1. Introduction
Cancer is a medical condition in which somatic cells
undergo genetic or epigenetic changes, causing aberrant cell
proliferation that can spread to other parts of the body (1). Breast cancer (BC) is one of the most
detrimental and heterogeneous diseases in modern times, affecting a
large number of individuals globally. It is the second most common
type of cancer among females worldwide, accounting for
685,000-related deaths (16% of the total female cancer-related
deaths) (2,3). In India, BC has become the most
frequent type of cancer among women, particularly in younger age
groups (4). BC is a heterogeneous
disease with various clinical, molecular and biological features;
some tumors exhibit a high estrogen receptor expression, which
influences the methods of treatment, as well as the prognosis of
patients (5). Numerous factors
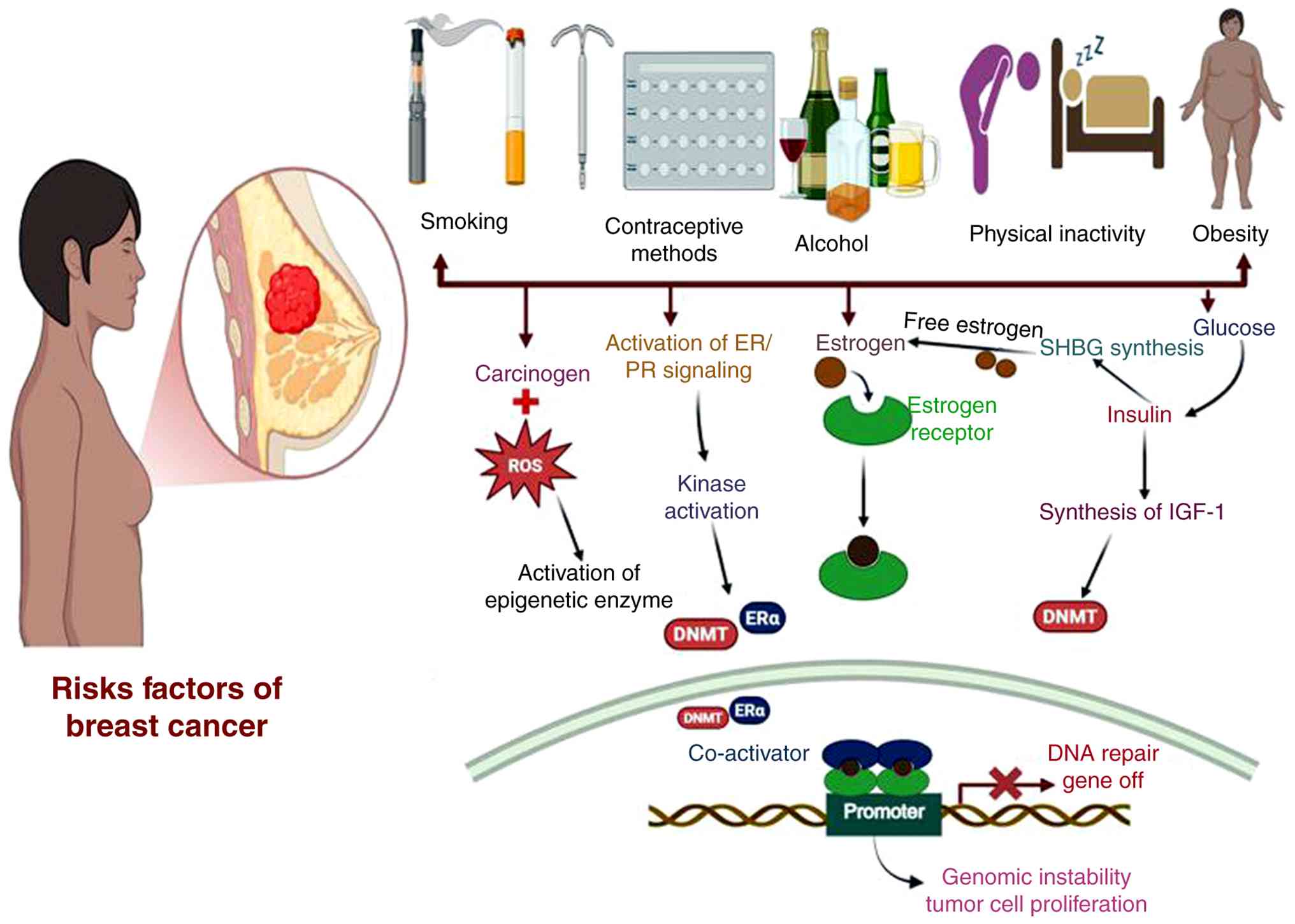
influence the development of breast tumors, as illustrated in
Fig. 1.
The processes of epigenetic modification [such as
DNA methylation, the post-transcriptional modification of histone
proteins and regulation through non-coding RNAs (ncRNAs)] are
critical in controlling the normal functions of a cell throughout
the developmental and differentiation states (6). These processes also help cells adapt
to environmental changes, including dietary changes or exposures to
chemicals or radiation. It is known that the action of cigarette
smoke and hormones is responsible for inducing epigenetic changes.
However, aberrant epigenetic changes lead to the development of
cancer (7). A key mechanism of
gene inactivation in breast cancer is promoter CpG island
hypermethylation. which targets CG-rich regions associated with
promoters of protein-coding genes (8). This epigenetic damage inactivates
several cellular processes, such as DNA repair, leading to mutator
pathways. Notably, patients with breast cancer frequently exhibit
hypermethylation of DNA repair genes, including breast cancer gene
1 (BRCA1), breast cancer gene 2 (BRCA2),
Abraxas1/BRCA1 A complex subunit (ABRA1), MRE11,
BRCA1-associated RING domain 1 (BARD1), mediator of DNA
damage checkpoint 1 (MDC1), ring finger protein (RNF)168,
UBC13, partner and localizer of BRCA2 (PALB2), RAD50,
RAD51, RAD51C, RNF8, NBS1, CtIP and ATM. The ability of
cells to respond to DNA damage is disrupted by epigenetic
alterations, resulting in increased tumor growth. Given that
epigenetic alterations are potentially reversible, they are an
attractive target for the development of treatments (9). Surgical intervention, chemotherapy
and radiation therapy continue to form the backbone of treatment
options for breast cancer; however, there are limitations to their
effectiveness, including dose-dependent toxicity, limited
selectivity, the potential for treatment resistance and significant
side-effects leading to the damage of non-cancerous cells.
Metastatic disease remains one of the key challenges of treatment
and is the principal cause of mortality in women with breast cancer
(10). Dietary phytochemicals
(natural compounds derived from plants) are being recognized as
potentially effective chemopreventive and therapeutic agents for
the treatmetn of breast cancer due to their low toxicity levels,
minimal side effects, affordability, and ease of access in
comparison to synthetic pharmaceuticals (11). Natural plant extracts, such as
Piper longum, Curcuma longa, Withania somnifera, Nigella sativa,
Amora rohituka and Dimocarpus longan contain anticancer
compounds with demonstrated anti-BC characteristics (12). This approach is particularly
promising as epigenetic changes, unlike genetic mutations, are
reversible and influence early cancer progression. Although this
exhibits immense potential, a knowledge gap currently exists in the
research of phytochemicals and their role in epigenetics in BC. The
present review thus aimed to fill this gap in the current
literature. The present review discusses the findings from the most
contemporary literature available with regard to the reversals of
hypermethylation in DNA repair genes by phytochemicals and their
mechanisms of action in BC.
