Introduction
Rheumatoid arthritis (RA) is a systemic chronic
inflammatory joint disease, which is characterized by persistent
synovitis, systemic inflammation and autoantibodies (1). Methotrexate (MTX) is the most widely
used and is regarded as the anchor drug in the treatment of RA.
Despite the advent of newer biologic therapies, MTX retains its
central role since it is relatively inexpensive, broad experience
with its use exists, and it is widely used in combination regimens
with other disease-modifying antirheumatic drugs (DMARDs) (2). In the US and European countries, the
recommended general dose of MTX is 15–20 mg/week, but individual
optimal dose is in the range of 5–25 mg/week. In China, the
conventional dose of MTX is 10 mg/week, but in practice 5–20
mg/week is prescribed based on individual sensitivity to and
tolerance of MTX (3,4). Although well proven, it is recognized
that there are large individual differences in the optimal dose of
MTX for RA patients (5). The
reasons for those individual differences are thought to be
different concentrations of intracellular MTX-polyglutamates
(MTX-PGs) and different enzyme activities at MTX-active sites
(6). Levels of various cytokines
and other mediators of inflammation, such as tumor necrosis
factor-α (TNF-α) and interleukin-10 (IL-10), may also correlate
with the efficacy of MTX (7,8).
However, rapid clinical assays to measure these such as MTX-PGs,
enzyme and cytokines are technically difficult and not available in
most clinical facilities (9,10).
To date, there are no useful and reliable clinical or molecular
markers to predict the efficacy of MTX treatment.
Recent advances in metabolomics offer us a chance to
identify changes in the concentration of small endogenous
metabolites in biological samples caused by drug therapy.
Metabolomics refers to the total metabolites present in biofluids
(e.g., blood and urine) and chemometric techniques combining
1H nuclear magnetic resonance (1H NMR) has
been applied for metabolomic profiling and characterization of
individually tailored therapeutic approaches (11,12).
Various studies have been performed to identify the major factors
predicting the response to MTX therapy in RA based on the
traditional method through evaluating the concentration of MTX-PGs
(13). In addition, 1H
NMR metabolomic study of rat urine was used to predict the risk of
adverse effects caused by nonsteroid anti-inflammatory drugs
(NSAIDs). A few endogenous metabolites of allantoin, taurine and
dimethylamine were selected as putative biomarkers for gastric
damage (14). Very recently, an
alternative approach to understanding such inter-subject
variability has been developed using metabolomics, and has been
used to predict the metabolism and toxicity of a dosed substance,
based solely on the analysis and modeling of a predose metabolic
profile (15).
In this study, we categorized the subsets of early
RA patients receiving MTX treatment for 24 weeks into two groups
according to the clinic assessment criterion, i.e., effective group
and non-effective group. Furthermore, 11 endogenous metabolites of
the effective group were significant different than the
non-effective group based on 1H NMR metabolomic analysis
and multivariate statistical analysis (p<0.05). As a potential
strategy and a more convenient technique, the 1H
NMR-based metabolomic approach deserves further evaluation for
assessing the efficacy of MTX treatment in patients with early
RA.
Patients and methods
Patient background and medical
records
This cohort study was conducted from January 2009 to
September 2010 at a single investigational site, the Department of
Integrated Chinese Traditional and Western Medicine, Tongji
Hospital (Wuhan, China). Patient demographic and clinical details
were collected on standardized data collection forms. All patients
underwent physical examination, electrocardiography and routine
blood testing. The duration of RA, age at which MTX administration
was initiated, gender, body weight, laboratory data; clinical data
including anti-cyclic citrullinated peptide (anti-CCP), serum
C-reactive protein (CRP) concentration, rheumatoid factor (RF),
erythrocyte sedimentation rate (ESR), serum creatinine clearance
(SCR) and alanine transaminase (ALT) were obtained from the medical
records.
Patients and study protocol
Patients 18 years of age and older with early
(<0.5 year) active RA, as defined by the American College of
Rheumatology (ACR; formerly, the American Rheumatism Association)
(16), were included in the study
and were scheduled to receive MTX monotherapy for 24 weeks. In all
cases, the starting dose of oral MTX was 10 mg/week. Active disease
was defined as following: ≥8 swollen joints, ≥10 tender joints and
an ESR of ≥28 mm/h or a CRP concentration of ≥7.5 mg/l. Exclusion
criteria were the following: previous use of glucocorticoids and/or
DMARDs within a period of three months before inclusion in the
study, alcohol abuse, serious comorbidity or recent major surgery.
A total of 20 healthy age-matched volunteers, who had no rheumatic
disease, were used as normal controls. Blood samples were collected
at baseline (before starting MTX treatment) and 24 weeks after MTX
monotherapy.
Ethical approval was obtained from the Ethics
Committee of Tongji Medical College, China. Written informed
consent was obtained from each participant in accordance with the
principles outlined in the Declaration of Helsinki.
Clinical assessment
Treatment efficacy was evaluated in early RA
patients receiving MTX monotherapy for 24 weeks, according to the
European League Against Rheumatism (EULAR) response criteria
(17). To obtain a better
analysis, the patients were categorized into the ‘effective’ or
‘non-effective’ group based on the amount of change in the DAS28
and the level of DAS28 reached. The effective group was defined as
patients who had a decrease in DAS28 from baseline (ΔDAS28) of
>1.2 and a DAS28 at the sixth month of <3.2; and had either
ΔDAS28 of >1.2 and a DAS28 at the sixth month of ≥3.2 or ΔDAS28
of 0.6 to 1.2 and a DAS28 at the sixth month of <5.1. The
non-effective group consisted of those who had either ΔDAS28 of
<0.6 or a DAS28 at the sixth month of ≥5.1.
Sample preparation
Venous blood samples were collected in the morning
preprandial, after overnight fasting, using vacuum tubes (BD, 8.5
ml, USA) and left at room temperature to coagulate for 45 min.
After this, samples were centrifuged at 1000 x g for 10 min, and
serum was collected and immediately stored at −80°C until being
used for metabolomic analysis (18).
Preparation of serum samples for metabolomic
analysis by 1H NMR was performed manually according to
Beckonert et al (19).
After defrosting at room temperature, 200 μl of serum was mixed
with 400 μl of 0.9% saline solution (wt/vol, NaCl in 90%
H2O/10%/D2O) and then centrifuged at 12000 x
g for 5 min at 4°C. After that, 550 μl of the supernatant was added
to a 5-mm NMR tube and NMR acquisition was performed
immediately.
