Introduction
Multiple myeloma (MM) is the second most common
hematological malignancy. Although patient survival has
significantly improved with the development of new treatments, such
as bortezomib and lenalidomide, some MM patients experience
long-term remission (1–3). Therefore, the development of novel
anticancer strategies for the effective treatment of this disease
is required.
DEP domain containing mammalian target of rapamycin
(mTOR)-interacting protein (DEPTOR) is an mTOR binding protein that
normally functions to inhibit the mTOR complex 1 (mTORC1) and 2
(mTORC2) pathways (4). The
expression of DEPTOR has been investigated in a number of human
tumors; low expression levels have been observed in the majority of
tumors (5). However, DEPTOR has
been found to be overexpressed in a subset of MM cells, and can
mediate the activation of the phosphoinositide 3-kinase (PI3K)/Akt
pathway in these cells (5,6).
This indirect mode of PI3K/Akt activation is crucial to the
survival of myeloma cells.
The PI3K/Akt signaling pathway is frequently
activated in many types of cancer and is therefore a major cell
survival pathway. Downstream effectors of the PI3K/Akt pathway
include caspase-3 and caspase-9 (7). Their activation has long been
associated with proliferation, differentiation and apoptosis
(8,9).
On the other hand, mTORC1 and PI3K/Akt have been
reported to inhibit the induction of autophagy (10–12). In addition, the connection between
autophagy and apoptosis has been extensively investigated over the
past decade (13–15). Certain studies have found that the
inhibition of autophagy induces apoptosis (16–20).
Therefore, in the present study, we aimed to clarify
the role of DEPTOR in the proliferation, apoptosis and autophagy in
MM cells, and to elucidate the mechanisms by which DEPTOR
contributes to the chemosensitivity of MM cells. We used the
RPMI-8226 cell line in which DEPTOR is highly expressed and treated
the cells with doxorubicin. In our study, we investigated the role
of DEPTOR using RNA interference (RNAi) technology in vitro.
RNAi is a sequence-specific, post-transcriptional gene silencing
technique induced by double-stranded RNA, homologous to the target
gene (21).
Materials and methods
Cell culture and reagents
RPMI-8226 cells (Wuhan University, Wuhan, China)
were cultured in RPMI-1640 medium (Gibco BRL, Carlsbad, CA, USA)
supplemented with 10% fetal calf serum (Gibco) at 37°C in a
humidified atmosphere containing 5% CO2. The following
reagents were used: anti-cleaved caspase-3 (Asp175), anti-cleaved
poly(ADP-ribose) polymerase (PARP; Asp214), anti-Akt,
anti-phosphorylated Akt (p-Akt; Ser 473), anti-phosphorylated
P70S6K (p-P70S6K) (Thr421/Ser424), anti-phosphorylated
eIF4E-binding protein-1 (p-4Ebp-1; Thr70) (Cell Signaling
Technology Inc., Danvers, MA, USA), anti-DEPTOR (Millipore,
Billerica, MA, USA), anti-autophagy-related 5 (Atg5), anti-light
chain (LC)-3 and anti-GAPDH (Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA) antibodies, as well as doxorubicin (Sigma-Aldrich,
St. Louis, MO, USA).
Lentivirus-mediated gene knockdown
To directly identify the biological function of
DEPTOR in MM, an effective RNAi sequence targeting the DEPTOR gene
was designed and screened by Genechem (Shanghai, China). Short
hairpin RNA (shRNA) was designed using DEPTOR RefSeq cDNA sequence
(GenBank accession no. NM_022783.2). The primer sequence was as
follows: 5′-CATGACAATCGGAAATCTA-3′. cDNA containing both sense and
antisense oligo DNA of the targeting sequence was designed,
synthesized and inserted into a GV115-EGFP vector to construct a
lentiviral vector that expressed DEPTOR shRNA. Lentiviral DEPTOR
shRNA and negative control shRNA were arrested and co-transfected
in 293T packaging cells. The negative control sequences have
previously been used in a number of studies (22,23), and have no significant homology to
any human gene sequences. The lentivirus in the supernatant was
collected and filtered and then used to transiently transfect the
RPMI-8226 MM cells. Lentivirus production and lentiviral infection
were performed by Genechem.
Cell proliferation assay
Cell viability was assessed by
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma-Aldrich) assays following lentiviral infection. Briefly, the
cells were seeded at a density of 2×103 cells/well in
96-well plates in 200 μl of medium and cultured for 24, 48 and 72
h. After incubation for a designated period of time, 20 μl of MTT
were added to each well and incubated for 4 h. The supernatant was
carefully removed and 200 μl DMSO were added to each well. The
optical density (OD) of each well was measured at 490 nm. The
inhibition rate (%) was calculated as follows: [1 − (OD of the
experimental samples/OD of the control)] ×100%. The concentration
of doxorubicin required to inhibit the growth by 50%
(IC50) was calculated. All of the experiments were
performed in triplicate.
Flow cytometric analysis of
apoptosis
Cells were washed twice with ice-cold PBS and fixed
with 70% ethanol at 4°C overnight. After washing with PBS, cells
were incubated in 0.5 ml PBS containing 50 μg/ml RNase A for 30 min
at 37°C, and then propidium iodide (PI) was added to a final
concentration of 50 μg/ml and incubated for 30 min in the dark. The
resultant cell suspension was then subjected to flow cytometric
analysis using a Coulter Epics XL flow cytometer (Beckman Coulter,
Inc., Miami, FL, USA). The percentage of apoptotic cells was
calculated.
RT-PCR and quantitative RT-PCR
(qRT-PCR)
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA). RNA (4 μg) was reverse
transcribed into cDNA using the Thermoscript RT-PCR System reagent
(Gibco) according to the manufacturer’s instructions. For
quantitative RT-PCR analysis, each 25 μl volume of qRT-PCR was
performed using the Applied Biosystems PRISM 7300 sequence
detection system (Applied Biosystem, Foster City, USA) in 96-well
plates. All reactions were conducted in triplicate. All the
threshold cycle (Ct) values were normalized to GAPDH. The
2−ΔΔCT method was used to relative quantify the
transcriptional level of DEPTOR. Primers were designed for PCR. The
primer sequences for human DEPTOR were:
5′-CCTACCCAAACTGTTTTGTCGC-3′ (sense) and
5′-CGGTCTGCTAATTTCTGCATGAG-3′ (antisense). Primers for the control
(GAPDH) were: 5′-TGACTTCAACAGCGACACCCA-3′ (sense) and
5′-CACCCTGTTGCTGTAGCCAAA-3′ (antisense). The PCR amplification
consisted of 35 cycles: 15 sec at 95°C for denaturation, 30 sec at
60°C for annealing and 45 sec at 72°C for elongation, and a final
extension of 72°C for 10 min. PCR was performed for GAPDH as a
control to ascertain the amount of the samples. The results were
expressed in relation to the control value.
