Introduction
Tuberculosis (TB) is one of the public health
emergencies all over the world and severely affects human health.
TB is especially epidemic in China, which accounted for 12% of the
global total cases in 2012 (1).
According to 2010 National Technical Steering Group of the
Epidemiological Sampling Survey for TB, the weighted prevalence of
active, smear-positive, bacteriological positive pulmonary TB were
459/100,000, 66/100,000, 119/100,000, respectively (2). The geographical distribution of TB
prevalence presented that it was relatively low in the eastern
parts and high in the western parts, and the prevalence rates of
active pulmonary TB, Mycobacterium-positive pulmonary TB and
smear-positive pulmonary TB in Xinjiang Uygur autonomous region
were all higher than that in other provinces (3,4), which
demonstrated that Xinjiang bore a heavy burden of TB.
TB is caused by Mycobacterium tuberculosis
(MTB), which has infected around a third of the world population
(5), but only 10% of those infected
progress to active disease in their lifetime, and up to 90% of
infected people are asymptomatic with a latent infection (6). Susceptibility to TB varies between
different people. Since 1890s, how genetic factors affect clinical
outcomes with MTB infection was illustrated by a series of
research. For example, several twin studies have found that
concordance of TB was higher in monozygotic twins compared to
dizygotic twins (7–9). Beyond that, adoption research,
Genome-wide association studies, and case-control analysis have
been performed to demonstrate that the associations between
individual genetics and susceptibility to TB (10–12).
These indicated that host genetic factors are important
determinants of TB susceptibility.
The ASAP1 gene (known as AMAP1 or DDEF1), located at
8q24.1–8q24.2, encodes an Arf GTPase-activating protein (Arf GAP),
which is a multifunctional scaffold protein that induces hydrolysis
of GTP bound to the ADP ribosylation factor family GTP-binding
(Arf) proteins (13–15). ASAP1 has been implicated in
regulating cell motility and invasion (16,17). It
was found to functionally link with the progression and metastasis
of tumor cells, including ovary cancer (18), prostate cancer (19), and breast cancer (20,21).
Curtis et al (14) found that
the expression of ASAP1 gene in MTB-infected dendritic cells was
dramatically reduced, which may impair DC migration, suggesting a
potential mechanism that predisposes to TB.
Genome-wide association study is an effective way
(9) to screen for the genes exerting
the best population-wide impact on susceptibility to a
multifactorial disease (22). By
using this method, Curtis et al identified a novel ASAP1
gene which was associated with susceptibility to TB (14). Eleven single-nucleotide polymorphisms
(SNP)s were identified with significant association with
susceptibility to MTB (P<5×10−8). Seven out of the
most significantly associated ASAP1 SNPs were individually
genotyped to replicate their discovery, the most significant
association was at rs4733781 (P=2.6×10−11). Then the
associations between ASAP1 SNPs and susceptibility to MTB were
studied in Chinese population, but not in Xinjiang minorities
(23,24). As far as we know, it is the first
study which explored association between ASAP1 SNPs and
susceptibility to MTB in Xinjiang Muslim populations.
Owing to population heterogeneity, different races
have different causative polymorphisms (25). In this study, we selected a set of
SNPs within the entire ASAP1 gene and used case-control
analysis in Xinjiang Muslim populations to investigate whether
ASAP1 SNP was associated with TB risk.
Materials and methods
Subject
In this study all the participants, including 400 TB
patients and 380 control subjects, were of Xinjiang Muslim
ethnicity. For TB patients, there were 322 Uyghur patients, 46
Kazak patients and 32 Hui nationality patients. Correspondingly,
the Uyghur patients, Kazak patients and Hui nationality patients
were 306, 44 and 30, respectively in the control group.
Eligible cases were adult patients who were newly
diagnosed with active TB. These patients have evident lesions of TB
through simple computed tomography, X-ray, and positive results of
sputum smears and cultures for mycobacteria. Patients with
HIV-infection, hepatitis virus infection, immunodeficiency disease,
and other lung diseases were excluded from this cohort. Healthy
controls were nationality-, age- and sex-matched Xinjiang
minorities from Department of Respiratory Medicine, Xinjiang Uygur
Autonomous Region Chest Hospital, the Xinjiang Uygur Autonomous
Region, Urumqi, Xinjiang, China. The controls were negative both
for history of TB and T-SPOT assay. All participants were BCG
vaccinated.
Each patient and control enrolled in this study
provided a written informed consent. The study protocol conformed
to the ethical guidelines of the 1975 Declaration of Helsinki and
was approved by the Ethics Committee of Department of Respiratory
Medicine, Xinjiang Uygur Autonomous Region Chest Hospital.
Blood sample collection, DNA
isolation, purification and quality test
Peripheral blood samples (10 ml) was collected from
each participants and stored at −80°C. Genomic DNA was extracted
from peripheral blood collected from 400 TB patients and 380 non-TB
controls using a Genomic DNA Mini Preparation kit (Beyotime,
Shanghai, China) according to the manufacturer's instructions. Then
a reference gene ASAP1 was used for qualifying the extracted
samples by polymerase chain reaction (PCR) and electrophoresis.