NMR acquisition
The NMR spectra were acquired using a Bruker Avance
III 600 spectrometer (Bruker BioSpin, Rheinstetten, Germany),
operating at a 1H frequency of 600.13 MHz, and equipped
with a 5-mm 1H TXI probe. Samples were afforded 5 min
within the spectrometer to equilibrate before acquisition. Standard
one-dimensional (1D) 1H NMR spectra were acquired using
a single 90º pulse experiment with water presaturation using a
recycle delay of 3 sec. Each data set was averaged over 64
transients using 32 K time domain points. The data were Fourier
transformed, and spectra were referenced to the TSP signal at 0
ppm.
Each NMR spectrum was reduced to 0.04 ppm wide
segments between 0.00 and 10.0 ppm using Chenomx NMR Suite
Professional software version 4.6 (Chenomx Inc., Edmonton, Canada),
giving a total of 230 integrated regions per NMR spectrum. The
spectrum regions of water (δ=2.6–3.0 ppm) and urea (δ=4.3–4.7 ppm)
were removed from the analysis for all groups in order to prevent
variation in each sample. Each NMR variable was normalized to the
total area in order to allow a spectrum-to-spectrum comparison.
Metabolites were assigned based on comparisons with the chemical
shifts of standard compounds in the Chenomx software. Custom
library entries were created for unidentified resonances in order
to carry them through the analysis for relative concentration
comparison. Typically, the concentrations of serum metabolites were
reported as ratios with creatinine.
Multivariate data analysis
The spectral data were converted to Microsoft Excel
format and imported into SIMCA-P software (version 11.5, Umetrics,
Umeå, Sweden) for multivariate analysis in which all spectral data
were mean-centered with no scaling. Initially, principal component
analysis (PCA) was applied to the clustering of all identified
outliers. Partial least squares-discrimination analysis (PLS-DA)
was then advanced for pattern recognition analysis from the
converted spectral data (20). All
the statistical analyses were carried out using binned bucket data
and the identified metabolites for spectral binning and targeted
profiling. PLS-DA made it possible to rotate the projection to
latent variables that focus on class separation. Thus, PLS-DA aimed
to find a model that separated classes of observations based on
their x variables. The PLS-DA model was validated by permutation
method by describing R2Y and Q2Y values. In
addition, the choice of input variables for ANOVA analysis was
based on magnitude of the variable influence on projection (VIP)
from SIMCA-P where a VIP >1 indicated contribution to the
model.
Statistical analysis
Clinic data are expressed as the means ± standard
deviation (SD), and the data were analyzed for statistical
significance by ANOVA test using SPSS version 17.0. Post-hoc tests
were applied to perform all pairwise comparisons between group
means by least significant difference (LSD) t-test. A value of
p<0.05 was considered to indicate a statistically significant
difference.
Results
Characteristics of the patients and
metabolite identification
Demographics and characteristics of the patients at
baseline and at the end of 24 weeks of MTX treatment are provided
in Table I. Overall, 38 patients
with early RA were enrolled in this study, and 20 healthy
age-matched volunteers (15 females and 5 males, 55.3±3.6 years of
age) were used as healthy controls. According to the EULAR DAS28
response criteria, the individuals treated with MTX montherapy were
divided into two subsets, i.e., effective and non-effective group.
Furthermore, the patients in the effective group showed a reduction
in RA signs and symptoms, inhibited progression of structural
damages, and improved physical function as compared with that of
non-effective group (Table I).
 | Table I.Demographics and clinical
characteristics of the RA patients at baseline and after MTX
treatment (10 mg/week) for 24 weeks. |
Table I.
Demographics and clinical
characteristics of the RA patients at baseline and after MTX
treatment (10 mg/week) for 24 weeks.
| Group | Healthy controls
(n=20) | RA patients
(n=38) | Early RA patients
receiving MTX treatment
|
|---|
| Non-effective
(n=13) | Effective
(n=25) |
|---|
| Female/male | 15/5 | 26/5 | 10/1 | 16/4 |
| Age (years) | 55.3±3.6 | 56.4±2.8 | 54.7±3.4 | 54.6±2.5 |
| Disease duration
(years) | - | 4.5±1.9 | 4.4±1.2 | 4.2±0.8 |
| DAS28 | - | 5.9±1.4 | 4.6±1.2 |
2.3±0.8a,b |
| Anti-CCP
(RU/ml) | 6.3±1.9 | 38.3±5.1 | 36.3±7.8 |
15.3±3.4a,b |
| CRP (mg/l) | 1.3±0.6 | 30.2±2.7 | 28.2±3.6 |
3.5±1.2a,b |
| RF (IU/ml) | 40.7±5.2 | 378.8±80.6 | 336.8±97.6 |
166.7±62.1a,b |
| ESR (mm/h) | 8.3±4.2 | 57.3±9.6 | 52.8±5.3 |
18.2±2.1a,b |
| SCR (mg/dl) | 53.6±9.5 | 58.6±7.4 | 68.2±9.8 | 60.7±8.2 |
| ALT (U/l) | 15.8±5.1 | 16.2±5.4 | 21.3±8.2 | 23.9±6.3 |
A 600 MHz 1D 1H NMR spectrum of a typical
human serum sample is shown in Fig.
1. This revealed extensive complexity and significant
information of biofluid. Regions of most significant metabolite
signals usually fell between δ1–5.6 ppm, including alanine
(δ1.3–1.35 ppm), acetate (δ1.35–1.4 ppm), citrate (δ3.0–3.2 ppm),
cysteine (δ3.2–3.4 ppm), taurine (δ3.5–3.6 ppm), methionine
(δ3.8–4.0 ppm), hypoxanthine (δ4.0–4.2 ppm) and uracil (δ5.4–5.6
ppm). Research in relation to the locations of the different
chemical groups and various molecular-weight metabolites has
already been reported (21,22).
To accurately detect the difference between the two groups,
specific software was used in the chemometric analysis.
