Introduction
Myocardial infarction (MI), a common presentation
for ischemic heart disease/coronary artery disease, is a major
contributor to mortality rates worldwide (1). The major trigger of MI is generally
owing to thrombus formation in a coronary artery. Accordingly, the
current treatments mainly include medical therapy (e.g.
anticoagulant medications, antiplatelets) and elective
catheterization. However, some disadvantages can not be ignored,
such as high risk of bleeding in medical therapy and high cost of
treatment in elective catheterization (2). In order to effectively prevent and
treat MI, seeking the exact pathogenesis of MI at various aspects
is necessary. Currently research directions mainly include
improvement of coronary blood flow, inhibition of apoptosis,
reduction of oxygen consumption, and revascularization procedures
(3). However, few studies have been
conducted in the pathogenesis of MI at the non-coding RNA (ncRNA)
level.
ncRNAs, RNA molecules, are not translated into
proteins, which mainly include ribosomal RNAs (rRNAs), transfer
RNAs (tRNAs), short non-coding RNAs such as microRNAs (miRNAs) and
the long non-coding RNAs (lncRNAs). lncRNAs are a set of RNAs
longer than 200 nucleotides, and participate in many fundamental
biological processes mainly including genomic imprinting and
chromatin modification (4). Although
the effects of lncRNAs on various cancers (5) and neuronal diseases (6,7) have
been widely investigated, the study number of lncRNA function in
cardiovascular diseases is very limited. Currently, some
transcripts have been investigated for their potential role as
biomarkers of cardiovascular diseases. The mitochondrial lncRNA
LIPCAR, as a novel biomarker, can predict future death in heart
failure patients (8). lncRNA
CoroMarker is a diagnostic biomarker for coronary artery disease
(9). In addition, lncRNA MIAT might
regulate MI via functioning as a competing endogenous RNA for
various targets (10,11).
miRNAs are short non-coding RNAs (approximately 20
nucleotides) and negatively regulate target genes. Different from
lncRNAs, miRNAs have been largely investigated in the context of MI
(12). The relationship between
miRNAs and lncRNAs has been extensively demonstrated, among which
lncRNAs can indirectly compete with mRNAs through binding to miRNAs
(13,14). For instance, lncRNA CHRF could
regulate cardiac hypertrophy by targeting miR-489 (15). Recognizing lncRNA competitively
regulated subpathway can reveal the pathogenesis of disease and the
molecular mechanism of lncRNAs in the disease context. However, the
relevant regulated mechanism of lncRNAs in MI remains unclear.
Subpathway-LNCE, as a novel method, can effectively integrate
lncRNA-mRNA expression profile and identify lncRNA competitively
regulated subpathway (16).
Moreover, subpathway-LNCE method is more accurate, advanced and its
calculation results are more relevant to disease. Accordingly, we
used the subpathway-LNCE method to investigate pathogenesis of MI
at genetic level.
Materials and methods
Recruitment and pretreatment of gene
expression data
Gene expression profiles with accession number
(GSE34198) for MI were recruited from the Gene Expression Omnibus
(GEO) (http://www.ncbi.nlm.nih.gov/geo/) database. GEO is a
public functional genomics data repository that freely distributes
high throughput gene expression profiles and other functional
genomics data sets. GSE34198 consists of 48 healthy controls and 49
patients with MI, and is deposited on GPL6102 data platform.
For purpose of controlling the quality of GSE34198,
GSE34198 was pretreated as follows: Background correction was
performed by Robust Multi-array Average (RMA) algorithm (17), the normalization was evaluated by
quantiles algorithm (18), perfect
match and mismatch was revised by Micro Array Suite (MAS) algorithm
(19), and all expression values
were summarized by medianpolish method (17). After removing invalid or duplicated
probes, we converted them into gene symbols by the annotate package
(20). Thus, 19,027 genes were
obtained in the pretreated GSE34198 of MI for further
application.