2. Epigenetic modifications in breast
cancer
The term ‘epigenetics’, originally used by
Waddington in 1942, describes genetic, reversible modifications in
gene expression without changing the DNA sequence (13). Breast cancer often involves
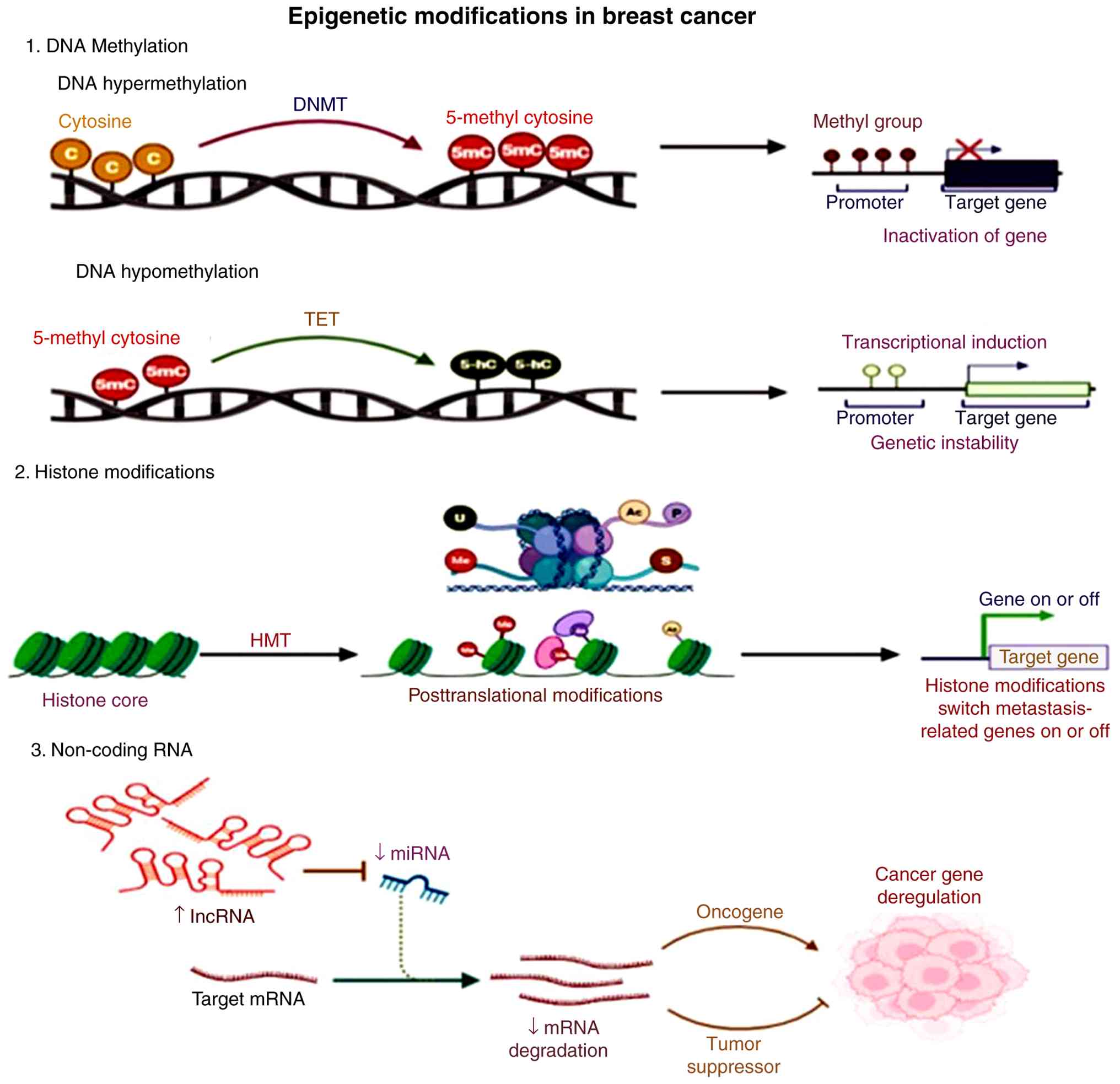
epigenetic reprogramming despite its genetic origin (14). These epigenetic alterations include
DNA methylation, histone modifications and ncRNA expression
(15), as illustrated in Fig. 2.
DNA methylation
DNA methylation is an epigenetic process linked to
cell development and various critical activities. Aberrant
methylation patterns have been observed in cancer cell genomes
(16). It is a reversible process
in which methyl groups are introduced to the fifth carbon position
of the cytosine from S-adenosyl methionine. Of note, two types of
methylation exist: Maintenance methylation (when CpG dinucleotides
on a single strand of DNA are methylated) and de novo
methylation (when CpG dinucleotides on both of the strands are
unmethylated) (17). DNA
methylation is a reversible process facilitated by DNA
methyltransferases (DNMTs). DNMT1, DNMT2, DNMT3A and
DNMT3B encode proteins with different functional
specificities. As previously demonstrated, two patterns of aberrant
methylation have been identified: Global hypomethylation throughout
the genome and hypermethylation at CpG islands within promoter
regions (18,19).
DNA hypomethylation
Global DNA hypomethylation, a cancer hallmark
coexisting with focal CpG island hypermethylation, involves
genome-wide loss of cytosine methylation (20). Mechanistically driven by an
impaired DNMT1 maintenance, deregulated DNMT3A/3B
activity and aberrant TET-mediated demethylation, and reduced
5-methylcytosine levels are associated with tumor progression and a
poor survival (21). Functionally,
hypomethylation destabilizes heterochromatin, causing chromosomal
instability. It reactivates transposable elements, inducing
mutagenesis, and aberrantly activates oncogenes and
metastasis-promoting genes (22).
DNA hypermethylation
The aberrant addition of methyl groups to cytosines
in CpG dinucleotides, particularly within promoter-associated CpG
islands, is a hallmark epigenetic alteration in cancer, causing
stable gene silencing without altering the DNA sequence. In cancer,
~5-10% of promoter CpG islands become hypermethylated, repressing
tumor suppressor genes involved in cell cycle control, DNA repair
and apoptosis (e.g., CDKN2A, MLH1 and BRCA1),
contributing to oncogenesis (23,24).
These reversible changes provide targets for epigenetic therapy, as
well as biomarkers for cancer diagnosis, prognosis and therapeutic
stratification (25). Both
modifications have commonly been documented in BC and are greatly
affected by environmental factors such as aging, stress, alcohol
consumption and air pollution (26).
Histone modifications
DNA wraps around a histone octamer in the nucleus.
The histone tails are sensitive to several covalent
posttranslational modifications (PTMs) that regulate chromatin
state. Changes in histone PTM patterns have been widely associated
with cancer (27). Key
modifications include the following:
Histone phosphorylation. The addition of
phosphate groups promoting chromatin relaxation, critical for DNA
damage responses (notably γ-H2AX at Ser139), transcriptional
regulation and mitotic chromosome condensation (28).
Histone acetylation. Acetylation by histone
acetyltransferases leads to decreased histone-DNA interaction,
creating euchromatin. This is strongly associated with gene
transcription activation. Histone deacetylases remove acetyl
groups, compacting chromatin and inhibiting transcription (29).
Histone ubiquitylation. H2A ubiquitination
(Lys119) usually causes gene repression, while H2B ubiquitination
(Lys120) aids transcription and is required prior to methylation
(30).
Histone sumoylation. SUMO conjugation drives
transcriptional repression and chromatin compaction, often
antagonizing acetylation and ubiquitination (31).
Histone methylation. This occurs on lysine
and arginine residues with variable outcomes. Activating markers
include H3K4me3; repressive markers include H3K9me3 and H3K27me3.
Histone methyltransferases and demethylases control methylation
dynamics (32).
ncRNA expression
Non-coding RNAs lack protein-coding ability, and
~52% of the human genome has been encoded, although only 1.2%
codify proteins. ncRNAs are categorized into ‘housekeeping ncRNAs’
(tRNAs, rRNAs, snRNAs and snoRNAs) and ‘regulatory ncRNAs’
(lncRNAs, >200 nucleotides; and short ncRNAs, <200
nucleotides) (33,34). They play a critical role in
controlling signaling pathways linked to tumor development, spread
and therapeutic resistance (35).
Short non-coding RNAs (sncRNAs). snRNAs are a
diverse group under ~200 nucleotides, including microRNAs (miRNAs),
small interfering RNAs (siRNAs) and PIWI-interacting RNAs (piRNAs),
that regulate gene expression primarily at the post-transcriptional
level. These molecules contribute to chromatin modulation, cell
differentiation, stress responses and disease processes (36,37).
