Introduction
Ornithine decarboxylase antizyme 1 (OAZ1) is a
member of the ornithine decarboxylase (ODC) antizyme family, which
targets ODC for ubiquitin-independent proteasome degradation,
thereby inhibiting the synthesis of polyamines and cell
proliferation (1). OAZ1 can also
inhibit cell growth through ODC-independent mechanisms, including
the inhibition of cellular uptake of polyamines by inactivating the
polyamine uptake transporter, promoting cyclin D1 degradation and
preventing ubiquitin-independent msp1 degradation (2–5).
Studies of cancer have also proved that OAZ1 has tumor suppressor
activities and it can affect the apoptosis and proliferation of
multiple tumor cell lines (6–8).
Several investigators have studied the involvement
of OAZ1 in tongue squamous cell carcinoma (TSCC). In p53-knockout
mice, overexpression of the OAZ1 protein inhibited the production
of keratinocyte proliferation marker, keratin 6 (K6),
whereas it upregulated the expression of keratinocyte
differentiation marker, loricrin (LOR) (9). In hamster malignant oral
keratinocytes, ectopic expression of OAZ1 induced epithelial
differentiation with the overexpression of involucrin (IVL)
(10). In several human oral
cancer cell lines, the expression level of the OAZ1 gene was
downregulated (11,12). A previous study has shown that
OAZ1 induces the conversion of the human tongue squamous cancer
cell line, UM1, to the less metastatic type, UM2, with the
hypomethylation of genome DNA and histone H3 lysine 9 dimethylation
(12). However, the overall
effects of OAZ1 on cellular proliferation and differentiation in
human oral cancer cells and the underlying mechanism remain to be
studied.
In the present study, the lentiviral vector
containing the OAZ1 gene was constructed and transfected
into the human tongue cancer cell line, SCC15, to evaluate the
effects of OAZ1 expression on proliferation and
differentiation of the cells. The results showed that the stable
expression of OAZ1 in SCC15 inhibited the cell proliferation
rate and induced G0/G1 arrest. OAZ1
expression also induced the formation of epithelial islands with
elevation of several differentiation marker genes (K10,
FLG and LOR). OAZ1 was also found to inhibit Smad
nuclear interacting protein 1 (SNIP1) and silencing of SNIP1
increased the expression of LOR in SCC15 cells. The results
of the study proved that OAZ1 can simultaneously inhibit
proliferation and induce differentiation of oral cancer cells in
humans.
Materials and methods
Cell culture and transfection
The human oral cancer cell line, SCC15, was obtained
from the American Type Culture Collection (ATCC, Manassas, VA, USA)
and maintained in Dulbecco’s modified Eagles medium/F12 medium
supplemented with 10% (v/v) fetal bovine serum at 37°C in 5%
CO2. The lentiviral vector (pLVX-Neo-IRES-ZsGreen)
containing the green fluorescent protein (GFP) gene,
ZsGreen, was ligated to OAZ1 cDNA (loss of the
frameshift site) to construct the stable expression vector of
OAZ1 (13). The cells were
transferred to 6-well plates and the OAZ1-containing vector
or control vector was transfected. When GFP was expressed, G418
(1,000 μg/ml) and flow cytometry were used to screen the stably
transfected clones. SCC15 cells transfected with the
OAZ1-expressed vector or the negative-control vector are
referred as SCC15/OAZ1 and SCC15/GFP.
Cell proliferation assay
Cells (1×104/well) were transferred in 3
replicates to 96-well plates and cultured on standard conditions.
At the time-points of 24, 48 and 72 h, the viability of the cells
was analyzed after the addition of 20 μl MTT (5 mg/ml) and dimethyl
sulfoxide. Absorbance was determined using the Multiskan MK3
Microplate Reader (Thermo Scientific, Waltham, MA, USA) at a
wavelength of 490 nm.
Cell cycle assay
Cells (2×105/well) were transferred in 3
replicates to 6-well plates. After 24 h of starvation, the cells
were recovered for 36 h before analysis. The cells were treated
according to the manufacturer’s instructions for the cell cycle
detection kit (Nanjing KeyGen Biotech, Nanjing, China) and
submitted to flow cytometric analysis.
Morphological observation
Cells (1×103/well) were plated in 100×20
mm plates and observed under an inverted microscope after 1
week.
RNA interference
SNIP1-specific small hairpin RNA (shRNA) (GeneChem
Co., Ltd., Shanghai, China) including sh-400
(5′-ATGCTGCTTTGAACAAGAC-3′), sh-401 (5′-TCGATGTATGTACATGACT-3′),
sh-402 (5′-TTAGAC GTTCTCCTTTGGT-3′) and the negative control, sh-NC
(5′-TTCTCCGAACGTGTCACGT-3′), were synthesized. SCC15 cells were
transferred to 6-well plates. When the cells reached 80% confluent,
shRNA was transfected with Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA) in 250 μl Opti-MEM (Gibco, Carlsbad, CA, USA).
