Introduction
The past decade has seen a rapid increase in the
incidence in melanoma, it is estimated that the annual increase in
the incidence rate of melanoma has been ~3–7% per year worldwide
for Caucasians (1). Melanoma is one
of the most lethal of all adult malignancies due to its extremely
high tendency to metastasize towards multiple human organs, such as
the liver, brain and lung (2,3). The
median survival following the onset of distant metastases is 6–9
months, and the 5-year survival rate is >5% (2,3). Surgery
resection remains the primary curative treatment for melanoma at
early stages (4). However, treatment
of melanoma at late stages is difficult and less effective
(5). Therefore, the development of
new molecular targeted therapies for melanoma is urgently
required.
MicroRNAs (miRs) are small non-coding RNAs (22–26
nucleotides), which inhibit the expression of target genes
primarily by binding to the 3′-untranslated region (3′-UTR) of
mature mRNAs, leading to mRNA degradation or inhibition of
translation (6,7). Aberrant expression of miRs has been
reported in various human cancer types. miRs have been reported to
function as oncogenes or tumor suppressor genes in tumor
progression as well as development, encompassing apoptosis,
proliferation, migration, invasion, metastasis and resistance to
therapy (7–10). Notably, numerous miRs serve pivotal
roles in melanoma development and progression (11–13).
miR-373 is among the most commonly upregulated miRs in melanoma
(14). However, the detailed
mechanism of miR-373 in melanoma is yet to be elucidated.
The serine/threonine kinase salt-inducible kinase 1
(SIK1) is a member of the SIK family, which consists of SIK1, SIK2
and SIK3 (15). SIK1 has been
reported to be involved in multiple biological processes, including
regulation of PKA activity (16) and
p53-dependent anoikis through the use of a kinome-wide
loss-of-function screen (17).
Attenuated SIK1 gene expression is identified to promote various
human cancer types. In non-small cell lung cancer, downregulation
of SIK1 enhances epithelial-mesenchymal transition (EMT) and
radioresistance (18). In addition,
induced SIK1 expression has been reported to inhibit gastric cancer
cell migration (19). However, the
functional role of SIK1 in melanoma remains largely unknown.
In this study, to clarify the vital role of miR-373
in human melanoma, reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) and western blot analyses were used to
detect the expression of miR-373 in clinical samples, wound healing
assays were used to detect cell migration and Transwell assays were
also used to detect cell migration. Based on the results of the
current study, miR-373 was demonstrated to serve as an oncomiR in
melanoma, by enhancing melanoma cell migration. Furthermore,
miR-373 was observed to attenuate SIK1 protein level through
directly binding its 3′-UTR. Notably, it was also identified that
downregulation of SIK1 enhanced melanoma cell migration.
Materials and methods
Clinical sample collection
A total of 16 melanoma tissues and normal skin
samples were collected from patients recruited from the Department
of Plastic Surgery, Central Hospital of Wuhan, Tongji Medical
College, Huazhong University of Science and Technology (Wuhan,
China) between February 2015 and June 2016. The mean age of the
recruited patients (10 male and 6 female) were 56.46 years
(standard deviation, 2.63 years). The fresh tissues were frozen in
liquid nitrogen (−196°C) to protect the protein or RNA from
degradation. The use of human tissues was approved by the Ethics
Committee of the Central Hospital of Wuhan and all patients
provided written, informed consent.
Cell culture and transfection
The human melanoma cell lines A375, WM115 and WM75
were purchased from the Shanghai Institute of Biochemistry and Cell
Biology (Shanghai, China), and were maintained in Dulbecco's
modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% (v/v)
fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.),
penicillin (25 U/ml), streptomycin (25 g/ml) and 1% L-glutamine.
Normal human melanocytes were purchased from Lifeline Cell
Technology, LLC (Frederick, MD, USA; cat. no. FC-0030) and grown in
LL-0027 medium (Lifeline Cell Technology, LLC). All cell lines were
cultured in a 5% CO2 and 37°C incubator. For the
upregulation of miR-373 expression, a synthesized miR-373 mimic
(5′-GAAGUGCUUCGAUUUUGGGGUGU-3) and a miR-NC
(5′-UUCUCCGAACGUGUCACGUTT-3′ (both 10 nM; Shanghai GenePharma Co.,
Ltd., Shanghai, China) was transfected into A375 or WM115 cells
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. The medium
containing the transfection reagents was removed 6 h after
transfection and 48 h later the cells were harvested for subsequent
assays. The transfection efficiency and changes in miR-373
expression were determined by RT-qPCR. For the downregulation of
SIK1 expression, short hairpin (sh)RNA against SIK1 (sh-SIK1;
Shanghai GenePharma Co., Ltd.) was stably transfected in A375 or
WM115 cells using Lipofectamine 2000, according to the
manufacturer's protocol. The sh-SIK1 sequence was
5-UGAACAAGAUCAAAGGGUU-3 and negative control shRNA (sh-NC) sequence
was 5-AATTCTCCGAACGTGTCACGT-3 (Hanyin Biotechnology, Shanghai,
China). The transfection efficiency and changes in SIK1 expression
were determined by western blotting. The medium containing the
transfection reagents was removed 6 h after transfection and 48 h
later the cells were harvested for the following assays.
