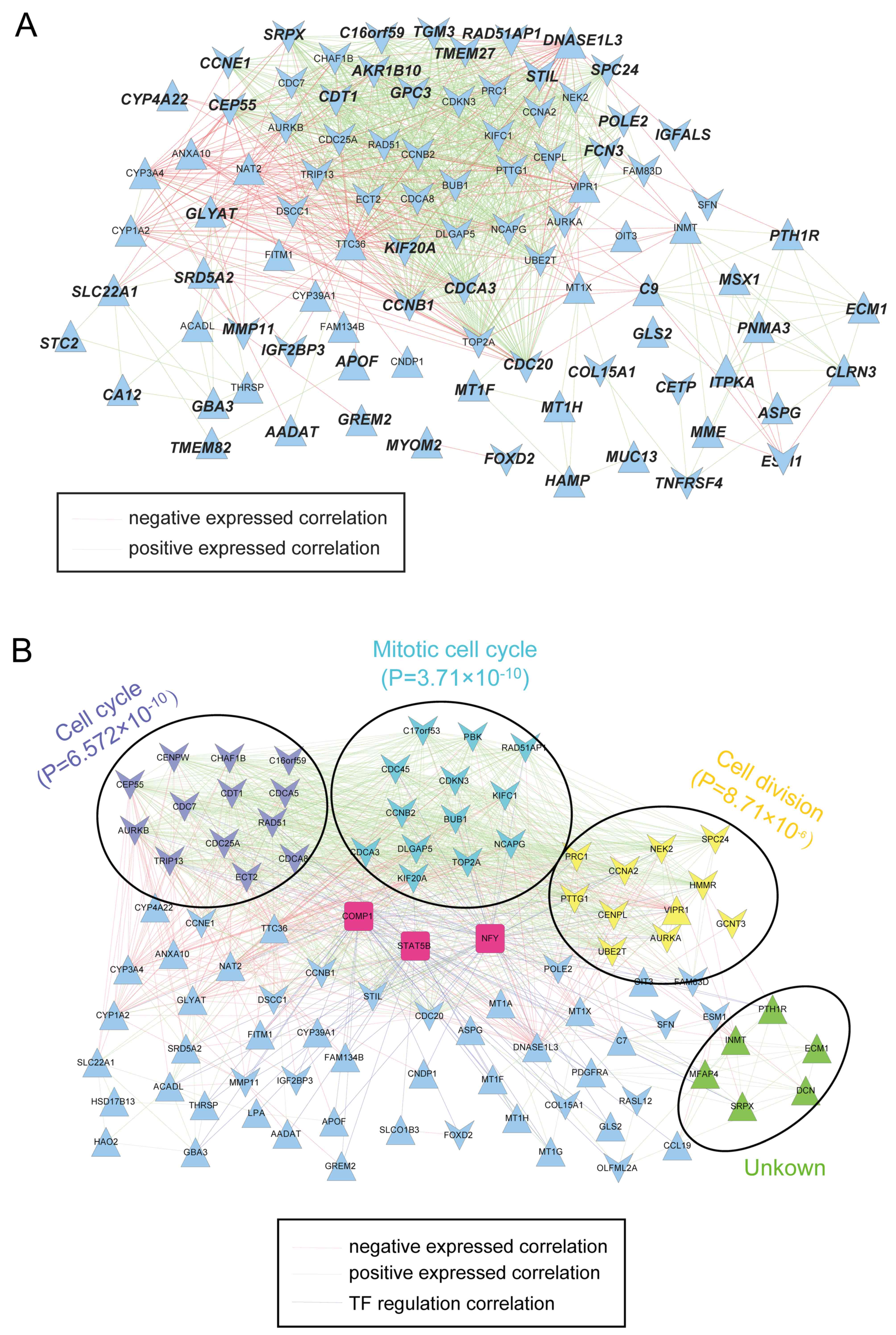
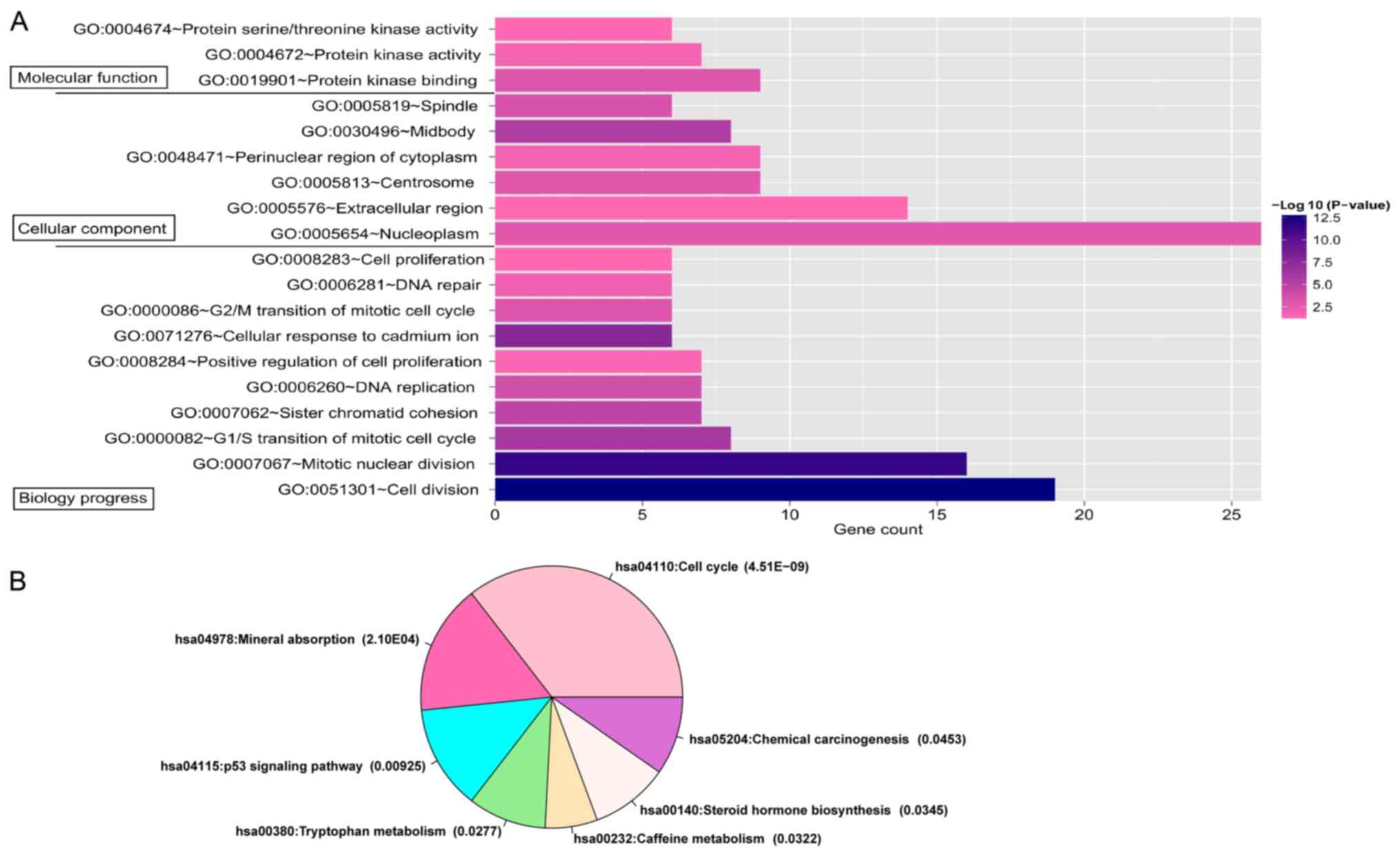
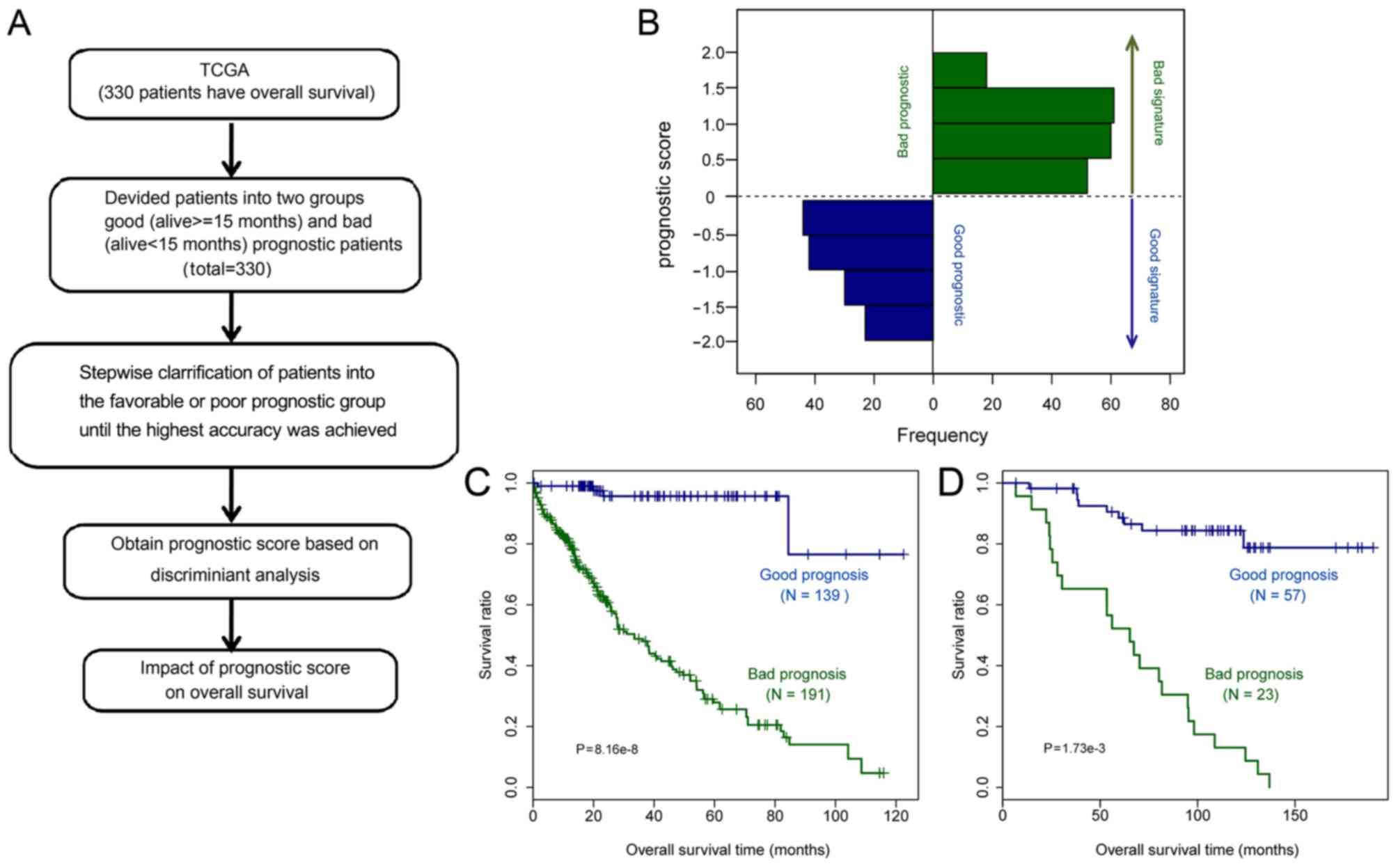
|
1
|
Zhu RX, Seto WK, Lai CL and Yuen MF:
Epidemiology of hepatocellular carcinoma in the asia-pacific
region. Gut and liver. 10:332–339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGlynn KA and London WT: Epidemiology and
natural history of hepatocellular carcinoma. Best Pract Res Clin
Gastroenterol. 19:3–23. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ayuso C, Rimola J, Vilana R, Burrel M,
Darnell A, García-Criado Á, Bianchi L, Belmonte E, Caparroz C,
Barrufet M, et al: Diagnosis and staging of hepatocellular
carcinoma (HCC): Current guidelines. Eur J Radiol. 101:72–81. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Laca L, Dedinska I, Miklusica J, Janik J,
Palkoci B and Pindura M: Surgical treatment of hepatocellular
