Introduction
Neurons and glial cells constitute most of the
central and peripheral nervous systems in the vertebrate that
regulate and control a wide range of thinking and behaviors; their
malfunction can cause serious nervous system diseases. Embryonic
stem cells (ESCs) provide significant therapeutic promise as they
can be induced to differentiate in vitro into various
tissues and organs originated from all three germ layers, including
neural cell lineages. Significant issues such as immunorejection,
ethical concerns and safety have inhibited the advancement of ESCs
toward clinical treatments, while induced pluripotent stem (iPS)
cells have become an attractive option for regenerative medicine
(1) and personalized medicine.
Therapeutic use of iPS cells will require greater understanding and
control of the reprogramming process, and demonstrations of safety
such as lack of tumorigenesis, through studies that cannot be
risked in humans. Mouse iPS cells are therefore a suitable model
for studying basic mechanisms of development and differentiation
and for evaluating the similarities between ES and iPS cells.
Several directed neuronal differentiation methods
have been developed, including via embryoid bodies (EBs) (2,3),
monolayer cultures (4), and
stromal cell-derived inducing activity (SDIA) (1,5).
In our study, we utilized a modified neuronal differentiation
method (2) to induce a tetraploid
complementation competent iPS cell line and two different ES cell
lines to differentiate into neurons. Neurons derived from all three
sources exhibited nearly the same differentiation patterns during
approximately 20 days of in vitro culture. Derived cell
populations are mixtures, and most cells are positive for the
neuron-specific marker protein MAP2. Several neural differentiation
stage marker genes were expressed by these cell mixtures as well,
including Blbp (Fabp7), Nestin and
Tuj1. Microarray profiling and single-cell PCR were employed
to further analyze the neural lineage differentiation process.
Following statistical comparisons and gene ontology analysis, 1,324
differentially expressed genes were identified, some of which are
involved in cell morphology, synaptic transmission, neurogenesis
and neuron recognition. The genes identified may be useful for
investigating important signaling factors and pathways regulating
neuronal differentiation.
Materials and methods
Cell culture
The mouse ES cell line CGR8.8 was a gift from Dr
Yanru Chen (Stanford University), and the mouse iPSC line IP14D-1
was derived from B6/DBA2F1 fetal fibroblasts and was confirmed to
be capable of developing into a complete embryo using the
tetraploid complementation assay (6,7).
Both R1 and IP14D-1 were cultured on mouse embryonic fibroblasts
(MEFs) with mitotic inactivation, while CGR8.8 was cultured under
feeder-free conditions using only 0.1% gelatin-coated culture
dishes. The complete culture medium utilized for mouse pluripotent
stem cells contained high glucose Dulbecco’s modified Eagle’s
medium with 15% fetal bovine serum tested for ES, 2 mM L-glutamine,
1 mM MEM sodium pyruvate, 1 mM MEM non-essential amino acids, 0.1
mM β-mercaptoethanol and 103 U/ml leukemia inhibitory
factor (LIF) (ESG1107; Millipore). Culture medium was changed daily
and cells were split every 2–3 days. E13.5 ICR mice were sterilely
dissected and harvested cortices were made into cell suspension.
They were cultured using Neurobasal culture medium containing 1%
B27 supplement in a 37°C, 5% CO2 incubator ~7–10 days
for further usage.
Induction of neural-specific embryoid
body formation in conditioned culture medium
R1 and IP14D-1 were passaged on 0.1% gelatin-coated
culture dishes prior to EB formation to avoid any effect of MEF
cells. EBs of all three pluripotent stem cell clones were digested
into single cells and cultured in suspension for the first four
days in pluripotent stem cell culture medium without LIF. For
further neural-specific induction, EBs were then transferred into
neural induction culture medium (NIM) for the following 3 or 4
days. EBs were then transferred into culture plates coated with
poly-D-lysine (PDL) and laminin for further adhesion culture with
EB culture medium using the following method: coating with a 100
μg/ml PDL solution on clean coverslips in 6-well plates
overnight at 37°C, then washing twice with water to remove PDL and
adding laminin solution (5 μg/ml in Hank’s media) for a few
hours to overnight, and rinsing once with Hank’s media prior to
use.
Twelve hours later, EB culture medium was replaced
with NIM and cultured for one week. NIM was a condition culture
medium which included Neurobasal Medium (21103-049) with
L-glutamine, NEAA, N2 (17502-048), B27 (17504-044) (Gibco), bFGF,
EGF and all-trans retinoic acid (R2625; Sigma) for another week.
bFGF and EGF were removed from NIM for subsequent culturing.
Samples of the original R1, CGR8.8, IP14D-1 lines,
derived neuronal cells (R1_Ne, CGR8.8_Ne and IP14D-1_Ne), and
primary cultured neurons as positive control were harvested in
TRIzol for RNA isolation. Reverse transcription of 0.5 μg
total RNA produced cDNA for PCR of Pax6, Sox1,
Blbp, Nestin, Tuj1, Ncam, Map2,
which test whether the pluripotent stem cells differentiate into
neurons. All PCR primers used are listed in Table I.
 | Table I.Primers for pluripotent stem cell
directed neuronal differentiation. |
Table I.
Primers for pluripotent stem cell
directed neuronal differentiation.
