Introduction
The sinoatrial node (SAN), located between the
superior vena cava and right atrium, is a natural cardiac
pacemaker. Understanding the mechanisms of pacemaker cell
development during embryogenesis is important for treating SAN
defects and constructing pacemakers that capable to adapt to
physiological requirements. Previous studies have confirmed that
T-box 18 (Tbx18) has key roles in heart development, particularly
in the formation of the SAN (1–4).
Multiple studies investigating SAN head dysplasia in Tbx18
knock-out models indicated that Tbx18 functions in SAN structure
formation (2,3,5).
Multipotent epicardial progenitor cells (EPCs) originate from the
proepicardial organ, a temporary structure outside the embryonic
heart that expresses transcription factors including Tbx18, Wnt1
and transcription factor 21 (2,6–8). Cells migrate from the pro-epicardial
organ to cover the surface of the embryonic heart and form the
epicardium. Studies have confirmed that most of the epicardial
cells express the Tbx18 transcription factor (9). Tbx18-positive (Tbx18+)
pro-epicardium develops into the SAN as a process of epicardium
formation (1,3,10,11).
Therefore, Tbx18+ EPCs may be among the best candidates
to investigate the mechanisms of SAN differentiation and generate
biological pacemakers. However, the differentiation process of
Tbx18+ EPCs into pacemaker cells has not been
elucidated.
Bone morphogenetic protein (Bmp), a member of the
transforming growth factor β superfamily, regulates various
processes during embryonic development. Disruption of Bmp4
(homozygous mutant embryos following homologous recombination in
embryonic stem cells) in mice is lethal to embryos in the early
gastrulation period [embryonic day (E)6.5-E9.5] (12). In addition to its roles in the
development of bone tissues, Bmp4 has a key role in embryonic heart
development. Changes in Bmp4 localization affect heart patterning
and looping (13). Furthermore, loss
of Bmp4 expression may lead to the development of abnormal cardiac
structures (14). In addition to its
influence on the development of cardiac structures, Bmp4 promotes
fibroblast reprogramming to cardiomyocytes that have spontaneous
pacemaker activity in embryonic mice (15). Furthermore, Bmp4 is a direct target
of Shox2 (short stature homeobox 2), which has a key role in SAN
development, and the expression patterns of Bmp4 and Shox2 overlap
in the embryonic SAN (16). Bmp4 has
an important role in the differentiation of pacemaker cells;
however, the role of Bmp4 in the differentiation of
Tbx18+ EPCs to pacemaker cells has remained to be
explored. The aim of the present study was to determine whether
Bmp4 regulates the differentiation of Tbx18+ EPCs into
pacemaker cells using Tbx18:Cre/Rosa26Renhanced yellow
fluorescence protein (EYFP) lineage tracing models in
vitro.
Materials and methods
Transgenic mice and primary culture of
Tbx18+ EPCs
All animal experiments were approved by the
Institutional Animal Care and Use Committee of Chongqing Medical
University (Chongqing, China) and were in compliance with the
‘Legislation for the Protection of Animals used for Scientific
Purposes’ of the P.R. China. Tbx18:Cre knock-in transgenic mice
(Evans Laboratory; University of California, San Diego, CA, USA)
and Rosa26REYFP (Jackson Laboratory; Bar Harbor, ME,
USA) mice were bred on a C57BL/6 background obtained from the
animal center of Chongqing Medical University. E11.5
double-transgenic embryos were isolated from Tbx18:Cre female mice
that were mated with Rosa26REYFP male mice. Atria and
outflow tract tissues were removed, and EPCs were allowed to grow
out from the retained ventricles. After the ventricles were
removed, EPCs were cultured in Dulbecco's modified Eagle's medium
containing 10% fetal bovine serum (both Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The procedure used to obtain
the EPCs was in accordance with previous studies (9,17).
Tbx18+ EPCs were separated into the following four
treatment groups: Control, Bmp4 (60 ng/ml; cat. no. P5958; Abnova,
Taipei, Taiwan), Bmp4+LDN193189 [final concentration of Bmp4, 60
ng/ml and LDN193189 (final concentration, 0.5 µmol/l; cat. no.
HY-12071; MedChem Express, Monmouth Junction, NJ, USA)] and
LDN193189 (0.5 µmol/l). The Bmp4+LDN193189 group was pre-treated
with LDN193189 for 30 min prior to simultaneous treatment with Bmp4
and LDN193189. The culture medium, which contained the drugs used
to treat each group, was changed every two days over a total of 6
days.
Knockdown of Gata4 in EPCs
Knockdown experiments were performed using EPCs from
wild-type C57BL/6 mice since almost all EPCs were Tbx18+
cells, as demonstrated by the current study and a previous study
(17). Tbx18+, which was
conjugated to YFP in the mice, was identified in EPCs using
immunofluorescence. Cells were transfected with Gata4-specificsmall
interfering (si)RNA (siGata4; cat. no. MSS247225) or control siRNA
(siControl; cat. no. 12935-300) with Lipofectamine RNAiMAX (cat.
no. 13778-150; all Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocols, and cultured for 72 h
without any additional media changes. Cells were divided into the
following five treatment groups: Control, siControl, Bmp4,
Bmp4+siGata4 and siGata4.
