfitting, and the
lack of standardized performance evaluation across studies, which
may limit the reproducibility of biomarker discovery. The ML-based
identification of molecular and cellular biomarkers in NDDs is
summarized in Table II (97-105).
 | Table IIMachine learning-based discovery of
molecular and cellular biomarkers in NDDs. |
Table II
Machine learning-based discovery of
molecular and cellular biomarkers in NDDs.
| First author, year
of publication | Biomarker type | Dataset | ML method | Accuracy | Key findings | Key
limitations | (Refs.) |
|---|
| Bellomo, 2021 | Core CSF
biomarkers: Aβ42/40 ratio, p-tau, t-tau | Two large patient
cohorts from AD biomarker centers | Unsupervised
Gaussian mixture model clustering | Not specified | Classified patients
into six clusters (AD-like and non-AD profiles); enabled
computation of cluster-based cut-off values; improved data-driven
stratification and phenotyping. | Cut-off values
still influenced by group heterogeneity; external validation not
reported; limited to CSF biomarkers only | (97) |
| Hallqvist,
2024 | Blood protein panel
(8 proteins: GRN, MASP2, BiP, PTGDS, ICAM1, C3, DKK3,
SERPING1) | Recently diagnosed
PD (n=99), pre-motor RBD cohorts (n=18 and n=54), healthy controls
(n=36) | Discriminant
OPLS-DA model | Classified and
separated de novo PD or control samples with 100% accuracy based on
the expression of eight proteins | A panel of eight
blood protein biomarkers, using machine learning, differentiated
early PD from controls, identified prodromal cases up to seven
years before symptom onset, and showed promise for early risk
stratification. | Relatively small
pre-motor cohorts; needs external validation for clinical use. | (98) |
| Xu, 2022 | Blood miRNA (serum
and plasma profiles) | miRPathDB and
GeneCards | Multilayer
Perceptron (MLP) classifier, the Naive Bayes (NB) classifier, the
Random Tree (RT) classifier, the Random Forest (RF) classifier, and
the ZeroR (ZR) classifier in WEKA | The ZR and NB
classifiers achieved an average accuracy of 80% in the
cross-validation test, whereas RT achieved 82%, RF 86%, and MLP
92%. | By analyzing miRNA
associated with AD, thousands of descriptors based on target genes
and pathways were generated, which may subsequently be used to
uncover new biomarkers and enhance disease detection. | Needs larger
prospective validation; translational application not
confirmed. | (99) |
| Kumar, 2024 | Blood miRNAs (112
miRNAs: 56 PD biomarkers, 56 non-PD) | miRNAs were
extracted from the miRpathDB database | Hoeffding Tree,
Naive Bayes, Multilayer Perceptron, Sequential Model (Keras); best=
Sequential Model | Identified miRNA
biomarkers with 95.65% accuracy. | The created machine
learning model using miRNAs their genomic route descriptors
attained great accuracy in predicting Parkinson's disease. | Details limited to
algorithm performance; requires larger independent validation; risk
of overfitting from feature reduction. | (100) |
| Lin, 2020 | Plasma protein
biomarkers: Aβ42, Aβ40, total Tau, p-Tau181, α-synuclein | Plasma samples
(n=377) from healthy controls, patients with AD spectrum
(including mild cognitive impairment (MCI)), PD spectrum with
variable cognitive severity [including PD with dementia (PDD)], and
FTD. | 7 deep-learning
classifiers (SVM, CART, C4.5, NB, LogReg, kNN, and RF and
leave-one-out cross-validation (LOOCV) model. | 76% (overall
classification), 83% (AD subgroup severity), 63% (PD subgroup
severity) | The constructed LDA
model with the RF classifier may aid physicians in differentiating
various NDDs. | The majority of
patients were on pharmacological treatment, potentially influencing
plasma protein profiles and impacting model precision; the control
group was younger than the AD/PDD patients, so constraining
comparability. | (101) |
| Khorsand, 2025 | Molecular
biomarkers: neprilysin, alpha-secretase, beta-secretase, amyloid
plaques, urinary formic acid | 191 AD patients and
59 non-AD subjects | Naive Bayes (NB),
Random Forest (RF), Decision Tree (DT), Support Vector Machine
(SVM), and K-Nearest Neighbors (KNN) | KNN, SVM, RF, and
DT achieved high sensitivity (94%) and accuracy (92%). | Targeted feature
selection enhances diagnostic precision; biomarker-driven
approaches distinguish AD from non-AD effectively. | Future research
with bigger, longitudinal cohorts is crucial to better clarify
these links and improve our comprehension of Alzheimer's processes,
eventually seeking novel treatment methods. | (102) |
| Lam, 2022 | Clinical
bio-markers (alanine amino-transferase, alkaline phosphatase,
bilirubin); Genetic biomarkers (SNPs) | 1,223 UK Biobank
participants (AD, PD, MND, MG) | Machine learning
with Monte Carlo randomization; multinomial model | 88.3% for NLD
diagnosis using clinical markers | This research
illustrates the efficacy of data-driven methodologies in
discovering new biomarkers when no existing or potential biomarkers
are available. | The multinomial
model yielded results that contradicted current literature,
including a negative coefficient for LDL in MND, suggesting reduced
serum LDL levels in MND patients, which is inconsistent with prior
findings. So, further research is required. | (103) |
| Yu, 2020 | Protein-protein
interaction (hub proteins) | Human interactome
datasets from the I2D database | Random forest
model, clustering algorithm MCODE | Prediction accuracy
of 0.77 ± 0.01, AUC=0.86, and the validation set showed 77%
accuracy. | Identified hub
proteins essential in PPIN; potential NDD-related proteins;
provides insights into disease pathogenesis. | Results need
experimental validation. | (104) |
| Yang, 2024 | Aging-related
biomarkers (whole-blood RNA-Seq) | Training: 11 PD
patients, 13 healthy controls; Validation: 3 GEO datasets + qRT-PCR
on PBMCs (10 PD, 10 HC) | LASSO, Random
Forest (RF), Support Vector Machine (SVM), Ridge Regression
(RR) | Combined model
AUC=0.98 (training); validation AUCs=0.833, 0.792, 0.725. | Found four
aging-related genes that are strong diagnostic biomarkers; tested
them in external datasets and PBMC samples; two biomarkers were
linked to immune cell infiltration. | The training sample
size is small (11 PD compared. 13 HC), and further testing is
required in bigger groups. | (105) |
6. Applications of machine learning in
neuroimaging for early diagnosis
Developments in neuroimaging and ML have shown the
ensured early detection of NDDs. CNNs have exhibited notale
efficacy in detecting AD, with a 94.7% accuracy rate in
distinguishing between early-stage AD and normal aging (106). Furthermore, DL and multimodal
imaging analysis have created new avenues for using ML in different
forms of dementia (107).
Additionally, the ML-based analysis of single-photon emission
computed tomography images has improved diagnostic precision and
outperformed conventional techniques in identifying dopaminergic
degradation in PD (108). Vieira
et al (109) investigated
ML and DL methods for identifying first-episode psychosis using
neuroimaging data. Their findings revealed the variations in
accuracy ranging from 50 to 70%, depending on the feature set. When
DL was used with surface-based features, the greatest accuracy of
70% was obtained (109). It has
been demonstrated that SVM and logistic regression are the optimal
schizophrenia classifiers. More accurate than surface area,
cortical thickness and subcortical volume align with the clinical
severity and neurobiological patterns of schizophrenia (110). It has been demonstrated that ML
can differentiate between AD, mild cognitive impairment (MCI) and
healthy individuals by focusing on key brain areas such as the
hippocampus. The accuracy rates are 66% for patients with MCI and
76% for AD compared to healthy controls (111). The ensemble transfer learning
approach achieved an AUC of 90.2%, accurately differentiating AD
from healthy individuals. Conversely, the lack of training images
in the custom DL model led to its low performance. These results
suggest that the use of transfer learning with neuroimages can
enhance the early diagnosis and prognosis of AD, even when models
are pre-trained on general images (112). The ML framework can be used to
predict future cognitive categories in non-demented older adults.