| Forward primer | Reverse primer | Size (bp) | Cycles | Parameters |
|---|
| Pax6 |
5′-GAAATCCGAGACAGATTATTATCCGAG-3′ |
5′-CCATTTGGCCCTTCGATTAGA-3′ | 495 | 35 | 94°C-40 sec;
58°C-30 sec; 72°C-40 sec |
| Sox1 |
5′-CCAAGAGACTGCGCGCGCTG-3′ |
5′-GGGTGCGCCGGGTGTGCGTG-3′ | 381 | 35 | 94°C-40 sec;
62°C-30 sec; 72°C-40 sec |
| Blbp |
5′-TGAGTACATGAAAGCTCTGGGCGT-3′ |
5′-TGAGCTTGTCTCCATCCAACCGAA-3′ | 224 | 35 | 94°C-40 sec;
58°C-30 sec; 72°C-30 sec |
| Nestin |
5′-CTGGAACAGAGATTGGAAGGCCGCT-3′ |
5′-GGATCCTGTGTCTTCAGAAAGGCTGTCAC-3′ | 403 | 35 | 94°C-40 sec;
59°C-30 sec; 72°C-40 sec |
| Tuj1 |
5′-ATCCACCTTCATTGGCAACAGCAC-3′ |
5′-ACTCGGACACCAGGTCATTCATGT-3′ | 173 | 35 | 94°C-40 sec;
58°C-30 sec; 72°C-30 sec |
| Ncam |
5′-TTCCTGTGTCAAGTGGCAGGAGAT-3′ |
5′-AGATCTTCACGTTGACAGTGGCCT-3′ | 229 | 35 | 94°C-40 sec;
60°C-30 sec; 72°C-30 sec |
| Map2 |
5′-AGCCGCAACGCCAATGGATT-3′ |
5′-TTTGTTCCGAGGCTGGCGAT-3′ | 313 | 35 | 94°C-40 sec;
58°C-30 sec; 72°C-40 sec |
| Gapdh |
5′-GCAAATTCAACGGCACAGTC-3′ |
5′-TCTTCTGGGTGGCAGTGATG-3′ | 399 | 35 | 94°C-40 sec;
59°C-30 sec; 72°C-40 sec |
Immunocytochemistry confirmation
Mouse pluripotent stem cells grown on coverslips
were washed with PBS twice and fixed with 4% paraformaldehyde for
15 min at room temperature. Cells were washed again with PBS and
permeabilized with 0.5% Triton X-100 in PBS for 15 min. The cells
were then blocked with 1% BSA, 0.5% Triton X-100 in PBS for 1 h at
room temperature. Mouse anti-mouse MAP2 primary antibodies were
diluted 1:200 in 1% BSA, 0.5% Triton X-100 in PBS and incubated
with fixed cells at 4°C overnight. After triple washing with PBS,
cells were incubated with Alexa Fluor 488 goat anti-mouse IgG
(1:1,000) secondary antibody for 1 h at room temperature. DAPI
counter-staining was used for cell nuclei.
Microarray assays of gene
expression
Total RNA was extracted from three replicates of
each cell population, including primary cultured neurons, three
pluripotent stem cells and corresponding neuron populations. cDNA
was hybridized to Affymetrix Mouse Gene 1.0 ST Arrays following
reverse transcription, in vitro transcription amplification
and quality control using the manufacturer’s standard
protocols.
Single cell PCR
Single cell isolation and cDNA
synthesis
A micromanipulator was employed to transfer single
cells into individual RNase-free EP tubes with 12 μl reverse
transcriptase mixture on ice. Reverse transcription reactions were
immediately performed to synthesize cDNA. Glycogen, ammonium
acetate and cold ethanol were added and stored at −80°C overnight
for precipitation.
In vitro transcription and cDNA
synthesis
Single-cell cDNA was amplified by in vitro
transcription (8,9), and the resulting aRNA was isolated
using high-speed low-temperature centrifugal sedimentation. The
amplified RNA was converted to single-stranded cDNA for PCR assays
of gene expression. PCR conditions were as described above.
Microarray data analysis
We applied the RMA algorithm in Affymetrix
Expression Console with default parameters to normalize and
summarize probe signals. We utilized the online NIA Array Analysis
Tool (http://lgsun.grc.nia.nih.gov/ANOVA/index.html) for
hierarchical clustering and statistical testing (6,10,11). For identification of significantly
different gene expression levels between pluripotent cell
populations and differentiated cells, we set cutoff thresholds at
