Introduction
Overall survival after liver transplantation has
significantly improved in recent years. However, early graft
failure remains a serious concern with high morbidity and
mortality. Primary graft non-function (PNF) is a rare, but serious
condition, of unknown pathophysiology developing in 2–6% cases
following liver transplantation (1–3).
Although PNF was discovered more than 25 years ago, a clear
consensus regarding the definition of PNF has not been reached
(4). The United Network for Organ
Sharing (UNOS) defines PNF as an irreversible graft function
requiring emergency liver replacement within the first 10 days of
liver transplantation. It is characterized by an aspartate
aminotransferase (AST) of ≥5,000 UI/l, international normalized
ratio (INR) of ≥3.0, and acidosis.
Other researchers have proposed a variety of
definitions for graft non-function. Silberhumer et al
suggested 4 grades of initial graft dysfunction, where PNF was
defined as a clinical patient status requiring retransplantation
within 7 days of primary transplantation (5). A further definition of PNF was
presented by the European Liver Transplant Registry (ELTR), where
PNF was restricted to retransplantation requirement within 1 month
(30 days) of primary transplantation. Yet another interpretation
was proposed by Amin et al, who altered the definition used
by ELTR to ‘a non-functioning graft within 1 month of receiving
liver from the deceased donor’ (6).
It is difficult to assess the incidence of PNF as it
depends on the definition used. According to Kemmer et al,
the incidence of PNF is similar in Europe and USA (7). The analysis conducted by Burroughs
et al (8) on the data
obtained from the ELTR (9) showed
the incidence of PNF to be 6% and 3%, where the cut-off time was 90
days and 30 days, respectively. Using the analysis of the 30-day
time period, Kemmer et al demonstrated that although the
utilization of extended criteria donors (ECD) over the last years
had increased, the incidence of PNF has decreased (7).
A failing graft has been shown to lead to
multi-organ instability, especially renal hemodynamic failure or
generalized sepsis (10). Liver
transplantation is the only treatment of choice for non-functioning
grafts (11). The decision for
retransplantation is mainly driven by clinical judgment and
experience, partly because of the variance in the definitions of
PNF. High perioperative recipient mortality is associated with the
lack of instant availability of new grafts. In such cases, rescue
hepatectomy has been proposed as a treatment, which aims to improve
patient conditions while awaiting a retransplantation (12,13). A large percentage of patients pass
away because of systemic complications, which lead to multi-organ
failure before a new graft becomes available.
The risk factors associated with PNF are related to
the organ donor as well as to the organ recipient (14). In many studies, a donor over the
age of 40 was associated with a significantly higher risk of graft
failure (15,16). In addition, hepatic steatosis of
graft has also become a major issue, and susceptibility to
steatosis increases with donor age, history of obesity,
dyslipidemia, and diabetes. Macrovesicular steatosis is considered
one of the most important risk factors for graft non-function
(17). Other donor-related risk
factors include >5-day stay in the intensive care unit (ICU),
warm ischemia time of >40 min, cold ischemia time (CIT) of
>10 h, and hypernatremia (18–21). In addition, research studies are
constantly adding new risk factors and questioning the significance
of the old risk factors for PNF (22–24).
Recent shortage in the available donor livers has
led to the development of ECD (25–27). These include donors over the age
of 70, as well as donors with a history of hepatitis C, inactive
hepatitis B, or liver steatosis. ECD carry a high risk of PNF
(28); consequently, a decreased
survival has been observed among high-risk recipients receiving
organs from ECD (29).
Currently, the only effective strategy of prevention
against primary graft non-function is donor and recipient selection
according to known risk factors which does not fully prevent PNF.
Little is known about the pathway of metabolic events leading to
PNF, although interrupted microcirculation may play an important
role (30). In contrast to
previous studies focusing on factors involved in the event of PNF
we hypothesized that a proteomic approach may lead to a better
understanding of the biological aspect of graft non-function. Our
10-year experience of 1,000 transplantations at the Department of
General, Transplant and Liver Surgery, (Medical University of
Warsaw) support the relevance of this topic in patient
survival.
In this study, we have used a proteomic approach to
identify the proteins associated with PNF following liver
transplantation. Our goal was to identify biomarkers that could be
measured before liver transplantation to predict the grafts that
will be susceptible to non-function. Clinical measurement of these
biomarkers, in donors could potentially reduce the likelihood of
PNF occurrence.
Materials and methods
Patient selection
Liver fragments were collected from 96 consecutive
liver transplantations. All 96 pairs of samples were subjected to
proteomic examination. Follow-up of the patients revealed that 3 of
them (3.1%) developed PNF within 10 days of transplantation
(Table I). All other potential
risk factors (HAT, acute rejection) were excluded in these 3 cases,
where the grafts were considered to meet the criteria of PNF
according to the UNOS criteria. All recipients received liver
grafts from ECD, defined in our study as heart beating deceased
donors over the age of 60, with steatosis of >30%, >5-day
stay in the ICU, CIT of >8 h, or [Na+] of >170
mmol/l at least at one point in time. In each case, the following
data were collected: cause of death, CIT, warm ischemia time, and
post-reperfusion syndrome. Two patients were listed for, and
subsequently underwent emergency retransplantation. The overall
mortality in this group was 100%. As a control group, we used 6
samples from donors with optimal grafts, who recovered after liver
transplantation with a good synthetic liver function. On the basis
of clinical, biochemical, and radiological parameters, the grafts
were considered as either a normal/optimal or a suboptimal/EDC.