This suggests that using a baseline neuropsychiatric symptoms and
mild behavioral impairment framework can improve the results
(113). This approach drives
research into dementia detection, optimizes resource utilization
and improves clinical practice sensitivity.
In 2021, Murugan et al (114) introduced the DEMentia NETwork
(DEMNET) for detecting dementia stages using MRI images. The model
outperformed existing approaches on the Kaggle dataset with 95.23%
accuracy, 97% AUC and 0.93 Cohen's Kappa. Additionally, the ability
of the model to identify AD phases was tested using the ADNI
dataset (114). In 2020, Jo et
al (115) found that Tau PET
images may be used to classify AD using a DL system that
incorporates 3D CNN and LRP algorithms. This framework will also be
useful for early identification during the prodromal stages of AD
(115). The resting-state
functional magnetic resonance imaging (fMRI) and DL approaches
identify and diagnose AD throughout six phases. The FT network
exhibited good accuracy throughout all phases, but the OTS network
had the highest average accuracy of 97.92% (116). A summary of the performance
metrics and clinical applications of FDA-approved AI/ML algorithms
used in diagnosing NDDs is presented in Table III (117-125).
These results indicate that combining fMRI with DL can enhance
early diagnosis and improve the identification of risk factors and
prognostic indicators.
 | Table IIIFDA-Approved AI/ML algorithms and
neuroimaging-based studies for the diagnosis of NDDs. |
Table III
FDA-Approved AI/ML algorithms and
neuroimaging-based studies for the diagnosis of NDDs.
| Algorithm | Developer | Diseases | Modality used | Performance
matrices | FDA approval
date | Function of the
algorithm | (Refs.) |
|---|
| Aidoc BriefCase-CSF
triage | Aidoc Medical,
Ltd. | Cervical Spine
Fractures | cervical spine CT
scans | Detection of
Cervical Spine Fractures Sensitivity-54.9 Specificity-94.1 PPV-38.7
NPV-96.8 | 5/31/19 | The system
automatically alerts clinicians when a CT scan of the neck shows
potential signs of a broken neck bone. | (117) |
| Vitrea CT Brain
Perfusion | Vital Images,
Inc. | Ischemic
stroke | CT images | Detection of
Ischemic Stroke Sensitivity-70.8 Specificity-80.0 PPV-98.8
NPV-10.2 | 11/20/18 | Automatically
computes quantitative brain perfusion metrics (rCBV, MTT, rCBF,
TTP) from CT perfusion scans. | (118) |
| Health VCF | Zebra Medical
Vision Ltd. | Vertebral
fractures | CT images | Detection of VCFs
Sensitivity-54.0 Specificity-92.0 PPV-69.0 NPV-87.0 | 5/12/20 | Automatically
detects and alerts on suspected intracranial hemorrhage (ICH) in CT
scans; analyzes CT perfusion scans for stroke detection. | (119) |
| Syngo.CT Neuro
Perfusion | Siemens
Healthineers | Ischemic
stroke | CT images | Detection of
Ischemic Core Volumes Sensitivity-93.0-97.0
Specificity-97.0-100.0 | 10/11/20 | | (120) |
| Brainance MD | Advantis Medical
Imaging | Major white matter
tracts | MRI images | Detection of ICH
Sensitivity-91.4 Specificity-97.5 PPV-80.2-97.3 NPV-91.9-99.0 | 10/14/21 | Performs diffusion
tensor imaging (DTI), dynamic susceptibility contrast (DSC)
perfusion, and functional MRI (fMRI) analyses. | (121) |
| NeuroQuant | cortechs.ai | Alzheimer's
disease | MRI images | Identification of
Alzheimer's Sensitivity-63.0-88.5 Specificity-66.0-92.0 | 9/7/17 | Processes
volumetric MRI scans. | (122) |
| NeuroQuant | cortechs.ai | Mild cognitive
impairment | MRI images | Identification of
MCI Sensitivity-48.9-60.2 Specificity-80.0-80.6 | 9/7/17 | Processes
volumetric MRI scans. | (123,124) |
| Quantib Brain | Quantib BV | Dementia | MRI images | Diagnosis of
Dementia Sensitivity-95.0-97.5 Specificity-60.0 | 3/9/18 | Processes
volumetric MRI scans. | (125) |
7. Machine learning approaches in cognitive
and behavioral assessment
ML is promising for cognitive and behavioral
testing. In cognitive workload assessment, artificial neural
networks and SVM accurately mimic physiological data (126). ML models based on previous
functional analysis can improve the accuracy of indirect
assessments such as the Questions About Behavioral Function (QABF),
enhancing the identification of behavioral functions (127). Moreover, ML methods have been
used to create robust personality assessment instruments using
digital records and social media data, potentially enhancing
personality theory when included in a thorough construct validation
framework (128). Javed et
al (129) designed the
Cognitive Assessment of Smart Home Residents (CA-SHR) to assess
daily functional health of elderly or cognitively impaired
individuals using the internet of things. They used predetermined
ratings and supervised classification to detect early cognitive
impairment (129). Research has
employed smart devices to automate test administration, speech
transcription and clinical state prediction for frequent remote
neuropsychological assessments, allowing for accurate evaluations
of cognitive and emotional states and enabling continuous mental
health monitoring (130). An
active superior temporal sulcus predicted stop-signal reaction time
well, accounting for 12% of the variation in multivariate ML
research. This indicates how multivariate methods can boost brain
function and performance knowledge (131). A supervised ML algorithm was
previously used to predict the response to working memory training
in patients with PD using demographic, clinical, cognitive and
learning data. The use of training-inherent learning parameters
improved the precision of the prediction models, potentially
maximizing training benefits following cognitive interventions
(132). Research has demonstrated
that transdiagnostic factors strongly affect psychotic cognitive
function. Psychosis-related cognitive impairment may reflect
overall cognitive performance. A diagnosis-agnostic,
symptom-targeted strategy may be suitable for evaluating therapies
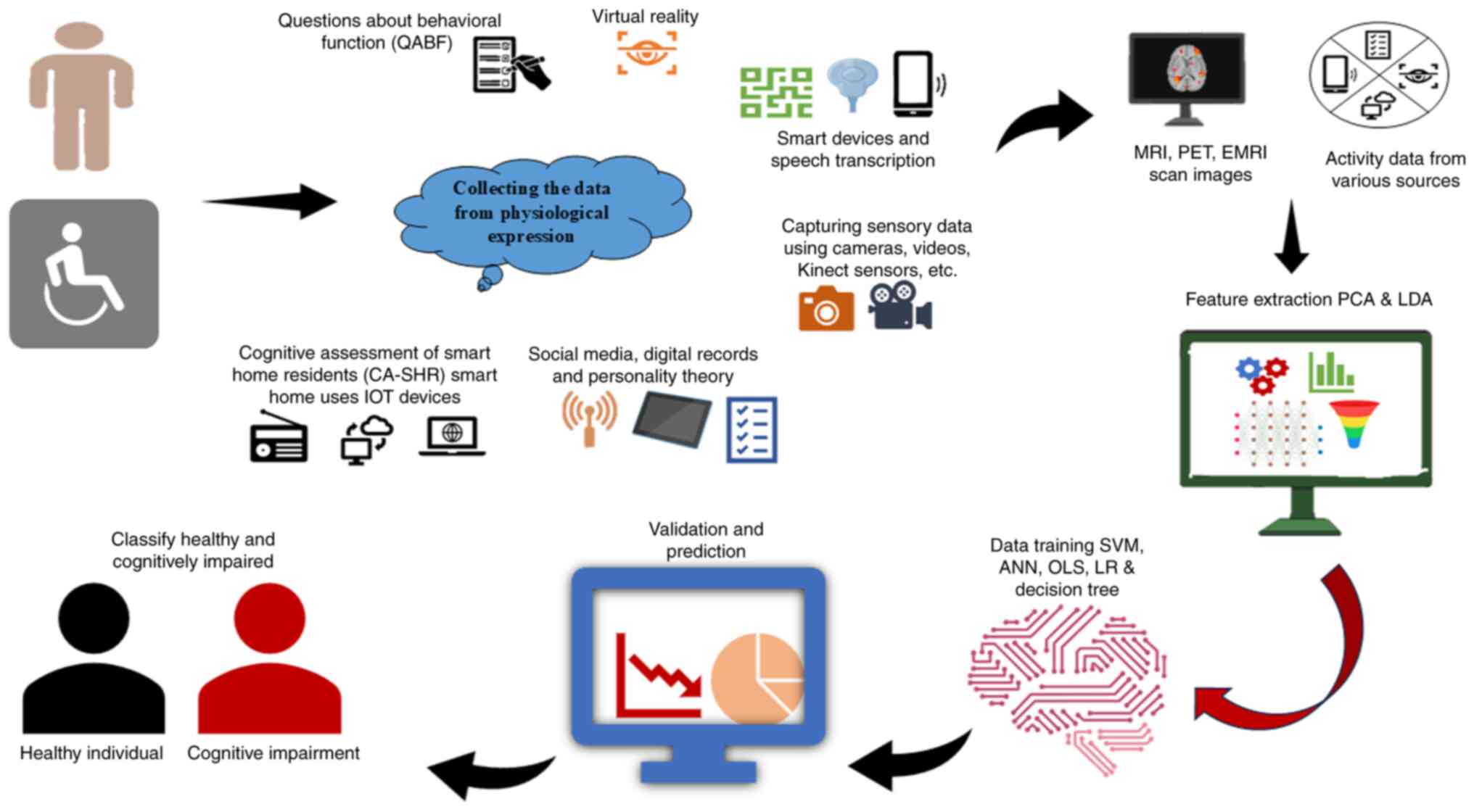
(133). The first validation
research by Kim et al (134) revealed that virtual reality (VR)