Long non-coding RNAs (lncRNAs). These RNA
species are longer than 200 nt without protein-coding potential
(33). The human transcriptome has
~10,000 lncRNAs. They function as oncogenes or tumor suppressors,
affecting cancer cell growth, apoptosis, metabolic processes,
epithelial-mesenchymal transition (EMT), metastasis and treatment
resistance. lncRNAs are usually exclusive to a particular tumor
subtype, and BC exhibits abnormal expression levels of several
lncRNAs (34,38).
3. Hypermethylated DNA repair genes in
breast cancer
DNA repair processes play a crucial role in
preserving genomic stability and limiting the growth of mutations
that may lead to cancer (39). In
BC, these mechanisms are frequently affected not only by genetic
mutations, but also by epigenetic alterations, particularly
promoter hypermethylation, that can silence essential DNA repair
genes without affecting their DNA sequence (40). In BC, promoter hypermethylation is
known to silence certain key DNA repair genes, as discussed below
and presented in Table I (41-67).
 | Table IRole of hypermethylated DNA repair
genes in breast cancer. |
Table I
Role of hypermethylated DNA repair
genes in breast cancer.
| Gene | Function in DNA
repair | Impact of
hypermethylation in breast cancer | Locus | (Refs.) |
|---|
| MSH6 | Forms MutSα complex
with MSH2 to repair base mismatches and small
insertion/deletion loops, preserving genomic stability. | Promoter
hypermethylation in both DCIS and IDC leads to early gene
silencing, impairing mismatch repair and promoting genomic
instability. | 2p16 | (41,42) |
| MDC1 | Scaffold protein in
DNA damage response, binds γH2AX at DSBs, recruits MRN complex,
ATM, RNF8, BRCA1, 53BP1, amplifies ATM signaling |
Hypermethylation/downregulation linked to
radioresistance, nodal failure, poor prognosis, and genomic
instability | 6p21.3 | (43,44) |
| RNF168 | E3 ubiquitin
ligase; ubiquitinates H2A at K13/K15, recruits 53BP1, BRCA1, PALB2,
promotes homologous recombination (HR) | Hypermethylation
may reduce expression, impair DNA repair, increase sensitivity to
DNA damage, or drive endocrine resistance | 3q29 | (45,46) |
| RAD51 | Key protein in
homologous recombination repair (HRR), mediates HR intermediate
formation and resolution | Promoter
hypermethylation causes gene silencing, seen in TNBC, linked to HR
deficiency, and poor prognosis | 17q23 | (47,48) |
| MLH1 | Key component of
the mismatch repair (MMR) system, forms a heterodimer with
PMS2 to correct DNA replication errors | Promoter
hypermethylation silences the gene, seen in ~43.5% of breast
cancers; linked to advanced stages, genomic instability, and
disease progression | 3p22.2 | (49,50) |
| RAD50 | key element of the
MRN complex that enables checkpoint activation, telomere
maintenance, and DSB repair through HR and Non-Homologous End
Joining (NHEJ) | Promoter
hypermethylation reduces expression, linked to impaired DNA repair
and increased breast malignancy risk | 5q31.1 | (51,52) |
| MRE11 | MRN complex member;
detects DSBs, initiates end resection, activates ATM, supports
HR/NHEJ | Reduced expression
(~31%) impairs repair; linked to BRCA1/ATM deregulation and
increased risk | 11q21 | (53,54) |
| BRCC36 | BRCA1-A complex
DUB; removes K63-linked ubiquitin, essential for BRCA1 activation
and 53BP1 recruitment | Overexpressed in
breast tumors; may disrupt BRCA1 function; silencing increases
radiosensitivity | Xq28 | (55,56) |
| BRCA1 | HR repair; RAD51
recruitment | Well-established
promoter hypermethylation in sporadic breast cancer → BRCA1
silencing & HRD | 17q12-21 | (57-59) |
| BRCA2 | HR mediator; loads
RAD51 | Promoter
hypermethylation in subsets, including DCIS → reduced HR | 13q12 | (41,59,60) |
| PALB2 | Bridges
BRCA1 and BRCA2; facilitates RAD51 loading, thereby
enabling HR repair of DSBs. | Rare but reported
promoter methylation → reduced HR competence in subsets | 16p12.2 | (61,62) |
| ATM | Serine/threonine
kinase that senses DSBs and activates DDR: phosphorylates key
effectors(e.g., p53, BRCA1, CHK2), triggering cell-cycle
checkpoints, DNA repair, or apoptosis. | Hypermethylation →
downregulation, larger tumors and an advanced stage | 11q22.3 | (63-65) |
| RNF8 | E3 ligase promotes
K63-Ub chains to recruit repair proteins | Direct methylation
is rare; aberrant expression, mostly upregulation, contributes to
EMT and chemoresistance | 6p21.3 | (66,67) |
MutS homolog 6 (MSH6)
MSH6 partners with MSH2 to form the
MutSα complex, which recognizes and repairs base-base mismatches
during DNA replication. In BC, promoter hypermethylation silences
MSH6 expression in both ductal carcinoma in situ
(DCIS) and invasive carcinomas, compromising mismatch repair (MMR)
fidelity and elevating mutation rates (41,42).
TCGA bioinformatic analyses have demonstrated inverse correlations
between promoter methylation and gene expression, establishing
MSH6 loss as an early driver of genomic instability in
breast tumorigenesis (42,68).
MDC1
MDC1 functions as a molecular scaffold at DNA
double-strand breaks (DSBs), binding phosphorylated histone H2AX
(γH2AX) and orchestrating the recruitment of the MRN complex,
ATM kinase, RNF8, BRCA1 and 53BP1.
Promoter-associated transcriptional downregulation in BC is
associated with genomic instability, radioresistance, nodal
metastasis and adverse clinical outcomes. In silico network
analyses position MDC1 as a central signaling hub within the
ATM-BRCA1-RNF8 axis, where its functional loss accelerates
DNA damage accumulation and impairs checkpoint responses (43,44).
RNF168
RNF168 (3q29), an E3 ubiquitin ligase,
catalyzes histone H2A ubiquitination at lysines 13 and 15,
facilitating the recruitment of 53BP1, BRCA1 and
PALB2 to DSB sites for homologous recombination (HR).
TCGA-BRCA and METABRIC dataset analyses reveal that promoter
hypermethylation suppresses RNF168 expression, resulting in
HR deficiency, heightened sensitivity to DNA-damaging agents and
potential endocrine therapeutic resistance (45,46).
TCGA data indicate RNF168 overexpression in
BRCA1-mutant BCs and in BCs with HR deficiency (HRD), driven
mainly by copy-number amplification and enriched in basal-like
tumors. A high expression of RNF168 is associated with HRD
mutational signature 3 and a worse survival, while lower
RNF168 levels are associated with improved outcomes and a
reduced risk of developing BC in BRCA1 mutation carriers
(69).
RAD51
RAD51 (17q23) is the central recombinase in
homologous recombination repair, mediating strand invasion and the
resolution of HR intermediates. Integrative analyses of TCGA and
DNA methylation arrays identify promoter hypermethylation-mediated
RAD51 silencing predominantly in triple-negative breast
cancer (TNBC), where it is associated with HR-deficiency molecular
signatures and reduced transcript abundance (47,48).
TCGA and drug-response datasets demonstrate that a low expression
of RAD51 is associated with a poor overall survival, but
increased sensitivity to DNA-damaging chemotherapy, particularly in
BC, reflecting homologous recombination deficiency that can be
therapeutically exploited (70).
MLH1
MLH1 heterodimerizes with PMS2 to
execute mismatch repair, correcting replication errors including
base mismatches and insertion-deletion loops. Promoter
hypermethylation silences MLH1 in ~43.5% of BCs, as
demonstrated by multi-platform in silico analyses
demonstrating strong inverse methylation-expression correlations
(49,50). The loss of MLH1 precipitates
MMR deficiency, microsatellite instability, elevated tumor
mutational burden, and the progression toward advanced, genomically
unstable disease states (71).
