Introduction
Over 170 million individuals worldwide are infected
with hepatitis C virus (HCV), which is a major causative agent of
liver disease. HCV infection persists after primary infection in
the majority of infected individuals, often leading to fibrosis,
cirrhosis or hepatocellular carcinoma (HCC). However, no effective
prophylactic vaccines or drugs against HCV are currently available
(1,2).
HCV is an enveloped virus that belongs to the genus
Hepacivirus in the Flaviviridae family, with a
positive single-stranded 9.5 kilobase (kb) RNA genome. HCV genomic
RNA encodes a polyprotein precursor of ~3,000 amino acid (aa)
residues, which is cleaved by a combination of host and viral
proteases into at least 10 proteins: the core, envelope proteins
(E1 and E2), p7, and non-structural 2 (NS2), NS3, NS4A, NS4B, NS5A
and NS5B proteins (3). Envelope
proteins are type 1 transmembrane proteins with N-terminal
ectodomains and C-terminal hydrophobic anchors (4). The proteins E1 and E2, especially
E2, mediate viral entry into target cells, which is an obligatory
step in virus replication and the maintenance of infection
(5,6). The frequency of HCV genomic mutation
is relatively high, and the virus is divided into six major
genotypes and multiple subtypes (7). In general, despite strong amino acid
variability, almost all viruses enter cells via specific cell
receptors, which are distinct for different genotypes of viruses
(8–11). Drugs targeting the viral entry
step may provide a potential candidate for anti-HCV therapy, with
the HCV E2 protein serving as an ideal target.
Full-length E2 protein extends from amino acids 384
to 746 of the HCV polyprotein and contains regions of extreme
variability. The C-terminal truncation of E2 at residue 661
(E2661) or 715 (E2715) leads to the secretion
of E2, as this deletion has been suggested to remove the
hydrophobic transmembrane anchor sequence (12,13). It has been shown that E2 truncated
at residue 661 has a higher folding efficiency compared to E2
truncated at 715 (12,13). In a previous study, we constructed
a recombinant prokaryotic plasmid expressing the truncated HCV E2
gene (E2661), and expressed and purified the protein. We
found that E2661 retained the ability to react with the
sera of HCV-infected individuals using western blotting (14). In the present study, we used the
phage-display random peptide library, a useful tool for obtaining
short peptides that specifically bind to a target ligand, to
identify novel peptides that specifically bind to HCV E2.
HCV-pseudotyped particles (HCVpp) harboring HCV envelope
glycoproteins (15,16) and cell-culture-produced HCV
(HCVcc) (17,18) were used as surrogate models to
investigate the ability of selected identified peptides to inhibit
the entry of HCV into cells in vitro.
Materials and methods
Cells and proteins
Huh7.5 cells (a kind gift from Professor Charles M.
Rice, The Rockefeller University, New York, NY, USA) and 293T cells
(ATCC, Manassas, VA, USA) were maintained in Dulbecco’s modified
Eagle’s medium (DMEM; Gibco, Gaithersburg, MD, USA) supplemented
with 10% fetal calf serum (FCS; Gibco), 1% L-glutamine and 1%
penicillin-streptomycin. The HCV proteins E2661, NS3
(19) and NS5B were
prokaryotically expressed with 6X His tags and purified using
Ni-NTA affinity chromatography (Qiagen, Valencia, CA, USA).
Phage display peptide biopanning
The Ph.D.-7 phage display peptide library was
obtained from New England Biolabs (Beverly, MA, USA). The original
library revealed a wide diversity of sequences with no obvious
positional biases. The E. coli host strain ER2738 provided
in the library kit was used for M13 phage propagation. Phage
panning procedures were performed according to the manufacturer’s
instructions.
Briefly, 35 mm-diameter polystyrene plates were
coated with HCV E2661 (100 μg) in 1.5 ml
NaHCO3 (pH 8.6, 0.1 mol/l) at 4°C overnight with gentle
agitation in a humidified container. The coating solution was
completely removed and each plate was filled with blocking buffer
(PBS containing 5% BSA) for 2 h at 4°C. Approximately
2×1011 phages were added to the blocked wells and mixed
gently for 1 h at 37°C. The plates were washed 10 times with Tris
buffer solution, pH 7.5, containing 0.1% (v/v) Tween-20 (TBST). The
adherent phages were eluted with 1 ml 0.2 mol/l glycine-HCl (pH
2.2) containing 1 mg/ml bovine serum albumin (BSA) and neutralized
with 150 μl 1 mol/l Tris-HCl (pH 9.1). The eluted phage titer was
subsequently evaluated by a blue plaque-forming assay on agar
plates containing tetracycline. The phages were amplified by mixing
the eluate with 20 ml of E. coli ER2738 culture and then
incubation at 37°C with vigorous agitation for 7 h. The titer of
the amplified eluates was determined as described in the kit
protocol, and the eluates were used for the following round of
biopanning. To enrich for phages that displayed the target peptide
on their surface, the phages were screened a total of three times,
as described above. The panning stringency was gradually increased
stepwise in each round, by decreasing the amount of input HCV
E2661 protein (100, 50, then 20 μg) and reducing the
binding time (60, 30, then 20 min).
Binding assays
The reactivity of the third round peptide population
for HCV E2661 protein was estimated using an ELISA.
Incubations were performed in a volume of 100 μl at 37°C.
Microtiter wells were coated with HCV E2661 protein
overnight at 4°C, and then blocked with BSA (15 mg/ml) for 4 h. The
third round peptide population was added [1×1011
plaque-forming units (PFU)/well] and incubated for 1 h at room
temperature with gentle agitation. The plate was washed 10 times
with TBST containing 0.5% Tween-20. Subsequently, horseradish
peroxidase (HRP)-conjugated anti-M13 phage antibody (Pharmacia,
Peapack, NJ, USA) diluted 1:5000 in phosphate-buffered saline (PBS,
pH 7.4) was added, incubated for 1 h, and the wells were washed 10
times with TBST. Peroxidase activity was determined at 37°C using
the substrate o-phenylenediamine-H2O2
in phosphoric acid buffer. The reaction was stopped using 25 ml of
2 mol/l H2SO4 for 15 min, and the absorbance
values were measured at 490 nm (A490). HCV proteins NS3 and NS5B,
and BSA and PBS were used as negative controls. Triplicate
determinations were generated for each data point.