Immunofluorescence
The culture medium was removed and cells, which were
grown on cover slips without coated, were fixed with 4%
paraformaldehyde at room temperature for 15 min, followed by
permeabilization with 0.25% Triton X-100 in PBS (0.01 mol/l) at
37°C for 10 min. Cells were blocked with 10% goat serum (Boster
Biological Technology, Pleasanton, CA, USA) at 37°C for 10 min, and
then incubated with the following primary antibodies at 4°C
overnight: Hyperpolarization-activated cyclic nucleotide gated
potassium channel 4 (Hcn4; 1:200 dilution; cat. no. ab69054; Abcam,
Cambridge, UK), Bmp4 (1:200 dilution; cat. no. MAB1049; EMD
Millipore, Billerica, MA, USA), Gata4 (1:200 dilution; ab134057)
and NK2 homeobox 5 (Nkx2.5; 1:150 dilution; cat. no. ab91196; both
Abcam). The cells were then incubated with cyanine 3-conjugated
goat anti-rabbit immunoglobulin G (CWBIO, Beijing, China) at 37°C
for 45 min, followed by DAPI at room temperature for 10 min.
Subsequently, the cells were subjected to confocal microscopy
imaging (original magnification, ×400). Imaging conditions for each
antibody were kept consistent across all samples.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Cells were collected and lysed with TRIzol (Takara
Bio Inc., Otsu, Japan) to extract total RNA, which was
reverse-transcribed to complementary DNA using a PrimeScript
Reverse Transcriptase reagent kit (cat. no. RR047A; Takara Bio
Inc.) according to the manufacturer's protocols. qPCR was performed
with SYBR premix Ex Taq (cat. no. RR820A; Takara Bio Inc.) on a
C1000 thermal cycler (BioRad Laboratories, Hercules, CA, USA) using
the following thermocycling conditions: 95°C for 35 sec, and 40
cycles of 95°C for 35 sec, 60°C for 30 sec and 72°C for 30 sec.
qPCR for Nkx2.5 was performed under similar conditions with 46
cycles due to its low mRNA expression levels. GAPDH was used as an
internal reference for each gene. Primers were provided by Sangon
Biotech Co. Ltd. (Shanghai, China) and their sequences are listed
in Table S1. The relative gene
expression levels were calculated using the
2−∆∆Cq method and normalized to GAPDH
(18).
Statistical analysis
All experiments were repeated three times. Values
are expressed as the mean ± standard deviation. The data were
analyzed using SPSS (version 20.0; IBM Corp., Armonk, NY, USA), and
one-way analysis of variance followed by a Tukey's test was applied
for comparison between groups. P<0.05 was considered to indicate
a statistically significant difference.
Results
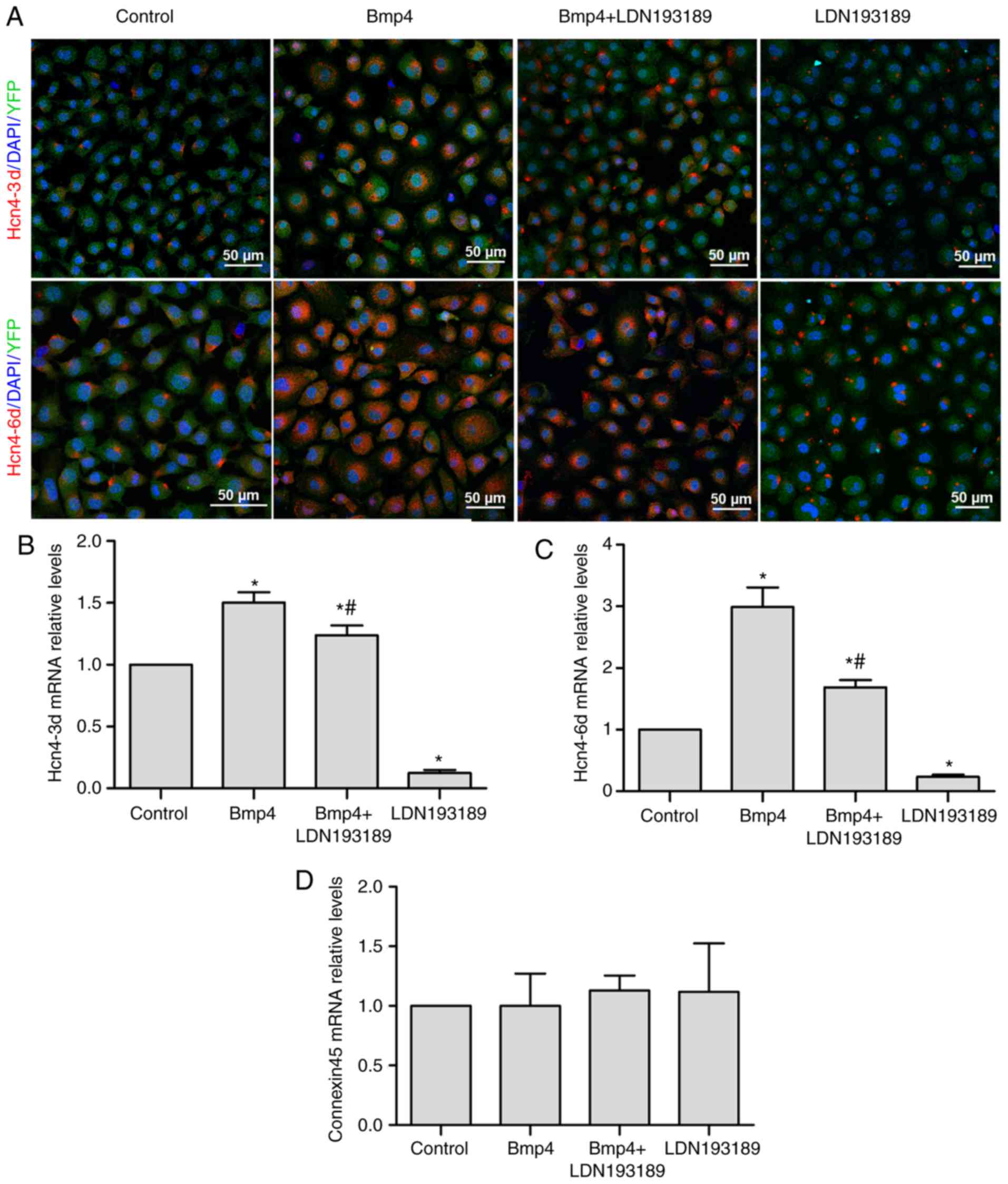
Bmp4 promotes the differentiation of
Tbx18+ EPCs to pacemaker-like cells
YFP expression in the EPCs, via which Tbx18
expression can be identified, was evident during immunofluorescence
under confocal microscope (Figs. 1
and 2). Tbx18+ EPCs were
treated with 60 ng/ml Bmp4 for 3 or 6 days. The expression of Hcn4,
a marker of pacemaker cells, was upregulated in Bmp4-treated
Tbx18+ EPCs that were isolated from
Tbx18:Cre/Rosa26REYFP mice and confirmed by
immunofluorescence analysis (Fig.