hand and eye motions may screen for MCI. SVM trained on virtual
kiosk test data effectively discriminated patients with MCI from
healthy controls, correlating these motions to cognitive domains
and facilitating VR for MCI screening (Fig. 4) (134). These studies demonstrate the
potential of ML in the enhancement of the accuracy and efficiency
of cognitive and behavioral assessments.
8. Prediction of neurodegenerative diseases
using multi-modal integration
Multi-modal integration techniques have exhibited
significant advantages in detecting NDDs. A novel methodology
employs graph neural networks (GNNs) to integrate image and
phenotypic data. Research has demonstrated the construction of
brain networks from structural MRI (sMRI) or PET images within a
multi-modal GNN framework. Experiments reveal that this method
improves the diagnosis of AD, underscoring the need for
comprehensive multi-modal diagnostic techniques (135). Lee et al (136) developed a multimodal recurrent
neural network combining neuroimaging, CSF and cognitive data to
predict MCI progression to AD. Using longitudinal, multi-domain
data, the model achieved 81% accuracy, aiding early risk
identification and clinical trial selection (136). In their study, Liu et al
(137) revealed the hierarchical
attention-based multi-task multi-modal fusion model (HAMMF)
designed to enhance AD diagnosis using multi-modal neuroimaging
data, including MRI and PET images. Their results achieved an
overall accuracy of 93.15% in differentiating between AD and
healthy cases (137). Wang et
al (138) introduced the
hypergraph-regularized multimodal learning by the graph diffusion
(HMGD) technique for the diagnosis of complex brain diseases. This
method improves similarity metrics across participants by including
imaging and genetic data (138).
Employing a consolidated graph and a multi-kernel support vector
machine (MK-SVM), HMGD exceeds current methodologies on ADNI data,
uncovering substantial correlations and critical areas associated
with genetic risk biomarkers for disease predictions.
The study by Zhu et al (139) developed a dynamic hyper-graph
learning framework for multi-modal imaging-based computer-assisted
diagnosis. The model estimates data representations and performs
classification and regression tasks, promising to identify
diagnostic labels and predict MCI and AD clinical scores (139). Castellano et al (140) examined multimodal models for 2D
and 3D MRI and amyloid PET scan-based AD diagnosis. Volumetric data
models outperform 2D images, and integrating imaging modalities
increases prediction accuracy by focusing on Alzheimer's-related
areas (140). By merging sMRI
with resting-state functional MRI (rs-fMRI) data, the localized
region extraction and multi-modal fusion (LRE-MMF) technique
improves PD diagnosis. PCA separates imaging data into localized
areas, identifies features, and decreases dimensionality, then
processes them via a neural network to reach 75% accuracy, possibly
enhancing diagnostic tools (141). Chen et al (142) described AD diagnosis using
neuroimage-MED multimodal image feature fusion. This method
improves classification and prediction, classifying AD, MCI and NC
with 84.1% accuracy and predicting MCI development with 93.9%.
Clinical diagnosis and neuroimaging bring the technique closer to
clinical practice. This method is relatively new, with 86.95% of
studies published over the past 5 years using data from biomedical
imaging, cognitive assessments, speech and language evaluations,
gait analysis, hand and eye movement tests, EEG and genetic
evaluations (142). The study
found that multimodal data categorization rates are sufficiently
enough to distinguish AD, PD and MCI from healthy controls
(143). Researchers use CNNs to
extract features from MRI and PET brain imaging data to improve
automated detection.
9. Conclusion and future directions
The present review emphasized the revolutionary
potential of ML for the early identification and treatment of NDDs.
The combination of numerous data sources, such as neuroimaging,
genetic profiling, and biomarker analysis, has shown encouraging
outcomes for improving diagnostic precision, recognizing disease
risk factors, and facilitating personalized treatment approaches.
The developments in ML approaches, particularly in processing
high-dimensional data, represent a major leap forward in the
capacity to predict disease progression and consequences. CNNs and
multilayered models have made significant progress. This
demonstrates that these technologies can accurately distinguish
between different stages of illness and help clinicians to make
decisions. However, despite these achievements, several problems
persist.
Issues related to data standards, privacy, and the
ethical implications of genetic testing require careful
consideration and regulation. There is a greater need for
longitudinal multimodal datasets that can more effectively document
disease progression and diversity across various groups.
Furthermore, the advancement of explainable ML techniques is
crucial for enhancing transparency, interpretability, and clinical
confidence in model predictions. The absence of defined biomarker
techniques persists in hindering reproducibility and comparability
across research, underscoring the need to create universal
standards. Further study is required to verify the robustness and
generalizability of ML models across varied demographics and
clinical situations. Additionally, to fully utilize ML approaches
in combating NDDs, multidisciplinary support among healthcare
professionals, data scientists, and ethicists is necessary. A
significant research gap exists in the application of machine
learning discoveries from controlled research settings to practical
clinical situations, necessitating collaboration among healthcare
providers, data scientists, and ethicists. By bridging the gap
between technology innovation and clinical application, researchers
may advance toward a future of more precise, efficient and
customized healthcare.