Selection of individual positive
clones
Twenty-five clones were randomly selected from the
eluates of the third round of biopanning and individually added to
E. coli ER2738 cultures for amplification and titration. The
relative binding affinities of the individual clones to HCV
E2661 protein were assayed using an ELISA, as described
above. Triplicate determinations were generated for each data
point.
DNA sequencing and peptide synthesis
The single-stranded DNA from the positive phages was
purified using an M13 purification kit (Beijing Sunbiotech Co.,
Ltd., Beijing, China). DNA sequencing was performed by Beijing Aoke
Biotechnology Ltd. (Beijing, China). The heptapeptide sequences
were deduced from the DNA sequences and synthesized by
Chinapeptides Co., Ltd. (Shanghai, China). Homologous analysis was
performed using BLAST. The unrelated synthetic heptapeptide
(VLRSDFK), the sequence deduced from the clone with the weakest
affinity to HCV E2661 protein of the 25 randomly
selected positive clones, was used as the negative control.
Entry inhibition assay using HCV
pseudotype particles
HCV pseudotype particles (HCVpp) were generated as
previously described (15,16).
Briefly, 2.25 μg of the HCV envelope glycoprotein expression
plasmid PVRC-E1E2hebei (genotype 1b), 9 μg of the transfer plasmid
pCS-CG expressing the reporter gene enhanced green fluorescent
protein (EGFP), and 6.75 μg of the lentiviral packaging plasmid
pHR’CMVΔ8.2 were co-transfected into 2×106 293T cells
using Fugene HD transfection reagent (Roche, Penzberg, Germany) to
produce infectious HCVpp. The three plasmids were a generous gift
from Professor Wenjie Tan (National Institute for Viral Disease
Control and Prevention, China CDC, Beijing, China).
HCVpp-containing supernatant was collected 48 h after transfection,
pooled, filtered and condensed using a Lenti-X™ Concentrator
(Takara, Otsu, Japan).
Incorporation of HCV El-E2 glycoproteins into the
pseudoparticles was verified by western blotting using HCV-infected
sera obtained from patients at Tangdu Hospital (Fourth Military
Medical University, Xi’an, China) as a primary antibody. HCVpp
titer was quantified by the p24 content of the samples using the
HIV-1 p24 ELISA kit (PerkinElmer, Boston, MA, USA). Then, 1.2 ml
HCVpp (1 ng/ml) was incubated with the peptides at final
concentrations of 20, 50, 100 and 200 μg/ml for 1 h at room
temperature, and added to the Huh7.5 cells. At 16 h post-infection,
the media were removed, the cells were washed three times with PBS,
incubated with fresh media for an additional 32 h, and then
collected. EGFP activity, a reflection of the degree of entry of
the pseudoparticles into the host cells, was measured using flow
cytometry. HCVpp bound to the unrelated peptide and naïve HCVpp
were used as a negative and mock control, respectively.
Entry inhibition assay using HCVcc
The pFL-J6/JFH plasmid containing the full-length
HCV chimeric genome (genotype 2a) was kindly provided by Professor
Charles M. Rice. Cell-culture-produced HCV (HCVcc) was generated as
previously described (17,18),
without modifications. Full-length HCV RNA was transcribed in
vitro from the pFL-J6/JFH plasmid, and electroporated into
Huh7.5 cells. The copy number of HCVcc in the cell supernatants was
determined by quantitative reverse-transcription (RT)-PCR. Then,
1×107 copies of HCVcc were incubated with the peptides
(final concentration, 100 μg/ml) for 1 h at room temperature and
then added to Huh7.5 cells. HCVcc incubated with the unrelated
peptide and naïve HCVcc were used as a negative and mock control,
respectively.
For quantitative real-time RT-PCR, the media were
discarded at 6 h post-infection. Subsequently, the cells were
washed three times with PBS and collected. Total RNA was extracted
using TRIzol (Gibco) according to the manufacturer’s instructions
and quantitative real-time RT-PCR was conducted using the
SYBR-Green Real-time PCR Master Mix kit (Takara) with the primers
listed in Table I.
 | Table IPrimers used for quantitative
real-time RT-PCR. |
Table I
Primers used for quantitative
real-time RT-PCR.
| Target gene | Primer |
|---|
| HCV
5′-UTR | F:
5′-CGGGAGAGCCATAGTGGTC-3′
R: 5′-AGTACCACAAGGCCTTTCG-3′ |
| GAPDH | F:
5′-TGCACCACCAACTGCTTAG-3′
R: 5′-GATGCAGGGATGATGTTC-3′ |
For western blot analysis, the media were removed at
16 h post-infection, the cells were washed three times with PBS,
incubated with fresh medium at 37°C for an additional 32 h and then
collected. Mouse anti-HCV core antigen monoclonal antibody (1:20;
Thermo Scientific Pierce Antibodies, Rockford, IL, USA) and mouse
anti-HCV NS5A protein monoclonal antibody (1:100; generous gift of
Professor Charles M. Rice) were used to detect the expression of
HCV core protein and NS5A by western blotting, respectively.
Statistical analysis
Differences between groups were compared using the
one-way ANOVA. Statistical significance was defined as
P<0.05.
