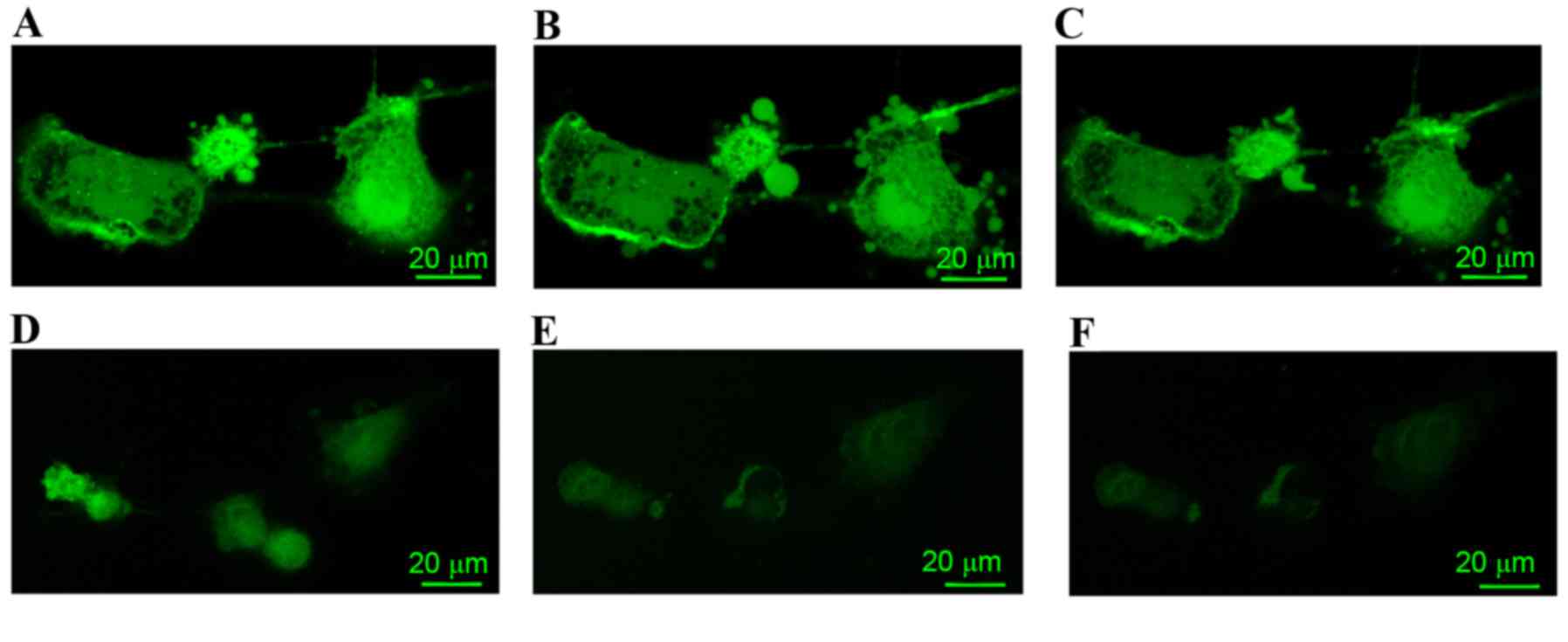
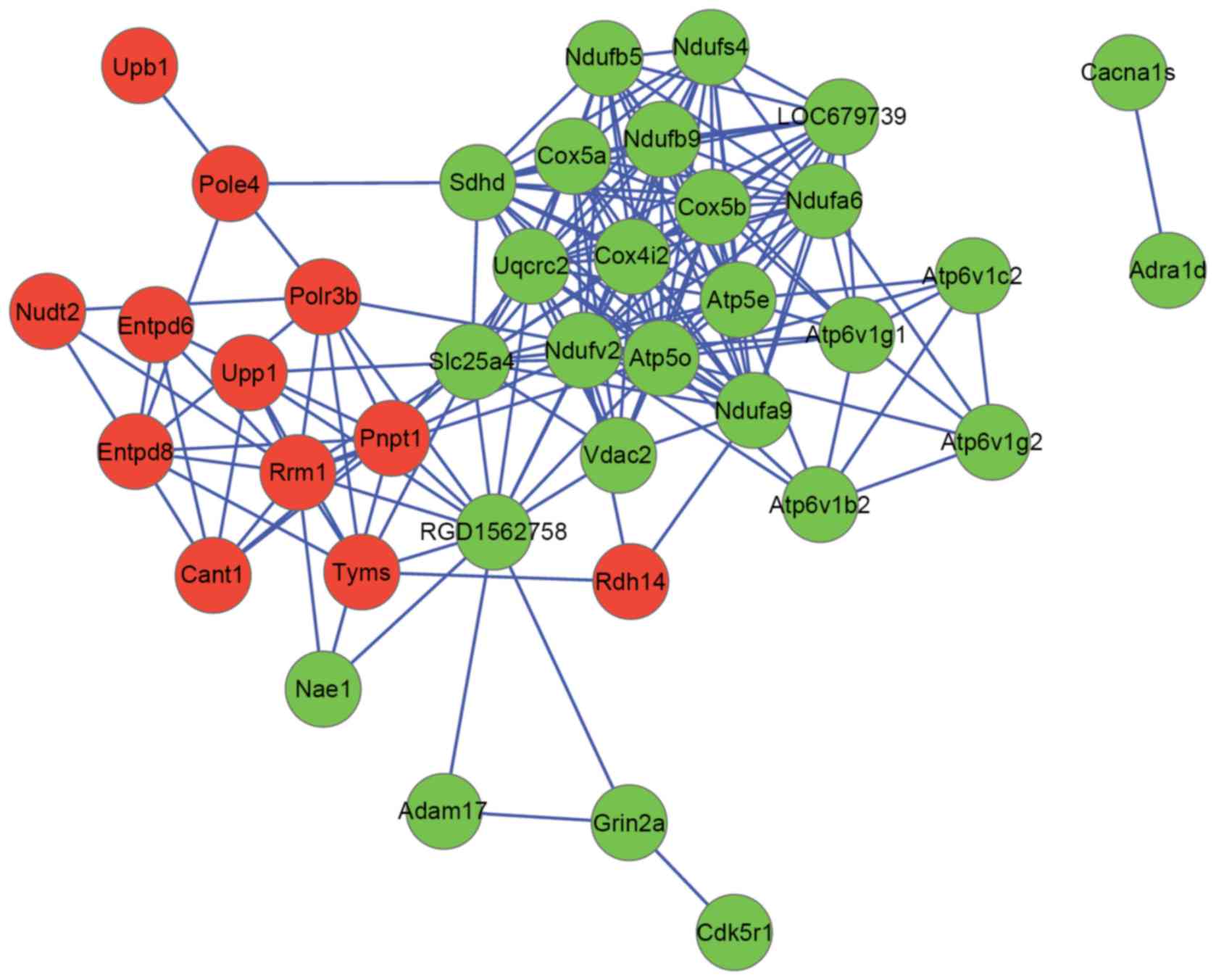
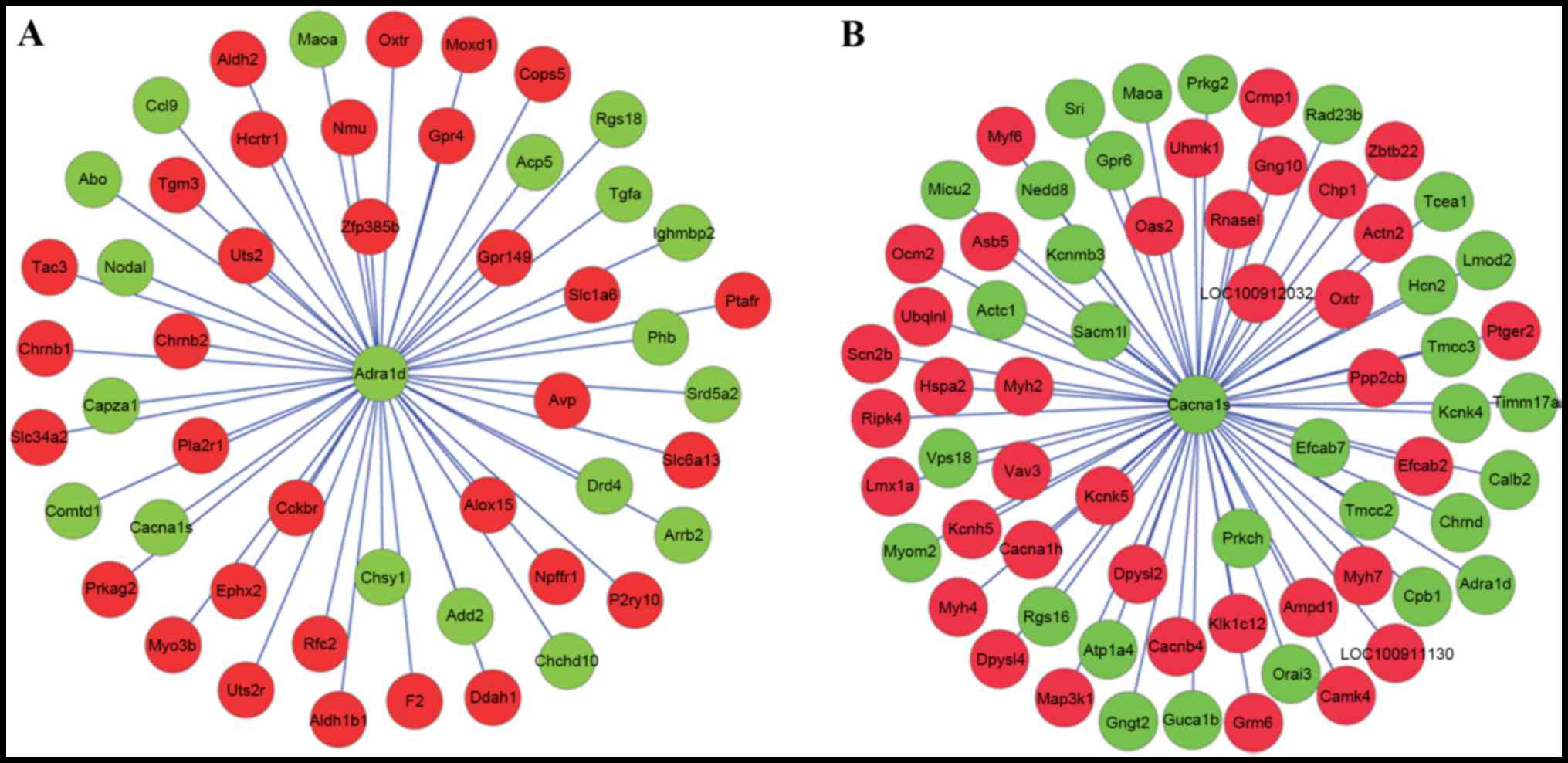
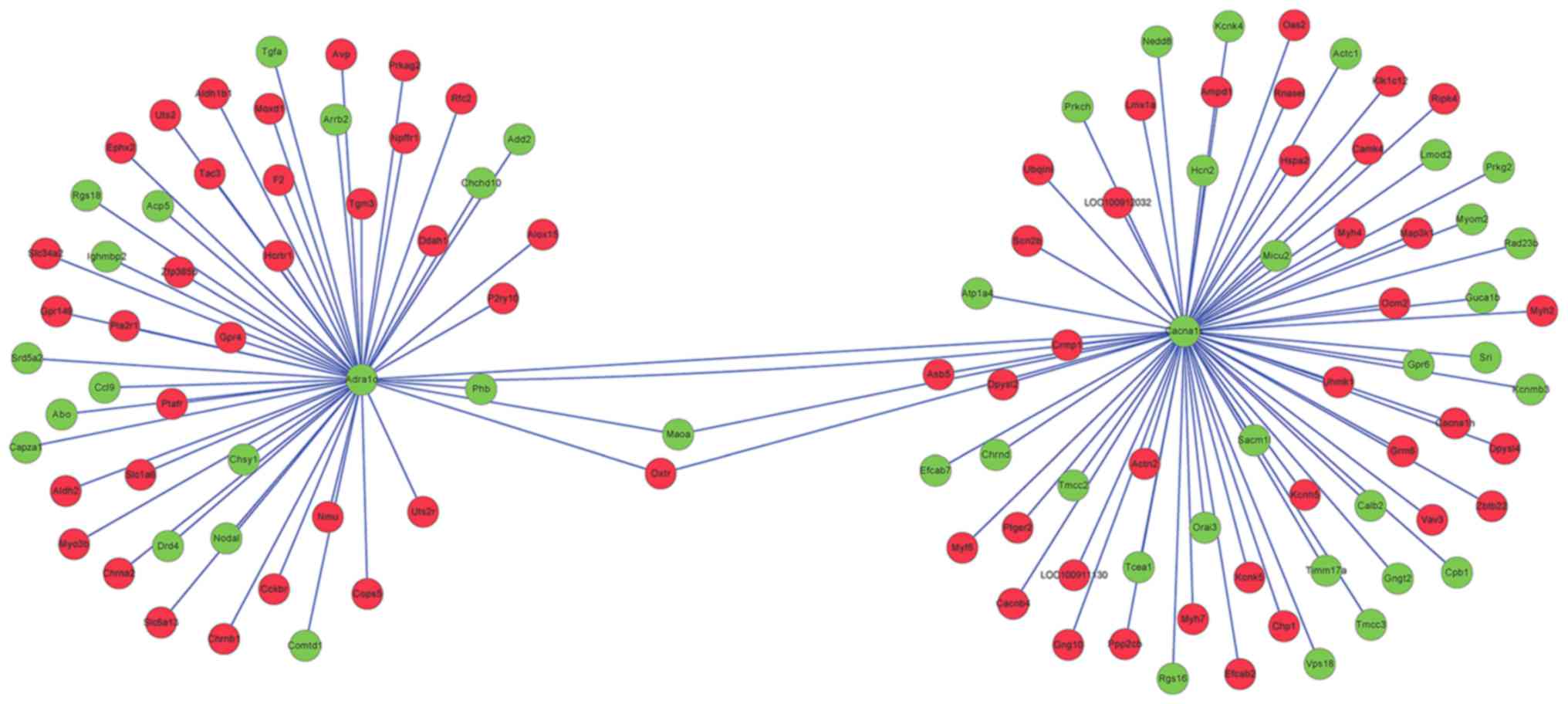
|
1
|
Owens GK: Regulation of differentiation of
vascular smooth muscle cells. Physiol Rev. 75:487–517.
1995.PubMed/NCBI
|
|
2
|
Fukata Y, Amano M and Kaibuchi K:
Rho-Rho-kinase pathway in smooth muscle contraction and
cytoskeletal reorganization of non-muscle cells. Trends Pharmacol
Sci. 22:32–39. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arita Y, Kihara S, Ouchi N, Maeda K,