However, challenges and limitations remain. The use
of ML in NDD research has considerable challenges. Limited and
diverse datasets, particularly in rare NDDs, restrict model
generalization and increase the risk of overfitting. Challenges
such as missing data, inconsistent formats (such as neuroimaging,
genetics and wearable sensor information), and difficulties in
integrating multiple data types render model development more
complex. A major obstacle is the lack of interpretability; many ML
algorithms act as ‘black boxes’, which can reduce confidence among
clinicians and patients. Additionally, the absence of standardized
evaluation methods and technical hurdles for clinical
implementation hinders real-world application. Ethical issues,
including patient privacy, algorithmic bias, and ensuring equitable
access, further contribute to these challenges. Addressing these
challenges requires larger, high-quality datasets, advancements in
explainable ML, the creation of standardized evaluation criteria,
and thorough validation across multiple centers to build trust and
ensure clinical use.
Acknowledgements
The authors would like to thank the management of
Chettinad Academy of Research and Education (Deemed to be
University), Chennai, India for providing the facilities to perform
the present review.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
SV conducted the literature search, collected data,
contributed to the writing of the manuscript, and created the
tables and figures. SW performed the validation and curation of the
data from the literature, and was involved in the revision process.
LK was involved in the preparation of the manuscript and provided
editing assistance. GKS conducted investigations, provided editing
assistance and supervision, and conceptualized the study. All
authors have thoroughly reviewed and approved the final manuscript.
Data authentication is not applicable.
Ethics and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
The ORCID IDs of the authors are as follows: SV
(0009-0004-1133-1646), SW (0000-0002-4703-0616), LK
(0000-0002-3154-1331 and GKS (0000-0002-0531-424X).
References
|
1
|
Newell ME, Babbrah A, Aravindan A, Rathnam
R, Kiernan R, Driver EM, Bowes DA and Halden RU: Prevalence rates
of neurodegenerative diseases versus human exposures to heavy
metals across the United States. Sci Total Environ.
928(172260)2024.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Adams JL, Myers TL, Waddell EM, Spear KL
and Schneider RB: Telemedicine: A valuable tool in
neurodegenerative diseases. Curr Geriatr Rep. 9:72–81.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hansson O: Biomarkers for
neurodegenerative diseases. Nat Med. 27:954–963. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Better MA: 2023 Alzheimer's disease facts
and figures. Alzheimers Dement. 19:1598–1695. 2023.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bhidayasiri R, Sringean J, Phumphid S,
Anan C, Thanawattano C, Deoisres S, Panyakaew P, Phokaewvarangkul
O, Maytharakcheep S, Buranasrikul V and Prasertpan T: The rise of
Parkinson's disease is a global challenge, but efforts to tackle
this must begin at a national level: A protocol for national
digital screening and ‘eat, move, sleep’ lifestyle interventions to
prevent or slow the rise of non-communicable diseases in Thailand.
Front Neurol. 15(1386608)2024.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Mead RJ, Shan N, Reiser HJ, Marshall F and
Shaw PJ: Amyotrophic lateral sclerosis: A neurodegenerative
disorder poised for successful therapeutic translation. Nat Rev
Drug Discov. 22:185–1212. 2023.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bloem BR, Okun MS and Klein C: Parkinson's
disease. Lancet. 397:2284–2303. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Baker E, Leonenko G, Schmidt KM, Hill M,
Myers AJ, Shoai M, de Rojas I, Tesi N, Holstege H, van der Flier WM
and Pijnenburg YA: What does heritability of Alzheimer's disease
represent? PLoS One. 18(e0281440)2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Nalls MA, Blauwendraat C, Vallerga CL,
Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A
and Bras J: Identification of novel risk loci, causal insights, and
heritable risk for Parkinson's disease: A Meta-analysis of
Genome-wide association studies. Lancet Neurol. 18:1091–1102.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sakowski SA, Koubek EJ, Chen KS, Goutman
SA and Feldman EL: Role of the exposome in neurodegenerative
disease: Recent insights and future directions. Ann Neurol.
95:635–652. 2024.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Rekatsina M, Paladini A, Piroli A, Zis P,
Pergolizzi JV and Varrassi G: Pathophysiology and therapeutic
perspectives of oxidative stress and neurodegenerative diseases: A
narrative review. Adv Ther. 37:113–139. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Cravello L, Di Santo S, Varrassi G,
Benincasa D, Marchettini P, de Tommaso M, Shofany J, Assogna F,
Perotta D, Palmer K and Paladini A: Chronic pain in the elderly
with cognitive decline: A narrative review. Pain Ther. 8:53–65.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Aldharman SS, Al-Jabr KH, Alharbi YS,
Alnajar NK, Alkhanani JJ, Alghamdi A, Abdellatif RA, Allouzi A,
Almallah AM and Jamil SF: Implications of early diagnosis and
intervention in the management of Neurodevelopmental Delay (NDD) in
children: A systematic review and Meta-analysis. Cureus.
15(e38745)2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Mobed A and Hasanzadeh M: Biosensing: The
best alternative for conventional methods in detection of
Alzheimer's disease biomarkers. Int J Biol Macromol. 161:59–71.
2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Selvam S and Ayyavoo V: Biomarkers in
neurodegenerative diseases: A broad overview. Exploration
Neuroprotective Ther. 4:119–147. 2024.
|
|
16
|
Rastogi S, Sharma V, Bharti PS, Rani K,
Modi GP, Nikolajeff F and Kumar S: The evolving landscape of
exosomes in neurodegenerative diseases: Exosomes characteristics
and a promising role in early diagnosis. Int J Mol Sci.
22(440)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Dubois B, von Arnim CA, Burnie N, Bozeat S
and Cummings J: Biomarkers in Alzheimer's disease: Role in early
and differential diagnosis and recognition of atypical variants.
Alzheimers Res Ther. 15(175)2023.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chudzik A, Śledzianowski A and
Przybyszewski AW: Machine learning and digital biomarkers can
detect early stages of neurodegenerative diseases. Sensors.
24(1572)2024.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Dorsey ER, Papapetropoulos S, Xiong M and
Kieburtz K: The first frontier: Digital biomarkers for
neurodegenerative disorders. Digital Biomarkers. 1:6–13.
2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Iftikhar M, Saqib M, Zareen M and Mumtaz
H: Artificial intelligence: Revolutionizing robotic surgery. Ann
Med Surg (Lond). 86:5401–5409. 2024.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bajwa J, Munir U, Nori A and Williams B:
Artificial intelligence in healthcare: Transforming the practice of
medicine. Future Healthc J. 8:e188–e194. 2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
García-Fonseca Á, Martin-Jimenez C,
Barreto GE, Pachón AF and González J: The emerging role of long
non-coding RNAs and microRNAs in neurodegenerative diseases: A
perspective of machine learning. Biomolecules.
11(1132)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Khaliq F, Oberhauser J, Wakhloo D and
Mahajani S: Decoding degeneration: The implementation of machine
learning for clinical detection of neurodegenerative disorders.
