Introduction
It is well known that adult female rats have heavier
adrenal glands than males of the same age. This difference appears
only after puberty and is dependent upon sex hormones. Earlier,
mainly morphologic, cytological and gravimetric data on this
subject had been extensively reviewed by Bachmann in 1954 (1). In the rat, adrenal sex dimorphism is
accompanied by functional differences, with females secreting
greater amounts of corticosterone than males (reviewed in refs.
2–5). Furthermore, female rats show greater
adrenocorticotropic hormone (ACTH) and corticosterone responses to
stress and these hormonal responses are modified by gonadectomy and
gonadal hormone replacement (6–8).
Numerous studies have suggested that observed sex differences in
the rat adrenal structure and function are dependent on the
inhibitory effects of testosterone on the
hypothalamo-pituitary-adrenal (HPA) axis, while estrogens exert
opposite effects (reviewed in refs. 4–5,9–12).
However, the molecular bases of the above outlined sex-related
differences in rat adrenal cortex structure and function have not
yet been fully elucidated.
The introduction of gene expression microarray
technology opens the possibility of discovering genes that may
contribute to various biological effects. As regards the adrenal
gland, an example of such a study is the identification of
genome-wide changes in gene expression following the treatment of
Y1 mouse adrenocortical cells with ACTH (13). In this cell line, ACTH affected
the levels of 1,275 different transcripts, and only 133 were
previously known as corticotropin-affected. Moreover, the authors
demonstrated that the cAMP/protein kinase A and protein kinase C
pathways appeared to account for approximately 60% of the effects
of ACTH (13). Furthermore, by
means of laser-capture microdissection, Nishimoto et al
(14,15) identified hundreds of transcripts
with differential expression in the zona glomerulosa (ZG) and zona
fasciculata (ZF) of adult male rats.
As far as sex-related differences in the adrenal
cortex are concerned, recently, El Wakil et al performed a
genomic analysis of gene expression in the mouse adrenal gland
(16). In their data, not
considering the transcription factors, nuclear receptor subfamily
5, group A, member 1 (Nr5a1) and nuclear receptor subfamily 0,
group B, member 1 (Nr0b1), none of the genes directly involved in
steroid hormone biosynthesis showed a differential expression in
the male and female mouse adrenal glands (16).
Therefore, in the present study, using the adrenal
glands of mature male and female rats, we performed whole
transcriptome analyses that allowed us to compare the expression
levels of approximately 27,000 genes by applying microarray
technology. Gene expression analyses were performed separately on
ZG and ZF/reticularis (ZF/R) samples. In the ZG, we revealed 32
differentially expressed genes, while 233 such genes were found in
the ZF/R. The functional profiles of the differentially
(male/female) expressed genes were characterized by means of the
GeneAnswers package of Bioconductor or by the Database for
Annotation, Visualization and Integrated Discovery (DAVID) tool, as
previously described (17,18).
Both methods revealed functional profiles of the differentially
(male/female) expressed genes and their role in the rat adrenal
gland was analyzed.
Materials and methods
Animals
Adult female and male Wistar rats (12 weeks old;
final body weight, 120–150 g) were obtained from the Laboratory
Animals Breeding Center, Department of Toxicology, Poznan
University of Medical Sciences, Poznan, Poland. The animals were
maintained under standardized conditions of light (14:10 h
light-dark cycle, illumination onset 06.00) at 23°C with free
access to standard food pellets and tap water. Female rats were
used in the estrous cycle phase, which was determined according to
the cell types observed in the vaginal smear. Following
decapitation (between 09:00 and 10:00), the adrenal glands were
promptly removed, freed of adherent fat and processed for analysis.
Briefly, under a stereomicroscope, the male and female (both
groups, n=6) rat adrenal glands were decapsulated to separate the
ZG from the ZF/R. The medulla of the adrenal gland was removed and
was not used in our analysis. The Local Ethics Committee for Animal
Studies at the Poznan University of Medical Sciences approved the
study protocol. Unless otherwise stated, all reagents were obtained
from Sigma-Aldrich (St. Louis, MO, USA) or from Avantor Performance
Materials Poland S.A. (Gliwice, Poland).
RNA isolation
RNA isolation from samples of adrenal glands was
carried out as previously described (19–23). Briefly, we used TRI reagent
(Sigma-Aldrich) and the isolated RNA was purified on columns
(RNeasy mini kit; Qiagen, Hilden, Germany). The quantity of the
total RNA was determined spectrophotometrically (optical density at
260 nm) and its purity was estimated by a 260/280 nm absorption
ratio (>1.8) (NanoDrop spectrophotometer; Thermo Scientific,
Waltham, MA, USA). RNA integrity and quality were examined using
the Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA,
USA). Evaluated RNA Integrity Numbers (RINs) were between 8.5 and
10 with the average of 9.2. The RNA concentration in each sample
was diluted to 100 ng/μl with an OD260/OD280 ratio of
1.8/2.0.