Bioinformatics analysis
To determine whether overexpression of miR-373
promoted melanoma through targeting of the 3′-UTR of its downstream
gene, three different algorithms [TargetScan (http://www.targetscan.org/vert_61/), miRTarBase
(http://mirtarbase.mbc.nctu.edu.tw/php/index.php) and
miRDB (http://mirdb.org/miRDB/)] were used to
select the potential target genes of miR-373.
Wound healing assay
A375 or WM115 cells were cultured in a 25
cm2 culture flask to 80–90% confluence in DMEM
supplemented with 10% FBS at 37°C, collected by digestion with
0.25% trypsin (Gibco; Thermo Fisher Scientific, Inc.) and
centrifuged at 4°C at 1,409 × g for 5 min, and then seeded into
6-well plates at 2.5×105 cells/well. miR-373 mimic,
miR-NC, sh-SIK1 and sh-NC were transfected into A375 or WM115 cells
as described above. Streaks were created in the monolayer with a
pipette tip. Progression of migration was observed and photographed
24 h after wounding. The floating cells were removed by washing
with PBS, and the width of scratch was observed at 0 and 24 h using
an inverted microscope (magnification, ×50). Each experiment was
repeated three times.
Transwell migration assay
The migration capability of A375 or WM115 cells was
determined using a Transwell assay. The two cell lines were
harvested and seeded with serum-free DMEM into the upper chambers
at 5×104 cells/well. The bottom chambers contained DMEM
with 10% FBS. The Transwells were incubated for 24 h at 37°C.
Following incubation, the migrated cells attached to the lower
surface of the membrane were fixed by 4% paraformaldehyde for 30
min at room temperature and stained with 1% toluidine blue for 15
min at room temperature. Cell numbers were counted in five randomly
chosen fields using an inverted microscope (magnification, ×100)
per membrane and counts were repeated three times.
RT-qPCR
Total RNA from tissues or cells was extracted using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. cDNA was synthesized
using a PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan),
according to the manufacturer's protocol. qPCR was performed with a
KAPA SYBR FAST qPCR kit (Kapa Biosystems, Inc., Wilmington, MA,
USA) using a 7900HT Fast Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The initial denaturation step was
for 10 sec at 95°C, followed by 40 cycles of denaturation for 5 sec
at 95°C, and annealing and extension for 20 sec at 60°C. The
expression levels of miRNA were normalized to endogenous small
nuclear RNA U6. The 2−ΔΔCq method was used to analyze
the expression level relative to the endogenous control (20). The primers used were as follows:
miR-373 forward, 5′-ATTTTGGTTAATACGGTGAAATTTC-3′ and reverse,
5′-CTATCGCCCAAACTAAAATACGAT-3′; SIK1 forward,
5-TGGACGTCTGGAGCCTCGGT-3′ and reverse, 5′-CTCGCGTTTTTCCTTAGCTG-3′;
U6, forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-ACGCTTCACGAATTTGCGT-3′.
Western blot analysis
Protein extraction and western blotting analysis
were conducted as described previously (21). The cells were incubated with the
primary antibodies, including anti-SIK1 (cat. no. ab64428; 1:1,000;
Abcam, Cambridge, MA, USA) or anti-GAPDH (cat. no. sc-25778;
1:5,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Then the
membranes were then washed and incubated with horseradish
peroxidase-conjugated goat anti-rabbit IgG secondary antibody (cat.
no. sc-2004; 1:5,000; Santa Cruz Biotechnology, Inc.).