Neural Regen Res. 18:1235–1242. 2023.PubMed/NCBI View Article : Google Scholar
|
|
24
|
DeTure MA and Dickson DW: The
neuropathological diagnosis of Alzheimer's disease. Mol
Neurodegener. 14(32)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sheppard O and Coleman M: Alzheimer's
disease: Etiology, neuropathology and pathogenesis. Exon
Publications. 19:1–21. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
García-Morales V, González-Acedo A,
Melguizo-Rodríguez L, Pardo-Moreno T, Costela-Ruiz VJ,
Montiel-Troya M and Ramos-Rodríguez JJ: Current understanding of
the physiopathology, diagnosis and therapeutic approach to
Alzheimer's disease. Biomedicines. 9(1910)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Tiwari S, Atluri V, Kaushik A, Yndart A
and Nair M: Alzheimer's disease: Pathogenesis, diagnostics, and
therapeutics. Int J Nanomedicine. 14:5541–5554. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Riederer P, Berg D, Casadei N, Cheng F,
Classen J, Dresel C, Jost W, Krüger R, Müller T, Reichmann H, et
al: α-Synuclein in Parkinson's disease: Causal or bystander? J
Neural Transm (Vienna). 126:815–840. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Masato A, Plotegher N, Terrin F, Sandre M,
Faustini G, Thor A, Adams S, Berti G, Cogo S, De Lazzari F and
Fontana CM: DOPAL Initiates αSynuclein-dependent impaired
proteostasis and degeneration of neuronal projections in
Parkinson's disease. NPJ Parkinsons Dis. 9(42)2023.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Simpson C, Vinikoor-Imler L, Nassan FL,
Shirvan J, Lally C, Dam T and Maserejian N: Prevalence of ten LRRK2
variants in Parkinson's disease: A comprehensive review.
Parkinsonism Relat Disord. 98:103–113. 2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhou ZD, Yi LX, Wang DQ, Lim TM and Tan
EK: Role of dopamine in the pathophysiology of Parkinson's disease.
Transl Neurodegener. 12(44)2023.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Calabresi P, Mechelli A, Natale G,
Volpicelli-Daley L, Di Lazzaro G and Ghiglieri V: Alpha-synuclein
in Parkinson's disease and other synucleinopathies: From overt
neurodegeneration back to early synaptic dysfunction. Cell Death
Dis. 14(176)2023.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhou W and Xu R: Current insights in the
molecular genetic pathogenesis of amyotrophic lateral sclerosis.
Front Neurosci. 17(1189470)2023.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liu J and Wang F: Role of
neuroinflammation in amyotrophic lateral sclerosis: Cellular
mechanisms and therapeutic implications. Front Immunol.
8(1005)2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Masrori P and Van Damme P: Amyotrophic
lateral sclerosis: A clinical review. Eur J Neurol. 27:1918–1929.
2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Semmler S, Gagné M, Garg P, Pickles SR,
Baudouin C, Hamon-Keromen E, Destroismaisons L, Khalfallah Y,
Chaineau M, Caron E and Bayne AN: TNF receptor-associated factor 6
interacts with ALS-linked misfolded superoxide dismutase 1 and
promotes aggregation. J Biol Chem. 295:3808–3825. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Farrawell NE and Yerbury JJ: Mutant Cu/Zn
superoxide dismutase (A4V) turnover is altered in cells containing
inclusions. Front Mol Neurosci. 14(771911)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Tedesco B, Ferrari V, Cozzi M,
Chierichetti M, Casarotto E, Pramaggiore P, Mina F, Galbiati M,
Rusmini P, Crippa V and Cristofani R: The role of small heat shock
proteins in protein misfolding associated motoneuron diseases. Int
J Mol Sci. 23(11759)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Maurel C, Dangoumau A, Marouillat S,
Brulard C, Chami A, Hergesheimer R, Corcia P, Blasco H, Andres CR
and Vourc'h P: Causative genes in amyotrophic lateral sclerosis and
protein degradation pathways: A link to neurodegeneration. Mol
Neurobiol. 55:6480–6499. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Bottero V, Santiago JA, Quinn JP and
Potashkin JA: Key disease mechanisms linked to amyotrophic lateral
sclerosis in spinal cord motor neurons. Front Mol Neurosci.
15(825031)2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Dokholyan NV, Mohs RC and Bateman RJ:
Challenges and progress in research, diagnostics, and therapeutics
in Alzheimer's disease and related dementias. Alzheimers Dement (N
Y). 8(e12330)2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Domínguez-Fernández C, Egiguren-Ortiz J,
Razquin J, Gómez-Galán M, De las Heras-García L, Paredes-Rodríguez
E, Astigarraga E, Miguélez C and Barreda-Gómez G: Review of
technological challenges in personalised medicine and early
diagnosis of neurodegenerative disorders. Int J Mol Sci.
24(3321)2023.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Shusharina N, Yukhnenko D, Botman S,
Sapunov V, Savinov V, Kamyshov G, Sayapin D and Voznyuk I: Modern
methods of diagnostics and treatment of neurodegenerative diseases
and depression. Diagnostics (Basel). 13(573)2023.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Anique M, Talib M, Ihsan A, Anwar I,
Zeeshan A and Ahsan N: Biomarker profiles in serum and CSF for
early diagnosis of selected neurodegenerative diseases: Serum and
CSF for early diagnosis of neurodegenerative diseases. Pakistan J
Health Sci. 5:166–70. 2024.
|
|
45
|
Kammeyer R, Chapman K, Furniss A, Hsieh E,
Fuhlbrigge R, Ogbu EA, Boackle S, Zell J, Nair KV, Borko TL, et al:
Blood-based biomarkers of neuronal and glial injury in active major
neuropsychiatric systemic lupus erythematosus. Lupus. 33:1116–1129.
2024.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Koníčková D, Menšíková K, Tučková L,
Hényková E, Strnad M, Friedecký D, Stejskal D, Matěj R and Kaňovský
P: Biomarkers of neurodegenerative diseases: Biology, taxonomy,
clinical relevance, and current research status. Biomedicines.
10(1760)2022.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hansson O, Lehmann S, Otto M, Zetterberg H
and Lewczuk P: Advantages and disadvantages of the use of the CSF
Amyloid β (Aβ) 42/40 ratio in the diagnosis of Alzheimer's Disease.
Alzheimers Res Ther. 11(34)2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Feng Y, Murphy MC, Hojo E, Li F and
Roberts N: Magnetic resonance elastography in the study of
neurodegenerative diseases. J Magn Reson Imaging. 59:82–96.
2024.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Young PN, Estarellas M, Coomans E,
Srikrishna M, Beaumont H, Maass A, Venkataraman AV, Lissaman R,
Jiménez D, Betts MJ, et al: Imaging biomarkers in
neurodegeneration: Current and future practices. Alzheimers Res
Ther. 12(49)2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Myrou A, Barmpagiannos K, Ioakimidou A and
Savopoulos C: Molecular biomarkers in neurological diseases:
Advances in diagnosis and prognosis. Int J Mol Sci.
26(2231)2025.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Ni A and Sethi A: Alzheimer's disease
Neuroimaging Initiative: Functional genetic biomarkers of
Alzheimer's disease and gene expression from peripheral blood.
bioRxiv. Jan 18, 2021 doi: 10.1101/2021.01.15.426891.
|
|
52
|
Abbas S, Asif M, Rehman A, Alharbi M, Khan
MA and Elmitwally N: Emerging research trends in artificial
intelligence for cancer diagnostic systems: A comprehensive review.
Heliyon. 10(e36743)2024.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Alowais SA, Alghamdi SS, Alsuhebany N,
Alqahtani T, Alshaya AI, Almohareb SN, Aldairem A, Alrashed M, Bin
Saleh K, Badreldin HA, et al: Revolutionizing healthcare: The role
of artificial intelligence in clinical practice. BMC Med Educ.
23(689)2023.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Sidey-Gibbons JA and Sidey-Gibbons CJ:
Machine learning in medicine: A practical introduction. BMC Med Res
Methodol. 19:1–8. 2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Mirnezami R, Nicholson J and Darzi A:
Preparing for precision medicine. N Engl J Med. 366:489–491.
2012.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ibrahim IM and Abdulazeez AM: The role of
machine learning algorithms for diagnosing diseases. Learning.