Western blot analysis
Total cell lysates were separated by 6–15% SDS-PAGE
gel electrophoresis and transferred onto polyvinylidene fluoride
(PVDF) membranes, which were blocked with TBST containing 5%
non-fat milk at 4°C overnight and then incubated with anti-cleaved
caspase-3, anti-cleaved PARP, anti-Akt, anti-p-Akt, anti-p-P70S6K,
anti-p-4Ebp-1, anti-DEPTOR, anti-Atg5, anti-LC-3 and anti-GAPDH
antibodies for 2 h. After washing with TBST, membranes were
incubated with HRP-labeled secondary antibodies for 2 h at room
temperature. The blots were detected using the enhanced
chemiluminescence (ECL) reagent kit (Beyotime, Shangshai,
China).
Transmission electron microscopy
The treated cells were collected by trypsinization
and fixed in 3% glutaraldehyde in 0.1 mol/l phosphate buffer for 1
h at 4°C. The samples were then fixed with 1% osmium tetroxide in
0.1 mol/l phosphate buffer for 1 h. Ultrathin sections (80 nm) were
prepared, stained with uranyl acetate for 15 min, followed by lead
citrate for 5 min, and then examined with a Philips EM 208
transmission electron microscope (Philips, Kassel, Germany) at an
accelerating voltage of 70 kV.
Visualization and quantification of
MDC-labeled autophagic vacuoles
Monodansylcadaverine (MDC) staining was also used to
detect autophagy and flow cytometry was used for quantification of
the autophagosomes. In addition, MDC has been proposed as a special
tracer for autophagic vacuoles (24). The autophagic vacuoles were filled
with MDC by incubating cell growth on cover-slips with 0.05 mmol/l
MDC in PBS at 37°C for 1 h. Following incubation, the cells were
washed twice with PBS and immediately analyzed by fluorescence
microscopy using an inverted microscope (Olympus IX-71; Olympus
Corp., Tokyo, Japan). The excitation wavelength was 380 nm and the
emission filter was 525 nm.
Sensitivity to doxorubicin
The cells were seeded in triplicate on 96-well
plates with 1×104 cells/well, then incubated for 24 h.
Subsequently, the medium was carefully removed and replaced with
fresh medium, containing doxorubicin in reasonable concentration,
and with the medium without doxorubicin as the control group. After
24 h incubation, the cells were treated with MTT as described
above. The inhibition rate was calculated as follows: 1 − OD490
(doxorubicin +)/OD490 (control) %.
Statistical analysis
The data are expressed as the means ± SD, and
one-way analysis of variance (ANOVA) was used to measure
statistical significance among the different groups, followed by
Student-Newman-Keuls analyses. A P-value <0.05 was considered to
indicate a statistically significant difference.
Results
shRNA mediates knockdown of DEPTOR in
RPMI-8226 MM cells
RNA and protein were harvested from the cells at 24
h post-transfection for the evaluation of DEPTOR knockdown. The
silencing effects of DEPTOR-specific shRNA in the RPMI-8226 cells
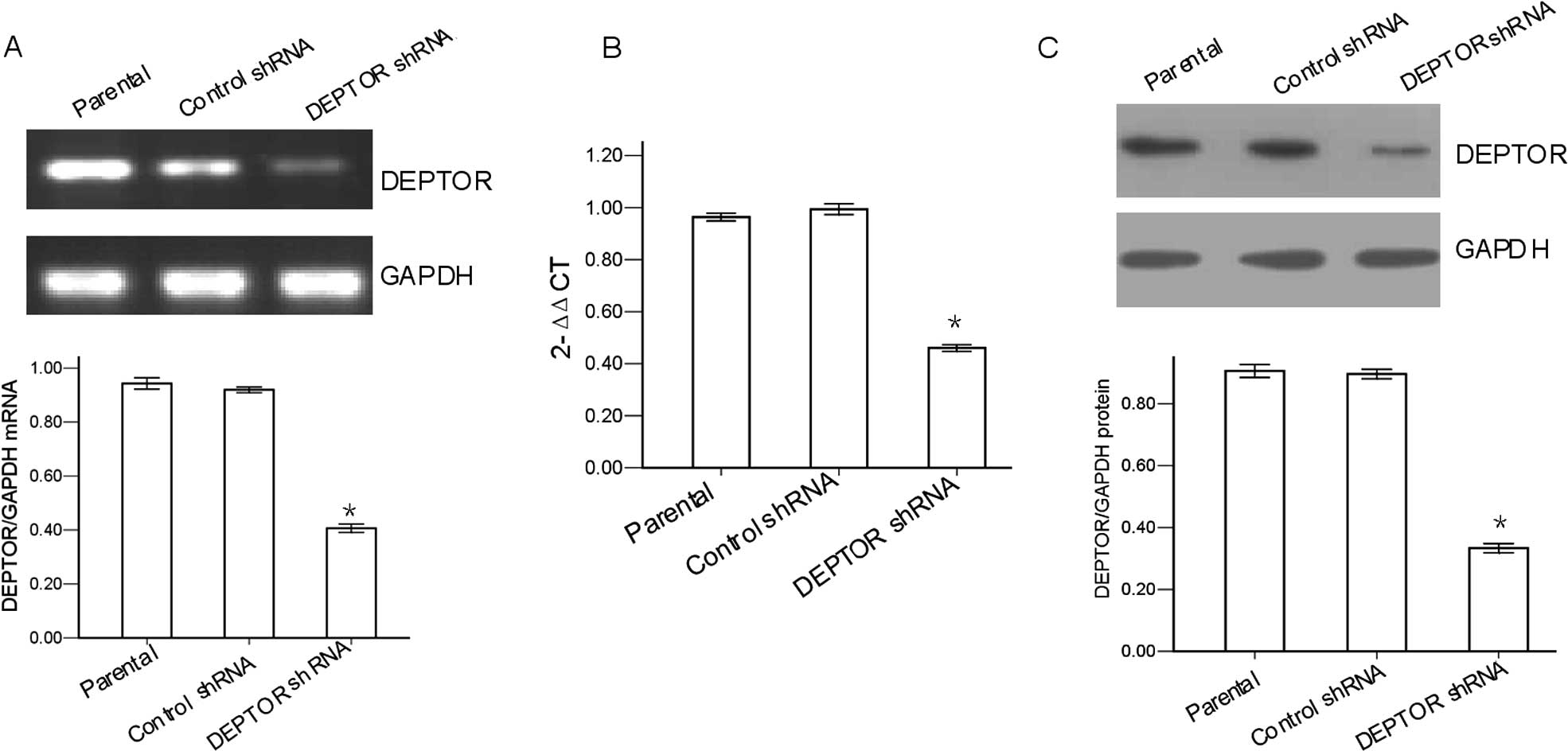
were first evaluated by RT-PCR. The results revealed that the ratio
of DEPTOR/GAPDH mRNA in the DEPTOR shRNA-transfected cells was
40.7±1.5%, significantly lower than that in the control
shRNA-transfected cells (92.0±1.0%) or in the parental cells
(94.3±2.1%; P<0.05; Fig. 1A).
