Introduction
Age-related thymic involution is manifested by a
progressive reduction in the size of the thymus with profound
architectural changes, the loss of thymic epithelial cells (TECs)
and the replacement of the thymic stroma with adipose tissue and
peripheral lymphocytes (1,2).
The atrophic thymus produces a reduced number of naïve T cells with
a diminished diversity of T cell receptors (3). Age-related thymic involution is
attributed to one of the main causes of immunosenescence: the
deterioration of immunocompetence in the elderly that is
characteristic of increased susceptibility to infections,
insufficient response to vaccinations and an increased propensity
for autoimmune diseases and cancer (4–6).
The development of T cells requires the importation
of T cell progenitors to the thymus from bone marrow (BM). The
early T cell progenitors (ETPs) residing in the
CD4−CD8− double negative 1 (DN1)
subpopulation proliferate and differentiate in response to various
thymic stromal signals. Following negative and positive selection,
the majority of thymocytes in the thymus undergo apoptosis. Only a
very small percentage of thymocytes differentiate into mature
CD4+ or CD8+ naïve T cells and are exported
to the periphery. Age-related thymic involution is accompanied by a
reduced number and proliferation of ETPs, as well as by the marked
deterioration of the thymic stroma. The change in the
microenvironment of the thymus, particularly the increased
apoptosis and decreased proliferation of TECs, is considered to be
the determining cause for defective T cell development in the aged
thymus (7–9).
The perturbation of gene expression levels in the
thymus can influence the process of thymic involution. For example,
the overexpression of the anti-apoptotic gene, myeloid cell
leukemia sequence 1 (MCL1), leads to an enlarged thymus in female
mice approaching advanced age (10). Targeting Ras homolog family member
B (RhoB), a member of the Rho subfamily of small GTPases, in the
thymic medullary epithelium results in an early thymic atrophy
(11). While the trigger of
thymic aging remains a mystery, the extensive pursuit of this topic
has revealed that aging is associated with changes in the
expression levels of several genes in thymocytes, including the E2A
and LIM domain only 2 (rhombotin-like 1) (LMO2) transcriptional
regulators (12). Gene array
analysis of aged thymocytes has revealed age-associated changes in
the mRNA levels of several genes, including those involved in
oxidative phosphorylation, T and B cell receptor signaling and
antigen presentation. Of note, some of the cancer-related genes and
immunoglobulin genes, such as immunoglobulin M (IgM) are
upregulated in 24-month-old mice. This may account for the high
incidence of cancer and increased thymic B cells observed in aged
humans (13). Alterations in the
expression levels of several genes have also been found in the
thymic stroma. These genes include forkhead box N1 (FoxN1),
interleukin (IL)-7, keratin 8 (12–14), Wnt4 and lamina-associated
polypeptide 2α (LAP2α) (15).
MicroRNAs (miRNAs) are a family of short (average
size, 22 bp), non-coding RNAs found in eukaryotic cells that affect
gene regulation in a sequence-specific manner (16). Emerging evidence has revealed that
mutations or alterations in miRNA expression correlate with the
development of various types of human cancer (17), as well as other diseases (18–20). miRNAs are also involved in the
regulation of the immune response, including T cell differentiation
and sensitivity (21,22). For example, miR-181a is highly
expressed in the thymus and miR-155 is upregulated in mature T
cells, both of which are required for T cell differentiation and
cell-mediated immune function (23,24). Knowledge of miRNA expression
profiles has been gained through the detailed examination of miRNA
expression at each individual developmental stage of thymocytes in
the thymus by microarray and quantitative polymerase chain reaction
(PCR) (25,26). However, miRNA expression in the
thymic stromal compartment is completely unknown. A number of
studies have shown the changes in miRNA levels in various tissues
in individuals of different age groups, in which miRNAs have been
suggested to be novel modulators that regulate the aging process
(27). Determining miRNA
expression in the thymic stroma, as well as changes in expression
during thymic aging would not only broaden our knowledge of T cell
development but would also provide a deep understanding of the
mechanisms underlying thymic aging.
In this study, we compared the transcriptional
levels of various miRNAs in TECs from young and aged mice using
microarrays. Quantitative PCR was performed to confirm the changes
in the expression of miRNAs in different age groups. Possible
downstream targets and pathways of these miRNAs were predicted by
performing bioinformatics analysis. To the best of our knowledge,
this is the first study to systematically analyze the expression of
miRNAs in mouse TECs and to demonstrate that miRNA expression is
altered with thymic aging.
Materials and methods
Mice and enrichment of TECs
C57BL/6 mice of different ages up to 10 months were
purchased from Charles River Laboratories (Wilmington, MA, USA).
Some of the 10-month-old mice were maintained in the animal
facility of China Medical University, Shenyang, China until they
began to age (≥19 months). All of the mice were maintained in a
specific, pathogen-free environment. All animal procedures were
performed according to the protocol approved by the Experiment
Animal Center of China Medical University in accordance with the
recommendations in the Guide for the Care and Use of Laboratory
Animals of the National Institutes of Health. The TECs were
purified according to a previously described method (7). In brief, the thymic sub-capsule was
gently torn apart with fine-tip forceps and shaken several times in
cold phosphate-buffered saline (PBS) to remove as many thymocytes
as possible. The remaining thymic tissue was further subjected to
enzymatic digestion with 1 mg/ml collagenase (Invitrogen, Carlsbad,
CA, USA) and 5 U/ml DNase I (Sigma-Aldrich, USA) for 10 min at 37°C
with intermittent shaking. The thymic dissociated cells were
stained by fluorescence-conjugated anti-CD45 (rat antibody),
followed by metal beads conjugated to anti-rat IgG (Miltenyi
Biotec, Bergisch Gladbach, Germany). The bead-labeled cells were
subjected to LS magnetic columns to obtain TEC-enriched cells by
negative selection, which were used for miRNA microarray and
quantitative PCR assays.
miRNA microarray
The microarray assay was performed by the service
provider Kangchen Biotech, China. Total RNA from enriched TECs of
young (2-month-old) and aged (20-month-old) mice was extracted
using TRIzol reagent (Invitrogen) and the RNeasy Mini kit (Qiagen,
Hilden, Germany), according to the manufacturer’s instructions. A
total of 0.25–1 μg RNA sample was labeled Hy3 using the miRCURY™
Array Power Labeling kit (Exiqon, Aarhus, Denmark). Hybridization
was performed using the miRCURY™ LNA Array (miRBase.14.0). Signal
scanning was performed using the Axon GenePix 4000B microarray
scanner. GenePix pro v6.0 was used to read the raw intensity of the
images.