1A). The level of Hcn4 expression increased with longer
durations of Bmp4 treatment (Fig.
1A-C). The effects of Bmp4 treatment were significantly blocked
by LDN193189 treatment at days 3 and 6, as demonstrated by RT-qPCR
analysis (both P<0.05; Fig. 1B and
C). In addition, it was indicated that the expression levels of
Hcn4 in the LDN193189-treated group were significantly lower
compared with those in the control group (P<0.05). The mRNA
levels of connexin45, a gap junction protein that is expressed
mainly in the SAN, did not differ between the groups (Fig. 1D).
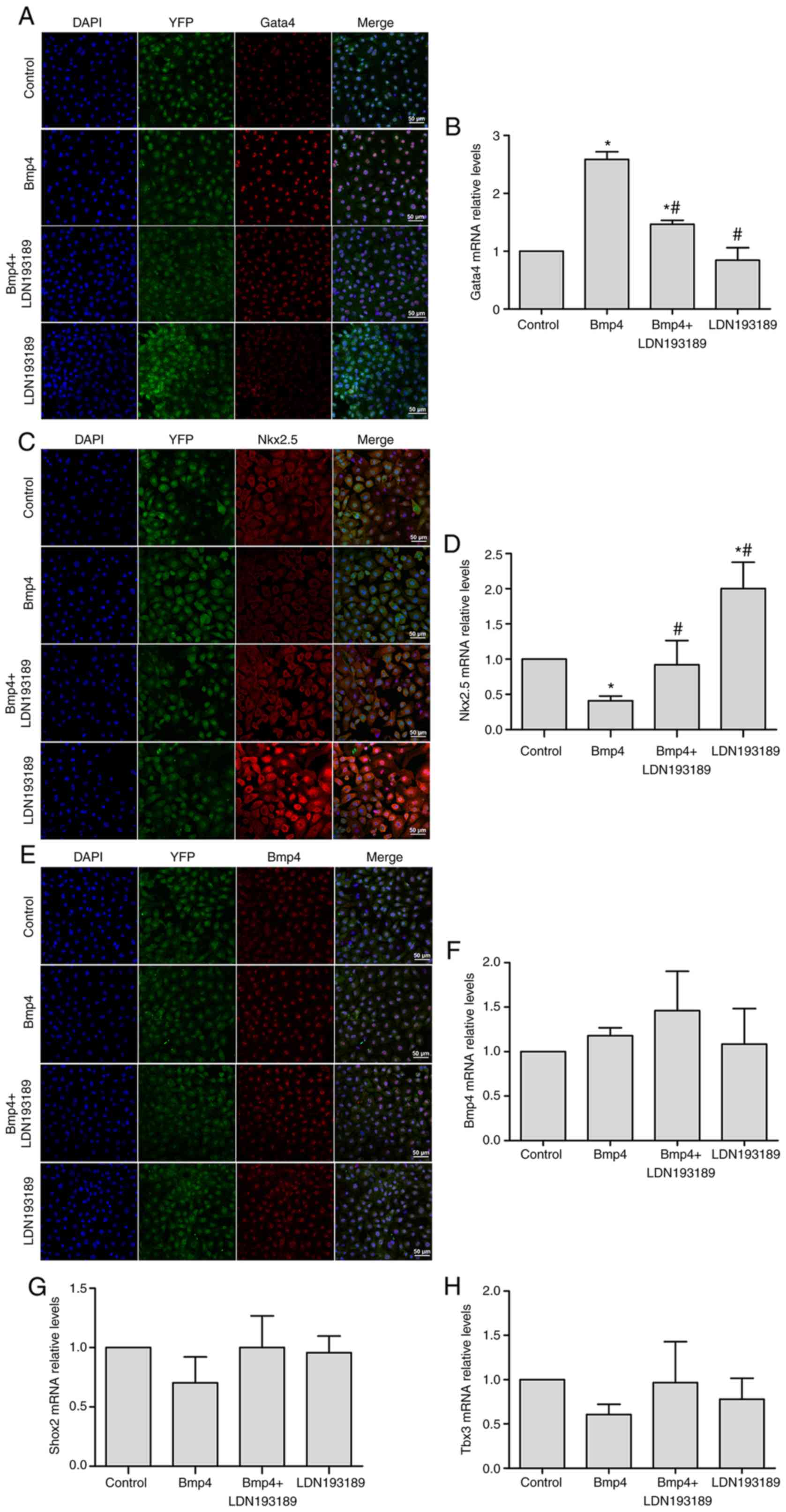 | Figure 2.Gata4 is a potential downstream
target of Bmp4. Bmp4 promotes the expression of Gata4 in
Tbx18+ EPCs, as assessed by (A) immunofluorescent
staining and (B) RT-qPCR. Inhibition of Bmp4 caused an upregulation
of Nkx2.5 expression, as assessed by (C) immunofluorescent staining
and (D) RT-qPCR. Treatment with exogenous Bmp4 had no significant
effect on the expression of Bmp4, as indicated by (E)
immunofluorescent staining and (F) RT-qPCR, and did not
significantly affect (G) Shox2 and (H) Tbx3 expression in
Tbx18+ EPCs. Representative immunofluorescence
microscopy images are presented with Gata4 or Nkx2.5 displaying in
red; the nuclei were counterstained with DAPI (blue) and the YFP
expressed by the cells is apparent (scale bar, 50 µm). Values are
expressed as the mean ± standard deviation. *P<0.05 vs. control
group; #P<0.05 vs. the Bmp4 group. EPCs, epicardial
progenitor cells; Bmp, bone morphogenetic protein; YFP, yellow
fluorescence protein; Hcn4, hyperpolarization-activated cyclic
nucleotide gated potassium channel 4; Tbx18, T-box 18; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction;
Gata4, GATA binding protein 4; Nkx2.5, NK2 homeobox 5; Shox2, short
stature homeobox 2. |
Gata4 is a potential downstream target
of Bmp4
Shox2 and Tbx3 are key regulatory factors that
mediate the patterning and pacemaker function of the SAN (3,19–24).
Gata4, a transcription factor critical in the development of the
epicardium, may interact with Bmp4 in specific signaling pathways
(25). By contrast, Nkx2.5, a
transcription factor that is not abundantly expressed in the SAN,
may prevent SAN development (3,26,27). In
the present study, the expression of these transcription factors
was determined by analyzing the downstream signaling molecules of