qRT-PCR showed that expression of DEPTOR mRNA was significantly
decreased in the DEPTOR shRNA-transfected group (Fig. 1B). The silencing effect of
DEPTOR-specific shRNA in the RPMI-8226 cells was also evaluated by
western blot analysis. The results revealed that the ratio of
DEPTOR/GAPDH protein in the DEPTOR shRNA-transfected cells was
33.3±1.5%, significantly lower than that in the control
shRNA-transfected cells (89.7±1.5%) or in the parental cells
(90.7±2.1%; P<0.05; Fig. 1C).
There was no significant difference observed between the control
shRNA-transfected cells and the parental cells (P>0.05). These
results demonstrated that DEPTOR was effectively knocked down in
the DEPTOR shRNA-transfected RPMI-8226 cells and could be used for
following experiments to characterize the role of DEPTOR in MM.
Knockdown of DEPTOR inhibits
proliferation and promotes apoptosis in RPMI-8226 cells
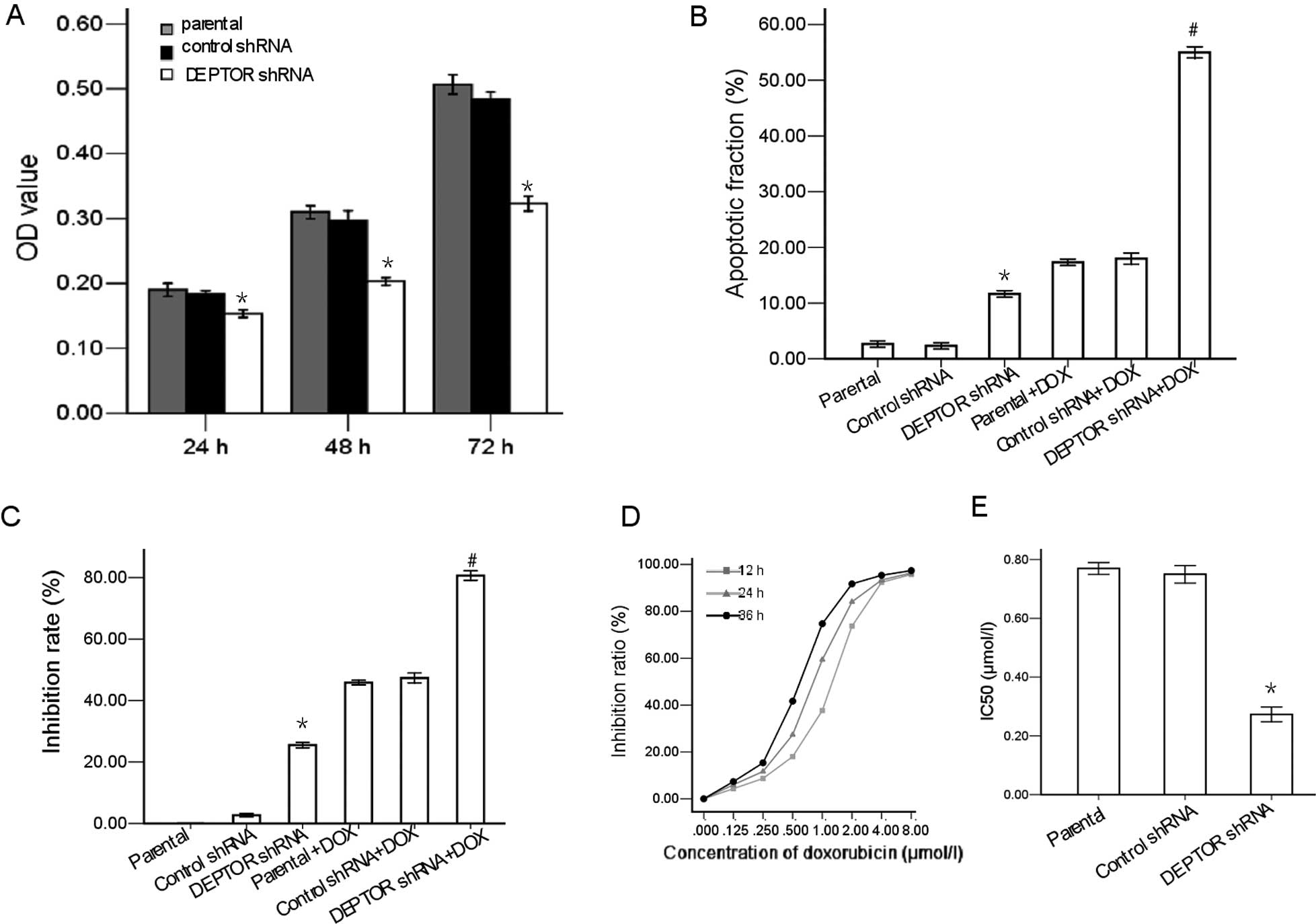
We then investigated whether DEPTOR shRNA decreases
the proliferation of RPMI-8226 cells. As indicated in Fig. 2A, compared to the parental cells,
the proliferation capacity of the DEPTOR shRNA-transfected cells
was inhibited by 67.3±1.32% (P<0.05), 61.6±1.35% (P<0.05) and
63.5±1.12% (P<0.05) at 24, 48 and 72 h, respectively. There was
no significant difference in the proliferation capacity between the
control shRNA-transfected cells and the parental cells (P>0.05).
To determine the apoptosis-inducing potential of DEPTOR shRNA in
the cells, flow cytometric analysis of the PI-stained cells was
performed. As shown in Fig. 2B,
the apoptotic rate in the DEPTOR shRNA-transfected group was
significantly higher than that in the parental or control
shRNA-transfected group. The results showed that the inhibition
rate in the DEPTOR shRNA-transfected cells was markedly higher than
that in the parental cells or in the control shRNA-transfected
RPMI-8226 cells (P<0.05; Fig.
