Introduction
Dental pulp cells (DPCs) have the potential to be
mineralized, which plays a key role in pulp repair and
dentinogenesis. Reparative dentin formation and pulp regeneration
after partial degradation are under the control of pulp progenitor
cells (1). Dentinogenesis is
regulated by tissue interactions associated with complicated signal
pathways. Dentin matrix protein 1 (DMP1), an essential
noncollagenous and acidic phosphorylated extracellular matrix
protein, is highly expressed in tooth odontoblasts and bone
osteocytes, with low level expression in osteoblasts and cartilage
(2–4). It is a multifunctional protein
involved in the biomineralization of bones and dentin (5), phosphate homeostasis, and
differentiation of odontoblasts and osteoblasts (6). The DMP1 gene has been mapped to
human chromosome 4q21:22 (7)
(chromosome 5q21 in mice) (8).
Dentinogenesis imperfecta type II, the autosomal dominant disorder
of dentin formation, has been also mapped to the same region of the
genome, indicating that DMP1 expression is tightly linked
genetically to its disease phenotype (9). Accordingly, DMP1 has been shown to
play a prime role in dentin mineralization (10,11). Dentin and bone extracellular
matrix (ECM) were shown to contain both the full length DMP1 as
well as its processed N-terminal (N-ter) (37 kDa) and C-terminal
(C-ter) (57 kDa) fragments. It was shown to regulate the
transcription of DSPP during early odontoblast differentiation
through binding of the promoter region through its carboxyl end
(residues 420–489) and was implicated in signaling functions
(12). This result led us to the
hypothesis that DMP1 may have functions other than regulating
mineralization. Recently, it was suggested that DMP1 belongs to the
SIBLING (small integrin binding ligand N-linked glycoprotein)
family of extracellular matrix proteins (13). The expression of SIBLING family
proteins such as osteopontin and bone sialoprotein in soft tissues
has been reported. DMP1 gene expression was also detected in fetal
bovine brain by northern analysis and in newborn mouse brain by
in situ hybridization (14,15). These reports raised the
possibility of DMP1 expression in other soft tissues, such as
liver, muscle, kidney, pancreas, salivary and eccrine sweat glands.
Together with results obtained in previous reports, it is suggested
that, in addition to its high affinity to calcium or hydroxyapatite
due to its acidic character and to its role in the mineralization
process, DMP1 may affect various cell activities.
Cells contain a variety of noncoding RNAs, such as
microRNAs (miRNAs), which are small (22-nt) endogenous noncoding
RNAs that anneal to the 3′ untranslated region (3′UTR) of target
mRNAs to inhibit translation and lower protein levels. It remains
to be established how specific miRNAs contribute to regulate the
onset of a tissue-specific phenotype in response to a
multifunctional morphogen. Let-7 was one of the first identified
miRNA families, consisting of 12 closely related genes in which
each isomer is usually located on a different chromosome and which
is highly conserved across animal species in sequence and timing of
expression (16,17). Let-7 plays a significant role in
cell proliferation, differentiation and oncogenesis; identification
of the target genes of miRNA may help to characterize its diverse
functions (17,18). By bioinformatic analysis, we have
found a potential binding site for let-7 within the 3′UTR of
DMP1.
It is important to understand the molecular
mechanism and specific genes during the developmental process. It
has been proven that DMP1 is regulated by many growth and
transcription factors in previous studies. However, the regulation
of DMP1 is not fully understood, particularly the understanding of
its post-transcriptional regulation. Here we present the probable
miRNA pathway of DMP1 in its post-transcriptional regulation.
Sophisticated computer-based prediction approaches of microRNAs and
of their targets, and effective biological validation techniques
for validating these predictions, now play a central role in
discovery of microRNAs and elucidating their functions. In this
study, we identified that let-7 may be the potential regulatory
miRNA of DMP1 gene through utilization of bioinformatics analysis
and a dual luciferase reporter assay. In addition, we assessed the
expression levels of let-7 at 10 and 21 days of differentiation in
odonto- and osteoblasts in dental pulp cells.
Materials and methods
Subjects and cell culture
Normal human impacted third molars were collected
from adults (16–24 years of age) at the Nanfang Hospital in the
Southern Medical University. The study protocol was approved by the
Institutional Ethics Committee, and informed consent was obtained
from all patients. DPCs were isolated and subjected to odontogenic
induction as previously reported (19). Tooth surfaces were cleaned and cut
around the cementum-enamel junction by using sterilized dental
fissure burs to reveal the pulp chamber. The pulp tissue was gently
separated from the crown and root and then digested in a solution
of 3 mg/ml collagenase type I and 4 mg/ml dispase for 30–60 min at
37°C. The cells were cultured in a growth medium containing
Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) with 15% fetal
bovine serum, 100 U/ml penicillin and 100,000 μg/ml
streptomycin, and then cultured at 37°C in 5% CO2. DPCs
at passage 3 were cultured in DMEM with 15% FBS until they reached
70–80% confluence. Cells were then induced in odontogenic medium
consisting of 50 mg/ml ascorbic acid, 10 mM β-glycerophosphate, and
0.01 mM dexamethasone (Sigma) in DMEM with 15% FBS for 7–21 days.
Control samples were DPCs grown in DMEM with 15% FBS and harvested
at 80% confluence. T293 cells were grown in DMEM (Gibco)
supplemented with 10% fetal bovine serum.
Mineralization staining
Mineralization of cultured DPCs was determined using
Alizarin Red (AR) staining. After Day 21, the cell layer was washed
with PBS and fixed in 10% formaldehyde (Sigma-Aldrich) at room
temperature for 15 min, then washed in duplicate with excess
dH2O prior to the addition of 1 ml of 40 mM AR (pH 4.1).