The intensity of the green signal was calculated
after background subtraction and 4 replicated spots of each probe
on the same slide were used to calculate the median. The median is
the 50% quantile of the miRNA intensity which is >50 in all
samples after background correction. The median normalization
method was used to obtain normalized data. Clustering and
statistical analysis were performed on the normalized data of
miRNAs.
Quantitative PCR
Total RNA (including small RNA) was extracted from
the TECs of young (1-month-old), young adults (6-month-old),
middle-aged (10-month-old) and aged (19-month-old) mice using the
RNeasy Mini kit (Qiagen). A poly(A) tail was added to 3′ end of
small RNA [E. coli Poly(A) Polymerase; New England Biolabs
(NEB), Ipswich, MA, USA] before the first-strand cDNA was
synthesized using the RT Reagent kit (Takara, Otsu, Japan). RNA
with no Poly(A) Polymerase was used as the internal control.
Instead of using a common oligo(dT) primer or a random primer, a
specific oligo(dT) primer with the following sequence was used:
5′-GCTGTCAACGATA CGCTACGTAACGGCATGACAGTG(T)24V-3′. cDNA (1 μl) was
used as the PCR template. The results were normalized to U6 snRNA
that was used as the internal reference (Ambion Inc., Foster City,
CA, USA). The forward primers for miRNAs are listed in Table I and every PCR reaction for miRNA
quantification shared the same reverse primer as follows:
5′-GCTGTCAACGATACGCTA CGTAACG-3′.
 | Table IPrimers of the 20 miRNAs used in
qPCR. |
Table I
Primers of the 20 miRNAs used in
qPCR.
| miRNA | Primer
sequence |
|---|
| miR-146a |
tgagaactgaattccatgggtt |
| miR-148b |
tcagtgcatcacagaactttgta |
| miR-150 |
ccaacccttgtaccagtgaaa |
| miR-154-3p |
cgaatcatacacggttgacctatt |
| miR-155 |
ttaatgctaattgtgatagggg |
| miR-181a |
cattcaacgctgtcggtgagt |
| miR-181b |
attcattgctgtcggtgggaa |
| miR-181c |
aacattcaacctgtcggtgagt |
| miR-192 |
gctgacctatgaattgacagcc |
| miR-194 |
tgtaacagcaactccatgtgga |
| miR-19a |
tgtgcaaatctatgcaaaactga |
| miR-19b |
gcaaatccatgcaaaactga |
| miR-22 |
aagctgccagttgaagaactgt |
| miR-24 |
tggctcagttcagcagga |
| miR-322 |
gcagcaattcatgttttgga |
| miR-382-3p |
ctctgtcattcacggacaaca |
| miR-431 |
caggccgtcatgcaaaa |
| miR-465a-3p |
gatcagggcctttctaagta |
| miR-93 |
aagtgctgttcgtgcaggtag |
| miR-96 |
tggcactagcacatttttgct |
Quantitative PCR was performed using the 7500 Real
Time PCR System (Applied Biosystems, Foster City, CA, USA) and the
SYBR-Green PCR master mix (Takara) with 40 cycles of 95°C for 15
sec and 62°C for 90 sec. Specificity of amplification was confirmed
by melting curve analyses.
Bioinformatics prediction of the targets
of miRNAs
The targets of the miRNAs were predicted using the
TargetScan, PicTar, miRanda and EIMMo databases. The predicted hits
from each algorithm were sorted as per the scores. The congruent
high score hits in all 4 of the algorithms were considered to be
reliable predictions.
Statistical analysis
A t-test was used for comparing the miRNA expression
levels in the 2- and 20-month-old groups. One-way analysis of
variance (ANOVA) was used for comparing the PCR results or the
weight of the thymus between the 1-, 6-, 10- and 19-month-old
groups. A linear correlation test was used for comparing miRNA
expression and changes in thymic weight change during aging. A
probability value (p-value) <0.05 was considered to indicate a
statistically significant difference. Each statistic and derived
figure was prepared using GraphPad Prism-5 software.
