Introduction
Clostridium difficile is a gram-positive
anaerobic gemma bacillus that becomes well established in human
colons, leading to diarrhea and inflammation in infected patients
(1). It has been estimated that ~30%
of antibiotic-associated diarrhea and 90% of pseudomembranous
enteritis result from C. difficile infection (CDI). CDI is
prevalent around the globe; it has been reported that €3.4 billion
is allocated to the diagnosis and treatment of CDI annually in the
UK (1). In the United States,
~300,000 cases of CDI are diagnosed every year, with an overall
medical cost of $1.1 billion (2).
However, the pathogenesis and prevalence of CDI in China is poorly
understood due to a lack of molecular and genetic data sources.
Pathogenic C. difficile strains produce
multiple toxins (1,2). The most well characterized are C.
difficile toxins A and B, which may induce diarrhea and
inflammatory diseases (3). These
potent toxins have been the focus of various investigations
(2,3). However, the specific secretion
procedures and relative contributions of C. difficile toxins
A and B remain to be elucidated, and this information is important
to determine the mechanism underlying CDI (3,4). In a
previous study by the present group (5), the existence of the type IV secretion
system (T4SS) in C. difficile was identified, and was
revealed to be potentially associated with the virulence of C.
difficile. T4SS is a secretion system that is associated with
the bacteria-binding mechanism (4).
It is capable of transferring toxins or proteins to the host cells
by forming an injector connecting to extracellular structures,
directly leading to infectious diseases (6). T4SS is associated with the virulence of
Helicobacter pylori, Bordetella pertussis,
Legionella pneumophila, Coxiella burnetii,
Escherichia coli and Bartonella (6–9). The
T4SS was previously identified in the genome-wide sequencing of
multiple C. difficile strains (9). However, the distribution of T4SS in
C. difficile colonies is poorly understood due to a lack of
epidemiological and experimental data. In the present study,
different sources and sequencing types of C. difficile were
preliminarily screened to investigate the distribution profiles of
three core genes of T4SS, including VirB4 m VirB6 and VirD4, in
C. difficile colonies. The aim of the present study was to
provide evidence for elucidating and establishing the molecular
polymorphism that results in T4SS in C. difficile.
Materials and methods
Materials
Sampling source
A total of 37 C. difficile strains of
different sources were obtained from patients treated at the
Department of Infectious diseases of different hospitals, 33 of
which were verified for subsequent experiment. Written informed
consent was obtained from all patients for inclusion in the present
study. One strain, BJ08, was isolated in Beijing (Department of
Clinical Laboratory, China-Japan Friendship Hospital; Beijing,
China), 13 were isolated in Guangdong Institute of Microbiology in
Guangzhou in the 1980s, 13 were isolated in Shanghai Institute of
Microbiology (Shanghai, China) between 2007 and 2011, 6 in Shandong
Institute of Microbiology (Jinan, China) between 2010 and 2012, 2
strains (UK1 and US1) were donated by Dr Feng Hanping from
University of Maryland in the United States, and ATCC9689 and CD630
strains were purchased from the American Type Culture Collection
(Manassas, VA, USA).
Reagents and instruments
Cycloserine-cefoxitin-fructose-egg yolk agar
substrate and additive (CCFA) medium, brain heart infusion (BHI)
agar, egg yolk emulsion and a BioMerieux kit were supplied by Oxoid
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). Proline paper
was supplied by Remel, Inc. (San Diego, CA, USA), goat blood was
obtained from Beijing Laboratory Biology Technology Co., Ltd
(Beijing, China) and bacterial genomic DNA was purified from
bacterial cultures using a QIAamp DNA extraction mini kit (Qiagen,
Inc., Valencia, CA, USA) strictly according to the manufacturer's
instructions. Polymerase chain reaction (PCR) primers and product
sequences were provided by Shanghai Sangon Biotech Co., Ltd.
(Shanghai, China), DNA polymerase was from Toyobo Co., Ltd. (Osaka,
Japan) and markers were obtained from Takara Bio, Inc. (Otsu,
Japan).
Bacterial culture and genome extraction
C. difficile strains of different sources
were inoculated in CCFA culture medium in an anaerobic environment
at 37°C for 48 h. Single flat, yellow ground-glass like colonies
with a horse dung smell and gram-positive bacillus were selected
and inoculated in BHI culture medium under anaerobic conditions at
37°C for 24 h. All cultured strains were verified strictly
according to the manufacturers' instructions (REF20300l; apiR20A
system, BioMerieux, Inc., St. Louis, MO, USA). The verified strains
were inoculated on a BHI blood disk in a streak pattern at 37°C for
48 h. The genomic DNA of 37 strains was analyzed using a DP320
bacterial genomic DNA extraction kit (DP320; Qiagen GmbH; Hilden
Germany), dissolved in Tris-EDTA buffer solution and stored at
−20°C for subsequent use. The present study was approved by the
Ethics Committee of The Affiliated Hospital of Binzhou Medical
University (Binzhou, China).