RAD50
RAD50 is a core part of the ATPase-containing
MRN complex, which is critical for the recognition of DSBs, the
activation of ATM, telomere maintenance, and selection of DSB
repair pathways between HR and non-homologous end joining (NHEJ)
repair (51,52). Multi-omics analysis has
demonstrated that the hypermethylation of promoters suppresses
RAD50 transcription and disrupts the stability of the MRN
complex, resulting in improper DSB signaling, genomic instability
and an increased risk of developing BC. RAD50 suppression is
particularly implicated in TNBC carcinogenesis and chemotherapeutic
resistance (72).
MRE11
MRE11 provides the nuclease activity within the MRN
complex, initiating DNA end resection, activating ATM-dependent
checkpoints, and supporting both HR and NHEJ pathways (53,54).
Previously, TCGA-BRCA analyses demonstrated a reduced MRE11
expression in a subset of breast cancers, while copy-number
alterations affecting MRE11 were observed at a lower frequency. A
low expression of MRE11 was shown to be associated with
differential gene expression and pathway enrichment indicating
impaired ATM signaling and homologous recombination,
consistent with genomic instability and aggressive tumor phenotypes
(73).
BRCC36 (BRCC3)
BRCC36 (Xq28) is a K63-specific
deubiquitinase within the BRCA1-A complex that removes ubiquitin
chains to fine-tune BRCA1 activation and regulate 53BP1 recruitment
during DSB repair pathway selection (55,56).
TCGA-BRCA transcriptomic data have revealed the
dysregulation/overexpression of BRCC36 in BC (74). Network and pathway analyses have
linked altered BRCC36 levels to a perturbed BRCA1-A complex
function and ubiquitin-dependent DSB repair, with computational
models suggesting the modulation of DSB pathway choice and
potential radiosensitization upon BRCC36 inhibition
(74).
BRCA1
BRCA1 is a tumor suppressor orchestrating
homologous recombination, cell cycle checkpoints, and
transcriptional regulation. In sporadic BC, promoter
hypermethylation constitutes a major mechanism of BRCA1
silencing, producing HR deficiency, triple-negative phenotype
enrichment, and synthetic lethality with PARP inhibitors (57-59).
Integrated bioinformatics analyses of BC cohorts have demonstrated
that BRCA1 promoter hypermethylation is tightly linked to
reduced BRCA1 expression and
homologous-recombination-deficient genomic and transcriptomic
signatures, closely mirroring BRCA1-mutated tumors and
supporting epigenetic BRCA1 silencing as a functional driver
of HR deficiency (75).
BRCA2
BRCA2 functions as a tumor suppressor that
regulates RAD51 nucleoprotein filament assembly during
HR-mediated DSB repair. Promoter hypermethylation represents an
early and frequent event in breast carcinogenesis, detected even in
DCIS, where it induces gene silencing, genomic instability and
repair incompetence (41,60). TCGA-based methylation and
integrative bioinformatic analyses link BRCA2 promoter
hypermethylation in pre-invasive breast lesions to homologous
recombination deficiency signatures and increased genomic
instability (76).
PALB2
PALB2 serves as a molecular bridge connecting
BRCA1 and BRCA2, facilitating RAD51 loading
and enabling efficient HR. While germline PALB2 mutations
confer hereditary breast cancer susceptibility, somatic promoter
hypermethylation also suppresses expression in a subset of sporadic
cases, resulting in HR deficiency and disease progression (61,62).
Bioinformatics methylome profiling in early-onset BC samples has
revealed increased methylation at PALB2 promoter-associated
CpG probes in tumor DNA compared with matched blood DNA, consistent
with potential epigenetic modulation of PALB2 expression in
a subset of cases (77).
ATM
ATM encodes a master checkpoint kinase that
phosphorylates numerous substrates, including p53, BRCA1 and
CHK2 in response to DSBs, orchestrating cell cycle arrest
and repair pathway activation. Promoter hypermethylation reduces
ATM expression in BC, and is associated with a larger tumor
size, advanced stage and an older patient age (63-65).
TCGA-based repair signature analyses have associated ATM
silencing with checkpoint deficiency and impaired DSB repair
capacity, supporting its utility as a prognostic and predictive
biomarker in sporadic disease (78).
RNF8
RNF8 (6p21.3) is an E3 ubiquitin ligase
initiating the ubiquitin cascade required for BRCA1 and
53BP1 recruitment to DSBs, promoting high-fidelity HR, while
suppressing mutagenic NHEJ. Paradoxically, RNF8 is
frequently overexpressed in BC, where it drives EMT, metastasis and
chemoresistance through the activation of the Twist and β-catenin
signaling pathways (66,67). Bioinformatics analyses of BC
datasets has demonstrated that RNF8 promoter methylation is rare;
however, RNF8 is transcriptionally dysregulated and embedded
in DDR networks linked to BRCA1-RAD51-ATM signaling,
indicating an altered homologous recombination and DDR pathway
balance (79).
4. Phytochemicals causing the reversal of
hypermethylation in DNA repair genes
The hypermethylation of functionally significant
genes has been observed in breast carcinoma; silencing of these
genes is crucial for carcinogenesis and tumor development (40). Treatment options for BC include
surgery, chemoradiotherapy, hormone treatments, monoclonal
antibodies, immunotherapy, nanomedicines and small molecule
inhibitors. Conventional medicines have limitations, including
resistance, a low efficacy and adverse effects, which limit their
clinical uses. Anticancer medications derived from plants that have
limited or no adverse effects may be a beneficial alternative to
chemotherapy (12). The ‘phyto’ in
the word phytochemicals is derived from the Greek word, meaning
‘plant’. Therefore, phytochemicals are plant compounds (80). Phytochemicals are plant-based
bioactive molecules that plants manufacture to protect themselves.
Over a thousand phytochemicals have been identified to date and can
be obtained from various foods, including whole grains, fruits,
vegetables, nuts, and herbs (81).
Phytochemicals primarily inhibit BC by reducing the
proliferation of cells, inducing apoptosis, limiting cancer cell
dissemination, inhibiting angiogenesis and impairing cancerous cell
migration. These chemicals have also been shown to improve the
curative effectiveness of other anticancer medications, sensitize
cells to radiation, and prevent resistance to therapy in malignant
tissue (82). Epigenetic
alterations are modulated by phytochemicals as inhibiting DNMTs
(83), modulating histone
acetylation and methylation, and affecting ncRNAs (84). Some of the phytochemicals with
targeted genes and their mechanisms are presented in Table II (85-97).
 | Table IIPhytochemicals targeting DNA repair
genes through epigenetic modulation in breast cancer. |
Table II
Phytochemicals targeting DNA repair
genes through epigenetic modulation in breast cancer.