2C).
Doxorubicin inhibits the proliferation of
RPMI-8226 cells
MTT assay was employed to detect the cytotoxic
effects of various concentrations of doxorubicin (0, 0.125, 0.25,
0.5, 1, 2, 4 and 8 μmol/l) on RPMI-8226 cells for 12, 24 and 36 h.
As shown in Fig. 2D, doxorubicin
induced a marked inhibition of cell proliferation in a time- and
dose-dependent manner with an IC50 of 0.77 μmol/l in the
RPMI-8226 cells at 24 h.
DEPTOR knockdown enhances the
doxorubicin-induced growth inhibitory effect and promotes apoptosis
in RPMI-8226 MM cells
We investigated whether the inhibition of DEPTOR by
shRNA affected the sensitivity of RPMI-8226 cells to the antitumor
drug, doxorubicin. The results demonstrated that the inhibition
rate in the DEPTOR shRNA-transfected cells treated with doxorubicin
was markedly higher than that in the parental cells treated with
doxorubicin or in the control shRNA-transfected cells treated with
doxorubicin (P<0.05). There was no significant difference
observed between the control shRNA-transfected cells treated with
doxorubicin and the parental cells treated with doxorubicin
(P>0.05; Fig. 2C).
We performed flow cytometry to evaluate apoptosis in
the RPMI-8226 cells treated with doxorubicin. The results showed
that upon exposure to doxorubicin, apoptosis was significantly
increased in the RPMI-8226 cells in which the expression of DEPTOR
had been knocked down compared with the control cells (Fig. 2B).
The RPMI-8226 cells were exposed to 0–8 μmol/l
doxorubicin for 24 h. The IC50 calculated based on the
data from MTT cytotoxicity assay showed that DEPTOR knockdown
enhanced the sensitivity of RPMI-8226 cells to doxorubicin. The
DEPTOR knockdown decreased the IC50 of doxorubicin. The
IC50 of doxorubicin in the RPMI-8226 cells decreased
from 0.77 μmol/l to 0.27 μmol/l (Fig.
2E).
Taken together, these data suggest that DEPTOR
knockdown enhances the doxorubicin-induced growth inhibitory
effect, promotes apoptosis, and increases the chemosensitivity of
RPMI-8226 human multiple myeloma cells to doxorubicin.
DEPTOR knockdown leads to changes in the
expression of apoptosis-associated proteins in RPMI-8226 MM
cells
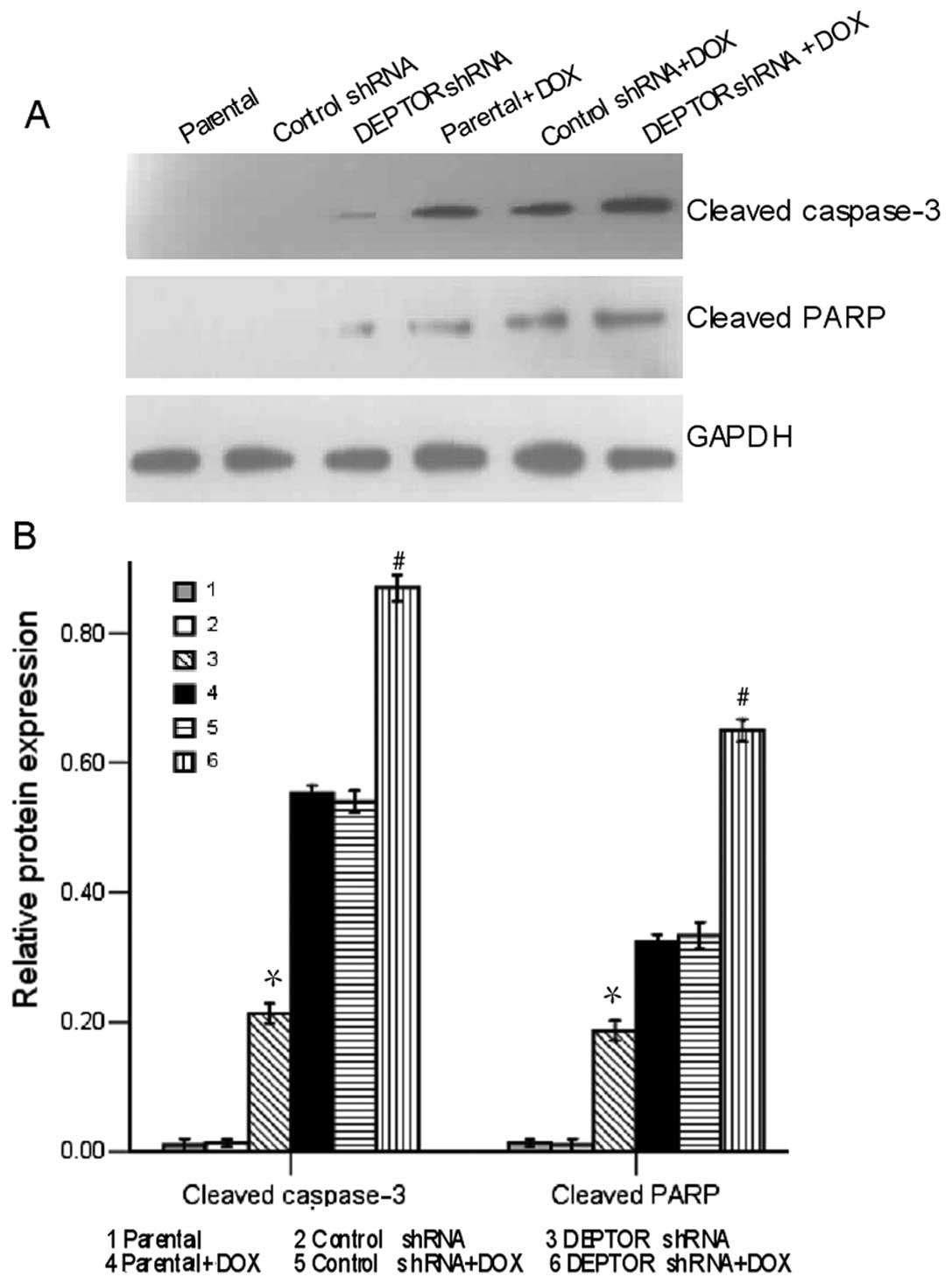
To determine the apoptosis-inducing potential of
DEPTOR shRNA in RPMI-8226 cells, we performed western blot analysis
to detect the expression of apoptosis-associated proteins. As shown
in Fig. 3, the expression levels
of cleaved caspase-3 and cleaved PARP in the DEPTOR
shRNA-transfected group were significantly higher than those
observed in the control shRNA-transfected and in the parental
group. Following exposure to doxorubicin for 24 h, the expression
levels of both proteins were markedly increased in the DEPTOR
shRNA-transfected cells, but not in the control shRNA-transfected
cells. These results suggest that DEPTOR knockdown induces the
upregulation of caspases, which then leads to apoptosis.