After transplantation, clinical follow-up was carried out, and all
the data were stored in a database. To minimize individual
variability, we compared PNF and healthy liver protein lysates in 3
different sets, as described below.
 | Table ICharacteristics of the PNF
patients. |
Table I
Characteristics of the PNF
patients.
| Patient no. | Extended criteria
donor | Non-optimal
recipient | Indication for
OLTxa | Recipient
gender | Primary
non-function | Decision of
reOLTx | reOLTxb | Survival |
|---|
| 1 | Yes | No | Cryptogenic
cirrhosis | F | + | − | − | 2 days after index
transplantation |
| 2 | Yes | Yes (reOLTx) | Metastases of
NETc | M | + | − | − | 5 days after index
transplantation |
| 3 | Yes | No | HCV positive
cirrhosis | F | + | + | + | 6 days after
reOLTx |
This study was approved by the independent Ethics
Committee of Warsaw Medical University in accordance with the
ethics guidelines of the ‘World Medical Association Declaration of
Helsinki - Ethical Principles for Medical Research Involving Human
Subjects’ adopted by the 18th WMA General Assembly, Helsinki,
Finland. Donor retrieval was performed under Polish law of organ
donation supervised by the Polish Transplant Coordinating Centre
‘Poltransplant’ (www.poltransplant.org.pl). All recipients enrolled in
this study provided written informed consent.
Sample preparation for protein
extraction
Tissues were snap-frozen in nitrogen and stored at
−80°C until use. The frozen liver specimens (100–200 mg) were
crushed to a fine powder by a ceramic mortar in liquid nitrogen.
The fine powder was suspended in 1 ml of rehydratation/sample
buffer 1 (RSB1) containing 7 M urea, 2 M thiourea, 1% ASB-14, 40 mM
Tris-Base, and trace bromophenol blue. Samples were then sonicated
and centrifuged at 16,000 rcf for 30 min at 20°C. The supernatants
were stored at −80°C for further analyses. Protein disulfide bonds
were reduced and alkylated using the ReadyPrep™
Reduction-Alkylation kit (Bio-Rad, Hercules, CA, USA), according to
the manufacturer’s protocol. Impure protein lysates were cleaned
using the ReadyPrep™ cleanup kit (Bio-Rad), according to the
manufacturer’s protocol. Protein concentrations were measured by a
modified Lowry assay using the RC DC Protein Assay (Bio-Rad).
Two-dimensional polyacrylamide gel
electrophoresis (2D-PAGE)
Each 100 μg of protein sample to be processed
by isoelectric focusing (IEF) using PROTEAN® IEF cell
(Bio-Rad) was diluted to a final volume of 500 μl with the
rehydratation/sample buffer 1+ (RSB1 enriched with 2 mM
TBP and 0.2% Bio-Lyte® 3/10 Ampholytes). The precast IPG
strips (pH 3–10, linear pH gradient, 24-cm long), used for the
first dimension, were actively rehydrated at 50 V and loaded with
the sample at 20°C for 12 h in mineral oil. During the pause after
rehydration, paper wicks soaked in ultrapure water were placed
between each electrode and IPG strip. IEF was immediately initiated
according to the following protocol: 10,000 V for 70,000 Vh, and
then, at 500 V. Strips were then equilibrated once for 30 min with
gentle shaking in an equilibration solution containing 6 M urea, 2%
SDS, 0.375 M Tris-HCl pH 8.8, and 20% glycerol. Separation by
protein molecular mass was performed in a ProteanPlus™ Dodeca Cell
(Bio-Rad) on homogeneous 10%, 1-mm-thick polyacrylamide gels.
Briefly, equilibrated IPG strips were placed onto gels and overlaid
with ReadyPrep™ overlay agarose to remove any residual air bubbles
from between the IPG strip and gel, and to add a trace of
bromophenol blue for electrophoresis control. Second dimension
SDS-PAGE was carried out according to the following protocol: 50 V
for 5 min, 100 V for 10 min, 150 V for 15 min, 200 V for 20 min,
and 250 V until the blue dye reached the bottom of each gel.
Immediately after electrophoresis, separated protein spots were
visualized using Silver Stain Plus kit (Bio-Rad), according to the
manufacturer’s protocol.
Gel scanning and image analysis
Stained 2D gels were scanned and analyzed to compare
matching spots. Gels were scanned using a Calibrated Densitometer
(GS-800; Bio-Rad). Gel images were processed for spot detection,
background subtraction, and matching by using the Quantity One and
PDQuest software (Bio-Rad). For image analysis, the proteomic
pattern of the control healthy liver was used as a reference
pattern, and the primary non-function liver protein patterns were
matched to this reference pattern. Spots were considered to be
differentially expressed if they were either present in a different
amount or absent in comparison with the reference gel. A
quantitative difference was determined when the normalized total
volume values differed significantly (p<0.01, Student’s t-test).
The ratio of expression intensity of PNF to that of the control of
≥2 and ≤0.5 were set as thresholds indicating significant change.
Protein spots selected by 2D-PAGE were excised from the gels,
washed with 1 ml of ultrapure water, and identified by liquid
chromatography followed by mass spectrometry (LC-MS). Peptide mass
fingerprinting was performed with the MASCOT engine (Matrix
Science, UK) against NCBI’s non-redundant human genome database
(NCBInr). The criteria for protein identification were based on
probability-based MOWSE scoring algorithm with a 95% confidence
level in MASCOT.
Results
Proteome differential expression in
functional and PNF livers
Total liver proteins were extracted from PNF and
healthy samples, and separated by 2D-PAGE using IPG strips with a
linear pH gradient (pH 3–10). The proteins were resolved in
homogeneous 10% acrylamide gels in the second dimension. After
processing the 2D-PAGE gels of PNF livers and paired healthy
tissues, well-resolved gels were obtained with the silver-stained
spots both sharply focused and widely distributed along pH 3–10.
Using Quantity One and PDQuest software, about 1,300 spots per gel
were detected, and their numbers were automatically determined.
Analysis of differentially expressed
proteins
Samples processed under identical conditions can be
compared by 2D-PAGE. Therefore, tandem experiments were performed
to compare pairs of samples prepared from functional livers (FL),
those that performed adequately in subsequent follow-up assessments
after transplantations, to that of PNF livers, that failed to
regain normal synthetic function after transplantation.