5% false discovery rate (FDR) and 2-fold magnitude of difference
and conducted pairwise comparisons using ANOVA with multiple
testing correction (6,10,11). Finally, the DAVID online database
and tools (http://david.abcc.ncifcrf.gov/) were employed to
annotate the differentially expressed genes by Gene Ontology
categories, and affected pathways were examined using Ingenuity
Pathway Analysis (http://www.ingenuity.com/products/pathways_analysis.html).
Results
Pluripotent stem cell lines remain
viable with shared culture conditions
The ES cell line R1 was established in 1991 from a
blastocyst produced by crossing two 129 substrains (129S1/SvImJ and
129X1/SvJ) (12). R1 cells were
cultured on inactivated MEF cells as recommended by the ATCC. The
CGR8.8 mouse ES cell line was derived from the 129/Ola mouse
strain. The induced pluripotent stem cell line IP14D-1 was induced
from C57/DBA2F1 MEFs and can produce embryos by tetraploid
complementation as previously reported (6). All three pluripotent stem cell lines
were cultured using the same conditions before testing the
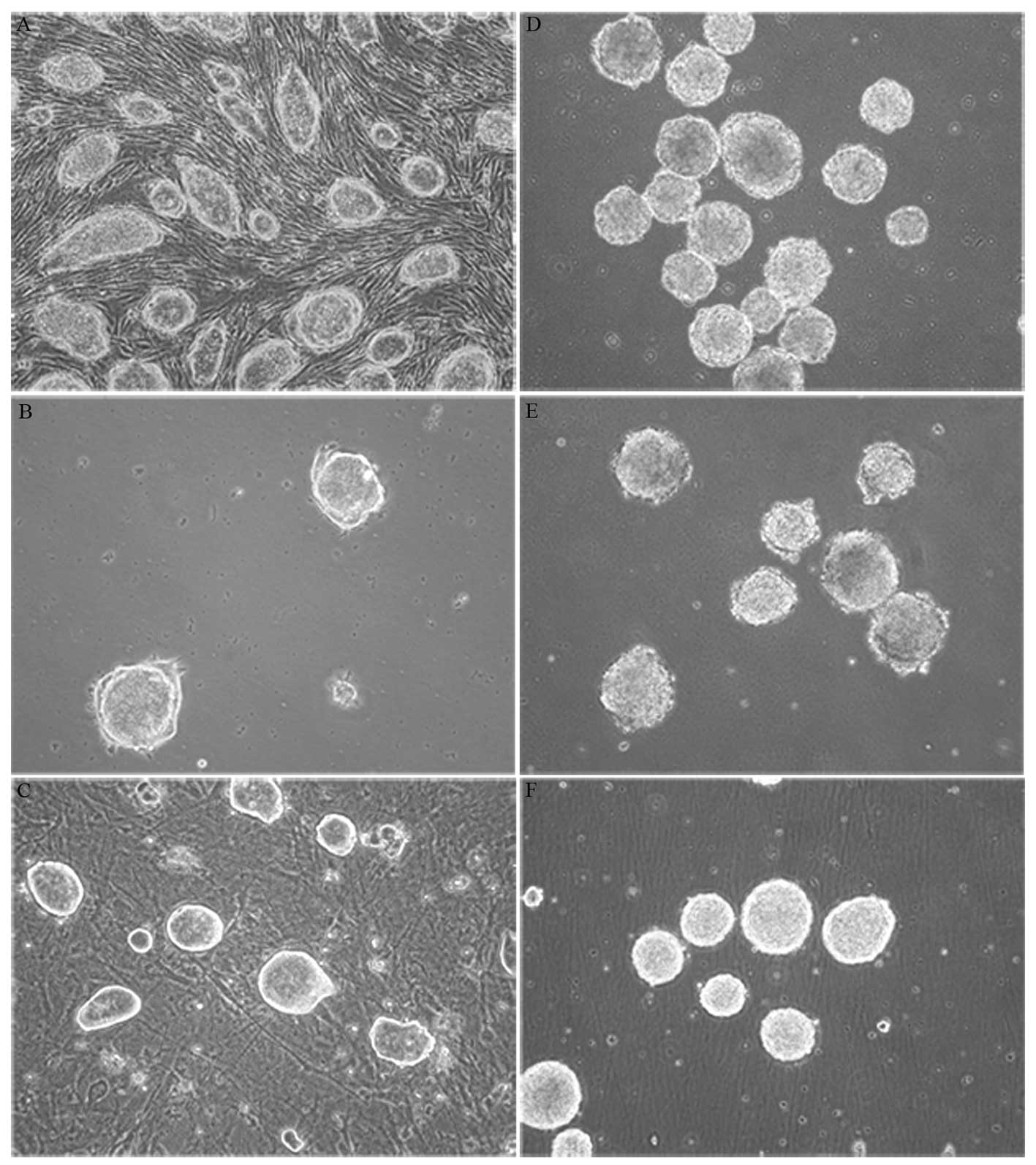
differentiation procedure. Each showed classic stem cell
characteristics such as colony growth with rapid proliferation,
smooth edges and strong refraction. There were no obvious cell
boundaries within stem cell colonies (Fig. 1A–C). Passages 20, 21 and 28 of R1,
CGR8.8 and IP14D-1, respectively, were employed for neuronal
differentiation.
Conditioned NIM is beneficial for
neural induction from EBs
EB formation is the first stage of differentiation
and allows pluripotent stem cells to be primed for further lineage
specific development. We adapted a standard neural differentiation
method with slight modifications to produce neurons via EB
formation. EBs formed after 4 days of suspension culture and then
0.5 μM ATRA was added into NIM for another 4 days of
suspension culture (Fig. 1D–F).
We used a modified ‘4+4’ induction method developed by Bain et
al (2). In the second 4 days
of EB suspension culture, we replaced EB medium with Neurobasal
containing some supplements, growth factors and RA without serum,
which was more suitable for promoting neuronal differentiation as
serum may have a negative effect on neural induction via RA
(13).
Pluripotent stem cells are induced
into MAP2 positive neurons
Following suspension culture induction, EBs were
transferred into PDL and laminin co-coated plates and cultured with
EB culture medium. After 12 h of adhesion, numerous EB cells
proliferated and extended from EBs. A week later, cells were
cultured in serum-free culture conditioned medium (NIM) for
neuronal differentiation. Total EB extension occurred after 2
weeks, and cells showed neuron-like morphology (Fig. 2A).
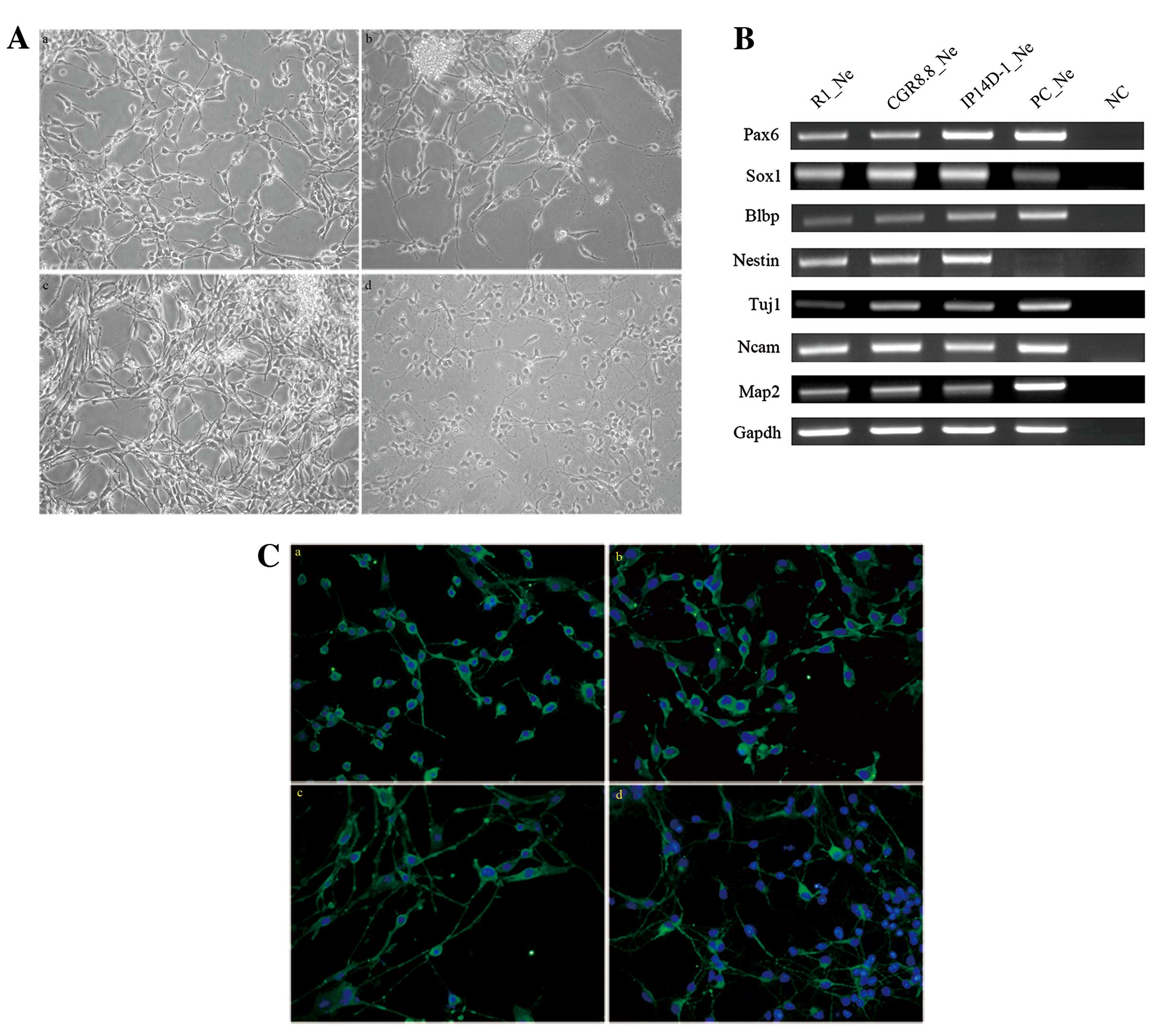
Neurons derived from each of the three pluripotent
stem cell lines were harvested to test for neuronal gene expression
patterns. Several genes specifically expressed in neurons and
during neural development processes were also expressed by the cell
populations differentiated from all three pluripotent cell lines,
such as Blbp (Fabp7), Nestin, Tuj1 and
Map2 (Fig. 2B). MAP2
protein is a marker for mature neurons, and the majority of cells
(~75%) from all three populations are positive for MAP2 antigen
expression (Fig. 2C).