Sh-NC was used as the negative control.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated using the RNAiso plus kit
(Takara Bio, Inc., Shiga, Japan). Full-length cDNA was generated
using the PrimeScript RT reagent kit (Takara, Dalian, China). cDNA
was subjected to RT-qPCR analysis using SYBR-Green I (Takara) and
results are reported as the ratio alteration among groups. The
primers used are presented in Table
I.
 | Table IPrimers used for RT-qPCR. |
Table I
Primers used for RT-qPCR.
| Primer | Sequence
(5′→3′) | Product length
(bp) |
|---|
| OAZ1
(NM_004152) | F:
TCTCCCTCCACTGCTGTAGTAACC
R: GTTGAGAATCCTCGTCTTGTCGTT | 198 |
| E-cadherin
(NM_004360) | F:
CTACAATGCCGCCATCGCTTA
R: CACTGATGACTCCTGTGTTCCTGTT | 98 |
| K1 (NM_006121) | F:
CGGAACTGAAGAACATGCAG
R: CATATAAGCACCATCCACATCC | 128 |
| K6A
(NM_005554) | F:
CAAGGCCCAATATGAGGAGA
R: GCAATCTCCTGCTTGGTGTT | 135 |
| K10
(NM_000421) | F:
AAACCATCGATGACCTTAAAAATC
R: GCGCAGAGCTACCTCATTCT | 134 |
| K19
(NM_002276) | F:
GCCACTACTACACGACCATCC
R: CAAACTTGGTTCGGAAGTCAT | 126 |
| FLG
(NM_002016) | F:
TTTCGGCAAATCCTGAAGAATCC
R: ACTGTGCTTTCTGTGCTTGTG | 195 |
| IVL
(NM_005547) | F:
CTGCCTCAGCCTTACTGTGA
R: TGGGTATTGACTGGAGGAGG | 133 |
| LOR
(NM_000427) | F:
GCACCGATGGGCTTAGAG
R: AGAAACCAAAGAGGCTAAACAG | 130 |
| SNIP1
(NM_024700) | F:
AAGAAGCAAGTCTCCTCGCAG
R: GTTCTGATGGTTCCCTGTGCT | 128 |
| GAPDH
(NM_002046) | F:
CGCTGAGTACGTCGTGGAGTC
R: GCTGATGATCTTGAGGCTGTTGTC | 172 |
Immunoblotting
The cells were washed twice in phospate-buffered
saline and lysed in radio-immunoprecipitation assay lysis buffer
(with phenylmethanesulfonyl fluoride). Equal amounts of total
protein were separated by 12% SDS-PAGE, transferred to a
polyvinylidene fluoride membrane (Millipore, Bedford, MA, USA) and
probed with OAZ1 (Abcam, Hong Kong, China); K1, K10, IVL, LOR,
filaggrin (FLG) and SNIP1 (Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA); and GAPDH (Cell Signaling Technology, Shanghai,
China) antibodies and subsequently with the horseradish
peroxidase-conjugated secondary antibody. The blots were evaluated
with the ECL detection system (Advansta, Menlo Park, CA, USA). The
protein bands of interest were quantified using FluorChem 8900
image analysis software (Alpha Innotech, San Leandro, CA, USA), and
the integrated signal densities were normalized to GAPDH first, and
subsequently expressed in terms of the fractional abundance
relative to the control cells. These experiments were replicated 3
times.
Statistical analysis
SPSS 16.0 (SPSS, Inc., Chicago, IL, USA) was used
for statistical analysis. Data are presented as means ± standard
deviation. One-way analysis of variance was used and P<0.05 was
considered to indicate a statistically significant difference.
Results
Overexpression of OAZ1 induces
morphological changes in SCC15 cells
The lentiviral vector expressing OAZ1 and GRF
(ZsGreen), and the control vector expressing GFP were
transfected into SCC15 cells separately and stable expression of
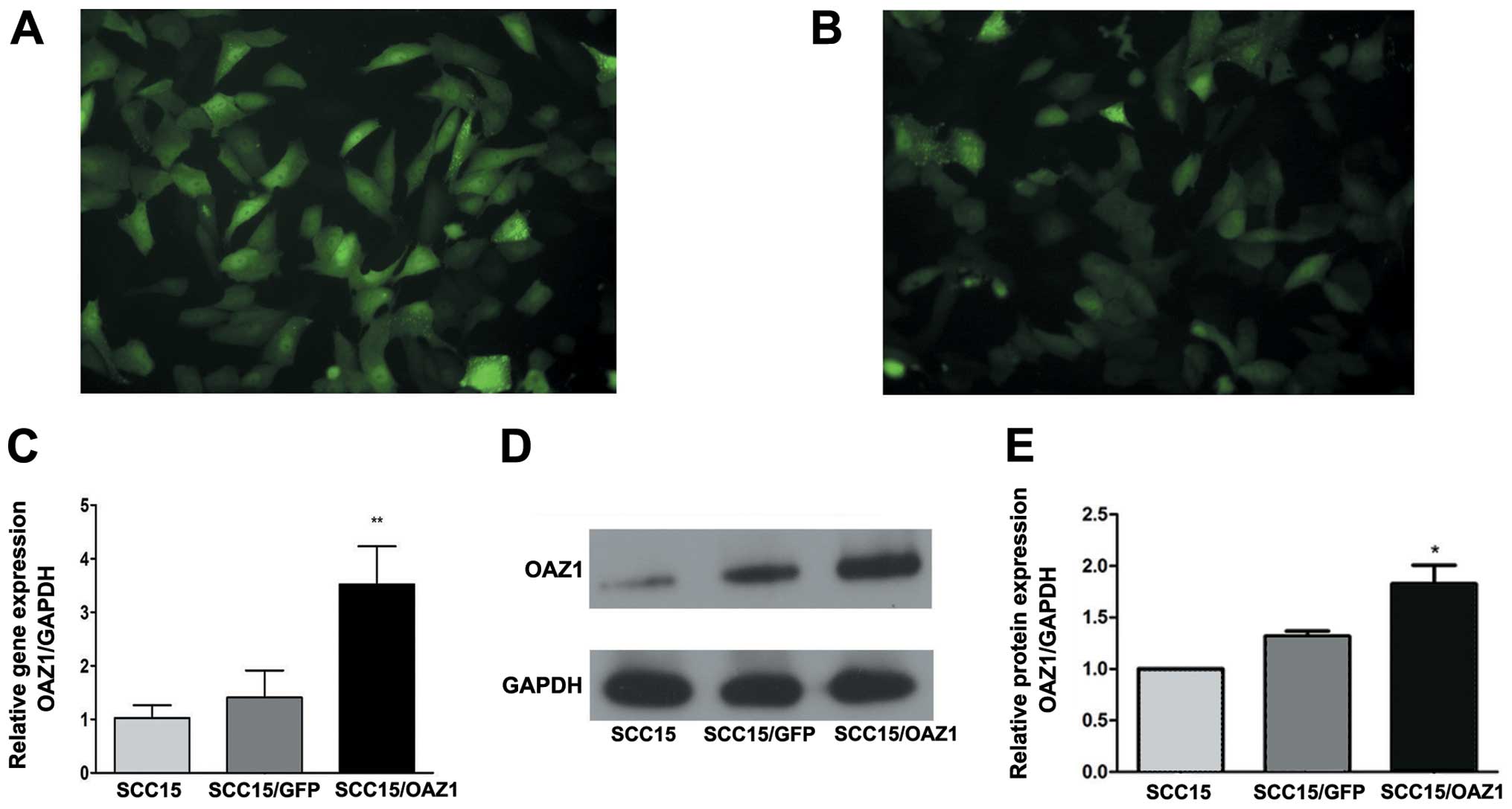
GFP was observed in the two groups (Fig. 1A and B). Quantitative analysis
showed that the mRNA level of OAZ1 was significantly
elevated in the OAZ1-transfected group (Fig. 1C). The OAZ1 protein expression was
verified by western blotting (Fig.
