Introduction
Dysregulation of hematopoietic cellular
differentiation contributes to leukemogenesis (1). The use of differentiation agents can
force malignant cells to undergo terminal differentiation, which is
viewed as a promising and revolutionary approach for the therapy of
leukemia diseases (2). The drug
12-O-Tetradecanoylphorbol-13-acetate (TPA) is a phase I clinical
therapeutic agent for patients with relapsed/refractory myelocytic
leukemia due to its capacity to induce differentiation or apoptosis
in malignant cells (3). Further
molecular and genetic analyses of the effects of TPA could promote
a better understanding of its mechanisms contributing to
differentiation and therapeutic response.
A new class of small non-coding RNA molecules known
as microRNAs (miRNAs) has introduced a whole new layer of gene
regulation in eukaryotes (4). The
miRNAs are transcribed by RNA polymerase II as large primary
non-coding transcripts or from regions of a known gene, and the
transcribed primary miRNAs are processed through the concerted
actions of biogenesis machineries, including their sequential
cleavage, export, and incorporation into the RNA-induced silencing
complex (RISC) to mediate the expression of target genes (5,6).
miRNA expression profiling in TPA-treated leukemia cells has
previously been performed in several studies (7–9).
However, considering the number of miRNAs that exist in cells and
the various types of myeloid leukemia, additional
differentiation-related miRNAs need to be identified (10). Moreover, miRNA expression
profiling should be conducted and compared among various leukemia
cell lines to identify the specifically induced miRNAs that respond
to TPA treatment.
The differentiation of leukemia cells induced by TPA
is highly dependent on the involvement of multiple signaling
pathways, including the mitogen-activated protein kinase (MAPK)
pathways, involving MEK/ERK/MAP kinase and c-Jun NH2-terminal
kinase, as well as MAPK-independent pathways such as the PI3K
pathway (11). These signaling
pathways are responsible for appropriately mediating gene
transcription with respect to cellular behavior (12). To better understand the mechanism
of TPA action, it is useful to investigate which pathways are
activated and how they mediate the induction of miRNAs in response
to TPA treatment in leukemia cells.
Here, we present the results of a microarray-based
screen for miRNAs that respond to TPA treatment in various leukemia
cell lines. We identified a series of specific
differentiation-induced miRNAs and analyzed their responses to
signal transduction by using pharmacological inhibitors, showing
the essential role of MEK/ERK signaling in miRNA induction.
Moreover, the regulation of both miRNA biogenesis machineries and
primary transcripts was analyzed in the same context, revealing the
major mechanisms for the induction of miRNA products.
Materials and methods
Cell culture and reagents
The NB4, HL-60, K562 and U937 cell lines were
obtained from the American Type Culture Collection (ATCC, Richmond,
MD, USA) and cultured in RPMI-1640 medium (Gibco, Grand Island, NY,
USA) supplemented with 10% fetal bovine serum (PAA Laboratories Pty
Ltd, Morningside, Australia), 2 mM L-glutamine and antibiotics in a
humidified incubator containing 5% CO2 at 37°C. TPA,
U0126 and LY294002 were purchased from Sigma (St. Louis, MO, USA)
and dissolved in dimethylsulfoxide (DMSO) according to the
supplier’s instructions. To induce cell differentiation, TPA was
added to the medium to a final concentration of 30 nM (13). To inhibit signal transduction,
cells were pretreated with specific inhibitors for 30 min prior to
TPA treatment (14,15). Antibodies against phospho-ERK1/2,
ERK1/2, Dicer, Drosha, Ago1, and Ago2 were purchased from Cell
Signaling Technology (Danvers, MA, USA). Antibodies against
phospho-Akt, Akt and β-actin were obtained from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA). The horseradish
peroxidase (HRP)-conjugated secondary antibodies were also obtained
from Santa Cruz Biotechnology, Inc.
Assessment of surface antigen
expression
Surface antigen expression was measured by flow
cytometry according to our previously published protocol (13). Briefly, cells were harvested at
the indicated times, washed twice with PBS, and then incubated with
20 μl FITC-labeled anti-CD14 antibody working solution
(eBioscience, San Diego, CA, USA) for 30 min at 4°C in the dark.
For every sample, separate aliquots of cells were also incubated
with isotype antibody controls (eBioscience) to determine
non-specific staining. Following incubation, cells were washed
twice with PBS and analyzed by flow cytometry with a 488 nm argon
laser. For each sample, a total of 10,000 cells were analyzed.
Assessment of cell cycle
Cell cycle was profiled by flow cytometry as
previously described (14).
Briefly, cells were harvested and fixed in 70% ethanol at 4°C
overnight. After washing with PBS, the fixed cells were
re-suspended in PBS containing 100 μg/ml RNase A and incubated for
30 min at 37°C. Finally, the cells were collected by centrifugation
and incubated in PBS containing 50 μg/ml of propidium iodide (PI)
(Sigma) for 20 min in the dark, and then analyzed using flow
cytometry with a 488 nm argon laser. A minimum of 20,000 cells was
analyzed for each sample.
miRNA microarray analysis
For miRNA microarray analysis, total RNA was
extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer’s instructions. CapitalBio Mammalian
miRNA Array V3.0 (CapitalBio, Beijing, China) microarrays were then
probed, which include 924 probes for human, mouse and rat miRNAs.
The microarray analyses were performed according to our previously
published protocols (13,16). Significance Analysis of
Microarrays (SAM; Stanford University, CA, USA) software was used
to determine the differentially expressed miRNAs, and the selection
criteria included a fold change ≥1.5 or ≤0.65, a q-value ≤5%, and a
SAM score >2 or <−2 (17).
The entire dataset described here is available from the Gene
Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) through accession
number GSE33537.