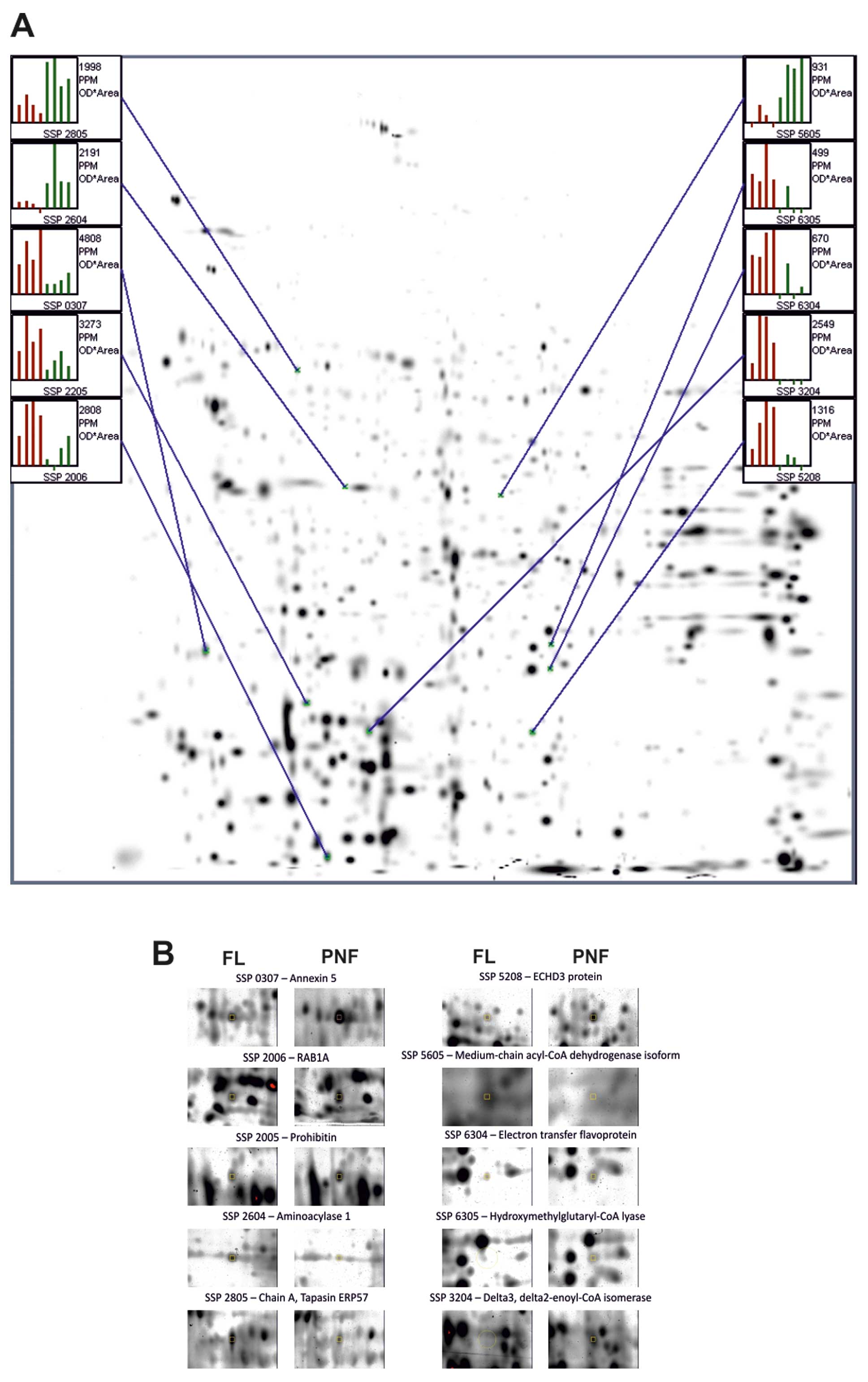
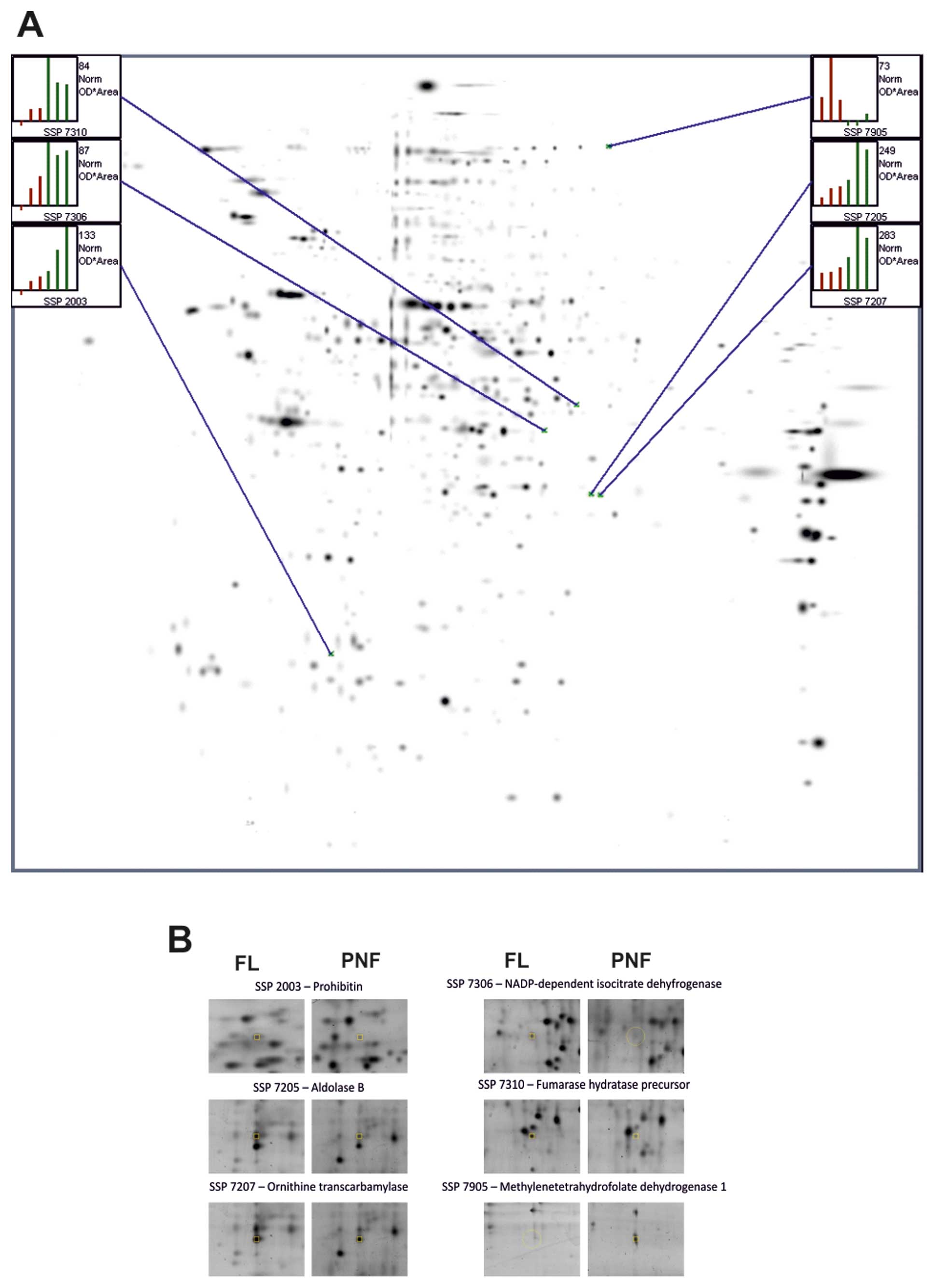
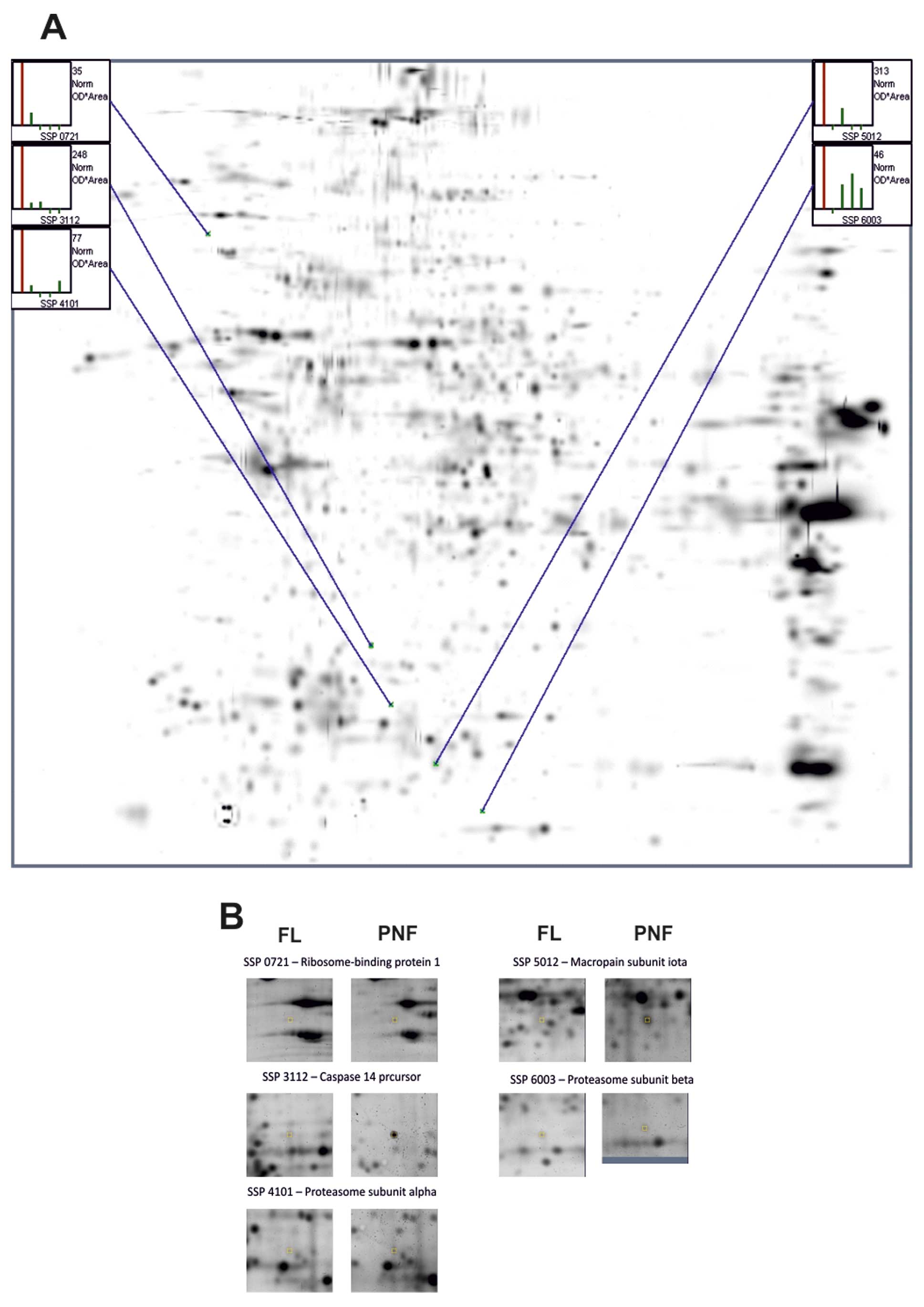
Representative 2D-PAGE maps obtained from 3
independent separations are shown in Figs. 1, 2 and 3. For reliable analysis of protein
expression, 2D gel maps were constructed and analyzed in
replicates. In the first comparison, both PNF and FL samples were
separated on 4 independent gels. The master gel, an average of 4
replicate runs of the gels, calculated and averaged in
silico, was selected as the reference gel. The Student’s t-test
showed that the volume of 10 protein spots was significantly
altered in these gels (Fig. 1,
p<0.01). In the second separation, all the samples were
separated on 2 independent gels, and the master gel was selected as
the reference gel.The Student’s t-test showed that the volume of 6
protein spots was significantly altered on these gels (Fig. 2, p<0.01). In the third
separation, one PNF sample was separated on 4 independent gels,
while 4 healthy samples were separated on 1 gel each, and the
master gel was selected as the reference gel. The student’s t-test
showed that the volume of the 5 protein spots was significantly
altered in the gels (Fig. 3,
p<0.01). Altogether, using the procedures described above, we
found 21 differential 2D-PAGE spots in the PNF livers: 13 that were
upregulated and 8 that were downregulated, compared to the control
livers.