Kuriyama H, Okamoto Y, Kumada M, Hotta K, Nishida M, Takahashi M,
et al: Adipocyte-derived plasma protein adiponectin acts as a
platelet-derived growth factor-BB-binding protein and regulates
growth factor-induced common postreceptor signal in vascular smooth
muscle cell. Circulation. 105:2893–2898. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Heo SK, Yun HJ, Noh EK, Park WH and Park
SD: LPS induces inflammatory responses in human aortic vascular
smooth muscle cells via Toll-like receptor 4 expression and nitric
oxide production. Immunol Lett. 120:57–64. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Irani K: Oxidant signaling in vascular
cell growth, death, and survival a review of the roles of reactive
oxygen species in smooth muscle and endothelial cell mitogenic and
apoptotic signaling. Circ Res. 87:179–183. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eltzschig HK and Carmeliet P: Hypoxia and
inflammation. N Engl J Med. 364:656–665. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Deten A, Volz HC, Holzl A, Briest W and
Zimmer HG: Effect of propranolol on cardiac cytokine expression
after myocardial infarction in rats. Mol Cell Biochem. 251:127–137.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Davel AP, Fukuda LE, De Sá LL, Munhoz CD,
Scavone C, Sanz-Rosa D, Cachofeiro V, Lahera V and Rossoni LV:
Effects of isoproterenol treatment for 7 days on inflammatory
mediators in the rat aorta. Am J Physiol Heart Circ Physiol.