Microarray analysis
Microarray analysis was carried out as previously
described (21–23). Isolated total RNA (100 ng) was
mixed with 1.5 μl of Poly-A RNA control solution and
subjected to reverse transcription. The obtained cDNA was used for
in vitro transcription to prepare antisense RNA (aRNA) by
incubation at 40°C for 16 h. Following purification, the aRNA was
applied for the second round of sense cDNA synthesis using the WT
Expression kit (Ambion, Austin, TX, USA). All subsequent steps were
performed using the Affimetrix microarray system (Affymetrix, Santa
Clara, CA, USA). The obtained cDNA was used for biotin labeling and
fragmentation by Affymetrix GeneChip® WT Terminal
Labeling and Hybridization. Biotin-labeled fragments of cDNA (5.5
μg) were hybridized to the Affymetrix® Rat Gene
1.1 ST Array Strip (45°C/24 h). Each array comprised of >720,000
unique 25-mer oligonucleotide probes, which included >27,000
genes. Up to 25 unique probes sequences were hybridized to a single
transcript. Following hybridization, each array strip was washed
and stained using the Fluidics Station of GeneAtlas system
(Affymetrix). The array strips were scanned using the Imaging
Station of the GeneAtlas system. A preliminary analysis of the
scanned chips was performed using Affymetrix GeneAtlas™ Operating
Software. The quality of gene expression data was examined
according to the quality control criteria provided with the
software. The intensity of fluorescence was converted to numerical
data by generating CEL files. The obtained CEL files were imported
into downstream data analysis software. Unless otherwise stated,
all presented analyses and graphs were performed using Bioconductor
and R programming language, as previously described (24). Each CEL file was merged with a
description file (downloaded from the Affymetrix webpage). In order
to perform background correction, normalization and summarization
of the results, we used the Robust Multiarray Averaging (RMA)
method. The statistical significance of the analyzed genes was
examined by moderated t-statistics from the empirical Bayes method.
The obtained p-values were corrected for multiple comparisons using
the Benjamini and Hochberg’s false discovery rate (statistical
method incorporated into Bioconductor calculations) (25). The selection of genes with a
significant change in expression was based on a p-value <0.05
and an expression fold change ≥2. Fold change calculations were
performed either for appropriate zones of male and female rats or
for the ZG and ZF/R of adrenal glands in both genders (Fig. 1).
Functional analysis by GeneAnswer
Singular enrichment analysis (SEA) was performed as
previously described (15,26,27).
Selected sets of differentially expressed genes were applied to
functional analysis using the GeneAnswers package of Bioconductor
which, among other, allows us to interpret a list of genes in the
context of their participation in particular biological processes
(GO.BP) (28). Gene ontology
analyses were also performed using the web deposited manual
(http://www.bioconductor.org/packages/release/bioc/vignettes/GeneAnswers/inst/doc/geneAnswers.pdf).
Lists of differentially expressed genes were combined as tables and
were subjected to further analyses. Since our dataset comprised 2
comparison sets (male vs. female ZG and male vs. female ZF/R) we
performed multigrup gene analyses. The GeneAnswers package allowed
to test the enrichment of each GO.BP category in a gene list using
a well-defined hypergeometric statistical test. The p-value was
determined based on the number of genes differentially expressed in
the investigated GO category. This test was performed separately
for the ZG and ZF/R and the results are presented in the respective
figures.
Functional analysis by DAVID
The other analysis was performed by functional
annotation clustering using DAVID. This database provides
functional annotation tools for understanding the biological
meaning behind a large list of genes (www.david.abcc.ncifcrf.gov). Among the many functions,
DAVID allows us to discover enriched function-related gene groups
and to cluster similar annotation terms (17,18).
All procedures were performed according to the web
provided manual (http://david.abcc.ncifcrf.gov/content.jsp?file=functional_annotation.html).
The official symbols of genes which were differentially expressed
(both in male and female ZG and ZF/R) were loaded separately into
DAVID as a gene list. Annotations and background (total number of
genes in the rat) were limited only to Rattus norvegicus. In
order to obtain the most meaningful clusters, the threshold of EASE
score (a modified Fisher exact p-Value) for gene enrichment
analysis was set ≤0.05. Again, the obtained p-values were corrected
for multiple comparisons using the Benjamini and Hochberg’s false
discovery rate (25). Analyses
were performed separately for the ZG and ZF/R zones of the adult
male and female rats, as indicated in the descriptions provided
with the figures and tables.