| Phytochemical | Source (plant
part) | Target gene | Mechanism | (Refs.) |
|---|
| Curcumin | Turmeric
(rhizome) | BRCA2 ↓,
RAD51 ↓, PARP1 modulation | In vitro
studies report reduced BRCA2 and RAD51 stability,
suggesting homologous recombination deficiency and increased
PARP-inhibitor sensitivity | (85-87) |
| Genistein | Soybeans
(seeds) | BRCA1 ↑,
BRCA2 ↑, ATM ↑ (transcriptional); MLH1
demethylation | In vitro and
limited in vivo data suggest DNMT inhibition and partial
promoter demethylation, leading to reactivation of BRCA1 and
MLH1 | (88,89) |
| Resveratrol | Grapes, berries,
red wine (skin, seeds) | RAD51 | In vitro
evidence indicates RAD51 downregulation, suggesting impaired
homologous recombination and increased DNA-damage sensitivity | (90) |
| Quercetin | Apples, onions,
grapes (fruits, peels) | RAD51,
Ku70, XRCC1 (DNA repair mediators) | In vitro
studies report reduced expression of multiple DNA repair genes,
suggesting global suppression of repair capacity | (91) |
| Thymoquinone | Black cumin
(seeds) | RAD51, Ku70,
XRCC1 (DNA repair) | In vitro
findings suggest decreased repair gene expression and increased DNA
damage markers, indicating impaired DNA repair | (91) |
| Berberine | Barberry
(roots/bark) |
XRCC1-mediated BER; BRCA1
axis (sensitization) | Preclinical studies
report inhibition of XRCC1-mediated BER, sensitizing cells
to DNA-damaging agents; BRCA1 effects are indirect. | (92) |
| Epigallocatechin
gallate | Green tea
(leaves) | DNMT1 ↓ →
MLH1 re-expression, BRCA1 promoter demethylation
reported | In vitro
studies demonstrate DNMT1 inhibition and re-expression of
MLH1; BRCA1 demethylation has been reported in select
models | (93,94) |
| Withaferin-A | Ashwagandha
(leaves, roots) | BRCA1 ↓,
ATR/ATM-DDR modulation | Preclinical
evidence suggests BRCA1 degradation and ATM/ATR-DDR
modulation, resulting in reduced homologous recombination | (95-97) |
5. Critical analysis, limitations and future
directions
Although DNA repair gene hypermethylation is widely
reported in BC, much of the evidence remains correlative as it
relies heavily on in vitro studies and bulk bioinformatic
datasets that fail to reflect the true biological complexity of
tumors. Large-scale methylation analyses demonstrate extensive
inter- and intra-tumoral epigenetic heterogeneity, indicating that
simplified models cannot fully capture patient-specific variation
(98). Moreover, the BC
microenvironment, including stromal and immune components, strongly
influences epigenetic states, yet this context is largely absent
from current experimental systems (99). Methodological variability across
methylation assays, histone modification profiling, and ncRNA
detection platforms further contributes to inconsistent and
difficult-to-compare findings in the epigenetics literature
(100). Notably, hypermethylation
in several DNA repair genes, including BRCA1, BRCA2, RAD51,
MLH1, MSH6, ATM, and components of the MRN complex
(MRE11-RAD50-NBS1), is frequently reported in BC; yet,
direct evidence demonstrating that these epigenetic alterations
consistently translate into functional protein loss or impaired DNA
repair activity remains limited (101). A number of published studies rely
on simplified experimental systems, including in vitro cell
line models, simplified animal models and bulk bioinformatic
analyses, which do not adequately capture the cellular
heterogeneity, tumor microenvironment and overall biological
complexity of primary breast tumors (14,26,78).
For this reason, determining the functional impact of
hypermethylation may be challenging as it remains unclear how this
process will affect cellular behavior. In vitro examples
have demonstrated that promoter hypermethylation can be reversed,
for example, with genes such as BRCA1, XRCC1, ATM and
RAD51; however, the majority of these studies are not
supported by comprehensive functional assays, either in terms of
confirming successful restoration of DNA repair function or
establishing the stability, reproducibility and long-term
biological impact of such reversions. Furthermore, in the majority
of cases, reversal of hypermethylation, in addition to being
unstable, may not have any biological significance in the long
term.
Such uncertainties in scientific research are
inclusive of the challenges related to the developing a medicinal
approach targeting hypermethylation reversal, particularly those
which are phytochemical-based. Variability in phytochemical
composition with respect to species, environmental factors, storage
and processing is very high, thus rendering standardization
difficult (102). Moreover, study
designs for establishing direct causality between phytochemical
consumption and subsequent cancer repression are not very feasible
due to confounding variables, such as dietary habits (103). The limited understanding of
phytochemical pharmacokinetics/pharmacodynamics, in addition, adds
complexity to predictive models of safety, efficacy and interaction
with conventional therapies (92).
Bioavailability is an issue with a number of phytochemicals, in
which a large absorption, low solubility, rapid metabolism and poor
tissue biodistribution frequently require supraphysiological
concentrations in vitro, which are not attainable in
vivo (104). The precise
biochemical mechanisms for their actions on epigenetic targets have
not yet been fully understood, and numerous compounds have
pleotropic or context-dependent effects that limit their
reliability as targeted epigenetic modulators (82).
Collectively, these limitations suggest more
integrated and rigorous research is required. Future studies
should, whenever possible, evaluate not only methylation and gene
expression, but also quantify endogenous protein levels, protein
abundance and functional DNA repair restoration in clinically
relevant models. In addition, enhancing the phytochemical
pharmacokinetics and developing standardized extraction protocols,
the use of combinatorial therapeutic approaches might help in
overcoming current barriers (105). Ultimately, well-designed clinical
trials with clearly defined molecular clinical endpoints become the
key to unlocking the true therapeutic potential of DNA repair gene
reversal and hypermethylation in breast cancer.
6. Conclusion
BC is a major concern that arises due to both
hereditary and epigenetic modifications, with hypermethylation of
DNA repair genes playing a critical role in carcinogenesis in BC.
Such epigenetic modifications lead to the inactivation of tumor
suppressor genes, resulting in DNA repair systems and promoting
genomic instability. Patients with cancer usually undergo
unconventional treatment methods, which have weaknesses, including
toxicity, resistance and negative side-effects. However, the use of
phytochemicals appears to be a promising strategy to reverse such
aberrant epigenetic modifications in BC, having multiple
advantages, including low toxicity, cost-effectiveness and
potential synergy with standard treatments. Phytochemicals are
natural molecules found in fruits, vegetables and medicinal plants
that are essential components of the human diet and have a large
impact in the direction of therapy of cancer. The present review
discussed some phytochemicals, such as curcumin, that have shown a
committing effect in targeting BC hypermethylated DNA repair
genes.
Despite their potential however, several
restrictions and challenges related to variability in the compound
composition of phytochemicals, low bioavailability, difficult
standardization and reproducibility, and a lack of clinical
validation prevent them from being widely used. Future studies are
thus required to elucidate the epigenetic effects of BC in order to
overcome these obstacles. This will aid in the development of more
individualized targets for phytochemical-based therapies, which
also provide a low-toxicity method of reactivating genes involved
in DNA repair and improving patient outcomes.
Acknowledgements
The authors acknowledge the support provided by the
School of Biosciences and Technology, Galgotias University, Greater
Noida, India, in terms of access to library resources, online
databases, and research facilities for the preparation of this
review.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
TG wrote the main manuscript, and AKJ provided
conceptual guidance. Both authors reviewed, and have read and
approved the final version of the manuscript. Data authentication
is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Saini A, Kumar M, Bhatt S, Saini V and
Malik A: Cancer causes and treatments. Int J Pharm Sci Res.
11:3121–3134. 2020.
|
|
2
|
Fatima N, Liu L, Hong S and Ahmed H:
Prediction of breast cancer, comparative review of machine learning
techniques, and their analysis. IEEE Access. 8:150360–150376.
2020.
|
|
3
|
Mathur R, Jha NK, Saini G, Jha SK, Shukla
SP, Filipejová Z, Kesari KK, Iqbal D, Nand P, Upadhye VJ, et al:
Epigenetic factors in breast cancer therapy. Front Genet.
13(886487)2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mehrotra R and Yadav K: Breast cancer in
India: Present scenario and the challenges ahead. World J Clin
Oncol. 13(209)2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mohanad M, Hamza HM, Bahnassy AA, Shaarawy
S, Ahmed O, El-Mezayen HA, Ayad EG, Tahoun N and Abdellateif MS:
Molecular profiling of breast cancer methylation pattern in triple
negative versus non-triple negative breast cancer. Sci Rep.
15(6894)2025.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Buocikova V, Rios-Mondragon I, Pilalis E,
Chatziioannou A, Miklikova S, Mego M, Pajuste K, Rucins M, Yamani
NE, Longhin EM, et al: Epigenetics in breast cancer therapy-new
strategies and future nanomedicine perspectives. Cancers (Basel).
12(3622)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Klibaner-Schiff E, Simonin EM, Akdis CA,
Cheong A, Johnson MM, Karagas MR, Kirsh S, Kline O, Mazumdar M,
Oken E, et al: Environmental exposures influence multigenerational
epigenetic transmission. Clin Epigenetics. 16(145)2024.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kang Z, Wang J, Liu J, Du L and Liu X:
Epigenetic modifications in breast cancer: From immune escape
mechanisms to therapeutic target discovery. Front Immunol.