Identification and multilocus sequence typing
(MLST) of toxins A and B
The tcdA gene, which encodes enterotoxin A (9), was amplified according to the protocol
described by Lemme et al (10). The tcdB gene, which encodes cytotoxin
B, was amplified according to the protocol described by Kato et
al (11). The products were
cultured on 1% agarose gel at 37°C for 24 h, subjected to 100 V
electrophoresis for 20–30 min and finally observed using a gel
imaging system (ChemiDoc MP Imaging System; Bio-Rad Laboratories,
Inc., Hercules, CA, USA) to verify the expression of toxins A and
B. According to the protocol by Griffiths et al (12), 7 housekeeping genes including
adenosine kinase, ATP synthase subunit alpha, 1-deoxy-D-xylulose
5-phosphate reductoisomerase, serine hydroxymethyltransferase,
recA, superoxide dismutase and triosephosphateisomerase were
subject to PCR amplification as follows: 94°C for 5 min, 94°C for
30 sec, 53°C for 30 sec, 72°C for 30 sec for 35 cycles and 72°C for
10 min. DNA polymerase (20–80 ng/µl; 3 µl) was used (Takara Bio,
Inc.). DNA sample was collected from the strains. The amplified
products were delivered for sequencing (Shanghai Bioengineering
Co., Ltd., Shanghai, China) and analyzed using Chromas software
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The sequence
obtained was subsequently submitted to a database (http://pubmlst.org/cdifficile). The allele of toxins A
and B was obtained and sequence typing of strains was verified, as
illustrated in Table I.
 | Table I.Toxins A and B primer and MLST of 7
housekeeping genes. |
Table I.
Toxins A and B primer and MLST of 7
housekeeping genes.
| Toxin | Gene | Direction | Sequence (5′-3′) | Fragment size
(bp) |
|---|
| tcdA | tcdA | F |
AGATTCCTATATTTACATGACAATAT | 369 |
|
|
| R |
GTATCAGGCATAAAGTAATATA CTTT |
|
| tcdB | NK104 | F |
GTGTAGCAATGAAAGTCCAAGTTTACGC | 204 |
|
| NK105 | R |
CACTTAGCTCTTTGATTGCTGCACCT |
|
|
| Adk | F |
TTACTTGGACCTCCAGGTGC | 635 |
|
|
| R |
TTTCCACTTCCTAAGGCTGC |
|
| MSLT | atpA | F |
TGATGATTTAAGTAAACAAGCTG | 674 |
|
|
| R |
AATCATGAGTGAAGTCTTCTCC |
|
|
| dxr | F |
GCTACTTTCCATTCTATCTG | 525 |
|
|
| R |
CCAACTCTTTGTGCTATAAA |
|
|
| glyA | F |
ATAGCTGATGAGGTTGGAGC | 625 |
|
|
| R |
TTCTAGCCTTAGATTCTTCATC |
|
|
| recA | F |
CAGTAATGAAATTGGGAGAAGC | 705 |
|
|
| R |
ATTCAGCTTGCTTAAATGGTG |
|
|
| sodA | F |
CCAGTTGTCAATGTATTCATTTC | 585 |
|
|
| R |
ATAACTTCATTTGCTTTTACACC |
|
|
| tpi | F |
ATGAGAAAACCTATAATTGCAG | 640 |
|
|
| R |
TTGAAGGTTTAACACTTCCACC |
|
PCR amplification and sequencing of T4SS
gene
T4SS was detected in the C. difficile 630
strain containing three core genes; VirB4, VirB6 and VirD4. DNA
sample was collected from the strains. DNA polymerase (20–80 ng/µl;
3 µl) was used (Takara Bio, Inc.). In the present study, the
sequences of VirB4, VirB6 and VirD4 in C. difficile 630 were
used as templates. To avoid false-positive or false-negative
results, three pairs of primers were designed for each core gene,
as illustrated in Table II.
 | Table II.Primer sequences of three core genes
of T4SS. |
Table II.