The plates were incubated at room temperature for 30 min under
gentle shaking. Following aspiration of the unincorporated dye, the
plates were washed twice with dH2O and visualized using
phase microscopy (Nikon).
Bioinformatics
DMP1 was identified as a potential target to search
for miRNAs. Three different miRNA target prediction programs were
used, TargetScan (http://www.targetscan.org/), the UCSC genome browser
tract for PicTar4 (http://genome.ucsc.edu/), and the miRBase for miRanda
(http://microrna.sanger.ac.uk/sequences/) (20–22). Each of these programs were
searched for complementarity to the miRNA seed region in the 3′UTRs
of DMP1. miRNAs were chosen based upon their targeted prediction by
all three programs, conser vation of the binding region, and
strength of the predicted interaction.
Dual luciferase reporter gene
construct
A 1041-bp fragment of the DMP1 3′UTR containing the
predicted binding site for let-7 was amplified from human genomic
DNA using primers with a short extension containing cleavage sites
for XhoI (5′ end) and NotI (3′ end): DMP1 forward
(5′-CCGCTCGAGC ATCAGCTGTCCTAAGAAGCAGTT-3′) and DMP1 reverse
(5′-ATAAGAATGCGGCCGCTTTCTTCTGGGTATTATAA TCTTTATTAC-3′). Amplicons
were cleaved with XhoI and NotI and cloned in between
the XhoI and NotI cleavage sites of the psi-CHECK2
luciferase vector (Promega) downstream of the Renilla luciferase
reporter gene.
Dual luciferase reporter assay
Luciferase assays were performed using the
Dual-Luciferase assay kit as previously described (Promega). For
let-7 target validation, 293-T cells were grown to a cell density
of 60–70% and co-transfected in 24-well plates with the indicated
psiCHECK2 luciferase construct (0.5 μg/well) and miRNA (20
μM) using Lipofectamine 2000. Following 48 h, the cells were
harvested in passive lysis buffer, and luciferase activity was
measured with a GloMax™ 20/20 luminometer (Promega). Renilla
luciferase activity was normalized to corresponding firefly
luciferase activity and plotted as a percentage of the control
between samples. Luciferase experiments were measured in triplicate
using independent samples, as indicated.
qRT-PCR
Dental pulp cells cultured in a mineralizing medium
were able to differentiate into odontoblast-like cells. Samples
were harvested for the isolation of RNA at 10 and 21 days of
differentiation to detect quantitative changes in gene DMP1 and
three miRNAs. Cells were cultured in a growth medium that served as
the control. Total-RNA was extracted from cells using the TRIzol
reagent (Invitrogen, Carlsbad, CA) per the manufacturer’s
instructions. RNA was analyzed for integrity, purity and
concentration by gel electrophoresis and spectrophotometry and
stored at −80°C until analysis.
For quantitative RT-PCR analysis, 0.1 μg of
RNA per reaction was used with the QuantiTech SYBR-Green RT-PCR kit
and primers specific for DMP1. To quantify miRNA expression,
total-RNA was reverse transcribed for use in two-step quantitative
RT-PCR using the stem-loop method. The resulting cDNA was subjected
to real-time qRT-PCR using the universal reverse primer in
conjunction with a sequence-specific forward primer for hsa-let-7a,
hsa-let-7c, hsa-let-7d, hsa-let-7f, hsa-let-7g and hsa-let-7i. Each
sample was performed in triplicate, and the results were normalized
using primers to 18S rRNA (for DMP1) or U6 (for miRNA analysis)
(Table I). The reactions were
incubated at 95°C for 5 min for one cycle and then 95°C/15 sec,
65°C/15 sec, 72°C/32 sec for 40 cycles, with a final 10-min
extension at 65°C. Results are expressed as fold change in
expression relative to the control sample calculated using the
equation RQ=2−ΔΔCt.
 | Table IOligonucleotides used in this
study. |
Table I
Oligonucleotides used in this
study.
| Primer name | Sequence-specific
primer |
|---|
| hsa-let-7a-RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATATCAA-3′ |
| hsa-let-7c-RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATACCAA-3′ |
| hsa-let-7d-RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTATCAA-3′ |
| hsa-let-7f-RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATATCAA-3′ |
| hsa-let-7g-RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATGTVAA-3′ |
| hsa-let-7i-RT |
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACGACAA-3′ |
| U6-RT |
5′-AACGCTTCACGAATTTGCGT-3′ |
| hsa-let-7a-F |
5′-TGAGGTAGTAGGTTGTATAGTT-3′ |
| hsa-let-7c-F |
5′-TGAGGTAGTAGGTTGTATGGTT-3′ |
| hsa-let-7d-F |
5′-AGAGGTAGTAGGTTGCATAGTT-3′ |
| hsa-let-7f-F |
5′-GCTGAGGTAGTAGATTGTATAGTT-3′ |
| hsa-let-7g-F |
5′-TGAGGTAGTAGTTTGTACAGTT-3′ |
| hsa-let-7i-F |
5′-GCATGAGGTAGTAGTTTGTGCTGTT-3′ |
| miRNA-R |
5′-GTGCAGGGTCCGAGGT-3′ |
| U6-F |
5′-CTCGCTTCGGCAGCACA-3′ |
| U6-R |
5′-AACGCTTCACGAATTTGCGT-3′ |
| DMP1-F |
5′-CCCTTGGAGAGCAGTGAGTC-3′ |
| DMP1-R |
5′-CTCCTTTTCCTGTGCTCCTG-3′ |
| 18s rRNA-F |
5′-CCTGGATACCGCAGCTAGGA-3′ |
| 18s rRNA-R |
5′-GCGGCGCAATACGAATGCCCC-3′ |
Statistical analyses
Statistical analysis was performed with SPSS for
Windows (version 13.0) using the ANOVA test. All data are presented
as the mean ± SD (n≥3). P≤0.01 was considered statistically
significant.