Vector construction and luciferase
reporter assay
For the luciferase reporter assay, A375 cells were
seeded on 24-well plates and co-transfected using Lipofectamine
2000 with 100 ng/well firefly luciferase UTR-reporter vector, 2
ng/well Renilla pRLCMV vector (internal control; all Promega
Corporation, Madison, WI, USA) and 20 ng/well of miR-373 mimics or
miR-NC (Applied Biosystems; Thermo Fisher Scientific, Inc.),
following the manufacturer's protocol. In brief, luciferase
reporters were constructed through cloning of the 3′-UTR of SIK1
wild-type (wt) as well as SIK1 mutant-type (mut) via a deletion at
position 1,905–1,911. The 3UTR of SIK1 mRNA containing the
wild-type or mutant miR-373 recognition sequences were
PCR-amplified and subcloned into the SacI and SalI
sites of the pmirGLO vector (Promega Corporation). A375 cells
transfected with miR-373 or miR-NC and cultured in 48-well plates
were co-transfected with 1.5 µg of firefly luciferase reporter and
0.35 ng Renilla luciferase reporter with Lipofectamine 2000
regent. After 24 h the relative luciferase activity was assessed
with the Dual-Luciferase Assay Reporter system (cat. no. E1910;
Promega Corporation) and normalized to Renilla luciferase
activity.
Statistical analysis
Data are presented as the mean ± standard deviation
from at least three independent experiments. All data were analyzed
using GraphPad 6.0 statistical software (GraphPad Software, Inc.,
La Jolla, CA, USA). Differences between two groups were analyzed
using two-independent-sample Student's t-test or non-parametric
Mann-Whitney U test. Differences between multiple groups were
analyzed using one-way analysis of variance with Dunnett's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-373 is upregulated in melanoma
tissues and cell lines
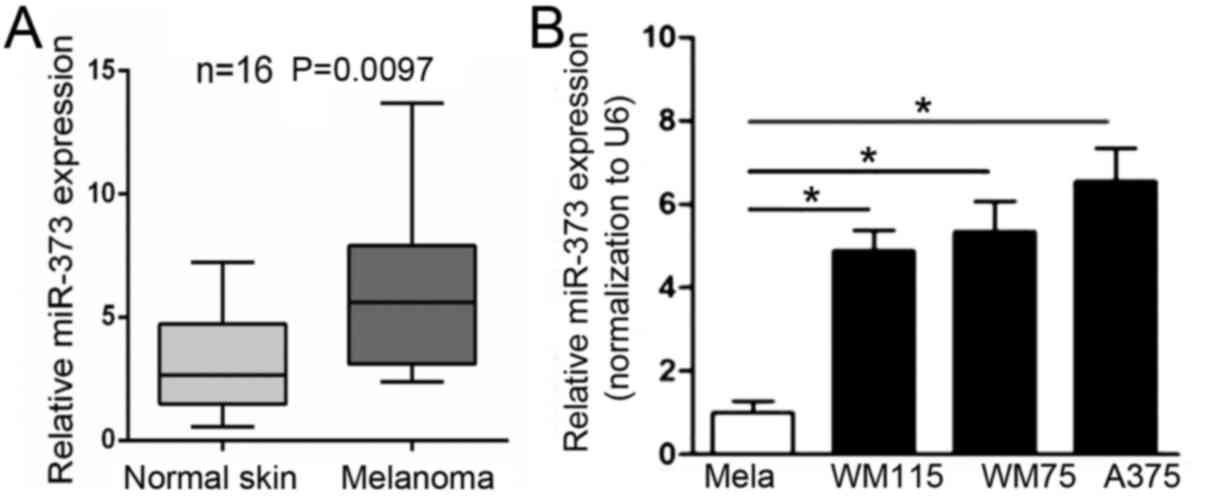
To investigate the expression of miR-373 in
melanoma, RT-qPCR was performed. The results indicated that the
mRNA level of miR-373 was significantly upregulated in melanoma
tissues compared with nevus (Fig.
1A). Next, the expression of miR-373 was evaluated in a panel
of normal melanocytes and human melanoma cell lines. The results
indicated that miR-373 was significantly upregulated in A375, WM115
and WM75 melanoma cell lines compared with normal melanocytes
(Fig. 1B). The results indicated
that the expression of miR-373 is increased in melanoma tissues and
cell lines.