Primer sequences of three core genes
of T4SS.
| Gene | Primer | Direction | Sequence
(5′-3′) | Fragment size
(bp) |
|---|
| VirB6 | Primer 1 | F |
CTACTGGGCGGTATTCAAGC | 288 |
|
|
| R |
CCATACAGCAATCCACATCTTG |
|
|
| Primer 2 | F |
GGAGAGCTTGTCATGATACTCTTTG | 317 |
|
|
| R |
ACCGCATATCCAAGTATCGT |
|
|
| Primer 3 | F |
GATGTGGATTGCTGTATGGTT | 304 |
|
|
| R |
TGGAATGGCTGAAATGGATG |
|
| VirB4 | Primer 1 | F |
CGGTAGAAGATACCATTCCCT | 257 |
|
|
| R |
TTTATCCGGTATCTGAATTGCC |
|
|
| Primer 2 | F |
GCGGATAATTTAGAACAGGCA | 442 |
|
|
| R |
CCGATGGAAGAATGTCCATA |
|
|
| Primer 3 | F |
CCATTTACCACAGAGGAGCTTT | 359 |
|
|
|
| R |
CTACCGCCTACTACAAGCTCAA |
|
| VirD4 | Primer 1 | F |
GAGTATGGCTCGGCAAGATG | 431 |
|
|
| R |
GCTTTTTCTCCCTCTCCTTTAG |
|
|
| Primer 2 | F |
TGCAAGATAAGGCAAAGTTTC | 760 |
|
|
| R |
ACTTCTGAAGCGTCTATCATATC |
|
|
| Primer 3 | F |
TCTTGCTAACGCAAACAGAAC | 1,300 |
|
|
| R |
AGTCCTCAAGGAGCTTGTAAT |
|
The PCR reaction system (30 µl) underwent 35 cycles
of 95°C for 3 min, 94°C for 30 sec, 55°C for 30 sec, 72°C for 120
sec, and extension at 72°C for 7 min. PCR products were subject to
1.5% agarose gel electrophoresis and positive results were verified
for sequencing. The sequencing results were analyzed using Segman
version 7.1 software (Beijing Genomics Institute, Beijing,
China).
Results
PCR analysis
Of the 37 strains, 25 (67.6%) were identified to be
positive for toxin A and toxin B. The remaining 12 (32.4%) were
negative for toxin A and positive for toxin B. MLST detected 7
types of stains including 11 strains with sequence type (ST) 37, 10
strains with ST2, 6 strains with ST35, 7 strains with ST3, 1 strain
with ST54, 1 strain with ST1 and 1 strain with ST119, respectively
(Table III).
 | Table III.Polymerase chain reaction results of
multilocus sequence typing of VirB4, VirB6 and VirD4 of 37 strains
of Clostridium difficile. |
Table III.
Polymerase chain reaction results of
multilocus sequence typing of VirB4, VirB6 and VirD4 of 37 strains