Validation by RT-qPCR
The methods applied for RT-qPCR were as described in
previous studies (19–23). Reverse transcription was performed
using AMV reverse transcriptase (Promega Corp., Madison, WI, USA)
with Oligo(dT) (PE Biosystems, Warrington, UK) as primers in the
temperature of 42°C for 60 min (Thermocycler UNO II; Biometra,
Goettingen, Germany). The primers used were designed by Primer 3
software (Whitehead Institute for Biomedical Research, Cambridge,
UK) (Table I). The primers were
purchased from the Laboratory of DNA Sequencing and Oligonucleotide
Synthesis, Institute of Biochemistry and Biophysics, Polish Academy
of Sciences, Warsaw, Poland. qPCR was performed using the
Lightcycler 2.0 instrument (Roche Diagnostics Corp., Indianapolis,
IN, USA) with the 4.05 software version. Using the above-mentioned
primers, the SYBR-Green detection system was applied. Each 20
μl reaction mixture contained 4 μl template cDNA
(standard or control), 0.5 μM of each gene-specific primer
and a previously determined optimum MgCl2 concentration
(3.5 μM for one reaction). LightCycler FastStart DNA Master
SYBR-Green I mix (Roche Diagnostics Corp.) was used. The real-time
PCR program included a 10-min denaturation step to activate the
TaqDNA polymerase, followed by a 3-step amplification program:
denaturation at 95°C for 10 sec, annealing at 56°C for 5 sec, and
extension at 72°C for 10 sec. The specificity of the reaction
products was examined by the determination of the melting points
(0.1°C/sec transition rate).
 | Table IPrimers used for the RT-qPCR
validation of selected genes. |
Table I
Primers used for the RT-qPCR
validation of selected genes.
| Gene symbol | Gene name | GenBank accession
no. | Primer sequences
(5′→3′) | Position | PCR product size
(bp) |
|---|
| Star | Steroidogenic acute
regulatory protein | NM_031558 |
CCTGAGCAAAGCGGTGTCATGCAAGTGGCTGGCGAACTCTA | 745–764
911–931 | 187 |
| Cyp11a1 | Cytochrome P450,
family 11, subfamily a, polypeptide 1 | NM_017286 |
GATGACCTATTCCGCTTTGCGTTGGCCTGGATGTTCTTG | 592–611
930–948 | 357 |
| Cyp11b1 | Cytochrome P450,
family 11, subfamily b, polypeptide 1 | NM_012537.3 |
TCATATCCGAGATGGTAGCACCTTCTGGGGATTAGCAACGA | 863–883
1049–1068 | 206 |
| Cyp11b2 | Cytochrome P450,
family 11, subfamily b, polypeptide 2 | NM_012538.2 |
TGGCAGCACTAATAACTCAGGAAAAGCCACCAACAGGGTAG | 875–895
1131–1150 | 276 |
| Lipe | Hormone-sensitive