carcinoma. Bratisl Lek Listy. 116:539–541. 2015.PubMed/NCBI
|
|
5
|
Ikeguchi M, Ueda T, Sakatani T, Hirooka Y
and Kaibara N: Expression of survivin messenger RNA correlates with
poor prognosis in patients with hepatocellular carcinoma. Diagn Mol
Pathol. 11:33–40. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakanishi K, Sakamoto M, Yamasaki S, Todo
S and Hirohashi S: Akt phosphorylation is a risk factor for early
disease recurrence and poor prognosis in hepatocellular carcinoma.
Cancer. 103:307–312. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu MZ, Yao TJ, Lee NP, Ng IO, Chan YT,
Zender L, Lowe SW, Poon RT and Luk JM: Yes-associated protein is an
independent prognostic marker in hepatocellular carcinoma. Cancer.
115:4576–4585. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fujii T, Nomoto S, Koshikawa K, Yatabe Y,
Teshigawara O, Mori T, Inoue S, Takeda S and Nakao A:
Overexpression of pituitary tumor transforming gene 1 in HCC is
associated with angiogenesis and poor prognosis. Hepatology.
43:1267–1275. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu YF, Yi Y, Qiu SJ, Gao Q, Li YW, Dai CX,
Cai MY, Ju MJ, Zhou J, Zhang BH and Fan J: PEBP1 downregulation is
associated to poor prognosis in HCC related to hepatitis B
infection. J Hepatol. 53:872–879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ding Y, Chen B, Wang S, Zhao L, Chen J,
Ding Y, Chen L and Luo R: Overexpression of Tiam1 in hepatocellular
carcinomas predicts poor prognosis of HCC patients. Int J Cancer.
124:653–658. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun JC, Liang XT, Pan K, Wang H, Zhao JJ,
Li JJ, Ma HQ, Chen YB and Xia JC: High expression level of EDIL3 in
HCC predicts poor prognosis of HCC patients. World J Gastroenterol.
16:4611–4615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamashita T, Forgues M, Wang W, Kim JW, Ye
Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, et al: EpCAM and
alpha-fetoprotein expression defines novel prognostic subtypes of
hepatocellular carcinoma. Cancer Res. 68:1451–1461. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calvisi DF, Ladu S, Gorden A, Farina M,
Lee JS, Conner EA, Schroeder IS, Factor VM and Thorgeirsson SS:
Molecular pathogenesis of human hepatocellular carcinoma:
Mechanistic and prognostic significance of aberrant methylation.
Cancer Res. 66:7632006.PubMed/NCBI
|
|
14
|
Gao Q, Qiu SJ, Fan J, Zhou J, Wang XY,
Xiao YS, Xu Y, Li YW and Tang ZY: Intratumoral balance of
regulatory and cytotoxic T cells is associated with prognosis of
hepatocellular carcinoma after resection. J Clin Oncol.