Obvious imaging was regarded as qualified, or genomic DNA would be
extracted again.
Gene polymorphism detection
SNP selection
According to Curtis et al (14), 7 polymorphic sites in ASAP1 gene were
adopted in our study, including rs1017281, rs10956514, rs1469288,
rs17285138, rs2033059, rs4733781, rs12680942. The details of
primers are presented in Table
I.
 | Table I.Clinical characteristics of patients
and controls. |
Table I.
Clinical characteristics of patients
and controls.
| Characteristics | TB patients | Control | χ2 | P-value |
|---|
| Number | 400 | 380 |
|
|
| Age (years) | 55.4±12.4 | 55.7±12.1 |
| 0.675 |
| Nationality |
|
| 0.281 | 0.869 |
|
Uyghur | 322 (79.8%) | 306 (81.2%) |
|
|
|
Kazakhs | 46 (11.5%) | 44 (11.0%) |
|
|
| Hui | 32 (8.8%) | 30 (7.9%) |
|
|
| Sex |
|
| 0.666 | 0.415 |
| Male | 237 (59.2%) | 236 (62.1%) |
|
|
|
Female | 163 (40.8%) | 144 (37.9%) |
|
|
| Family history of
TB |
|
| 1.319 | 0.251 |
| Yes | 38 (9.5%) | 29 (7.2%) |
|
|
| No | 362 (90.5%) | 371 (92.8%) |
|
|
Primer design
After searching the whole ASAP1 gene sequence in
Genebank, 7 primers pairs were designed and synthetized by Shanghai
Biological Engineering Company (Table
II).
 | Table II.SNPs with their primers. |
Table II.
SNPs with their primers.
| SNP ID | Gene | SNP | Primer sequence
(5′-3′) | Annealing
temperature (°C) | Fragment size
(bp) |
|---|
| rs10956514 | ASAP1 | A/G |
GGCCACTGGCAAAAATAAGC | 55 | 320 |
|
|
|
|
AGTTGTCCAACTGCGGATAC |
|
|
| rs4733781 | ASAP1 | A/C |
CAAATGAACCCCCATAAAGG | 55 | 238 |
|
|
|
|
CCAGTGGCTGCATCCTACAT |
|
|
| rs1017281 | ASAP1 | C/T |
TATCTAATGTGCAGGGGATTG | 55 | 298 |
|
|
|
|
TCTCCCTTTTGCAGCTCACA |
|
|
| rs1469288 | ASAP1 | C/T |
TCCACACTGCTGAAAAATCTG | 55 | 521 |
|
|
|
|
AAGGATGTGGGGAGTTGAGG |
|
|
| rs2033059 | ASAP1 | C/T |
ACATACGTGGTGGTTGACTG | 54 | 393 |
|
|
|
|
TCCCAAAGCACAGAGGAAGA |
|
|
| rs12680942 | ASAP1 | A/G |
GCTGCTATAAAGACCCAGAAG | 56 | 207 |
|
|
|
|
GGCCATTTCTCCAAAGCCTCT |
|
|
| rs17285138 | ASAP1 | A/T |
CTGACTTGGTGCCAGCCTAC | 54 | 319 |
|
|
|
|
TGCTTTCCCAGAGCTTTCAG |
|
|
Multiplex PCR amplification and
product purification
SNPs were genotyped by multiplex PCR reaction using
AmpliTaq Gold® 360 Master Mix (Applied Biosystems,
Carlsbad, CA, USA), according to its protocol. The PCR reaction was
designed to amplify fragments covering all 7 SNP loci. The PCR
product was purified using PCR Clean Up kit (Beyotime) according to
the protocol.
SNP detection
Polymorphic loci genotypes were detected using
Sanger sequencing.
Statistical analysis
Statistical power was calculated by a post hoc power
analysis by G*Power 3.1.9.2 software (Program written,
conceptualized and designed by Franz, Universitat Kiel, Germany;
freely available windows application software). The data were
analyzed by SPSS 18.0 software (SPSS Inc., Chicago, IL). Continuous
variables and categorical data were compared by χ2 test.
Hardy-Weinberg equilibrium test was used to detect whether the two
groups were in genetic equilibrium. Genotype frequency comparisons
between groups were presented as odds ratio (OR) and 95% confidence
interval (CI). Haploview 4.2 (26)
was used to performed linkage disequilibrium analysis. The tests
were 2-sided and P<0.05 was considered to indicate a
statistically significant difference.