Results
Specific enrichment of HCV
E2661 protein-bound phages
Phages specifically binding to HCV E2661
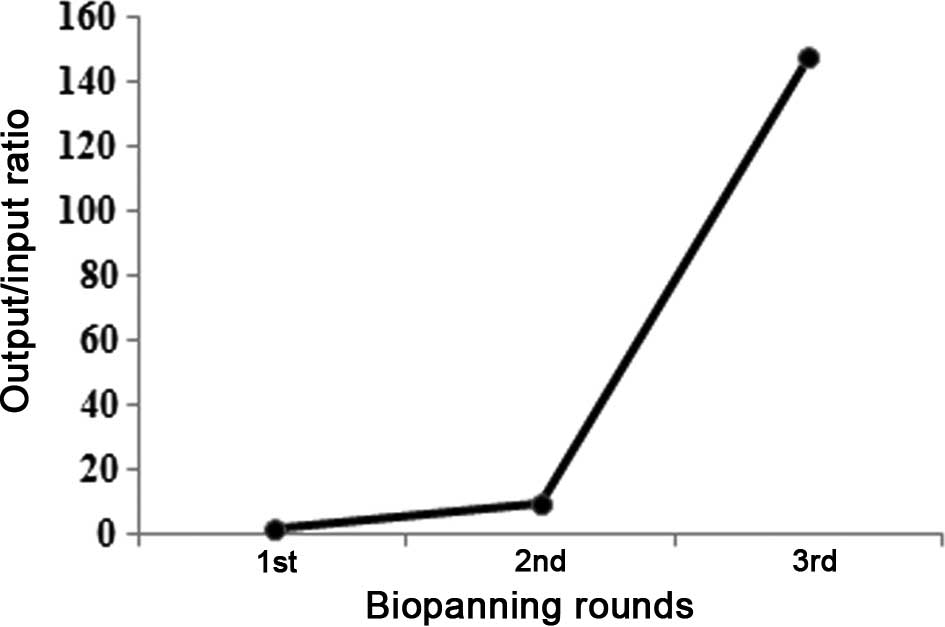
protein were enriched through three rounds of panning. The
output/input ratio of phages after each round of panning was used
to determine the phage recovery efficiency. After three rounds of
panning, the output/input ratio in the third round had increased by
almost 150-fold, compared with the first round, demonstrating that
effective biopanning of phages specifically binding to HCV
E2661 protein had occurred (Fig. 1).
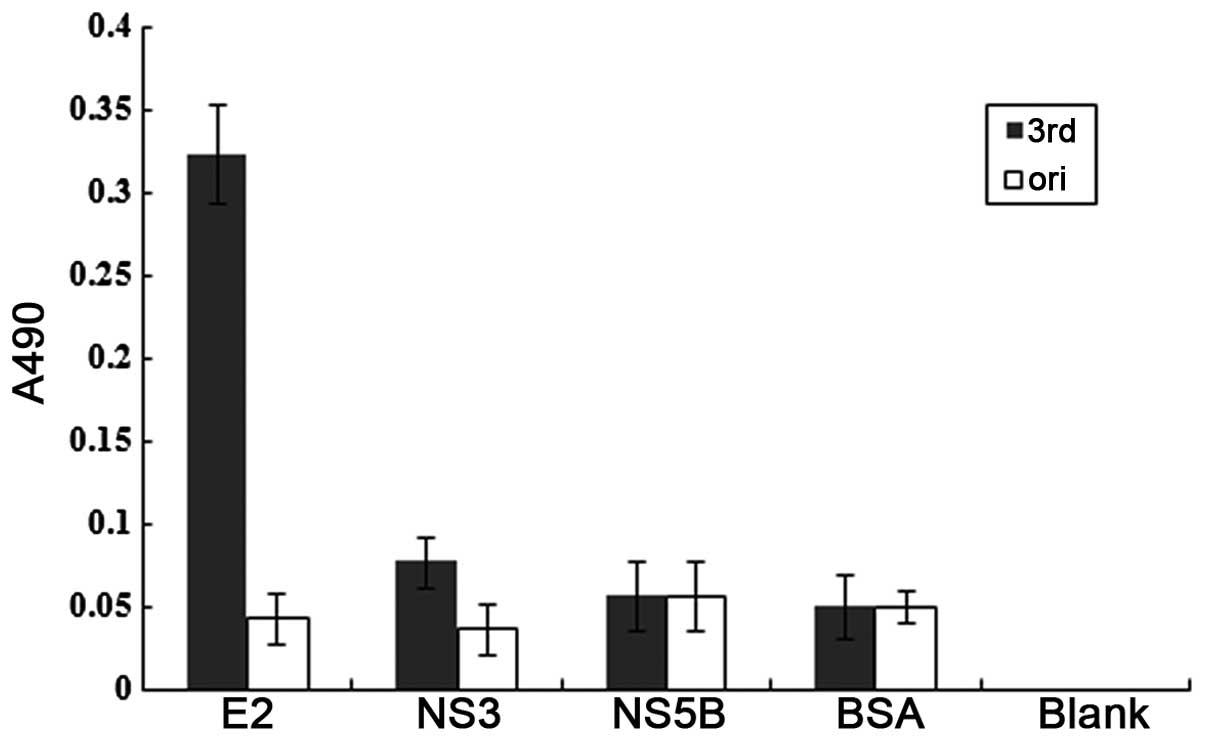
Binding assays
As shown in Fig.
2, the binding reactivity of the third round phages was much
higher in the HCV E2661 protein as compared to other
unrelated proteins. The original phages had similar reactivity in
all the proteins.
