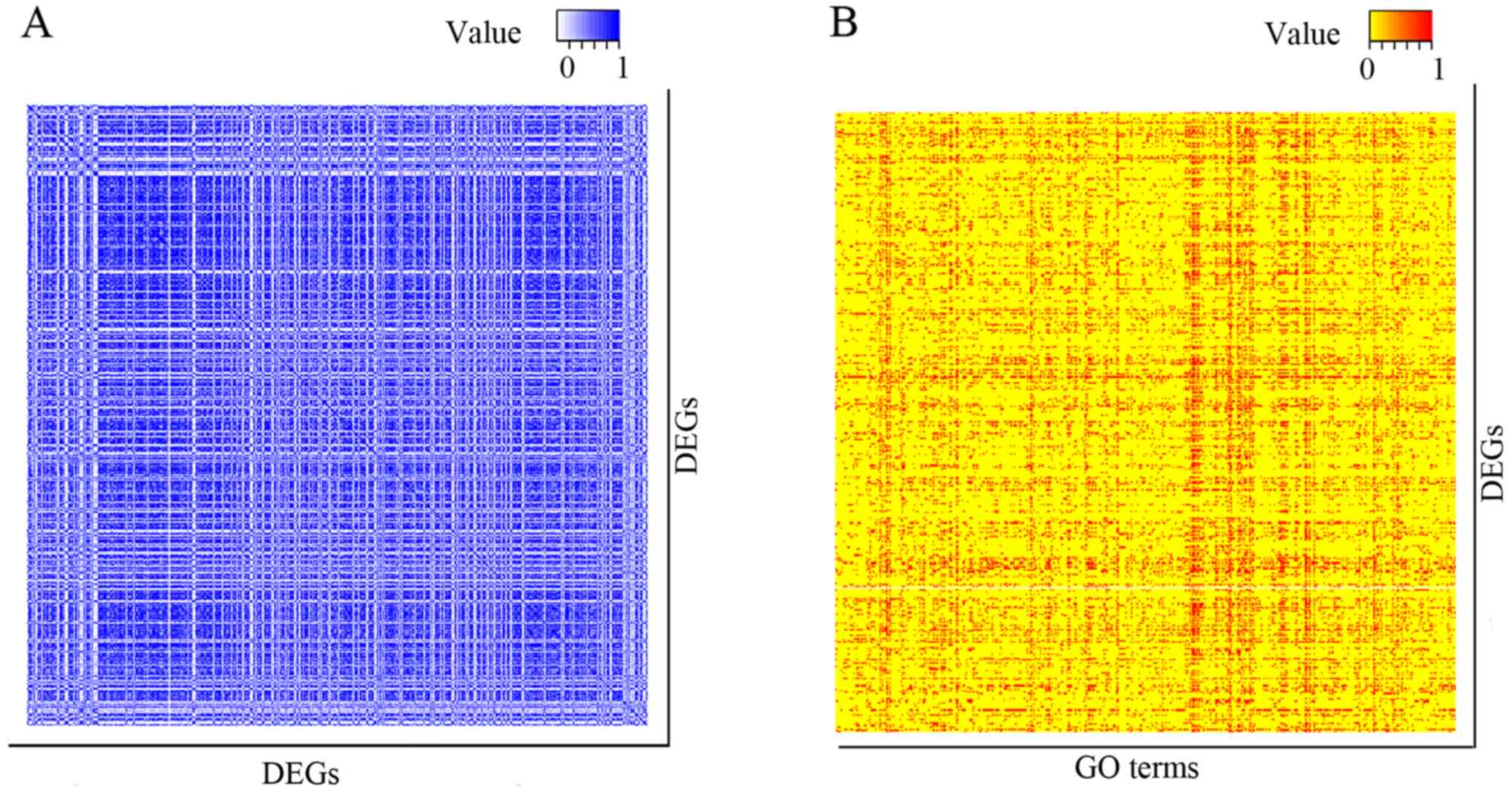
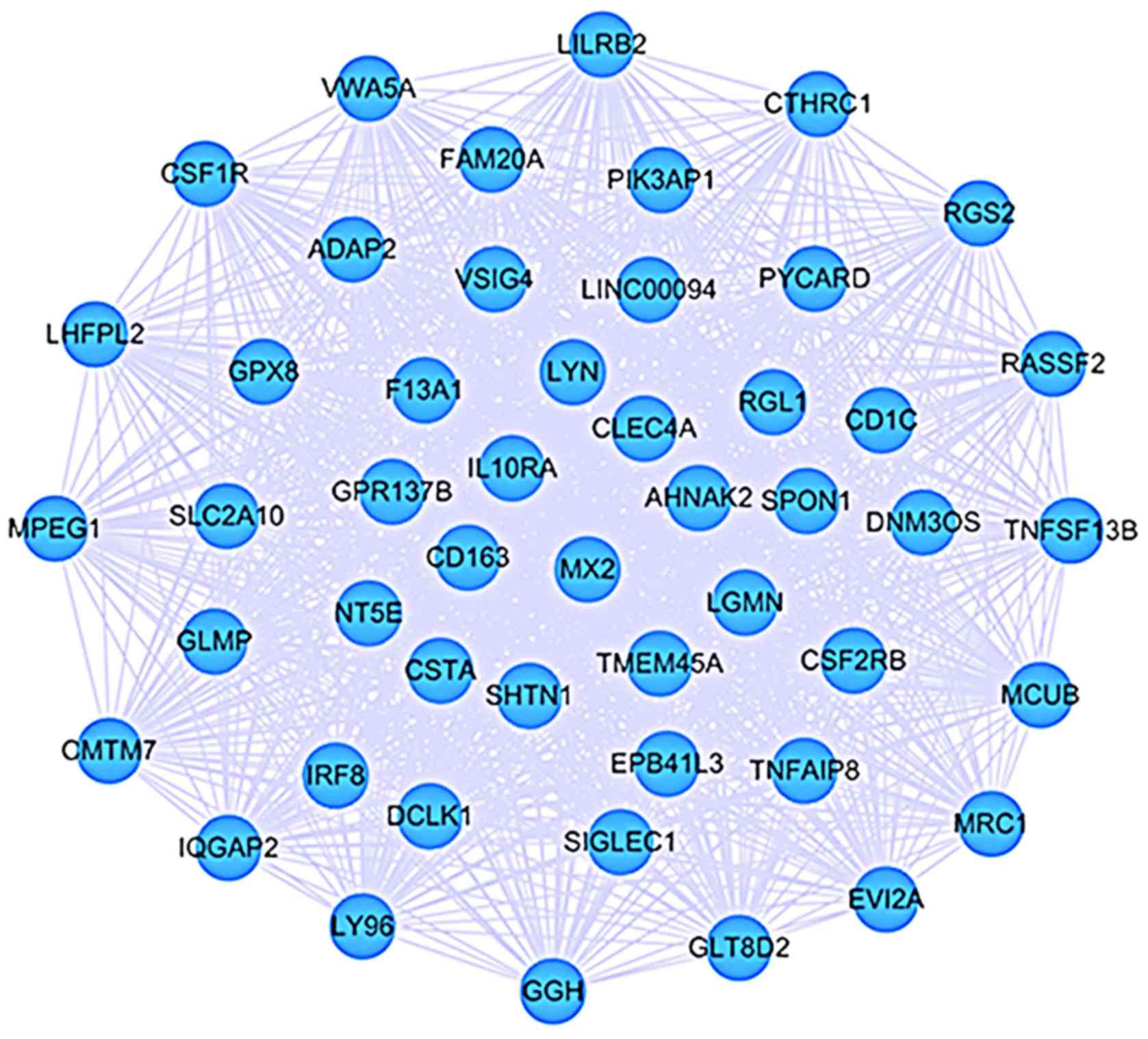
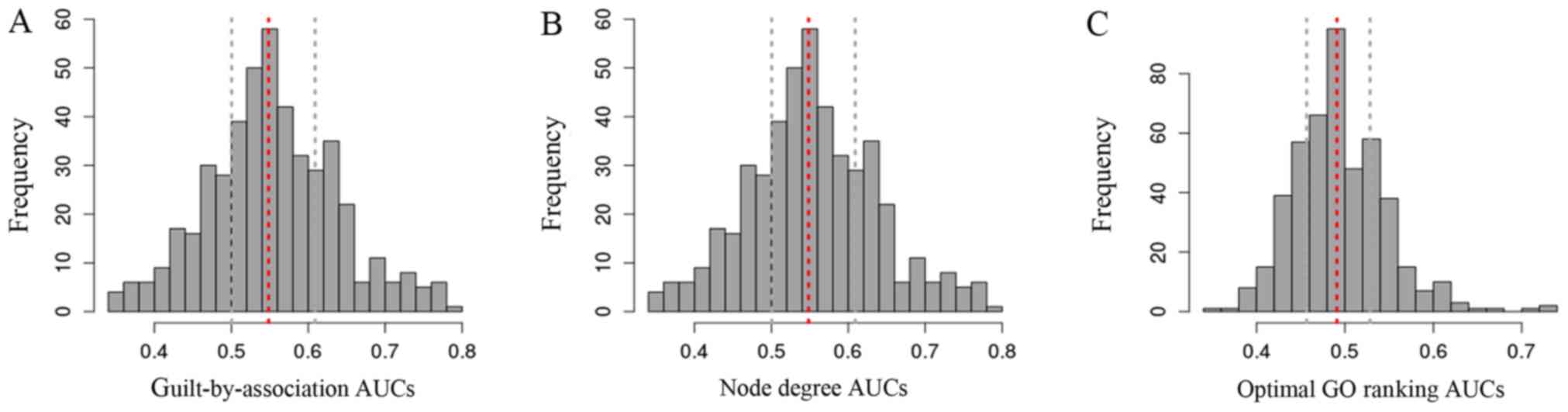
|
1
|
Nurputra DK, Lai PS, Harahap NI, Morikawa
S, Yamamoto T, Nishimura N, Kubo Y, Takeuchi A, Saito T, Takeshima
Y, et al: Spinal muscular atrophy: From gene discovery to clinical
trials. Ann Hum Genet. 77:435–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grunseich C, Zukosky K, Kats IR, Ghosh L,
Harmison GG, Bott LC, Rinaldi C, Chen KL, Chen G, Boehm M, et al:
Stem cell-derived motor neurons from spinal and bulbar muscular
atrophy patients. Neurobiol Dis. 70:12–20. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kirschner J, Schorling D, Hauschke D,
Rensing-Zimmermann C, Wein U, Grieben U, Schottmann G, Schara U,
Konrad K, Müller-Felber W, et al: Somatropin treatment of spinal
muscular atrophy: A placebo-controlled, double-blind crossover
pilot study. Neuromuscul Disord. 24:134–142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gillis J and Pavlidis P: The impact of
multifunctional genes on ‘guilt by association’ analysis. PLoS One.
6:e172582011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gillis J and Pavlidis P: ‘Guilt by
Association’ is the exception rather than the rule in gene
networks. PLoS Comput Biol. 8:e10024442012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zeng X, Liao Y, Liu Y and Zou Q:
Prediction and validation of disease genes using HeteSim Scores.
IEEE/ACM Trans Comput Biol Bioinformatics. 14:687–695. 2017.
View Article : Google Scholar
|
|
7
|
Wang S, Huang G, Hu Q and Zou Q: A
network-based method for the identification of putative genes
related to infertility. Biochim Biophys Acta 1860 (11 Pt B).
2716–2724. 2016.
|
|
8
|
Molet M, Stagner JP, Miller HC, Kosinski T
and Zentall TR: Guilt by association and honor by association: The
role of acquired equivalence. Psychon Bull Rev. 20:385–390. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vey G: Metagenomic guilt by association:
An operonic perspective. PLoS One. 8:e714842013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mulcahy PJ, Iremonger K, Karyka E,
Herranz-Martín S, Shum KT, Tam JK and Azzouz M: Gene therapy: A
promising approach to treating spinal muscular atrophy. Hum Gene
Ther. 25:575–586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Raleigh DP: Guilt by association: The
physical chemistry and biology of protein aggregation. J Phys Chem
Lett. 5:2012–2014. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Verleyen W, Ballouz S and Gillis J:
Measuring the wisdom of the crowds in network-based gene function
inference. Bioinformatics. 31:745–752. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Levy PD: Guilt by association - A closer
look at calcium, heart failure, and mortality. J Card Fail.
21:628–629. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: The Gene Ontology Consortium. Gene ontology: Tool for the
unification of biology. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huntley RP, Harris MA, Alam-Faruque Y,
Blake JA, Carbon S, Dietze H, Dimmer EC, Foulger RE, Hill DP,
Khodiyar VK, et al: A method for increasing expressivity of Gene
Ontology annotations using a compositional approach. BMC
Bioinformatics. 15:1552014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gene Ontology Consortium: Gene Ontology
Consortium: Going forward. Nucleic Acids Res 43 (D1). D1049–D1056.
2015. View Article : Google Scholar
|
|
17
|
Zou M, Liu Z, Zhang XS and Wang Y:
NCC-AUC: An AUC optimization method to identify multibiomarker
panel for cancer prognosis from genomic and clinical data.
Bioinformatics. 15:3330–3338. 2015. View Article : Google Scholar
|
|
18
|
Gillis J and Pavlidis P: The role of
indirect connections in gene networks in predicting function.
Bioinformatics. 27:1860–1866. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Willson DF: Outcomes and risk factors in
pediatric ventilator-associated pneumonia: Guilt by association.