Results
General characteristic of TB patients
and healthy controls
The clinical characteristics of the total TB
patients and healthy controls are summarized in Table III. TB patients were age matched
with healthy controls. The age distribution between the patients
(55.4±12.4 years) and controls (55.7±12.1 years) was not
significantly different based on Mann-Whitney U test (P=0.675), as
the data were non-normally distributed (Table I). Patients were also gender-matched
with controls, no significant difference of gender was found
between the two groups (χ2=0.666, P=0.415). Nationality
of objects display no difference between patients and controls
(χ2=0.281, P=0.869). Among 400 TB patients, 38 persons
had family history of TB, while there were 29 persons with family
history of TB in control group, no significance was found between
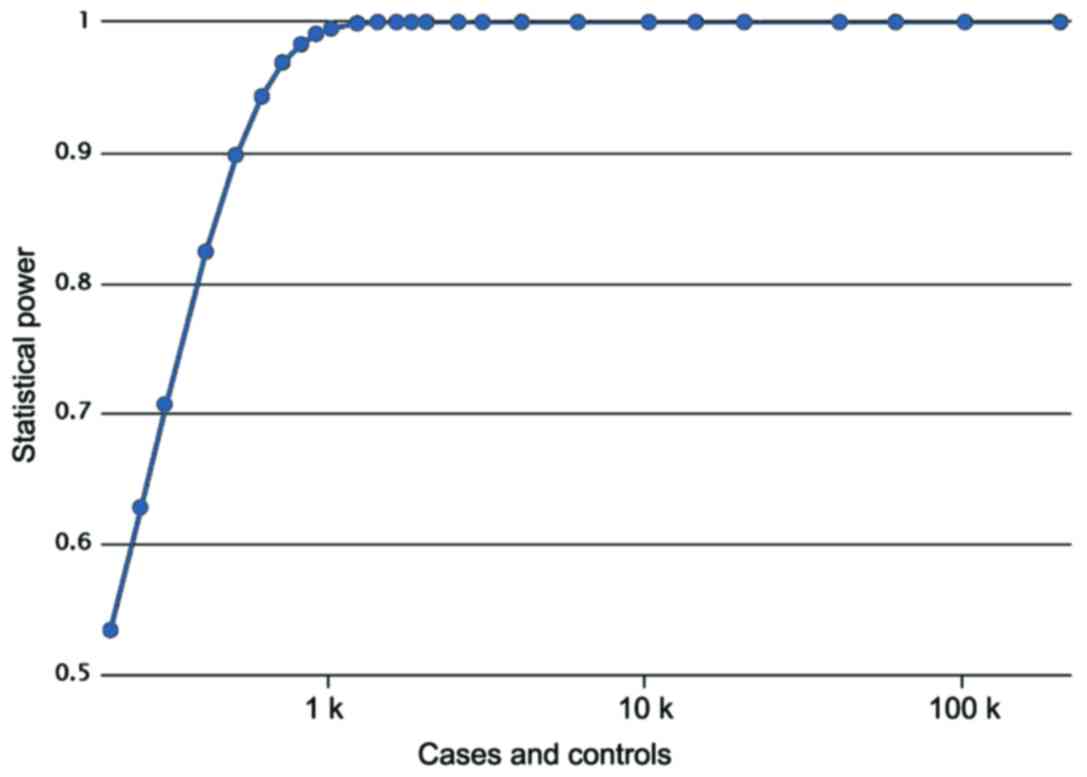
two groups (χ2=1.319, P=0.251). A post hoc power
analysis showed that Power value equals 0.8620, meaning a high
statistical effect (Fig. 1). The
parameters used in power analysis were as follows: cases, 400;
controls, 380; prevalence, 0.00178; odds ratio, 1.5070; minor
allele frequency, 0.3536.
 | Table III.Genotyping of rs1469288, rs2033059,
rs12680942, Rs17285138 in TB and control group in Xinjiang Muslin
population. |
Table III.
Genotyping of rs1469288, rs2033059,
rs12680942, Rs17285138 in TB and control group in Xinjiang Muslin
population.