Quantitative real-time PCR analysis
To quantify the mature miRNAs, stem-loop RT-PCR
assays were performed according to the method of Chen et al
(18); snRNA U6 was used as an
internal standard. To quantify the pri-miRNAs, total RNA was
subjected to DNase I treatment and reverse-transcribed into cDNA
with M-MLV (Promega, Wisconsin, WI, USA) (19). Following reverse transcription,
PCR reactions were performed using a SuperGreen quantitative PCR
kit II (CapitalBio) according to the manufacturer’s instructions.
ACTB was used as an internal standard. Relative expression levels
were calculated using the formula: Q.rel. = 2−ΔCT, where
ΔCT = average CT test gene - average CT internal standard, and CT
is the cycle threshold. All primers used for quantitative RT-PCR
are listed in Table I.
 | Table IPCR primers designed for the
amplification of the investigated microRNAs and primary
transcripts. |
Table I
PCR primers designed for the
amplification of the investigated microRNAs and primary
transcripts.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| hsa-miR-21 |
GCCGCTAGCTTATCAGACTGATGT | GTGCAGGGTCCGAGGT |
| hsa-miR-22 |
CGAAGCTGCCAGTTGAAGAA | GTGCAGGGTCCGAGGT |
| hsa-miR-23a |
GATATCACATTGCCAGGGATT | GTGCAGGGTCCGAGGT |
| hsa-miR-24 |
GGTGGCTCAGTTCAGCAGGA | GTGCAGGGTCCGAGGT |
| hsa-miR-23b |
GCATCACATTGCCAGGGATT | GTGCAGGGTCCGAGGT |
| hsa-miR-29a |
CGTAGCACCATCTGAAATCGG |
GTGCAGGGTCCGAGGT |
| hsa-miR-29b |
GCATAGCACCATTTGAAATAGT |
GTGCAGGGTCCGAGGT |
| hsa-miR-146b |
CGGCTGAGAACTGAATTCCATAG |
GTGCAGGGTCCGAGGT |
| hsa-miR-638 |
AGGGATCGCGGGCGG |
GTGCAGGGTCCGAGGT |
| SnRNA U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
| Pri-miR-21 |
TTTTGTTTTGCTTGGGAGGA |
AGCAGACAGTCAGGCAGGAT |
| Pri-miR-22 |
GCAGAAAGCCTTGGGTTG |
CGAACAGCAGGGTGGATGA |
| Pri-23a-24-2 |
TCACCCCTGTGCCACG |
CAAACCAACTGTGTTTCAGCT |
| Pri-miR-146b |
GAGCAGCGTCCAGGCTG |
CCGGGCACCAGAACTGAGT |
| ACTB |
CATGTACGTTGCTATCCAGGC |
CTCCTTAATGTCACGCACGAT |
Immunoblotting analysis
The cells were quickly lysed on ice using lysis
buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 10%
glycerol, 2 mM EDTA, 1 mM Na3VO4, and
protease inhibitors) and clarified by centrifugation at 14,000 rpm
for 10 min; the supernatants represented the cellular protein
extracts. Equal quantities of protein extracts were resolved by
SDS-PAGE and transferred to nitrocellulose membranes. The membranes
were blocked in TBST (10 mM Tris pH 7.6, 150 mM NaCl, and 1%
Tween-20) containing 5% low-fat milk for 1 h. Subsequently, the
membranes were incubated with primary antibody dilutions at 4°C