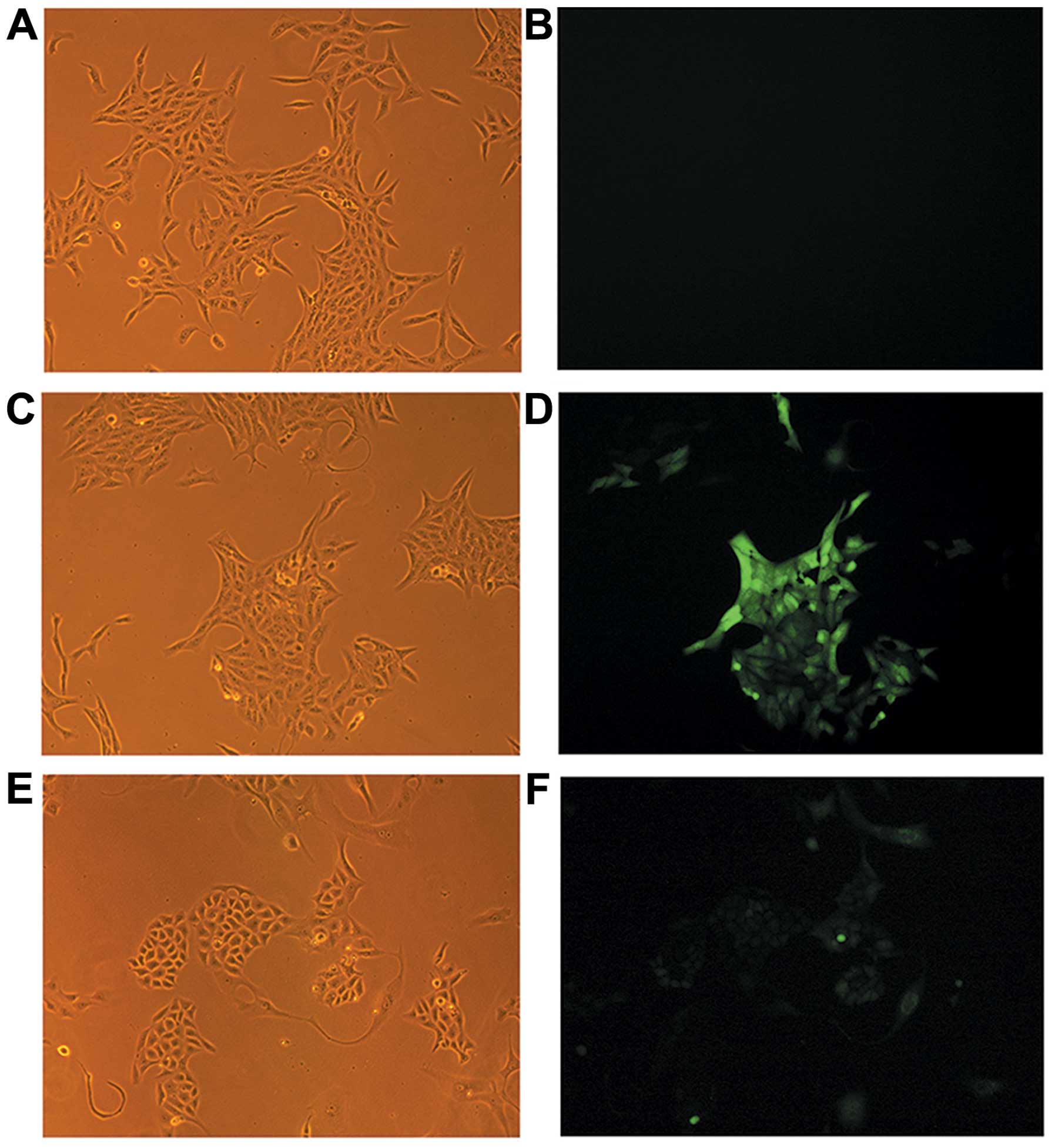
1D). The cells transfected with the control vector (Fig. 2C and D) demonstrated the typical
appearance of SCC15 (Fig. 2A and
B). However, the appearance of the OAZ1 transfectants
exhibited evidence of epithelial island formation and increased
cellular junctions indicating the terminal differentiation of the
cells (Fig. 2E and F).