4(6)2021.
|
|
57
|
Saputra NA, Riza LS, Setiawan A and
Hamidah I: A systematic review for classification and selection of
deep learning methods. Decision Analytics J. 12(100489)2024.
|
|
58
|
Labory J, Njomgue-Fotso E and Bottini S:
Benchmarking feature selection and feature extraction methods to
improve the performances of machine-learning algorithms for patient
classification using metabolomics biomedical data. Comput Struct
Biotechnol J. 23:1274–1287. 2024.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Jia W, Sun M, Lian J and Hou S: Feature
dimensionality reduction: A review. Complex Intelligent Systems.
8:2663–2693. 2022.
|
|
60
|
Sarder MA, Maniruzzaman M and Ahammed B:
Feature selection and classification of leukemia cancer using
machine learning techniques. Machine Learning Res. 5(18)2020.
|
|
61
|
Remeseiro B and Bolon-Canedo V: A review
of feature selection methods in medical applications. Comput Biol
Med. 112(103375)2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Harrison CJ and Sidey-Gibbons CJ: Machine
learning in medicine: A practical introduction to natural language
processing. BMC Med Res Methodol. 21(158)2021.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Pfob A, Lu SC and Sidey-Gibbons C: Machine
learning in medicine: A practical introduction to techniques for
data pre-processing, hyperparameter tuning, and model comparison.
BMC Med Res Methodol. 22(282)2022.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Samala RK, Chan HP, Hadjiiski L and Helvie
MA: Risks of feature leakage and sample size dependencies in deep
feature extraction for breast mass classification. Medical Physics.
48:2827–2837. 2021.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Varoquaux G and Colliot O: Evaluating
machine learning models and their diagnostic value. In: Machine
Learning for Brain Disorders [Internet]. New York, NY, Humana,
2023.
|
|
66
|
Erickson BJ and Kitamura F: Magician's
corner: 9. Performance metrics for machine learning models. Radiol
Artif Intell. 3(e200126)2021.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Handelman GS, Kok HK, Chandra RV, Razavi
AH, Huang S, Brooks M, Lee MJ and Asadi H: Peering into the black
box of artificial intelligence: Evaluation metrics of machine
learning methods. Am J Roentgenol. 212:38–43. 2019.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Hicks SA, Strümke I, Thambawita V, Hammou
M, Riegler MA, Halvorsen P and Parasa S: On evaluation metrics for
medical applications of artificial intelligence. Sci Rep.
12(5979)2022.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Ledesma D, Symes S and Richards S:
Advancements within modern machine learning methodology: Impacts
and prospects in biomarker discovery. Curr Med Chem. 28:6512–6531.
2021.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Lam S, Arif M, Song X, Uhlen M and
Mardinoglu A: Machine learning analysis reveals biomarkers for the
detection of neurodegenerative diseases. medRxiv. Feb 15, 2022.
|
|
71
|
Arisi I, D'Onofrio M, Brandi R, Sonnessa
M, Campanelli A, Florio R, Sposato V, Malerba F, Cattaneo A,
Mecocci P and Bruno G: Mining clinical and laboratory data of
neurodegenerative diseases by machine learning: Transcriptomic
biomarkers. In 2018 IEEE International Conference on Bioinformatics
and Biomedicine (BIBM), IEEE, pp2735-2737, Dec 3, 2018.
|
|
72
|
Li Z, Guo W, Ding S, Chen L, Feng K, Huang
T and Cai YD: Identifying key MicroRNA signatures for
neurodegenerative diseases with machine learning methods. Front
Genet. 13(880997)2022.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Huseby CJ, Delvaux E, Brokaw DL and
Coleman PD: Blood transcript biomarkers selected by machine
learning algorithm classify neurodegenerative diseases including
Alzheimer's disease. Biomolecules. 12(1592)2022.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Ren J, Zhang B, Wei D and Zhang Z:
Identification of methylated gene biomarkers in patients with
Alzheimer's disease based on machine learning. Biomed Res Int.
2020(8348147)2020.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Abd El Hamid MM, Mabrouk MS and Omar YM:
Developing an early predictive system for identifying genetic
biomarkers associated to Alzheimer's disease using machine learning
techniques. Biomed Engineering Applications Basis Communications.
31(1950040)2019.
|
|
76
|
Kelly J, Moyeed R, Carroll C, Luo S and Li
X: Blood biomarker-based classification study for neurodegenerative
diseases. Sci Rep. 13(17191)2023.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Abdelwahab MM, Al-Karawi KA and Semary HE:
Deep learning-based prediction of Alzheimer's disease using
microarray gene expression data. Biomedicines.
11(3304)2023.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Wang Y, Wu D, Zheng M and Yang T: An
integrated bioinformatics and machine learning approach to
identifying biomarkers connecting Parkinson's disease with purine
metabolism-related genes. BMC Neurol. 25(161)2025.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Huang Y, Sun X, Jiang H, Yu S, Robins C,
Armstrong MJ, Li R, Mei Z, Shi X, Gerasimov ES and De Jager PL: A
machine learning approach to brain epigenetic analysis reveals
kinases associated with Alzheimer's disease. Nat Commun.
12(4472)2021.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Alamro H, Thafar MA, Albaradei S, Gojobori
T, Essack M and Gao X: Exploiting machine learning models to
identify novel Alzheimer's disease biomarkers and potential
targets. Sci Rep. 13(4979)2023.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Madar IH, Sultan G, Tayubi IA, Hasan AN,
Pahi B, Rai A, Sivanandan PK, Loganathan T, Begum M and Rai S:
Identification of marker genes in Alzheimer's disease using a
machine-learning model. Bioinformation. 17:348–355. 2021.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Lin RH, Wang CC and Tung CW: A machine
learning classifier for predicting stable MCI patients using gene
biomarkers. Int J Environ Res Public Healt. 19(4839)2022.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Sharma A and Dey P: A machine learning
approach to unmask novel gene signatures and prediction of
Alzheimer's disease within different brain regions. Genomics.
113:1778–1789. 2021.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Augustine J and Jereesh AS: Blood-based
gene-expression biomarkers identification for the non-invasive
diagnosis of Parkinson's disease using two-layer hybrid feature
selection. Gene. 823(146366)2022.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Sekaran K, Alsamman AM, George Priya Doss
C and Zayed H: Bioinformatics investigation on blood-based gene
expressions of Alzheimer's disease revealed ORAI2 gene biomarker
susceptibility: An explainable artificial intelligence-based
approach. Metabolic Brain Disease. 38:1297–1310. 2023.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Bhandari N, Walambe R, Kotecha K and
Kaliya M: Integrative gene expression analysis for the diagnosis of
Parkinson's disease using machine learning and explainable AI.
Comput Biol Med. 163(107140)2023.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Yu WY, Sun TH, Hsu KC, Wang CC, Chien SY,
Tsai CH and Yang YW: Comparative analysis of machine learning
algorithms for Alzheimer's disease classification using EEG signals
and genetic information. Comput Biol Med.
176(108621)2024.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Shi K, Lin W and Zhao XM: Identifying
molecular biomarkers for diseases with machine learning based on
integrative omics. IEEE/ACM Trans Comput Biol Bioinform.
18:2514–2525. 2020.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Brar A, Zhu A, Baciu C, Sharma D, Xu W,
Orchanian-Cheff A, Wang B, Reimand J, Grant R and Bhat M:
Development of diagnostic and prognostic molecular biomarkers in
hepatocellular carcinoma using machine learning: A systematic
review. Liver Cancer International. 3:141–161. 2022.
|
|
90
|
Mertins SD: Capturing biomarkers and
molecular targets in cellular landscapes from dynamic reaction
network models and machine learning. Front Oncol.