Identification of proteins by LC-MS
All of the 21 differentially expressed protein spots
were excised from the gels and identified by liquid chromatography
followed by mass spectrometry (LC-MS). Peptide mass fingerprinting
performed with the MASCOT engine against NCBInr identified the
proteins listed in Table II. In
Table III, the 21 proteins shown,
are grouped according to function and localization.
 | Table IIMass spectrometry-based
identification of the 2D-PAGE protein spots differently expressed
in primary non-functional human livers. |
Table II
Mass spectrometry-based
identification of the 2D-PAGE protein spots differently expressed
in primary non-functional human livers.
| Protein number | Spot number | Protein name | Scorea | Theoretical pI;Mr
(kDa) | PNF vs. FL |
|---|
| 1 | 0307 | Annexin 5 | 148 | 4.94; 36 | Upregulation |
| 2 | 2006 | RAB1A, member Ras
oncogene family isoform 1 | 335 | 5.93; 23 | Upregulation |
| 3 | 2205 | Prohibitin | 362 | 5.57; 29.8 | Upregulation |
| 4 | 2604 | Aminoacylase 1 | 169 | 5.77; 46 | Downregulation |
| 5 | 2805 | Chain A, tapasin
ERP57 heterodimer | 432 | 5.61; 54.5 | Downregulation |
| 6 | 3204 | Δ3,
Δ2-enoyl-CoA isomerase | 416 | 6.39; 29.7 | Upregulation |
| 7 | 5208 | ECHDC3 protein | 129 | 8.81; 31.5 | Upregulation |
| 8 | 5605 | Medium-chain
acyl-CoA dehydrogenase isoform α precursor | 124 | 8.61; 47 | Downregulation |
| 9 | 6304 | Electron transfer
flavoprotein | 393 | 8.62; 35.4 | Upregulation |
| 10 | 6305 |
Hydroxymethylglutaryl-CoA lyase | 137 | 7.01; 32.6 | Upregulation |
| 11 | 2003 | Prohibitin | 595 | 5.57; 29.8 | Downregulation |
| 12 | 7205 | Aldolase B | 456 | 8.00; 40 | Downregulation |
| 13 | 7207 | Ornithine
transcarbamylase | 183 | 8.84; 40 | Downregulation |
| 14 | 7306 | NADP-dependent
isocitrate dehydrogenase | 146 | 6.34; 46.9 | Downregulation |
| 15 | 7310 | Fumarate hydratase
precursor | 396 | 8.85; 54.8 | Downregulation |
| 16 | 7905 |
NADP+-dependent
methylenetetrahydrofolate dehydrogenase 1 | 189 | 6.75; 102.2 | Upregulation |
| 17 | 0721 | Ribosome-binding
protein 1 | 630 | 5.45; 109 | Upregulation |
| 18 | 3112 | Caspase 14
precursor | 137 | 5.44; 28 | Upregulation |
| 19 | 4101 | Proteasome subunit
α type-3 isoform 1 | 108 | 5,19; 28.6 | Upregulation |
| 20 | 5012 | Macropain subunit
ι | 98 | 5.58; 25.1 | Upregulation |
| 21 | 6003 | Proteasome subunit
β type-2 isoform 1 | 142 | 6.51; 23 | Upregulation |
 | Table IIIList of proteins grouped according to
function and localization. |
Table III
List of proteins grouped according to
function and localization.
| Protein number | Protein name | Subcellular
localization | Function | PNF vs. FL |
|---|
| 4 | Aminoacylase 1 | Cytoplasmic | Amino acid
metabolism | Downregulation |
| 5 | Chain A, tapasin
ERP57 heterodimer | Cytoplasmic | Antigen
presentation/endoplasmic reticulum stress | Downregulation |
| 8 | Medium-chain
acyl-CoA dehydrogenase isoform α precursor | Mitochondrial | Lipid
metabolism | Downregulation |
| 11 | Prohibitin | Mitochondrial | Cell
proliferation | Downregulation |
| 12 | Aldolase B | Cytoplasmic | Energy
metabolism | Downregulation |
| 13 | Ornithine
transcarbamylase | Mitochondrial | Energy
metabolism | Downregulation |
| 14 | NADP-dependent
isocitrate dehydrogenase |
Mitochondrial/Cytoplasmic | Energy
metabolism | Downregulation |
| 15 | Fumarate hydratase
precursor |
Mitochondrial/Cytoplasmic | Cell
proliferation | Downregulation |
| 1 | Annexin 5 | Cytoplasmic | Cell apoptosis,
cell membrane scaffolds | Upregulation |
| 2 | RAB1A, member Ras
oncogene family isoform 1 | Cytoplasmic | Intracellular
vesicle traffic | Upregulation |
| 3 | Prohibitin | Mitochondrial | Cell
proliferation | Upregulation |
| 6 | Δ3,
Δ2-enoyl-CoA isomerase | Cytoplasmic | Metabolism of amino
acids | Upregulation |
| 7 | ECHDC3 protein | Mitochondrial | Energy
metabolism | Upregulation |
| 9 | Electron transfer
flavoprotein | Mitochondrial | Energy
metabolism | Upregulation |
| 10 |
Hydroxymethylglutaryl-CoA lyase | Mitochondrial | Energy
metabolism | Upregulation |
| 16 |
NADP+-dependent
methylenetetrahydrofolate dehydrogenase 1 | Mitochondrial | Energy
metabolism | Upregulation |
| 17 | Ribosome-binding
protein 1 | Cytoplasmic | Intracellular
vesicle traffic | Upregulation |
| 18 | Caspase 14
precursor | Cytoplasmic | Cell apoptosis | Upregulation |
| 19 | Proteasome subunit
α type-3 isoform 1 | Cytoplasmic | Peptide
cleavage/cell trafficking | Upregulation |
| 20 | Macropain subunit
ι | Cytoplasmic | Peptide
cleavage/cell trafficking | Upregulation |
| 21 | Proteasome subunit
β type-2 isoform 1 | Cytoplasmic | Peptide
cleavage/cell trafficking | Upregulation |
Discussion
The prevalence of PNF has remained steady over the
past 2 decades, since the syndrome has been discovered.