Constructing the candidate lncRNA-mRNA
interactions
As far as we known, small non-coding RNAs target
Base version 2.0 (starBase v2.0, http://starbase.sysu.edu.cn/) has been used to
identify the RNA-RNA and protein-RNA interaction networks. Hence,
miRNA-lncRNA interactions and miRNA-mRNA interactions were
collected using StarBase v2.0. The miRNA-mRNA interactions were
acquired based on miRecords (21),
mir2Disease (22), mirTarBase
(23) and TarBase (24). Then candidate lncRNA-mRNA
competitively regulated relationships were constructed using their
shared miRNAs between miRNA-mRNA and miRNA-lncRNA interactions. To
ensure the data reliability, we used two criteria to identify the
candidate competing miRNA of each lncRNA as follows: i)
hypergeometric test of shared miRNAs under a threshold of P=0.05
and ii) Jaccard Coefficient of shared miRNAs rank at top 20%.
To make the candidate lncRNA-mRNA interactions
involved in MI, all the genes in GSE34198 were mapped on them and
the intersections were selected for further analysis.
Obtaining the lncRNA and mRNA
co-expression relationship pairs
To screen the candidate lncRNA-mRNA interactions, we
evaluated co-expression for any pair of relations in the candidate
lncRNA-mRNA network using Pearson's correlation coefficient.
Subsequently, the significance of Pearson's correlation coefficient
was evaluated by Fisher's Z-transform, which converts the values
into the normally distributed variable Z. Then the Z-transform test
utilizes the one-to-one mapping of the standard normal curve to the
P-value of a one tailed test. When the calculation value exceeded a
significant positive threshold (P<0.05), the lncRNA-mRNA
co-expression relationship pairs (LncGenePairs) were retained.
Reconstructing condition-specific
lncRNA competitively regulated signal pathways
We used Fisher's test to identify the gene
enrichment pathways in the mRNA-gene expression profiles, and the
pathways were obtained from KEGG database. The gene enrichment
pathways were collected when the P-value of gene enrichment pathway
was no longer than 0.01, and considered as the candidate difference
pathways. We put lncRNAs in the LncGenePairs into the candidate
difference pathway graphs as nodes, then we acquired
condition-specific lncRNA regulated signal pathways (csLncRPs). The
lncRNA nodes were considered as signature nodes.
Located subpathways within pathways
according to signature nodes
Signature nodes represent information on the
competing regulation and genes of interest, which can help to
efficiently locate subpathways through further considering their
topologies within pathways. Moreover, distances are usually similar
between certain nodes in a subpathway. We utilized ‘lenient
distance’ similarity of signature nodes to locate subpathways
competing for regulation by lncRNAs. We computed the shortest path
between any two signature nodes as follows: If the molecule number
between two signature nodes was less than n, they were combined
into a single node. Finally, the node number in the molecule sets
within pathway was longer than s, and defined as subpathway
regions. The n parameter conducts the intensity of regulated
signals, and the s parameter regulates the size of candidate
subpathways. Thus, n=1 and s=8 were used as default parameters.
Statistical significance of candidate
subpathways
Wallenius approximation methods were applied to
evaluate the statistical significance of each subpathway. The
needed values were shown as follows: i) the number of interesting
mRNAs (x) submitted for analysis; ii) background mRNAs (n) number;
iii) the number of background mRNAs (m1) annotated to each
subpathway; iv) the number of interesting mRNAs (m2) annotated to
each subpathway and v) the weight of each subpathway (w), which
indicated the intensity of competitively regulated lncRNAs involved
in this subpathway. The formula of the subpathway weight is as
follows:
W=1+β(−log2(GLPG))
According to the formula above, PG
represents the number of mRNAs in the subpathways. GL
represents the number of mRNAs competing regulation by lncRNAs in
this subpathway. Moreover, β is the control parameter (β =1). The
Wallenius approximation methods were carried out by R package
BiasedUrn (25).