lipase | NM_012859.1 |
GCCCTCCAAACAGAAACCCAAATCCATGCTGTGTGAGAA | 967–985
1082–1101 | 135 |
| Nr0b1 | Nuclear receptor
subfamily 0, group B, member 1 | NM_053317.1 |
AGAGTACGCCTATCTGAAGATCGGTGTTGATGAATCTC | 1141–1159
1321–1339 | 199 |
| Hcrtr2 | Hypocretin (orexin)
receptor 2 | NM_013074.1 |
GGCTTATCTCCAAATATTCCGTAACTCTGAACCACAGAAGAAGTTCC | 782–806
828–850 | 69 |
| Hprt | Hypoxanthine
phosphoribosyltransferase | NM_012583 |
ATTTTGGGGCTGTACTGCTTGACAGTCAACGGGGGACATAAAAG | 391–412
515–536 | 146 |
Statistical analysis
The RT-qPCR data are expressed as the means ± SE,
and the statistical significance of the differences between the
control and experimental groups was estimated using the Student’s
t-test. A p-value <0.05 was considered to indicate a
statistically signficiant difference.
Results
To obtain more precise data on sexually dimorphic
gene expression in the adrenal glands of adult male and female
rats, we performed analyses of the samples of the ZG and ZF/R of
the glands. Due to the nature of the applied experiment, all data
were analyzed in relation to the adrenal glands of the male rats.
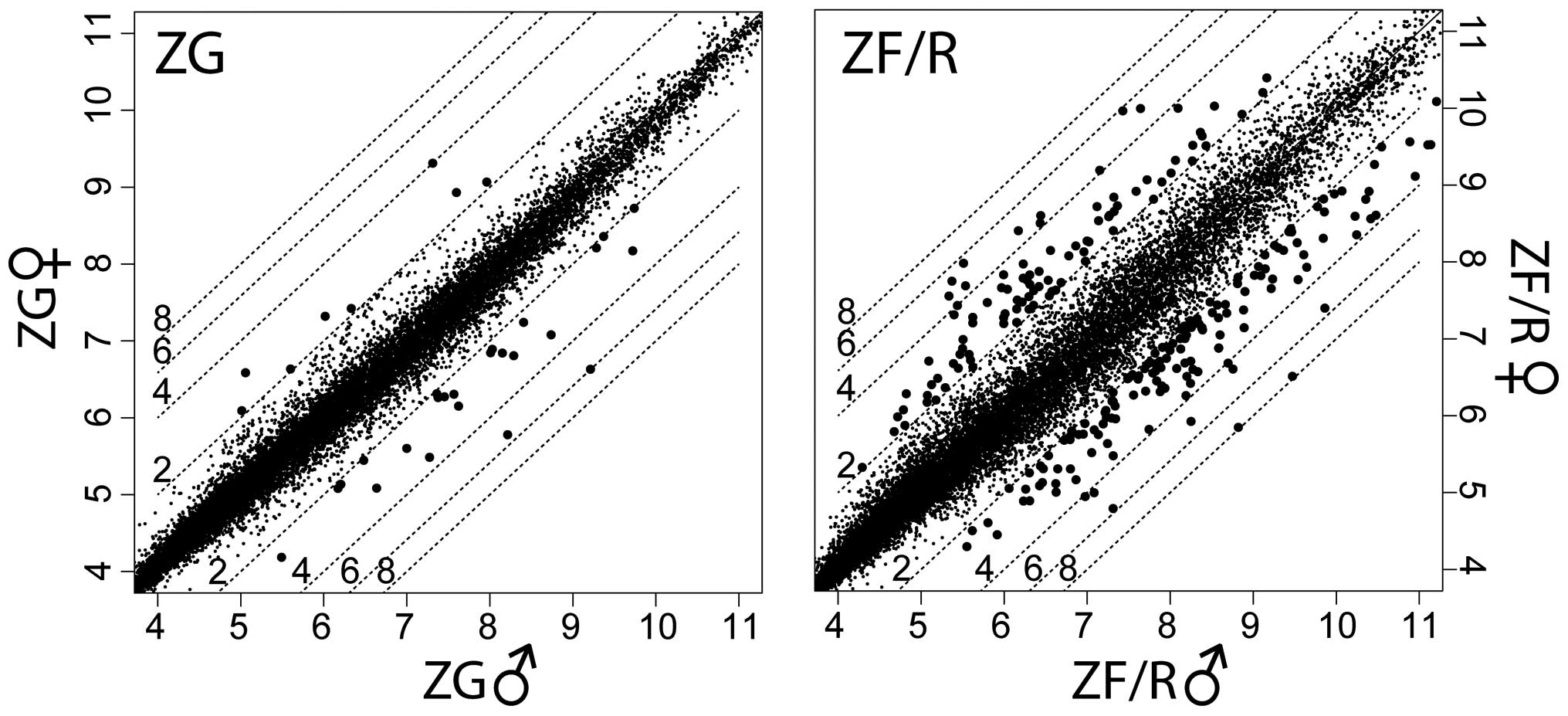
The mean expression value of each gene was presented in scatter
plot graphs (Fig. 1). The left
upper part of the graphs shows genes, the expression levels of
which were higher in the female than in the male adrenal glands. In
the right lower part of the graphs, genes are also shown, the
expression levels of which were lower in the female than in the
male adrenal glands. By applying the previously mentioned cut-off
parameters (fold change ±2; p<0.05), the Affymetrix Rat Gene 1.1
ST Array data revealed 32 differentially expressed genes in the ZG,
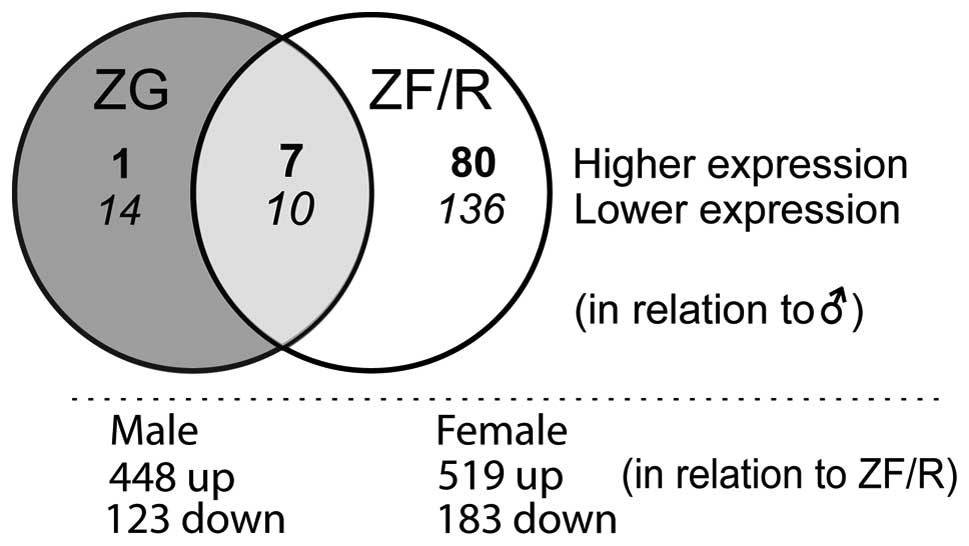
and 233 genes in the ZF/R. A Venn diagram demonstrated their
localization to the adrenocortical zones examined (Fig. 2). Of these, in the ZG only 15,
while in the ZF/R 216 genes were exclusively expressed; 17 genes
were regulated both in the ZG and ZF/R. In the ZG, the expression
levels of 24 genes were lower and 8 were higher in the female rats.
More pronounced sex-related differences were observed in the ZF/R.
In this compartment of the rat adrenal cortex, the expression
levels of 146 genes were lower and those of 87 genes were higher in
the female rats. In both the ZG and ZF/R, the expression levels of
10 genes were lower and 7 were higher in the female rats.