16(1584087)2025.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Watanabe Y, Maeda I, Oikawa R, Wu W,
Tsuchiya K, Miyoshi Y, Itoh F, Tsugawa KI and Ohta T: Aberrant DNA
methylation status of DNA repair genes in breast cancer treated
with neoadjuvant chemotherapy. Genes Cells. 18:1120–1130.
2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chiou YS, Li S, Ho CT and Pan MH:
Prevention of breast cancer by natural phytochemicals: Focusing on
molecular targets and combinational strategy. Mol Nutr Food Res.
62(e1800392)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Abbasi BA, Iqbal J, Mahmood T, Khalil AT,
Ali B, Kanwal S, Shah SA and Ahmad R: Role of dietary
phytochemicals in modulation of miRNA expression: Natural swords
combating breast cancer. Asian Pac J Trop Med. 11:501–509.
2018.
|
|
12
|
Sohel M, Aktar S, Biswas P, Amin MA,
Hossain MA, Ahmed N, Mim MIH, Islam F and Mamun AA: Exploring the
anti-cancer potential of dietary phytochemicals for the patients
with breast cancer: A comprehensive review. Cancer Med.
12:14556–14583. 2023.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Thuy LHA, Thuan LD and Phuong TK: DNA
hypermethylation in breast cancer. In: Breast Cancer-From Biology
to Medicine. IntechOpen, 2017.
|
|
14
|
Flores-García LC, Rubio K, Ibarra-Sierra
E, Silva-Cázares MB, Palma-Flores C and López-Camarillo C:
Epigenetic and transcriptional reprogramming in 3D culture models
in breast cancer. Cancers (Basel). 17(3830)2025.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Park M, Kim D, Ko S, Kim A, Mo K and Yoon
H: Breast cancer metastasis: Mechanisms and therapeutic
implications. Int J Mol Sci. 23(6806)2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Smith ZD, Hetzel S and Meissner A: DNA
methylation in mammalian development and disease. Nat Rev Genet.
26:7–30. 2025.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Karami Fath M, Azargoonjahromi A, Kiani A,
Jalalifar F, Osati P, Akbari Oryani M, Shakeri F, Nasirzadeh F,
Khalesi B, Nabi-Afjadi M, et al: The role of epigenetic
modifications in drug resistance and treatment of breast cancer.
Cell Mol Biol Lett. 27(52)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Jha AK, Nikbakht M, Jain V, Sehgal A,
Capalash N and Kaur J: Promoter hypermethylation of p73 and p53
genes in cervical cancer patients among north Indian population.
Mol Biol Rep. 39:9145–9157. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Agarwal N and Jha AK: DNA hypermethylation
of tumor suppressor genes among oral squamous cell carcinoma
patients: A prominent diagnostic biomarker. Mol Biol Rep.
52(44)2024.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Fu J, Qin T, Li C, Zhu J, Ding Y, Zhou M,
Yang Q, Liu X, Zhou J and Chen F: Research progress of LINE-1 in
the diagnosis, prognosis, and treatment of gynecologic tumors.
Front Oncol. 13(1201568)2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chen C, Wang Z, Ding Y, Wang L, Wang S,
Wang H and Qin Y: DNA methylation: From cancer biology to clinical
perspectives. Front Biosci (Landmark Ed). 27(326)2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Besselink N, Keijer J, Vermeulen C,
Boymans S, de Ridder J, van Hoeck A, Cuppen E and Kuijk E: The
genome-wide mutational consequences of DNA hypomethylation. Sci
Rep. 13(6874)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jung I, An J and Ko M: Epigenetic
regulators of DNA cytosine modification: Promising targets for
cancer therapy. Biomedicines. 11(654)2023.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Heery R and Schaefer MH: DNA methylation
variation along the cancer epigenome and the identification of
novel epigenetic driver events. Nucleic Acids Res. 49:12692–12705.
2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Dunican DS, Mjoseng HK, Duthie L, Flyamer
IM, Bickmore WA and Meehan RR: Bivalent promoter hypermethylation
in cancer is linked to the H327me3/H3K4me3 ratio in embryonic stem
cells. BMC Biol. 18(25)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Vietri MT, D'elia G, Benincasa G, Ferraro
G, Caliendo G, Nicoletti GF and Napoli C: DNA methylation and
breast cancer: A way forward (Review). Int J Oncol.
59(98)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Duan X, Xing Z, Qiao L, Qin S, Zhao X,
Gong Y and Li X: The role of histone post-translational
modifications in cancer and cancer immunity: Functions, mechanisms
and therapeutic implications. Front Immunol.
15(1495221)2024.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Gong P, Guo Z, Wang S, Gao S and Cao Q:
Histone phosphorylation in DNA damage response. Int J Mol Sci.
26(2405)2025.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Fang Z, Wang X, Sun X, Hu W and Miao QR:
The role of histone protein acetylation in regulating endothelial
function. Front Cell Dev Biol. 9(672447)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Baweja L and Wereszczynski J: Mechanistic
basis for the opposing effects of H2A and H2B ubiquitination on
nucleosome stability and dynamics. bioRxiv [Preprint]:
2025.02.13.638112, 2025.
|
|
31
|
Ryu HY and Hochstrasser M: Histone
sumoylation and chromatin dynamics. Nucleic Acids Res.
49:6043–6052. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Millán-Zambrano G, Burton A, Bannister AJ
and Schneider R: Histone post-translational modifications-cause and
consequence of genome function. Nat Rev Genet. 23:563–580.
2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
George TP, Subramanian S and Supriya MH: A
brief review of noncoding RNA. Egypt J Med Hum Genet.
25(98)2024.
|
|
34
|
Amelio I, Bernassola F and Candi E:
Emerging roles of long non-coding RNAs in breast cancer biology and
management. Semin Cancer Biol. 72:36–45. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Keyvani-Ghamsari S, Khorsandi K, Rasul A
and Zaman MK: Current understanding of epigenetics mechanism as a
novel target in reducing cancer stem cells resistance. Clin
Epigenetics. 13(120)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Jouravleva K and Zamore PD: A guide to the
biogenesis and functions of endogenous small non-coding RNAs in
animals. Nat Rev Mol Cell Biol. 26:347–370. 2025.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Xiong Q and Zhang Y: Small RNA
modifications: Regulatory molecules and potential applications. J
Hematol Oncol. 16(64)2023.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Lu C, Wei D, Zhang Y, Wang P and Zhang W:
Long non-coding RNAs as potential diagnostic and prognostic
biomarkers in breast cancer: progress and prospects. Front Oncol.
11(710538)2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Hopkins JL, Lan L and Zou L: DNA repair
defects in cancer and therapeutic opportunities. Genes Dev.
36:278–293. 2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kundu S, Sarkar S, Ghosh S and Chowdhury
AA: DNA methylation as an oncogenic driver in breast cancer:
Therapeutic targeting via epigenetic reprogramming of DNA
methyltransferases. Biochem Pharmacol. 242(117313)2025.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Moelans CB, Verschuur-Maes AH and Van
Diest PJ: Frequent promoter hypermethylation of BRCA2, CDH13, MSH6,
PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast
cancer. J Pathol. 225:222–231. 2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Aravind Kumar M, Naushad SM, Narasimgu N,
Nagaraju Naik S, Kadali S, Shanker U and Lakshmi Narasu M: Whole
exome sequencing of breast cancer (TNBC) cases from India:
Association of MSH6 and BRIP1 variants with TNBC risk and oxidative
DNA damage. Mol Biol Rep. 45:1413–1419. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zou R, Zhong X, Wang C, Sun H, Wang S, Lin
L, Sun S, Tong C, Luo H, Gao P, et al: MDC1 enhances estrogen
receptor-mediated transactivation and contributes to breast cancer
suppression. Int J Biol Sci. 11:992–1005. 2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Torres Esteban M: Structural and
Functional Characterization of Two Phosphorylation Site Clusters in
the DNA Damage Response Adaptor Protein MDC1. Doctoral
Dissertation, University of Zurich, 2024.
|
|
45
|
Kelliher J, Ghosal G and Leung JWC: New
answers to the old RIDDLE: RNF168 and the DNA damage response
pathway. FEBS J. 289:2467–2480. 2022.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Xie T, Qin H, Yuan Z, Zhang Y, Li X and
Zheng L: Emerging roles of RNF168 in tumor progression. Molecules.