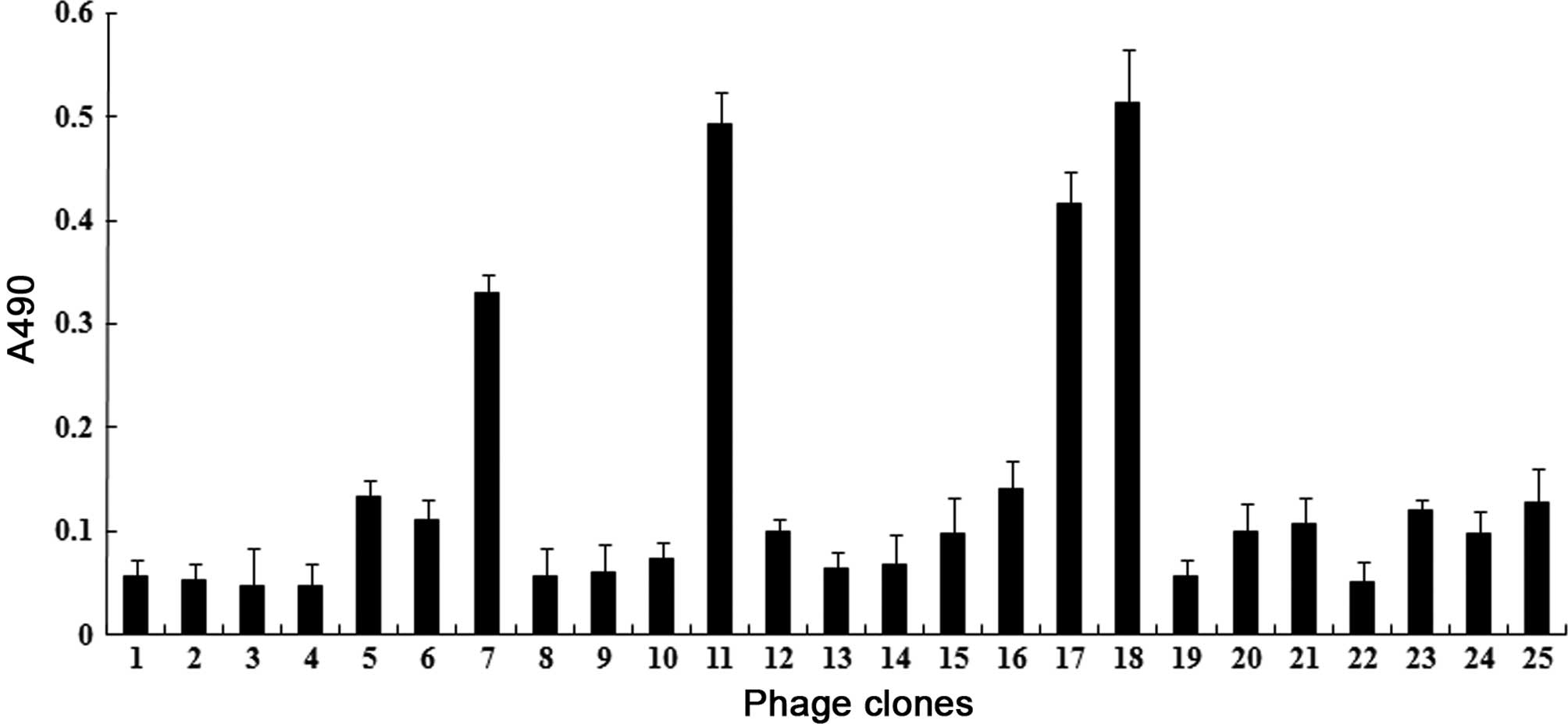
Positive clones and their sequences
Twenty-five plaques were randomly selected from the
third round phages, and their binding reactivity to the HCV
E2661 protein was determined individually using an
ELISA. Clone no. 3 had the lowest affinity to E2661
protein. Four clones with the highest affinities (nos. 7, 11, 17
and 18) were selected for subsequent analysis (Fig. 3).
Following DNA sequencing, the amino acid sequences
of the selected clones were deduced and designated as C7, C11, C17
and C18, corresponding to the respective clone numbers (Fig. 4). The four peptides harbored the
conserved motif WPWXXXR, where X is any residue, with a bias for H.
This similarity suggested that the four peptides were capable of
binding to the same site of the target protein E2661.
Furthermore, the four sequences were analyzed using BLAST; however,
no homologous proteins were identified. The sequence VLRSDFK
deduced from clone no. 3 was synthesized as an unrelated
peptide.
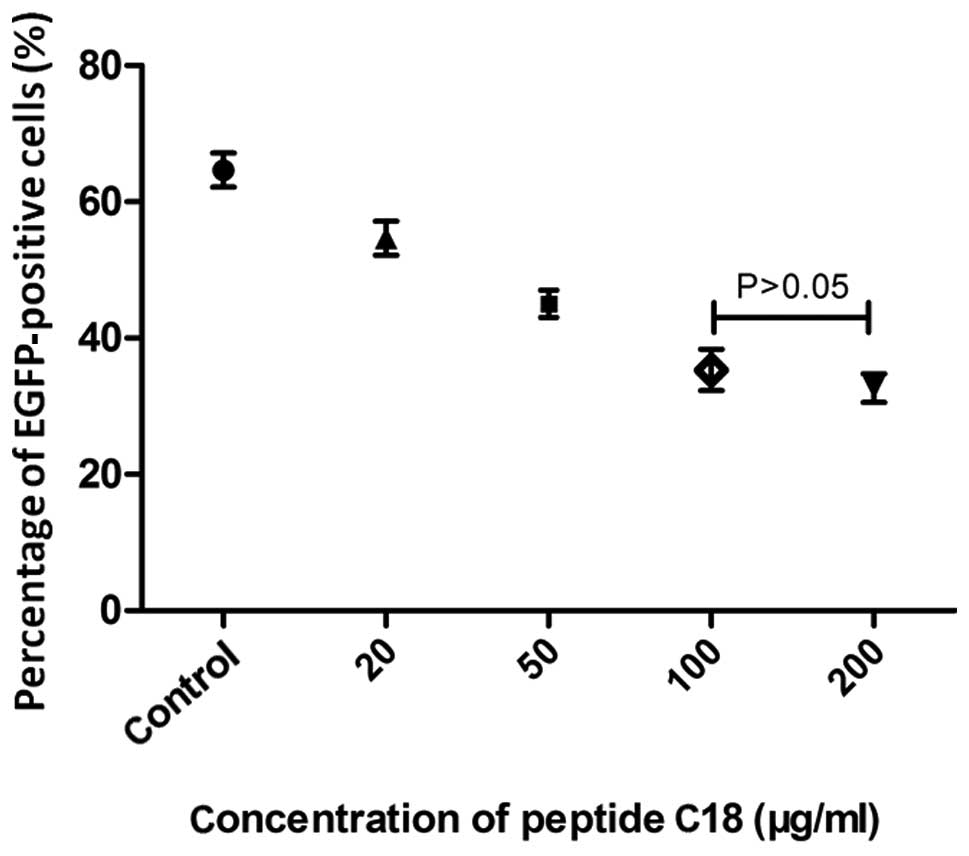
Evaluation of the antiviral activity of
peptide C18
Due to the high binding reactivity between clone no.