|
Site/genotype/allele | SNP | TB patients | Control | OR (95% CI) | P-value |
|---|
| rs1469288 | HWE(P) |
|
|
| 0.20 |
|
Genotype | AA | 203 (50.8%) | 194 (51.1%) |
|
|
|
| AG | 169 (42.4%) | 160 (42.2%) |
|
|
|
| GG | 28 (6.8%) | 26 (6.7%) |
| 0.994 |
|
Allele | A | 575 (71.9%) | 548 (72.1%) |
|
|
|
| G | 225 (28.1%) | 212 (27.9%) | 0.989
(0.793–1.233) | 0.919 |
| rs2033059 | HWE(P) |
|
|
| 0.45 |
|
Genotype | CC | 130 (32.5%) | 108 (28.5%) |
|
|
|
| CT | 192 (48.0%) | 203 (53.3%) |
|
|
|
| TT | 78
(19.5%) | 69
(18.2%) |
| 0.293 |
|
Allele | C | 452 (56.5%) | 419 (55.1%) |
|
|
|
| T | 348 (43.5%) | 341 (44.9%) | 1.507
(0.866–1.291) | 0.586 |
| rs12680942 | HWE(P) |
|
|
| 0.99 |
|
Genotype | AA | 132 (33.0%) | 110 (29.0%) |
|
|
|
| AG | 183 (45.9%) | 202 (53.0%) |
|
|
|
| GG | 85
(21.2%) | 68
(17.9%) |
| 0.116 |
|
Allele | A | 447 (55.9%) | 422 (55.5%) |
|
|
|
| G | 353 (44.1%) | 338 (44.5%) | 1.014
(0.83–1.239) | 0.89 |
| rs17285138 | HWE(P) |
|
|
| 0.09 |
|
Genotype | AA | 126 (31.6%) | 105 (27.7%) |
|
|
|
| AT | 204 (51.0%) | 204 (53.6%) |
|
|
|
| TT | 70
(17.4%) | 71
(18.7%) |
| 0.496 |
|
Allele | A | 456 (57.0%) | 414 (54.5%) |
|
|
|
| T | 344 (43.0%) | 346 (45.5%) | 1.108
(0.907–1.353) | 0.315 |
Hardy-Weinberg equilibrium test
The rs10956514, rs1469288, rs2033059, rs4733781,
rs1017281, rs17285138, rs12680942 SNPs were investigated in 400
pulmonary TB cases and 380 healthy controls in the Xinjiang Muslim
population. rs10956514 site was not in Hardy-Weinberg equilibrium
(P<0.05), so it was excluded from our research. The last six
SNPs were in Hardy-Weinberg equilibrium in the control group and
the pulmonary TB group (P>0.05) (Tables III and IV).
 | Table IV.Genotyping of rs10956514, rs4733781,
rs1017281 in TB and control group in Xinjiang Muslin
population. |
Table IV.
Genotyping of rs10956514, rs4733781,
rs1017281 in TB and control group in Xinjiang Muslin
population.
|
Site/genotype/allele | SNP | TB patients | Control | OR (95% CI) | P-value |
|---|
| rs10956514 | HWE(P) |
|
|
| 0.013 |
|
Genotype | GG | 136 (34.0%) | 133 (35.0%) |
|
|
|
| GA | 207 (51.8%) | 199 (52.4%) |
|
|
|
| AA | 57
(14.2%) | 48
(12.6%) |
| 0.799 |
|
Allele | G | 479 (59.9%) | 465 (61.2%) |
|
|
|
| A | 321 (40.1%) | 295 (38.8%) | 0.947
(0.773–1.160) | 0.597 |
| rs4733781 | HWE(P) |
|
|
| 0.12 |
|
Genotype | AA | 194 (48.6%) | 174 (45.8%) |
|
|
|
| AC | 173 (43.2%) | 149 (39.2%) |
|
|
|
| CC | 33 (8.2%) | 57 (15%) |
| 0.012 |
|
Allele | A | 561 (70.1%) | 497 (65.4%) |
|
|
|
| C | 239 (29.9%) | 263 (34.6%) | 1.242
(1.004–1.537) | 0.046 |
| rs1017281 | HWE(P) |
|
|
| 0.56 |
|
Genotype | AA | 165 (41.2%) | 171 (45.0%) |
|
|
|
| AG | 187 (46.8%) | 177 (46.6%) |
|
|
|
| GG | 58 (12.0%) | 32 (8.4%) |
| 0.034 |
|
Allele | A | 517 (64.6%) | 519 (67.3%) |
|
|
|
| G | 283 (35.4%) | 241 (31.7%) | 0.792
(0.643–0.976) | 0.028 |
Genotype frequency distribution of
ASAP1 gene SNPs in the Xinjiang Muslim population
We used the case-control analysis to examine whether
7 polymorphisms in the ASAP1 gene were associated with
susceptibility to TB in Xinjiang Muslim population. The genotype
and allelic frequencies of ASAP1 7 SNPs are summarized in Tables III and IV. Two polymorphisms were associated with
TB (P<0.05), while the other five SNPs showed no significance.
For SNP rs4733781 the frequency of allele A in the pulmonary TB
group was higher than that in the control group, and there was a
significant difference between the two groups (P=0.046). While SNP
rs1017281 was lower than that in the control group (Tables III and IV). The direction of effect for the
associated alleles of rs4733781 and rs1017281 was the same as in
Curtis et al (14).
Associations between genetic model of
SNPs and TB risk
In order to find the optimal genetic model, we built
codominant, dominant, recessive and overdominant model of ASAP1
gene polymorphisms. rs4733781 site was found related to the
occurrence of TB in the recessive model (CC vs. AA+AC: OR, 0.51;
95% CI: 0.324–0.802; P=0.003) and co-dominant model (AA vs. CC: OR,
1.926; 95% CI: 1.198–3.097; P=0.006). rs1017281 site was associated
with TB in the recessive model (GG vs. AA+AG: OR, 1.792; 95% CI:
1.135–2.828; P=0.011) and co-dominant model (AA vs. GG: OR, 0.532;
95% CI: 0.329–0.826; P=0.01) (Tables
V and VI).
 | Table V.Association of ASAP1 SNP genotypes
with pulmonary TB under different genotype models. |
Table V.