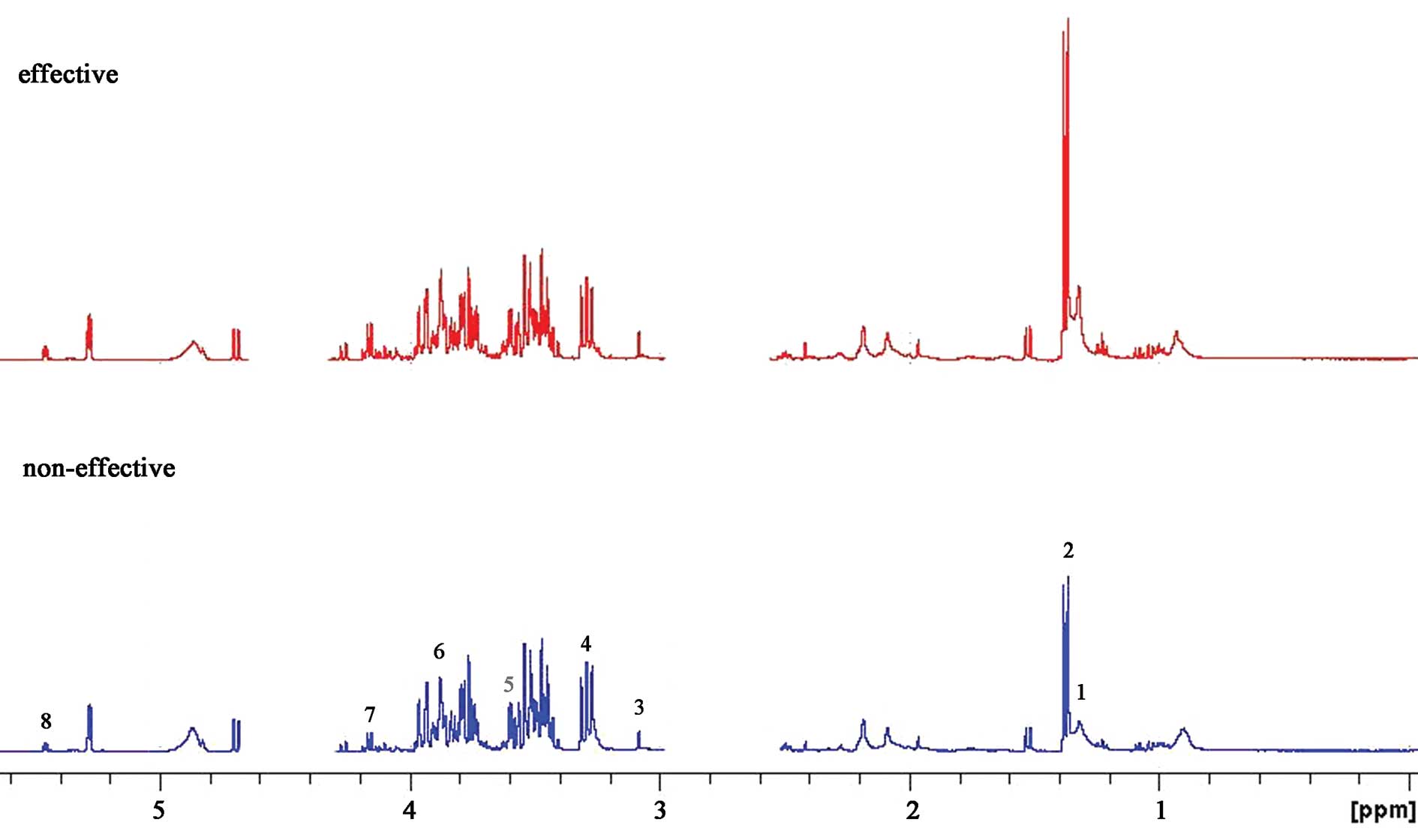 | Figure 1.Representative 600-MHz 1H
NMR spectrum of serum samples from the ‘effective’ group (red, top)
and ‘non-effective’ group (blue, bottom). Typical serum samples are
shown and peak assignments of major metabolites on 1H
NMR spectra. The spectrum regions of water (δ=2.6–3.0 ppm) and urea
(δ=4.3–4.7 ppm) were removed from the analysis for all groups in
order to prevent variation in each sample. 1, alanine; 2, acetate;
3, citrate; 4, cysteine; 5, taurine; 6, methionine; 7,
hypoxanthine; 8, uracil. |
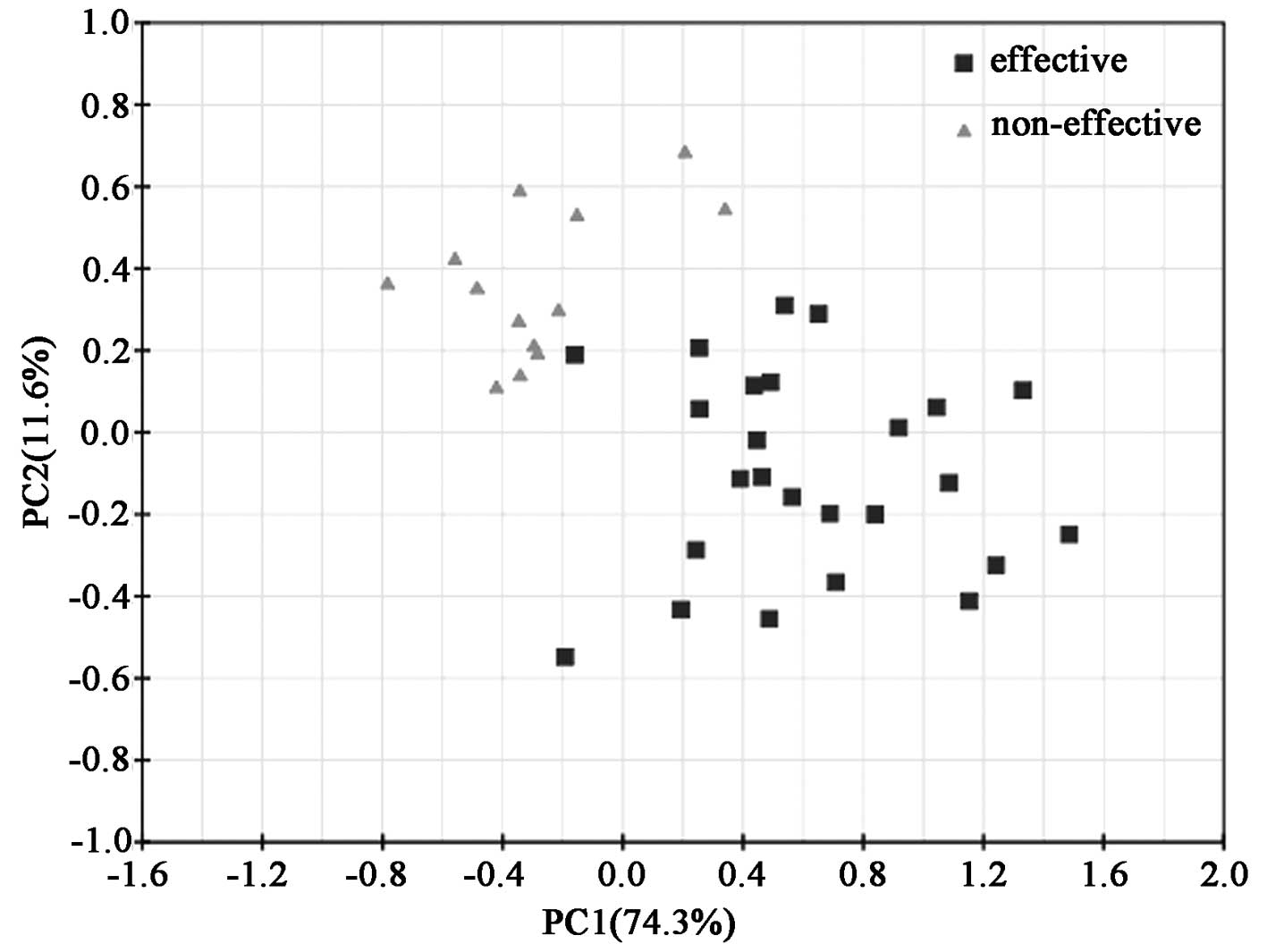
Principal component analysis of the serum
samples in the patients receiving MTX treatment
The mean-centered data from 1D 1H NMR
spectra were input into the SIMCA-P software and processed by the
unsupervised method of the principal component analysis (PCA), and
the resulting score loading plots for both the effective and the
non-effective group are shown (Fig.