Nonetheless, increasing incidence of PNF has already been
documented and can be expected to continue an increasing number of
ECD donors are accepted for transplantation (7). Regular surveillance of high-risk
grafts is carefully carried out each time a donor is accepted by a
transplant center for recovery. Despite all efforts, even an
experienced retrieval team and accepted means of preventive
treatment may not protect the recipient from PNF in every case.
Often, the combination of risk factors associated with each graft
can be responsible for the problem. When evaluated independently,
their impact on PNF is not obvious, but when they coexist, the
graft dysfunction is clinically more significant. Therefore, we
believe that the results of a proteomic approach will provide
valuable novel data to evaluate grafts for PNF.
In our experience, >50% of the donors fulfill the
criteria of ECD. All patients included in the study developed graft
non-function following transplantation of grafts received from ECD.
The currently extended donor criteria used by the Department of
General, Transplant and Liver Surgery are generally consistent with
the literature. The donor was considered ECD if at least 2 of the
criteria were fulfilled. If steatosis is encountered during organ
recovery, the other variables of ECD graft are then considered.
Although liver retransplantation is the only
definitive treatment for PNF, it carries a high mortality rate
(2). However, successful
retransplantations with long-term follow-up showed that the patient
and graft survival rates after retransplantation following PNF were
not different compared to retransplantation for alternative causes
(31). In contrast, Yoo et
al showed inferior results of retransplantation for PNF
(2). Nevertheless, in spite of
high perioperative mortality, we are in favor of emergency
retransplantation. Therefore, 2 of 3 patients were listed for
reOLTx. In our group, the survival was 0%. In all cases,
multi-organ failure was considered the cause of death. The general
critical condition of the remaining 1 recipient made it impossible
to offer retransplantation as a treatment. One patient passed away
because of systemic complications, which led to multi-organ failure
before a new graft was available. One patient passed away 6 days
after liver retransplanation reOLTx.
To the best of our knowledge, this is the first
attempt to utilize a proteomic approach in the study of PNF in
order to identify novel predictive biomarkers. To do this, the
protein expression profile of the liver grafts transplanted to
recipients who developed PNF were compared to those with optimal
liver grafts, which showed normal primary function after
transplantation. Proteomics-based technologies can identify and
quantify novel proteins that can function as biomarkers of the
presence or severity of a disease state. In general, human liver
proteome profiling is quite challenging. A minority of proteins,
including albumin, immunoglobulins, transferrin, and fibrinogen,
are highly abundant, and typically constitute >90% of the total
protein mass. However, the low abundant proteins are most likely to
be biologically relevant as the markers of a disease state. In this
study, we present the liver proteome analysis of patients with PNF
and functional liver after transplantation. We identified changes
in 21 proteins (13 upregulated and 8 downregulated) when comparing
the samples from the PNF grafts to those from the grafts with
normal function after transplantation.
Tables II and
III list the proteins that were
either upregulated or downregulated. The characteristic functions
of the differentially expressed proteins can be classified into two
major categories: one category is proteins associated with
mitochondrial oxidative phosphorylation, while the other category
is proteins essential for the ATP-dependent turnover of proteins.
Among the upregulated proteins, 4 are related to energy metabolism:
ECHDC3 protein, electron transfer flavoprotein,
hydroxymethylglutaryl-CoA lyase, and NADP+-dependent
ehydrogenase 1.
Only 2 of the upregulated proteins are associated
with cell apoptosis: Annexin 5 and caspase 14 precursor. This
observation indicates that hepatocyte apoptosis is not a major
factor for PNF. One may also speculate that even a suboptimal graft
is not prone to dysfunction prior to harvesting simply on the basis
cell death pathways. It may also imply that events leading to PNF
are multifactorial and are rather metabolic in origin and thereby
potentially reversible. Indirect proof of that hypothesis is a
relative rarity of the PNF in contrast to the increasing number of
ECD.
Only one among the upregulated proteins, an electron
transfer flavoprotein, is associated with cellular protection
against oxidative stress (ischemia/reperfusion [I/R] injury).
Vascotto et al reported the upregulation of flavoproteins
when they compared the changes in liver proteome profile upon
reperfusion (32), but this is
the only reported occurrence of such a finding. This may mean that
the grafts were not subjected to a major ischemic stress, which
might impair the metabolic function after reperfusion.
The 8 proteins that were downregulated in the PNF
livers are all involved in various metabolic pathways, mostly in
energy and energy substrate metabolism. Vascotto et al also
reported an increase in the hepatic levels of aldolase B, a protein
responsible for energy metabolism, after reperfusion (32). In our data, aldolase B was
downregulated in only one case, indirectly suggesting the lack of a
major ischemic insult to the graft before donation. Considering
that tissues were collected from deceased donors, we can
hypothesize that the alterations in these protein levels are not
directly related to ischemic tissue responses.