OAZ1 affects the expression of marker
genes in cell differentiation
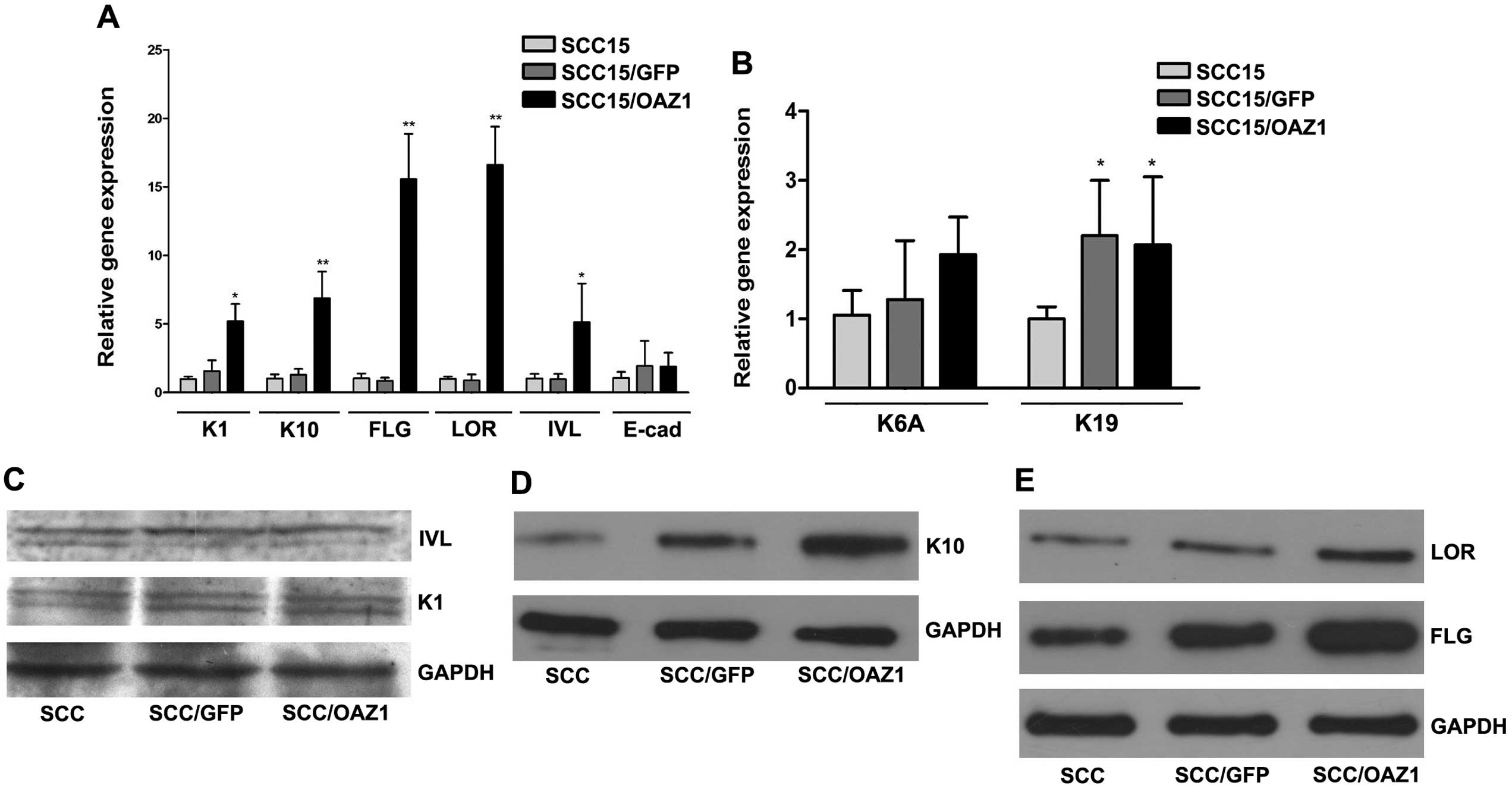
OAZ1-transfected SCC15 cells showed
morphological alterations indicating terminal differentiation. To
further confirm the effects of OAZ1 on the differentiation of
SCC15, the expression of several reported differentiation marker
genes were examined. Elevation of early terminal differentiation
genes, K1, K10 and IVL, was observed in
OAZ1-expression cells. As for late terminal differentiation
genes, the elevation of FLG and LOR was also
observed, whereas the mRNA level of E-cadherin remained stable
(Fig. 3A). When detected by
western blot analysis, K10, LOR and FLG showed increased expression
on protein level (Fig. 3C–E).