2). PCA score plots showed clustering of serum samples in the
first and second principal components. The diverged score plots
signified a separation between the effective and the non-effective
group along the axes corresponding to PC 1 and 2 with these two
principal components accounting for 74.3 and 11.6% of the variation
in the data, respectively. A tendency for grouping according to the
effects of MTX treatment was noted along the first principal
components. For further elucidation of the effective group
following MTX treatment and providing validation, supervised
analysis of the pattern recognition such as PLS and PLS-DA was
performed.
Partial least squares (PLS) and
PLS-discriminant analysis (PLS-DA)
The spectral data of the serum samples were
collected from the effective (n=25) and non-effective group (n=13),
respectively, and the metabolic profiles of the serum samples are
depicted in Fig. 3. The loading
plots of the PCA demonstrated that the outer subset of compounds
was important in the discrimination of the two groups. Furthermore,
several features in the NMR metabolite profile were identified by
the Chenomx NMR Suite (Table
II).
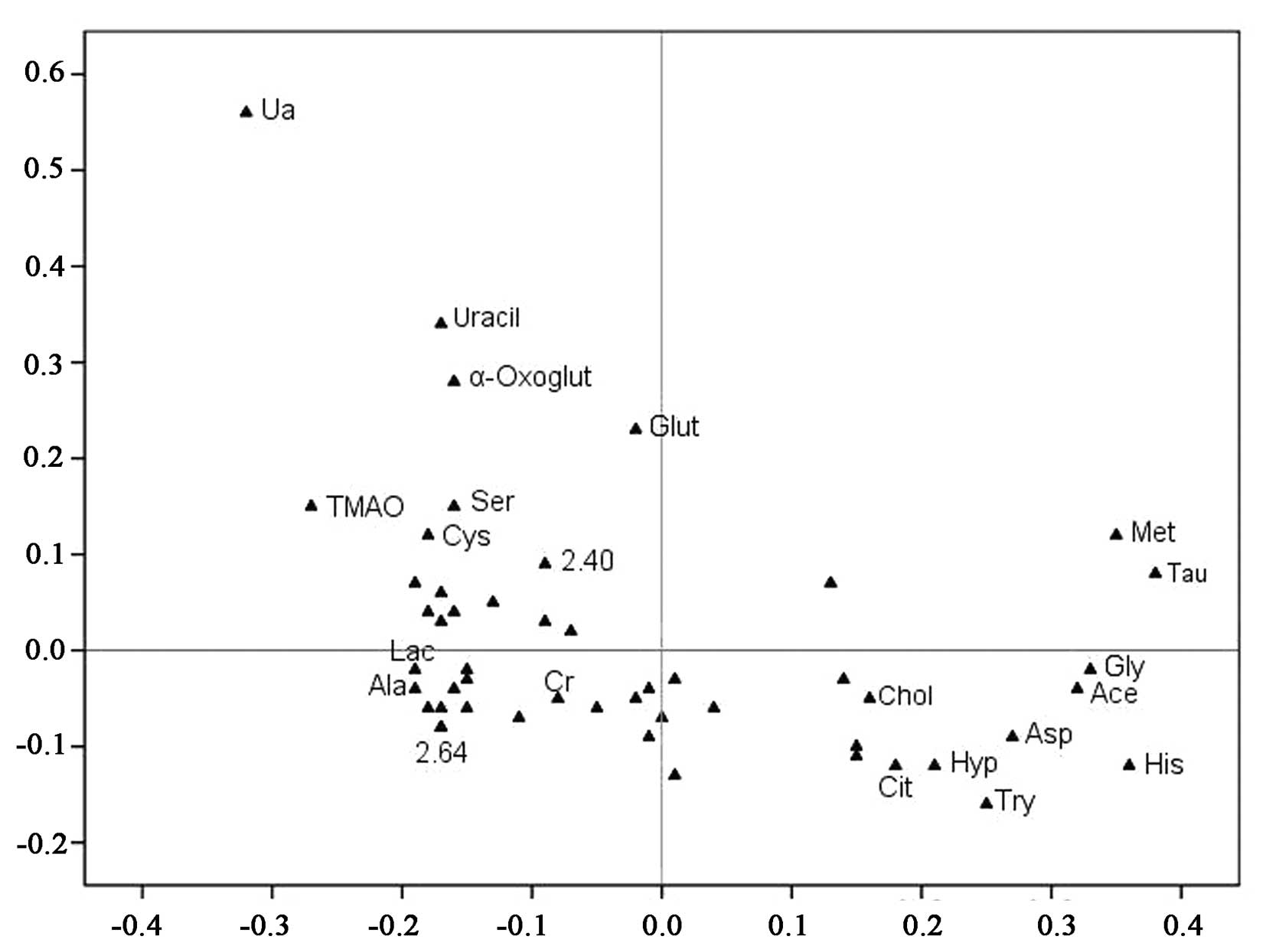 | Figure 3.Loading plot from the PLS-DA analysis.