Results
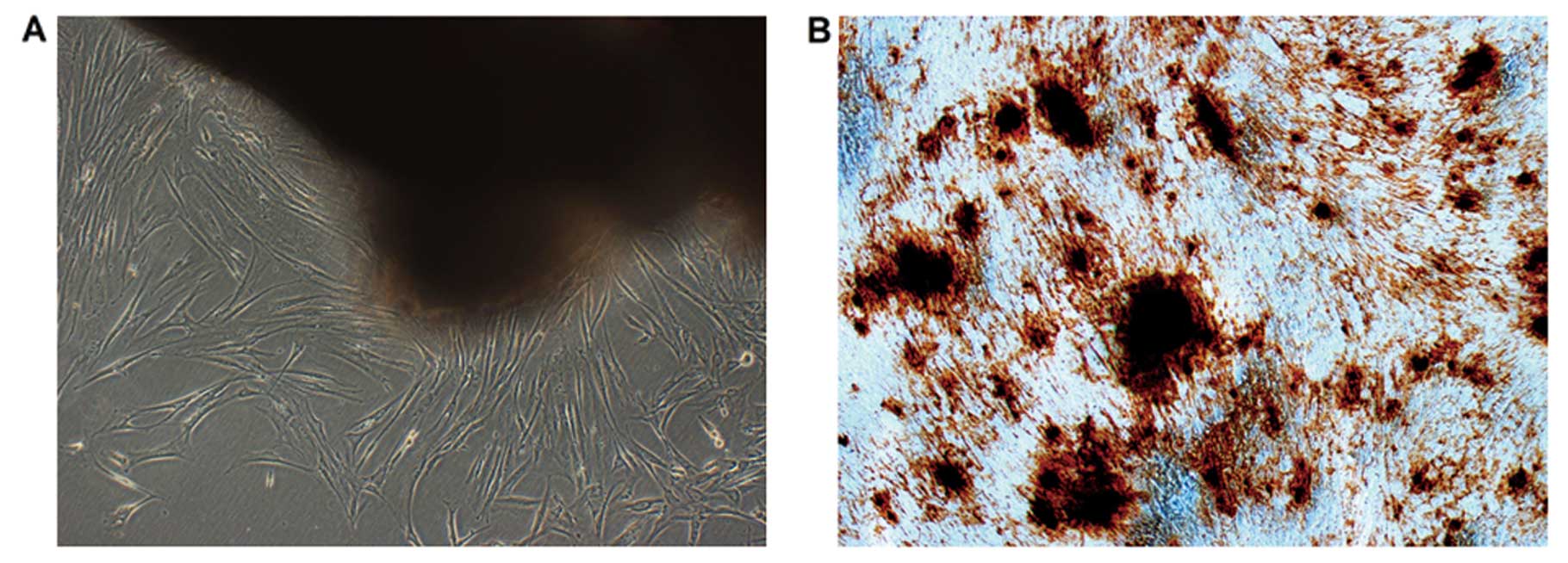
Characteristics of dental pulp cells
It has been demonstrated that dental pulp cells can
be expanded as a dentin-pulp-like structure. Our results show that
DPCs have the potential to differentiate into odontoblast-like
cells when cultured in the culture medium contain dexamethasone
(Dex) and/or β-glycerophosphate (β-GP) (23). Ten days later, mineralized nodules
formed and became more condensed. Alizarin Red staining of
mineralized nodules in representative cell cultures is demonstrated
at 21 days in Fig. 1B.
Bioinformatic analyses identify let-7 as
a regulator of DMP1
With DMP1 being identified as a preferential target,
miRNA target prediction was carried out using a combination of the
following computational algorithms: TargetScan, miRanda, and
PicTar. Based on the stem-loop feature of the miRNA and
cross-species comparison, a number of computational algorithms have
been developed to predict miRNAs from the genome. Among miRNAs
being predicted to target DMP1, let-7 was complementary to multiple
sequences of potential miRNAs with a high probability of binding to
the 3′UTR of DMP1. To increase our probability of identifying
miRNAs capable of regulating DMP1 post-transcriptionally, we
selected to further examine eight miRNAs that were identified by
all three search algorithms.