overnight, followed by three washes with TBST at room temperature.
The membranes were then incubated with secondary antibody dilutions
for 1 h at room temperature, followed by four washes with TBST. The
enhanced chemiluminescence (ECL) system (Pierce, Rockford, IL, USA)
was used to detect reactive proteins.
Statistical analysis
Experiments were performed in duplicate or
triplicate and independently repeated at least three times. Data
are presented as the means ± standard deviation (SD). Statistical
significance was determined using two-tailed t-tests, and P<0.05
was considered to indicate statistically significant
differences.
Results
Analysis of the miRNA expression pattern
of myeloid leukemia cell lines following TPA treatment
NB4 (acute promyelocytic leukemia), HL-60 (acute
myeloblastic leukemia), U937 (monoblastic leukemia), and K562
(chronic myelogenous leukemia) cell lines are the typical models
for studies of human leukemia cell differentiation in vitro.
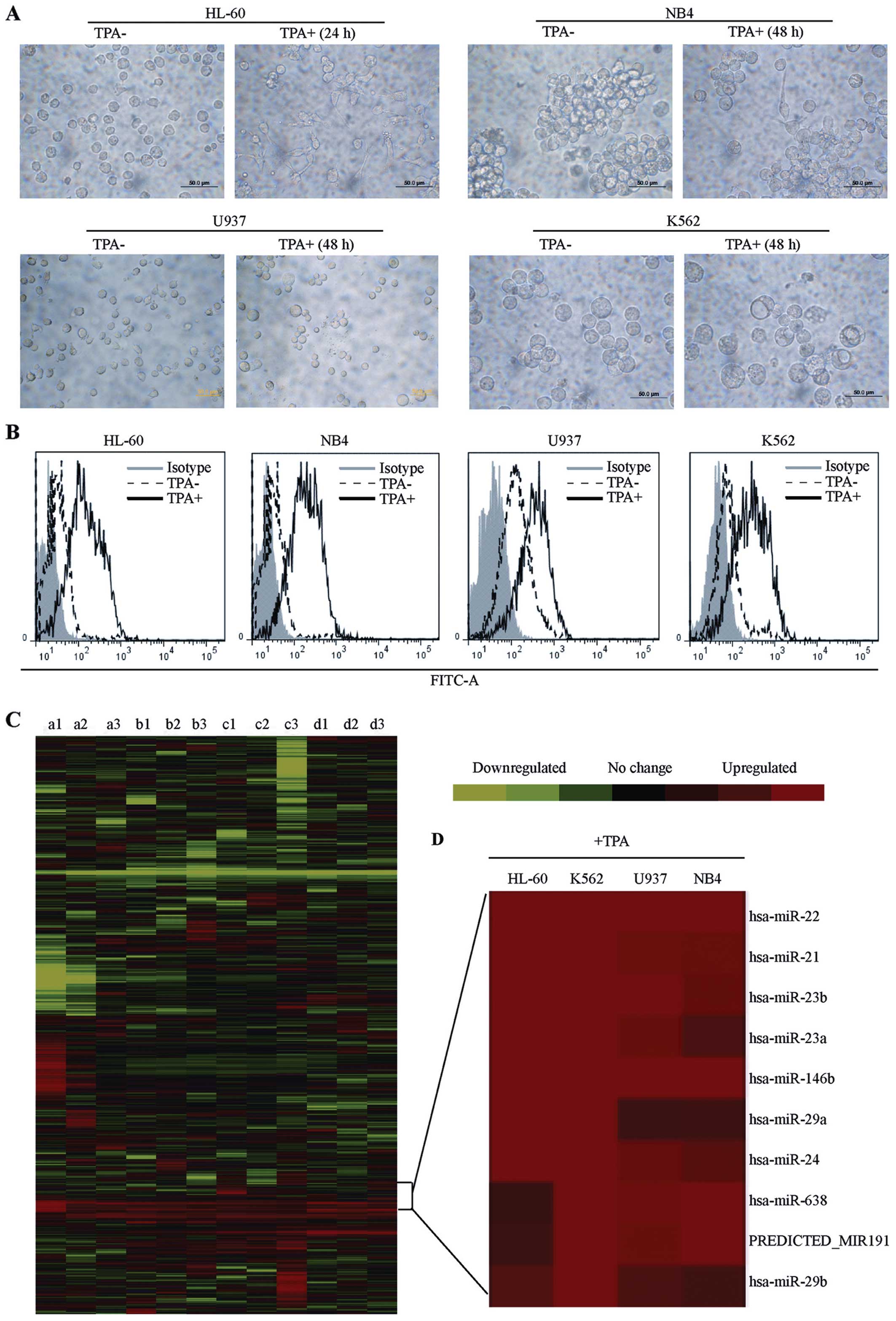
The differentiation of these cells induced by TPA was determined by
assessing cell morphology and the expression of a differentiation
marker. After TPA treatment, the cells spread and adhered to the
culture dishes, and some cells displayed pseudopod-like protrusions
(Fig. 1A). A significant increase
in CD14 expression was also observed in leukemia cells treated with
TPA (Fig. 1B). These results
suggest that cellular differentiation was induced.
Using microarrays, the global changes in miRNA
expression were analyzed after treatment of the myeloid leukemia
cell lines with TPA. The microarray chips contained 924 probes,
allowing a survey of 802 mature human, mouse and rat miRNAs after
discarding redundant sequences, and a further 122 predicted miRNAs
from published data. To reduce individual variability, replicate
array analysis used independently treated samples, and a technical
replicate was also performed for each sample.
Employing hierarchical cluster analysis, the global
expression patterns of miRNAs in the four leukemia cell lines were
obtained (Fig. 1C). Through SAM
statistics (17), significantly
regulated miRNAs in differentiated cells were identified. TPA
induction resulted in the upregulation of 21 miRNAs in the NB4 cell
line, 17 miRNAs in the HL-60 cell line, 15 miRNAs in the K562 cell
line, and 35 miRNAs in the U937 cell line (Table II). To identify specific
differentiation-induced miRNAs, the overlaps of upregulated miRNAs
were evaluated between these four myeloid leukemia lines (20). The cell lines each exhibited