Unknown resonances are indicated by the numerical ppm value or
unmarked if not significant between the two groups. Each point
represents the number of a metabolite included in the PLS-DA
analysis (total, 20 metabolic data points are shown), and the axes
are associated with the score plot of Fig. 2. Met, methionine; His, histidine;
Gly, glycine; Ala, alanine; Cys, cysteine; Ser, serine; Tau,
taurine; Glut, glutamine; Chol, cholesterol; Try, tryptophan; Hyp,
hypoxanthine; Asp, aspartate; α-Oxoglut, α-oxoglutarate; Ace,
acetate; TMAO, trimethylamine-N-oxide. |
 | Table II.List of the metabolites identified by
the Chenomx NMR Suite in the serum samples. |
Table II.
List of the metabolites identified by
the Chenomx NMR Suite in the serum samples.
| Metabolites | Chemical shift
(ppm) and multiplicity |
|---|
| α-oxoglutarate | δ2.44 (t), 3.07
(t) |
| Glycine | δ3.55 (s) |
| Citrate | δ2.55 (d), 2.65
(d) |
| Aspartate | δ3.56 (s), 6.74
(m), 7.08 (m) |
| Acetate | δ1.92 (s) |
| Alanine | δ1.32 (d), 3.72
(q) |
| Cholesterol | δ3.60 (s) |
| Creatinine | δ3.00 (s), 3.96
(s) |
| Cysteine | δ2.72 (s) |
| Histidine | δ3.96 (d), 6.49
(t), 7.73–7.76 (m) |
| Hypoxanthine | δ3.14 (d), 4.05
(t), 5.31–5.37 (m), 6.77 (d) |
| Lactate | δ1.32 (s), 4.23
(s) |
| Glutamine | δ2.04 (d) |
| Methionine | δ1.36 (s), 2.42
(t), 3.40–3.42 (q), 3.56 (d), 3.98 (s) |
| Serine | δ2.40 (s) |
| Taurine | δ3.36 (t), 3.68
(t) |
| Tryptophan | δ4.43 (s), 7.47
(t) |
|
Trimethylamine-N-oxide | δ3.18 (s) |
| Uracil | δ5.81 (d), 7.53
(d) |
| Uric acid | δ3.13 (t), 5.46
(m) |
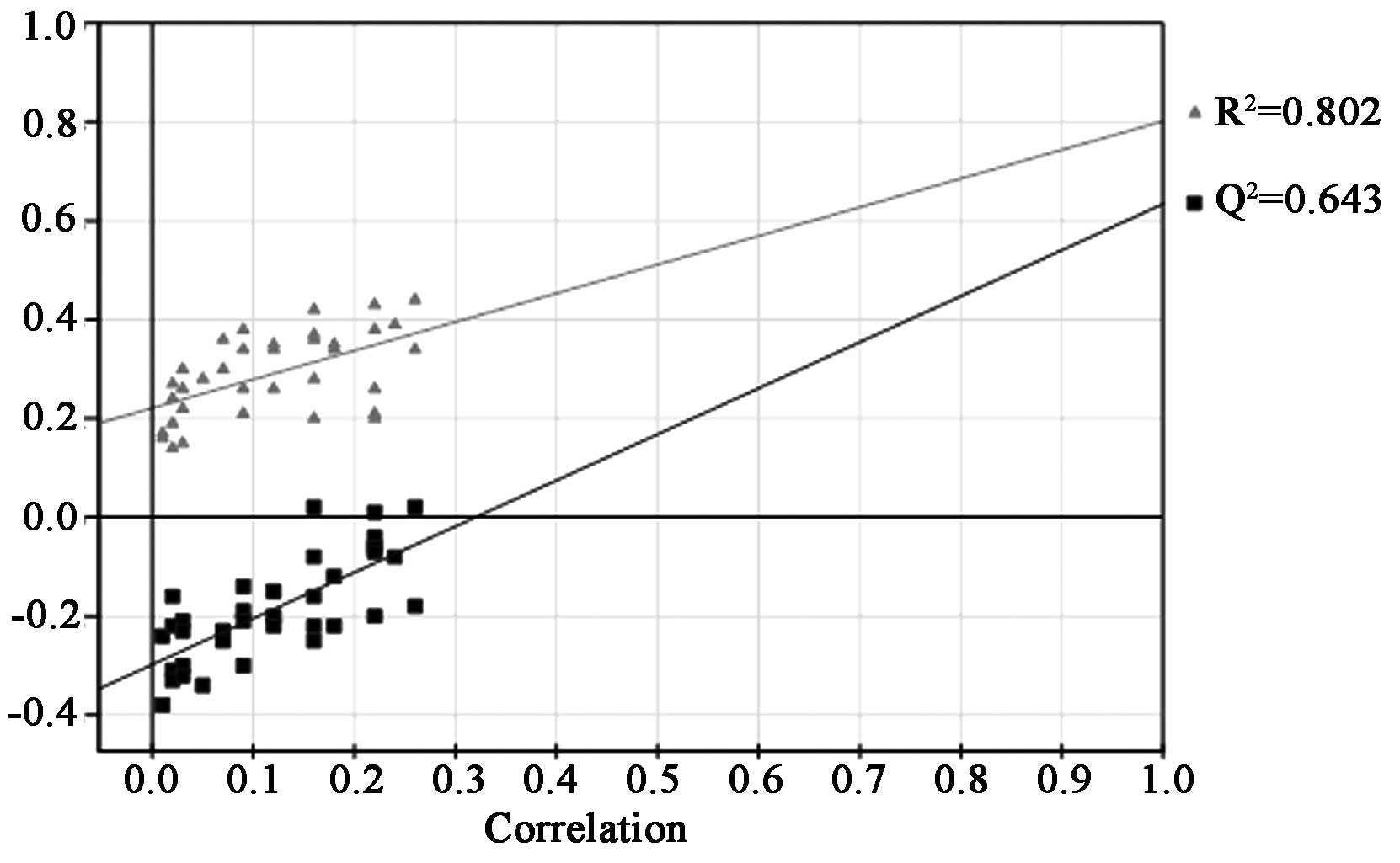
The quality of the principal component models was
evaluated with the parameters R2 and Q2, and the
goodness of fit was quantified by R2 while the
predictive ability was indicated by Q2. Generally,
R2Y and Q2Y explained variation in the data,
where 0 was no variation explained, and 1, for which 100% variation
was accounted for. The results indicated that this PLS-DA model had
a high R2Y value of 0.802 and Q2Y value of
0.643, which suggested that although each patient in the two groups
was treated with the same concentration of MTX, and it was possible
to metabolically discriminate with different physical constitutions
(Fig. 3).