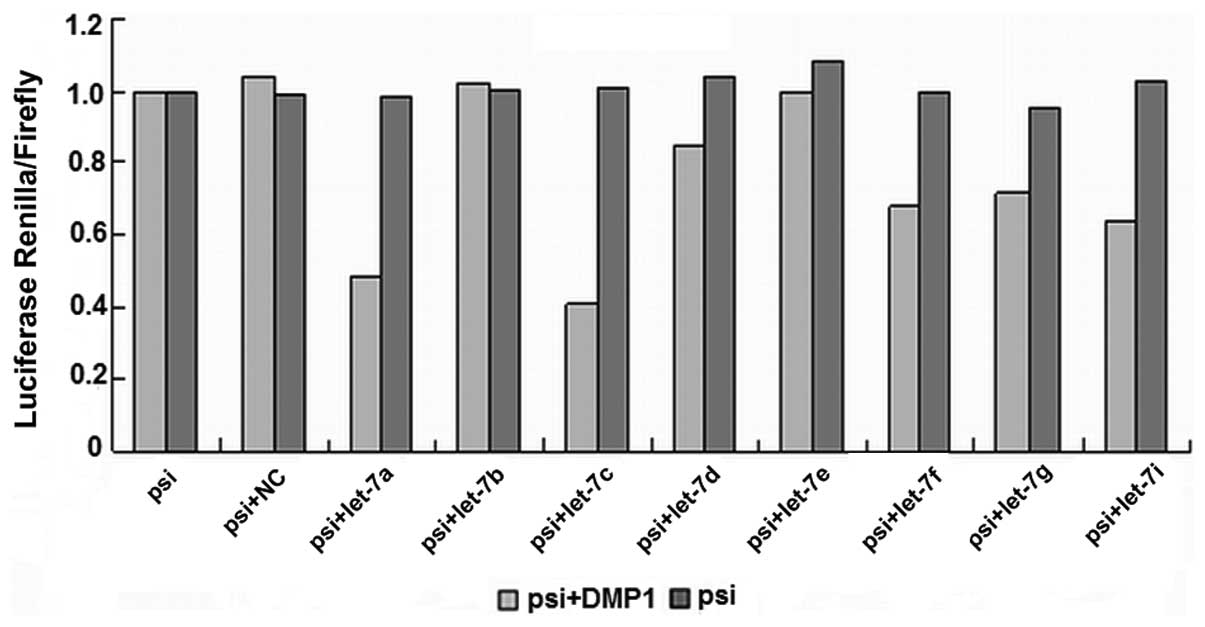
Let-7 regulates DMP1
We used a luciferase reporter assay to test whether
the 3′UTR of DMP1 contained sequences capable of interacting with
let-7. The potential binding sites of let-7 within DMP1 3′UTR were
obtained by TargetScan and PicTar. Synthetic oligos including
predicted binding sites were annealed then cloned into
XhoI/NotI site of psiCHECK-2. 293-T cells were
transiently transfected with the relevant DMP1 3′UTR fragment and
microRNA using Lipofectamine 2000 (Invitrogen) at the indicated
concentrations.
To test whether the luciferase assay responds to
let-7, we then tested reporters in which the 3′UTR of DMP1 was
inserted downstream of firefly luciferase. The 3′UTR of DMP1
contains conserved let-7 seed matches at positions 988–994. When
adding a miRNA that is able to interact with DMP1 3′UTR, the
bioluminescence reaction of Photinus pyralis luciferase was