Bmp4 in Tbx18+ EPCs. Gata4 was expressed in >99% of
Tbx18+ EPCs and was mainly localized in the nucleus
(Fig. 2A). The addition of Bmp4 for
6 days upregulated nuclear expression of Gata4 protein in
Tbx18+ EPCs (Fig. 2A) and
significantly increased Gata4 mRNA levels compared with the control
group (P<0.05; Fig. 2B); this
upregulation was partially blocked by the co-administration of Bmp4
and LDN193189 (P<0.05; Fig. 2A and
B). The addition of the Bmp4 inhibitor caused an upregulation
of Nkx2.5 expression (P<0.05; Fig. 2C
and D), particularly in the cytoplasm (Fig. 2C). Furthermore, immunofluorescence
staining demonstrated that Bmp4 is expressed in the nuclei of
Tbx18+ EPCs and that treatment with exogenous Bmp4 did
not change the intracellular Bmp4 protein localization (Fig. 2E) and did not significantly affect
the mRNA levels (Fig. 2F). In
addition, Bmp4 had no significant effect on the expression of Shox2
or Tbx3, as indicated by RT-qPCR analysis (Fig. 2G and H). Overall, these results
indicated that Bmp4 may promote the differentiation of
Tbx18+ EPCs to pacemaker-like cells via upregulation of
Gata4 and downregulation of Nkx2.5.
Knockdown of Gata4 inhibits the
differentiation of Tbx18+ EPCs into pacemaker-like
cells
To determine whether Gata4 has a role in the
Bmp4-dependent differentiation of Tbx18+ EPCs to
pacemaker-like cells, a knockdown experiment with Gata4 was
performed. The effectiveness of Gata4 knockdown was assessed by
immunofluorescence and RT-qPCR. The results indicated that the
protein and mRNA expression of Gata4 was significantly decreased at
72 h after Gata4-siRNA transfection compared with the control group
(P<0.05; Fig. 3A and B,
respectively). No difference in Gata4 expression was identified
between the control group and the negative control group (Fig. 3A and B), indicating that knockdown of
Gata4 was caused by Gata4-siRNA and no off-target effects. The
expression of Hcn4, Nkx2.5 and Bmp4 after knockdown of Gata4 was
then assessed. Not only did knockdown of Gata4 significantly
downregulate Hcn4 mRNA expression compared with the control group
(P<0.05; Fig. 3C and D), but
Nkx2.5 mRNA expression was significantly and Nkx2.5protein
expression was markedly upregulated (P<0.05; Fig. 3E and F). However, there was no
difference in Bmp4 expression between the groups (Fig. 3G and H). The expression of Bmp4 did
not differ in the Bmp4 and Bmp4+siGATA4 groups (Fig. 3G and H).