Association of ASAP1 SNP genotypes
with pulmonary TB under different genotype models.
|
| Model | Genotype | Case | Control | OR (95% CI) | P-value |
|---|
| rs4733781 |
| AA | 194 (48.6%) | 174 (45.8%) | 1 |
|
|
| Codominant | AC | 173 (43.2%) | 149 (39.2%) | 0.96
(0.712–1.296) | 0.791 |
|
|
| CC | 33 (8.2%) | 57 (15%) | 1.926
(1.198–3.097) | 0.006 |
|
| Dominant | AA | 194 (48.5%) | 174 (45.8%) |
|
|
|
|
| AC+CC | 206 (51.5%) | 206 (54.2%) | 1.115
(0.841–1.477) | 0.448 |
|
| Recessive | CC | 33 (8.2%) | 57
(15.0%) |
|
|
|
|
| AC+AA | 367 (91.8%) | 323 (85.0%) | 0.51
(0.324–0.802) | 0.003 |
|
| Overdominant | AA+CC | 227 (56.8%) | 231 (60.8%) |
|
|
|
|
| AC | 173 (43.2%) | 149 (39.2%) | 0.846
(0.636–1.126) | 0.252 |
| rs1017281 |
| AA | 165 (41.2%) | 171 (45.0%) | 1 |
|
|
| Codominant | AG | 187 (46.8%) | 177 (46.6%) | 0.913
(0.679–1.229) | 0.549 |
|
|
| GG | 58
(12.0%) | 32 (8.4%) | 0.532
(0.329–0.862) | 0.01 |
|
| Dominant | AA | 165 (41.2%) | 171 (45.0%) |
|
|
|
|
| AG+GG | 235 (58.8%) | 209 (55.0%) | 0.858
(0.646–1.140) | 0.29 |
|
| Recessive | GG | 48
(12.0%) | 32 (8.4%) |
|
|
|
|
| AA+AG | 352 (88.0%) | 348 (91.0%) | 1.792
(1.135–2.828) | 0.011 |
|
| Overdominant | AA+GG | 213 (53.2%) | 203 (53.4%) |
|
|
|
|
| AG | 187 (46.8%) | 177 (46.6%) | 1.04
(0.786–1.375) | 0.785 |
 | Table VI.The data of D' and r2 among 7 SNPs of
ASPAP1 gene in the Chinese Xinjiang Muslim population. |
Table VI.
The data of D' and r2 among 7 SNPs of
ASPAP1 gene in the Chinese Xinjiang Muslim population.
| L1 | L2 | D' | r^2 |
|---|
| rs1469288 | rs1017281 | 1 | 1 |
| rs1469288 | rs10956514 | 1 | 0.979 |
| rs1469288 | rs12680942 | 1 | 1 |
| rs1469288 | rs2033059 | 1 | 0.959 |
| rs1469288 | rs4733781 | 1 | 1 |
| rs1469288 | rs17285138 | 1 | 0.979 |
| rs1017281 | rs10956514 | 1 | 0.979 |
| rs1017281 | rs12680942 | 1 | 1 |
| rs1017281 | rs2033059 | 1 | 0.959 |
| rs1017281 | rs4733781 | 1 | 1 |
| rs1017281 | rs17285138 | 1 | 0.979 |
| rs10956514 | rs12680942 | 1 | 0.979 |
| rs10956514 | rs2033059 | 1 | 0.979 |
| rs10956514 | rs4733781 | 1 | 0.979 |
| rs10956514 | rs17285138 | 0.979 | 0.958 |
| rs12680942 | rs2033059 | 1 | 0.959 |
| rs12680942 | rs4733781 | 1 | 1 |
| rs12680942 | rs17285138 | 1 | 0.979 |
| rs2033059 | rs4733781 | 1 | 0.959 |
| rs2033059 | rs17285138 | 1 | 0.979 |
| rs4733781 | rs17285138 | 1 | 0.979 |
Linkage disequilibrium analysis
Haploview 4.2 was used to performed linkage
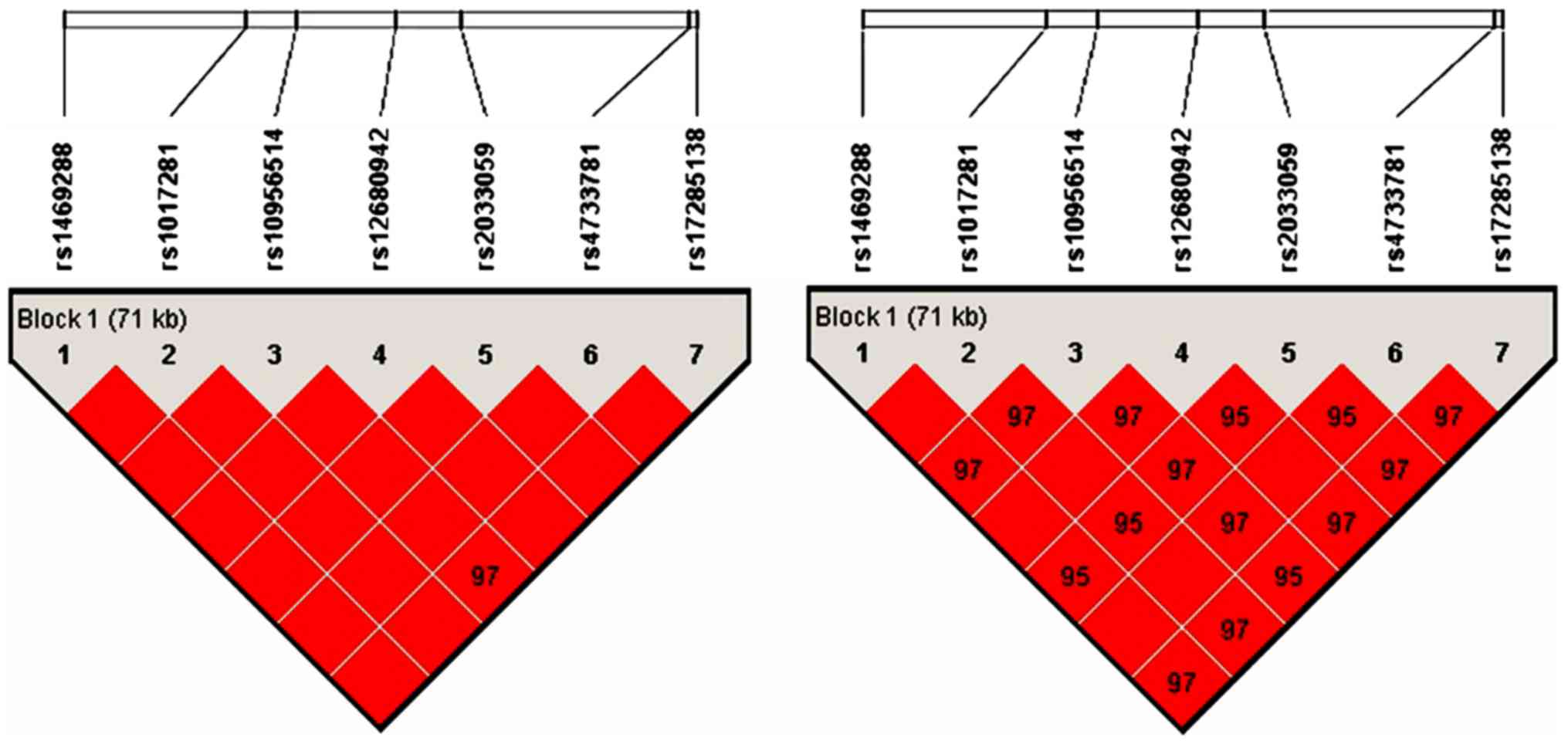
disequilibrium analysis. As shown in Fig. 2, linkage disequilibrium patterns for