annihilated, as miRNA inhibits translation of vector mRNA. This
construct allowed us to quickly and quantitatively evaluate the
miRNA’s effect on the 3′UTR of DMP1. The DMP1 3′UTR fragment was
inserted into the psiCHECK2 luciferase vector. The relative
luciferase activity in 293-T cells transfected with the luciferase
vector alone was set at 100% for comparison. The binding ability of
the eight predicted let-7 miRNAs with DMP1 is shown in Fig. 2.
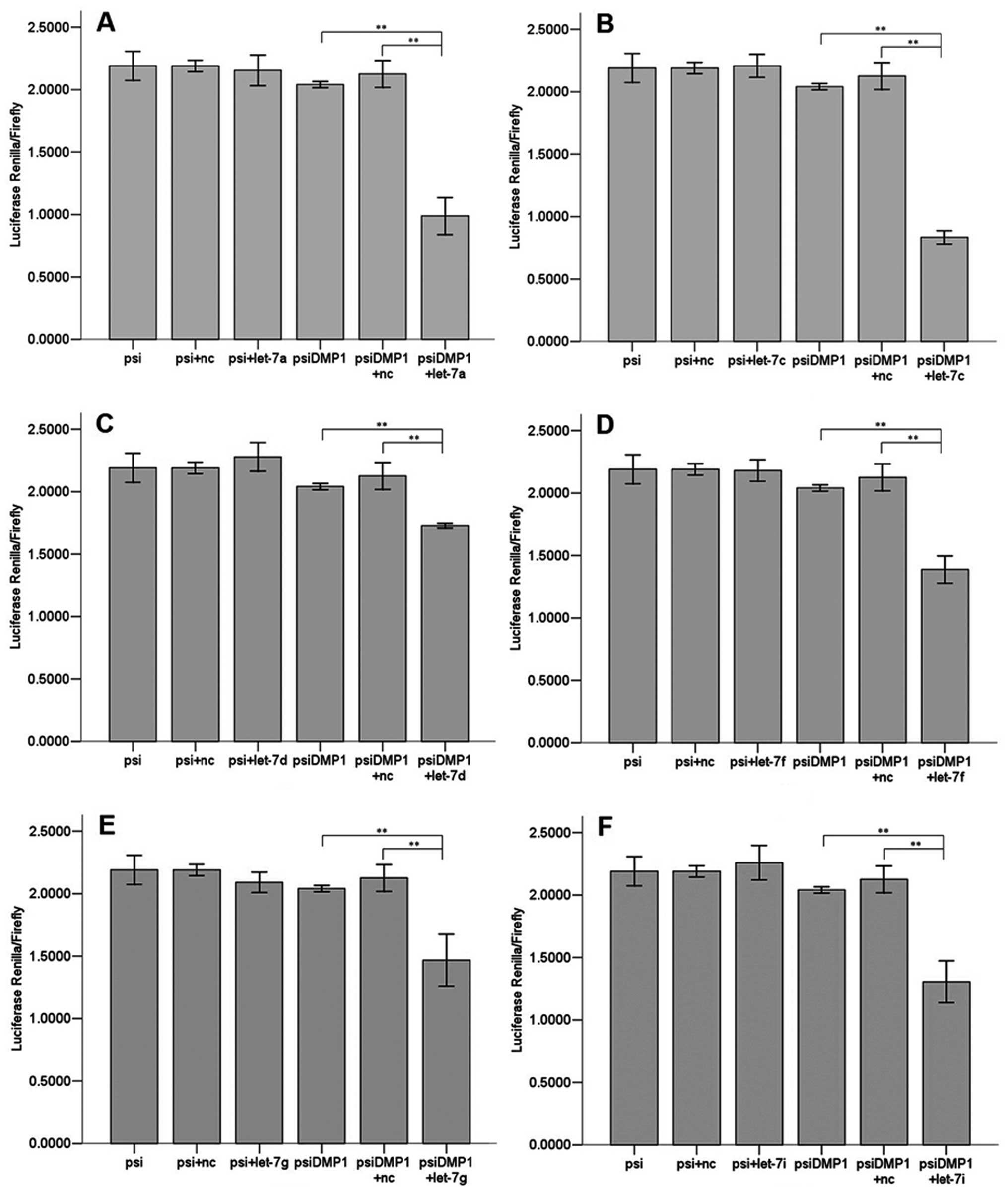
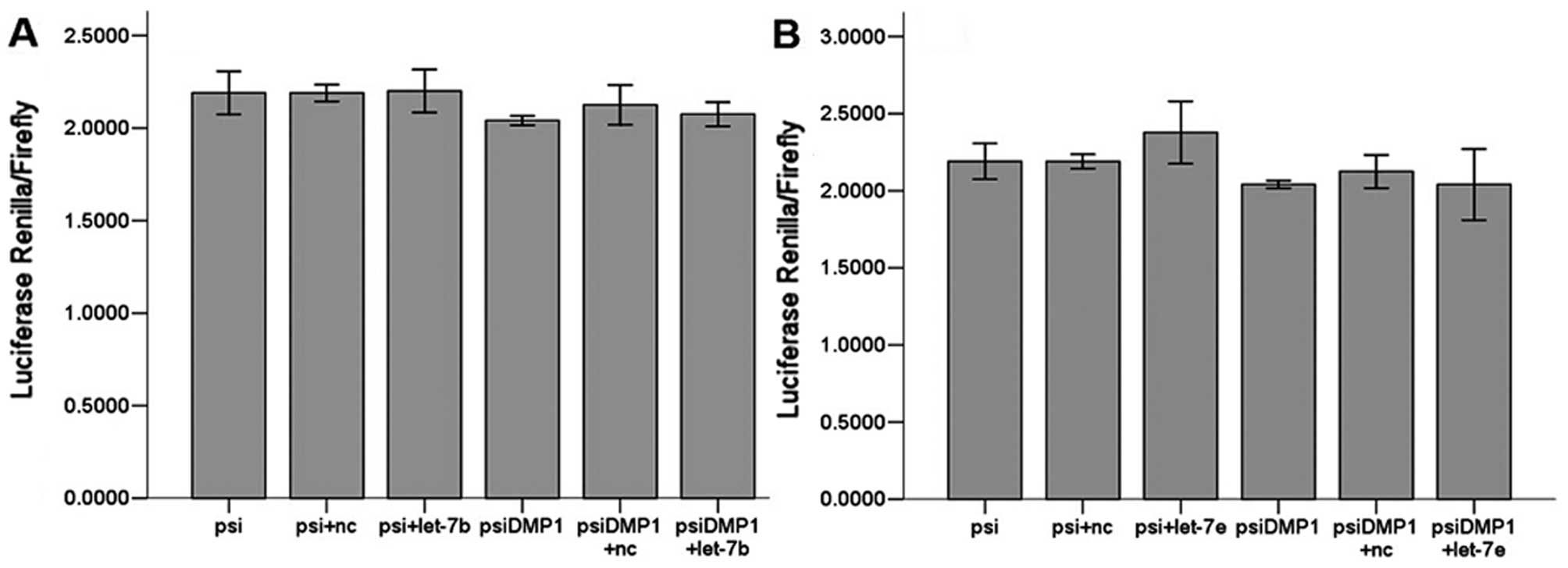
Let-7a, let-7c, let-7d, let-7f, let-7g and let-7i
(Fig. 3) all significantly
(P<0.01) reduced luciferase activity when compared to the
negative scrambled miRNA and the luciferase vector alone, while
let-7b and let-7e (Fig. 4) did
not. There are 6 members of let-7 family miRNAs predicted by the
biology software TargetScan and miRanda, and 5 by PicTar4. Let-7
family is considered a potentially important regulator of DMP1
3′UTR.