characteristic miRNA profiling due to their different origins.
However, 10 unique miRNAs were consistently induced in all four
leukemia cell lines after exposure to TPA (Fig. 1D). We support that the
differentially regulated miRNAs may represent individual
characteristics of each cell line, while the commonly regulated
miRNAs may be ‘key players’ in the differentiation of leukemia
cells.
 | Table IISignificantly upregulated miRNAs
obtained by microarray analysis. |
Table II
Significantly upregulated miRNAs
obtained by microarray analysis.
| Cell lines | miRNAs (fold
changesa ≥1.5-folds;
q-valueb ≤0.05) |
|---|
| HL-60 | hsa-miR-21,
hsa-miR-22, hsa-miR-146a, hsa-miR-27a, hsa-miR-222, hsa-miR-27b,
hsa-miR-24, hsa-miR-146b-5p, hsa-miR-23b, hsa-miR-29a, hsa-miR-23a,
hsa-miR-221, hsa-miR-509-3p, hsa-miR-17, hsa-miR-29b,
PREDICTED_MIR191, hsa-miR-638 |
| NB4 | hsa-miR-146a,
hsa-miR-222, hsa-miR-22, hsa-miR-23b, hsa-miR-146b-5p, hsa-miR-221,
hsa-miR-638, hsa-miR-21, PREDICTED_MIR191, hsa-miR-23a, hsa-miR-24,
hsa-miR-27b, hsa-miR-663, hsa-miR-509-3p, hsa-miR-155, hsa-miR-124,
mmu-miR-193b, hsa-miR-193a-3p, hsa-miR-29b, hsa-let-7g,
hsa-miR-29a |
| U937 | hsa-miR-638,
hsa-miR-486-3p, hsa-miR-508-5p, PREDICTED_MIR191, hsa-miR-663,
hsa-miR-22, hsa-miR-584, hsa-miR-146b-5p, hsa-miR-487b,
hsa-miR-888, hsa-miR-21, hsa-miR-23b, hsa-miR-146a, hsa-miR-27b,
PREDICTED_MIR255, hsa-miR-222, hsa-miR-381, hsa-miR-24,
hsa-miR-658, hsa-miR-221, hsa-miR-509-3p, hsa-miR-27a, hsa-miR-424,
hsa-miR-23a, hsa-miR-28-5p, hsa-miR-193a-3p, hsa-miR-29b,
hsa-miR-218, hsa-miR-410, hsa-miR-216a, hsa-miR-29a, hsa-miR-26a,
PREDICTED_MIR160, hsa-miR-192, hsa-miR-125b |
| K562 | PREDICTED_MIR191,
hsa-miR-638, hsa-miR-663, mmu-miR-762, hsa-miR-22, hsa-miR-21,
hsa-miR-146b-5p, hsa-miR-24, hsa-miR-29b, hsa-miR-23b, hsa-miR-27a,
hsa-miR-29a, hsa-miR-23a, hsa-miR-17, hsa-miR-16 |
Validation of differentiation-specific
miRNAs by quantitative RT-PCR
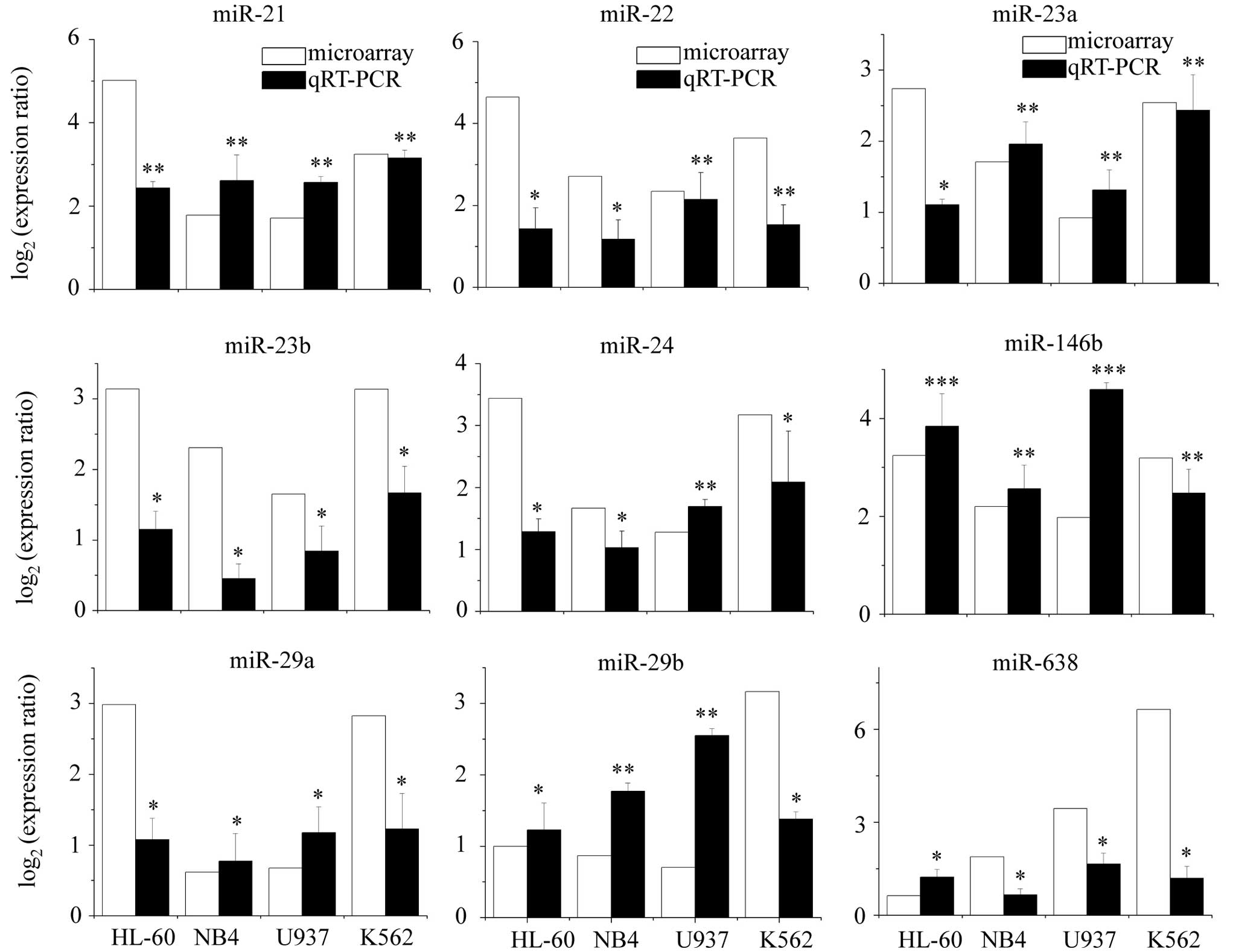
To confirm the accuracy of our microarray analysis,
stem-loop RT-PCR assays were performed on the identified
differentiation-specific miRNAs (except predicted-miR191) using
independent samples. The results of qRT-PCR were consistent with
those of microarray analysis in all four cell lines (Fig. 2). These nine miRNAs were confirmed
to be differentiation-specific in leukemia cells induced by TPA, of
which a subset (miR-21, miR-22, miR-146b, miR-23a and miR-24) was
selected for further investigation largely based on the magnitude
of the detected induction.