miR-373 promotes melanoma cell
migration
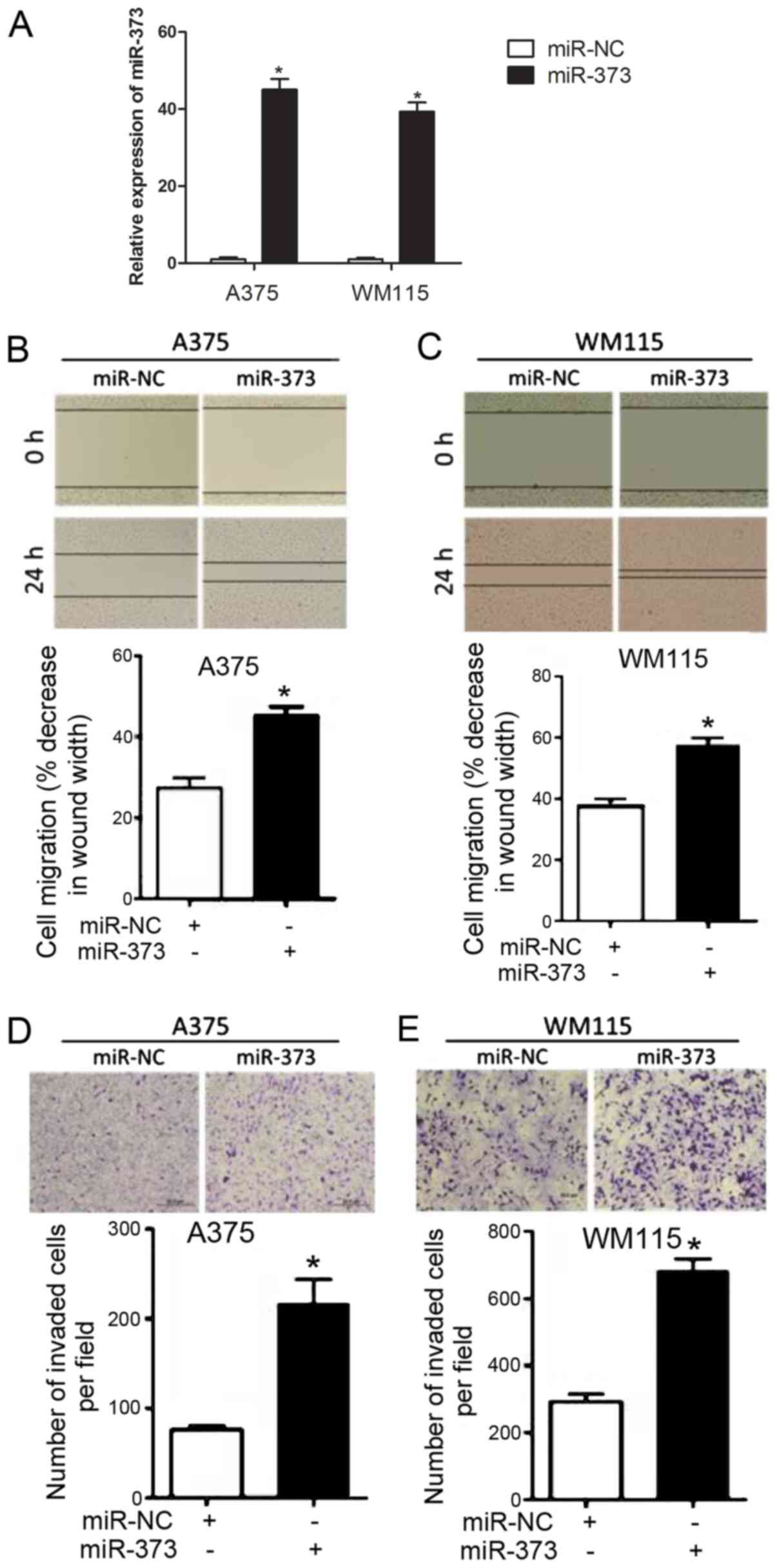
To explore the functional role of miR-373 in
melanoma progression, melanoma cells [A375 (relatively highest
level of miR-373 among the three cell groups from Fig. 1B) and WM115 (relatively lowest level
of miR-373 among the three cell groups from Fig. 1B)] were stably transfected with
miR-373 mimics. It was confirmed that the expression of miR-373 was
upregulated in A375 and WM115 cells following miR-373 mimic
transfection compared with cells transfected with miR-NC (Fig. 2A). As indicated in Fig. 2B and C, a wound healing assay
demonstrated that transfection with miR-373 mimic significantly
enhanced melanoma cell migration in A375 and WM115 cells compared
with transfection with miR-NC. The effects of miR-373 on melanoma
cell migration were evaluated further using a Transwell assay
(Fig. 2D and E). A375 and WM115
cells transfected with miR-373 mimic were observed to have a
significantly higher migration capability compared with cells
transfected with miR-NC. These results suggested that miR-373
elevates melanoma cell migration.