Identifying hub lncRNAs
According to the values of the subpathways, the
subpathways competing for regulation by lncRNAs were obtained and
constructed, that is, lncRNA-mRNA networks were constructed. Then,
we collected the hub lncRNAs when the lncRNA degree was longer than
the average lncRNA degree in the lncRNA-mRNA networks.
Results
Identifying the relationship between
lncRNAs and mRNAs for MI
In this study, 19,027 genes were selected from
GSE34198 of MI after standard corrections and normalizations. To
understand the relationship between lncRNA and mRNA, we extracted
lncRNA-mRNA interactions by their shared miRNAs based on mRNA-miRNA
interactions and lncRNA-miRNA interactions. All genes in GSE34198
were mapped on these lncRNA-mRNA interactions and the intersections
were selected. We then collected these intersections which
satisfied P<0.05. The results revealed that 7,693 lncRNA-mRNA
interactions were obtained, which included 835 lncRNAs and 1,749
mRNAs. Next, 1,681 mRNAs and 112 lncRNAs were indentified for MI
after taking intersections with 19,027 genes in gene expression
data. Subsequently, Pearson's correlation coefficient was performed
to evaluate co-expression for any pair of relations in the
intersections. We obtained 300 lncRNA-mRNA co-expression
relationship pairs (LncGenePairs), among which include 58 lncRNAs,
and 243 mRNAs. The top 6 LncGenePairs are illustrated in Table I.
 | Table I.The diagram of the top 6
LncGenePairs. |
Table I.
The diagram of the top 6
LncGenePairs.
| Lnc | Gene | corValue | P-value |
|---|
| TDRG1 | HENMT1 |
0.220560501443871 |
0.0299373311676897 |
| LIMD1-AS1 | LRRC56 |
0.246484637523062 |
0.0149413386406672 |
| LEF1-AS1 | MAGEH1 |
0.454787669314928 |
2.87221540632211×10−6 |
| SNHG11 | MRPL53 |
0.680510717664986 |
1.75464015432401×10−14 |
| LINC00485 | NME7 |
0.249365243302853 |
0.0137715015048636 |
| LEF1-AS1 | SLC27A5 |
0.424727193889112 |
0.0000145102562949063 |
Reconstruction of condition-specific
lncRNA competitively regulated signal pathways
In order to obtain the lncRNA competitively
regulated signal pathways (csLncRPs), we firstly annotated the
mRNAs in the mRNAs-gene expression profiles into KEGG pathways. A
total of 62 candidate difference pathways were obtained, among
which the top 6 pathways were selected as shown in Table II. We then put lncRNAs based on the
LncGenePairs in the candidate difference pathways as nodes through
linking to their regulated mRNAs. Hence, a total of 1,241 csLncRPs
were acquired, among which the top 14 pathways were selected as
shown in Table III.
 | Table II.The diagram of the top 6 candidate
different pathways. |
Table II.
The diagram of the top 6 candidate
different pathways.
| Index | Pathway_id | Pathway_pvalue | p.adjust (FDR) |
|---|
| 05215 | hsa05215: Prostate
cancer |
9.99732401124532×10−12 |
9.99732401124532×10−12 |
| 05200 | hsa05200: Pathways
in cancer |
8.45066918278111×10−14 |
8.45066918278111×10−14 |
| 05220 | hsa05220: Chronic
myeloid leukemia |
7.81664671803157×10−17 |
7.81664671803157×10−17 |
| 04910 | hsa04910: Insulin
signaling pathway |
0.0000771758111031205 |
0.0000771758111031205 |
| 04510 | hsa04510: Focal
adhesion |
6.65828057822821×10−06 |
6.65828057822821×10−06 |
| 05166 | hsa05166: HTLV–I
infection |
6.59501272205171×10−07 |
6.59501272205171×10−07 |
 | Table III.The diagram of the top 14
csLncRPs. |
Table III.