Global gene expression profiles of
cells derived from mouse ESCs and iPSCs are similar
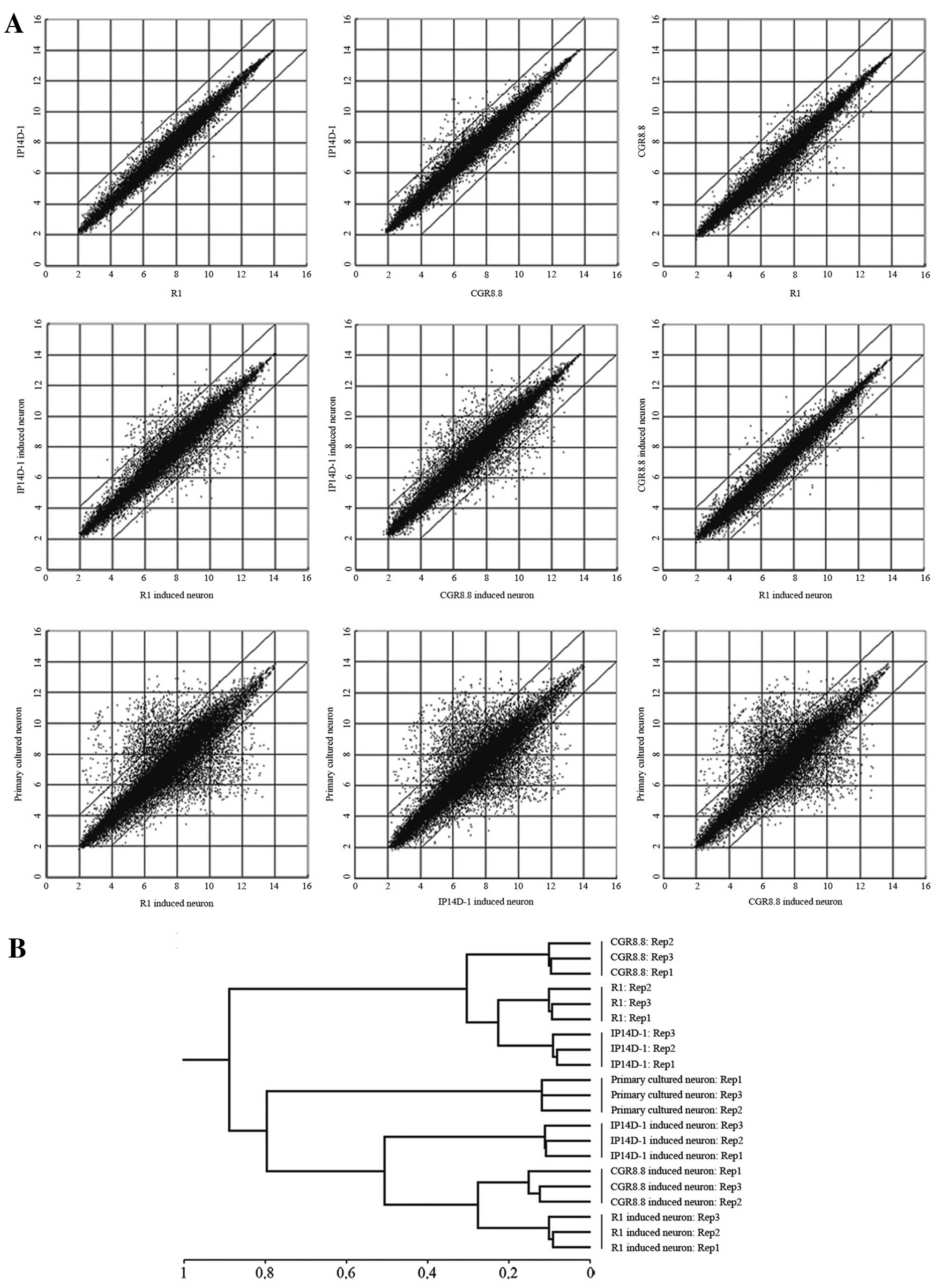
Microarrays were employed to analyze directed neural
differentiation of ESCs and iPSCs at a global RNA expression level
(Fig. 3A). Gene expression
profiles of induced neurons from ES and iPS cells are nearly
identical, particularly between derivatives of the two ES cell
populations. However, there are some differences between induced
neurons from ESCs and iPSCs, despite the confirmation that the iPS
cell line IP14D-1 has a developmental capacity equivalent to ES
cells. Moreover, the expression profiles of all these induced
neurons are to some extent not consistent with primary cultured
neurons. Hierarchical clustering of expression patterns grouped the
three induced neuron populations as more related to each other than
to primary cultured neurons, but all neurons were distinct from the
precursor pluripotent stem cells (Fig. 3B).