18 and HCV E2661 protein, peptide C18 with the sequence
WPWHNHR was synthesized. The most appropriate concentration of the
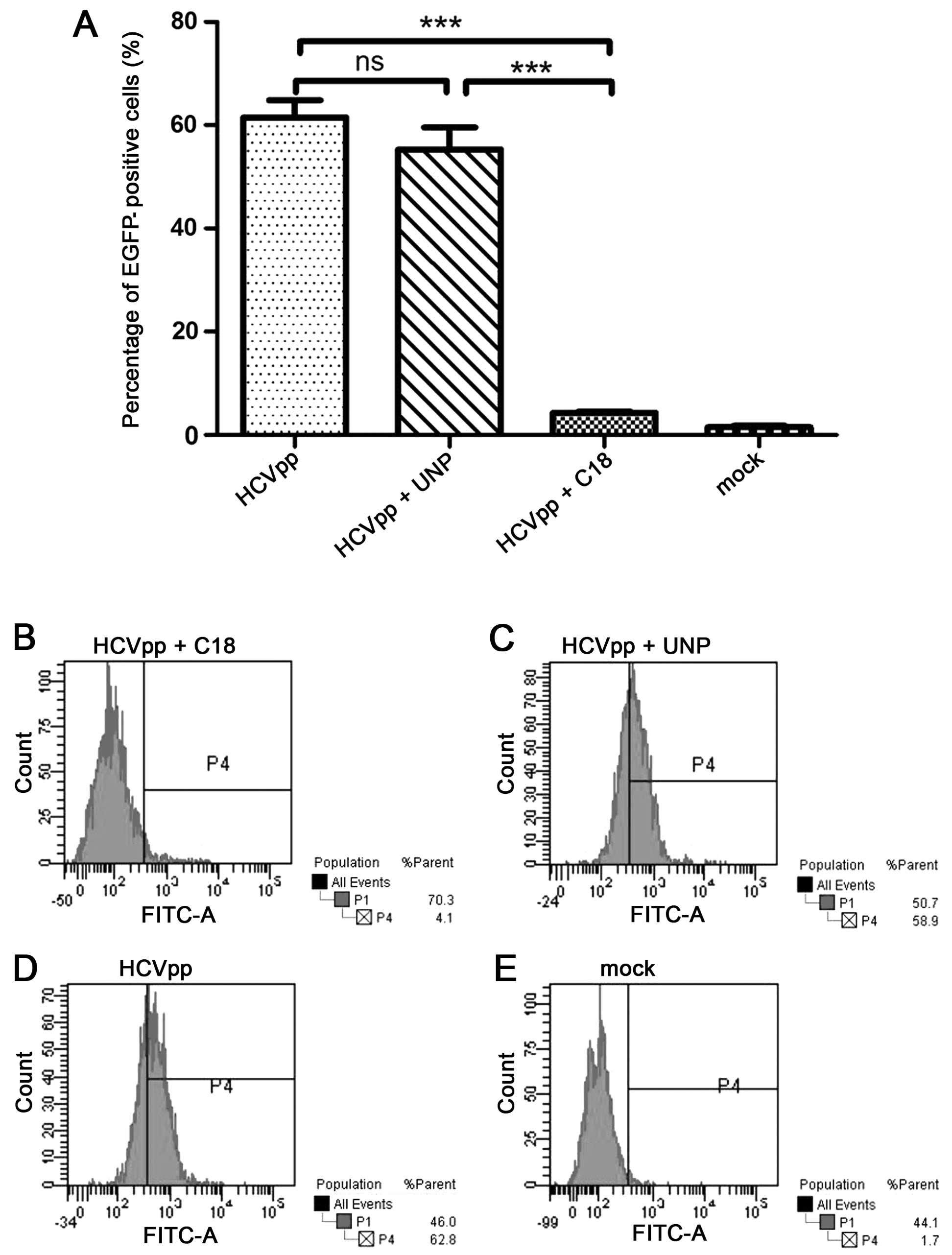
peptide used in the inhibition assay was 100 μg/ml (Fig. 5). It is indicated that peptide C18
significantly inhibited HCVpp from entering Huh7.5 cells detected
by flow cytometry. The HCVpp entry was notably inhibited in the
presence of peptide C18, compared to the group of HCVpp and the
group of the unrelated peptide (Fig.
6).
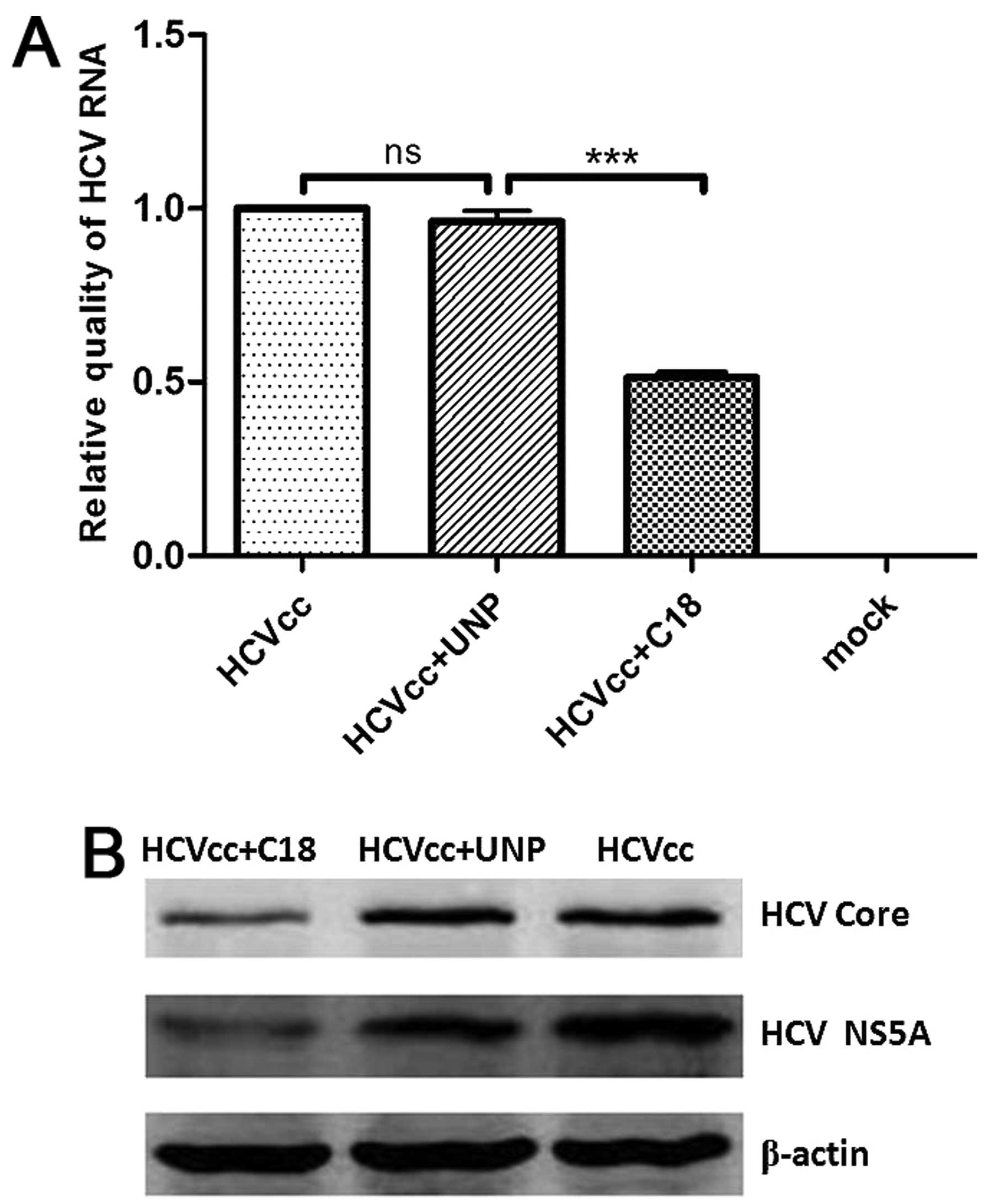
Furthermore, quantitative real-time RT-PCR
demonstrated that the level of HCV RNA in HCVcc-infected Huh7.5
cells was significantly lower in the presence of peptide C18,
compared to Huh7.5 cells incubated with HCVcc and the unrelated
peptide (Fig. 7A). Western
blotting revealed that the expression of the HCV core and NS5A
proteins decreased in HCVcc-infected Huh7.5 cells incubated with
peptide C18, compared to cells incubated with HCVcc and the
unrelated peptide (Fig. 7B).
These results demonstrated that peptide C18 is able to block both
HCVpp and HCVcc entering the host cells.
Discussion
Entry of hepatitis C virus (HCV) into cells may
involve the cellular components CD81 tetraspanin (20,21), the scavenger receptor class B type
I (SR-BI) (22,23), low-density lipoprotein receptor
(LDLR) (24), the mannose-binding
lectins DC-SIGN and L-SIGN (25),
glycosaminoglycans (GAGs) (26),
and the junction proteins claudin-1 (CLDN1) (27) and occluding (OCLN) (28). The HCV envelope glycoprotein E2 is
also thought to play a major role in virus-cell attachment.
Inhibition of the process by which viruses bind to their target
cells may provide an effective way to block viral entry and
infection (10).
The emergence of entry inhibitors has derived from
anti-HIV infection studies (29–31). Enfuvirtide, licensed in 2003, is
the first drug in this class and provides significant clinical
anti-HIV activity. Enfuvirtide is a 36 amino acid peptide that is
based on the stem region of HIV gp41, a viral envelope protein that
mediates the fusion of HIV with host cells (29). This new type of viral inhibitor
may also provide a novel mechanism to treat other types of viral
infection.
There have been some successful examples in the
study of HCV entry inhibitors. Receptor-mimics or neutralizing
antibodies targeting HCV candidate cell receptors such as CD81,
SR-BI and CLDN1 may be potential novel antiviral strategies
(10,32). The CD81-like small peptide
ATWVCGPCT, which aligns with 153–161 of the hCD81 sequence, is able