of Clostridium difficile.
|
|
|
|
| VirB4 | VirB6 | VirD4 |
|---|
|
|
|
|
|
|
|
|
|---|
| No. | Strain | Toxin
genotyping | ST | F1R1 | F2R2 | F3R3 | F1R1 | F2R2 | F3R3 | F1R1 | F2R2 | F3R3 |
|---|
| 1 | CD630 | A+B+ | 54 | + | + | + | + | + | + | + | + | + |
| 2 | UK1 | A+B+ |
1 | + | + | + | + | + | + | + | + | − |
| 3 | ATCC9689 | A+B+ |
3 | − | − | − | − | − | − | − | − | − |
| 4 | BJ08 | A-B+ | 37 | + | + | + | + | + | + | + | + | − |
| 5 | US1 | A-B+ | 37 | + | + | + | + | + | + | + | + | + |
| 6 | GZ15 | A-B+ | 119 | +a | + | + | − | +a | + | +a | + | + |
| 7 | GZ1 | A+B+ | 35 | + | + | + | + | − | + | + | − | − |
| 8 | GZ2 | A-B+ | 37 | + | + | + | + | + | + | + | + | + |
| 9 | GZ3 | A-B+ | 37 | +a | + | + | + | +a | + | +a | + | + |
| 10 | GZ5 | A+B+ |
2 | −/+b | − | −/+ | −/+ | −/+b | − | −/+b | −/+ | −/+ |
| 11 | GZ6 | A-B+ | 37 | +a | + | + | + | +a | + | +a | + | + |
| 12 | GZ7 | A+B+ |
2 | −/+ | − | −/+ | + | + | −/+ | −/+ | −/+ | − |
| 13 | GZ8 | A-B+ | 37 | + | + | + | + | + | + | + | + | + |
| 14 | GZ9 | A-B+ | 37 | + | + | + | + | + | + | + | + | + |
| 15 | GZ11 | A-B+ | 37 | + | + | + | + | + | + | + | + | + |
| 16 | GZ12 | A-B+ | 37 | + | + | + | + | + | + | + | + | + |
| 17 | GZ13 | A-B+ | 37 | + | + | + | + | + | + | + | + | + |
| 18 | GZ14 | A-B+ | 37 | + | + | + | + | + | + | + | + | + |
| 19 | SH1 | A+B+ | 35 | +a | + | + | + | +a | + | +a | +c | − |
| 20 | SH2 | A+B+ | 35 | +a | + | + | + | +a | + | +a | +c | − |
| 21 | SH3 | A+B+ |
2 | −/+ | −/+ | −/+ | − | −/+ | −/+ | −/+ | −/+ | − |
| 22 | SH4 | A+B+ |
2 | −/+b | −/+ | −/+ | − | −/+b | −/+ | −/+b | −/+ | − |
| 23 | SH6 | A+B+ |
2 | −/+ | −/+ | − | − | −/+ | −/+ | − | −/+ | − |
| 24 | SH7 | A+B+ |
2 | −/+ | −/+ | − | − | −/+ | −/+ | − | −/+ | − |
| 25 | SH8 | A+B+ |
2 | −/+ | −/+ | − | − | −/+ | −/+ | −/+ | −/+ | − |
| 26 | SH9 | A+B+ |
2 | −/+ | −/+ | − | − | −/+ | −/+ | −/+ | −/+ | − |
| 27 | SH10 | A+B+ | 35 | + | + | + | + | + | + | + | +c | − |