Knockdown of DEPTOR inhibits cell
autophagy in RPMI-8226 MM cells
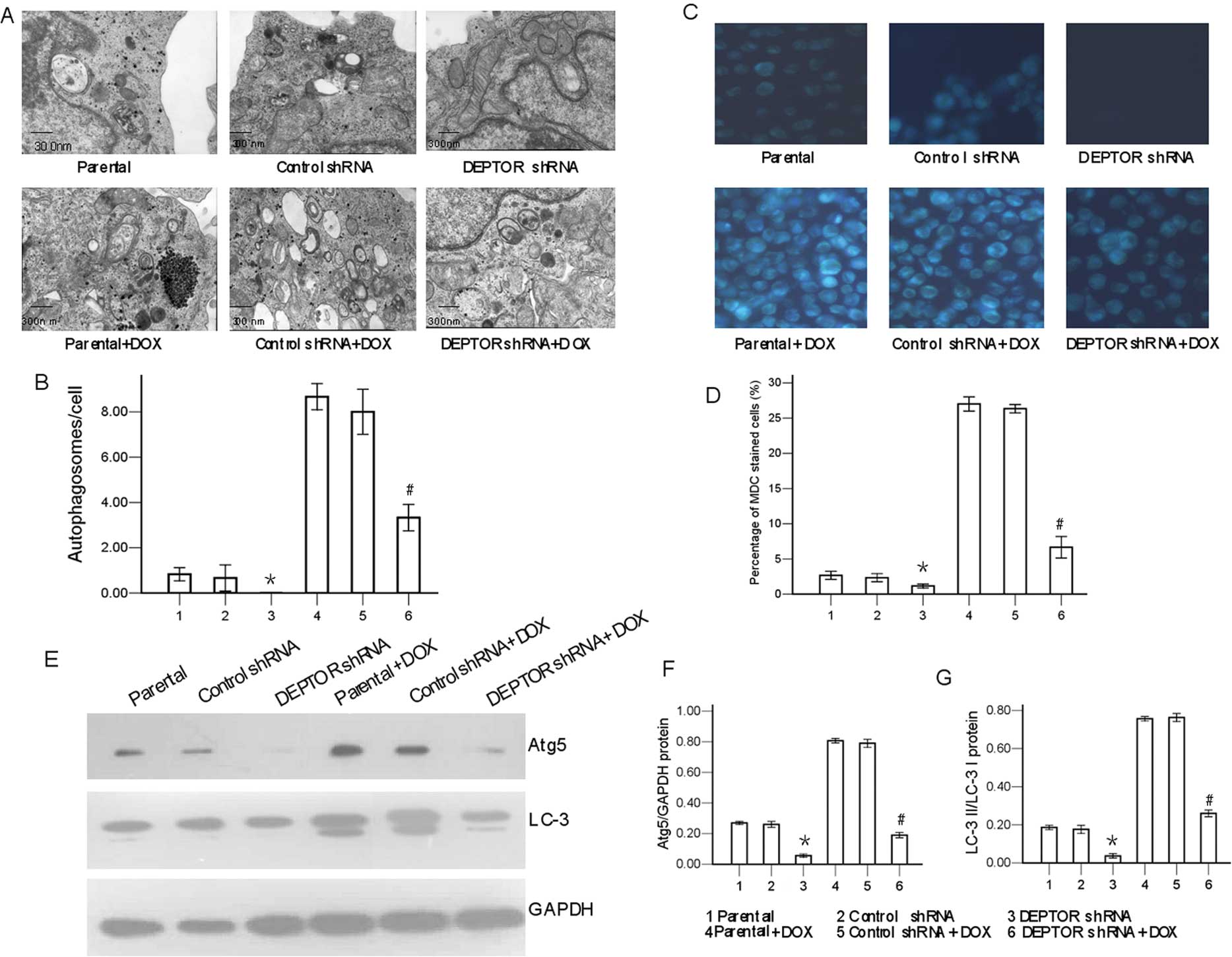
Evidence indicates that autophagy can be detected
morphologically and biochemically (25,26). In this study, transmission
electron microscopy and the fluorescence of MDC observations
revealed that the number of autophagic vacuoles in the DEPTOR
shRNA-transfected cells was markedly lower than that in the
parental or the control shRNA-transfected cells (P<0.05;
Fig. 4). Consistent with these
findings, the different treatments also induced the conversion of
LC-3 I to LC-3 II.
To determine the autophagy-inducing potential of
DEPTOR shRNA in RPMI-8226 cells, we performed western blot analysis
to detect the expression of autophagy-associated proteins. As shown
in Fig. 4, the expression levels
of Atg5 and LC-3 in the DEPTOR shRNA-transfected group were
significantly lower than those observed in the control
shRNA-transfected group and the parental group. Following exposure
to doxorubicin for 24 h, the expression levels of Atg5 and LC-3
II/LC-3 I, which indicated the activation of autophagy, were
markedly reduced in the DEPTOR shRNA-transfected cells, but not in
the control shRNA-transfected cells.
Thus, taken together, these data indicate the
inhibition of autophagic response in the DEPTOR shRNA-transfected
RPMI-8226 cells. Our data suggest that the knockdown of DEPTOR
inhibits autophagy in MM cells.