We also identified transcripts with differential
expression in the ZG and ZF/R of adult male and female rats
(Fig. 2). In male rats, compared
to the ZF/R, in the ZG 448 genes were upregulated and 123
downregulated. In females, these figures were 519 and 183,
respectively.
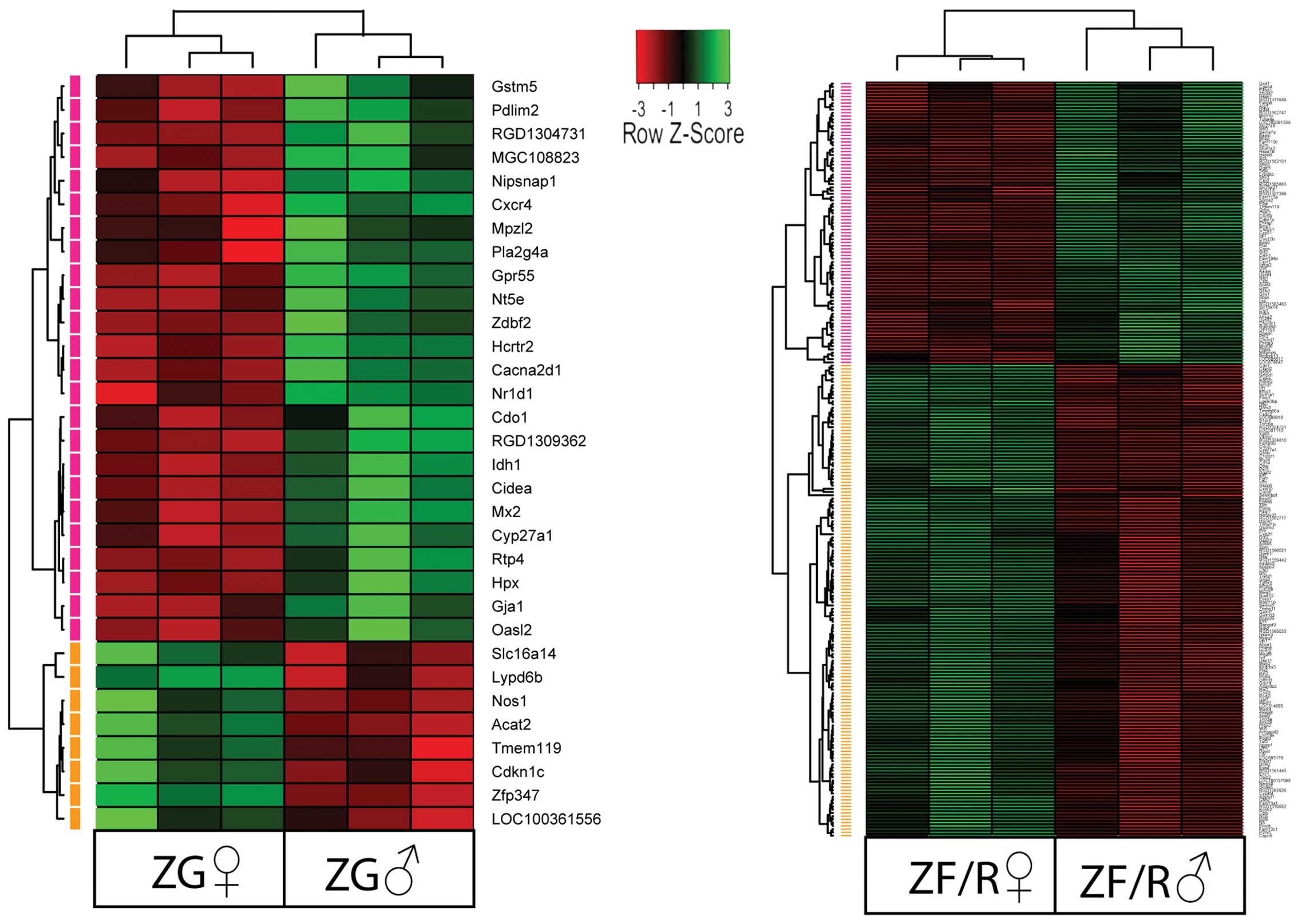
Each of the raw expression values from
gender-specific genes was grouped using a hierarchical clustering
algorithm. The results of this analysis are presented as a heat map
(Fig. 3). The clustering
confirmed that in the ZG, the expression levels of 24 genes were
lower and 8 were higher in the female rats and the symbols of these
genes are shown. The same applied to the ZF/R; however, in this
compartment, the expression levels of 146 genes were lower and
those of 87 genes were higher in the female rats.