25:2586–2593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Budhu A, Forgues M, Ye QH, Jia HL, He P,
Zanetti KA, Kammula US, Chen Y, Qin LX, Tang ZY and Wang XW:
Prediction of venous metastases, recurrence, and prognosis in
hepatocellular carcinoma based on a unique immune response
signature of the liver microenvironment. Cancer Cell. 10:99–111.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaposi-Novak P, Lee JS, Gòmez-Quiroz L,
Coulouarn C, Factor VM and Thorgeirsson SS: Met-regulated
expression signature defines a subset of human hepatocellular
carcinomas with poor prognosis and aggressive phenotype. J Clin
Invest. 116:1582–1595. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bruix J and Llovet JM: Prognostic
prediction and treatment strategy in hepatocellular carcinoma (p
519–524). Hepatology. 35:519–524. 2010. View Article : Google Scholar
|
|
18
|
Smyth GK: Limma: Linear models for
microarray data. Bioinformatics Comput Biol Solut Using R
Bioconductor. pp397–420. 2005. View Article : Google Scholar
|
|
19
|
Rao Y, Lee Y, Jarjoura D, Ruppert AS, Liu
CG, Hsu JC and Hagan JP: A comparison of normalization techniques
for microRNA microarray data. Stat Appl Genet Mol Biol.
7:Article222008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pollard KS, Gilbert HN, Ge Y, Taylor S and
Dudoit S: multtest: Resampling-based multiple hypothesis testing.
2018.
|
|
21
|
Liu H, Xu R, Liu X, Sun R and Wang Q:
Bioinformatics analysis of gene expression in peripheral blood
mononuclear cells from children with type 1 diabetes in 3 periods.
Exp Clin Endocrinol Diabetes. 122:477–483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang X, Xu S, Chen L, Shen D, Cao Y, Tang
R, Wang X, Ji C, Li Y, Cui X and Guo X: Profiling analysis reveals
the potential contribution of peptides to human adipocyte
differentiation. Proteomics Clin Appl. 16:e17001722018. View Article : Google Scholar
|
|
23
|
Varešlija D, Priedigkeit N, Fagan A,
Purcell S, Cosgrove N, O'Halloran PJ, Ward E, Cocchiglia S,
Hartmaier R, Castro CA, et al: Transcriptome characterization of
matched primary breast and brain metastatic tumors to detect novel
actionable targets. J Natl Cancer Inst. 28:2018.
|
|
24
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H and Wang Y: RNA-seq analyses of
multiple meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gosling C: Encyclopedia of Distances. Ref
Rev. 24:342010.
|
|
26
|
Wang P, Wang Y, Hang B, Zou X and Mao JH:
A novel gene expression-based prognostic scoring system to predict
survival in gastric cancer. Oncotarget. 7:55343–55351.
2016.PubMed/NCBI
|
|
27
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Reimand J, Tooming L, Peterson H, Adler P
and Vilo J: GraphWeb: Mining heterogeneous biological networks for
gene modules with functional significance. Nucleic Acids Res 36
(Web Server Issue). W452–W459. 2008. View Article : Google Scholar
|
|
31
|
Joutsijoki H, Haponen M, Rasku J,
Aalto-Setälä K and Juhola M: Error-correcting output codes in
classification of human induced pluripotent stem cell colony
images. Biomed Res Int. 2016:30250572016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dimitriadou E, Hornik K, Leisch F and
Meyer D: The e1071 package. Ethnos J Anthropol. 23:55–56. 2006.
|
|
33
|
Sun J and Zhao H: The application of
sparse estimation of covariance matrix to quadratic discriminant
analysis. BMC Bioinformatics. 16:482015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jellinek DA, Chang AC, Larsen MR, Wang X,
Robinson PJ and Reddel RR: Stanniocalcin 1 and 2 are secreted as
phosphoproteins from human fibrosarcoma cells. Biochem J.