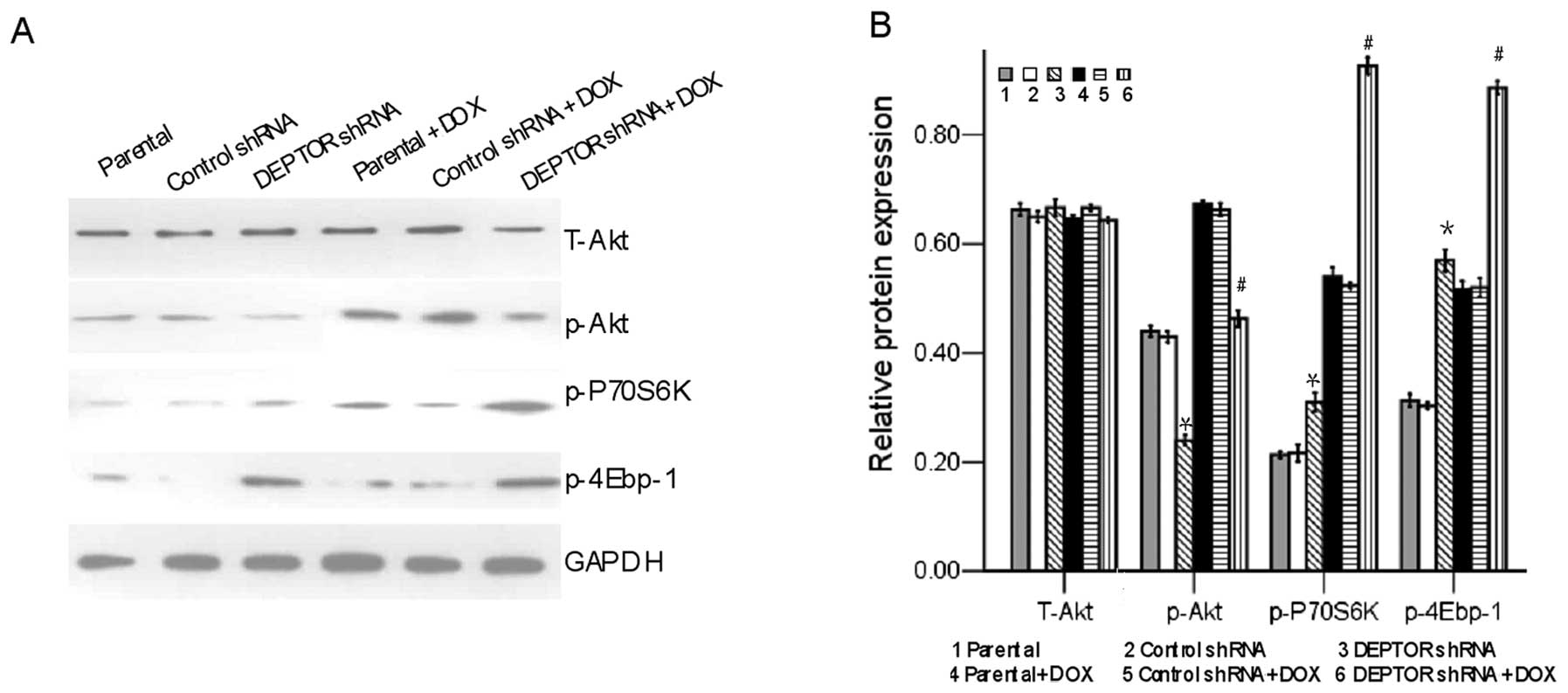
DEPTOR knockdown reduces PI3K/Akt
activity in RPMI-8226 MM cells
We detected the PI3K/Akt activity in the RPMI-8226
MM cells unexposed or exposed to doxorubicin. Western blot analysis
showed that the level of p-Akt was markedly reduced in the
RPMI-8226 cells in which the expression of DEPTOR had been knocked
down, but not in the control RPMI-8226 cells; however, the level of
total Akt remained unaltered between the cells in which DEPTOR
expression had been knocked down and the control RPMI-8226 cells
(Fig. 5). In addition,
doxorubicin markedly activated the mTOR complex 1 targets, p-P70S6K
and p-4Ebp-1, in the RPMI-8226 cells in which DEPTOR expression had
been knocked down, but had no such effects on the control
shRNA-transfected RPMI-8226 cells (Fig. 5). Collectively, these results
demonstrate that DEPTOR knockdown reduces the PI3K/Akt activity in
the RPMI-8226 MM cells.
Discussion
Our results demonstrated the the knockdown of
DEPTOR, a recently identified inhibitor of mTOR complexes (5), induces apoptosis, increases the
chemosensitivity to doxorubicin, and suppresses autophagy and
PI3K/Akt signaling in RPMI-8226 cells. Our data have demonstrated
that targeting DEPTOR can be used for the treatment of MM. shRNA is
an effective and efficient technique for the study of tumors, as
well as treatment (27). In our
study, we successfully transfected shRNA targeting the DEPTOR gene
into the human MM cell line, RPMI-8226, and DEPTOR expression was
effectively inhibited at the protein level.
A recent study found that high expression levels of
DEPTOR are predictive of response to thalidomide in myeloma.
However, there was no survival benefit for thalidomide in the low
DEPTOR expression group (28).
The results showed that the level of DEPTOR expression is crucial
to the survival of myeloma patients. The results from our study
also indicate that the level of DEPTOR expression is crucial to the
survival of myeloma cells.
We examined the effect of DEPTOR silencing on the
autophagic capacity of RPMI-8226 cells. The autophagic capacity of
DEPTOR shRNA-transfected cells was suppressed compared to the
control shRNA-transfected cells or the parental cells. At the same
time, mTORC1 and the PI3K/Akt pathway plays an important role in
cell autophagy (10–12). Since DEPTOR is an mTOR-interacting
protein that normally functions to inhibit the mTORC1 pathway, the
knockdown of DEPTOR inhibits cell autophagy.
Certain evidence indicates that the inhibition of
autophagy may induce apoptosis (16–20). An earlier study suggested a role
for autophagy as a potential pro-survival mechanism in MM cells
(29). Recently, another study
suggested that the suppression of autophagy significantly augments
the in vitro and in vivo antimyeloma activity of
DNA-damaging chemotherapy (30).
Thus, apoptosis may be enhanced by the inhibition of autophagy
using shRNA targeting the DEPTOR gene in MM cells.
Active caspases play a vital role in the induction
of apoptosis. Following the activation of caspase-3, PARP was
cleaved (31). The cleavage of
PARP has often been viewed as an indicator of apoptosis. In our
study, DEPTOR shRNA-transfected RPMI-8226 cells demonstrated a
higher level of cleaved caspase-3 fragments and cleaved PARP. These
results reveal that the DEPTOR knockdown by shRNA is sufficient to
trigger caspase-dependent apoptosis, which could be the reason for
the decrease in cell viability. We found that the inhibition of
DEPTOR expression led to increased levels of cleaved caspase-3 and
cleaved PARP, which contributed to doxorubicin-induced apoptosis in
RPMI-8226 cells. These results are consistent with those from a
previous study, reporting that doxorubicin-induced apoptosis in MM
cells was associated with the activation of caspase-3 and PARP
(30,32).