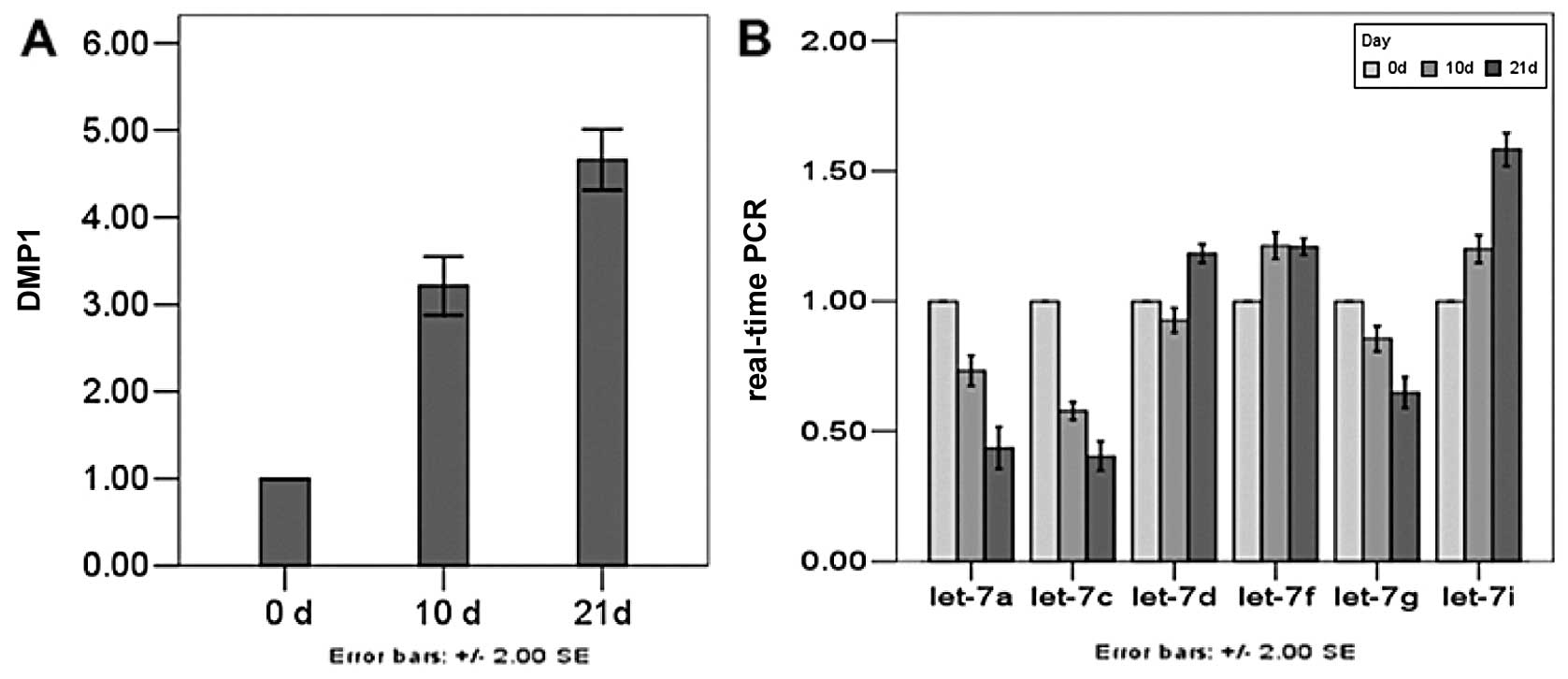
Expression levels of DMP1, let-7a,
let-7c, let-7d, let-7f, let-7g and let-7i miRNA genes determined
using qRT-PCR analysis
To validate the above data, the expression of 6
miRNA genes in the process of dental pulp cell differentiation was
determined using qRT-PCR analysis. The DMP1 transcript was
predominantly expressed in odontoblasts and transiently in
preameloblasts along with an involvement with odontoblast
differentiation and mineralization, which is used as an indicator
of odontoblastic differentiation. Dental pulp cells cultured in
mineral izing medium exhibited odontoblastic features, including
increasing DMP1. In our study, expression of DMP1 was weak before
Day 0, but the amount increased by Day 10. After Day 21, DMP1 was
strongly expressed. As a regulator of DMP1, the members of let-7
family need to exist in undifferentiated and differentiated dental
pulp cells. Expression of 3 miRNAs on Day 10 was decreased,
especially that of let-7a and let-7c. On Day 21, expression level
of the 3 miRNAs was significantly decreased when DMP1 was at its
peak. The expression of the other 3 miRNAs was higher in
differentiated than undifferentiated cells, but the relative
expression of let-7i rose higher than that of the other two miRNAs
(Fig. 5).
Discussion
miRNAs constitute a type of endogenous
post-transcriptional regulatory gene expression. miRNAs provide
important regulatory functions as key negative regulators of
diverse biological and pathological processes, including
development, organogenesis, apoptosis, cell proliferation,
differentiation and in the control of tumorigenesis. Some miRNAs
can control gene expression during mesenchymal differ entiation and
in modulating osteogenic differentiation (24). A group of miRNAs can be
co-regulatory when they target genes in common and therefore can
involve a complex process. The conserved let-7 miRNA was originally
discovered in Caenorhabditis elegans as a switch gene
induced as cells exit the cell cycle when C. elegans reach
their adult stage (25–27). In humans and mouse, like C.
elegans, the expression of let-7 is barely detectable in
embryonic developmental stages but increases after differentiation
and in mature tissue (28). A
previous study has implicated let-7 as a tumor suppressor (29). Let-7 family members may play a
role in cancer progression, as they map onto genomic regions
altered or deleted, such as HMGA2. This target was identified by a
different let-7, which contributes to differentiation during
mammary epithelial cell differentiation affecting self-renewal
(30,31). Previous studies on the functional
effects of let-7 have focused on the targets Ras, HMGA2, and c-Myc
(32–35).
Two approaches are used to identify candidate genes
targeted by miRNAs: computational target prediction algorithms and
experimental target identification strategies. Prediction tools
such as TargetScan and miRanda can be used to identify potential
target genes for all miRNAs. Current miRNA target prediction tools
have the common problem that their false positive rate is
unavoidable, so experimental target identification is necessary. A
dual luciferase reporter assay is a quick, sensitive and direct
method to verify these predicted miRNAs (36). In this study, we predicted let-7
targeting DMP1 using computational analyses and observed various
let-7 isoforms identified concurrently, let-7a, let-7c, let-7d,
let-7f, let-7g and let-7i being the most predominant. Combined with
qRT-PCR, we have shown for the first time that let-7 expression is
differentially expressed on the marker gene DMP1 during
differentiation of dental pulp cells to odontoblast-like cells. The
results showed that on Day 21 the expression levels of DMP1 were
highest, and that of let-7a, let-7c and let-7g were lowest. When
the level of DMP1 were low, let-7a, let-7c and let-7g were
relatively high, which demonstrated that there was some link
between DMP1 and miRNAs. But it remains unclear whether let-7
regulates other target genes and what are the key roles of the
complex regulatory network between miRNA and mRNA in this molecular
pathway for odonto-differentiation.