The permutation testing was performed to validate
PLS-DA models. It provided the statistical significance of the
estimated predicted power of the models by comparing R2Y and
Q2Y values of the original model with those of the
re-ordered model, which was created newly whenever y data was
permutated at random. It is known that models with the R2Y
intercept <0.4 and a Q2Y intercept <0.05 indicate
valid models. As shown in Fig. 4,
the PLS-DA model had a proper R2Y intercept value of 0.208 and a
Q2Y intercept value of −0.285.
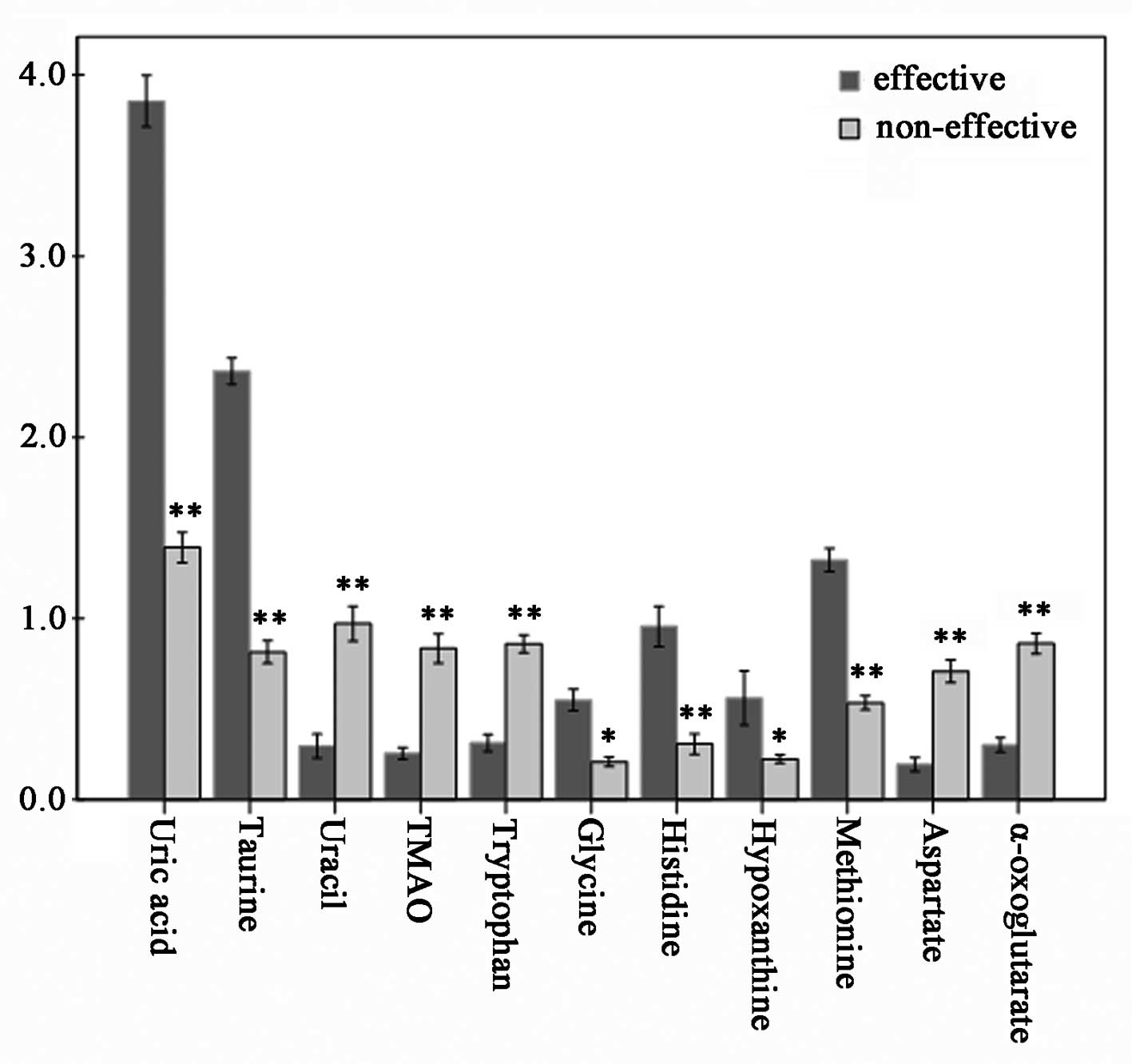
VIP value of the major contributing
metabolites
VIP values of several metabolites assigned by using
SIMCA-P are listed in Table III.
Usually, the metabolites with VIP >1 were considered to be
influential for separating each sample in these PLS-DA models.
Consequently, we performed an ANOVA test to determine the relative
intensities of the important metabolites contributing to the
separation of the two groups. Eleven metabolites were selected as
key metabolites for the classification of each group. VIP analysis
showed the order of metabolites according to their critical
influence on clustering, with metabolites on the x-axis arranged
according to their VIP values (VIP >1), and the y-axis
illustrating the relative intensities of the metabolites (Table III). The mean uric acid, taurine,
histidine, methionine, glycine and hypoxanthine concentrations in
the effective group were elevated (p<0.05), while uracil, TMAO,
α-oxoglutarate, aspartate and tryptophan were statistically
significant decreased (p<0.05) (Fig. 5).
 | Table III.VIP values of the major contributing
metabolites for the separation in the score plots derived from
PLS-DA. |
Table III.