OAZ1 inhibits the proliferation of SCC15
cells
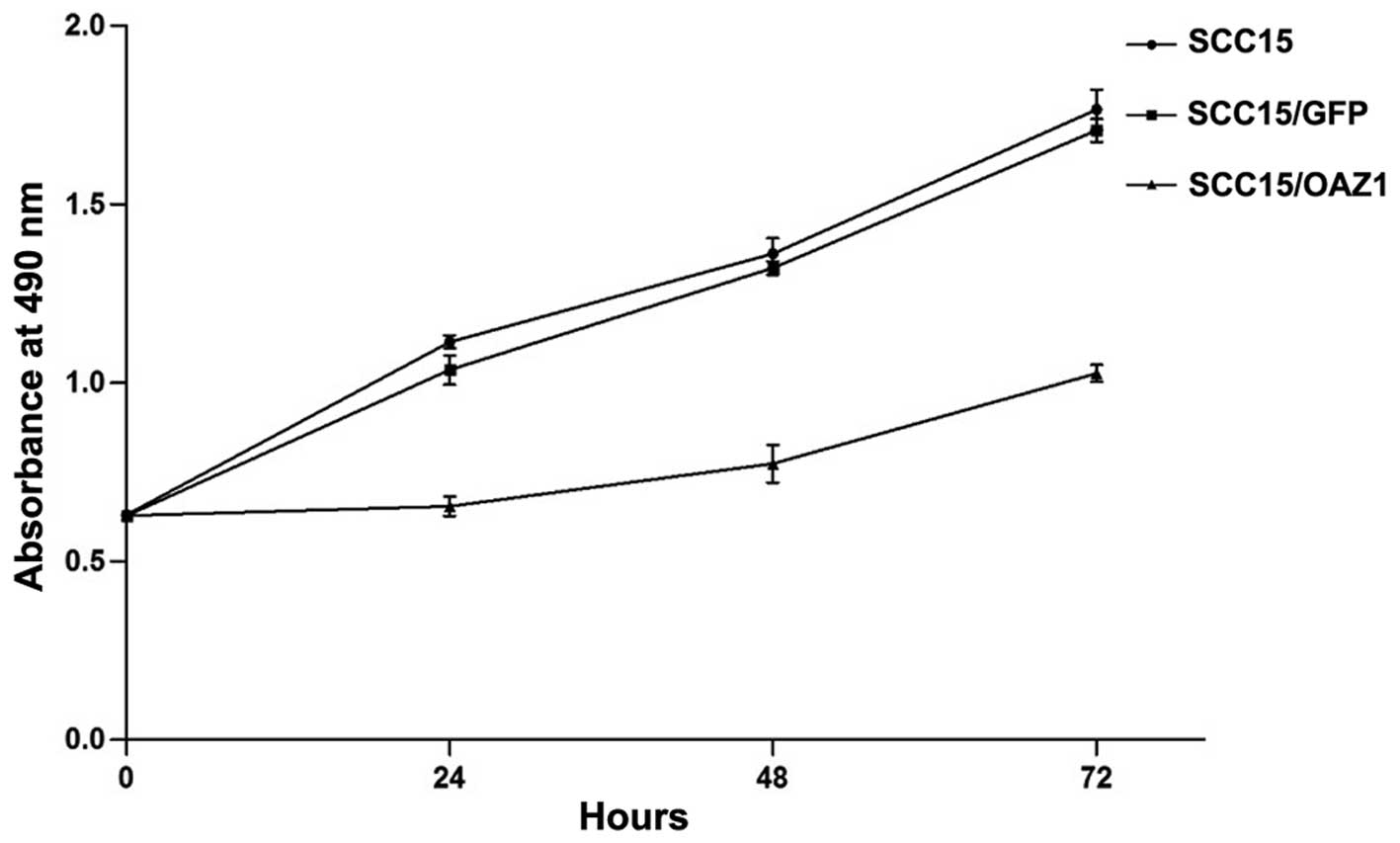
MTT assay was performed to detect the effects of
OAZ1 expression on the proliferation of SCC15. Compared to
the control, SCC15 cells stably expressing OAZ1 showed a
significant reduction in the cell growth rate (P<0.001 compared
to the other groups) (Fig. 4),
indicating that OAZ1 expression inhibited the proliferation
of SCC15.
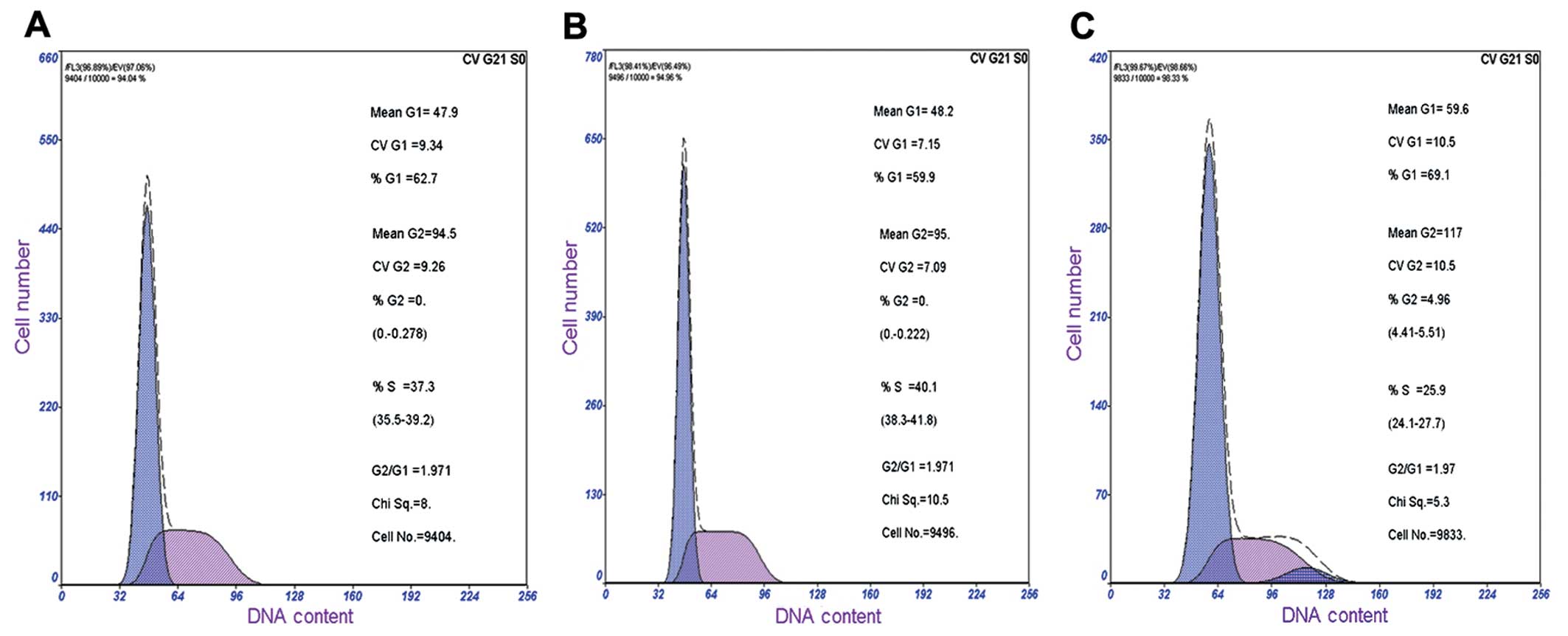
The cell cycle profiles of the
OAZ1-transfected SCC15 cells and the control cells were
further examined (Fig. 5). The
flow cytometry analysis showed that the cells in the G1
phase were increased in OAZ1-expressed SCC15 cells (65.26%)
compared to the control (55.83%). Whereas cells in the S phase were
decreased to 30.07% compared to the control (42.47%) (Table II). These results indicate that
OAZ1 may inhibit the cell proliferation through induction of
G0/G1 arrest.