295:H211–H219. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu Y, Tuerxun A, Hui Y and Abliz P:
Effects of propranolol and isoproterenol on infantile hemangioma
endothelial cells in vitro. Exp Ther Med. 8:647–651.
2014.PubMed/NCBI
|
|
10
|
Romacho T, Azcutia V, Vázquez-Bella M,
Matesanz N, Cercas E, Nevado J, Carraro R, Rodríguez-Mañas L,
Sánchez-Ferrer CF and Peiró C: Extracellular PBEF/NAMPT/visfatin
activates pro-inflammatory signalling in human vascular smooth
muscle cells through nicotinamide phosphoribosyltransferase
activity. Diabetologia. 52:2455–2463. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cuff CA, Kothapalli D, Azonobi I, Chun S,
Zhang Y, Belkin R, Yeh C, Secreto A, Assoian RK, Rader DJ and Puré
E: The adhesion receptor CD44 promotes atherosclerosis by mediating
inflammatory cell recruitment and vascular cell activation. J Clin
Invest. 108:10312001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dichtl W, Dulak J, Frick M, Alber HF,
Schwarzacher SP, Ares MP, Nilsson J, Pachinger O and Weidinger F:
HMG-CoA reductase inhibitors regulate inflammatory transcription
factors in human endothelial and vascular smooth muscle cells.
Arterioscler Thromb Vasc Biol. 23:58–63. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Christodoulou DC, Gorham JM, Herman DS and
Seidman J: Construction of normalized RNA-seq libraries for
next-generation sequencing using the crab duplex-specific nuclease.
Curr Protoc Mol Biol Chapter. 4:Unit4.12. 2011. View Article : Google Scholar
|
|
14
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
D'Antonio M, Picardi E, Castrignanò T,
D'Erchia AM and Pesole G: Exploring the RNA editing potential of
RNA-Seq data by ExpEdit. Methods Mol Biol. 1269:327–328. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tarazona S, Furio-Tari P, Ferrer A and
Conesa A: NOISeq: Exploratory analysis and differential expression
for RNA-seq data. R package version 200. 2012.
|
|
18
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nature genetics. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Altermann E and Klaenhammer TR:
PathwayVoyager: Pathway mapping using the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
da W Huang, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2008. View Article : Google Scholar
|
|
21
|
Snel B, Lehmann G, Bork P and Huynen MA:
STRING: A web-server to retrieve and display the repeatedly
occurring neighbourhood of a gene. Nucleic Acids Res. 28:3442–3444.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Willoughby D, Dunn C, Yamamoto S, Capasso
F, Deporter D and Giroud J: Calcium pyrophosphate-induced pleurisy
in rats: A new model of acute inflammation. 1975. Agents Actions.
43:221–224. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nishioka K, Vilariño-Güell C, Cobb SA,
Kachergus JM, Ross OA, Hentati E, Hentati F and Farrer MJ: Genetic
variation of the mitochondrial complex I subunit NDUFV2 and
Parkinson's disease. Parkinsonism Relat Disord. 16:686–687. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Washizuka S, Kametani M, Sasaki T, Tochigi
M, Umekage T, Kohda K and Kato T: Association of mitochondrial
complex I subunit gene NDUFV2 at 18p11 with schizophrenia in the
Japanese population. Am J Med Genet B Neuropsychiatr Genet.