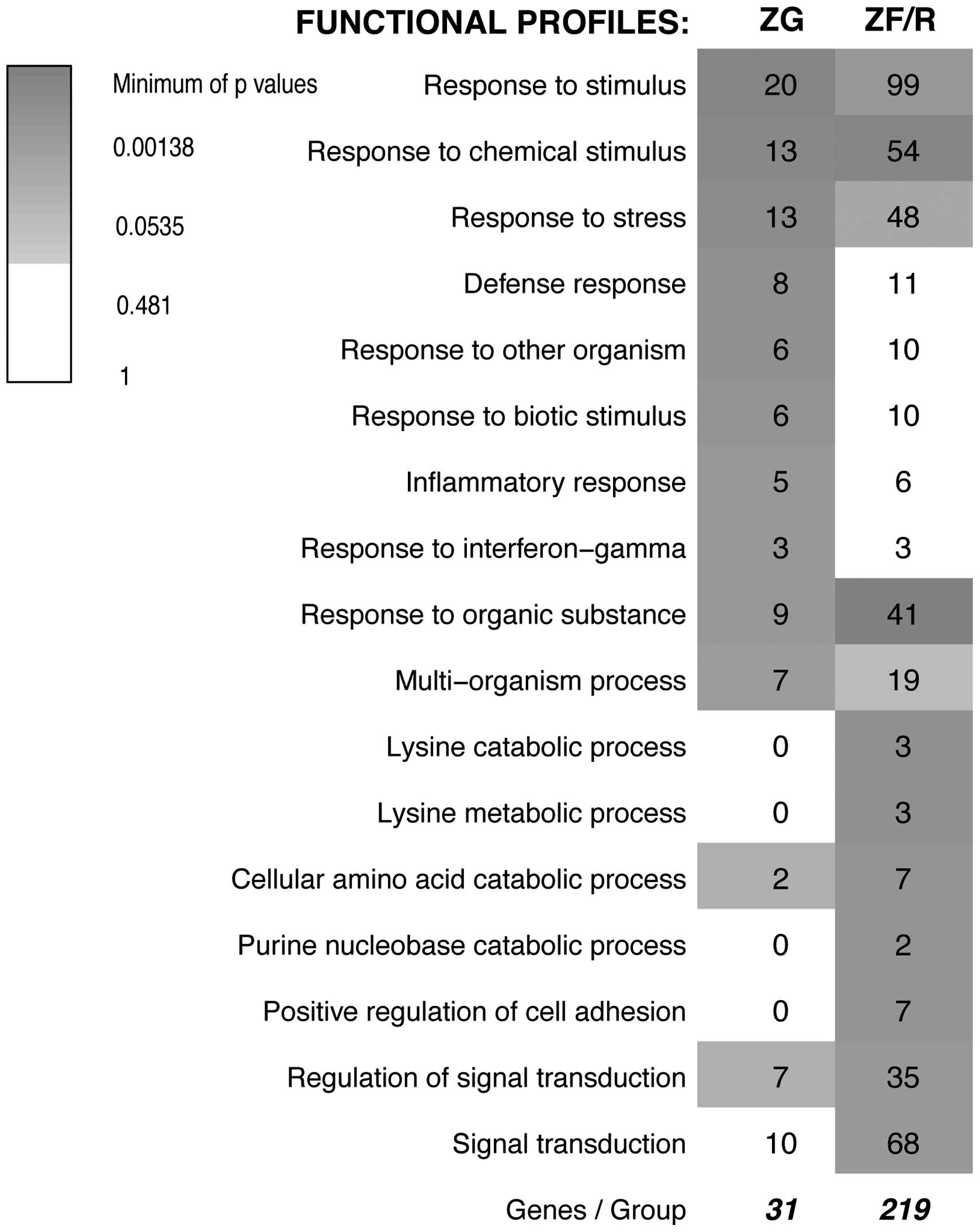
Subsequently, we performed SEA followed by
functional analysis using the GeneAnswers package of Bioconductor
which, among other things, allows us to interpret a list of genes
in the context of their participation in a certain particular
biological process (GO.BP). The original results of GeneAnswer
analysis (as of December 16, 2013) revealed 17 groups of genes with
different functional profiles. These profiles were rather general
(for example ‘response to stimulus’, ‘response to chemical
stimulus’, ‘response to organic substances’) and the prevailing
number of the obtained profiles were out of our interest (Fig. 4). It should be emphasized that the
GO database is composed of some general, as well as specific
categories with a similar meaning and, therefore, a single gene may
be mapped to several GO terms and may be counted more than once.
Moreover, GO functional annotations of genes are still in the
developing stage and are far from complete. Since this analysis
yielded rather unsatisfactory results, we decided to analyze our
results using the DAVID system.
The DAVID system is a powerful tool that allows us
to discover enriched functionally related gene groups and to
cluster similar annotation terms. This system extracts data from
numerous databases and, as evidenced by the Science Citation Index,
this system is gaining wide popularity among molecular biologists.