350:453–461. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu F, Li TY, Su SC, Yu JS, Zhang HL, Tan
GQ, Liu JW and Wang BL: STC2 as a novel mediator for
Mus81-dependent proliferation and survival in hepatocellular
carcinoma. Cancer Lett. 388:177–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang ZH, Wu YG, Qin CK, Rong ZH, Su ZX
and Xian GZ: Stanniocalcin 2 expression predicts poor prognosis of
hepatocellular carcinoma. Oncol Lett. 8:2160–2164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fung TK and Poon RY: A roller coaster ride
with the mitotic cyclins. Semin Cell Dev Biol. 16:335–342. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Weinstein J, Jacobsen FW, Hsu-Chen J, Wu T
and Baum LG: A novel mammalian protein, p55CDC, present in dividing
cells is associated with protein kinase activity and has homology
to the Saccharomyces cerevisiae cell division cycle proteins Cdc20
and Cdc4. Mol Cell Biol. 14:3350–3363. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li J, Gao JZ, Du JL, Huang ZX and Wei LX:
Increased CDC20 expression is associated with development and
progression of hepatocellular carcinoma. Int J Oncol. 45:1547–1555.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zeng JD, Zhang N, Zhao GJ, Xu LX, Yang Y,
Xu XY, Chen MK, Wang HY, Zheng SX and Li XX: MT1G is silenced by
DNA methylation and contributes to the pathogenesis of
hepatocellular carcinoma. J Cancer. 9:2807–2816. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kroemer G and Pouyssegur J: Tumor cell
metabolism: Cancer's Achilles' heel. Cancer Cell. 13:472–482. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Neri D and Supuran CT: Interfering with pH
regulation in tumours as a therapeutic strategy. Nat Rev Drug
Discov. 10:767–777. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hsieh MJ, Chen KS, Chiou HL and Hsieh YS:
Carbonic anhydrase XII promotes invasion and migration ability of
MDA-MB-231 breast cancer cells through the p38 MAPK signaling
pathway. Eur J Cell Biol. 89:598–606. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shi G, Abbott KN, Wu W, Salter RD and
Keyel PA: Dnase1L3 regulates inflammasome-dependent cytokine
secretion. Front Immunol. 8:5222017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang L, Seino J, Tomotake H, Funakoshi Y,
Hirayama H and Suzuki T: Co-expression of NEU2 and GBA3 causes a
drastic reduction in cytosolic sialyl free N-glycans in human MKN45
stomach cancer cells-evidence for the physical interaction of NEU2
and GBA3. Biomolecules. 5:1499–1514. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhu P, Jin J, Yan L, Li J, Yu XZ, Liao W
and He S: A novel prognostic biomarker SPC24 up-regulated in
hepatocellular carcinoma. Oncotarget. 6:41383–41397. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
He J, Ding C, He G and Huang Q:
Relationship between expression of ESM-1 and MMP-3 and invasion and
metastasis of human hepatocellular carcinoma. Med Sci J Central
South China. 40:368–372. 2012.(In Chinese).
|
|
48
|
Zhang X, Hu S, Zhang X, Wang L, Zhang X,
Yan B, Zhao J, Yang A and Zhang R: MicroRNA-7 arrests cell cycle in
G1 phase by directly targeting CCNE1 in human hepatocellular
carcinoma cells. Biochem Biophys Res Commun. 443:1078–1084. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Karavias D, Maroulis I, Papadaki H, Gogos
C, Kakkos S, Karavias D and Bravou V: Overexpression of CDT1 Is a
Predictor of Poor Survival in Patients with Hepatocellular
Carcinoma. J Gastrointest Surg. 20:568–579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shi C, Huang D, Lu N, Chen D, Zhang M, Yan
Y, Deng L, Lu Q, Lu H and Luo S: Aberrantly activated Gli2-KIF20A
axis is crucial for growth of hepatocellular carcinoma and predicts
poor prognosis. Oncotarget. 7:26206–26219. 2016.PubMed/NCBI
|
|
51
|
Tennant DA, Durán RV and Gottlieb E:
Targeting metabolic transformation for cancer therapy. Nat Rev
Cancer. 10:267–277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jin B, Wang W, Du G, Huang GZ, Han LT,
Tang ZY, Fan DG, Li J and Zhang SZ: Identifying hub genes and
dysregulated pathways in hepatocellular carcinoma. Eur Rev Med
Pharmacol Sci. 19:592–601. 2015.PubMed/NCBI
|