Validation of microarray data
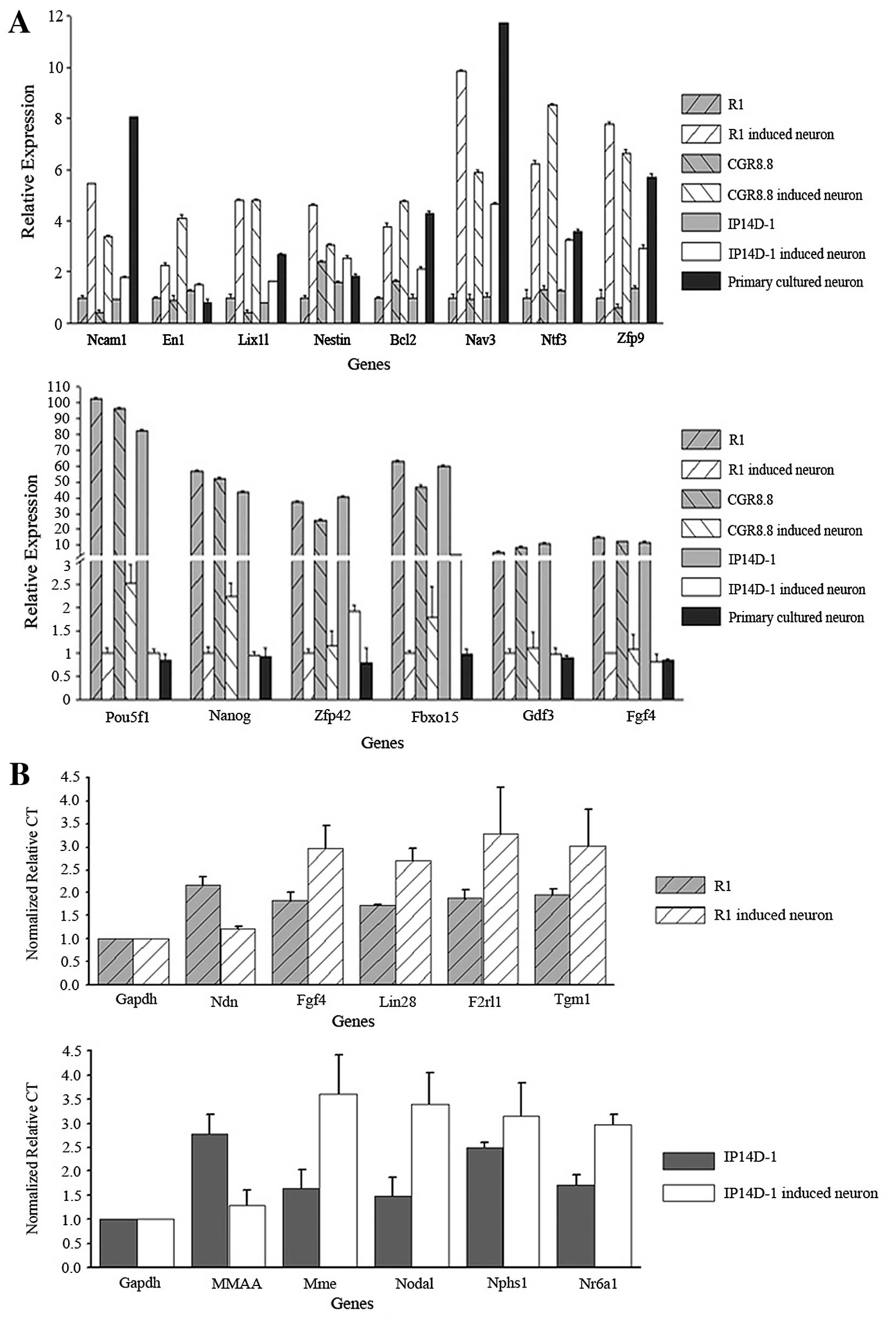
We selected differentially expressed candidates from
the microarray data for genes related to neurogenesis and
pluripotency. Ncam (neural cell adhesion molecule),
En1 and Lix1l were selected as neuron marker genes,
and Nestin, Bcl2, Nav3, Ntf3 and
Zfp9 were selected as genes related to neural
differentiation. Each of the neurogenesis-related genes was
upregulated during direct neural differentiation as assayed by
microarrays and quantitative PCR. RNA expression differences for
the neuron marker genes were also confirmed. Additionally, the
three well-known pluripotency genes Oct4, Nanog and
Klf4 were downregulated during the differentiation process
(Fig. 4A).
We also used single cell PCR to validate gene
expression levels in individual neurons derived both from R1 and
IP14D-1. We randomly chose five genes from the microarray candidate
lists for each cell population, as well as selected genes with
known relevance in pluripotency or neural differentiation.
Expression of genes involved in pluripotency maintenance, such as
Fgf4 and Lin28, decreased during neural
differentiation. Genes involved in neural lineage development,
including Ndn and Nr6a1, were confirmed to be
upregulated (Fig. 4B).
Differential expression between ES/iPS
and derived neurons
At an FDR <5% and a difference threshold greater
than 2-fold, almost 5,000 differentially expressed genes were
identified by pairwise comparisons between pluripotent stem cells
and their corresponding induced neurons. There are 1,126
differentially expressed genes common to all three pairwise
comparisons (Table II), including
824 upregulated genes and 302 downregulated genes during neural
lineage development.
 | Table II.Number of differentially expressed
genes detected after induced neural differentiation. |
Table II.
Number of differentially expressed
genes detected after induced neural differentiation.
| Pairwise
comparisons | Upregulated genes
(pluripotent > neurons) | Downregulated genes
(pluripotent < neurons) |
|---|
| IP14D-1 vs.
IP14D-1_neuron | 2,278 | 2,103 |
| IP14D-1 vs. primary
neuron | 4,574 | 3,684 |
| Shared set | 1,634 | 613 |
| R1 vs.
R1_neuron | 2,684 | 2,707 |
| R1 vs. primary
neuron | 4,033 | 3,778 |
| Shared set | 1,829 | 962 |
| CGR8.8 vs.
CGR8.8_neuron | 2,599 | 2,378 |
| CGR8.8 vs. primary
neuron | 4,099 | 3,680 |
| Shared set | 1,791 | 934 |
| Common among 3
inductions | 824 | 302 |
| Fold-change >2,
FDR <0.05 | | |
Annotation analysis of regulated genes
in ES/iPS cell differentiation
We investigated functional categories within sets of
differentially expressed genes using Ingenuity Pathway Analysis
tools. Among the 1,126 differentially expressed genes, nearly 700
can be classified into annotated categories that include
transcription regulators, growth factors and ion channel formation
(Table III). We also tested for
statistically significant overrepresentation of Gene Ontology (GO)
categories, and classified 302 upregulated genes into biological
processes involved in synaptic transmission, regulation of membrane
potential, axonogenesis, and neuron recognition, as well as basic
functions such as transcription, metabolism and biosynthesis
(Table IV). We ranked the GO
categories according to overrepresentation P-value, and with the
exception of some basic biological processes, the most important
categories involved in neural development are focused on neuron
projection morphogenesis and axonogenesis.
 | Table III.Molecular function categories for
differentially expressed genes. |
Table III.