| 28 | SH11 | A+B+ | 35 | + | + | + | + | + | + | + | +c | − |
| 29 | SH12 | A+B+ | 35 | + | + | + | + | + | + | + | +c | − |
| 30 | SH13 | A+B+ |
2 | −/+ | −/+ | − | − | −/+ | −/+ | −/+ | − | − |
| 31 | SH14 | A+B+ |
2 | −/+ | −/+ | − | − | −/+ | −/+ | −/+ | − | − |
| 32 | JN09 | A+B+ |
3 | + | + | + | + | + | + | + | + | + |
| 33 | JN012 | A+B+ |
3 | + | + | + | + | + | + | + | + | + |
| 34 | JN31 | A+B+ |
3 | + | + | + | + | + | + | + | + | + |
| 35 | JN33 | A+B+ |
3 | + | + | + | + | + | + | + | + | + |
| 36 | JN43 | A+B+ |
3 | +a | + | + | + | +a | + | +a | + | + |
| 37 | JN159 | A+B+ |
3 | +a | + | + | + | +a | + | +a | + | + |
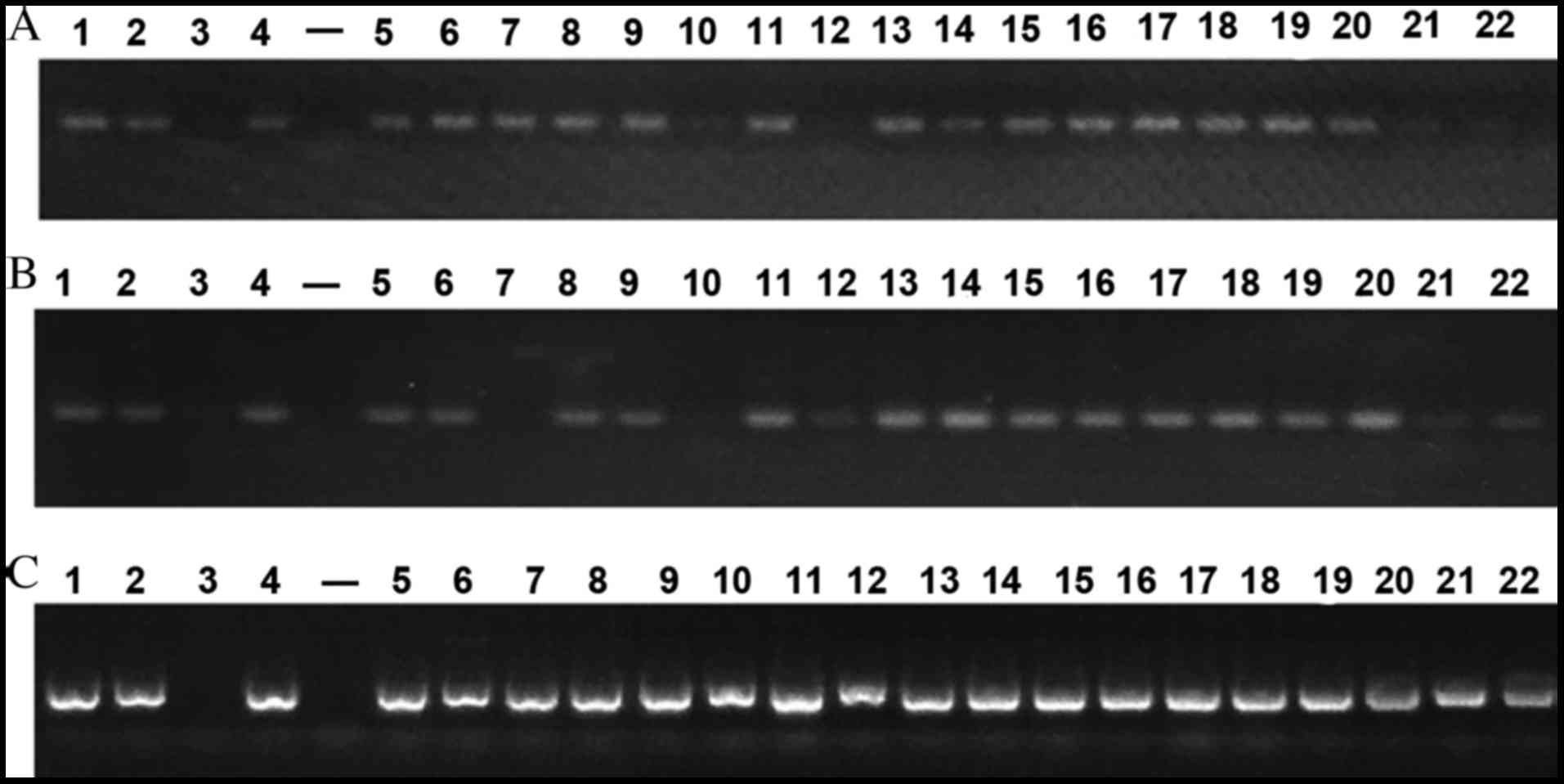
PCR outcomes of VirB4, VirB6 and VirD4 of 37 strains
are presented in Fig. 1. Strains of
the same ST shared similar outcomes. Positive results were observed
in three core genes of 11 ST37 strains and 7 ST3 strains. A
positive result was also noted in ST119 strains. Negative outcomes
were observed in 3 core genes of the ATCC9689 strain, however
positive results were detected in the other 3 standard strains. A
total of 10 ST2 stains were observed to be weakly positive and 3
core genes of 6 ST35 stains were positive. However, double bands
located at the target gene and at 2,000 bp were noted in the
VirD4-F2R2 strain. This requires further verification in subsequent
studies analyzing T4SS function. Based on the preliminary PCR
outcomes, 14 representative strains of different sources and ST
were selected for repeat PCR analysis. VirB4-F1R1, VirB6-F2R2 and
VirD4-F1R1 primers with the highest positive rate were chosen for
subsequent PCR. Following one cycle of PCR analysis, the results of
7 strains including ST6 (GZ15), ST9 (GZ3), ST11 (GZ6), ST19 (SH1),
ST20 (SH2), ST36 (JN43) and ST37 (JN159) were selected for
sequencing. Following a second PCR analysis, the results of ST10
(GZ5) and ST22 (SH4) strains were chosen for sequencing
analysis.