None of the proteins appeared more than once in
separate analyses. The absence of similar proteomic patterns among
the cases studied may indicate that PNF is a heterogeneous
multifactor phenomenon, and that a graft is not deemed to primary
dysfunction on a proteomic basis. A relatively small number of
upregulated and downregulated proteins in our study may also
indicate that PNF grafts were in a stable metabolic homeostasis
before retrieval and not much different from optimal grafts.
Additional factors may have also influenced the outcome. These
factors may be linked with cold and warm ischemia time, liver
perfusion, or possibly, post-reperfusion syndrome. We may conclude
that it is not the graft itself, but other factors leading to I/R
syndrome that function as the determinants of primary dysfunction
(32). Therefore, our data may be
useful in determining the likelihood of I/R syndrome rather than
primary graft non-function. At present, there is not enough data to
support the notion that a proteomic study alone is a useful tool
for diagnosing primary PNF.
Our study has demonstrated that grafts developing
PNF demonstrate modification of the liver proteome. Considering the
complexity of the problem, which can be influenced by many external
and internal factors, finding one specific pattern of biomarkers
will be difficult. Therefore, rather than focusing on the
expression of each individual protein, we focused our attention on
the global aspects of variations in protein expression with respect
to a functional response. The majority of protein expression
changes were noted among proteins associated with energy
metabolism. The other major groups of proteins found in PNF were
proteins involved in ATP-dependent turnover of proteins and lipids,
as well as cell trafficking control. These changes were noted only
in single cases, so they may represent an alteration of their
normal involvement in metabolic pathways typical for the liver, the
site of amino acids and lipid metabolism. In our opinion, the
alteration of these protein levels was not directly related to the
ischemic tissue response or other insult to the potential
graft.
Our study has certain limitations. The definition of
PNF used in our study was provided for graft non-function occurring
within 10 days of transplantation. In addition, the study analyzed
only a small number of cases. We think that an increased sample
size could potentially alter the results. Another limitation of the
study was the difficulty involved in collecting a similar group of
samples in a controlled and repetitive manner. Quite a few
variations were beyond the control of the researchers. For example,
the grafts were recovered and implanted by different teams of
surgeons. However, the research protocol was very strict, and a
single surgeon performed the entire procedure of sample freezing
and preparation for further research. There are some variables in
the 2D-PAGE procedure that could not be overcome. For example, the
temperature at which silver staining is performed has tremendous
impact on the intensity of stained protein spots (33). Another considerable problem that
occurs during the preparation of protein samples is the lack of
protein-loading control. To address these technical problems, all
tandem analyses were performed under strictly controlled
conditions, including simultaneous staining and multiple
independent measurements of sample protein concentration.
In summary, this study identified 21 proteins
differentially expressed in a PNF graft sample compared to normal
liver grafts. The individual protein associations identified in the
proteomic study validate the technique. The data also led us to
hypothesize that a unique profile for PNF may not be possible
because of many donor-associated factors. A further explanation of
the problem is that PNF is a complex phenomenon. Therefore, in our
opinion, it would be worthwhile to search for a global aspect of a
unique proteomic profile that would be clinically relevant to the
discovery of diagnostic, prognostic, and therapeutic biomarkers to
advance the knowledge and treatment of PNF.
Abbreviations:
|
PNF
|
primary graft non-function;
|
|
LC-MS
|
liquid chromatography-mass
spectrometry;
|
|
ICU
|
intensive care unit;
|
|
UNOS
|
United Network for Organ Sharing;
|
|
HAT
|
hepatic artery thrombosis;
|
|
INR
|
international normalized ratio;
|
|
AST
|
aspartate aminotransferase;
|
|
ELTR
|
European Liver Transplant
Registry;
|
|
ECD
|
extended criteria donors;
|
|
CIT
|
cold ischemia time;
|
|
2D-PAGE
|
two-dimensional polyacrylamide gel
electrophoresis;
|
|
IEF
|
isoelectric focusing;
|
|
FL
|
functional liver;
|
|
NCBInr
|
NCBI non-redundant human genome
database;
|
|
I/R
|
ischemia/reperfusion
|
Acknowledgements
This study was supported by grants N
40306732/3612 (to M.K.) from the Polish Ministry of Science. J.G.
is supported by the European Union within European Regional
Development Fund through Innovative Economy grant
POIG.01.01.02-00-008/08. J.G. is a recipient of the Mistrz Award
from the Foundation for Polish Science. J.G. is a member of TEAM
Programme co-financed by the Foundation for Polish Science and the
EU European Regional Development Fund.
References
|
1.
|
RJ PloegAM D’AlessandroSJ KnechtleRisk
factors for primary dysfunction after liver transplantation-a
multivariate
analysisTransplantation55807813199310.1097/00007890-199304000-000248475556
|
|
2.
|
HY YooA MaheshwariPJ
ThuluvathRetransplantation of liver: primary graft nonfunction and
hepatitis C virus are associated with worse outcomeLiver
Transpl9897904200310.1053/jlts.2003.5017612942450
|
|
3.
|
T UemuraHB RandallEQ SanchezLiver
retransplantation for primary nonfunction: analysis of a 20-year
single-center experienceLiver Transpl13227233200717256780
|
|
4.
|
BW Shaw JrRD GordonS IwatsukiTE
StarzlRetransplantation of the liverSemin Liver
Dis5394401198510.1055/s-2008-1040638
|
|
5.
|
GR SilberhumerH PokornyH HetzCombination
of extended donor criteria and changes in the Model for End-Stage
Liver Disease score predict patient survival and primary
dysfunction in liver transplantation: a retrospective
analysisTransplantation83588592200710.1097/01.tp.0000255319.07499.b7
|
|
6.
|
MG AminMP WolfJA TenBrook JrExpanded
criteria donor grafts for deceased donor liver transplantation
under the MELD system: a decision analysisLiver
Transpl1014681475200410.1002/lt.20304
|
|
7.