28(1417)2023.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Pettitt S, Proszek P, Torga G, Gulati A,
Llop-Guevara A, Jank P, Felder B, Rodriguez A, Cooke S,
Garcia-Murillas I, et al: 435 (PB423): Persistence of BRCA1 and
RAD51C methylation after neoadjuvant chemotherapy in high risk
TNBC. Eur J Cancer. 211 (Suppl 1)(S114944)2024.
|
|
48
|
Tabano S, Azzollini J, Pesenti C, Lovati
S, Costanza J, Fontana L, Peissel B, Miozzo M and Manoukian S:
Analysis of BRCA1 and RAD51C promoter methylation in Italian
families at high-risk of breast and ovarian cancer. Cancers
(Basel). 12(910)2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Malik SS, Zia A, Mubarik S, Masood N,
Rashid S, Sherrard A, Khan MB and Khadim MT: Correlation of MLH1
polymorphisms, survival statistics, in silico assessment and gene
downregulation with clinical outcomes among breast cancer cases.
Mol Biol Rep. 47:683–692. 2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Dasgupta H, Islam S, Alam N, Roy A,
Roychoudhury S and Panda CK: Hypomethylation of mismatch repair
genes MLH1 and MSH2 is associated with chemotolerance of breast
carcinoma: Clinical significance. J Surg Oncol. 119:88–100.
2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Heng J, Zhang F, Guo X, Tang L, Peng L,
Luo X, Xu X, Wang S, Dai L and Wang J: Integrated analysis of
promoter methylation and expression of telomere related genes in
breast cancer. Oncotarget. 8(25442)2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Abad E, Civit L, Potesil D, Zdrahal Z and
Lyakhovich A: Enhanced DNA damage response through RAD50 in triple
negative breast cancer resistant and cancer stem-like cells
contributes to chemoresistance. FEBS J. 288:2184–2202.
2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Borde V: The multiple roles of the Mre11
complex for meiotic recombination. Chromosome Res. 15:551–563.
2007.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lu R, Zhang H, Jiang YN, Wang ZQ, Sun L
and Zhou ZW: Post-translational modification of MRE11: Its
implication in DDR and diseases. Genes (Basel).
12(1158)2021.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Block-Schmidt AS, Dukowic-Schulze S,
Wanieck K, Reidt W and Puchta H: BRCC36A is epistatic to BRCA1 in
DNA crosslink repair and homologous recombination in Arabidopsis
thaliana. Nucleic Acids Res. 39:146–154. 2011.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Rabl J, Bunker RD, Schenk AD, Cavadini S,
Gill ME, Abdulrahman W, Andrés-Pons A, Luijsterburg MS, Ibrahim
AFM, Branigan E, et al: Structural basis of BRCC36 function in DNA
repair and immune regulation. Mol Cell. 75:483–497.e9.
2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Rosen EM, Fan S, Pestell RG and Goldberg
ID: BRCA1 gene in breast cancer. J Cell Physiol. 196:19–41.
2003.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Tarsounas M and Sung P: The
antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication.
Nat Rev Mol Cell Biol. 21:284–299. 2020.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Stefansson OA and Esteller M: Epigenetic
modifications in breast cancer and their role in personalized
medicine. Am J Pathol. 183:1052–1063. 2013.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Fishchuk L, Rossokha Z, Lobanova O,
Cheshuk V, Vereshchako R, Vershyhora V, Medvedieva N, Dubitskaa O
and Gorovenko N: Hypermethylation of the BRCA2 gene promoter and
its co-hypermethylation with the BRCA1 gene promoter in patients
with breast cancer. Cancer Biomark. 40:275–283. 2024.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Pauty J, Rodrigue A, Couturier A, Buisson
R and Masson JY: Exploring the roles of PALB2 at the crossroads of
DNA repair and cancer. Biochem J. 460:331–342. 2014.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Potapova A, Hoffman AM, Godwin AK,
Al-Saleem T and Cairns P: Promoter hypermethylation of the PALB2
susceptibility gene in inherited and sporadic breast and ovarian
cancer. Cancer Res. 68:998–1002. 2008.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Khanna KK: Cancer risk and the ATM gene: A
continuing debate. J Natl Cancer Inst. 92:795–802. 2000.PubMed/NCBI View Article : Google Scholar
|
|
64
|
He C, Kawaguchi K and Toi M: DNA damage
repair functions and targeted treatment in breast cancer. Breast
Cancer. 27:355–362. 2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Vo QN, Kim WJ, Cvitanovic L, Boudreau DA,
Ginzinger DG and Brown KD: Erratum: The ATM gene is a target for
epigenetic silencing in locally advanced breast cancer. Oncogene.
24(1964)2005.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Zhou T, Yi F, Wang Z, Guo Q, Liu J, Bai N,
Li X, Dong X, Ren L, Cao L and Song X: The functions of DNA damage
factor RNF8 in the pathogenesis and progression of cancer. Int J
Biol Sci. 15:909–918. 2019.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Lee HJ, Li CF, Ruan D, Powers S, Thompson
PA, Frohman MA and Chan CH: The DNA damage transducer RNF8
facilitates cancer chemoresistance and progression through twist
activation. Mol Cell. 63:1021–1033. 2016.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Hacking S, Chou C, Baykara Y, Wang Y, Uzun
A and Gamsiz Uzun ED: MMR deficiency defines distinct molecular
subtype of breast cancer with histone proteomic networks. Int J Mol
Sci. 24(5327)2023.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Patel PS, Abraham KJ, Guturi KK, Halaby
MJ, Khan Z, Palomero L, Ho B, Duan S, St-Germain J, Algouneh A, et
al: RNF168 regulates R-loop resolution and genomic stability in
BRCA1/2-deficient tumors. J Clin Invest.
131(e140105)2021.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Liu H and Weng J: A pan-cancer
bioinformatic analysis of RAD51 regarding the values for diagnosis,
prognosis, and therapeutic prediction. Front Oncol.
12(858756)2022.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Yan Y, Wang Y, Tang J, Liu X, Wang J, Song
G and Li H: Comprehensive analysis of oncogenic somatic alterations
of mismatch repair gene in breast cancer patients. Bioengineering
(Basel). 12(426)2025.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Karamat U and Ejaz S: Overexpression of
RAD50 is the marker of poor prognosis and drug resistance in breast
cancer patients. Current Cancer Drug Targets. 21:163–176.
2021.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Alblihy A, Shoqafi A, Toss MS, Algethami
M, Harris AE, Jeyapalan JN, Abdel-Fatah T, Servante J, Chan SYT,
Green A, et al: Untangling the clinicopathological significance of
MRE11-RAD50-NBS1 complex in sporadic breast cancers. NPJ Breast
Cancer. 7(143)2021.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Mohamed RI, Bargal SA, Mekawy AS,
El-Shiekh I, Tuncbag N, Ahmed AS, Badr E and Elserafy M: The
overexpression of DNA repair genes in invasive ductal and lobular
breast carcinomas: Insights on individual variations and precision
medicine. PLoS One. 16(e0247837)2021.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Glodzik D, Bosch A, Hartman J, Aine M,
Vallon-Christersson J, Reuterswärd C, Karlsson A, Mitra S, Niméus
E, Holm K, et al: Comprehensive molecular comparison of BRCA1
hypermethylated and BRCA1 mutated triple negative breast cancers.
Nat Commun. 11(3747)2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Pang JMB, Deb S, Takano EA, Byrne DJ, Jene
N, Boulghourjian A, Holliday A, Millar E, Lee CS, O'Toole SA, et
al: Methylation profiling of ductal carcinoma in situ and its
relationship to histopathological features. Breast Cancer Res.