Results
Microarray data reveal the difference in
the expression levels of miRNAs in TECs from young and aged
thymus
To determine the age-related changes in miRNA
expression in thymic stromal tissue, we compared miRNA expression
in enriched TECs from the thymi of 2- and 20-month-old mice by
miRNA microarray analysis. Hierarchical clustering analysis
partitioned the samples into 2 groups, 2-month-old (young; sample 1
and 2) and 20-month-old (aged; sample 3 and 4), which suggested
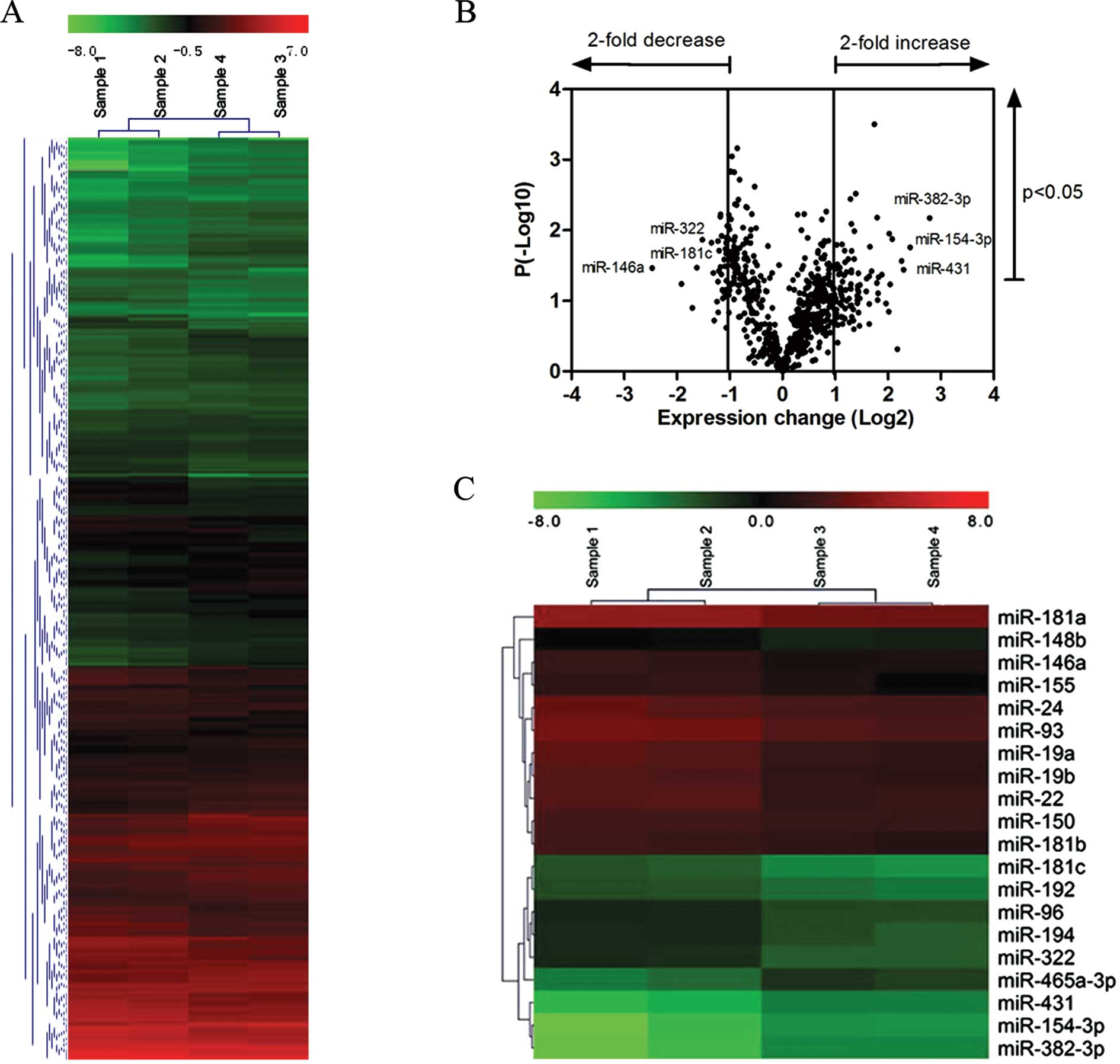
that each sample had a distinct miRNA expression profile (Fig. 1A). Among the 678 mouse miRNA
probes in the miRNA arrays, 111 (16.4%) miRNAs were expressed and
detected in the TECs from the 2 groups (fold change >2 or
<0.5). From the data analysis based on the p-values (t-test), 31
miRNAs were upregulated and 22 were downregulated significantly
(p<0.05) in the TECs from the aged thymus (Fig. 1B). The 20 miRNAs that had a
greater fold change in expression than the other 33 were selected
for quantification by quantitative PCR (Fig. 1C). Among the 20 miRNAs that were
analyzed, 4 (miR-382-3p, miR-154-3p, miR-431 and miR-465a-3p) were
upregulated in the aged TECs, whereas the other 16 miRNAs were
downregulated (Table II). The
upregulated miRNAs as measured in the microarray assay increased by
at least 4-fold and the downregulated miRNAs were decreased by at
least 0.5-fold in the aged TECs compared with those from the
2-month-old (young) mice.
 | Table IIExpression pattern of 20 miRNAs as
shown by microarray analysis. |
Table II
Expression pattern of 20 miRNAs as
shown by microarray analysis.
| Normalization | | |
|---|
|
| | |
|---|
| miRNA | 2-month-old
mice | 20-month-old
mice | Folda | p-valueb |
|---|
| miR-382-3p | 0.0165 | 0.1139 | 6.8919 | 0.0067 |