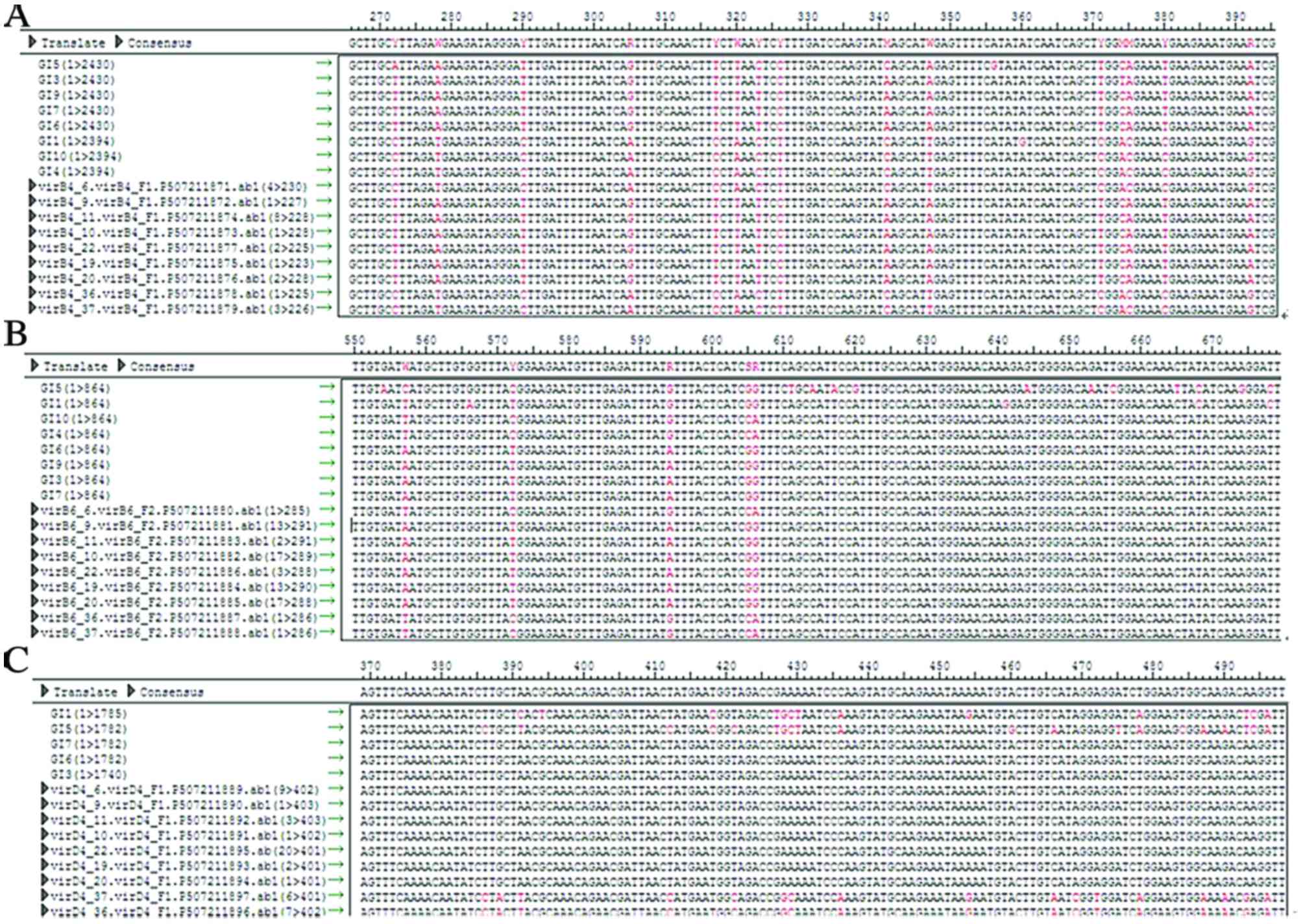
Sequencing analysis
The PCR results of 9 strains were sequenced and
matched with the T4SS sequence verified by Segman software. SNPs
were detected in a minority of strains. MLST revealed SNPs in 11
ST37 strains, 10 ST2 strains, 6 ST35 strains, 7 ST3 strains, 1 ST54
strain, 1 ST1 strain and 1 ST119 strain. Among 37 strains, 25 were
positive for toxins A and B and 12 were negative for A and positive
for B. The genome sequence of ATCC9689 strain was retrieved from
the link below. (http://www.ncbi.nlm.nih.gov/nuccore/484228681?report=fasta).
The Gene Prediction System For Type IV Secretion Systems
(http://www.secretion.org/navigateT4SP.action) invented
by our study group, was used and T4SS was not detected in the
ATCC9689 strain, which was consistent with the PCR results.
However, T4SS was detected in another 3 standard strains.
Considering varying geographic and ST factors, 9 strains of 3
regions and 5 STs were selected for PCR sequencing. The positive
rate of T4SS was up to 100%.
The VirB4, VirB6 and VirD4 sequences of ST2, ST37
and ST35 were observed to be identical. Strains with identical ST
shared the same SNP loci of T4SS, whereas these loci differed in
strains with different ST. This suggests that there is heredity
disparity of T4SS in strains with different ST, as illustrated in
Fig. 2.