VIP values of the major contributing
metabolites for the separation in the score plots derived from
PLS-DA.
| Metabolites | VIP value |
|---|
| α-oxoglutarate | 1.328 |
| Glycine | 1.942 |
| Citrate | 0.704 |
| Aspartate | 1.213 |
| Acetate | 2.348 |
| Alanine | 0.841 |
| Cholesterol | 0.536 |
| Cysteine | 0.964 |
| Histidine | 2.815 |
| Hypoxanthine | 1.224 |
| Lactate | 0.872 |
| Glutamine | 1.116 |
| Methionine | 2.478 |
| Serine | 0.836 |
| Taurine | 3.624 |
| Tryptophan | 1.462 |
|
Trimethylamine-N-oxide | 2.367 |
| Uracil | 2.437 |
| Uric acid | 3.837 |
Discussion
Metabolomics is a novel scientific discipline
focused on the association between disease and metabolic profile in
tissue and biofluids, as determined by techniques including
1H NMR spectroscopy, mass spectroscopy and liquid
chromatography mass spectroscopy. Moreover, metabolomic analysis
can also be utilized to identify biomarkers of drug treatment
(23). From a therapeutic point of
view, it is well accepted that MTX is effective in only 60–75% of
RA patients (24). In this study,
we performed an 1H NMR-based metabolomic approach in
early RA patients with MTX treatment to identify potential
biomarkers for therapeutic evaluation. Using PCA and PLS-DA
analysis, we detected the relationships between the individual
response of 38 patients with early RA receiving MTX mono-therapy
and various associated factors in the effective group. Furthermore,
11 endogenous metabolites as the major predictive factors for the
response to MTX were identified in the metabolic profile.
Significantly, the current study demonstrates the feasibility of
using serum for diagnostic or prognostic testing in patients with
early RA receiving MTX treatment.
Biological fluids, such as blood and urine, contain
a large number of metabolites that may provide valuable information
on the metabolism of an organism, and thus concerning its health
status. In the present study, 11 metabolites were found to vary
with the differential response of the early RA patients receiving
MTX treatment, which suggest they could be used to predict the
therapeutic effects of MTX. It appeared that nucleic acid
metabolism may be highly interrelated to the therapeutic effects,
with uric acid and uracil being two of the most prominent
metabolites noted. In addition, the changes in levels of TMAO and
hypoxanthine suggest an alteration in purine and pyrimidine ratios,
and in turn may affect cell metabolism, energy conservation and
biosynthetic pathways, even signal transduction and translation
(23). Glycine has been suggested
to have anti-inflammatory properties by acting on macrophage
chloride channels to blunt cytokine release, and methionine is
associated with SAM synthesis that may influence metabolism of
nucleic acid (24,25). Both glycine and methionine are
important metabolites in homocysteine metabolism, and higher levels
of glycine and methionine in the effective group implied a
favorable response to MTX in the present study. As far as we know,
the main physiological function of the one-carbon unit is to
synthesize purines and pyrimidines, and it is closely linked to
amino acid metabolism and nucleic acid metabolism (26). The one-carbon unit mainly arises
from serine, glycine, histidine and tryptophan, and the three
compounds were found to be altered in the serum of the effective
group (27). Increased levels of
hypoxanthine in the serum of the effective group may be attributed
to a decrease in the levels of oxidative stress following MTX
therapy. Taurine is also known to have antioxidant effects which
appear to downregulate pro-inflammatory cytokine production, and
was present at increased levels in the effective group (28). Furthermore, the Krebs cycle is also
likely to play an important role in the course of treatment since
the concentrations of α-oxoglutarate and aspartate were decreased
in the effective group when compared with those of the
non-effective group (29,30).
The potential metabolic pathways highlight the
complexity of the metabolic response to MTX treatment. Although
conclusively defining roles for each metabolite were not feasible
based on the data from this study, 11 metabolites presented here
may be considered useful biomarkers for their discriminatory power
to distinguish the effectiveness of MTX treatment in patients with
early RA. We therefore hypothesized that several metabolic pathways
are related to the therapeutic effects of MTX, such as nucleic acid
metabolism, homocysteine metabolism, one-carbon metabolism, and
metabolite analysis may be used to predict the effects of MTX
treatment in RA patients.
One of the ultimate goals of the current method was
to enable comparisons between RA patients with a favorable response
to MTX and those who respond poorly in clinical settings. According
to the different situations, we could rapidly adjust the therapy
regimen in the clinic. But one problem faced in clinical
investigations is the inherently greater variability in a human
population compared with that seen in our study. Thus, the study
factors such as age, gender, diet, individual difference and other
environmental influences, can be more extensively controlled by
investigators than when the enrolled human study populations are
investigated. Although the exact metabolites identified as
potential ‘biomarkers’ may vary among different individuals, more
studies could use the methodology applied here to investigate the
efficacy of MTX treatment in RA patients. Metabolomic analysis is
undoubtedly applicable to the search for biological predictors of
response to drug treatment in RA, but future studies should employ
larger patient cohorts, more discriminatory analyses, and a less
equivocal clinical phenotype.
In conclusion, the monitoring of entire sets of
metabolic features is critical to evaluate the efficacy of drug
treatments. A metabolic ‘bioprofile’, consisting of predictive
serum metabolite features from 1H NMR spectral data of
early RA patients receiving MTX treatment are presented in this
study. Significantly, our study demonstrated that various serum
biomarkers can be used to evaluate MTX treatment in patients with
early RA, and the metabolomic approach including 1H NMR
spectroscopy coupled with multivariate statistical analysis may be
useful to predict the outcome of drug response and treatment.
Abbreviations:
|
1H NMR
|
1H nuclear magnetic
resonance
|
|
RA
|
rheumatoid arthritis
|
|
MTX
|
methotrexate
|
|
DAS
|
disease activity score
|
|
PCA
|
principal component analysis
|
|
PLS-DA
|
partial least squares discriminant
analysis
|
|
DMARDs
|
disease-modifying anti-rheumatic
drugs
|
|
NSAIDs
|
non-steroid anti-inflammatory
drugs
|
Acknowledgements
The authors specially thank Dr Ming
Jiang of the School of Pharmacy, Tongji Medical College for his
technical assistance.
References
|
1
|
Scott DL, Wolfe F and Huizinga TW:
Rheumatoid arthritis. Lancet. 376:1094–1108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rath T and Rubbert A: Drug combinations
with methotrexate to treat rheumatoid arthritis. Clin Exp
Rheumatol. 28:S52–S57. 2010.PubMed/NCBI
|
|
3
|
Saag KG, Teng GG, Patkar NM, et al:
American College of Rheumatology 2008 recommendations for the use
of nonbiologic and biologic disease-modifying antirheumatic drugs
in rheumatoid arthritis. Arthritis Rheum. 59:762–784. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Braun J: Optimal administration and dosage
of methotrexate. Clin Exp Rheumatol. 28:46–51. 2010.
|
|
5
|
Ranganathan P and McLeod HL: Methotrexate
pharmacogenetics: the first step toward individualized therapy in
rheumatoid arthritis. Arthritis Rheum. 54:1366–1377. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dervieux T, Furst D, Lein DO, Capps R,
Smith K, Walsh M and Kremer J: Polyglutamation of methotrexate with
common polymorphisms in reduced folate carrier, aminoimidazole
carboxamide ribonucleotide transformylase, and thymidylate synthase
are associated with methotrexate effects in rheumatoid arthritis.