| miR-154-3p | 0.0166 | 0.0887 | 5.3488 | 0.0175 |
| miR-431 | 0.0377 | 0.1847 | 4.8985 | 0.0361 |
| miR-465a-3p | 0.0361 | 0.1522 | 4.2228 | 0.0133 |
| miR-146a | 0.1065 | 0.0192 | 0.1805 | 0.0343 |
| miR-181c | 0.3194 | 0.1035 | 0.3241 | 0.0338 |
| miR-322 | 0.7906 | 0.2762 | 0.3494 | 0.0136 |
| miR-194 | 0.8985 | 0.3531 | 0.3930 | 0.0150 |
| miR-19b | 9.8918 | 4.2293 | 0.4276 | 0.0142 |
| miR-19a | 11.2469 | 4.8793 | 0.4338 | 0.0378 |
| miR-181a | 0.6530 | 0.2839 | 0.4348 | 0.0195 |
| miR-96 | 0.9457 | 0.4177 | 0.4417 | 0.0064 |
| miR-155 | 2.2002 | 0.9730 | 0.4422 | 0.0059 |
| miR-24 | 10.5154 | 4.7114 | 0.4480 | 0.0422 |
| miR-181b | 0.2506 | 0.1127 | 0.4496 | 0.0120 |
| miR-148b | 0.1819 | 0.0845 | 0.4646 | 0.0260 |
| miR-150 | 1.9059 | 0.8968 | 0.4705 | 0.0324 |
| miR-192 | 0.3752 | 0.1773 | 0.4725 | 0.0184 |
| miR-22 | 3.8834 | 1.8492 | 0.4762 | 0.0213 |
| miR-93 | 1.0102 | 0.4813 | 0.4764 | 0.0151 |
Quantitative PCR confirms the difference
in miRNA expression in young and aged TECs
To confirm our results obtained from the microarray
assay, we performed a more thorough investigation of miRNA
expression in mouse TECs by quantitative PCR, a method with better
sensitivity than microarray analysis. To be able to observe the
trend in the changes in miRNA expression with thymic aging, we
compared the miRNA expression levels in TECs from mice of various
ages, including 1-month-old (young), 6-month-old (young adults),
10-month-old (middle-aged) and 19-month-old (aged) mice. The
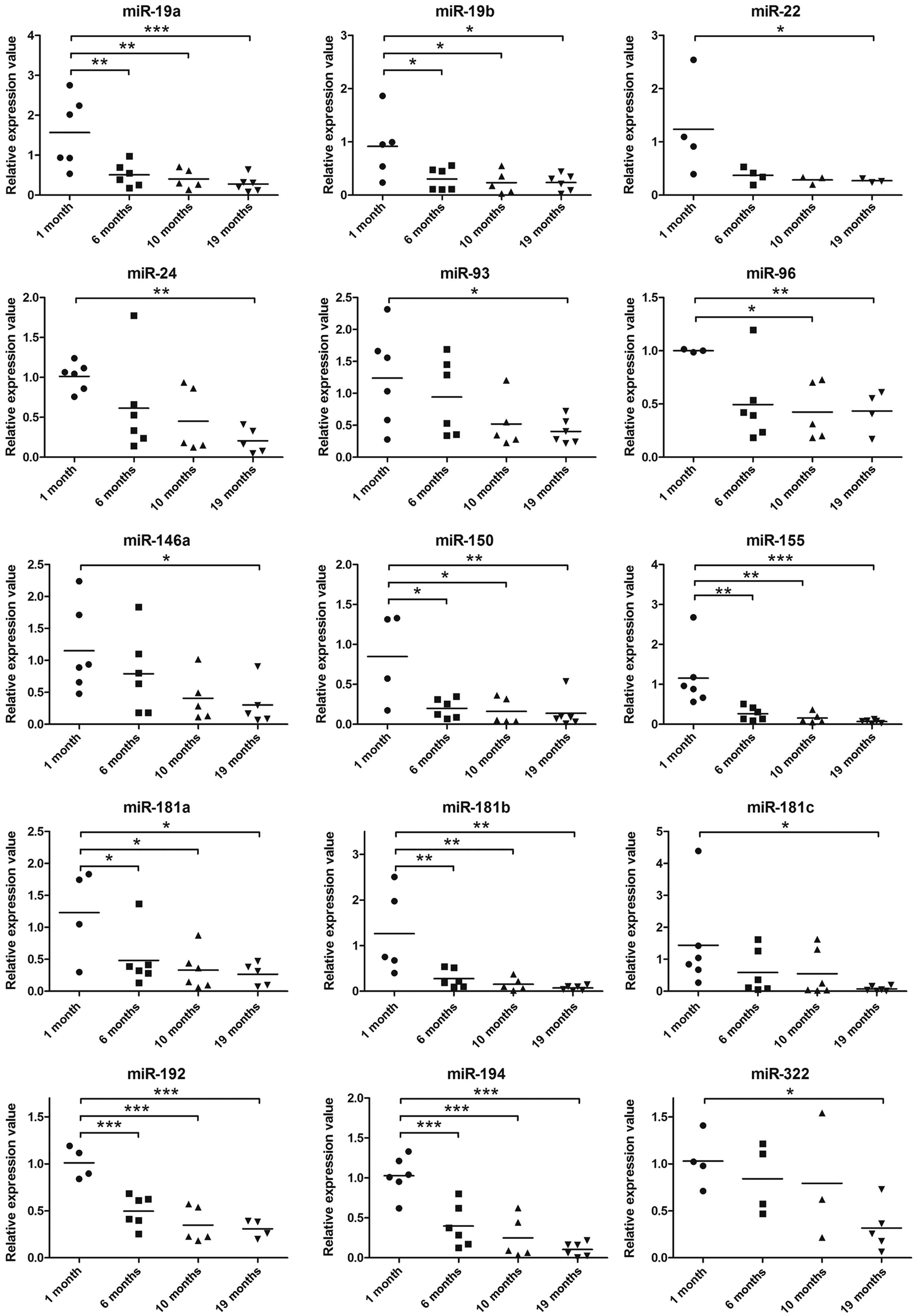
results of quantitative PCR were in agreement with the majority of
the results obtained from microarray analysis. The expression of 17
out of 20 miRNAs in the 19-month-old group decreased significantly,
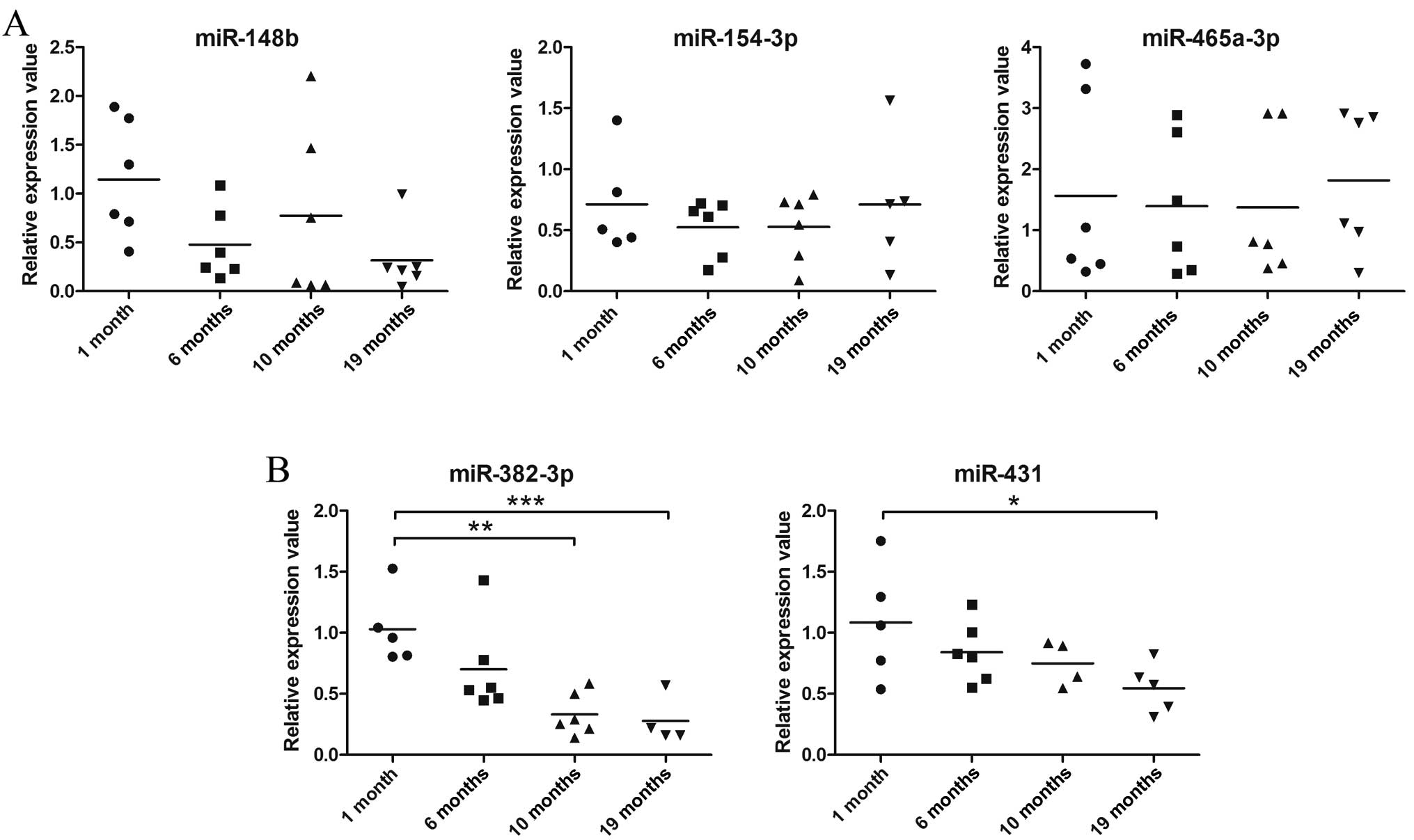