to block the CD81 binding site of the HCV E2 protein (33). A CD81-binding peptide and mimotope
of the HCV E2 protein, SPQYWTGPA, can competitively inhibit the
binding of HCV E2 to native CD81-expressing MOLT-4 cells (34). However, cell receptor-based
antagonists potentially affect the expression of these cell
receptors and disrupt receptor associations with other cell surface
proteins. Therefore, the identification of viral protein-based
inhibitory compounds may be a superior and safer strategy.
In the present study, we expressed truncated HCV
E2661 protein in E. coli, and selected the
peptides which specifically bound to HCV E2661 protein
from a phage display peptide library. Phage display technology has
been used in a number of studies for epitope mapping and the
identification of peptides which bind to target proteins (33–36). The heptapeptide library used in
this study contained a complexity of 2.7×109 individual
clones, representing the entire obtainable repertoire of 7-mer
peptide sequences (207=1.28×109). As a
result, effective panning was conducted after three rounds
(Fig. 1). Although HCV
E2661 protein was prokaryotically expressed with a 6X
His tag, the negative control HCV proteins NS3 and NS5B were also
expressed with 6X His tags. Therefore, the third round phages did
not bind to the 6X His tags, as they had a higher binding
reactivity for E2661 compared to the negative control
proteins with the same tags (Fig.
2). Eventually, four peptides which were able to bind
specifically to the HCV E2661 protein were obtained. Of
these peptides, C18, the candidate peptide with the greatest
affinity for E2661, was selected for subsequent
analysis.
Investigation of virus-cell interactions were
blocked until the development of HCVpp and HCVcc, which enabled
efficient growth of HCV in cell culture and provided valuable tools
to understand the viral life cycle and in the search for new
antiviral compounds. Baldick et al screened a small molecule
library and identified a potent HCV-specific triazine inhibitor,
EI-1, which blocked the cellular entry of a series of HCVpp and
HCVcc with E1–E2 sequences prepared from various HCV isolates
(genotype 1a, 1b and 2a) (37).
Additionally, a 16-residue polypeptide containing a portion of the
E2 transmembrane domain inhibited HCVpp infection at concentrations
up to 50 mM (38). Hong et
al prokaryotically expressed a 50 amino acid C-terminal
region-truncated HCV E2 protein (genotype 2a) and performed
biopanning using a phage display peptide against HCV-truncated E2.