Pediatr Crit Care Med. 16:299–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Becker D, Otto M, Ammann P, Keller I,
Drögemüller C and Leeb T: The brown coat colour of Coppernecked
goats is associated with a non-synonymous variant at the TYRP1
locus on chromosome 8. Anim Genet. 46:50–54. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rousseau B, Larrieu-Lahargue F, Javerzat
S, Guilhem-Ducléon F, Beermann F and Bikfalvi A: The
tyrp1-Tag/tyrp1-FGFR1-DN bigenic mouse: a model for selective
inhibition of tumor development, angiogenesis, and invasion into
the neural tissue by blockade of fibroblast growth factor receptor
activity. Cancer Res. 64:2490–2495. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Veiga-da-Cunha M, Hadi F, Balligand T,
Stroobant V and Van Schaftingen E: Molecular identification of
hydroxylysine kinase and of ammoniophospholyases acting on
5-phosphohydroxy-L-lysine and phosphoethanolamine. J Biol Chem.
287:7246–7255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vives-Rodriguez A, Sivaraju A and Louis
ED: Drop attacks: A clinical manifestation of LGI1 encephalitis.
Neurol Clin Pract. 7:442–443. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nikolaus M, Jackowski-Dohrmann S, Prüss H,
Schuelke M and Knierim E: Morvan syndrome associated with CASPR2
and LGI1 antibodies in a child. Neurology. 90:183–185. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Y, Huang Q, Zhou H, Wang Y, Hu X and
Li T: Inhibition of canonical WNT/β-catenin signaling is involved
in leflunomide (LEF)-mediated cytotoxic effects on renal carcinoma
cells. Oncotarget. 7:50401–50416. 2016.PubMed/NCBI
|
|
26
|
Chen Z, Gao Y, Yao L, Liu Y, Huang L, Yan
Z, Zhao W, Zhu P and Weng H: LncFZD6 initiates Wnt/β-catenin and
liver TIC self-renewal through BRG1-mediated FZD6 transcriptional
activation. Oncogene. 37:3098–3112. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wan Y, Rogers MB and Szabo-Rogers HL: A
six-gene expression toolbox for the glands, epithelium and
chondrocytes in the mouse nasal cavity. Gene Expr Patterns.
27:46–55. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cui S, Zhang W, Xiong L, Pan F, Niu Y, Chu
T, Wang H, Zhao Y and Jiang L: Use of capture-based next-generation
sequencing to detect ALK fusion in plasma cell-free DNA of patients
with non-small-cell lung cancer. Oncotarget. 8:2771–2780.
2017.PubMed/NCBI
|
|
29
|
Shojaati G, Khandaker I, Sylakowski K,
Funderburgh ML, Du Y and Funderburgh JL: Compressed collagen
enhances stem cell therapy for corneal scarring. Stem Cells Transl
Med. 7:487–494. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Drilon A, Somwar R, Mangatt BP, Edgren H,
Desmeules P, Ruusulehto A, Smith RS, Delasos L, Vojnic M,
Plodkowski AJ, et al: Response to ERBB3-directed targeted therapy
in NRG1-rearranged cancers. Cancer Discov. 8:686–695. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Varner VD and Nelson CM: Cellular and
physical mechanisms of branching morphogenesis. Development.
141:2750–2759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu M and Li J: Numb family proteins: Novel
players in cardiac morphogenesis and cardiac progenitor cell
differentiation. Biomol Concepts. 6:137–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kolb SJ and Kissel JT: Spinal muscular
atrophy. Neurol Clin. 33:831–846. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Anderton RS and Mastaglia FL: Advances and
challenges in developing a therapy for spinal muscular atrophy.
Expert Rev Neurother. 15:895–908. 2015.PubMed/NCBI
|
|
35
|
Takahashi K, Satoh M, Takahashi Y, Osaki
T, Nasu T, Tamada M, Okabayashi H, Nakamura M and Morino Y:
Dysregulation of ossification-related miRNAs in circulating
osteogenic progenitor cells obtained from patients with aortic
stenosis. Clin Sci (Lond). 130:1115–1124. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Jiang D, Zhou H, Li F, Yang J, Hong
L, Fu X, Li Z, Liu Z, Li J, et al: Expression of
RNA-interference/antisense transgenes by the cognate promoters of
target genes is a better gene-silencing strategy to study gene
functions in rice. PLoS One. 6:e174442011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Klie S, Nikoloski Z and Selbig J:
Biological cluster evaluation for gene function prediction. J
Comput Biol. 21:428–445. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zanetta C, Nizzardo M, Simone C, Monguzzi
E, Bresolin N, Comi GP and Corti S: Molecular therapeutic
strategies for spinal muscular atrophies: Current and future
clinical trials. Clin Ther. 36:128–140. 2014. View Article : Google Scholar : PubMed/NCBI
|