 | Table IIEffect of OAZ1 on cell cycle in SCC15
cells. |
Table II
Effect of OAZ1 on cell cycle in SCC15
cells.
| Groups | G1,
% | S, (%) | G2,
% |
|---|
| SCC15 | 55.83±5.95 | 42.47±4.49 | 1.73±1.57 |
| SCC15/GFP | 54.83±4.43 | 44.17±3.69 | 1.36±1.52 |
| SCC15/OAZ1 | 65.26±3.32a | 30.07±4.42b | 4.64± 2.58 |
The expression of two genes reported to be elevated
during epithelial cell proliferation, K6A and K19,
were also investigated. RT-qPCR results showed that K19
expression was significantly elevated (P<0.05), and the level of
K6A remained stable (Fig.
3B).
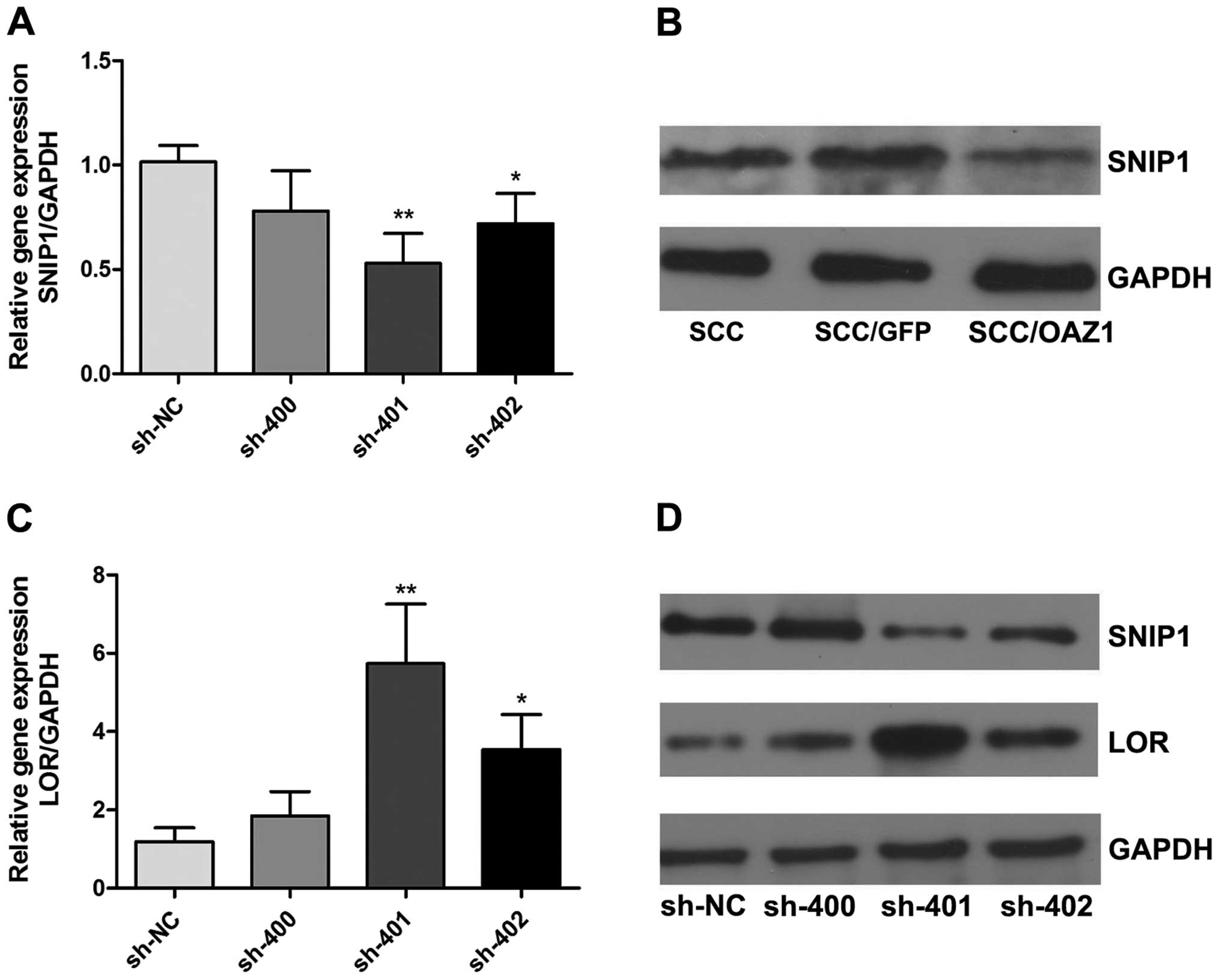
OAZ1 inhibits LOR by promoting SNIP1
degradation
OAZ1 can induce the degradation of SNIP1, leading to
the release of CREB-binding protein (CBP), a key factor in the
regulation of LOR transcription (14,15). To further explore the mechanism of
LOR upregulation by OAZ1, the expression of SNIP1 in
OAZ1-transfected cells was examined. The SNIP1 protein level
was downregulated (Fig. 6B) in
OAZ1 transfectants. The RNA interference experiment showed
that inhibition of SNIP1 in the SCC15 cells induces the
expression of LOR (Fig. 6A, C
and D). These results indicate that OAZ1 may increase
LOR expression through the degradation of SNIP1.