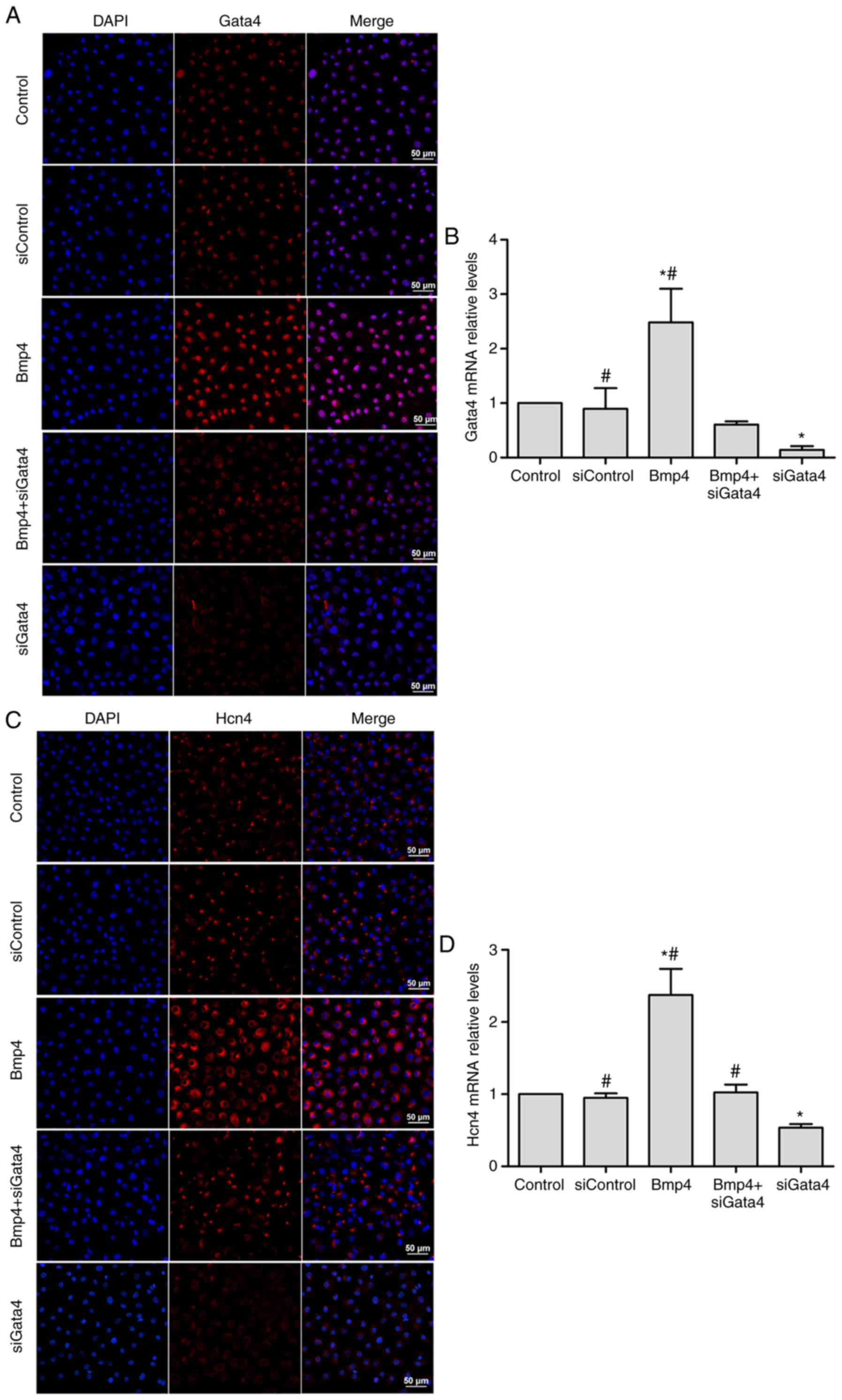 | Figure 3.Effects of Gata4 knockdown in
Tbx18+ epicardial progenitor cells. (A and B)
Confirmation of the efficacy of Gata4 knockdown by siGata4.
Knockdown of Gata4 (C and D) decreased Hcn4 expression.
Representative immunofluorescence microscopy images in (A and C)
are presented with Gata4 and Hcn4, respectively. Gata4 and Hcn4
expression is displayed in red; the nuclei were counterstained with
DAPI (blue; scale bar, 50 µm). The mRNA expression shown in (B and
D) was assessed by reverse transcription-quantitative polymerase
chain reaction. Values are expressed as the mean ± standard
deviation. *P<0.05 vs. the control group; #P<0.05
vs. the siGata4 group. Effects of Gata4 knockdown in
Tbx18+ epicardial progenitor cells. Knockdown of Gata4
(E and F) increased Nkx2.5 expression, but (G and H) did not affect
Bmp4 expression levels. Representative immunofluorescence
microscopy images in (E and G) are presented with Nkx2.5 and Bmp4,
respectively. Nkx2.5 and Bmp4 expression is displayed in red; the
nuclei were counterstained with DAPI (blue; scale bar, 50 µm). The
mRNA expression shown in (F and H) was assessed by reverse
transcription-quantitative polymerase chain reaction. Values are
expressed as the mean ± standard deviation. *P<0.05 vs. the
control group; #P<0.05 vs. the siGata4 group. Gata4,
GATA binding protein 4; siGata4, small interfering RNA targeting
Gata4; Bmp, bone morphogenetic protein; YFP, yellow fluorescence
protein; Hcn4, hyperpolarization-activated cyclic nucleotide gated
potassium channel 4; Tbx18, T-box 18; Nkx2.5, NK2 homeobox 5. |
Discussion
In the present study, a primary culture of
Tbx18+ EPCs isolated from
Tbx18:Cre/Rosa26REYFP mice was established. YFP
fluorescence in the cells of Tbx18:Cre/Rosa26REYFP mice
was used to identify the cells expressing Tbx18. It was indicated
that Bmp4 promotes the differentiation of Tbx18+ EPCs
into pacemaker-like cells in vitro, and that Gata4 is a
downstream target of Bmp4.