Molecular function categories for
differentially expressed genes.
| Category | No. of mapped genes
overexpressed in pluripotent cell vs. neurons | No. of mapped genes
underexpressed in pluripotent cell vs. neurons | Sum |
|---|
| Enzyme | 1190 | 35 | 154 |
| Transcription
regulator | 67 | 26 | 93 |
| Kinase | 23 | 22 | 45 |
| Transporter | 28 | 13 | 41 |
| Peptidase | 12 | 10 | 22 |
| Phosphatase | 8 | 6 | 14 |
| Growth factor | 8 | 3 | 11 |
| Transmembrane
receptor | 5 | 4 | 9 |
| Ion channel | 1 | 7 | 8 |
| Ligand-dependent
nuclear receptor | 3 | 4 | 7 |
| Translation
regulator | 5 | 0 | 5 |
| G-protein coupled
receptor | 1 | 3 | 4 |
| Other | 271 | 113 | 384 |
| Sum | 551 | 246 | 797 |
 | Table IV.Gene ontology analysis of upregulated
genes. |
Table IV.
Gene ontology analysis of upregulated
genes.
| Category | Biological
process | Count | P-value | Fold
enrichment |
|---|
| Transcription
related | Positive regulation
of transcription, DNA-dependent | 19 | 2.8E-05 | 3.2 |
| Positive regulation
of transcription from RNA polymerase II promoter | 17 | 5.6E-05 | 3.3 |
| Regulation of
transcription from RNA polymerase II promoter | 22 | 1.9E-04 | 2.5 |
| Regulation of
transcription, DNA-dependent | 35 | 1.8E-03 | 1.7 |
| Regulation of
transcription | 48 | 2.0E-03 | 1.5 |
| Metabolism
related | Positive regulation
of macromolecule metabolic process | 27 | 9.8E-07 | 3.0 |
| Positive regulation
of RNA metabolic process | 19 | 3.1E-05 | 3.2 |
| Phosphorus
metabolic process | 25 | 1.3E-03 | 2.0 |
| Regulation of RNA
metabolic process | 35 | 2.3E-03 | 1.7 |
| Regulation of
cellular protein metabolic process | 9 | 4.4E-02 | 2.3 |
| Biosynthesis
related | Positive regulation
of macromolecule biosynthetic process | 22 | 2.1E-05 | 2.9 |
| Positive regulation
of cellular biosynthetic process | 22 | 3.8E-05 | 2.8 |
| Positive regulation
of biosynthetic process | 22 | 4.2E-05 | 2.8 |
| Cell adhesion
related | Cell adhesion | 22 | 4.9E-05 | 2.7 |
| Homophilic cell
adhesion | 8 | 1.5E-03 | 4.7 |
| Cell-cell
adhesion | 11 | 2.0E-03 | 3.3 |
| Regulation of
cell-matrix adhesion | 3 | 1.7E-02 | 14.9 |
| Phosphorylation
related | Regulation of
phosphorylation | 14 | 2.5E-04 | 3.4 |
| Protein amino acid
phosphorylation | 22 | 3.1E-04 | 2.4 |
|
Phosphorylation | 22 | 1.3E-03 | 2.1 |
| Regulation of
protein amino acid phosphorylation | 7 | 7.0E-03 | 4.1 |
| Ion homeostasis
related | Ion
homeostasis | 13 | 9.2E-04 | 3.1 |
| Cellular calcium
ion homeostasis | 6 | 9.9E-03 | 4.6 |
| Homeostatic
process | 17 | 8.4E-03 | 2.1 |
| Cellular ion
homeostasis | 11 | 3.9E-03 | 3.0 |
| Cellular chemical
homeostasis | 11 | 4.7E-03 | 2.9 |
| Cation
homeostasis | 8 | 1.5E-02 | 3.1 |
| Di-, tri-valent
inorganic cation homeostasis | 7 | 1.6E-02 | 3.4 |
| Regulation of metal
ion transport | 4 | 3.1E-02 | 5.8 |
| Regulation of
sodium ion transport | 3 | 1.9E-02 | 13.9 |
| Cell morphogenesis
related | Neuron projection
morphogenesis | 13 | 7.7E-06 | 5.2 |
| Cell
morphogenesis | 16 | 3.5E-05 | 3.6 |
| Cell morphogenesis
involved in neuron differentiation | 12 | 5.7E-05 | 4.6 |
| Regulation of
neuron projection development | 4 | 3.0E-02 | 5.9 |
| Cell motion
related | Cell motion | 20 | 1.1E-06 | 3.8 |
| Cell migration | 14 | 4.0E-05 | 4.1 |
| Axon guidance | 6 | 1.3E-02 | 4.2 |
| Neural crest cell
migration | 3 | 4.2E-02 | 9.1 |
| Synaptic
transmission related | Regulation of
membrane potential | 7 | 7.3E-03 | 4.1 |
| Synaptic
transmission | 8 | 1.4E-02 | 3.2 |
| Transmission of
nerve impulse | 9 | 1.6E-02 | 2.8 |
| Regulation of
postsynaptic membrane potential | 3 | 4.9E-02 | 8.3 |
| Neural development
related | Axonogenesis | 12 | 2.0E-05 | 5.2 |
| Neuron
recognition | 4 | 5.8E-04 | 23.1 |
| Regulation of
neurogenesis | 9 | 5.9E-04 | 4.8 |
| Neuron
development | 13 | 9.2E-04 | 3.1 |
| Regulation of
nervous system development | 9 | 1.3E-03 | 4.2 |
| Axonal
fasciculation | 3 | 4.1E-03 | 29.7 |
| Neural crest cell
development | 4 | 1.2E-02 | 8.4 |
| Negative regulation
of neurogenesis | 4 | 1.6E-02 | 7.5 |
| Regulation of
axonogenesis | 4 | 1.8E-02 | 7.1 |
| Negative regulation
of axonogenesis | 3 | 2.1E-02 | 13.0 |
Discussion
During the last decade, mouse and human ES cells
have been induced to differentiate into several cell types to study
developmental potential in vitro and to develop valuable
therapeutics (1,14,15). Diseases of the nervous system have
a serious impact on human health, therefore, a number of methods
have been adapted to induce ESCs into neurons with the aim of
curing disease (1,16). We used a classic induction method
with slight modifications to cause mouse ES and iPS cells to
differentiate into neurons. RA, an efficient induction factor for
neural development, was added to the EB culture medium to promote
EBs to differentiate into the neural lineage as suggested by
previous in vitro research (17,18). With a low RA concentration in
culture, pluripotent cells, both ES and EC cells, could
differentiate into neurons (19).