The diagram of the top 14
csLncRPs.
| path_name | matched_lnc | matched_gene |
|---|
| 3008 | DLEU2 | BMS1 |
| 3008 | C14orf169 | BMS1 |
| 3008 |
ERVK13-1 | RCL1 |
| 3008 |
PCBP1-AS1 | POP1 |
| 3008 |
ERVK13-1 | MDN1 |
| 3008 |
ERVK13-1 | WDR75 |
| 3008 |
ERVK13-1 | UTP15 |
| 3008 | PDXDC2P | HEATR1 |
| 3008 |
ERVK13-1 | WDR43 |
| 3008 | ERVK13–1 | LSG1 |
| 3010 |
ERVK13-1 | RPS6 |
| 3010 |
ERVK13-1 | RPS27 |
| 3010 |
ERVK13-1 | RPS25 |
| 3010 |
ERVK13-1 | RPS3A |
Identifying the signal subpathways
competitively regulated by lncRNA for MI
The csLncRPs were calculated using lenient distance
similarity to obtain the lncRNA competitively regulated
subpathways. Then the Wallenius approximation methods were
performed to evaluate the significance of signal subpathways. A
total of 65 lncRNA competitively regulated subpathways were
obtained, among which the top rank 6 subpathways are listed in
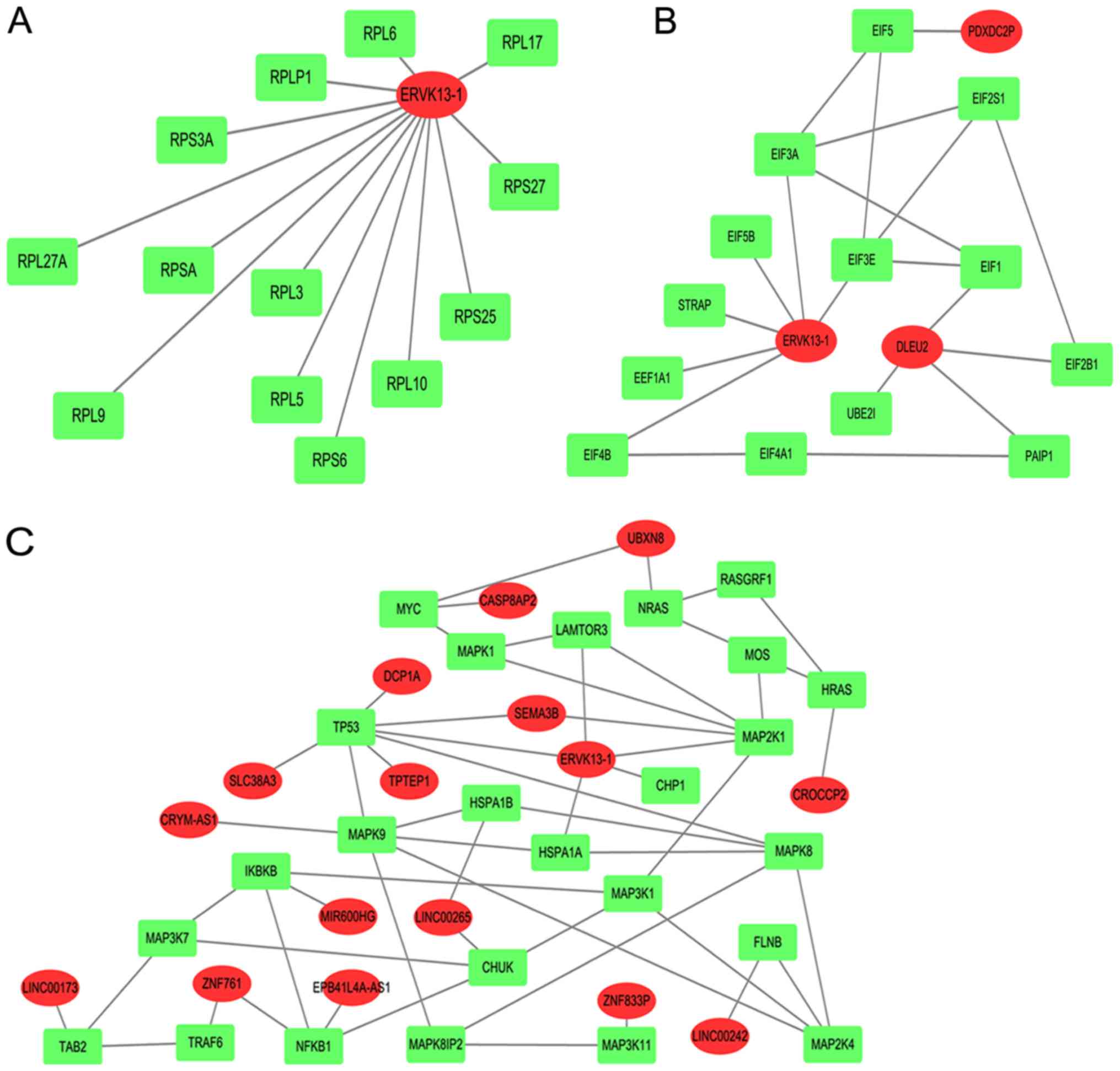
Table IV. We illustrated the
schematic diagram of top rank 3 lncRNA competitively regulated
subpathways, that is Ribosome, RNA transport and mitogen activated
protein kinase (MAPK) subpathways (Fig.
1). According to the value, the signal subpathways were
constructed into lncRNA-mRNA networks. Then 13 hub lncRNAs were
collected from the lncRNA-mRNA networks (Fig. 2).
 | Table IV.The diagram of the top six of the
lncRNA competitively regulated subpathways. |
Table IV.
The diagram of the top six of the
lncRNA competitively regulated subpathways.
| Pathway Id | Pathway name | Molecule ratio
(m2/x) | Bg ratio
(m1/n) | Weight | P-value | FDR |
|---|
| 03010_1 | Ribosome | 13/264 | 13/26232 | 1.000000 | <0.001 | 0.0000E+0000 |
| 03013_1 | RNA transport | 11/264 | 13/26232 | 1.241008 | <0.001 | 0.0000E+0000 |
| 04010_1 | MAPK signaling
pathway | 17/264 | 25/26232 | 1.556393 | <0.001 | 0.0000E+0000 |