3′-UTR of SIK1 is the direct target of
miR-373
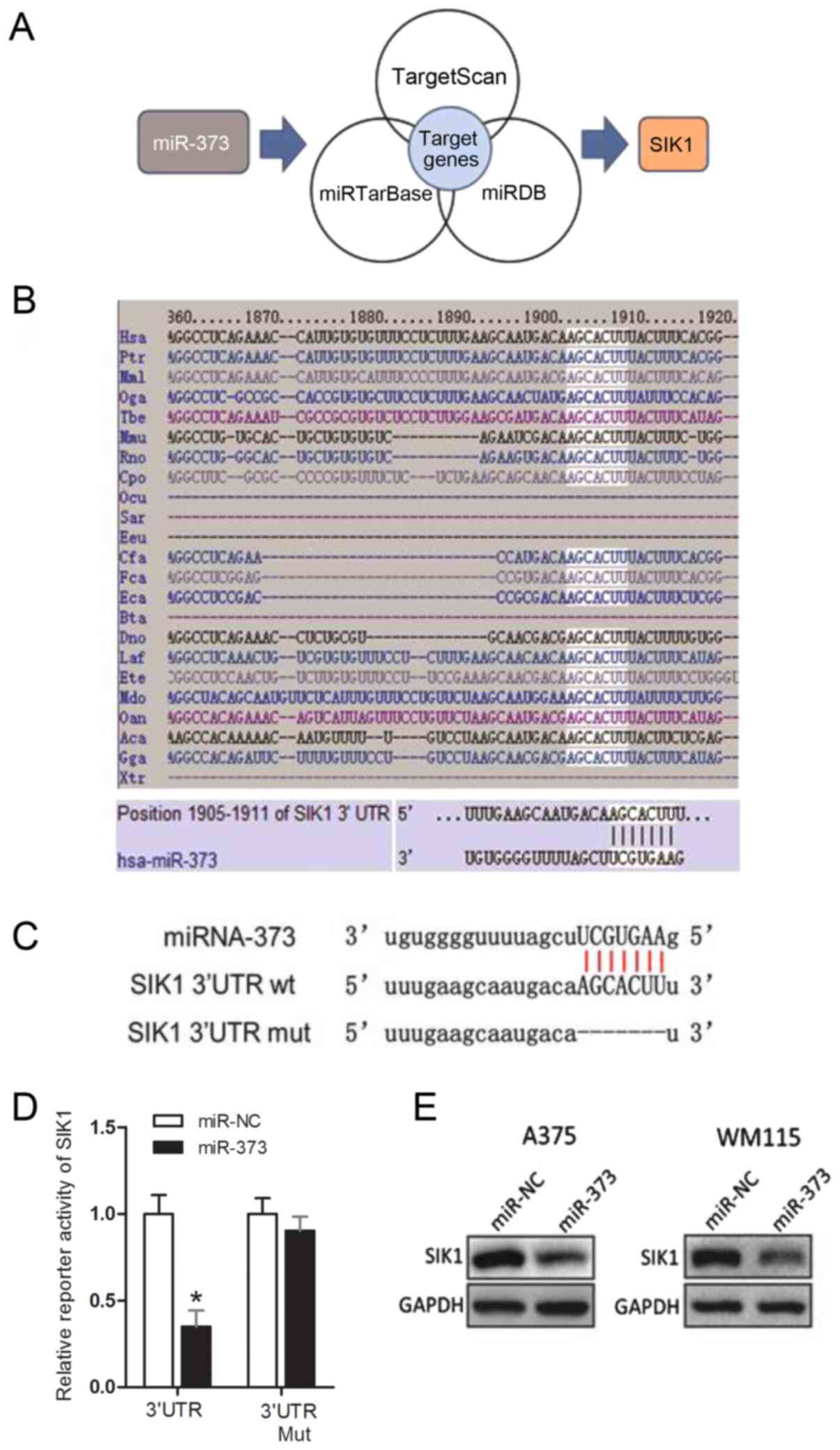
Among the potential candidate targets of miR-373 was
SIK1 (Fig. 3A). Fig. 3B indicates that the seed sequence of
miR-373 is complementary to the 3′-UTR of SIK1 and is highly
conserved among 18 different species. Dual-luciferase reporter
assays illustrated that the transient transfection of A375 cells
with SIK1 wt 3′-UTR significantly reduced reporter expression when
compared with the control vector (Fig.
3C and D). However, the firefly luciferase reporter activity of
the SIK1 mut 3′-UTR was not notably affected by
miR-373-transfection (Fig. 3D). This
result indicated that the 3′-UTR of SIK1 was targeted by miR-373.