RA is particularly critical for GABAergic neuron differentiation of
ESCs (3,20). To maintain cellular proliferation
and promote expansion from EBs, we pretreated cell culture dishes
with PDL and laminin at least 2 h (or, optimally, overnight) at
37°C. Both substances promote efficient cell adhesion and
stretching (21). We added growth
factors into the neural induction medium to increase cell
proliferation and directed differentiation (22,23). These factors were previously shown
to protect induced neurons by promoting resistance to apoptosis and
necrosis (24), perhaps via
regulation of genes such as Bcl-2 expressed during the
neural directed differentiation process (25).
By comparing RNA expression levels among induced
neurons from pluripotent stem cells and to primary cultured neurons
isolated from ICR E13.5 mice, we found ES and iPS cells follow
almost the same neural differentiation process, and the derived
neurons have marker gene expression patterns identical to primary
cultured neurons.
This level of similarity extended to genomic
expression patterns from microarray assays. Neurons derived from
all three types of pluripotent cells had mostly overlapping gene
expression by cluster analysis, and the differences between these
patterns and that of primary cultured neurons is likely due to a
heterogeneous mixture of cell types in the induced populations. The
number of differentiated cells never exceeded 75%, so samples taken
from induced cell populations contained a mixture of neuron and
non-neuron RNA.
As an independent validation of the microarray
results, we used quantitative RT-PCR of several well-known
pluripotency and neuron-relevant genes. As expected, pluripotency
genes were downregulated and neural genes were upregulated during
differentiation. We also employed single cell PCR in the validation
tests, which has become a relatively accurate technology for
assessing gene expression (26–28). Primary cultured neurons served as
positive control. Genes selected for the PCR assays showed
expression patterns consistent with the microarray results. After
confirming the concordance of selected microarray and qPCR assays,
we conducted a statistical analysis on the full microarray data set
to test for gene expression differences between ES/iPS cells and
their derived neurons. We identified 824 genes highly expressed in
pluripotent cells and 302 genes highly expressed in both induced
neurons and primary cultured neurons. The resulting lists of
candidate genes were used for pathway and gene ontology analyses
that highlighted several biological processes apparently operating
during induced neural differentiation. The cadherin super-family
contains calcium-dependent cell adhesion molecules including
protocadherins (Pcdh) that are important regulators of mouse
and human nervous system development (29–32). The Pcdh family is involved
in maintenance of spinal interneurons, axon convergence and
synaptic development, particularly connectivity between neuronal
cells (29). In the present
study, we detected increased expression of several members of this
family including Cdh2, Cdh10, Pcdh7, Pcdh16, Pcdh18 and
Pcdh22. The solute carrier gene family (Slc) encodes
another group of key proteins in neural development for maintaining
neuron functions such as ion transport, glutamate transport, and
neurotransmitter symporters. Genetic variants of Slc9a9
affect the development of attention-deficit/hyperactivity disorder,
a common behavioral disorder with over-activity and inattentiveness
(33–35). This gene, therefore, may play a
part in nervous system development, and we observed more than
2-fold upregulation during neuron differentiation.
We also detected differential expression of
Sema family genes including 3a, 3d, 4c and 6d which have
roles in nervous system development (36). The Sema3a and Npn1
genes were upregulated during the directed neuronal differentiation
process. Both genes are involved in a neural development regulatory
pathway for motor and sensory axon outgrowth (37). microRNA let-7 is known to promote
neural lineage differentiation (38,39) and to suppress expression of
several pluripotency genes (40).
The precursor transcript for let-7 was upregulated 20-fold on
average during induced neural differentiation.
Our analysis of in vitro directed neuronal
differentiation indicates that iPSCs follow almost the same
differentiation process as mouse ESCs. Neurons induced from iPSCs
and ESCs have similar morphology, neuron marker expression, and
global gene expression patterns. Several genes related to known
neuronal differentiation processes showed statistically significant
changes in expression, and these patterns may be useful for
optimizing induction methods, improving the efficiency of neural
differentiation from pluripotent stem cells, and understanding
neuronal differentiation mechanisms underlying nervous system
development.
Acknowledgements
This study was supported by grants
from the National Natural Science Foundation of China (81125003,
30900259), the Hi-Tech Research and Development Program of China
(2011AA020116), the China National Basic Research Program
(2010CB945200), the Science and Technology Committee of Shanghai
Municipality (10140900200), and the Shanghai Leading Academic
Discipline Project (S30201) to FZ.
References
|
1.
|
Kim DW: Efficient induction of
dopaminergic neurons from embryonic stem cells for application to
Parkinson’s disease. Yonsei Med J. 45(Suppl): S23–S27. 2004.
|
|
2.
|
Bain G, Kitchens D, Yao M, Huettner JE and
Gottlieb DI: Embryonic stem cells express neuronal properties in
vitro. Dev Biol. 168:342–357. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Chatzi C, Scott RH, Pu J, et al:
Derivation of homogeneous GABAergic neurons from mouse embryonic
stem cells. Exp Neurol. 217:407–416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Ying QL, Stavridis M, Griffiths D, Li M
and Smith A: Conversion of embryonic stem cells into
neuroectodermal precursors in adherent monoculture. Nat Biotechnol.
21:183–186. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Kim DW, Chung S, Hwang M, et al: Stromal
cell-derived inducing activity, Nurr1, and signaling molecules
synergistically induce dopaminergic neurons from mouse embryonic
stem cells. Stem Cells. 24:557–567. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Zhao XY, Li W, Lv Z, et al: iPS cells
produce viable mice through tetraploid complementation. Nature.
461:86–90. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Zhao XY, Lv Z, Li W, Zeng F and Zhou Q:
Production of mice using iPS cells and tetraploid complementation.
Nat Protoc. 5:963–971. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Sul JY, Wu CW, Zeng F, et al:
Transcriptome transfer produces a predictable cellular phenotype.