We then identified the signaling pathway through
which DEPTOR modulates the effects of chemotherapy in MM. Recently,
DEPTOR was identified as a regulator of the PI3K/Akt pathway.
DEPTOR appears to play a specific role in upregulating the PI3K/Akt
pathway in myeloma (5). The
PI3K/Akt signaling pathway plays an important role in cell
proliferation, development and apoptotic resistance and survival
(8,9,28,33). At the same time, PI3K/Akt
inhibition has been found to induce chemosensitization in MM cells
(34,35). The constitutive activation of the
PI3K/Akt pathway has been accepted as an important molecular event
that contributes to the malignant phenotype of MM cells (36).
Our results have demonstrated that DEPTOR shRNA
suppressed PI3K/Akt activity in the RPMI-8226 cells, indicating the
involvement of the PI3K/Akt signaling pathway downstream of DEPTOR.
Akt can also phosphorylate procaspase-3 to inhibit apoptosis
(37). Akt is a major mediator of
cell survival, either directly by inhibiting pro-apoptotic
proteins, such as caspase-9 and Bad, or indirectly by modulating
regulators of cell death including p53 and nuclear factor-κB
(NF-κB) (38–42). Activated Akt modulates the
function of many substrates involved in cell cycle progression,
cell growth and the regulation of cell survival (43–45). The major upstream regulator of Akt
is PI3K, which is activated by a variety of transmembrane receptors
(46). DEPTOR also acts as an
oncogene by relieving the feedback inhibition from S6 kinase 1
(S6K1) to PI3K, thus activating Akt (47).
Our study has demonstrated that the
doxorubicin-induced increase in the level of p-Akt level was
markedly reduced in the DEPTOR shRNA-transfected cells, but was
unaffected in the control shRNA-transfected cells or in the
parental RPMI-8226 cells. Moreover, Akt regulates cell
proliferation through its effects on the mTOR/P70S6 kinase pathway
(8,36,48). mTORC1 controls cell proliferation
partly by phosphoylating S6K1 and 4Ebp-1, key regulators of protein
synthesis (49). In our study, we
found that doxorubicin markedly activated the mTORC1 targets, S6K1
and 4Ebp-1, in the DEPTOR shRNA-transfected RPMI-8226 cells.
Collectively, these results suggest that the suppression of
PI3K/Akt activity following DEPTOR knockdown is responsible for the
increased sensitivity of RPMI-8226 cells to doxorubicin.
In conclusion, our study demonstrates that the
knockdown of DEPTOR by RNAi induces apoptosis, increases the
chemosensitivity to doxorubicin, suppresses autophagy and inhibits
the activation of the PI3K/Akt signaling pathway in RPMI-8226
cells. To the best of our knowledge, this the first study to
demonstrate a possible correlation between DEPTOR gene expression
and autophagy in MM cells. Our results provide evidence that DEPTOR
is an important therapeutic target for the treatment of MM. These
findings raise the possibility that DEPTOR inhibitors may be used
to enhance the effectiveness of doxorubicin in the treatment of
myeloma. Animal experiments should be performed to further confirm
the effects of DEPTOR knockdown on the proliferation, apoptosis and
autophagy of MM cells. The anticancer effects induced by DEPTOR
knockdown require further investigation.
Acknowledgements
This study was supported in part by grants from the
National Natural Science Foundation of China (no. 30871111), the
Provincial Education Department of Fujian Province (no. JB11056),
and the National Natural Science Foundation of China (no.
81272628).
References
|
1
|
Kyle RA and Rajkumar SV: Multiple myeloma.
N Engl J Med. 351:1860–1873. 2004. View Article : Google Scholar
|
|
2
|
Richardson PG, Mitsiades CS, Hideshima T
and Anderson KC: Novel biological therapies for the treatment of
multiple myeloma. Best Pract Res Clin Haematol. 18:619–634. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dimopoulos MA, San-Miguel JF and Anderson
KC: Emerging therapies for the treatment of relapsed or refractory
multiple myeloma. Eur J Haematol. 86:1–15. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Duan S, Skaar JR, Kuchay S, et al: mTOR
generates an auto-amplification loop by triggering the βTrCP- and
CK1α-dependent degradation of DEPTOR. Mol Cell. 44:317–324.
2011.PubMed/NCBI
|
|
5
|
Peterson TR, Laplante M, Thoreen CC, et
al: DEPTOR is an mTOR inhibitor frequently overexpressed in
multiple myeloma cells and required for their survival. Cell.
137:873–886. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao Y, Xiong X and Sun Y: DEPTOR, an mTOR
inhibitor, is a physiological substrate of SCF(βTrCP) E3 ubiquitin
ligase and regulates survival and autophagy. Mol Cell. 44:304–316.
2011.PubMed/NCBI
|
|
7
|
Morgensztern D and McLeod HL:
PI3K/Akt/mTOR pathway as a target for cancer therapy. Anticancer
Drugs. 16:797–803. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hennessy BT, Smith DL, Ram PT, Lu Y and
Mills GB: Exploiting the PI3K/AKT pathway for cancer drug
discovery. Nat Rev Drug Discov. 4:988–1004. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wullschleger S, Loewith R and Hall MN: TOR
signaling in growth and metabolism. Cell. 124:471–484. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Edinger AL and Thompson CB: Defective
autophagy leads to cancer. Cancer Cell. 4:422–424. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gozuacik D and Kimchi A: Autophagy as a
cell death and tumor suppressor mechanism. Oncogene. 23:2891–2906.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Maiuri MC, Zalckvar E, Kimchi A and
Kroemer G: Self-eating and selfkilling: crosstalk between autophagy
and apoptosis. Nat Rev Mol Cell Biol. 8:741–752. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim R, Emi M, Tanabe K, Murakami S, Uchida
Y and Arihiro K: Regulation and interplay of apoptotic and
non-apoptotic cell death. J Pathol. 208:319–326. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thorburn A: Apoptosis and autophagy:
regulatory connections between two supposedly different processes.
Apoptosis. 13:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Boya P, González-Polo RA, Casares N, et
al: Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol.
25:1025–1040. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Amaravadi RK, Yu D, Lum JJ, et al:
Autophagy inhibition enhances therapy-induced apoptosis in a
Myc-induced model of lymphoma. J Clin Invest. 117:326–336. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ravikumar B, Berger Z, Vacher C, O’Kane CJ
and Rubinsztein DC: Rapamycin pre-treatment protects against
apoptosis. Hum Mol Genet. 15:1209–1216. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Longo L, Platini F, Scardino A, Alabiso O,
Vasapollo G and Tessitore L: Autophagy inhibition enhances
anthocyanin-induced apoptosis in hepatocellular carcinoma. Mol
Cancer Ther. 7:2476–2485. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Herman-Antosiewicz A, Johnson DE and Singh
SV: Sulforaphane causes autophagy to inhibit release of cytochrome
c and apoptosis in human prostate cancer cells. Cancer Res.