In a previous study, we used a dual luciferase
reporter assay and qRT-PCR to confirm that mir885-5p, mir586 and
mir32-targeted DSPP were expressed during differentiation into
odontoblast-like cells (37).
These findings warrant additional studies to investigate whether
miRNA alterations are also involved in the process of
differentiation of dental pulp cells to odontoblast-like cells and
whether miRNA expression levels would manifest the biological and
biochemical consequences in the development of differentiation. The
results of the present study suggest the possibility of using
miRNAs for the development of cell differentiation. Further
experiments will be required to assess the differentiation effect
of let-7 miRNA at various stages of the process, and such
experiments are currently underway in our laboratory.
Accordingly, future identification of the targets
for miRNAs and the regularity for change of miRNAs may provide
clues to develop a novel marker during differentiation of dental
pulp cells and result in abnormal dentin formation. It is envisaged
that such future studies may ultimately provide a foundation for a
new paradigm of the involvement of noncoding small RNA species,
miRNA, in stem cell differentiation. Our findings suggest a
mechanistic link between the let-7 family of miRNAs and DMP1 gene
expression in dental pulp cells. We anticipate that unraveling the
molecular mechanisms by which miRNAs mediate effects during
odontoblast differentiation will allow us to decipher the central
regulatory role of miRNAs in many fundamental biological
processes.
Acknowledgements
This study was supported by a
grant-in-aid for the College intelligences funds from Guangdong
province (no. C1030270) and the Events of Science and Technology
Program of the Guangdong province (no. 2010B060900053).
References
|
1.
|
GT HuangS GronthosS ShiMesenchymal stem
cells derived from dental tissues vs. those from other sources:
their biology and role in regenerative medicineJ Dent
Res88792806200919767575
|
|
2.
|
A GeorgeB SabsayPA SimonianA
VeisCharacterization of a novel dentin matrix acidic
phosphoprotein. Implications for induction of biomineralizationJ
Biol Chem268126241263019938509401
|
|
3.
|
KL HirstK Ibaraki-O’ConnorMF YoungMJ
DixonCloning and expression analysis of the bovine dentin matrix
acidic phosphoprotein geneJ Dent
Res76754760199710.1177/002203459707600307019109824
|
|
4.
|
M MacDougallTT GuX LuanD SimmonsJ
ChenIdentification of a novel isoform of mouse dentin matrix
protein 1: spatial expression in mineralized tissuesJ Bone Miner
Res13422431199810.1359/jbmr.1998.13.3.4229525343
|
|
5.
|
C QinJC BrunnRG CookRS OrkiszewskiJP
MaloneA VeisWT ButlerEvidence for the proteolytic processing of
dentin matrix protein 1. Identification and characterization of
processed fragments and cleavage sitesJ Biol
Chem2783470034708200310.1074/jbc.M30531520012813042
|
|
6.
|
A AlmushaytK NarayananAE ZakiA
GeorgeDentin matrix protein 1 induces cytodifferentiation of dental
pulp stem cells into odontoblastsGene
Ther13611620200610.1038/sj.gt.330268716319946
|
|
7.
|
M MacDougallBR DuPontD SimmonsRJ
LeachAssignment of DMP1 to human chromosome 4 band q21 by in situ
hybridizationCytogenet Cell
Genet74189199610.1159/0001344108941370
|
|
8.
|
A GeorgeJ GuiNA JenkinsDJ GilbertNG
CopelandA VeisIn situ localization and chromosomal mapping of the
AG1 (Dmp1) geneJ Histochem
Cytochem4215271531199410.1177/42.12.79833537983353
|
|
9.
|
HM AplinKL HirstAH CrosbyMJ DixonMapping
of the human dentin matrix acidic phosphoprotein gene (DMP1) to the
dentinogenesis imperfecta type II critical region at chromosome
4q21Genomics30347349199510.1006/geno.1995.98678586437
|
|
10.
|
G HeT DahlA VeisA GeorgeDentin matrix
protein 1 initiates hydroxyapatite formation in vitroConnect Tissue
Res44Suppl 1240245200310.1080/71371363712952204
|
|
11.
|
G HeS GajjeramanD SchultzD CooksonC QinWT
ButlerJ HaoA GeorgeSpatially and temporally controlled
biomineralization is facilitated by interaction between
self-assembled dentin matrix protein 1 and calcium phosphate nuclei
in solutionBiochemistry441614016148200510.1021/bi051045l
|
|
12.
|
K NarayananA RamachandranJ HaoG HeKW ParkM
ChoA GeorgeDual functional roles of dentin matrix protein 1.
Implications in biomineralization and gene transcription by
activation of intracellular Ca2+ storeJ Biol
Chem2781750017508200310.1074/jbc.M21270020012615915
|
|
13.
|
LW FisherNS FedarkoSix genes expressed in
bones and teeth encode the current members of the SIBLING family of
proteinsConnect Tissue Res44Suppl
1S33S40200310.1080/71371364412952171
|
|
14.
|
RN D’SouzaA CavenderG SunavalaJ AlvarezT
OhshimaAB KulkarniM MacDougallGene expression patterns of murine
dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein
(DSPP) suggest distinct developmental functions in vivoJ Bone Miner
Res122040204919979421236
|
|
15.
|
KL HirstK Ibaraki-O’ConnorMF YoungMJ
DixonCloning and expression analysis of the bovine dentin matrix
acidic phosphoprotein geneJ Dent
Res76754760199710.1177/002203459707600307019109824
|
|
16.