Furthermore, western blot analysis results demonstrated that
overexpression of miR-373 inhibited SIK1 protein level in A375 and
WM115 cells (Fig. 3E). These results
supported the conclusion that SIK1 is the target gene of miR-373 in
melanoma.
sh-SIK1 augments cell migration in
melanoma
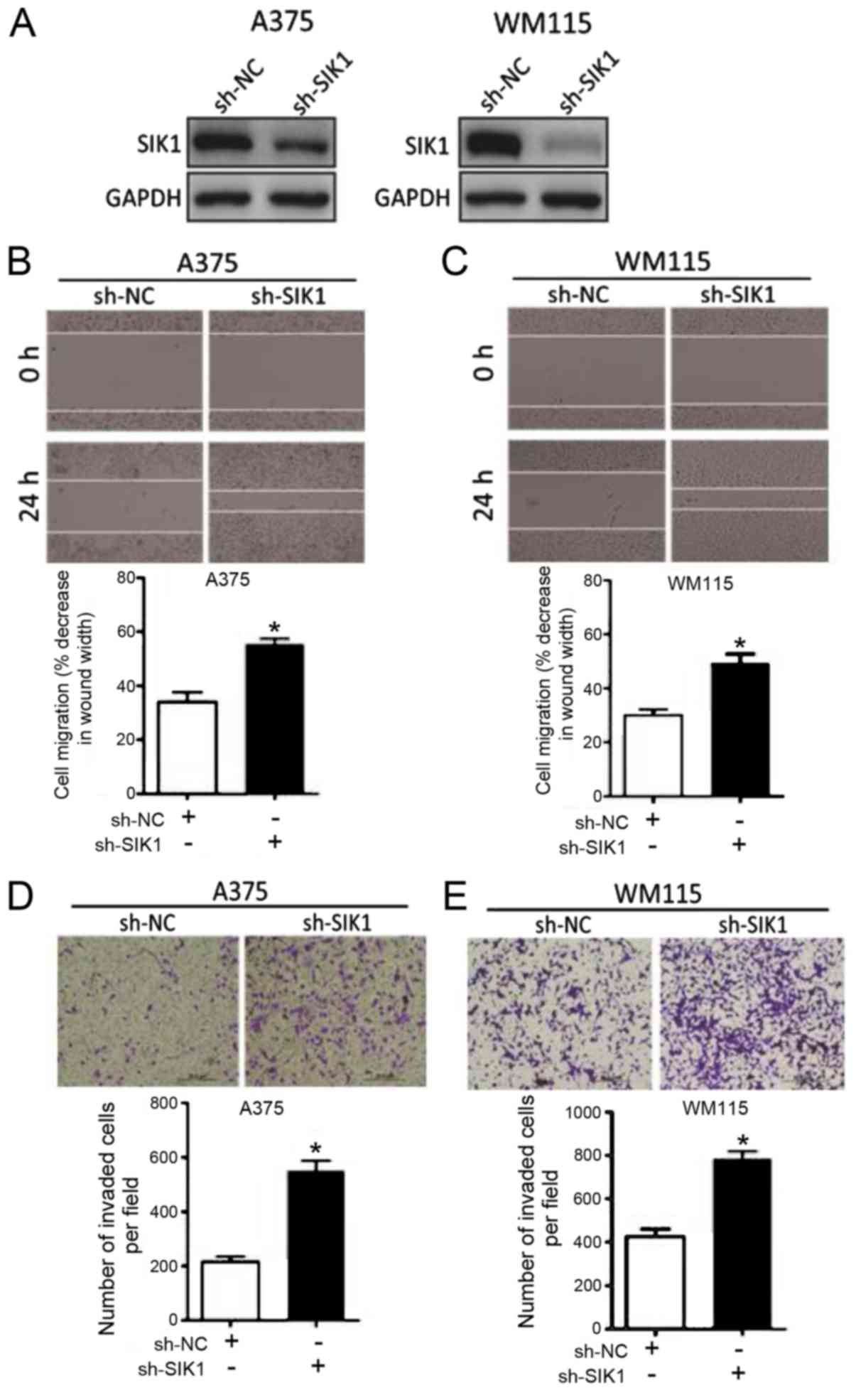
The functional role of SIK1 in melanoma was
evaluated. The effect of sh-SIK1 and sh-NC on SIK1 expression in
A375 and WM115 cell lines was observed via western blot analysis.
It was confirmed that transfection with sh-SIK1 resulted in
decreased SIK1 expression compared with sh-NC in the two cell lines
(Fig. 4A). A375 cells and WM115
cells transfected with sh-SIK1 exhibited significantly increased
cell migration compared with sh-NC Transfected cells, as
demonstrated by wound healing and Transwell assays (Fig. 4B-E). These results indicated that
downregulation of SIK1 promotes melanoma cell progression.