Discussion
OAZ1 is an important regulator of cellular
differentiation and proliferation. Studies have focused on its
unique frameshift regulation mechanism, function in regulating
polyamine metabolism and ability of inducing ubiquitin-independent
degradation of numerous large molecules. Previously, it has been
reported that OAZ1 has tumor suppressor activities and it can
affect the apoptosis and proliferation of multiple tumor cell lines
(6–8,16).
There is also evidence that overexpression of OAZ1 can
induce the differentiation of certain cancer cells (10,11,17). The mechanism underlying the tumor
suppressor activity of OAZ1 remains unclear and further exploration
may help with the understanding.
TSCC is one of the most common malignant oral
tumors. Previous studies of the oral cancer cell line UM1 have
proved that TSCC is an appropriate model to investigate the
detailed molecular mechanisms of differentiation and proliferation
regulation in oral cancer development (11,12). In the present study, the human
TSCC cell line SCC15 was studied and a lentivirial vector was
constructed that stably expresses the frame shift form of
OAZ1 to continuously trigger the degradation of ODC. The
expression of OAZ1 in SCC15 was verified by western blot
analysis. Morphological study showed that the cells overexpressing
OAZ1 exhibit the formation of more epithelial islands, which
is a marker of terminal differentiation of epithelial cells. This
is also consistent with the study on the hamster oral cancer cell
line, HCPC-1 (10).
To elucidate the mechanism of OAZ1-induced
cell differentiation in SCC15 cells, the expression of several
terminal differentiation-associated genes was examined, including
K1, K10, FLG, IVL and LOR. All these genes are
marker genes in epithelial differentiation that have decreased
expression in epithelial cancer cells, which is resumed during
differentiation (18–25). In hamster oral cancer cells,
OAZ1 has been proved to promote the expression of LOR
and IVL (9,10). In the present study,
OAZ1-expressed SCC15 cells showed significantly increased
expression of the majority of the marker genes assessed, confirming
the positive role of OAZ1 in epithelial differentiation. The gene
involved in cellular junction, E-cadherin, was also examined as
closer conjunction of cells expressing OAZ1 was observed and
E-cadherin is reduced in squamous carcinoma (26). However, the mRNA level of
E-cadherin showed no significant difference between the
OAZ1-transfected SCC15 and control cells, which is
consistent with a previous study (11). This suggests that factors other
than E-cadherin may be involved in the induction of closer cellular
junctions for this condition.
The mechanism underlying the induction of LOR
expression in OAZ1-expressed SCC15 cells was further
explored. LOR is the component of the cell membrane in terminal
differentiated keratinocytes and p300/CBP can function as a bridge
during its transcription-activator complex formation (15). SNIPI has been reported to
competitively inhibit the binding of CBP in the promotor region,
leading to the inhibition of transcription (14,27). The present study showed that
OAZ1 overexpression promoted the degradation of SNIP1 and
the silencing of SNIP1 by RNA interference resumed the level of LOR
in SCC15 cells. These results indicate that OAZ1 may induce
LOR expression partly through SNIP1 degradation.
For the study of cell proliferation in
OAZ1-expressing SCC15 cells, the results showed that
overexpression of the antizyme significantly inhibited cell
proliferation with G0/G1 arrest. Two genes,
K6A and K19, were further examined that are elevated
during epithelial proliferation (9,26).
However, the results of RT-qPCR showed that K19 expression
was elevated instead of reduced and the level of K6A
remained stable. OAZ1 is assumed to inhibit the proliferation of
SCC15 cells without the involvement of regulating K6A and
K19 expression.
In conclusion, the study showed that OAZ1 induced
the formation of epithelial islands with significant upregulation
of epithelial terminal differentiation marker genes in SCC15.
OAZ1 expression also inhibited cell proliferation and
induced G0/G1 arrest. OAZ1 simultaneously
inhibited proliferation and induced differentiation of oral cancer
cells in humans. This may be a novel perspective to consider in the
induction of tumor cell differentiation, particularly in human oral
cancer.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 30901757), the
Natural Science Foundation of Guangdong Province (grant no.
10151008004000028) and the Leading Talent Project of Guangdong
Province.