Pharmacological inhibition of MEK/EKR
activation suppresses the upregulation of differentiation-specific
miRNAs
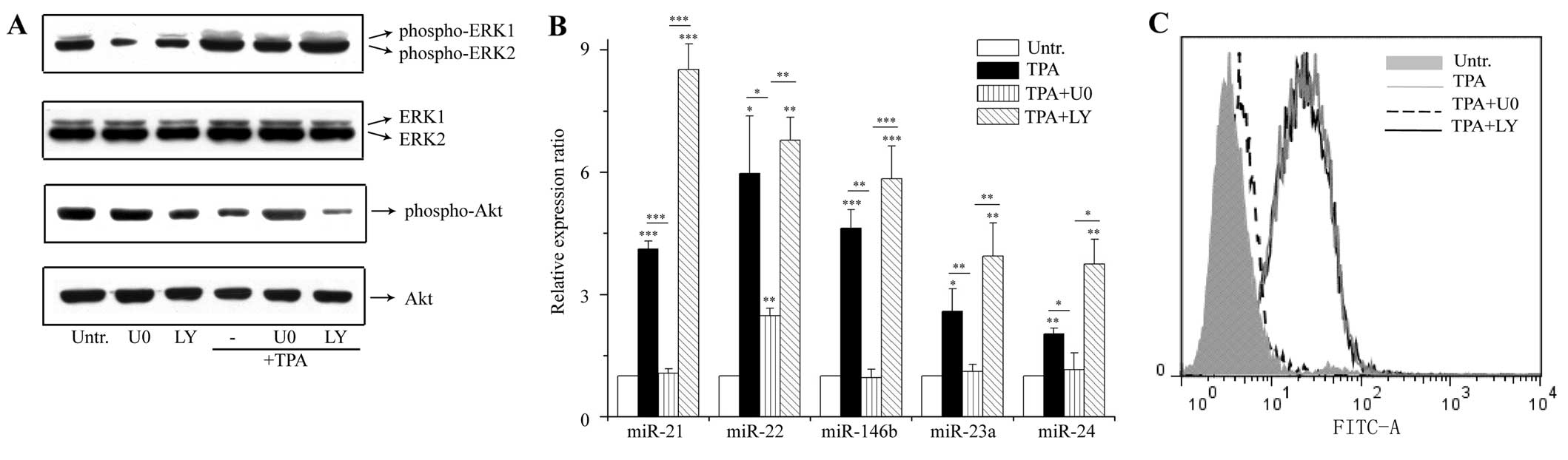
To ascertain how the expression of these
differentiation-specific miRNAs was induced, cell signaling
analysis was performed. Both the MEK/ERK and PI3K/Akt pathways are
associated with the induction of differentiation by TPA in leukemia
cells (11). To investigate the
effects of these pathways on miRNA induction, MEK/ERK and PI3K/Akt
signal transduction was blocked using pharmacological inhibitors:
U0126 (MEK1/2 inhibitor) and LY294002 (PI3K inhibitor). U0126
blocked the TPA-stimulated phosphorylation of ERK1/2 (Fig. 3A), and inhibited the induction of
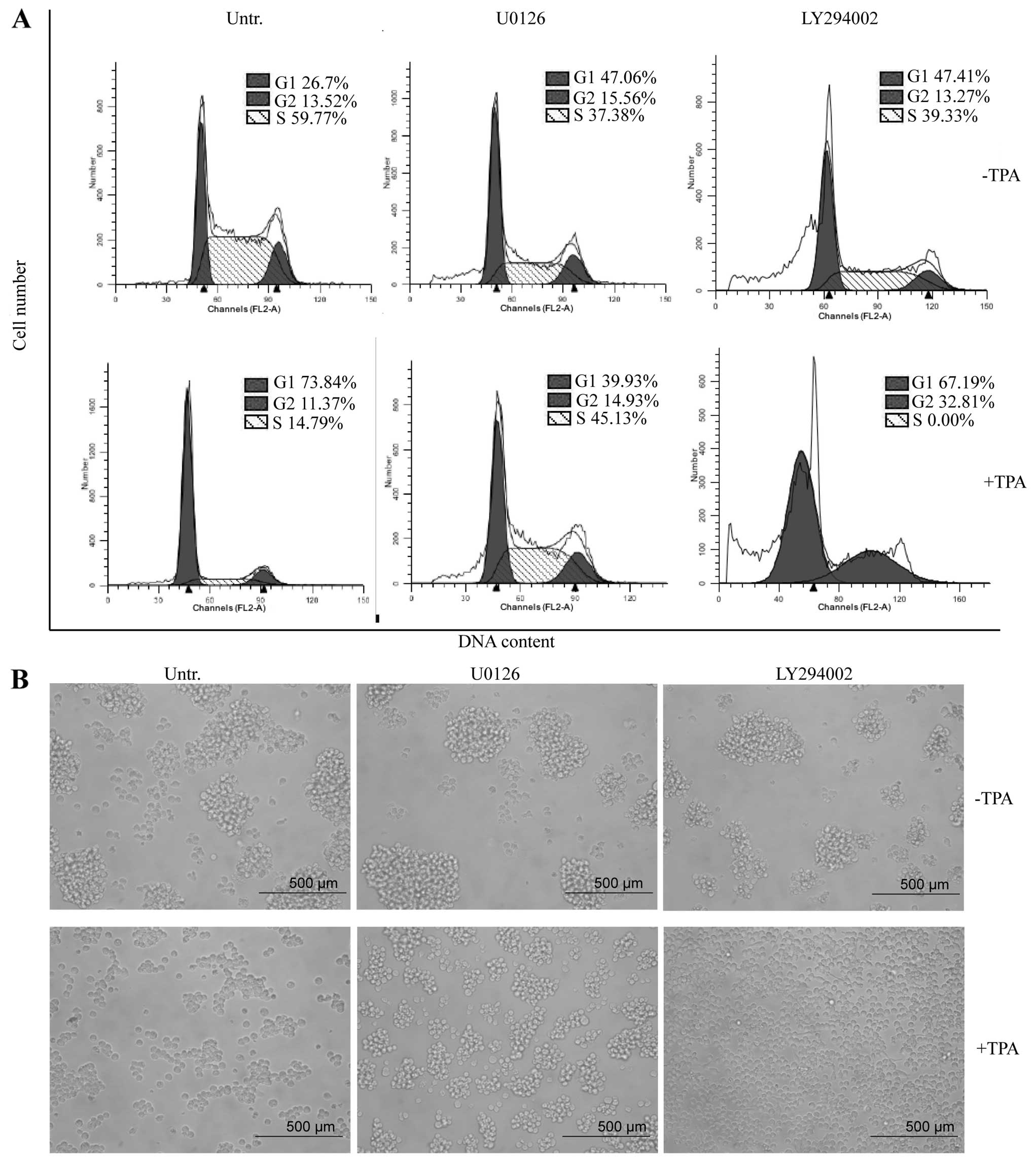
miR-21, miR-22, miR-146b, miR-23a and miR-24 (Fig. 3B). The changes in the expression
of the differentiation marker, growth arrest, and cell morphology
were also inhibited by pretreatment with U0126 (Figs. 3C and 4). By contrast, LY294002 failed to
suppress the TPA-induced miRNA expression and cellular
differentiation (Figs. 3B, C and
4). The reduction of Akt
phosphorylation proved the inhibitory effect of LY294002 on
PI3K/Akt signaling (Fig. 3A).