Discussion
Accumulating evidence indicates that miR-373 acts as
a tumor suppressor or oncogene in numerous human cancer types
(22). More specifically, miR-373
targeting CD44 and transforming growth factor-β receptor 2
suppressed tumor migration and invasion in glioma cells (23). In parallel, miR-373 inhibited ovarian
cancer cell invasion and metastasis by targeting RAB22A (24). On the other hand, miR-373 was
associated with aggressive human mucinous colorectal cancer
(25). Upregulated miR-373 promoted
cell migration in breast cancer through epigenetic silencing of
integrin subunit α-2 (26). miR-373
enhanced EMT transition and metastasis via targeting
thioredoxin-interacting protein in breast cancer (27). However, the association between
miR-373 and melanoma remains unclear.
In the present study, it was demonstrated that the
expression of miR-373 is upregulated in melanoma tissues and
certain melanoma cell lines compared with nevus and normal
melanocytes, respectively. Functionally, miR-373 mimics
significantly enhanced cell migration in wound healing and
Transwell assays in A375 and WM115 cell lines. miR-373 was
identified to target downstream gene SIK1 through directly binding
a complementary sequence. In addition, downregulated SIK1 was
indicated to be promote migration in A375 and WM115 cell lines.
Collectively, these findings suggest that miR-373 serves as an
oncomiR to promote melanoma progression and development via
targeting SIK1.
Previous studies have illustrated that SIK1 is
downregulated in multiple human tumor types. In human
hepatocellular carcinoma (HCC), SIK1 markedly inhibited EMT, tumor
growth and metastasis and provided a potential new candidate for
HCC therapy (28). Reduced
expression of SIK1 was also identified to be associated with poor
outcomes in gastric carcinoma (19).
Furthermore, SIK1 could be the potential target for numerous miRs.
In particular, miR-203 induces proliferation, migration and
invasion by targeting SIK1 in pancreatic cancer (29). In ovarian cancer, miR-141 promotes
cell growth through reversing SIK1-suppressed proliferation and
cancer stem cell-associated traits (30). However, the phenotypic role of SIK1
in melanoma remains largely unknown. In the present study, it was
identified that downregulation of SIK1, as a target of miR-373,
elevates cell migration of melanoma.
In summary, the present study identified that
miR-373 functions as an oncomiR to promote melanoma progression
through targeting SIK1 expression. This signaling pathway may
provide a new therapeutic approach for melanoma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
XB performed the total experiments and was a major
contributor in writing the manuscript. MY collected and analyzed
the clinical data regarding the melanoma. YX contributed to the
design of this project. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The use of human tissues was approved by the Ethics
Committee of the Central Hospital of Wuhan and all patients
provided written, informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cellerino P, Corsi F, Morandi E, Foschi D
and Trabucchi E: Metastatic melanoma of the gallbladder. Eur J Surg
Oncol. 26:815–816. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haskaraca MF, Ozsoy M, Ozsan I and Kurt K:
Primary malignant melanoma of the gallbladder: A case report and
review of the literature. Case Rep Surg. 2012:6935472012.PubMed/NCBI
|
|
4
|
Corrie P, Hategan M, Fife K and Parkinson
C: Management of melanoma. Br Med Bull. 111:149–162. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Burki TK: Defining the genetics of
melanoma progression. Lancet Oncol. 17:e72016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zeng Y and Cullen BR: Sequence
requirements for micro RNA processing and function in human cells.
Rna. 9:112–123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li YC, Li CF, Chen LB, Li DD, Yang L, Jin
JP and Zhang B: MicroRNA-766 targeting regulation of SOX6
expression promoted cell proliferation of human colorectal cancer.
Onco Targets Ther. 8:2981–2988. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cao J, Liu J, Xu R, Zhu X, Liu L and Zhao
X: MicroRNA-21 stimulates epithelial-to-mesenchymal transition and
tumorigenesis in clear cell renal cells. Mol Med Rep. 13:75–82.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Puglisi F, Minisini AM, De Angelis C and
Arpino G: Overcoming treatment resistance in HER2-positive breast
cancer: Potential strategies. Drugs. 72:1175–1193. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bennett PE, Bemis L, Norris DA and
Shellman YG: miR in melanoma development: miRNAs and acquired
hallmarks of cancer in melanoma. Physiol Genomics. 45:1049–1059.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
da Cruz AT and Jasiulionis MG: miRNAs and
melanoma: How are they connected? Dermatol Res Pract.