Arthritis Rheum. 50:2766–2774. 2004. View Article : Google Scholar
|
|
7
|
Kawabata T, Nishida K, Takasugi K, et al:
Increased activity and expression of histone deacetylase 1 in
relation to tumor necrosis factor-alpha in synovial tissue of
rheumatoid arthritis. Arthritis Res Ther. 12:R133–R145. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roybal JL, Endo M, Radu A, Zoltick PW and
Flake AW: Early gestational gene transfer of IL-10 by systemic
administration of lentiviral vector can prevent arthritis in a
murine model. Gene Ther. 18:719–726. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Drouin J and Haraoui B; 3e Initiative
Group: Predictors of clinical response and radiographic progression
in patients with rheumatoid arthritis treated with methotrexate
monotherapy. J Rheumatol. 37:1405–1410. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rudwaleit M, Yin Z, Siegert S, Grolms M,
Radbruch A, Braun J and Sieper J: Response to methotrexate in early
rheumatoid arthritis is associated with a decrease of T cell
derived tumour necrosis factor alpha, increase of interleukin 10,
and predicted by the initial concentration of interleukin 4. Ann
Rheum Dis. 59:311–314. 2000. View Article : Google Scholar
|
|
11
|
Nicholson JK and Lindon JC: Systems
biology: metabolomics. Nature. 455:1054–1056. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Malet-Martino M and Holzgrabe U: NMR
techniques in biomedical and pharmaceutical analysis. J Pharm
Biomed Anal. 55:1–15. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Inoue S, Hashiguchi M, Takagi K, Kawai S
and Mochizuki M: Preliminary study to identify the predictive
factors for the response to methotrexate therapy in patients with
rheumatoid arthritis. Yakugaku Zasshi. 129:843–849. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Um SY, Chung MW, Kim KB, et al: Pattern
recognition analysis for the prediction of adverse effects by
non-steroidal anti-inflammatory drugs using 1H NMR-based
metabolomics in rats. Anal Chem. 81:4734–4741. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lauridsen MB, Bliddal H, Christensen R, et
al: 1H NMR spectroscopy-based interventional metabolic
phenotyping: a cohort study of rheumatoid arthritis patients.
Proteome Res. 9:4545–4553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arnett FC, Edworthy SM, Bloch DA, et al:
The American Rheumatism Association 1987 revised criteria for the
classification of rheumatoid arthritis. Arthritis Rheum.
31:315–324. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fransen J, Creemers MC and van Riel PL:
Remission in rheumatoid arthritis: agreement of the disease
activity score (DAS28) with the ARA preliminary remission criteria.
Rheumatology. 43:1252–1255. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
MacIntyre DA, Jiménez B, Lewintre EJ, et
al: Serum metabolome analysis by 1H-NMR reveals
differences between chronic lymphocytic leukaemia molecular
subgroups. Leukemia. 24:788–797. 2010.PubMed/NCBI
|
|
19
|
Beckonert O, Keun HC, Ebbels TM, Bundy J,
Holmes E, Lindon JC and Nicholson JK: Metabolic profiling,
metabolomic and metabolomic procedures for NMR spectroscopy of
urine, plasma, serum and tissue extracts. Nat Protoc. 2:2692–2703.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Okada T, Afendi FM, Altaf-Ul-Amin M,
Takahashi H, Nakamura K and Kanaya S: Metabolomics of medicinal
plants: the importance of multivariate analysis of analytical
chemistry data. Curr Comput Aided Drug Des. 6:179–196. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bourré-Tessier J and Haraoui B:
Methotrexate drug interactions in the treatment of rheumatoid
arthritis: a systematic review. J Rheumatol. 37:1416–1421.
2010.PubMed/NCBI
|
|
22
|
Bathon JM, Martin RW, Fleischmann RM, et
al: A comparison of etanercept and methotrexate in patients with
early rheumatoid arthritis. N Engl J Med. 343:1586–1593. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Simoni RE, Gomes LN, Scalco FB, Oliveira
CP, Aquino Neto FR and de Oliveira ML: Uric acid changes in urine
and plasma: an effective tool in screening for purine inborn errors
of metabolism and other pathological conditions. J Inherit Metab
Dis. 30:295–309. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Weljie AM, Dowlatabadi R, Miller BJ, Vogel
HJ and Jirik FR: An inflammatory arthritis-associated metabolite
biomarker pattern revealed by 1H NMR spectroscopy. J
Proteome Res. 6:3456–3464. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rees WD, Wilson FA and Maloney CA: Sulfur
amino acid metabolism in pregnancy: the impact of methionine in the
maternal diet. J Nutr. 136:S1701–S1705. 2006.PubMed/NCBI
|
|
26
|
Jámbor A and Molnár-Perl I: Amino acid
analysis by high-performance liquid chromatography after
derivatization with 9-fluorenylmethyloxycarbonyl chloride.
Literature overview and further study. J Chromatogr A.
1216:3064–3077. 2009.
|
|
27
|
Carr DF, Whiteley G, Alfirevic A and
Pirmohamed M; FolATED study team: Investigation of inter-individual
variability of the one-carbon folate pathway: a bioinformatic and
genetic review. Pharmacogenomics J. 9:291–305. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schirra HJ, Anderson CG, Wilson WJ, Kerr
L, Craik DJ, Waters MJ and Lichanska AM: Altered metabolism of
growth hormone receptor mutant mice: a combined NMR metabolomics
and microarray study. PLoS One. 3:e27642008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sadykov MR, Zhang B, Halouska S, et al:
Using NMR metabolomics to investigate tricarboxylic acid
cycle-dependent signal transduction in Staphylococcus
epidermidis. J Biol Chem. 285:36616–36624. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lenz EM, Bright J, Wilson ID, Morgan SR
and Nash AF: A 1H NMR-based metabolomic study of urine
and plasma samples obtained from healthy human subjects. J Pharm
Biomed Anal. 33:1103–1115. 2003.
|