Proc Natl Acad Sci USA. 106:7624–7629. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Van Gelder RN, von Zastrow ME, Yool A,
Dement WC, Barchas JD and Eberwine JH: Amplified RNA synthesized
from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci
USA. 87:1663–1667. 1990.PubMed/NCBI
|
|
10.
|
Zeng F, Baldwin DA and Schultz RM:
Transcript profiling during preimplantation mouse development. Dev
Biol. 272:483–496. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Zeng F and Schultz RM: RNA transcript
profiling during zygotic gene activation in the preimplantation
mouse embryo. Dev Biol. 283:40–57. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Nagy A, Rossant J, Nagy R, Abramow-Newerly
W and Roder JC: Derivation of completely cell culture-derived mice
from early-passage embryonic stem cells. Proc Natl Acad Sci USA.
90:8424–8428. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Pachernik J, Bryja V, Esner M, Kubala L,
Dvorak P and Hampl A: Neural differentiation of pluripotent mouse
embryonal carcinoma cells by retinoic acid: inhibitory effect of
serum. Physiol Res. 54:115–122. 2005.PubMed/NCBI
|
|
14.
|
Caspi O, Huber I, Kehat I, et al:
Transplantation of human embryonic stem cell-derived cardiomyocytes
improves myocar-dial performance in infarcted rat hearts. J Am Coll
Cardiol. 50:1884–1893. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Laflamme MA, Chen KY, Naumova AV, et al:
Cardiomyocytes derived from human embryonic stem cells in
pro-survival factors enhance function of infarcted rat hearts. Nat
Biotechnol. 25:1015–1024. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Swistowski A, Peng J, Liu Q, et al:
Efficient generation of functional dopaminergic neurons from human
induced pluripotent stem cells under defined conditions. Stem
Cells. 28:1893–1904. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Maden M: Retinoic acid in the development,
regeneration and maintenance of the nervous system. Nat Rev
Neurosci. 8:755–765. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Lu J, Tan L, Li P, et al: All-trans
retinoic acid promotes neural lineage entry by pluripotent
embryonic stem cells via multiple pathways. BMC Cell Biol.
10:572009. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Okada Y, Shimazaki T, Sobue G and Okano H:
Retinoic-acid-concentration-dependent acquisition of neural cell
identity during in vitro differentiation of mouse embryonic stem
cells. Dev Biol. 275:124–142. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Addae C, Yi X, Gernapudi R, Cheng H, Musto
A and Martinez-Ceballos E: All-trans-retinoid acid induces the
differentiation of encapsulated mouse embryonic stem cells into
GABAergic neurons. Differentiation. 83:233–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Ma W, Tavakoli T, Derby E, Serebryakova Y,
Rao MS and Mattson MP: Cell-extracellular matrix interactions
regulate neural differentiation of human embryonic stem cells. BMC
Dev Biol. 8:902008. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Ratzka A, Baron O, Stachowiak MK and
Grothe C: Fibroblast growth factor 2 regulates dopaminergic neuron
development in vivo. J Neurochem. 122:94–105. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Schwindt TT, Motta FL, Gabriela FB, et al:
Effects of FGF-2 and EGF removal on the differentiation of mouse
neural precursor cells. An Acad Bras Cienc. 81:443–452. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Zucchini S, Buzzi A, Barbieri M, et al:
Fgf-2 overexpression increases excitability and seizure
susceptibility but decreases seizure-induced cell loss. J Neurosci.
28:13112–13124. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Bernier PJ and Parent A: Bcl-2 protein as
a marker of neuronal immaturity in postnatal primate brain. J
Neurosci. 18:2486–2497. 1998.PubMed/NCBI
|
|
26.
|
Esumi S, Wu SX, Yanagawa Y, Obata K,
Sugimoto Y and Tamamaki N: Method for single-cell microarray
analysis and application to gene-expression profiling of GABAergic
neuron progenitors. Neurosci Res. 60:439–451. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Bontoux N, Dauphinot L, Vitalis T, et al:
Integrating whole transcriptome assays on a lab-on-a-chip for
single cell gene profiling. Lab Chip. 8:443–450. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Takayama J, Faumont S, Kunitomo H, Lockery
SR and Iino Y: Single-cell transcriptional analysis of taste
sensory neuron pair in Caenorhabditis elegans. Nucleic Acids
Res. 38:131–142. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Junghans D, Heidenreich M, Hack I, Taylor
V, Frotscher M and Kemler R: Postsynaptic and differential
localization to neuronal subtypes of protocadherin beta16 in the
mammalian central nervous system. Eur J Neurosci. 27:559–571. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Yagi T: Clustered protocadherin family.
Dev Growth Differ. 50(Suppl 1): S131–S140. 2008. View Article : Google Scholar
|
|
31.
|
Han MH, Lin C, Meng S and Wang X:
Proteomics analysis reveals overlapping functions of clustered
protocadherins. Mol Cell Proteomics. 9:71–83. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Garrett AM and Weiner JA: Control of CNS
synapse development by {gamma}-protocadherin-mediated
astrocyte-neuron contact. J Neurosci. 29:11723–11731. 2009.
|
|
33.
|
Markunas CA, Quinn KS, Collins AL, et al:
Genetic variants in SLC9A9 are associated with measures of
attention-deficit/hyperactivity disorder symptoms in families.
Psychiatr Genet. 20:73–81. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Lasky-Su J, Anney RJ, Neale BM, et al:
Genome-wide association scan of the time to onset of attention
deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr
Genet. 147B:1355–1358. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Franke B, Neale BM and Faraone SV:
Genome-wide association studies in ADHD. Hum Genet. 126:13–50.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Mauti O, Domanitskaya E, Andermatt I,
Sadhu R and Stoeckli ET: Semaphorin6A acts as a gate keeper between
the central and the peripheral nervous system. Neural Dev.
2:282007. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Ben-Zvi A, Manor O, Schachner M, Yaron A,
Tessier-Lavigne M and Behar O: The Semaphorin receptor PlexinA3
mediates neuronal apoptosis during dorsal root ganglia development.
J Neurosci. 28:12427–12432. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Rybak A, Fuchs H, Smirnova L, et al: A
feedback loop comprising lin-28 and let-7 controls pre-let-7
maturation during neural stem-cell commitment. Nat Cell Biol.
10:987–993. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Zhao C, Sun G, Li S, et al: MicroRNA
let-7b regulates neural stem cell proliferation and differentiation
by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci
USA. 107:1876–1881. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Zhao C, Huang C, Weng T, Xiao X, Ma H and
Liu L: Computational prediction of MicroRNAs targeting GABA
receptors and experimental verification of miR-181, miR-216 and
miR-203 targets in GABA-A receptor. BMC Res Notes. 5:912012.
View Article : Google Scholar : PubMed/NCBI
|