Thus, MEK/ERK activation contributed to the induction of these
differentiation-specific miRNAs.
MEK/ERK activation triggers the
transcription of differentiation-specific miRNAs
To investigate the mechanism by which MEK/ERK
activation caused the upregulation of differentiation-specific
miRNAs, the expression of both miRNA biogenesis machineries and
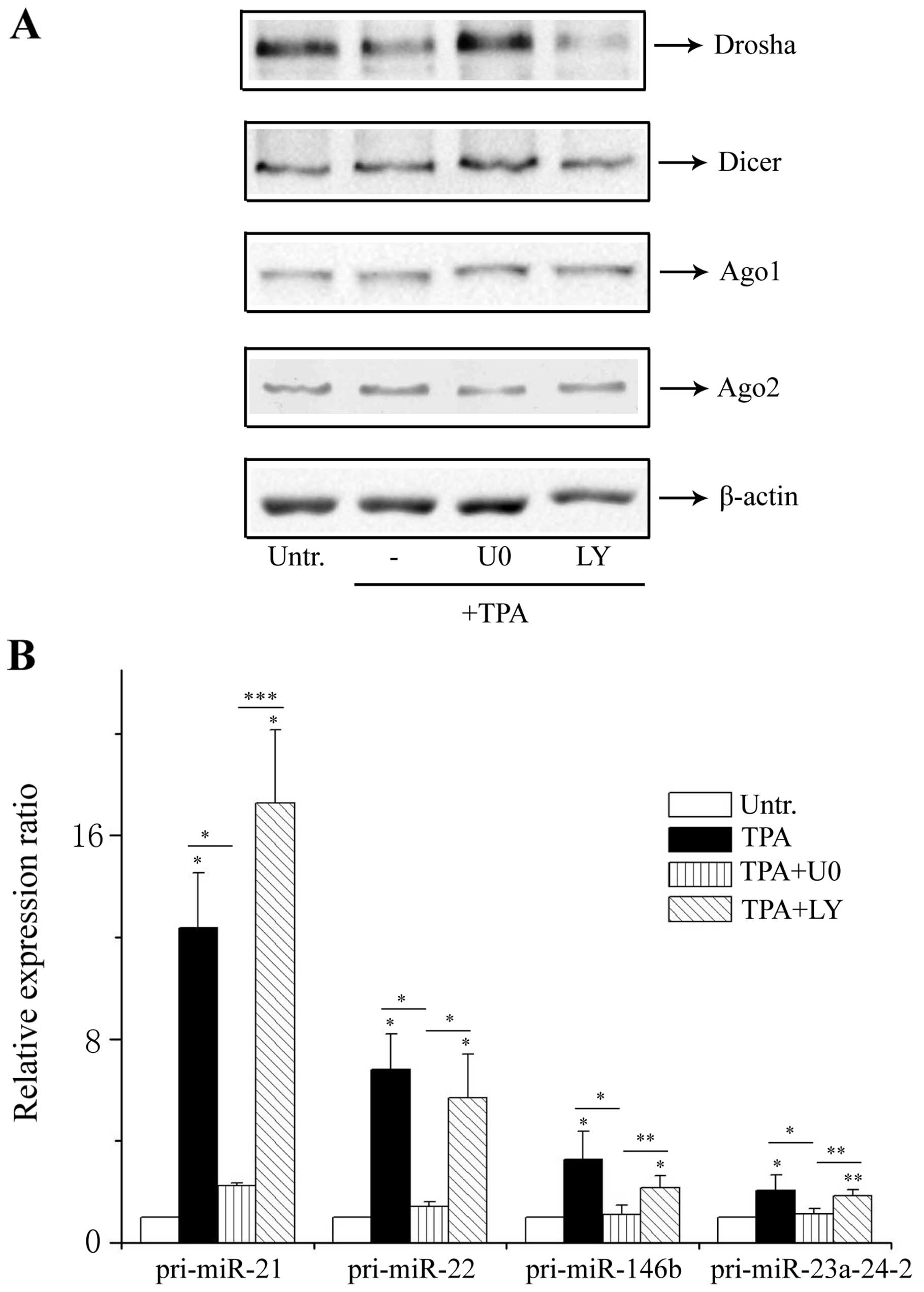
primary transcripts was examined. Using immunoblotting analysis,
four proteins that are the key machineries of miRNA biogenesis
pathways (Drosha, Dicer, Ago1 and Ago2) were investigated. Drosha
and Dicer are involved in successively cleaving primary transcripts
into mature miRNAs; Ago1 and Ago2 are the major components of RISC
(5,6). The expression of Dicer, Ago1 and
Ago2 was not significantly altered in differentiated cells. Only
Drosha was downregulated in cells after TPA treatment, and U0126
inhibited the downregulation of Drosha caused by TPA (Fig. 5A). These results suggest that the
expression levels of the key miRNA biogenesis machineries in
differentiated cells were not responsible for the increased
expression of the aforementioned miRNAs induced by TPA.
Next, we assessed if the MEK/ERK activation induced
the upregulation of the differentiation-specific miRNAs at the
transcriptional level. qRT-PCR was performed to examine the primary
transcripts of miR-21, miR-22, miR-146b, miR-23a and miR-24. Since
there was a coordinated effect of processing at the miR-23a and
miR-24 loci (21), a pair of
specific primers was designed to quantify the expression levels of
the pri-23a-24-2 transcript loci. The basal levels of these miRNA
primary transcripts were increased after TPA induction, and the
induction of these transcripts was significantly inhibited by U0126
but not by LY294002 (Fig. 5B).