|
LP LimNC LauEG WeinsteinA AbdelhakimS
YektaMW RhoadesCB BurgeDP BartelThe microRNAs of Caenorhabditis
elegansGenes Dev1799110082003
|
|
17.
|
S RoushFJ SlackThe let-7 family of
microRNAsTrends Cell Biol18505516200810.1016/j.tcb.2008.07.007
|
|
18.
|
XY HeJX ChenZ ZhangCL LiQL PengHM PengThe
let-7a microRNA protects from growth of lung carcinoma by
suppression of k-Ras and c-Myc in nude miceJ Cancer Res Clin
Oncol13610231028201010.1007/s00432-009-0747-520033209
|
|
19.
|
S GronthosJ BrahimW LiLW FisherN ChermanA
BoydeP DenBestenPG RobeyS ShiStem cell properties of human dental
pulp stem cellsJ Dent
Res81531535200210.1177/15440591020810080612147742
|
|
20.
|
BP LewisIH ShihMW Jones-RhoadesDP BartelCB
BurgePrediction of mammalian microRNA
targetsCell115787798200310.1016/S0092-8674(03)01018-314697198
|
|
21.
|
RM MarinJ VanicekEfficient use of
accessibility in microRNA target predictionNucleic Acids
Res391929201110.1093/nar/gkq76820805242
|
|
22.
|
SF MaddenSB CarpenterIB JefferyH
BjorkbackaKA FitzgeraldLA O’NeillDG HigginsDetecting microRNA
activity from gene expression dataBMC
Bioinformatics11257201010.1186/1471-2105-11-25720482775
|
|
23.
|
ML CoubleJC FargesF BleicherB
Perrat-MabillonM BoudeulleH MagloireOdontoblast differentiation of
human dental pulp cells in explant culturesCalcif Tissue
Int66129138200010.1007/PL0000583310652961
|
|
24.
|
LA GoffS BoucherCL RicuperoS
FenstermacherM SwerdelLG ChaseCC AdamsJ ChesnutU LakshmipathyRP
HartDifferentiating human multipotent mesenchymal stromal cells
regulate microRNAs: prediction of microRNA regulation by PDGF
during osteogenesisExp
Hematol3613541369200810.1016/j.exphem.2008.05.004
|
|
25.
|
AL AbbottUncovering new functions for
microRNAs in Caenorhabditis elegansCurr
Biol21R668R671201110.1016/j.cub.2011.07.02721920301
|
|
26.
|
SM JohnsonSY LinFJ SlackThe time of
appearance of the C. elegans let-7 microRNA is
transcriptionally controlled utilizing a temporal regulatory
element in its promoterDev Biol259364379200312871707
|
|
27.
|
AL AbbottE Alvarez-SaavedraEA MiskaNC
LauDP BartelHR HorvitzV AmbrosThe let-7 MicroRNA family members
mir-48, mir-84, and mir-241 function together to regulate
developmental timing in Caenorhabditis elegansDev
Cell9403414200510.1016/j.devcel.2005.07.00916139228
|
|
28.
|
H JinS LvJ YangX WangH HuC SuC ZhouJ LiY
HuangL LiX LiuM WuQ QianUse of microRNA Let-7 to control the
replication specificity of oncolytic adenovirus in hepatocellular
carcinoma cellsPLoS
One6e21307201110.1371/journal.pone.002130721814544
|
|
29.
|
Y LiTN VandenBoomD KongZ WangS AliPA
PhilipFH SarkarUp-regulation of miR-200 and let-7 by natural agents
leads to the reversal of epithelial-to-mesenchymal transition in
gemcitabine-resistant pancreatic cancer cellsCancer
Res6967046712200910.1158/0008-5472.CAN-09-129819654291
|
|
30.
|
AX ChenKD YuL FanJY LiC YangAJ HuangZM
ShaoGermline genetic variants disturbing the Let-7/LIN28
double-negative feedback loop alter breast cancer
susceptibilityPLoS
Genet7e1002259201110.1371/journal.pgen.100225921912531
|
|
31.
|
J YunCA FrankenbergerWL KuoMC BoelensEM
EvesN ChengH LiangWH LiH IshwaranAJ MinnMR RosnerSignalling pathway
for RKIP and Let-7 regulates and predicts metastatic breast
cancerEMBO J3045004541201110.1038/emboj.2011.31221873975
|
|
32.
|
L BoominathanThe guardians of the genome
(p53, TA-p73, and TA-p63) are regulators of tumor suppressor miRNAs
networkCancer Metastasis
Rev29613639201010.1007/s10555-010-9257-920922462
|
|
33.
|
YS LeeA DuttaThe tumor suppressor microRNA
let-7 represses the HMGA2 oncogeneGenes
Dev2110251030200710.1101/gad.154040717437991
|
|
34.
|
SM JohnsonH GrosshansJ ShingaraM ByromR
JarvisA ChengE LabourierKL ReinertD BrownFJ SlackRAS is regulated
by the let-7 microRNA
familyCell120635647200510.1016/j.cell.2005.01.01415766527
|
|
35.
|
S WangY TangH CuiX ZhaoX LuoW PanX HuangN
ShenLet-7/miR-98 regulate Fas and Fas-mediated apoptosisGenes
Immun12149154201110.1038/gene.2010.5321228813
|
|
36.
|
FE NicolasExperimental validation of
microRNA targets using a luciferase reporter systemMethods Mol
Biol732139152201110.1007/978-1-61779-083-6_1121431711
|
|
37.
|
X HuangS XuJ GaoF LiuJ YueT ChenB WumiRNA
expression profiling identifies DSPP regulators in cultured dental
pulp cellsInt J Mol Med28659667201121687927
|