16(423)2014.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Scott CM, Joo JE, O'Callaghan N, Buchanan
DD, Clendenning M, Giles GG, Hopper JL, Wong EM and Southey MC:
Methylation of breast cancer predisposition genes in early-onset
breast cancer: Australian breast cancer family registry. PLoS One.
11(e0165436)2016.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Knijnenburg TA, Wang L, Zimmermann MT,
Chambwe N, Gao GF, Cherniack AD, Fan H, Shen H, Way GP, Greene CS,
et al: Genomic and molecular landscape of DNA damage repair
deficiency across the cancer genome atlas. Cell Rep. 23:239–254.
2018.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Liu C, Kuang J, Wang Y, Duan T, Min L, Lu
C, Zhang T, Chen R, Wu Y and Zhu L: A functional reference map of
the RNF8 interactome in cancer. Biol Direct. 17(17)2022.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Jayakumari S: Phytochemicals and
pharmaceutical: Overview: Recent progress and future applications.
In: Advances in Pharmaceutical Biotechnology. Springer Singapore,
pp163-173, 2020.
|
|
81
|
Kumar A, P N, Kumar M, Jose A, Tomer V, Oz
E, Proestos C, Zeng M, Elobeid T, K S and Oz F: Major
phytochemicals: Recent advances in health benefits and extraction
method. Molecules. 28(887)2023.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Bhattacharya T, Dutta S, Akter R, Rahman
MH, Karthika C, Nagaswarupa HP, Murthy HCA, Fratila O, Brata R and
Bungau S: Role of phytonutrients in nutrigenetics and nutrigenomics
perspective in curing breast cancer. Biomolecules.
11(1176)2021.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Pop S, Enciu AM, Tarcomnicu I, Gille E and
Tanase C: Phytochemicals in cancer prevention: Modulating
epigenetic alterations of DNA methylation. Phytochem Rev.
18:1005–1024. 2019.
|
|
84
|
Hsieh HH, Kuo MZ, Chen IA, Lin CJ, Hsu V,
Huangfu WC and Wu TY: Epigenetic modifications as novel therapeutic
strategies of Cancer chemoprevention by phytochemicals. Pharm Res.
42:69–78. 2025.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Guney Eskiler G, Sahin E, Deveci Ozkan A,
Cilingir Kaya OT and Kaleli S: Curcumin induces DNA damage by
mediating homologous recombination mechanism in triple negative
breast cancer. Nutr Cancer. 72:1057–1066. 2020.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Choi YE and Park E: Curcumin enhances
poly(ADP-ribose) polymerase inhibitor sensitivity to chemotherapy
in breast cancer cells. J Nutr Biochem. 26:1442–1447.
2015.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Komar ZM, Ladan MM, Verkaik NS, Dahmani A,
Montaudon E, Marangoni E, Kanaar R, Nonnekens J, Houtsmuller AB,
Jager A and van Gent DC: Curcumin induces homologous recombination
deficiency by BRCA2 degradation in breast cancer and normal cells.
Cancers (Basel). 17(2109)2025.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Xie Q, Bai Q, Zou LY, Zhang QY, Zhou Y,
Chang H, Yi L, Zhu JD and Mi MT: Genistein inhibits DNA methylation
and increases expression of tumor suppressor genes in human breast
cancer cells. Genes Chromosomes Cancer. 53:422–431. 2014.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Donovan MG, Selmin OI, Doetschman TC and
Romagnolo DF: Epigenetic activation of BRCA1 by genistein in vivo
and triple negative breast cancer cells linked to antagonism toward
aryl hydrocarbon receptor. Nutrients. 11(2559)2019.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Sharma M and Tollefsbol TO: Combinatorial
epigenetic mechanisms of sulforaphane, genistein and sodium
butyrate in breast cancer inhibition. Exp Cell Res.
416(113160)2022.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Karimian A, Majidinia M, Moliani A, Alemi
F, Asemi Z, Yousefi B and Naghibi AF: The modulatory effects of two
bioflavonoids, quercetin and thymoquinone on the expression levels
of DNA damage and repair genes in human breast, lung and prostate
cancer cell lines. Pathol Res Pract. 240(154143)2022.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Gao X, Wang J, Li M, Wang J, Lv J, Zhang
L, Sun C, Ji J, Yang W, Zhao Z and Mao W: Berberine attenuates
XRCC1-mediated base excision repair and sensitizes breast cancer
cells to the chemotherapeutic drugs. J Cell Mol Med. 23:6797–6804.
2019.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H,
Welsh W and Yang CS: Tea polyphenol (-)-epigallocatechin-3-gallate
inhibits DNA methyltransferase and reactivates methylation-silenced
genes in cancer cell lines. Cancer Res. 63:7563–7570.
2003.PubMed/NCBI
|
|
94
|
Mirza S, Sharma G, Parshad R, Gupta SD,
Pandya P and Ralhan R: Expression of DNA methyltransferases in
breast cancer patients and to analyze the effect of natural
compounds on DNA methyltransferases and associated proteins. J
Breast Cancer. 16:23–31. 2013.PubMed/NCBI View Article : Google Scholar
|
|
95
|
Zhang X, Timmermann B, Samadi AK and Cohen
MS: Withaferin a induces proteasome-dependent degradation of breast
cancer susceptibility gene 1 and heat shock factor 1 proteins in
breast cancer cells. ISRN Biochem. 2012(707586)2012.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Hahm ER, Lee J, Abella T and Singh SV:
Withaferin A inhibits expression of ataxia telangiectasia and
Rad3-related kinase and enhances sensitivity of human breast cancer
cells to cisplatin. Mol Carcinog. 58:2139–2148. 2019.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Yu Y, Katiyar SP, Sundar D, Kaul Z, Miyako
E, Zhang Z, Kaul SC, Reddel RR and Wadhwa R: Withaferin-A kills
cancer cells with and without telomerase: Chemical, computational
and experimental evidences. Cell Death Dis. 8(e2755)2017.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Batra RN, Lifshitz A, Vidakovic AT, Chin
SF, Sati-Batra A, Sammut SJ, Provenzano E, Ali HR, Dariush A, Bruna
A, et al: DNA methylation landscapes of 1538 breast cancers reveal
a replication-linked clock, epigenomic instability and
cis-regulation. Nat Commun. 12(5406)2021.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Yang J, Xu J, Wang W, Zhang B, Yu X and
Shi S: Epigenetic regulation in the tumor microenvironment:
Molecular mechanisms and therapeutic targets. Signal Transduct
Target Ther. 8(210)2023.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Sherif ZA, Ogunwobi OO and Ressom HW:
Mechanisms and technologies in cancer epigenetics. Front Oncol.
14(1513654)2025.PubMed/NCBI View Article : Google Scholar
|
|
101
|
McCarthy-Leo C, Darwiche F and Tainsky MA:
DNA repair mechanisms, protein interactions and therapeutic
targeting of the MRN complex. Cancers (Basel).
14(5278)2022.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Gupta VK, Singh R and Sharma B:
Phytochemicals mediated signalling pathways and their implications
in cancer chemotherapy: Challenges and opportunities in
phytochemicals based drug development: A review. Biochem Comp.
5:1–5. 2017.
|
|
103
|
Alaouna M, Penny C, Hull R, Molefi T,
Chauke-Malinga N, Khanyile R, Makgoka M, Bida M and Dlamini Z:
Overcoming the challenges of phytochemicals in triple negative
breast cancer therapy: The path forward. Plants (Basel).
12(2350)2023.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Singh VK, Arora D, Ansari MI and Sharma
PK: Phytochemicals based chemopreventive and chemotherapeutic
strategies and modern technologies to overcome limitations for
better clinical applications. Phytother Res. 33:3064–3089.
2019.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Koklesova L, Jakubikova J, Cholujova D,
Samec M, Mazurakova A, Šudomová M, Pec M, Hassan STS, Biringer K,
Büsselberg D, et al: Phytochemical-based nanodrugs going beyond the
state-of-the-art in cancer management-targeting cancer stem cells
in the framework of predictive, preventive, personalized medicine.
Front Pharmacol. 14(1121950)2023.PubMed/NCBI View Article : Google Scholar
|