the cluster of 7 SNPs in the ASAP1 gene genotyped in a Chinese
Xinjiang Muslim population. The results showed that the whole areas
were strong linkage disequilibrium and they can be used as a block.
Therefore, rs4733781 and rs1017281 were linked together, they
should be considered as one locus.
Discussion
Xinjiang Uygur Autonomous Region bears heavier
tuberculosis (TB) burden than other areas in China (3). The selection of candidate genes and
detection of their polymorphism sites have been considered
breakthroughs of TB prevention and treatment. Curtis et al
(14) found single-nucleotide
polymorphisms of ASAP1 gene was associated with the susceptibility
to TB in the Russian population (14), while Hu et al (23) and Miao et al (24) found no associations. This may be
partly due to differences between populations and small statistical
power of the follow-up studies to detect weak genetic effects.
However, no case-control analysis has been reported in Xinjiang
Muslim population to date. Hence, investigating the susceptibility
genes in Xinjiang Muslim population may provide access to control
TB in Xinjiang Uygur autonomous region.
In the present study, we tested association of ASAP1
gene polymorphism and TB susceptibility in Xinjiang Muslim
population by SNP genotyping. Our data suggested that ASAP1
rs4733781 and rs1017281 polymorphisms were associated with TB
susceptibility in Xinjiang Muslim population (P=0.046 and P=0.028).
While no significant associations were found in rs10956514,
rs1469288, rs2033059, rs12680942 and rs17285138. In contrast to our
findings, Hu et al (23) and
Miao et al (24) found that
ASAP1 gene polymorphism was not associated with TB susceptibility
in Chinese population and Tibetan population. As known, there are
ethnic variations of the allelic frequency distribution in the
investigated polymorphism markers (25). In addition, numerous gene studies
indicated that the risk variants of genetic heterogeneity and
ethnicity specificity was associated with TB (27,28). An
explanation for these divergent results may involve whether
polymorphism itself acts functionally and confers a truly altered
susceptibility to TB disease or the associated allele was an
unknown disease susceptibility allele in linkage disequilibrium. In
this regard, it is necessary to note that excessive reduction of
ASAP1 expression results in the impaired migration of
Mycobacterium tuberculosis (MTB)-infected DCs, which may
contribute to TB pathogenesis (14).