Therefore, the transcription of these differentiation-specific
miRNAs was induced by MEK/ERK activation, suggesting a major
mechanism for the upregulation of these miRNAs in the
differentiation of leukemia cells induced by TPA.
Discussion
This study included: i) miRNA expression (induced by
TPA) profiling in four distinct leukemia cell lines; ii)
confirmation of the differentiation-specific miRNAs in this
process; and iii) investigation of the association of signaling
pathways with the induction of differentiation-specific miRNAs,
revealing the essential role of MEK/ERK signaling in regulating
miRNA transcription in response to TPA treatment.
Since miRNAs are expressed in a tissue-specific and
context-dependent manner, the large-scale profiling of miRNAs using
microarrays has aided drug research and disease diagnosis (22). Previous studies on miRNA profiling
of TPA-treated leukemia cells demonstrated heterogeneous results
due to the differences in platform, drug dose, and cell types
(7–9). In our study, miRNA expression
profiling was analyzed in four distinct leukemia cells using the
same experimental platform with the same concentration of drug,
which is a better approach for further data mining. Through our
comparison, 10 commonly upregulated miRNAs were identified in the
four cell lines, representing the differentiation-specific miRNAs.
Among them, miR-146b and miR-29a were previously reported to be
upregulated in TPA-induced HL-60 cells, and they were also
identified in K562 and NB4 cells by our analysis. In addition,
miR-23b and miR-24 are induced in K562 cells treated with TPA, and
these two miRNAs were also upregulated in TPA-induced U937, NB4 and
HL-60 cells by our analysis. Such specifically induced genes that
respond to drug treatment are largely regarded as potentially major
targets of drug action (20).
Therefore, the differentiation-specific miRNAs identified in this
study provide significant insights into the thorough understanding
of the mechanism of TPA action in leukemia cells.
Signal transduction is an important mechanism for
gene regulation in cells (12),
and a large number of miRNAs are under the control of various
important signal pathways (23).
Using specific signal transduction inhibitors, we demonstrated that
MEK/ERK activation contributes to the induction of several
differentiation-specific miRNAs (miR-21, miR-22, miR-23a, miR-146b
and miR-24). These miRNAs target several genes related to
differentiation, and most of them function as tumor suppressors
(24–26). Their expression patterns
correspond to cellular behaviors, i.e., induction in differentiated
cells and inhibition in undifferentiated cells. Moreover, these
MEK/ERK signaling-induced miRNAs (such as miR-21 and miR-24) can in
turn regulate the MEK/ERK signaling pathway by targeting the
components of this pathway or other related pathways, forming a
complex regulatory network in TPA-induced leukemia differentiation
(27,28). These differentiation-specific
miRNAs are an important molecular link between MEK/ERK signal
transduction and TPA-induced differentiation.
The common upregulation of differentiation-specific
miRNAs via MEK/ERK activation may indicate a uniform regulatory
program. To explore this possibility, we examined both miRNA
biogenesis machineries and primary transcripts. The expression
changes of miRNA biogenesis enzymes can affect the miRNA expression
in some cases (29–31). We found that the expression levels
of several key miRNA biogenesis machineries were not increased in
TPA-induced NB4 cells, and the same trend was also observed in
HL-60 and K562 cells (data not shown). Based on this, we
hypothesized that MEK/ERK activation may work on the
transcriptional level for miRNA production. Indeed, we found that
MEK/ERK activation contributed to the induction of the primary
transcripts of the differentiation-specific miRNAs by qRT-PCR.
Among these transcripts, the upregulation of miR-21 and miR-24 is
also observed in other biological processes induced by MEK/ERK
activation (32,33). In addition, the promoter regions
of these induced miRNAs also contain potential binding sites for
the transcription factors RUNX1, NF-κB and RREB-1 (34,35), all of which are downstream
effectors of Raf/MEK/ERK signaling. These previous findings and our
data indicate a causal effect of the MEK/EKR signaling pathway on
the induction of the investigated miRNAs, and further studies will
be conducted to confirm the direct targets of the MEK/ERK pathways
that contribute to the induction of the differentiation-specific
miRNAs.
In conclusion, elucidating the modulation of miRNA
expression related to signal transduction advanced our
understanding of an intracellular signaling network. Moreover, with
increasing clinical administration of differentiation therapy in
leukemia patients, the miRNA expression signature reported in this
study may facilitate the development of differentiation therapeutic
strategies and ultimately be predictive of response to therapy.
Acknowledgements
We thank Yonggui Wang and Xiaoyu Zhang for their
microarray technical assistance, Yu Liu for microarray data
analysis, and Chao Wang for flow cytometric analysis. We thank Dr
JunWei Guo for the helpful discussion and comments on the
manuscript. We also thank Dr M. Bochman for checking the
manuscript.
References
|
1
|
Kluiver J, Kroesen BJ, Poppema S and van
den Berg A: The role of microRNAs in normal hematopoiesis and
hematopoietic malignancies. Leukemia. 20:1931–1936. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nowak D, Stewart D and Koeffler HP:
Differentiation therapy of leukemia: 3 decades of development.
Blood. 113:3655–3665. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schaar D, Goodell L, Aisner J, et al: A
phase I clinical trial of 12- O-tetradecanoylphorbol-13-acetate for
patients with relapsed/refractory malignancies. Cancer Chemother
Pharmacol. 57:789–795. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chang SH and Hla T: Gene regulation by RNA
binding proteins and microRNAs in angiogenesis. Trends Mol Med.
17:650–658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Davis-Dusenbery BN and Hata A: Mechanisms
of control of microRNA biogenesis. J Biochem. 148:381–392.