In the present study, ventricular EPCs were less
difficult to use than atrial EPCs, particularly from the right
atrium. Hcn4 is a specific marker of SAN pacemaker cells, and a
majority of Tbx18+ EPCs express Hcn4 protein. The ion
channels formed by Hcn4 channel proteins are required to generate
the current of pacemaker cells, which has a critical role in
spontaneous depolarization. Knockout of Hcn4 in mice results in
embryonic death (28).
Tbx18+ EPCs have the potential to differentiate into
pacemaker cells. It has been indicated that exogenous Bmp4 promotes
the mRNA and protein expression of Hcn4, suggesting that Bmp4 is a
critical factor in the differentiation of Tbx18+ EPCs
into pacemaker-like cells. Indeed, extended treatment with Bmp4
amplified the effects on the expression of downstream targets.
LDN193189, an effective Bmp4 inhibitor, inhibits BMP type I
receptor kinases and subsequently the downstream signal effectors
(29,30). Immunofluorescence and RT-qPCR
analyses revealed that the expression levels of Hcn4 were lower in
the LDN193189-treated group than in the control group. As Bmp4 was
expressed in Tbx18+ EPCs, it was hypothesized that Bmp4
generated by Tbx18+ EPCs had an autocrine and a
paracrine effect that may be partially blocked by exogenous
LDN193189. Furthermore, Bmp4 treatment had no effect on the
expression of Bmp4 in Tbx18+ EPCs, suggesting that Bmp4
exerts its functions via a signaling mechanism through membrane
receptors. This is consistent with the classical mechanism of Bmp4
(31) and with the notion that Bmp4
does not stimulate its own expression. A previous study using mouse
embryonic stem cells (embryoid bodies) performed by Hashem et
al (23) demonstrated that
disruption of Shox2 downregulated Bmp4 and Hcn4, while addition of
Bmp4 partially rescued this effect. This is consistent with a
previous study indicating that Bmp4 directly affects the expression
of Hcn4 in the development of the dorsal mesenchymal protrusions
(24). Taken together, these results
are consistent with those of the present study, indicating that
Bmp4 is an upstream regulator of Hcn4 in Tbx18+ EPCs.
Hcn4 and Connexin45 are specific markers of pacemaker cells.
Connexin45, but not connexin 40 or connexin 43, is expressed in
pacemaker cells (32–34). In the present study, no changes in
connexin45 mRNA expression were observed after Bmp4 treatment,
indicating that the cells may have differentiated into
pacemaker-like cells lacking this feature. However, Hcn4 was
affected.
Tbx3 is expressed in the embryonic SAN (20). Loss of Tbx3 in the SAN leads to
expression of mature myocardium-specific genes, while abnormal
expression of Tbx3 upregulates the expression of Hcn4, forming a
pacemaker in the atria (21,35,36).
However, Tbx3 is not required for the formation of the SAN
structure (3). In addition, the
expression of Shox2 is restricted to the sinoatrial node and the
venous valves. Shox2-deficient embryos have markedly decreased SAN,
dysfunctional cardiac pacemaker activity and reduced Hcn4
expression (26,37,38). The
present results indicated that Bmp4 promotes Hcn4 expression via
upregulation of Gata4, while transcription of Shox2 and Tbx3 was
not affected by Bmp4. However, the present study also suggested
that Tbx18+ EPCs do not abundantly express the Nkx2.5
transcription factor, which is consistent with the results of other
studies (3,39). Taken together, it is indicated that
Nkx2.5 inhibits SAN differentiation, and its expression is
regulated by Bmp4 and Gata4.
In the present study, the mRNA and protein
expression of Gata4 was effectively silenced by siGata4. There was
no significant difference in the mRNA expression of Gata4 between
the Bmp4+siGata4 group and the siGata4 group even though Bmp4
upregulated Gata4, which may be attributed to transcriptional gene
silencing of Gata4. However, Hcn4 expression levels were higher in
the Bmp4+siGata4-treated group compared with those in the
siGata4-treated group, indicating that there may be other
transcription factors in the same regulatory network compensating
for Hcn4 expression. In addition, almost all of the EPCs isolated
from the Tbx18:Cre/Rosa26REYFP mice were
Tbx18+ according to the immunofluorescence analysis.
These results indicated the EPCs used in the Gata4 silencing
experiment were Tbx18 positive as well. The expression of Gata4 and
Nkx2.5 were affected by Bmp4, while Gata4 affected the expression
of Nkx2.5. Inhibition of Nkx2.5 is essential for the
differentiation of EPCs into pacemaker cells (26), and Nkx2.5 expression changes their
fate to form the SAN as a vital part of the working myocardium
(38,40). It may therefore be speculated that
high levels of Nkx2.5 in EPCs after knockdown of Gata4 downregulate
Hcn4 expression to promote their differentiation into atrial
myocytes. In addition, the low mRNA expression level of Nkx2.5
according to qPCR results in the current study may mean that these
results are dubious. In conclusion, the present study explored the
association between Bmp4, Gata4 and Hcn4 in Tbx18+ EPCs
and revealed that the expression of Nkx2.5 is regulated by Bmp4 and
Gata4, providing important information for further studies.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation (grant no. 81770251).