| 04012_1 | ErbB signaling
pathway | 10/264 | 15/26232 | 1.736966 | <0.001 | 0.0000E+0000 |
| 04062_1 | Chemokine signaling
pathway | 14/264 | 20/26232 | 1.514573 | <0.001 | 0.0000E+0000 |
| 04066_1 | HIF-1 signaling
pathway | 11/264 | 16/26232 | 1.540568 | <0.001 | 0.0000E+0000 |
Discussion
In our study, the subpathway-LNCE method was first
used to identify the lncRNAs competitively regulated subpathways
for MI. Then the lncRNA-mRNA network was constructed according to
these signal subpathways, in which the hub lncRNAs were
detected.
Plenty of literature has reported that lncRNAs
regulate many fundamental biological processes and play a key role
in various diseases (4,5). Besides, lncRNAs competitively regulate
mRNA expression levels through binding to miRNAs, so that lncRNAs
can maintain normal biological functions (13,14).
Hence, identifying the functional relationships between lncRNA and
disease relevant subpathways may help understand the pathogenesis
of diseases. However, the relevant regulated mechanism of lncRNAs
in MI remains unclear. Thus, it is very important to search for a
suitable method to examine the functions of lncRNAs in MI.
Therefore, we applied subpathway-LINCE with GEO data
set of MI and identified a total of 65 lncRNA competitively
regulated subpathways under the condition of P<0.01. Of which,
P-values of 36 subpathways were nearly zero, which showed the
significant difference between MI group and control group, such as
Ribosome, RNA transport and MAPK subpathways. Ribosome subpathways
are critical to ribosome assembly and protein synthesis. When using
some methods to regulate the ribosome biogenesis, the cell growth
will be extensively affected. For example, mTOR signaling can
regulate multiple steps in ribosome biogenesis, thus influencing
the cell proliferation and survival (26,27).