Other co-variables may not be excluded, such as socio-economic
factors, nutritional status, and interactions between genes
(29). Therefore, in-depth study on
the function of these sites is needed. In addition, in light of
marginal P-values of associations between ASAP1 SNPs and TB
susceptibility (P=0.046 and P=0.028), multiple corrections should
be considered in our later work, such as nationality, age, sex,
smoking and family history. An independent Uyghur sample will be a
good option to get converse results.
Regarding different genotype models, our results
demonstrated that a trend of higher rs4733781 A allele and
rs1017281 G allele in TB group compared to controls. Different
genotype models revealed that subjects with rs4733781 A and
rs1017281 G could be more susceptible to TB while subjects with
rs4733781 T and rs1017281 A could be more resistant. The recessive
model of rs4733781 (CC vs. AC+AA) in Xinjiang Muslim populations
tended to be related with a lower TB risk [P=0.003, OR=0.51
(0.324–0.802)], while the recessive model of rs1017281 (GG vs.
AA+AG) seemed to be related with a higher TB risk [P=0.011,
OR=1.792 (1.135–2.828)].Additionally, the linkage disequilibrium
analysis showed that rs4733781 and rs1017281 were linked together,
they should be considered one locus. Next, profound studies will be
performed to explore the potential functional roles of these two
SNPs to help understand major findings. Do they participate in the
immunoreaction caused by TB? What is the exact mechanism? Much work
remains to be done.
In conclusion, ASAP1 rs4733781 and rs1017281
polymorphism may be a genetic factor for susceptibility to MTB
among the Xinjiang Muslim populations. Further investigations of
the functional role of SNP rs4733781 and rs1017281 and the genomic
surrounding region are warranted.
Acknowledgements
Not applicable.
Funding
The study was supported by the National Natural
Science Foundation of China (81673160) and The Xinjiang Uygur
Autonomous Region Natural Science Foundation (2016D01B015) and The
Xinjiang Uygur Autonomous Region Natural Science Foundation
(2017D01A25).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW contributed to the conception of the study; AM
and XH contributed significantly to analysis and manuscript
preparation; AL performed the data analyses and wrote the
manuscript; FX helped perform the analysis with constructive
discussions. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study protocol conformed to the ethical
guidelines of the 1975 Declaration of Helsinki and was approved by
the Ethics Committee of Department of Respiratory Medicine,
Xinjiang Uygur Autonomous Region Chest Hospital. Each patient and
control enrolled in this study provided a written informed
consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zumla A, George A, Sharma V, Herbert RH,
Oxley A and Oliver M: Baroness Masham of Ilton: The WHO 2014 global
tuberculosis report - further to go. Lancet Glob Health. 3:e10–e12.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
National Technical Steering Group of the
Epidemiological Sampling Survey for Tuberculosis, . Implementing
Office of the Epidemiological Sampling Survey for Tuberculosis: The
prevalence of pulmonary tuberculosis in a national survey across
China in 2010. Zhonghua Jie He He Hu Xi Za Zhi. 35:665–668.
2012.(In Chinese). PubMed/NCBI
|
|
3
|
Li XX, Zhang H, Jiang SW, Liu XQ, Fang Q,
Li J, Li X and Wang LX: Geographical distribution regarding the
prevalence rates of pulmonary tuberculosis in China in 2010.
Zhonghua Liu Xing Bing Xue Za Zhi. 34:980–984. 2013.(In Chinese).
PubMed/NCBI
|
|
4
|
Wubuli A, Xue F, Jiang D, Yao X, Upur H
and Wushouer Q: Socio-demographic predictors and distribution of
pulmonary tuberculosis (TB) in Xinjiang, China: A spatial analysis.
PLoS One. 10:e01440102015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siroka A, Ponce NA and Lönnroth K:
Association between spending on social protection and tuberculosis
burden: A global analysis. Lancet Infect Dis. 16:473–479. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Png E, Alisjahbana B, Sahiratmadja E,
Marzuki S, Nelwan R, Balabanova Y, Nikolayevskyy V, Drobniewski F,
Nejentsev S, Adnan I, et al: A genome wide association study of
pulmonary tuberculosis susceptibility in Indonesians. BMC Med
Genet. 13:52012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Comstock GW: Tuberculosis in twins: A
re-analysis of the Prophit survey. Am Rev Respir Dis. 117:621–624.