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
QS and LW contributed to the conception of the study
and were major contributors in writing the manuscript. JD and XJ
made substantial contributions to the analysis and interpretation
of data and critical reading of the manuscript. YY, SD and ZH
contributed to the acquisition of data and helped perform the
analysis, including constructive discussions. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments in the present study were
approved by the Institutional Animal Care and Use Committee of
Chongqing Medical University and were in compliance with the
‘Legislation for the Protection of Animals used for Scientific
Purposes’ of the P.R. China.
Patients' consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Greulich F, Rudat C and Kispert A:
Mechanisms of T-box gene function in the developing heart.
Cardiovasc Res. 91:212–222. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cai CL, Martin JC, Sun Y, Cui L, Wang L,
Ouyang K, Yang L, Bu L, Liang X, Zhang X, et al: A myocardial
lineage derives from Tbx18 epicardial cells. Nature. 454:104–108.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wiese C, Grieskamp T, Airik R, Mommersteeg
MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF,
Kispert A and Christoffels VM: Formation of the sinus node head and
differentiation of sinus node myocardium are independently
regulated by Tbx18 and Tbx3. Circ Res. 104:388–397. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Christoffels VM, Grieskamp T, Norden J,
Mommersteeg MT, Rudat C and Kispert A: Tbx18 and the fate of
epicardial progenitors. Nature. 458:E8–E9; discussion E9-E10. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Grieskamp T, Rudat C, Lüdtke TH, Norden J
and Kispert A: Notch signaling regulates smooth muscle
differentiation of epicardium-derived cells. Circ Res. 108:813–823.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jenkins SJ, Hutson DR and Kubalak SW:
Analysis of the proepicardium-epicardium transition during the
malformation of the RXRalpha-/- epicardium. Dev Dyn. 233:1091–1101.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Witty AD, Mihic A, Tam RY, Fisher SA,
Mikryukov A, Shoichet MS, Li RK, Kattman SJ and Keller G:
Generation of the epicardial lineage from human pluripotent stem
cells. Nat Biotechnol. 32:1026–1035. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tandon P, Miteva YV, Kuchenbrod LM,
Cristea IM and Conlon FL: Tcf21 regulates the specification and
maturation of proepicardial cells. Development. 140:2409–2021.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jing X, Gao Y, Xiao S, Qin Q, Wei X, Yan
Y, Wu L, Deng S, Du J, Liu Y and She Q: Hypoxia induced the
differentiation of Tbx18-positive epicardial cells to CoSMCs. Sci
Rep. 6:304682016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Wijk B, van den Berg G, Abu-Issa R,
Barnett P, van der Velden S, Schmidt M, Ruijter JM, Kirby ML,
Moorman AF and van den Hoff MJ: Epicardium and myocardium separate
from a common precursor pool by crosstalk between bone
morphogenetic protein- and fibroblast growth factor-signaling
pathways. Circ Res. 105:431–441. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Norden J, Greulich F, Rudat C, Taketo MM
and Kispert A: Wnt/β-catenin signaling maintains the mesenchymal
precursor pool for murine sinus horn formation. Circ Res.
109:e42–e50. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Winnier G, Blessing M, Labosky PA and
Hogan BL: Bone morphogenetic protein-4 is required for mesoderm
formation and patterning in the mouse. Genes Dev. 9:2105–2116.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen JN, van Eeden FJ, Warren KS, Chin A,
Nüsslein-Volhard C, Haffter P and Fishman MC: Left-right pattern of
cardiac BMP4 may drive asymmetry of the heart in zebrafish.
Development. 124:4373–4382. 1997.PubMed/NCBI
|
|
14
|
McCulley DJ, Kang JO, Martin JF and Black
BL: BMP4 is required in the anterior heart field and its
derivatives for endocardial cushion remodeling, outflow tract
septation, and semilunar valve development. Dev Dyn. 237:3200–3209.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Efe JA, Hilcove S, Kim J, Zhou H, Ouyang
K, Wang G, Chen J and Ding S: Conversion of mouse fibroblasts into
cardiomyocytes using a direct reprogramming strategy. Nat Cell
Biol. 13:215–222. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Puskaric S, Schmitteckert S, Mori AD,
Glaser A, Schneider KU, Bruneau BG, Blaschke RJ, Steinbeisser H and
Rappold G: Shox2 mediates Tbx5 activity by regulating Bmp4 in the
pacemaker region of the developing heart. Hum Mol Genet.
19:4625–4633. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qin Q, Wang J, Yan Y, Jing X, Du J, Deng
S, Wu L, Liu Y and She Q: Angiotensin II induces the
differentiation of mouse epicardial progenitor cells into vascular
smooth muscle-like cells. Biochem Biophys Res Commun. 480:696–701.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Frank DU, Carter KL, Thomas KR, Burr RM,
Bakker ML, Coetzee WA, Tristani-Firouzi M, Bamshad MJ, Christoffels
VM and Moon AM: Lethal arrhythmias in Tbx3-deficient mice reveal
extreme dosage sensitivity of cardiac conduction system function
and homeostasis. Proc Natl Acad Sci USA. 109:E154–E163. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hoogaars WM, Tessari A, Moorman AF, de
Boer PA, Hagoort J, Soufan AT, Campione M and Christoffels VM: The
transcriptional repressor Tbx3 delineates the developing central
conduction system of the heart. Cardiovasc Res. 62:489–499. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bakker ML, Boukens BJ, Mommersteeg MT,
Brons JF, Wakker V, Moorman AF and Christoffels VM: Transcription
factor Tbx3 is required for the specification of the
atrioventricular conduction system. Circ Res. 102:1340–1349. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Boogerd CJ, Wong LY, van den Boogaard M,
Bakker ML, Tessadori F, Bakkers J, 't Hoen PA, Moorman AF,
Christoffels VM and Barnett P: Sox4 mediates Tbx3 transcriptional
regulation of the gap junction protein Cx43. Cell Mol Life Sci.