Moreover, mTOR is activated in MI and mTOR inhibition could reduce
cardiac dilation and infract size and improve cardiac function
(28). Hence, clarifying the effect
of ribosome subpathways on MI may offer opportunities for new
therapies. RNA transport is the process where specific RNA
molecules are transported from one cellular region to another via
different sorting and transport mechanisms (29). In the present study, RNA transport
subpathway in MI was associated with lncRNAs, which might assist in
better understanding of the pathogenesis of MI. In particular, MAPK
influences mitochondria mediated cell functions, mainly including
proliferation, apoptosis and gene expression, because mitochondria
are great power providers and gate-keepers of cell life and death.
Moreover, MAPK can significantly affect cellular signaling
underlying cardiac compensation and decompensation via interacting
with the mitochondria (30). Our
study showed that MAPK signaling subpathways participate in the
pathological process of MI and are involved in lncRNAs. This could
help to clarify the pathogenesis of MI.
In order to explore hub lncRNAs in the signal
subpathways, the lncRNA-mRNA network was constructed through
selecting the high degree of the subpathways. Then, a total of 13
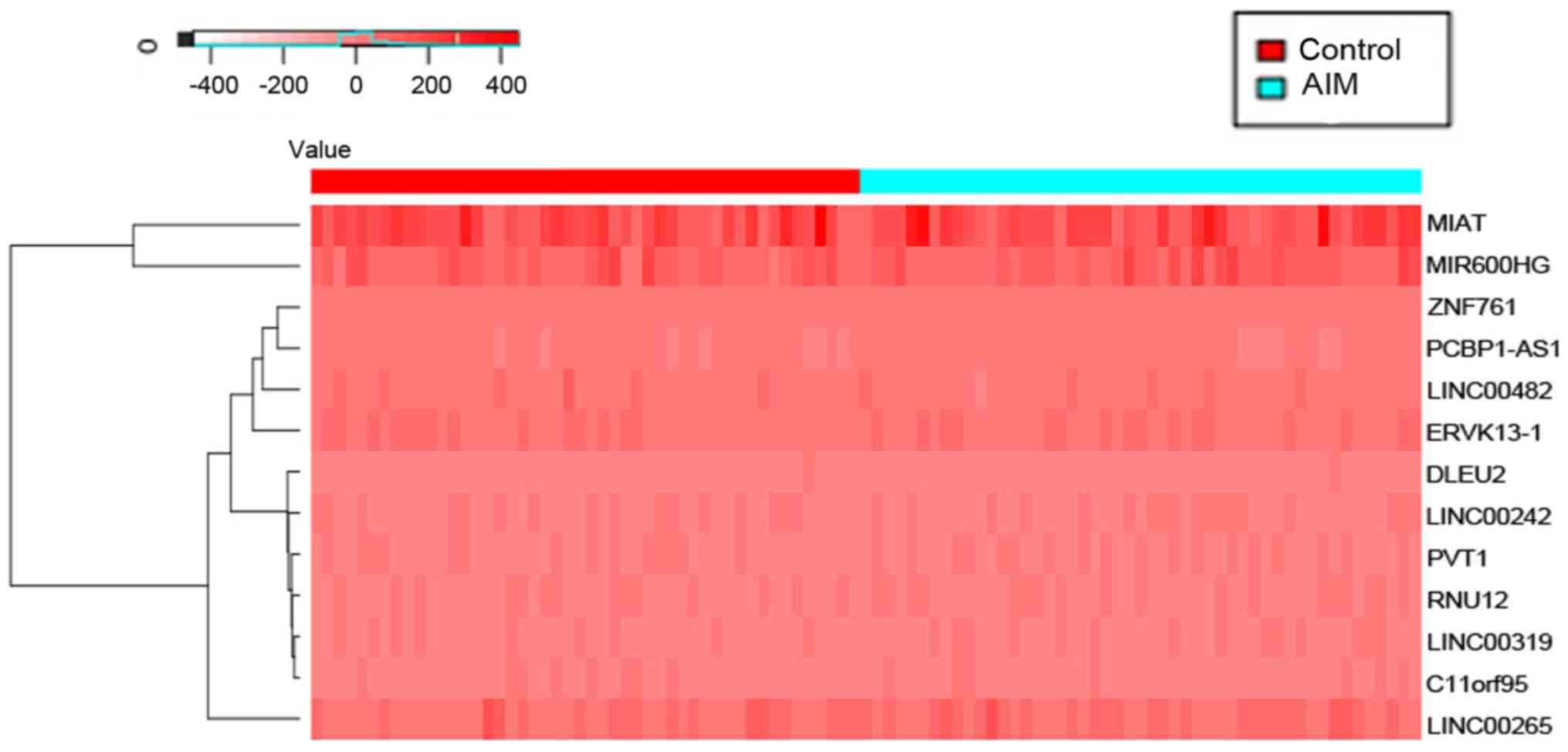
hub lncRNAs were collected from the lncRNA-mRNA networks. Among
these hub lncRNAs, MI associated transcript (MIAT), as an lncRNA,
has been associated with a risk of MI (10,31). It
has been reported that MIAT expression levels are found to change
in peripheral blood cells in patients who have suffered from MI,
and smoking as a cardiovascular risk factor is found to be
positively associated with MIAT (32). Apart from MIAT, other new hub lncRNAs
for MI were found using subpathway-LINCE in this study. These hub
lncRNAs could become potential diagnostic and therapeutic targets
for MI.
In conclusion, using subpathway-LINCE to study MI