unlike that in the 1-month-old group (all 15 miRNAs in Fig. 2 and miR-382-3p and miR-431 in
Fig. 3). The decrease in the
expression levels of miR-382-3p and miR431 based on quantitative
PCR (Fig. 3) is not consistent
with the results obtained from microarray analysis, as the
expression of these 2 miRNAs was shown to be upregulated by
microarray analysis (Table II).
The expression of miR-154-3p and miR-465a-3p, which was also
upregulated in the aged mice as shown by the results of microarray
analysis, displayed no change in the different age-group samples as
shown by the results of quantitative PCR (Fig. 3). The inconsistency between
microarray and quantitative PCR may be the result of the variation
in miRNA expression in each individual mouse. By retrospective
examination of the microarray data, we found that the absolute
values of the fluorescence intensity of these 4 upregulated miRNAs
(miR-382-3p, miR-431, miR-154-3p and miR-465a-3p) belong to the
lower end of the detection limit in the microarray. In other words,
the microarray data for these miRNAs may be less reliable than that
obtained by quantitative PCR (Table
II). In addition, even though miR-148b expression showed an
age-associated decrease by quantitative PCR, which conforms to the
trend observed in the microarray data, the difference in the
expression in each age group was not significant (Fig. 3).
Changes in miRNA expression in TECs with
age closely correlate with age-related thymic atrophy
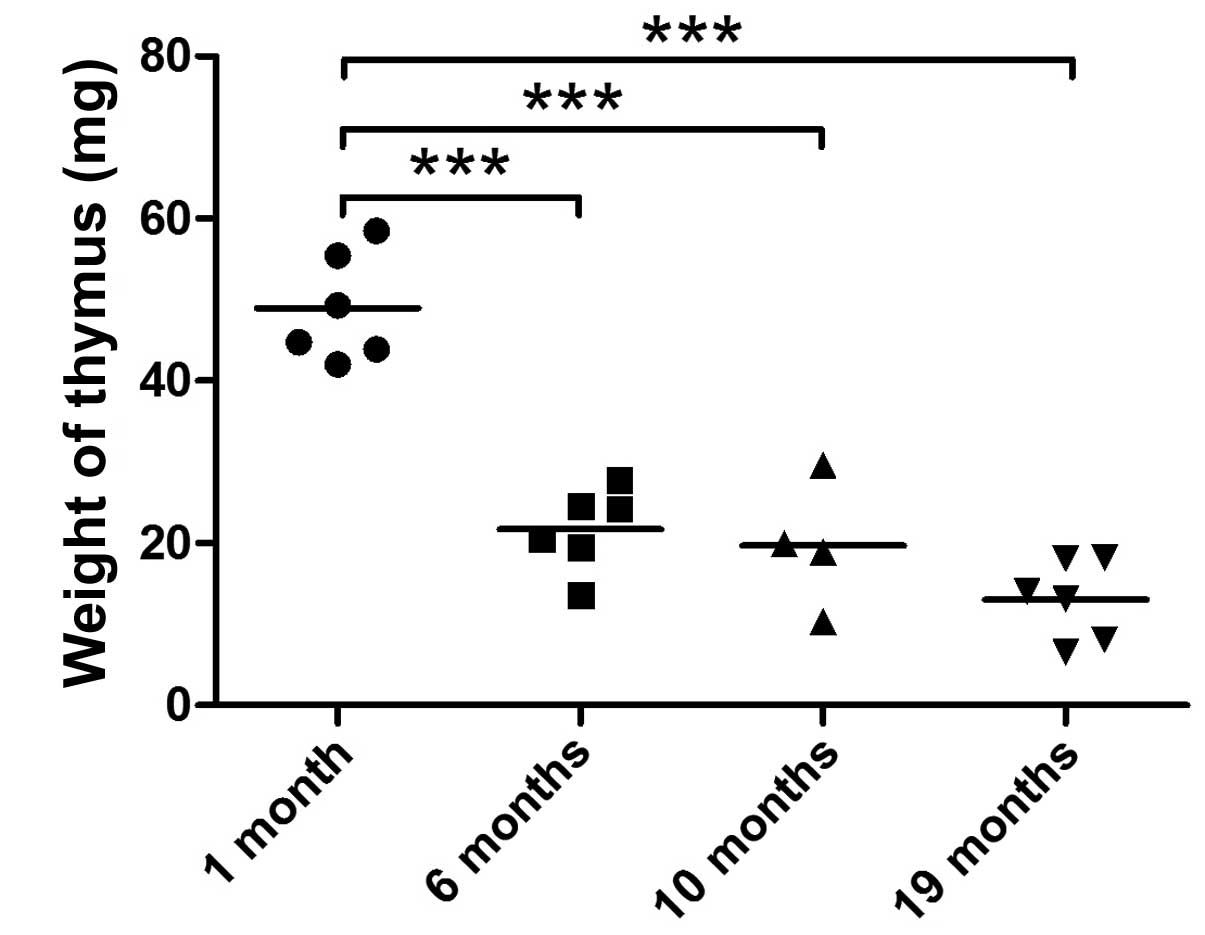
Thymic weight loss is a reliable index of thymic
aging and diminished thymopoiesis. By measuring the weight of the
thymus of mice with different ages ranging from 1 to 19 months, we
found that the thymus loses its weight in a biphasic pattern.