The peptide pep7-1 was found to notably decrease the infectivity of
HCVcc in the cell culture (39).
Similarly, in the present study, it was shown that
short peptides with a high affinity for E2661 were
capable of inhibiting the entry of HCV into cells, and serve as
potential antiviral agents. The conserved motif WPWXXXR was
identified from the HCV E2 protein with genotype 1a (Fig. 4). The representative synthetic
peptide C18 demonstrated a good ability to inhibit the cell entry
of HCVpp (with the E2 sequence from genotype 1b, Fig. 6) and HCVcc (with the E2 sequence
from genotype 2a, Fig. 7). These
results confirm the assumption that, viruses with different
genotypes enter cells via a similar mechanism.
Notably, if the solution of peptide and HCVpp or
HCVcc were not removed from the cells within 16 h, the entry of HCV
(as indicated by the expression of the viral reporter EGFP, or HCV
RNA and viral protein expression) was not significantly different
in HCVpp- or HCVcc-infected cells incubated with C18 or the
unrelated peptide (data not shown). This may be explained by the
fact that if the HCVpp or HCVcc and peptide solution were not
removed, the peptide may undergo degradation and/or endocytosis in
the cells; thereby, weakening the inhibitory effect of peptide C18
on viral entry to the cells. Entry inhibitors block virus
attachment and entry, and cannot exert an effect once the viruses
have entered the cells.
Peptide C18 with the sequence WPWHNHR was identified
in the present study and is a potential candidate for HCV
therapeutic intervention. Entry inhibitors, a novel type of viral
inhibitor, may be an effective anti-HCV strategy, regardless of the
genotype of the virus, and may eventually provide a valuable
component of the optimal therapy for HCV infection. A novel
cocktail therapy, using a combination of entry inhibitors, HCV
proteases and polymerase inhibitors, with a backbone of pegIFN/RBV,
may have preferable, distinct modes of action and lead to
resistance to HCV infection.
Acknowledgements
We gratefully acknowledge Professor Charles M. Rice
for generously providing the Huh7.5 cell line and the HCV
full-length clones and Professor Wenjie Tan for the kind gift of
plasmids for construction of the HCVpp. This study was supported by
grants from the National Natural Science Foundation of China (nos.
30600021 and 81171637).
References
|
1
|
McGivern DR and Lemon SM: Virus-specific
mechanisms of carcinogenesis in hepatitis C virus associated liver
cancer. Oncogene. 30:1969–1983. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bacon BR and McHutchison JG: Into the
light: strategies for battling hepatitis C. Am J Manag Care.
13(Suppl 12): S319–S326. 2007.PubMed/NCBI
|
|
3
|
Berwyn C: Molecular virology of hepatitis
C virus. J Gen Virol. 78:2397–2410. 1997.
|
|
4
|
Penin F, Dubuisson J, Rey FA, Moradpour D
and Pawlotsky JM: Structural biology of hepatitis C virus.
Hepatology. 39:5–19. 2004. View Article : Google Scholar
|
|
5
|
Reed KE and Rice CM: Overview of hepatitis
C virus genome structure, polyprotein processing, and protein
properties. Curr Top Microbiol Immunol. 242:55–84. 2000.PubMed/NCBI
|
|
6
|
Flint M and McKeating JA: The role of the
hepatitis C virus glycoproteins in infection. Rev Med Virol.
10:101–117. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Simmonds P: Genetic diversity and
evolution of hepatitis C virus - 15 years on. J Gen Virol.
85:3173–3188. 2004.PubMed/NCBI
|
|
8
|
Bartosch B, Verney G, Dreux M, Donot P,
Morice Y, Penin F, Pawlotsky JM, Lavillette D and Cosset FL: An
interplay between hypervariable region 1 of the hepatitis C virus
E2 glycoprotein, the scavenger receptor BI, and high-density
lipoprotein promotes both enhancement of infection and protection
against neutralizing antibodies. J Virol. 79:8217–8229. 2005.
View Article : Google Scholar
|
|
9
|
Callens N, Ciczora Y, Bartosch B, Vu-Dac
N, Cosset FL and Pawlotsky JM: Basic residues in hypervariable
region 1 of hepatitis C virus envelope glycoprotein E2 contribute
to virus entry. J Virol. 79:15331–15341. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zeisel MB, Fofana I, Fafi-Kremer S and
Baumert TF: Hepatitis C virus entry into hepatocytes: molecular
mechanisms and targets for antiviral therapies. J Hepatol.
54:566–576. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Owsianka A, Clayton RF, Loomis-Price LD,
McKeating JA and Patel AH: Functional analysis of hepatitis C virus
E2 glycoproteins and virus-like particles reveals structural
dissimilarities between different forms of E2. J Gen Virol.
82:1877–1883. 2001.
|
|
12
|
Lavillette D, Tarr AW, Voisset C, Donot P,
Bartosch B, Bain C, Patel AH, Dubuisson J, Ball JK and Cosset FL:
Characterization of host-range and cell entry properties of the
major genotypes and subtypes of hepatitis C virus. Hepatology.
41:265–274. 2005. View Article : Google Scholar
|
|
13
|
Flint M, Dubuisson J, Maidens C, Harrop R,
Guile GR, Borrow P and McKeating JA: Functional characterization of
intracellular and secreted forms of a truncated hepatitis C virus
E2 glycoprotein. J Virol. 74:702–709. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yao M, Fang HL, Yin W, Lei YF, Yang J, Sun
MN, Tian JH and Lü X: Prokaryotic expression and purification of
truncated hepatitis c virus envelope glycoprotein E2. Sci Technol
Eng. 9:2296–2298. 2009.
|
|
15
|
Bartosch B, Dubuisson J and Cosset FL:
Infectious hepatitis C virus pseudo-particles containing functional
E1–E2 envelope protein complexes. J Exp Med. 197:633–642.