lncRNA competitively regulated subpathways were gained, and the hub
lncRNAs for MI in lncRNA-mRNA network were also obtained.
Identifying the lncRNAs competitively regulated subpathways could
help us to understand the pathogenesis of MI. The hub lncRNAs might
represent novel regulators of MI and become new diagnostic and
therapeutic targets for MI. Although the results in this study
still need to be verified by experiments, these findings can help
understand the roles of lncRNAs in MI.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW and LS conceived the study and drafted the
manuscript. XW acquired the data. LS and ZW analyzed the data and
revised the manuscript. All authors read and approved the final
study.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Frangogiannis NG: Pathophysiology of
myocardial infarction. Compr Physiol. 5:1841–1875. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boateng S and Sanborn T: Acute myocardial
infarction. Dis Mon. 59:83–96. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Heyn J, Hinske C, Möhnle P, Luchting B,
Beiras-Fernandez A and Kreth S: MicroRNAs as potential therapeutic
agents in the treatment of myocardial infarction. Curr Vasc
Pharmacol. 9:733–740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang K, Liu CY, Zhou LY, Wang JX, Wang M,
Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, et al: APF lncRNA
regulates autophagy and myocardial infarction by targeting
miR-188-3p. Nat Commun. 6:67792015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clark BS and Blackshaw S: Long non-coding
RNA-dependent transcriptional regulation in neuronal development
and disease. Front Genet. 5:1642014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iyengar BR, Choudhary A, Sarangdhar MA,
Venkatesh KV, Gadgil CJ and Pillai B: Non-coding RNA interact to
regulate neuronal development and function. Front Cell Neurosci.