2012:5283452012.PubMed/NCBI
|
|
13
|
Mannavola F, Tucci M, Felici C, Stucci S
and Silvestris F: miRNAs in melanoma: A defined role in tumor
progression and metastasis. Expert Rev Clin Immunol. 12:79–89.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mueller DW, Rehli M and Bosserhoff AK:
miRNA expression profiling in melanocytes and melanoma cell lines
reveals miRNAs associated with formation and progression of
malignant melanoma. J Invest Dermatol. 129:1740–1751. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bertorello AM and Zhu JK: SIK1/SOS2
networks: Decoding sodium signals via calcium-responsive protein
kinase pathways. Pflugers Arch. 458:613–619. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee J, Tong T, Takemori H and Jefcoate C:
Stimulation of StAR expression by cAMP is controlled by inhibition
of highly inducible SIK1 via CRTC2, a co-activator of CREB. Mol
Cell Endocrinol. 408:80–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng H, Liu P, Wang ZC, Zou L, Santiago
S, Garbitt V, Gjoerup OV, Iglehart JD, Miron A, Richardson AL, et
al: SIK1 couples LKB1 to p53-dependent anoikis and suppresses
metastasis. Sci Signal. 2:ra352009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yao YH, Cui Y, Qiu XN, Zhang LZ, Zhang W,
Li H and Yu JM: Attenuated LKB1-SIK1 signaling promotes
epithelial-mesenchymal transition and radioresistance of non-small
cell lung cancer cells. Chin J Cancer. 35:502016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Selvik LK, Rao S, Steigedal TS, Haltbakk
I, Misund K, Bruland T, Prestvik WS, Lægreid A and Thommesen L:
Salt-inducible kinase 1 (SIK1) is induced by gastrin and inhibits
migration of gastric adenocarcinoma cells. PLoS One. 9:e1124852014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhai W, Sun Y, Jiang M, Wang M, Gasiewicz
TA, Zheng J and Chang C: Differential regulation of LncRNA-SARCC
suppresses VHL-mutant RCC cell proliferation yet promotes
VHL-normal RCC cell proliferation via modulating androgen
receptor/HIF-2α/C-MYC axis under hypoxia. Oncogene. 35:4866–4880.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei F, Cao C, Xu X and Wang J: Diverse
functions of miR-373 in cancer. J Transl Med. 13:1622015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei F, Wang Q, Su Q, Huang H, Luan J, Xu X
and Wang J: miR-373 inhibits glioma cell U251 migration and
invasion by down-regulating CD44 and TGFBR2. Cell Mol Neurobiol.
36:1389–1397. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Zhao FJ, Chen LL, Wang LQ, Nephew
KP, Wu YL and Zhang S: MiR-373 targeting of the Rab22a oncogene
suppresses tumor invasion and metastasis in ovarian cancer.
Oncotarget. 5:12291–12303. 2014.PubMed/NCBI
|
|
25
|
Eyking A, Reis H, Frank M, Gerken G,
Schmid KW and Cario E: MiR-205 and MiR-373 are associated with
aggressive human mucinous colorectal cancer. PLoS One.
11:e01568712016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ding W, Fan XL, Xu X, Huang JZ, Xu SH,
Geng Q, Li R, Chen D and Yan GR: Epigenetic silencing of ITGA2 by
MiR-373 promotes cell migration in breast cancer. PLoS One.
10:e01351282015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen D, Dang BL, Huang JZ, Chen M, Wu D,
Xu ML, Li R and Yan GR: MiR-373 drives the
epithelial-to-mesenchymal transition and metastasis via the
miR-373-TXNIP-HIF1α-TWIST signaling axis in breast cancer.
Oncotarget. 6:32701–32712. 2015.PubMed/NCBI
|
|
28
|
Qu C, He D, Lu X, Dong L, Zhu Y, Zhao Q,
Jiang X, Chang P, Jiang X, Wang L, et al: Salt-inducible kinase
(SIK1) regulates HCC progression and WNT/β-catenin activation. J
Hepatol. 64:1076–1089. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ren ZG, Dong SX, Han P and Qi J: miR-203
promotes proliferation, migration and invasion by degrading SIK1 in
pancreatic cancer. Oncol Rep. 35:1365–1374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen JL, Chen F, Zhang TT and Liu NF:
Suppression of SIK1 by miR-141 in human ovarian cancer cell lines
and tissues. Int J Mol Med. 37:1601–1610. 2016. View Article : Google Scholar : PubMed/NCBI
|