141B:301–304. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu HY, Liao PC, Chuang KT and Kao MC:
Mitochondrial targeting of human NADH dehydrogenase (ubiquinone)
flavoprotein 2 (NDUFV2) and its association with early-onset
hypertrophic cardiomyopathy and encephalopathy. J Biomed Sci.
18:292011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Teoh J, Boulos S, Chieng J, Knuckey NW and
Meloni BP: Over-expressing prohibitin (PHB) in neuronal cultures
exacerbates cell death following hydrogen peroxide and L-glutamic
acid induced injury. Neurosci Med. 5:149–160. 2014. View Article : Google Scholar
|
|
28
|
Muraguchi T, Kawawa A and Kubota S:
Prohibitin protects against hypoxia-induced H9c2 cardiomyocyte cell
death. Biomed Res. 31:113–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsutsumi T, Matsuda M, Aizaki H, Moriya K,
Miyoshi H, Fujie H, Shintani Y, Yotsuyanagi H, Miyamura T, Suzuki T
and Koike K: Proteomics analysis of mitochondrial proteins reveals
overexpression of a mitochondrial protein chaperon, prohibitin, in
cells expressing hepatitis C virus core protein. Hepatology.
50:378–386. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Amrani Y, Syed F, Huang C, Li K, Liu V,
Jain D, Keslacy S, Sims MW, Baidouri H, Cooper PR, et al:
Expression and activation of the oxytocin receptor in airway smooth
muscle cells: Regulation by TNFalpha and IL-13. Respir Res.
11:1042010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kimura T, Tanizawa O, Mori K, Brownstein
MJ and Okayama H: Structure and expression of a human oxytocin
receptor. Nature. 356:3561992. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Heinrichs M, Baumgartner T, Kirschbaum C
and Ehlert U: Social support and oxytocin interact to suppress
cortisol and subjective responses to psychosocial stress.
Biological Psychiatry. 54:1389–1398. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Thompson RJ, Parker KJ, Hallmayer JF,
Waugh CE and Gotlib IH: Oxytocin receptor gene polymorphism
(rs2254298) interacts with familial risk for psychopathology to
predict symptoms of depression and anxiety in adolescent girls.
Psychoneuroendocrinology. 36:144–147. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li K, Qi Y, Xia T, Yao Y, Zhou L, Lau KM
and Ng HK: CRMP1 inhibits proliferation of medulloblastoma and is
regulated by HMGA1. PLoS One. 10:e01279102015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yao L, Liu YH, Li X, Ji YH, Yang XJ, Hang
XT, Ding ZM, Liu F, Wang YH and Shen AG: CRMP1 interacted with Spy1
during the collapse of growth cones induced by Sema3A and acted on
regeneration after sciatic nerve crush. Mol Neurobiol. 53:879–893.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Indraswari F, Wong PT, Yap E, Ng YK and
Dheen ST: Upregulation of Dpysl2 and Spna2 gene expression in the
rat brain after ischemic stroke. Neurochem Int. 55:235–242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tanaka H, Morimura R and Ohshima T: Dpysl2
(CRMP2) and Dpysl3 (CRMP4) phosphorylation by Cdk5 and DYRK2 is
required for proper positioning of Rohon-Beard neurons and neural
crest cells during neurulation in zebrafish. Dev Biol. 370:223–236.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xiong T, Tang J, Zhao J, Chen H, Zhao F,
Li J, Qu Y, Ferriero D and Mu D: Involvement of the
Akt/GSK-3β/CRMP-2 pathway in axonal injury after hypoxic-ischemic
brain damage in neonatal rat. Neuroscience. 216:123–132. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang CY, Feng SJ, Xu L, Bu XN, Zhang N,
Zheng YX, Yuan XW, Li XG and Li JF: CRMP-2 is involved in hypoxic
preconditioning-induced neuroprotection against cerebral ischemic
injuries of mice. Chin J Basic Clin Med. 11:0072009.(In
Chinese).
|