References
|
1
|
Kahana C: Antizyme and antizyme inhibitor,
a regulatory tango. Cell Mol Life Sci. 66:2479–2488. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Newman RM, Mobascher A, Mangold U, et al:
Antizyme targets cyclin D1 for degradation. A novel mechanism for
cell growth repression. J Biol Chem. 279:41504–41511. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kasbek C, Yang CH, Yusof AM, Chapman HM,
Winey M and Fisk HA: Preventing the degradation of mps1 at
centrosomes is sufficient to cause centrosome reduplication in
human cells. Mol Biol Cell. 18:4457–4469. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kasbek C, Yang CH and Fisk HA: Mps1 as a
link between centrosomes and genomic instability. Environ Mol
Mutagen. 50:654–665. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kasbek C, Yang CH and Fisk HA: Antizyme
restrains centrosome amplification by regulating the accumulation
of Mps1 at centrosomes. Mol Biol Cell. 21:3878–3889. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dulloo I, Gopalan G, Melino G, et al: The
antiapoptotic DeltaNp73 is degraded in a c-Jun-dependent manner
upon genotoxic stress through the antizyme-mediated pathway. Proc
Natl Acad Sci USA. 107:4902–4907. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feith DJ, Origanti S, Shoop PL, Sass-Kuhn
S and Shantz LM: Tumor suppressor activity of ODC antizyme in
MEK-driven skin tumorigenesis. Carcinogenesis. 27:1090–1098. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsuji T, Todd R, Meyer C, et al: Reduction
of ornithine decarboxylase antizyme (ODC-Az) level in the
7,12-dimethylbenz(a)anthracene-induced hamster buccal pouch
carcinogenesis model. Oncogene. 16:3379–3385. 1998. View Article : Google Scholar
|
|
9
|
Feith DJ, Pegg AE and Fong LY: Targeted
expression of ornithine decarboxylase antizyme prevents upper
aerodigestive tract carcinogenesis in p53-deficient mice.
Carcinogenesis. 34:570–576. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsuji T, Usui S, Aida T, et al: Induction
of epithelial differentiation and DNA demethylation in hamster
malignant oral keratinocyte by ornithine decarboxylase antizyme.
Oncogene. 20:24–33. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tsuji T, Katsurano M, Ibaragi S, Shima K,
Sasaki A and Hu GF: Ornithine decarboxylase antizyme upregulates
DNA-dependent protein kinase and enhances the nonhomologous
end-joining repair of DNA double-strand breaks in human oral cancer
cells. Biochemistry. 46:8920–8932. 2007. View Article : Google Scholar
|
|
12
|
Yamamoto D, Shima K, Matsuo K, et al:
Ornithine decarboxylase antizyme induces hypomethylation of genome
DNA and histone H3 lysine 9 dimethylation (H3K9me2) in human oral
cancer cell line. PLoS One. 5:e125542010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu BP, Wang X, Ma WL, Zhang SY, Zheng WL
and Jiang L: Effect of ornithine decarboxylase antizyme 1 on the
erythroid differentiation in human leukemia cell line K562. Chin J
Biochem Mol Biol. 29:361–367. 2013.
|
|
14
|
Lin Y, Martin J, Gruendler C, et al: A
novel link between the proteasome pathway and the signal
transduction pathway of the bone morphogenetic proteins (BMPs). BMC
Cell Biol. 3:152002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jang SI and Steinert PM: Loricrin
expression in cultured human keratinocytes is controlled by a
complex interplay between transcription factors of the Sp1, CREB,
AP1, and AP2 families. J Biol Chem. 277:42268–42279. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu GY, Liao YF, Hsu PC, et al: Antizyme,
a natural ornithine decarboxylase inhibitor, induces apoptosis of
haematopoietic cells through mitochondrial membrane depolarization
and caspases’ cascade. Apoptosis. 11:1773–1788. 2006.PubMed/NCBI
|
|
17
|
Suzuki J, Murakami Y, Samejima K, Ohtani
KK and Oka T: Antizyme is necessary for conversion of pancreatic
tumor cells into glucagon-producing differentiated cells. Endocr
Relat Cancer. 16:649–659. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu N, Sulpice E, Obeid P, Benzina S,
Kermarrec F, Combe S and Gidrol X: The miR-17 family links p63
protein to MAPK signaling to promote the onset of human
keratinocyte differentiation. PLoS One. 7:e457612012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Woelfle U, Laszczyk MN, Kraus M, et al:
Triterpenes promote keratinocyte differentiation in vitro, ex vivo
and in vivo: a role for the transient receptor potential canonical
(subtype) 6. J Invest Dermatol. 130:113–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Robertson ED, Weir L, Romanowska M, Leigh
IM and Panteleyev AA: ARNT controls the expression of epidermal
differentiation genes through HDAC- and EGFR-dependent pathways. J
Cell Sci. 125:3320–3332. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chan KS, Sano S, Kataoka K, et al: Forced
expression of a constitutively active form of Stat3 in mouse
epidermis enhances malignant progression of skin tumors induced by
two-stage carcinogenesis. Oncogene. 27:1087–1094. 2008. View Article : Google Scholar
|
|
22
|
Kawachi Y, Fujisawa Y, Furuta J, Nakamura
Y, Ishii Y and Otsuka F: Superficial epithelioma with sebaceous
differentiation: immunohistochemical study of keratinocyte
differentiation markers. Eur J Dermatol. 21:1016–1017. 2011.
|
|
23
|
Tu CL, Chang W and Bikle DD: The
calcium-sensing receptor-dependent regulation of cell-cell adhesion
and keratinocyte differentiation requires Rho and filamin A. J
Invest Dermatol. 131:1119–1128. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cohen I, Birnbaum RY, Leibson K, Taube R,
Sivan S and Birk OS: ZNF750 is expressed in differentiated
keratinocytes and regulates epidermal late differentiation genes.
PLoS One. 7:e426282012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Blander G, Bhimavarapu A, Mammone T, et
al: SIRT1 promotes differentiation of normal human keratinocytes. J
Invest Dermatol. 129:41–49. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gasparoni A, Fonzi L, Schneider GB, Wertz
PW, Johnson GK and Squier CA: Comparison of differentiation markers
between normal and two squamous cell carcinoma cell lines in
culture. Arch Oral Biol. 49:653–664. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim RH, Flanders KC, Birkey Reffey S,
Anderson LA, Duckett CS, Perkins ND and Roberts AB: SNIP1 inhibits
NF-kappa B signaling by competing for its binding to the C/H1
domain of CBP/p300 transcriptional co-activators. J Biol Chem.
276:46297–46304. 2001. View Article : Google Scholar : PubMed/NCBI
|