66:5828–5835. 2006. View Article : Google Scholar
|
|
21
|
Bernstein E, Caudy AA, Hammond SM and
Hannon GJ: Role for a bidentate ribonuclease in the initiation step
of RNA interference. Nature. 409:363–366. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zielske SP and Stevenson M: Importin 7 may
be dispensable for human immunodeficiency virus type 1 and simian
immunodeficiency virus infection of primary macrophages. J Virol.
79:11541–11546. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pullmann R Jr, Juhaszova M, López de
Silanes I, et al: Enhanced proliferation of cultured human vascular
smooth muscle cells linked to increased function of RNA-binding
protein HuR. J Biol Chem. 280:22819–22826. 2005. View Article : Google Scholar
|
|
24
|
Munafo DB and Colombo MI: A novel assay to
study autophagy: regulation of autophagosome vacuole size by amino
acid deprivation. J Cell Sci. 114:3619–3629. 2001.PubMed/NCBI
|
|
25
|
Mizushima M: Methods for monitoring
autophagy. Int J Biochem Cell Biol. 36:2491–2502. 2004. View Article : Google Scholar
|
|
26
|
Klionsky DJ, Abeliovich H, Agostinis P, et
al: Guidelines for the use and interpretation of assays for
monitoring autophagy in higher eukaryotes. Autophagy. 4:151–175.
2008. View Article : Google Scholar
|
|
27
|
Brummelkamp TR, Bernards R and Agami R: A
system for stable expression of short interfering RNAs in mammalian
cells. Science. 296:550–553. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boyd KD, Walker BA, Wardell CP, et al:
High expression levels of the mammalian target of rapamycin
inhibitor DEPTOR are predictive of response to thalidomide in
myeloma. Leuk Lymphoma. 51:2126–2129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hideshima T, Bradner JE, Wong J, et al:
Small-molecule inhibition of proteasome and aggresome function
induces synergistic antitumor activity in multiple myeloma. Proc
Natl Acad Sci USA. 102:8567–8572. 2005. View Article : Google Scholar
|
|
30
|
Pan Y, Gao Y, Chen L, et al: Targeting
autophagy augments in vitro and in vivo antimyeloma activity of
DNA-damaging chemotherapy. Clin Cancer Res. 17:3248–3258. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kaufman SH: Induction of endonucleolytic
DNA cleavage in human acute myelogenous leukemia cell by etoposide,
camptothecin and other cytotoxic anticancer drugs: a cautionary
note. Cancer Res. 49:5870–5878. 1989.PubMed/NCBI
|
|
32
|
Cheriyath V, Kuhns MA, Kalaycio ME and
Borden EC: Potentiation of apoptosis by histone deacetylase
inhibitors and doxorubicin combination: cytoplasmic cathepsin B as
a mediator of apoptosis in multiple myeloma. Br J Cancer.
104:957–967. 2011. View Article : Google Scholar
|
|
33
|
Carnero A, Blanco-Aparicio C, Renner O,
Link W and Leal JF: The PTEN/PI3K/Akt signaling pathway in cancer,
therapeutic implications. Curr Cancer Drug Targets. 8:187–198.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Que W, Chen J, Chuang M and Jiang D:
Knockdown of c-Met enhances sensitivity to bortezomib in human
multiple myeloma U266 cells via inhibiting Akt/mTOR activity.
APMIS. 120:195–203. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
McMillin DW, Ooi M, Delmore J, et al:
Antimyeloma activity of the orally bioavailable dual
phosphatidylinositol 3-kinase/mammalian target of rapamycin
inhibitor NVP-BEZ235. Cancer Res. 69:5835–5842. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
de la Rubia J and Such E: DEPTOR
expression and response to thalidomide: toward a new therapeutic
target in multiple myeloma? Leuk Lymphoma. 51:1960–1961. 2010.
|
|
37
|
Allan LA and Clarke PR: Apoptosis and
autophagy: regulation of caspase-9 by phosphorylation. FEBS J.
276:6063–6073. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Akiyama M, Hideshima T, Hayashi T, et al:
Cytokines modulate telomerase activity in a human multiple myeloma
cell line. Cancer Res. 62:3876–3882. 2002.PubMed/NCBI
|
|
39
|
Cantley LC: The phosphoinositide 3-kinase
pathway. Science. 296:1655–1657. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Brunet A, Bonni A, Zigmond MJ, et al: Akt
promotes cell survival by phosphorylating and inhibiting a Forkhead
transcription factor. Cell. 96:857–868. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Datta SR, Brunet A and Greenberg ME:
Cellular survival: a play in three Akts. Genes Dev. 13:2905–2927.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim D and Chung J: Akt: versatile mediator
of cell survival and beyond. J Biochem Mol Biol. 35:106–115. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kang HY, Shim D, Kang SS, Chang SI and Kim
HY: Protein kinase B inhibits endostatin-induced apoptosis in
HUVECs. J Biochem Mol Biol. 39:97–104. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Han B, Wei W, Hua F, et al: Requirement
for ERK activity in sodium selenite-induced apoptosis of acute
promyelocytic leukemia-derived NB4 cells. J Biochem Mol Biol.
40:196–204. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Que WZ and Chen JM: Knockdown of c-Met
inhibits cell proliferation and invasion and increases
chemosensitivity to doxorubicin in human multiple myeloma U266
cells in vitro. Mol Med Rep. 4:343–349. 2011.PubMed/NCBI
|
|
46
|
Knobloch J, Schmitz I, Götz K,
Schulze-Osthoff K and Rüther U: Thalidomide induces limb anomalies
by PTEN stabilization, Akt suppression, and stimulation of
caspase-dependent cell death. Mol Cell Biol. 28:529–538. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Efeyan A and Sabatini DM: mTOR and cancer:
many loops in one pathway. Curr Opin Cell Biol. 22:169–176. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zeng X and Kinsella TJ: Mammalian target
of Rapamycin and S6 Kinase 1 positively regulate
6-thioguanine-induced autophagy. Cancer Res. 68:2384–2390. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Guertin DA and Sabatini DM: Defining the
role of mTOR in cancer. Cancer Cell. 12:9–22. 2007. View Article : Google Scholar
|