2010.
|
|
7
|
Kasashima K, Nakamura Y and Kozu T:
Altered expression profiles of microRNAs during TPA-induced
differentiation of HL-60 cells. Biochem Biophys Res Commun.
322:403–410. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lal A, Pan Y, Navarro F, et al:
miR-24-mediated downregulation of H2AX suppresses DNA repair in
terminally differentiated blood cells. Nat Struct Mol Biol.
16:492–498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Forrest AR, Kanamori-Katayama M, Tomaru Y,
et al: Induction of microRNAs, mir-155, mir-222, mir-424 and
mir-503, promotes monocytic differentiation through combinatorial
regulation. Leukemia. 24:460–466. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang XS and Zhang JW: The microRNAs
involved in human myeloid differentiation and
myelogenous/myeloblastic leukemia. J Cell Mol Med. 12:1445–1455.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matsumoto E, Hatanaka M, Bohgaki M and
Maeda S: PKC pathway and ERK/MAPK pathway are required for
induction of cyclin D1 and p21Waf1 during 12-o-tetradecanoylphorbol
13-acetate-induced differentiation of myeloleukemia cells. Kobe J
Med Sci. 52:181–194. 2006.PubMed/NCBI
|
|
12
|
Schaar DG, Liu H, Sharma S, et al:
12-O-tetradecanoylphorbol-13-acetate (TPA)-induced dual-specificity
phosphatase expression and AML cell survival. Leuk Res.
29:1171–1179. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang J, Xiang G, Mitchelson K and Zhou Y:
Microarray profiling of monocytic differentiation reveals
miRNA-mRNA intrinsic correlation. J Cell Biochem. 112:2443–2453.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim S, Lee HS, Lee SK, et al:
12-O-Tetradecanoyl phorbol-13-acetate (TPA)-induced growth arrest
is increased by silibinin by the downregulation of cyclin B1 and
cdc2 and the upregulation of p21 expression in MDA-MB231 human
breast cancer cells. Phytomedicine. 17:1127–1132. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park MH, Park SY and Kim Y: Induction of
proline-rich tyrosine kinase2 (Pyk2) through C/EBPbeta is involved
in PMA-induced monocyte differentiation. FEBS Lett. 582:415–422.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo Y, Chen Z, Zhang L, et al: Distinctive
microRNA profiles relating to patient survival in esophageal
squamous cell carcinoma. Cancer Res. 68:26–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:5116–5121. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Liu J, Zhou R, Huang S and Chen XM:
Gene silencing of MIR22 in acute lymphoblastic leukaemia involves
histone modifications independent of promoter DNA methylation. Br J
Haematol. 148:69–79. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heller G, Schmidt WM, Ziegler B, et al:
Genome-wide transcriptional response to 5-aza-2′-deoxycytidine and
trichostatin a in multiple myeloma cells. Cancer Res. 68:44–54.
2008.
|
|
21
|
Cameron JE, Fewell C, Yin Q, et al:
Epstein-Barr virus growth/latency III program alters cellular
microRNA expression. Virology. 382:257–266. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nagpal JK, Rani R, Trink B and Saini KS:
Targeting miRNAs for drug discovery: a new paradigm. Curr Mol Med.
10:503–510. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ichimura A, Ruike Y, Terasawa K and
Tsujimoto G: miRNAs and regulation of cell signaling. FEBS J.
278:1610–1618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsuchiya N, Izumiya M, Ogata-Kawata H, et
al: Tumor suppressor miR-22 determines p53-dependent cellular fate
through post-transcriptional regulation of p21. Cancer Res.
71:4628–4639. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang S, He X, Ding J, et al: Upregulation
of miR-23a approximately 27a approximately 24 decreases
transforming growth factor-beta-induced tumor-suppressive
activities in human hepatocellular carcinoma cells. Int J Cancer.
123:972–978. 2008. View Article : Google Scholar
|
|
26
|
Wang P, Zou F, Zhang X, et al: microRNA-21
negatively regulates Cdc25A and cell cycle progression in colon
cancer cells. Cancer Res. 69:8157–8165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sayed D, Rane S, Lypowy J, et al:
MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol
Biol Cell. 19:3272–3282. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zaidi SK, Dowdy CR, van Wijnen AJ, et al:
Altered Runx1 subnuclear targeting enhances myeloid cell
proliferation and blocks differentiation by activating a
miR-24/MKP-7/MAPK network. Cancer Res. 69:8249–8255. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin RJ, Lin YC, Chen J, et al: microRNA
signature and expression of Dicer and Drosha can predict prognosis
and delineate risk groups in neuroblastoma. Cancer Res.
70:7841–7850. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vaksman O, Hetland TE, Trope CG, Reich R
and Davidson B: Argonaute, Dicer, and Drosha are upregulated along
tumor progression in serous ovarian carcinoma. Hum Pathol. May
29–2012.(Epub ahead of print).
|
|
31
|
Yan M, Huang HY, Wang T, et al:
Dysregulated expression of dicer and drosha in breast cancer.
Pathol Oncol Res. 18:343–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Frezzetti D, De Menna M, Zoppoli P, et al:
Upregulation of miR-21 by Ras in vivo and its role in tumor growth.
Oncogene. 30:275–286. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Takagi S, Nakajima M, Kida K, Yamaura Y,
Fukami T and Yokoi T: MicroRNAs regulate human hepatocyte nuclear
factor 4alpha, modulating the expression of metabolic enzymes and
cell cycle. J Biol Chem. 285:4415–4422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schmeier S, MacPherson CR, Essack M, et
al: Deciphering the transcriptional circuitry of microRNA genes
expressed during human monocytic differentiation. BMC Genomics.
10:5952009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou R, Hu G, Gong AY and Chen XM: Binding
of NF-kappaB p65 subunit to the promoter elements is involved in
LPS-induced transactivation of miRNA genes in human biliary
epithelial cells. Nucleic Acids Res. 38:3222–3232. 2010. View Article : Google Scholar : PubMed/NCBI
|