|
N KemmerM SecicV ZachariasT KaiserGW
NeffLong-term analysis of primary nonfunction in liver transplant
recipientsTransplant
Proc3914771480200710.1016/j.transproceed.2006.11.01217580166
|
|
8.
|
AK BurroughsCA SabinK Rolles3-month and
12-month mortality after first liver transplant in adults in
Europe: predictive models for
outcomeLancet367225232200610.1016/S0140-6736(06)68033-116427491
|
|
9.
|
R AdamP McMasterJG O’GradyEvolution of
liver transplantation in Europe: report of the European Liver
Transplant RegistryLiver
Transpl912311243200310.1016/j.lts.2003.09.01814625822
|
|
10.
|
J PitreO SoubraneB DoussetHow valid is
emergency liver transplantation for acute liver necrosis in
patients with multiple-organ failure?Liver Transpl
Surg217199610.1002/lt.5000201029346621
|
|
11.
|
HR DoyleF MorelliJ McMichaelHepatic
retransplantation - an analysis of risk factors associated with
outcomeTransplantation6114991505199610.1097/00007890-199605270-000168633379
|
|
12.
|
KJ OldhaferA BornscheuerNR FrühaufRescue
hepatectomy for initial graft non-function after liver
transplantationTransplantation6710241028199910.1097/00007890-199904150-0001510221488
|
|
13.
|
SK SoJA BarteauGA PerdrizetJW
MarshSuccessful retransplantation after a 48-hour anhepatic
stateTransplant Proc251962196319938385827
|
|
14.
|
H ChenCH PengBY ShenMulti-factor analysis
of initial poor graft function after orthotopic liver
transplantationHepatobiliary Pancreat Dis
Int6141146200717374571
|
|
15.
|
JR LakeJS ShorrBJ SteffenAH ChuRD GordonRH
WiesnerDifferential effects of donor age in liver transplant
recipients infected with hepatitis B, hepatitis C and without viral
hepatitisAm J
Transplant5549557200510.1111/j.1600-6143.2005.00741.x15707410
|
|
16.
|
S FengNP GoodrichJL
Bragg-GreshamCharacteristics associated with liver graft failure:
the concept of a donor risk indexAm J
Transplant6783790200610.1111/j.1600-6143.2006.01242.x16539636
|
|
17.
|
PA ClavienM SelznerHA RüdigerA prospective
randomized study in 100 consecutive patients undergoing major liver
resection with versus without ischemic preconditioningAnn
Surg238843852200310.1097/01.sla.0000098620.27623.7d
|
|
18.
|
SM StrasbergTK HowardEP MolmentiM
HertlSelecting the donor liver: risk factors for poor function
after orthotopic liver
transplantationHepatology20829838199410.1002/hep.18402004107927223
|
|
19.
|
E TotsukaF DodsonA UrakamiInfluence of
high donor serum sodium levels on early postoperative graft
function in human liver transplantation: effect of correction of
donor hypernatremiaLiver Transpl
Surg5421428199910.1002/lt.500050510
|
|
20.
|
H YersizA ShakedK OlthoffCorrelation
between donor age and the pattern of liver graft recovery after
transplantationTransplantation60790794199510.1097/00007890-199510270-000057482736
|
|
21.
|
AM CameronRM GhobrialH YersizOptimal
utilization of donor grafts with extended criteria: a single-center
experience in over 1000 liver transplantsAnn
Surg243748755200610.1097/01.sla.0000219669.84192.b316772778
|
|
22.
|
SR JohnsonS AlexopoulosM CurryDW
HantoPrimary nonfunction (PNF) in the MELD era: An SRTR database
analysisAm J
Transplant710031009200710.1111/j.1600-6143.2006.01702.x17286618
|
|
23.
|
J BusquetsX XiolJ FiguerasThe impact of
donor age on liver transplantation: influence of donor age on early
liver function and on subsequent patient and graft
survivalTransplantation7117651771200110.1097/00007890-200106270-0001111455256
|
|
24.
|
WK WashburnLB JohnsonWD LewisRL
JenkinsGraft function and outcome of older (> or = 60 years)
donor liversTransplantation61106210661996
|
|
25.
|
H PokornyT GruenbergerT SolimanS
RockenschaubF LängleR SteiningerOrgan survival after primary
dysfunction of liver grafts in clinical orthotopic liver
transplantationTranspl Int13Suppl
1S154S157200010.1007/s00147005031011111986
|
|
26.
|
B MüllhauptD DimitroulisJT GerlachPA
ClavienHot topics in liver transplantation: organ allocation -
extended criteria donor - living donor liver transplantationJ
Hepatol48Suppl 1S58S67200818308415
|
|
27.
|
HY ChungSC ChanCM LoST FanStrategies for
widening liver donor poolAsian J
Surg336369201010.1016/S1015-9584(10)60011-521029941
|
|
28.
|
RW BusuttilK TanakaThe utility of marginal
donors in liver transplantationLiver
Transpl9651663200310.1053/jlts.2003.5010512827549
|
|
29.
|
M GastacaExtended criteria donors in liver
transplantation: adapting donor quality and recipientTransplant
Proc41975979200910.1016/j.transproceed.2009.02.01619376402
|
|
30.
|
K HatsugaiN OhkohchiT FukumoriY AkamatsuS
SatomiMechanism of primary graft non-function in a rat model for
fatty liver transplantationTranspl Int13Suppl
1S583S590200010.1007/s00147005040811112079
|
|
31.
|
D AzoulayMM LinharesE HuguetDecision for
retransplantation of the liver: an experience- and cost-based
analysisAnn
Surg236713721200210.1097/00000658-200212000-0000312454509
|
|
32.
|
C VascottoL CesarattoC D’AmbrosioProteomic
analysis of liver tissues subjected to early ischemia/reperfusion
injury during human orthotopic liver
transplantationProteomics634553465200610.1002/pmic.200500770
|
|
33.
|
M ChevalletS LucheT RabilloudSilver
staining of proteins in polyacrylamide gelsNat
Protoc118521858200610.1038/nprot.2006.28817487168
|