11(805592)2022.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Macyszyn L, Akbari H, Pisapia JM, Da X,
Attiah M, Pigrish V, Bi Y, Pal S, Davuluri RV, Roccograndi L and
Dahmane N: Imaging patterns predict patient survival and molecular
subtype in glioblastoma via machine learning techniques. Neuro
Oncol. 18:417–425. 2015.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Wang J, Kang Z, Liu Y, Li Z, Liu Y and Liu
J: Identification of immune cell infiltration and diagnostic
biomarkers in unstable atherosclerotic plaques by integrated
bioinformatics analysis and machine learning. Front Immunol.
13(956078)2022.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Liang Y, Lin F and Huang Y: Identification
of biomarkers associated with diagnosis of osteoarthritis patients
based on bioinformatics and machine learning. J Immunol Res.
2022(5600190)2022.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Sinkala M, Mulder N and Martin D: Machine
learning and network analyses reveal disease subtypes of pancreatic
cancer and their molecular characteristics. Sci Rep.
10(1212)2020.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Zheng H, Zhang Q, Gong Y, Liu Z and Chen
S: Identification of prognostic biomarkers for stage iii non-small
cell lung carcinoma in female nonsmokers using machine learning
arXiv: Aug 28, 2024.
|
|
96
|
Rydzewski NR, Helzer KT, Bootsma M, Shi Y,
Bakhtiar H, Sjöström M and Zhao SG: Machine learning &
molecular radiation tumor biomarkers. Semin Radiat Oncol.
33:243–251. 2023.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Bellomo G, Indaco A, Chiasserini D,
Maderna E, Paolini Paoletti F, Gaetani L, Paciotti S, Petricciuolo
M, Tagliavini F, Giaccone G and Parnetti L: Machine learning driven
profiling of cerebrospinal fluid core biomarkers in Alzheimer's
disease and other neurological disorders. Front Neurosci.
15(647783)2021.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Hallqvist J, Bartl M, Dakna M, Schade S,
Garagnani P, Bacalini MG, Pirazzini C, Bhatia K, Schreglmann S,
Xylaki M and Weber S: Plasma proteomics identify biomarkers
predicting Parkinson's disease up to 7 years before symptom onset.
Nat Commun. 15(4759)2024.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Xu A, Kouznetsova VL and Tsigelny IF:
Alzheimer's disease diagnostics using mirna biomarkers and machine
learning. J Alzheimers Dis. 86:841–859. 2022.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Kumar A, Kouznetsova VL, Kesari S and
Tsigelny IF: Parkinson's disease diagnosis using miRNA biomarkers
and deep learning. Front Biosci (Landmark Ed). 29(4)2024.PubMed/NCBI View Article : Google Scholar
|
|
101
|
Lin CH, Chiu SI, Chen TF, Jang JS and Chiu
MJ: Classifications of neurodegenerative disorders using a
multiplex blood biomarkers-based machine learning model. Int J Mol
Sci. 21(6914)2020.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Khorsand B, Salehi S, Karimi S,
Karimipasand S, Fariborzi N, Houri H and Asri N: Investigating
Alzheimer's disease biomarkers by applying machine learning models
bioRxiv: Mar 21, 2025 doi: 10.1101/2025.03.19.643368.
|
|
103
|
Lam S, Arif M, Song X, Uhlén M and
Mardinoglu A: Machine learning analysis reveals biomarkers for the
detection of neurological diseases. Front Mol Neurosci.
15(889728)2022.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Yu X, Lai S, Chen H and Chen M:
Protein-protein interaction network with machine learning models
and multiomics data reveal potential neurodegenerative
disease-related proteins. Hum Mol Genet. 29:1378–1387.
2020.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Yang W, Xu S, Zhou M and Chan P:
Aging-related biomarkers for the diagnosis of Parkinson's disease
based on bioinformatics analysis and machine learning. Aging
(Albany NY). 16(12191)2024.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Mohammed EM, Fakhrudeen AM and Alani OY:
Detection of Alzheimer's disease using deep learning models: A
systematic literature review. Informatics Med Unlocked.
50(101551)2024.
|
|
107
|
Myszczynska MA, Ojamies PN, Lacoste AM,
Neil D, Saffari A, Mead R, Hautbergue GM, Holbrook JD and
Ferraiuolo L: Applications of machine learning to diagnosis and
treatment of neurodegenerative diseases. Nat Rev Neurol.
16:440–456. 2020.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Ahmed MR, Zhang Y, Feng Z, Lo B, Inan OT
and Liao H: Neuroimaging and machine learning for dementia
diagnosis: Recent advancements and future prospects. IEEE Rev
Biomed Eng. 12:19–33. 2018.PubMed/NCBI View Article : Google Scholar
|
|
109
|
Vieira S, Gong QY, Pinaya WH, Scarpazza C,
Tognin S, Crespo-Facorro B, Tordesillas-Gutierrez D, Ortiz-García
V, Setien-Suero E, Scheepers F, et al: Using machine learning and
structural neuroimaging to detect first episode psychosis:
Reconsidering the evidence. Schizophr Bull. 46:17–26.
2020.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Yassin W, Nakatani H, Zhu Y, Kojima M,
Owada K, Kuwabara H, Gonoi W, Aoki Y, Takao H, Natsubori T, et al:
Machine-learning classification using neuroimaging data in
schizophrenia, autism, ultra-high risk and first-episode psychosis.
Transl Psychiatry. 10(278)2020.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Salvatore C, Cerasa A, Battista P, Gilardi
MC, Quattrone A and Castiglioni I: Alzheimer's Disease Neuroimaging
Initiative. Magnetic resonance imaging biomarkers for the early
diagnosis of Alzheimer's disease: A machine learning approach.
Front Neurosci. 9(307)2015.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Nanni L, Interlenghi M, Brahnam S,
Salvatore C, Papa S, Nemni R and Castiglioni I: Alzheimer's Disease
Neuroimaging Initiative. Comparison of transfer learning and
conventional machine learning applied to structural brain MRI for
the early diagnosis and prognosis of Alzheimer's disease. Front
Neurol. 11(576194)2020.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Gill S, Mouches P, Hu S, Rajashekar D,
MacMaster FP, Smith EE, Forkert ND and Ismail Z: Alzheimer's
disease neuroimaging initiative. Using machine learning to predict
dementia from neuropsychiatric symptom and neuroimaging data. J
Alzheimers Dis. 75:277–288. 2020.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Murugan S, Venkatesan C, Sumithra MG, Gao
XZ, Elakkiya B, Akila M and Manoharan S: DEMNET: A deep learning
model for early diagnosis of Alzheimer diseases and dementia from
MR images. Ieee Access. 9:90319–90329. 2021.
|
|
115
|
Jo T, Nho K, Risacher SL and Saykin AJ:
Alzheimer's Neuroimaging Initiative. Deep learning detection of
informative features in tau PET for Alzheimer's disease
classification. BMC Bioinformatics. 21 (Suppl
21)(S496)2020.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Ramzan F, Khan MU, Rehmat A, Iqbal S, Saba
T, Rehman A and Mehmood Z: A deep learning approach for automated
diagnosis and multi-class classification of Alzheimer's disease
stages using resting-state fMRI and residual neural networks. J Med
Syst. 44(37)2019.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Voter AF, Larson ME, Garrett JW and Yu JP:
Diagnostic accuracy and failure mode analysis of a deep learning
algorithm for the detection of cervical spine fractures. AJNR Am J
Neuroradiol. 42:1550–1556. 2021.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Rava RA, Snyder KV, Mokin M, Waqas M,
Allman AB, Senko JL, Podgorsak AR, Bhurwani MS, Hoi Y, Siddiqui AH,
et al: Assessment of a Bayesian Vitrea CT perfusion analysis to
predict final infarct and penumbra volumes in patients with acute
ischemic stroke: A comparison with RAPID. AJNR Am J Neuroradiol.