Thymic weight markedly decreased from 1 to 6 months and it
decreased at a much slower rate from 6 to 19 months (Fig. 4). Of note, the expression of 13
miRNAs in the TECs (miR-194, miR-192, miR-155, miR-150, miR-19a,
miR-19b, miR-148b, miR-382-3p, miR-146a, miR-93, miR-181a, miR-181b
and miR-181c), as measured by quantitative PCR, rapidly decreased
from 1 to 6 months and was only slightly altered from 6 to 10 and
19 months (Figs. 2 and 3), which is reminiscent of the pattern
of thumic weight loss with age. These 13 miRNAs are possibly
closely involved in the process of age-related thymic involution,
whereas age-associated changes in the expression of the other 7
miRNAs did not follow the biphasic pattern of thymic weight loss
(Table III). miR-465a-3p and
miR-154-3p are the only 2 miRNAs in the TECs that showed an
increasing trend in expression with age; however, this was not
statistically significant.
 | Table IIILinear correlation analysis of the
expression of 20 miRNAs (by qPCR) and changes in thymic weight with
aging. |
Table III
Linear correlation analysis of the
expression of 20 miRNAs (by qPCR) and changes in thymic weight with
aging.
| Name | R-valuea | p-value |
|---|
| miR-155 | 0.6957 | 0.0002b |
| miR-181b | 0.632 | 0.0012b |
| miR-194 | 0.6077 | 0.0016b |
| miR-19b | 0.5993 | 0.002b |
| miR-181c | 0.5886 | 0.0025b |
| miR-150 | 0.5733 | 0.0053b |
| miR-19a | 0.5477 | 0.0056b |
| miR-181a | 0.5328 | 0.0129b |
| miR-192 | 0.5155 | 0.0168b |
| miR-148b | 0.4602 | 0.0236b |
| miR-382-3p | 0.4555 | 0.038b |
| miR-146a | 0.4505 | 0.0354b |
| miR-93 | 0.4168 | 0.0428b |
| miR-322 | 0.4428 | 0.0859c |
| miR-96 | 0.4193 | 0.0833c |
| miR-22 | 0.3682 | 0.1102c |
| miR-24 | 0.2783 | 0.188c |
| miR-431 | 0.2597 | 0.2688c |
| miR-465a-3p | −0.1506 | 0.4824c |
| miR-154-3p | −0.2017 | 0.4376c |
Possible targets of miRNAs predicted
possible pathways involved in age-related thymic involution
miRNAs exert their functions through multiple
downstream targets, including a number of transcription factors,
apoptosis control factors and other important factors for cell
function. In the present study, we predicted the targets of these
miRNAs by comparing the targets from 4 databases: Target Scan,
PicTar, Miranda and EIMMo. These miRNAs not only differed in
expression between the young and aged TECs (Fig. 2 and miR-382-3p and miR-431 in
Fig. 3), but also closely
correlated with changes in thymic weight with aging (Table III). We paid much attention to
the predictions of targets implicated to play a role in thymic
aging. We selected the output hits with a high score in all 4
databases for each of the analyzed miRNAs. The predicted targets
are listed in Table IV.
 | Table IVPredicted target genes of 12 miRNAs
found in 4 databases (TargetScan, PicTar, Miranda and EIMMo). |
Table IV
Predicted target genes of 12 miRNAs
found in 4 databases (TargetScan, PicTar, Miranda and EIMMo).