1978.PubMed/NCBI
|
|
8
|
Varpela E: Studies on tuberculosis in
twins. Duodecim. 73:242–250. 1957.(In Finnish). PubMed/NCBI
|
|
9
|
Rubinstein U, Schachter J, Sharon N,
Talnir R and Amir J: Tuberculosis in a pair of twins - the use of
molecular biology methods for the detection of the source of
infection. Harefuah. 146:170–172, 248. 2007.(In Hebrew). PubMed/NCBI
|
|
10
|
Khor CC, Vannberg FO, Chapman SJ, Guo H,
Wong SH, Walley AJ, Vukcevic D, Rautanen A, Mills TC, Chang KC, et
al: CISH and susceptibility to infectious diseases. N Engl J Med.
362:2092–2101. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Garzón MC, Angée DY, Llerena C, Orjuela DL
and Victoria JE: Surveillance of Mycobacterium tuberculosis
resistance to antituberculosis drugs. Biomedica. 28:319–326.
2008.(In Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ladefoged K, Rendal T, Skifte T, Andersson
M, Søborg B and Koch A: Risk factors for tuberculosis in Greenland:
Case-control study. Int J Tuberc Lung Dis. 15:44–49.
2011.PubMed/NCBI
|
|
13
|
Nie Z and Randazzo PA: Arf GAPs and
membrane traffic. J Cell Sci. 119:1203–1211. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Curtis J, Luo Y, Zenner HL,
Cuchet-Lourenço D, Wu C, Lo K, Maes M, Alisaac A, Stebbings E, Liu
JZ, et al: Susceptibility to tuberculosis is associated with
variants in the ASAP1 gene encoding a regulator of dendritic cell
migration. Nat Genet. 47:523–527. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tien DN, Kishihata M, Yoshikawa A,
Hashimoto A, Sabe H, Nishi E, Kamei K, Arai H, Kita T, Kimura T, et
al: AMAP1 as a negative-feedback regulator of nuclear factor-κB
under inflammatory conditions. Sci Rep. 4:50942014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Oda A, Wada I, Miura K, Okawa K, Kadoya T,
Kato T, Nishihara H, Maeda M, Tanaka S, Nagashima K, et al: CrkL
directs ASAP1 to peripheral focal adhesions. J Biol Chem.
278:6456–6460. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nie Z, Hirsch DS, Luo R, Jian X, Stauffer
S, Cremesti A, Andrade J, Lebowitz J, Marino M, Ahvazi B, et al: A
BAR domain in the N terminus of the Arf GAP ASAP1 affects membrane
structure and trafficking of epidermal growth factor receptor. Curr
Biol. 16:130–139. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hou T, Yang C, Tong C, Zhang H, Xiao J and
Li J: Overexpression of ASAP1 is associated with poor prognosis in
epithelial ovarian cancer. Int J Clin Exp Pathol. 7:280–287.
2013.PubMed/NCBI
|
|
19
|
Lin D, Watahiki A, Bayani J, Zhang F, Liu
L, Ling V, Sadar MD, English J, Fazli L, So A, et al: ASAP1, a gene
at 8q24, is associated with prostate cancer metastasis. Cancer Res.
68:4352–4359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sabe H, Hashimoto S, Morishige M, Ogawa E,
Hashimoto A, Nam JM, Miura K, Yano H and Onodera Y: The
EGFR-GEP100-Arf6-AMAP1 signaling pathway specific to breast cancer
invasion and metastasis. Traffic. 10:982–993. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Onodera Y, Hashimoto S, Hashimoto A,
Morishige M, Mazaki Y, Yamada A, Ogawa E, Adachi M, Sakurai T,
Manabe T, et al: Expression of AMAP1, an ArfGAP, provides novel
targets to inhibit breast cancer invasive activities. EMBO J.
24:963–973. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bellamy R: Susceptibility to mycobacterial
infections: The importance of host genetics. Genes Immun. 4:4–11.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu X, Peng W, Chen X, Zhao Z, Zhang J,
Zhou J, Cai B, Chen J, Zhou Y, Lu X, et al: No significant effect
of ASAP1 gene variants on the susceptibility to tuberculosis in
Chinese population. Medicine (Baltimore). 95:e37032016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miao R, Ge H, Xu L, Sun Z, Li C, Wang R,
Ding S, Yang C and Xu F: Genetic variants at 18q11.2 and 8q24
identified by genome-wide association studies were not associated
with pulmonary tuberculosis risk in Chinese population. Infect
Genet Evol. 40:214–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yim JJ and Selvaraj P: Genetic
susceptibility in tuberculosis. Respirology. 15:241–256. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: Analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Delgado JC, Baena A, Thim S and Goldfeld
AE: Ethnic-specific genetic associations with pulmonary
tuberculosis. J Infect Dis. 186:1463–1468. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
El Baghdadi J, Remus N, Benslimane A, El
Annaz H, Chentoufi M, Abel L and Schurr E: Variants of the human
NRAMP1 gene and susceptibility to tuberculosis in Morocco. Int J
Tuberc Lung Dis. 7:599–602. 2003.PubMed/NCBI
|
|
29
|
Perrin P: Human and tuberculosis
co-evolution: An integrative view. Tuberculosis (Edinb). 95 Suppl
1:S112–S116. 2015. View Article : Google Scholar : PubMed/NCBI
|