2003.PubMed/NCBI
|
|
16
|
Hsu M, Zhang J, Flint M, Logvinoff C,
Cheng-Mayer C, Rice CM and McKeating JA: Hepatitis C virus
glycoproteins mediate pH-dependent cell entry of pseudotyped
retroviral particles. Proc Natl Acad Sci USA. 100:7271–7276. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lindenbach BD, Evans MJ, Syder AJ, Wölk B,
Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR,
McKeating JA and Rice CM: Complete replication of hepatitis C virus
in cell culture. Science. 309:623–626. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lindenbach BD, Meuleman P, Ploss A,
Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM,
Major ME, Leroux-Roels G and Rice CM: Cell culture-grown hepatitis
C virus is infectious in vivo and can be recultured in vitro. Proc
Natl Acad Sci USA. 103:3805–3809. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang J, Lei YF, Yin W, Wei SH, An QX, Lv
X, Hu XB and Xu ZK: Production and characterization of monoclonal
antibody specific for NS3 helicase of hepatitis C virus. Hybridoma.
27:181–186. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pileri P, Uematsu Y, Campagnoli S, Galli
G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G
and Abrignani S: Binding of hepatitis C virus to CD81. Science.
282:938–941. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang J, Randall G, Higginbottom A, Monk
P, Rice CM and McKeating JA: CD81 is required for hepatitis C virus
glycoprotein-mediated viral infection. J Virol. 78:1448–1455. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Scarselli E, Ansuini H, Cerino R,
Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R
and Vitelli A: The human scavenger receptor class B type I is a
novel candidate receptor for the hepatitis C virus. EMBO J.
21:5017–5025. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bartosch B, Vitelli A, Granier C, Goujon
C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A and
Cosset FL: Cell entry of hepatitis C virus requires a set of
co-receptors that include the CD81 tetraspanin and the SR-B1
scavenger receptor. J Biol Chem. 278:41624–41630. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Agnello V, Abel G, Elfahal M, Knight GB
and Zhang QX: Hepatitis C virus and other flaviviridae viruses
enter cells via low density lipoprotein receptor. Proc Natl Acad
Sci USA. 96:12766–12771. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cormier EG, Durso RJ, Tsamis F, Boussemart
L, Manix C, Olson WC, Gardner JP and Dragic T: L-SIGN (CD209L) and
DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis
C virus. Proc Natl Acad Sci USA. 101:14067–14072. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Garson JA, Lubach D, Passas J, Whitby K
and Grant PR: Suramin blocks hepatitis C binding to human hepatoma
cells in vitro. J Med Virol. 57:238–242. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Evans MJ, von Hahn T, Tscherne DM, Syder
AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD and
Rice CM: Claudin-1 is a hepatitis C virus co-receptor required for
a late step in entry. Nature. 446:801–805. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ploss A, Evans MJ, Gaysinskaya VA, Panis
M, You H, de Jong YP and Rice CM: Human occludin is a hepatitis C
virus entry factor required for infection of mouse cells. Nature.
457:882–886. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cervia JS and Smith MA: Enfuvirtide
(T-20): a novel human immunodeficiency virus type 1 fusion
inhibitor. Clin Infect Dis. 37:1102–1106. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dorr P, Westby M, Dobbs S, Griffin P,
Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier
C, Webster R, Armour D, Price D, Stammen B, Wood A and Perros M:
Maraviroc (UK-427, 857), a potent, orally bioavailable, and
selective small-molecule inhibitor of chemokine receptor CCR5 with
broad-spectrum antihuman immunodeficiency virus type 1 activity.
Antimicrob Agents Chemother. 49:4721–4732. 2005. View Article : Google Scholar
|
|
31
|
Fätkenheuer G, Pozniak AL, Johnson MA,
Plettenberg A, Staszewski S, Hoepelman AI, Saag MS, Goebel FD,
Rockstroh JK, Dezube BJ, Jenkins TM, Medhurst C, Sullivan JF,
Ridgway C, Abel S, James IT, Youle M and van der Ryst E: Efficacy
of short-term monotherapy with maraviroc, a new CCR5 antagonist, in
patients infected with HIV-1. Nat Med. 11:1170–1172.
2005.PubMed/NCBI
|
|
32
|
Shimizu YK, Hijikata M, Iwamoto A, Alter
HJ, Purcell RH and Yoshikura H: Neutralizing antibodies against
hepatitis C virus and the emergence of neutralization escape mutant
viruses. J Virol. 68:1494–1500. 1994.PubMed/NCBI
|
|
33
|
Cao J, Zhao P, Miao XH, Zhao LJ, Xue LJ
and Qi ZT: Phage display selection on whole cells yields a small
peptide specific for HCV receptor human CD81. Cell Res. 13:473–479.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cao J, Liao XL, Wu SM, Zhao P, Zhao LJ, Wu
WB and Qi ZT: Selection of a phage-displayed peptide recognized by
monoclonal antibody directed blocking the site of hepatitis C virus
E2 for human CD81. J Microbiol Methods. 68:601–604. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Molek P, Strukelj B and Bratkovic T:
Peptide phage display as a tool for drug discovery: targeting
membrane receptors. Molecules. 16:857–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim MS, Park C, Lee JH and Myung H:
Selection and target-site mapping of peptides inhibiting HCV NS5B
polymerase using phage display. J Microbiol Biotechnol. 18:328–333.
2008.PubMed/NCBI
|
|
37
|
Baldick CJ, Wichroski MJ, Pendri A, Walsh
AW, Fang J, Mazzucco CE, Pokornowski KA, Rose RE, Eggers BJ, Hsu M,
Zhai W, Zhai G, Gerritz SW, Poss MA, Meanwell NA, Cockett MI and
Tenney DJ: A novel small molecule inhibitor of hepatitis C virus
entry. PLoS Pathog. 6:e10010862010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu R, Tewari M, Kong R, Zhang R,
Ingravallo P and Ralston R: A peptide derived from hepatitis C
virus E2 envelope protein inhibits a post-binding step in HCV
entry. Antiviral Res. 86:172–179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hong HW, Lee SW and Myung H: Selection of
peptides binding to HCV E2 and inhibiting viral infectivity. J
Microbiol Biotechnol. 20:1769–1771. 2010.PubMed/NCBI
|