| miRNA | Target genes |
|---|
| miR-155 | Myb, Hivep2,
Ptpn2 |
| miR-181b | Rnf34 |
| miR-194 | Ptpn12, Hbegf,
Sumo2 |
| miR-19b | Rin2, Nbea,
Dlx1 |
| miR-181c | Zfp14 |
| miR-150 | Myb |
| miR-19a | Zmynd11, Rnf11,
Phtf2 |
| miR-181a | Klf6, Fbxo33,
Zfp36l2 |
| miR-192 | Wnk1, Msn,
Cdc6 |
| miR-146a | Irak1 |
| miR-93 | Epha4, Zfp367,
Rab5b |
| miR-382-3p | none |
Discussion
The thymus is responsible for thymocyte development
and de novo T cell production. Age-related thymic involution
is one of the prominent causes of T cell-mediated immunodeficiency
in the elderly. Emerging evidence indicates that the deterioration
of the thymic microenvironment, particularly the loss of TECs,
largely contributes to the defect in T cell development in the
thymus (7–9). Changes in several critical genes
associated with age-related thymic involution have been extensively
investigated (12,15). To our knowledge, this is the first
study to explore miRNA expression in TECs and the correlation
between changes in miRNA expression with age-related thymic weight
loss. By microarray and quantitative PCR analyses, our findings
indicated that the majority of the 20 miRNAs that we examined had
decreased expression levels with thymic aging. These results are in
agreement with those from a study using peripheral blood
mononuclear cells, in which >800 miRNAs were profiled and the
majority of the studied miRNAs decreased in abundance with age
(28). In this study, the
fluorescence intensity of miR-431, miR-154-3p, miR-382-3p and
miR-465a-3p, as shown by the microarray analysis was very weak,
indicating that the expression of these miRNAs in the TECs of both
young and aged mice was very low. Given the better sensitivity and
larger sample size that was used in quantitative PCR compared with
the microarray analysis, we have more confidence in the results
from quantitative PCR for these 4 miRNAs than in the results from
the microarray analysis.
It should be noted that several of the miRNAs that
we analyzed have been implicated in important immune functions. A
number of studies have confirmed that miR-155, miR-181a and miRNAs
derived from the miR-17-92 cluster (miR-19a and miR-19b in the
present study) are involved in the development and differentiation
of lymphoid and myeloid cells (25, 29–31). To the best of our knowledge, this
is the first study to provide evidence that these miRNAs with
important immunological functions are expressed in TECs.
Another important finding in this study comes from
the correlation analysis between changes in the expression of
miRNAs with age and the age-related thymic weight loss. In total,
13 out of the 20 miRNAs showed a significant correlation. The
expression of all 13 miRNAs showed a positive correlation with the
thymic weight loss during aging. It is very likely that the
decrease in miRNA expression in the TECs precedes the age-induced
thymic involution. This may be particularly true for miRNAs, such
as miR-194, miR-192, miR-155, miR-19a, miR-19b, miR-181a and
miR-181b from the 13 miRNAs, as the decrease in the expression of
these miRNAs with age follows a more obvious biphasic pattern [1 to
6 months compared with 6 months onwards (Fig. 2)], which conforms to the pattern
of thymic weight loss during aging (Fig. 4).
We predicted 3 target genes for miRNAs that may be
involved in the process of age-related thymic involution (Table IV). Some of the targets have
previously been implicated in immune functions, as well as T cell
functions. For example, Myb regulates T cell differentiation
(32) and the development of
hematopoietic stem cells (33).
Hivep2, a target gene of miR-155, plays a critical role in signal
transduction (34,35), lymphocyte development (36) and the production of memory T
helper cells (37,38). Wnk1 is required for mitosis
(39,40), proliferation and migration
(41) and modulates of TGFβ-Smad
pathways (42). The IRAK family
of genes may be involved in the expression of inflammatory gene
(43). One of these genes, Irak1
may disrupt the balance of the generation of pro-inflammatory
cytokines and type I interferons in the inherent immune response
(44) and has been identified as
a danger gene in systemic lupus erythematosus (45).
Based on the targets of miRNAs, our belief is that
further analysis of these targets may aid in the understanding of
how these miRNAs control the thymic aging process. It can be
concluded that changes in the expression of these miRNAs in TECs
may at least partially regulate thymocyte development and T cell
maturation, considering the important role that TECs play in the
thymic microenvironment for thymocyte development and
differentiation.
Acknowledgements
The present study was supported by the National
Natural Scientific Foundation of China (grant no. 30872715,
81270430), by the Special Research Fund for Doctoral Program of
Education Department of China (grant no. 20112104110011) and by the
Free Research Program Fund of Shengjing Hospital (grant no. 200805)
to X.Z.
Abbreviations:
|
ANOVA
|
analysis of variance
|
|
BM
|
bone marrow
|
|
DN
|
double negative
|
|
ETPs
|
early T cell progenitors
|
|
PCR
|
polymerase chain reaction
|
|
PBS
|
phosphate-buffered saline
|
|
TECs
|
thymic epithelial cells
|
References
|
1
|
Aw D, Silva AB, Maddick M, et al:
Architectural changes in the thymus of aging mice. Aging Cell.
7:158–167. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aw D and Palmer DB: The origin and
implication of thymic involution. Aging Dis. 2:437–443.
2011.PubMed/NCBI
|
|
3
|
Naylor K, Li G, Vallejo A N, et al: The
influence of age on T cell generation and TCR diversity. J Immunol.
174:7446–7452. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maue AC, Yager EJ, Swain SL, et al: T-cell
immunosenescence: lessons learned from mouse models of aging.
Trends Immunol. 30:301–305. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dorshkind K and Swain S: Age-associated
declines in immune system development and function: causes,
consequences, and reversal. Curr Opin Immunol. 21:404–407. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Foster AD, Sivarapatna A and Gress RE: The
aging immune system and its relationship with cancer. Aging health.
7:707–718. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gui J, Zhu X, Dohkan J, et al: The aged
thymus shows normal recruitment of lymphohematopoietic progenitors
but has defects in thymic epithelial cells. Int Immunol.
19:1201–1211. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu X, Gui J, Dohkan J, et al:
Lymphohematopoietic progenitors do not have a synchronized defect
with age-related thymic involution. Aging Cell. 6:663–672. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gui J, Mustachio LM, Su DM and Craig RW:
Thymus size and age-related thymic involution: early programming,
sexual dimorphism, progenitors and stroma. Aging Dis. 3:280–290.
2012.PubMed/NCBI
|
|
10
|
Gui J, Morales AJ, Maxey SE, et al: MCL1
increases primitive thymocyte viability in female mice and promotes
thymic expansion into adulthood. Int Immunol. 23:647–659. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bravo-Nuevo A, O’Donnell R, Rosendahl A,
et al: RhoB deficiency in thymic medullary epithelium leads to
early thymic atrophy. Int Immunol. 23:593–600. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ortman CL, Dittmar KA, Witte PL and Le PT:
Molecular characterization of the mouse involuted thymus:
aberrations in expression of transcription regulators in thymocyte
and epithelial compartments. Int Immunol. 14:813–822. 2002.