Discussion
Previous studies have demonstrated that it is
difficult to identify multiple strains of C. difficile
(11–13), and researchers from the present study
group have previously conducted fundamental studies on C.
difficile (13). The gene
polymorphism of toxins A and B of C. difficile has
previously been studied in a clinical setting, as have the gene
polymorphism and evolutionary characteristics of toxin A-negative
and toxin B-positive C. difficile (13).
The genetic function, synthetic mechanism, receptor
factors and evolution of toxins A and B have previously been
intensively investigated (11–13).
Nevertheless, how these toxins are transmitted from C.
difficile to the outside environment remains poorly understood.
Govind and Dupuy (14) initially
proposed that toxins A and B may be transmitted to the external
environment through C. difficile toxin E (TcdE) protein.
However, TcdE deactivation in C. difficile 630 failed to
alter the secretion levels of toxins A and B (15). The specific underlying mechanism of
toxin secretion in C. difficile remains of interest in the
medical field. In 2013, Brouwer et al (16) demonstrated that the pathogenicity
locus of C. difficile 630 is capable of horizontally
transferring toxigenic genes via a conjugation-like mechanism to
non-toxigenic strains, which results in its conversion to a toxin
producer. This study suggested that non-toxigenic strains may be a
promising therapy for C. difficile-associated diarrhea
(16).
T4SS is a secretion system associated with the
mechanism underlying bacterial binding (17). It directly transports toxigenic
proteins, but also mediates transportation at the genetic level via
bacterial binding, transmits toxigenic genes, enhances pathogen
virulence and contributes to bacterial evolution (17–20). In
previous studies, a novel subtype of T4SS known as the type IVC
secretion system has been observed (17,20). In
porcine streptococcus, type IVC has been demonstrated to mediate
the horizontal transferring of 1 pathogenesis island of 89 K
(4). As three core genes of T4SS
subtype, VirB4, VirB6 and VirD4 serve synergistic effects mediating
DNA transfer (17). In a previous
study, similar T4SS subtypes were detected in Streptococcus
agalactiae, Streptococcus pneumonia and Pyogenic
streptococcus (5). In addition,
our study group first identified the existence of T4SS in C.
difficile and its association with C. difficile
virulence (5), suggesting that T4SS
may serve as the vital mechanism underlying the secretion of toxins
A and B.
In the present study, 37 strains of C.
difficile from different sources and with different STs were
preliminarily screened for VirB4, VirB6 and VirD4 to investigate
the distribution profile of T4SS in C. difficile. Of these
37 strains, 25 were positive for toxins A and B, and the remaining
12 were negative for toxin A and positive for toxin B. MLST
identified 7 strain types, including 11 strains with ST37, 10 with
ST2, 6 with ST35, 7 with ST3, 1 with ST54, 1 with ST1 and 1 with
ST119. Considering the variety of geographic and ST factors, 9
strains of 3 regions and 5 STs were selected for PCR sequencing.
The positive rate of T4SS was up to 100%, suggesting heredity
disparity in the T4SS of strains with different ST.
There are several limitations to the present study.
Firstly, T4SS sequencing analysis of the strains was restricted to
the genetic level, and so whether these genes are able to function
normally or express functional proteins remains unclear. Therefore,
the actual detection rate of T4SS with normal functionality may be
lower than reported here. Secondly, only A+B+ or A-B+ toxigenic
strains were screened in the present study; non-toxigenic strains
were not included. Consequently, the exact detection rate of T4SS
in C. difficile colonies should be further investigated.
Thirdly, only 37 C. difficile strains of 7 STs were
investigated. A larger sample-size should be used in future studies
to investigate the distribution profile of T4SS on a wider
scale.
In conclusion, the detection rate of VirB4, VirB6
and VirD4 is equally 100% in T4SS, and strains with identical STs
possess similar SNP loci. The results of the present study provide
a basis for subsequent identification of T4SS distribution,
epidemiological investigations, polymorphism analyses and
investigations into the association between T4SS, cytotoxicity and
enterotoxication in C. difficile.
Acknowledgements
The present study was supported by the National
Natural Science Foundation (grant no. 81301402).
References
|
1
|
Bauer MP, Notermans DW, van Benthem BH,
Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT and
Kuijper EJ: ECDIS Study Group: Clostridium difficile infection in
Europe: A hospital-based survey. The Lancet. 377:63–73. 2011.