41:206–212. 2020.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Kolanu N, Silverstone EJ, Ho BH, Pham H,
Hansen A, Pauley E, Quirk AR, Sweeney SC, Center JR and Pocock NA:
Clinical utility of computer-aided diagnosis of vertebral fractures
from computed tomography images. J Bone Miner Res. 35:2307–2312.
2020.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Koopman MS, Berkhemer OA, Geuskens RR,
Emmer BJ, van Walderveen MA, Jenniskens SF, van Zwam WH, van
Oostenbrugge RJ, van der Lugt A, Dippel DW, et al: Comparison of
three commonly used CT perfusion software packages in patients with
acute ischemic stroke. J Neurointerv Surg. 11:1249–1256.
2019.PubMed/NCBI View Article : Google Scholar
|
|
121
|
Christidi F, Karavasilis E, Samiotis K,
Bisdas S and Papanikolaou N: Fiber tracking: A qualitative and
quantitative comparison between four different software tools on
the reconstruction of major white matter tracts. Eur J Radiol Open.
3:153–161. 2016.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Persson K, Barca ML, Cavallin L, Brækhus
A, Knapskog AB, Selbæk G and Engedal K: Comparison of automated
volumetry of the hippocampus using NeuroQuant® and
visual assessment of the medial temporal lobe in Alzheimer's
disease. Acta Radiol. 59:997–1001. 2018.PubMed/NCBI View Article : Google Scholar
|
|
123
|
Kwon C, Kang KM, Byun MS, Yi D, Song H,
Lee JY, Hwang I, Yoo RE, Yun TJ, Choi SH, et al: Assessment of mild
cognitive impairment in elderly subjects using a fully automated
brain segmentation software. Invest Magnetic Resonance Imaging.
25:164–171. 2021.
|
|
124
|
Persson K, Selbæk G, Brækhus A, Beyer M,
Barca M and Engedal K: Fully automated structural MRI of the brain
in clinical dementia workup. Acta Radiol. 58:740–747.
2017.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Zaki LA, Vernooij MW, Smits M, Tolman C,
Papma JM, Visser JJ and Steketee RM: Comparing two artificial
intelligence software packages for normative brain volumetry in
memory clinic imaging. Neuroradiology. 64:1359–1366.
2022.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Elkin C, Nittala S and Devabhaktuni V:
Fundamental cognitive workload assessment: A machine learning
comparative approach. In: Advances in Neuroergonomics and Cognitive
Engineering: Proceedings of the AHFE 2017 International Conference
on Neuroergonomics and Cognitive Engineering, July 17-21,. 2017,
The Westin Bonaventure Hotel, Springer International Publishing,
Los Angeles, CA, pp275-284, 2018.
|
|
127
|
Bailey JD, Baker JC, Rzeszutek MJ and
Lanovaz MJ: Machine learning for supplementing behavioral
assessment. Perspect Behav Sci. 44:605–619. 2021.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Bleidorn W and Hopwood CJ: Using machine
learning to advance personality assessment and theory. Pers Soc
Psychol Rev. 23:190–203. 2019.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Javed AR, Fahad LG, Farhan AA, Abbas S,
Srivastava G, Parizi RM and Khan MS: Automated cognitive health
assessment in smart homes using machine learning. Sustainable
Cities Soc. 65(102572)2021.
|
|
130
|
Chandler C, Foltz PW, Cohen AS, Holmlund
TB, Cheng J, Bernstein JC, Rosenfeld EP and Elvevåg B: Machine
learning for ambulatory applications of neuropsychological testing.
Intelligence Based Med. 1(100006)2020.
|
|
131
|
Yuan D, Hahn S, Allgaier N, Owens MM,
Chaarani B, Potter A and Garavan H: Machine learning approaches
linking brain function to behavior in the ABCD STOP task. Hum Brain
Mapp. 44:1751–1766. 2023.PubMed/NCBI View Article : Google Scholar
|
|
132
|
Ophey A, Wenzel J, Paul R, Giehl K,
Rehberg S, Eggers C, Reker P, van Eimeren T, Kalbe E and
Kambeitz-Ilankovic L: Cognitive performance and learning parameters
predict response to working memory training in Parkinson's disease.
J Surg Case Rep. 12:2235–2247. 2022.PubMed/NCBI View Article : Google Scholar
|
|
133
|
McCutcheon RA, Keefe RS, McGuire PM and
Marquand A: Deconstructing cognitive impairment in psychosis with a
machine learning approach. JAMA Psychiatry. 82:57–65.
2025.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Kim SY, Park J, Choi H, Loeser M, Ryu H
and Seo K: Digital marker for early screening of mild cognitive
impairment through hand and eye movement analysis in virtual
reality using machine learning: First validation study. J Med
Internet Res. 25(e48093)2023.PubMed/NCBI View
Article : Google Scholar
|
|
135
|
Zhang Y, He X, Chan YH, Teng Q and
Rajapakse JC: Multi-modal graph neural network for early diagnosis
of Alzheimer's disease from sMRI and PET scans. Comput Biol Med.
164(107328)2023.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Lee G, Nho K, Kang B, Sohn KA and Kim D:
Predicting Alzheimer's disease progression using multi-modal deep
learning approach. Sci Rep. 9(1952)2019.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Liu X, Li W, Miao S, Liu F, Han K and
Bezabih TT: HAMMF: Hierarchical attention-based multi-task and
multi-modal fusion model for computer-aided diagnosis of
Alzheimer's disease. Comput Biol Med. 176(108564)2024.PubMed/NCBI View Article : Google Scholar
|
|
138
|
Wang M, Shao W, Huang S and Zhang D:
Hypergraph-regularized multimodal learning by graph diffusion for
imaging genetics based alzheimer's disease diagnosis. Med Image
Anal. 89(102883)2023.PubMed/NCBI View Article : Google Scholar
|
|
139
|
Zhu Y, Zhu X, Kim M, Yan J, Kaufer D and
Wu G: Dynamic hyper-graph inference framework for computer-assisted
diagnosis of neurodegenerative diseases. IEEE Trans Med Imaging.
38:608–616. 2018.PubMed/NCBI View Article : Google Scholar
|
|
140
|
Castellano G, Esposito A, Lella E,
Montanaro G and Vessio G: Automated detection of Alzheimer's
disease: A multi-modal approach with 3D MRI and amyloid PET. Sci
Rep. 14(5210)2024.PubMed/NCBI View Article : Google Scholar
|
|
141
|
Chatterjee I and Bansal V: LRE-MMF: A
novel multi-modal fusion algorithm for detecting neurodegeneration
in Parkinson's disease among the geriatric population. Exp
Gerontol. 197(112585)2024.PubMed/NCBI View Article : Google Scholar
|
|
142
|
Chen H, Guo H, Xing L, Chen D, Yuan T,
Zhang Y and Zhang X: Multimodal predictive classification of
Alzheimer's disease based on Attention-combined fusion network:
Integrated neuroimaging modalities and medical examination data.
IET Image Processing. 17:3153–3164. 2023.
|
|
143
|
Huang G, Li R, Bai Q and Alty J:
Multimodal learning of clinically accessible tests to aid diagnosis
of neurodegenerative disorders: A scoping review. Health Inf Sci
Syst. 11(32)2023.PubMed/NCBI View Article : Google Scholar
|