We performed separate analyses for the ZG and ZF/R. For the ZG,
with the selected cut-off value (p<0.05), 3 annotation clusters
were obtained (ordered by enrichment score) (Table II). On the other hand, for the
ZF/R 5 clusters were obtained. All clusters (ZG and ZF/R) combined
516 genes (counts). This figure indicated that numerous genes
participated in more than one annotation cluster. In the ZG 90
counts were either up- or downregulated, while in the ZF/R the
number of counts was notably higher, i.e., 426.
 | Table IIFunctional annotation clustering
report of differentially expressed genes in the ZG and ZF/R of
adult male and female rats, as revealed by DAVID analysis (separate
analyses for ZG and ZF/R). |
Table II
Functional annotation clustering
report of differentially expressed genes in the ZG and ZF/R of
adult male and female rats, as revealed by DAVID analysis (separate
analyses for ZG and ZF/R).
| Gene function | Up | Down |
|---|
|
|---|
| Zona
gromelurosa | ↓ | ↑ |
|---|
| Annotation cluster
1/enrichment score: 3.73 |
| Regulation of
calcium ion transport | 3 | 1 |
| Regulation of
metal ion transport | 3 | 1 |
| Regulation of ion
transport | 3 | 1 |
| Annotation cluster
2/enrichment score: 2.32 |
| Iron ion
binding | 4 | 1 |
| Metal binding | 8 | 1 |
| Transition metal
ion binding | 8 | 2 |
| Metal ion
binding | 10 | 2 |
| Cation
binding | 10 | 2 |
| Iron | 3 | 1 |
| Ion binding | 10 | 2 |
| Annotation cluster
3/enrichment score: 2.2 |
| Response to
hormone stimuli | 4 | 1 |
| Response to
endogenous stimuli | 4 | 1 |
| Response to
steroid hormone stimuli | 3 | 1 |
|
| Zona
fasciculata/reticularis | | |
|
| Annotation cluster
1/enrichment score: 4.66 |
| Steroid metabolic
process | 4 | 10 |
| Cholesterol
metabolic process | 3 | 6 |
| Sterol metabolic
process | 3 | 6 |
| Steroid
biosynthetic process | 0 | 8 |
| Lipid biosynthetic
process | 3 | 11 |
| Steroid
biosynthesis | 1 | 4 |
| Annotation cluster
2/enrichment score: 3.75 |
| Microsome | 8 | 6 |
| Vesicular
fraction | 8 | 6 |
| Insoluble
fraction | 15 | 7 |
| Membrane
fraction | 14 | 7 |
| Cell fraction | 17 | 8 |
| Annotation cluster
3/enrichment score: 2.99 |
| Oxidation
reduction | 14 | 9 |
|
Oxidoreductase | 12 | 7 |
| Monooxygenase | 5 | 3 |
| Electron carrier
activity | 7 | 4 |
| Cytochrome P450, C
terminal region | 4 | 3 |
| Cytochrome
P450 | 4 | 3 |
| Secondary
metabolites biosynthesis, transport and catabolism | 4 | 3 |
| Iron ion
binding | 7 | 4 |
| Iron | 7 | 4 |
| Cytochrome P450,
conserved site | 3 | 3 |
| Chromoprotein | 3 | 3 |
|
Metalloprotein | 4 | 3 |
| Cytochrome P450, E
class, group I | 4 | 1 |
| Zona
fasciculata/reticularis | ↓ | ↑ |
| Annotation cluster
4/enrichment score: 2.94 |
| Response to
extracellular stimulus | 7 | 7 |
| Response to
nutrient levels | 6 | 6 |
| Response to
nutrients | 4 | 6 |
| Response to
steroid hormone stimuli | 6 | 5 |
| Annotation cluster
5/enrichment score: 2.6 |
| Response to
organic substance | 17 | 11 |
| Response to
hormone stimuli | 10 | 7 |
| Response to
endogenous stimuli | 10 | 7 |
| Response to
steroid hormone stimuli | 6 | 5 |
| Response to
glucocorticoid stimuli | 4 | 3 |
| Response to
corticosteroid stimuli | 4 | 3 |
| Response to
peptide hormone stimuli | 5 | 4 |
For the ZG, annotation clusters 1 and 2 combined
genes involved in the regulation of ion transport, while cluster 3
contained transcripts linked to response to hormones (Table II). In the case of ZF/R, we
obtained 5 clusters. The first cluster combined genes regulating
steroid biosynthesis and metabolism. Cluster 2 incorporated genes
primarily connected with cell fractions, while annotation cluster 3
(with 13 subclusters) combined transcripts primarily connected with
oxidation/reduction processes. Cluster 4 was composed of genes
regulating responses to nutrients and to extracellular stimuli or
steroid hormone stimuli. Annotation cluster 5 also contained genes
involved in response to stimuli, among which were ‘response to
hormone stimulus’ (subcluster 2) and ‘response to peptide hormone
stimulus’ (subcluster 7).