68:3949–3961. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hashem SI, Lam ML, Mihardja SS, White SM,
Lee RJ and Claycomb WC: Shox2 regulates the pacemaker gene program
in embryoid bodies. Stem Cells Dev. 22:2915–2926. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun C, Yu D, Ye W, Liu C, Gu S, Sinsheimer
NR, Song Z, Li X, Chen C, Song Y, et al: The short stature homeobox
2 (Shox2)-bone morphogenetic protein (BMP) pathway regulates dorsal
mesenchymal protrusion development and its temporary function as a
pacemaker during cardiogenesis. J Biol Chem. 290:2007–2023. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nemer G and Nemer M: Transcriptional
activation of BMP-4 and regulation of mammalian organogenesis by
GATA-4 and −6. Dev Biol. 254:131–148. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Espinoza-Lewis RA, Yu L, He F, Liu H, Tang
R, Shi J, Sun X, Martin JF, Wang D, Yang J and Chen Y: Shox2 is
essential for the differentiation of cardiac pacemaker cells by
repressing Nkx2-5. Dev Biol. 327:376–385. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jay PY, Harris BS, Maguire CT, Buerger A,
Wakimoto H, Tanaka M, Kupershmidt S, Roden DM, Schultheiss TM,
O'Brien TX, et al: Nkx2-5 mutation causes anatomic hypoplasia of
the cardiac conduction system. J Clin Invest. 113:1130–1137. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Stieber J, Herrmann S, Feil S, Löster J,
Feil R, Biel M, Hofmann F and Ludwig A: The
hyperpolarization-activated channel HCN4 is required for the
generation of pacemaker action potentials in the embryonic heart.
Proc Natl Acad Sci USA. 100:15235–15240. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu PB, Deng DY, Lai CS, Hong CC, Cuny GD,
Bouxsein ML, Hong DW, McManus PM, Katagiri T, Sachidanandan C, et
al: BMP type I receptor inhibition reduces heterotopic
ossification. Nat Med. 14:1363–1369. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Katakawa Y, Funaba M and Murakami M:
Smad8/9 is regulated through the BMP pathway. J Cell Biochem.
117:1788–1796. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Miyazono K, Kusanagi K and Inoue H:
Divergence and convergence of TGF-beta/BMP signaling. J Cell
Physiol. 187:265–276. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Verheijck EE, van Kempen MJ, Veereschild
M, Lurvink J, Jongsma HJ and Bouman LN: Electrophysiological
features of the mouse sinoatrial node in relation to connexin
distribution. Cardiovasc Res. 52:40–50. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Christoffels VM, Smits GJ, Kispert A and
Moorman AF: Development of the pacemaker tissues of the heart. Circ
Res. 106:240–254. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yamamoto M, Dobrzynski H, Tellez J, Niwa
R, Billeter R, Honjo H, Kodama I and Boyett MR: Extended atrial
conduction system characterised by the expression of the HCN4
channel and connexin45. Cardiovasc Res. 72:271–281. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hoogaars WM, Engel A, Brons JF, Verkerk
AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P,
et al: Tbx3 controls the sinoatrial node gene program and imposes
pacemaker function on the atria. Genes Dev. 21:1098–1112. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bakker ML, Boink GJ, Boukens BJ, Verkerk
AO, van den Boogaard M, den Haan AD, Hoogaars WM, Buermans HP, de
Bakker JM, Seppen J, et al: T-box transcription factor TBX3
reprogrammes mature cardiac myocytes into pacemaker-like cells.
Cardiovasc Res. 94:439–449. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ye W, Wang J, Song Y, Yu D, Sun C, Liu C,
Chen F, Zhang Y, Wang F, Harvey RP, et al: A common Shox2-Nkx2-5
antagonistic mechanism primes the pacemaker cell fate in the
pulmonary vein myocardium and sinoatrial node. Development.
142:2521–2532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Blaschke RJ, Hahurij ND, Kuijper S, Just
S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J,
Hardt SE, et al: Targeted mutation reveals essential functions of
the homeodomain transcription factor Shox2 in sinoatrial and
pacemaking development. Circulation. 115:1830–1838. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Christoffels VM, Mommersteeg MT, Trowe MO,
Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler
K, Harvey RP, Moorman AF and Kispert A: Formation of the venous
pole of the heart from an Nkx2-5-negative precursor population
requires Tbx18. Circ Res. 98:1555–1563. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Espinoza-Lewis RA, Liu H, Sun C, Chen C,
Jiao K and Chen Y: Ectopic expression of Nkx2.5 suppresses the
formation of the sinoatrial node in mice. Dev Biol. 356:359–369.
2011. View Article : Google Scholar : PubMed/NCBI
|