View Article : Google Scholar
|
|
13
|
Lustig A, Carter A, Bertak D, et al:
Transcriptome analysis of murine thymocytes reveals age-associated
changes in thymic gene expression. Int J Med Sci. 6:51–64. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun L, Guo J, Brown R, et al: Declining
expression of a single epithelial cell-autonomous gene accelerates
age-related thymic involution. Aging Cell. 9:347–357. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kvell K, Varecza Z, Bartis D, et al: Wnt4
and LAP2alpha as pacemakers of thymic epithelial senescence. PLoS
One. 5:e107012010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Weatherall DJ: Thalassaemia: the long road
from bedside to genome. Nat Rev Genet. 5:625–631. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
18
|
Zhao Y, Ransom JF, Li A, et al:
Dysregulation of cardiogenesis, cardiac conduction, and cell cycle
in mice lacking miRNA-1-2. Cell. 129:303–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mencia A, Modamio-Høybjør S, Redshaw N, et
al: Mutations in the seed region of human miR-96 are responsible
for nonsyndromic progressive hearing loss. Nat Genet. 41:609–613.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng J, Sun G, Yan J, et al: Evidence for
X-chromosomal schizophrenia associated with microRNA alterations.
PLoS One. 4:e61212009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Muljo SA, Ansel KM, Kanellopoulou C, et
al: Aberrant T cell differentiation in the absence of Dicer. J Exp
Med. 202:261–269. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li QJ, Chau J, Ebert PJ, et al: miR-181a
is an intrinsic modulator of T cell sensitivity and selection.
Cell. 129:147–161. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rodriguez A, Vigorito E, Clare S, et al:
Requirement of bic/microRNA-155 for normal immune function.
Science. 316:608–611. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Neilson JR, Zheng GX, Burge CB and Sharp
PA: Dynamic regulation of miRNA expression in ordered stages of
cellular development. Genes Dev. 21:578–589. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Virts EL and Thoman ML: Age-associated
changes in miRNA expression profiles in thymopoiesis. Mech Ageing
Dev. 131:743–748. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen LH, Chiou GY, Chen YW, et al:
MicroRNA and aging: a novel modulator in regulating the aging
network. Ageing Res Rev. 9(Suppl 1): S59–S66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Noren Hooten N, Abdelmohsen K, Gorospe M,
et al: microRNA expression patterns reveal differential expression
of target genes with age. PLoS One. 5:e107242010.PubMed/NCBI
|
|
29
|
O’Connell RM, Rao DS, Chaudhuri AA, et al:
Sustained expression of microRNA-155 in hematopoietic stem cells
causes a myeloproliferative disorder. J Exp Med. 205:585–594.
2008.PubMed/NCBI
|
|
30
|
Ventura A, Young AG, Winslow MM, et al:
Targeted deletion reveals essential and overlapping functions of
the miR-17 through 92 family of miRNA clusters. Cell. 132:875–886.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiao C, Srinivasan L, Calado DP, et al:
Lymphoproliferative disease and autoimmunity in mice with increased
miR-17-92 expression in lymphocytes. Nat Immunol. 9:405–414. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sheiness D and Gardinier M: Expression of
a proto-oncogene (proto-myb) in hemopoietic tissues of mice. Mol
Cell Biol. 4:1206–1212. 1984.PubMed/NCBI
|
|
33
|
Schulz C, Gomez Perdiguero E, Chorro L, et
al: A lineage of myeloid cells independent of Myb and hematopoietic
stem cells. Science. 336:86–90. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shukla A and Yuspa SH: CLIC4 and
Schnurri-2: A dynamic duo in TGF-beta signaling with broader
implications in cellular homeostasis and disease. Nucleus.
1:144–149. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shukla A, Malik M, Cataisson C, et al:
TGF-beta signalling is regulated by Schnurri-2-dependent nuclear
translocation of CLIC4 and consequent stabilization of
phospho-Smad2 and 3. Nat Cell Biol. 11:777–784. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Staton TL, Lazarevic V, Jones DC, et al:
Dampening of death pathways by schnurri-2 is essential for T-cell
development. Nature. 472:105–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nakayama T and Kimura MY: Memory Th1/Th2
cell generation controlled by Schnurri-2. Adv Exp Med Biol.
684:1–10. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kimura MY, Iwamura C, Suzuki A, et al:
Schnurri-2 controls memory Th1 and Th2 cell numbers in vivo. J
Immunol. 178:4926–4936. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tu SW, Bugde A, Luby-Phelps K and Cobb MH:
WNK1 is required for mitosis and abscission. Proc Natl Acad Sci
USA. 108:1385–1390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jiang ZY, Zhou QL, Holik J, et al:
Identification of WNK1 as a substrate of Akt/protein kinase B and a
negative regulator of insulin-stimulated mitogenesis in 3T3-L1
cells. J Biol Chem. 280:21622–21628. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun X, Gao L, Yu RK and Zeng G:
Down-regulation of WNK1 protein kinase in neural progenitor cells
suppresses cell proliferation and migration. J Neurochem.
99:1114–1121. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee BH, Chen W, Stippec S and Cobb MH:
Biological cross-talk between WNK1 and the transforming growth
factor beta-Smad signaling pathway. J Biol Chem. 282:17985–17996.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gottipati S, Rao NL and Fung-Leung WP:
IRAK1: a critical signaling mediator of innate immunity. Cell
Signal. 20:269–276. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
An H, Hou J, Zhou J, et al: Phosphatase
SHP-1 promotes TLR- and RIG-I-activated production of type I
interferon by inhibiting the kinase IRAK1. Nat Immunol. 9:542–550.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jacob CO, Zhu J, Armstrong DL, et al:
Identification of IRAK1 as a risk gene with critical role in the
pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci
USA. 106:6256–6261. 2009. View Article : Google Scholar : PubMed/NCBI
|