8:472014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kumarswamy R, Bauters C, Volkmann I, Maury
F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F and Thum
T: Circulating long noncoding RNA, LIPCAR, predicts survival in
patients with heart failure. Circ Res. 114:1569–1575. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Y, Cai Y, Wu G, Chen X, Liu Y, Wang
X, Yu J, Li C, Chen X, Jose PA, et al: Plasma long non-coding RNA,
CoroMarker, a novel biomarker for diagnosis of coronary artery
disease. Clin Sci (Lond). 129:675–685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ishii N, Ozaki K, Sato H, Mizuno H, Saito
S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al:
Identification of a novel non-coding RNA, MIAT, that confers risk
of myocardial infarction. J Hum Genet. 51:1087–1099. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li
YJ, Tao ZF, Song YC, Chen Q and Jiang Q: lncRNA-MIAT regulates
microvascular dysfunction by functioning as a competing endogenous
RNA. Circ Res. 116:1143–1156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Busch A, Eken SM and Maegdefessel L:
Prospective and therapeutic screening value of non-coding RNA as
biomarkers in cardiovascular disease. Ann Transl Med. 4:2362016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoon JH, Abdelmohsen K and Gorospe M:
Functional interactions among microRNAs and long noncoding RNAs.
Semin Cell Dev Biol. 34:9–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang K, Liu F, Zhou LY, Long B, Yuan SM,
Wang Y, Liu CY, Sun T, Zhang XJ and Li PF: The long noncoding RNA
CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res.
114:1377–1388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi X, Xu Y, Zhang C, Feng L, Sun Z, Han
J, Su F, Zhang Y, Li C and Li X: Subpathway-LNCE: Identify
dysfunctional subpathways competitively regulated by lncRNAs
through integrating lncRNA-mRNA expression profile and pathway
topologies. Oncotarget. 7:69857–69870. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guzzi PH and Cannataro M: Micro-Analyzer:
Automatic preprocessing of Affymetrix microarray data. Comput
Methods Programs Biomed. 111:402–409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Allen JD, Wang S, Chen M, Girard L, Minna
JD, Xie Y and Xiao G: Probe mapping across multiple microarray
platforms. Brief Bioinform. 13:547–554. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res 37 (Database). D105–D110. 2009.
View Article : Google Scholar
|
|
22
|
Jiang Q, Wang Y, Hao Y, Juan L, Teng M,
Zhang X, Li M, Wang G and Liu Y: miR2Disease: A manually curated
database for microRNA deregulation in human disease. Nucleic Acids
Res 37 (Database). D98–D104. 2009. View Article : Google Scholar
|
|
23
|
Chou CH, Chang NW, Shrestha S, Hsu SD, Lin
YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al: miRTarBase
2016: Updates to the experimentally validated miRNA-target
interactions database. Nucleic Acids Res. 44(D1): D239–D247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vergoulis T, Vlachos IS, Alexiou P,
Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N,
Dalamagas T and Hatzigeorgiou AG: TarBase 6.0: Capturing the
exponential growth of miRNA targets with experimental support.
Nucleic Acids Res. 40(D1): D222–D229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Epstein MP, Duncan R, Jiang Y, Conneely
KN, Allen AS and Satten GA: A permutation procedure to correct for
confounders in case-control studies, including tests of rare
variation. Am J Hum Genet. 91:215–223. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gentilella A, Kozma SC and Thomas G: A
liaison between mTOR signaling, ribosome biogenesis and cancer.
Biochim Biophys Acta. 1849:812–820. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iadevaia V, Liu R and Proud CG: mTORC1
signaling controls multiple steps in ribosome biogenesis. Semin
Cell Dev Biol. 36:113–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sciarretta S, Volpe M and Sadoshima J:
Mammalian target of rapamycin signaling in cardiac physiology and
disease. Circ Res. 114:549–564. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nakielny S, Fischer U, Michael WM and
Dreyfuss G: RNA transport. Annu Rev Neurosci. 20:269–301. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Javadov S, Jang S and Agostini B:
Crosstalk between mitogen-activated protein kinases and
mitochondria in cardiac diseases: Therapeutic perspectives.
Pharmacol Ther. 144:202–225. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liao J, He Q, Li M, Chen Y, Liu Y and Wang
J: LncRNA MIAT: Myocardial infarction associated and more. Gene.
578:158–161. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vausort M, Wagner DR and Devaux Y: Long
noncoding RNAs in patients with acute myocardial infarction. Circ
Res. 115:668–677. 2014. View Article : Google Scholar : PubMed/NCBI
|