View Article : Google Scholar
|
|
2
|
Kuijper EJ, Coignard B and Tüll P: ESCMID
Study Group for Clostridium difficile; EU Member States; European
Centre for Disease Prevention and Control: Emergence of Clostridium
difficile-associated disease in North America and Europe. Clin
Microbiol Infect. 12 Suppl 6:S2–S18. 2006. View Article : Google Scholar
|
|
3
|
Huang H, Wu S, Wang M, Zhang Y, Fang H,
Palmgren AC, Weintraub A and Nord CE: Clostridium difficile
infections in a Shanghai hospital: Antimicrobial resistance, toxin
profiles and ribotypes. Int J Antimicrob Agents. 33:339–342. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wilkins TD and Lyerly DM: Clostridium
difficile testing: After 20 years, still challenging. J Clin
Microbiol. 41:531–534. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang W, Rong C, Chen C and Gao GF:
Type-IVC Secretion system: A novel subclass of type IV secretion
system (T4SS) common existing in gram-positive genus Streptococcus.
Plos One. 7:e463902012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rupnik M: How to detect Clostridium
difficile variant strains in a routine laboratory. Clin Microbiol
Infect. 7:417–420. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He M, Sebaihia M, Lawley TD, Stabler RA,
Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, et
al: Evolutionary dynamics of Clostridium difficile over short and
long time scales. Proc Natl Acad Sci USA. 107:7527–7532. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Janvilisri T, Scaria J, Thompson AD,
Nicholson A, Limbago BM, Arroyo LG, Songer JG, Gröhn YT and Chang
YF: Microarray identification of Clostridium difficile core
components and divergent regions associated with host origin. J
Bacteriol. 191:3881–3891. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan Q, Zhang J, Chen C, Zhou H, Du P, Cui
Z, Cen R, Liu L, Li W, Cao B, et al: Multilocus sequence typing
(MLST) analysis of 104 Clostridium difficile strains from China.
Epidemiol Infect. 4:195–199. 2013. View Article : Google Scholar
|
|
10
|
Lemme L, Halluin A, Pestel-Caron M,
Lemeland JF and Pons JL: Multilocus sequence typing analysis of
human and animal Clostridium difficile isolates of various
toxigenic types. J Clin Microbiol. 42:2609–2617. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kato H, Kato N, Watanabe K, Iwai N,
Nakamura H, Yamamoto T, Suzuki K, Kim SM, Chong Y and Wasito EB:
Identification of toxin A-negative, toxin B-positive Clostridium
difficile by PCR. J Clin Microbiol. 36:2178–2182. 1998.PubMed/NCBI
|
|
12
|
Griffiths D, Fawley W, Kachrimanidou M,
Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJ,
Jolley KA, et al: Multilocus sequence typing of Clostridium
difficile. J ClinMicrobiol. 48:770–778. 2010.
|
|
13
|
Cheng Y, Du P, Chen C, Yan S, Jia H, Wang
J, Yan Q, Feng H and Lu J: Toxin A-negative, toxin B-positive
Clostridium difficile infection diagnosed by polymerase chain
reaction. Infect Control Hosp Epidemiol. 32:520–522. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Govind R and Dupuy B: Secretion of
Clostridium difficile toxins A and B requires the holin-like
protein TcdE. PLoS Pathog. 8:e10027272012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Olling A, Seehase S, Minton NP, Tatge H,
Schröter S, Kohlscheen S, Pich A, Just I and Gerhard R: Release of
TcdA and TcdB from Clostridium difficile cdi 630 is not affected by
functional inactivation of the tcdE gene. Microb Pathog. 52:92–100.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brouwer MS, Roberts AP, Hussain H,
Williams RJ, Allan E and Mullany P: Horizontal gene transfer
converts non-toxigenic Clostridium difficile strains into toxin
producers. Nat Commun. 4:20612013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Segal G, Feldman M and Zusman T: The
Icm/Dot type-IV secretion systems of Legionella pneumophila and
Coxiella burnetii. FEMS Microbiol Rev. 29:65–81. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ninio S and Roy CR: Effector proteins
translocated by Legionella pneumophila: Strength in numbers. Trends
Microbiol. 15:372–380. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang P, Xiong Y, Lan R, Ye C, Wang H, Ren
J, Jing H, Wang Y, Zhou Z, Cui Z, et al: pO157_Sal, a novel
conjugative plasmid detected in outbreak isolates of escherichia
coli O157:H7. J Clin Microbiol. 49:1594–1597. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saenz HL, Engel P, Stoeckli MC, Lanz C,
Raddatz G, Vayssier-Taussat M, Birtles R, Schuster SC and Dehio C:
Genomic analysis of Bartonella identifies type IV secretion systems
as host adaptability factors. Nat Genet. 39:1469–1476. 2007.
View Article : Google Scholar : PubMed/NCBI
|