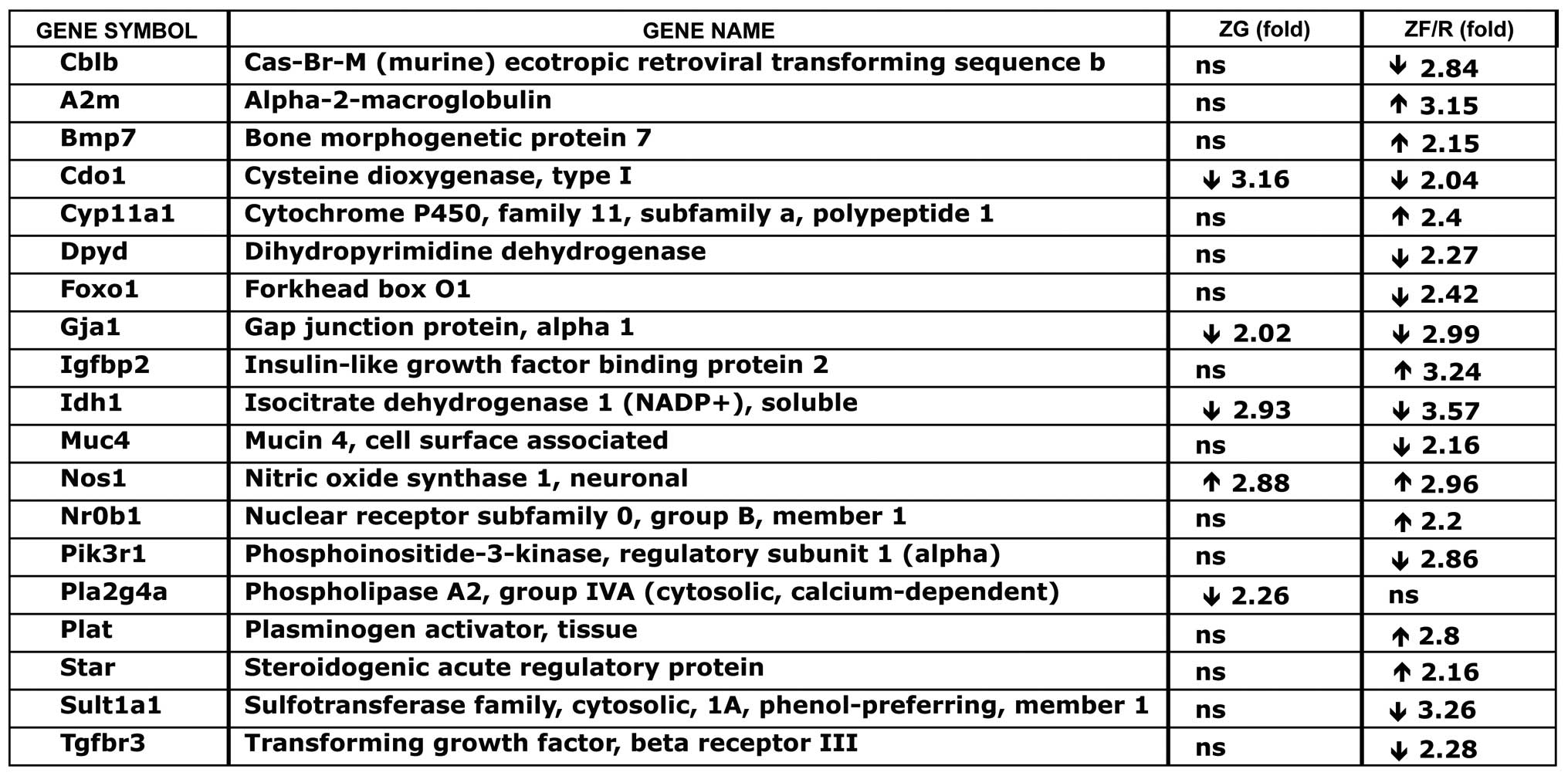
Subsequently, we analyzed a list of genes which were
differentially expressed in the ZG and ZF/R of the adrenal glands
in adult female rats, in comparison to adult male rats and
annotated as a ‘response to hormone stimulus’ (for the ZG from
cluster 3, and for ZF/R from cluster 5 shown in Table II). Of these genes, the
expression levels of cysteine dioxygenase type 1 (Cdo1), gap
junction protein, alpha 1 (Gja1), isocitrate dehydrogenase 1 (Idh1)
and phospholipase A2, group IVA (Pla2g4a) were lower and those of
nitric oxide synthase 1 (Nos1) were higher in the ZG of female
rats, when compared with the males (Fig. 5). More pronounced differences were
observed in the ZF/R zones of the rat adrenal cortex. Of the 19
genes forming the above-mentioned subcluster, the expression levels
of 7 of these genes were higher and those of 10 genes were lower in
the female adrenal glands. In the female ZF/R, higher expression
levels were observed in the following genes: alpha-2-macroglobulin
(A2m), bone morphogenetic protein 7 (Bmp7), cytochrome P450, family
11, subfamily A, polypeptide 1 (Cyp11a1), insulin-like growth
factor binding protein 2 (Igfbp2), Nos1, Nr0b1 and plasminogen
activator, tissue (Plat). On the contrary, in the female ZF/R of
the adrenal cortex, lower expression levels were observed in the
following genes: Cbl proto-oncogene B, E3 ubiquitin protein ligase
(Cblb), Cdo1, dihydropyrimidine dehydrogenase (Dpyd), forkhead box
O1 (Foxo1), Gja1, Idh1, mucin 4, cell surface associated (Muc4),
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) (Pik3r1),
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1
(Sult1a1) and transforming growth factor, beta receptor III
(Tgfbr3).
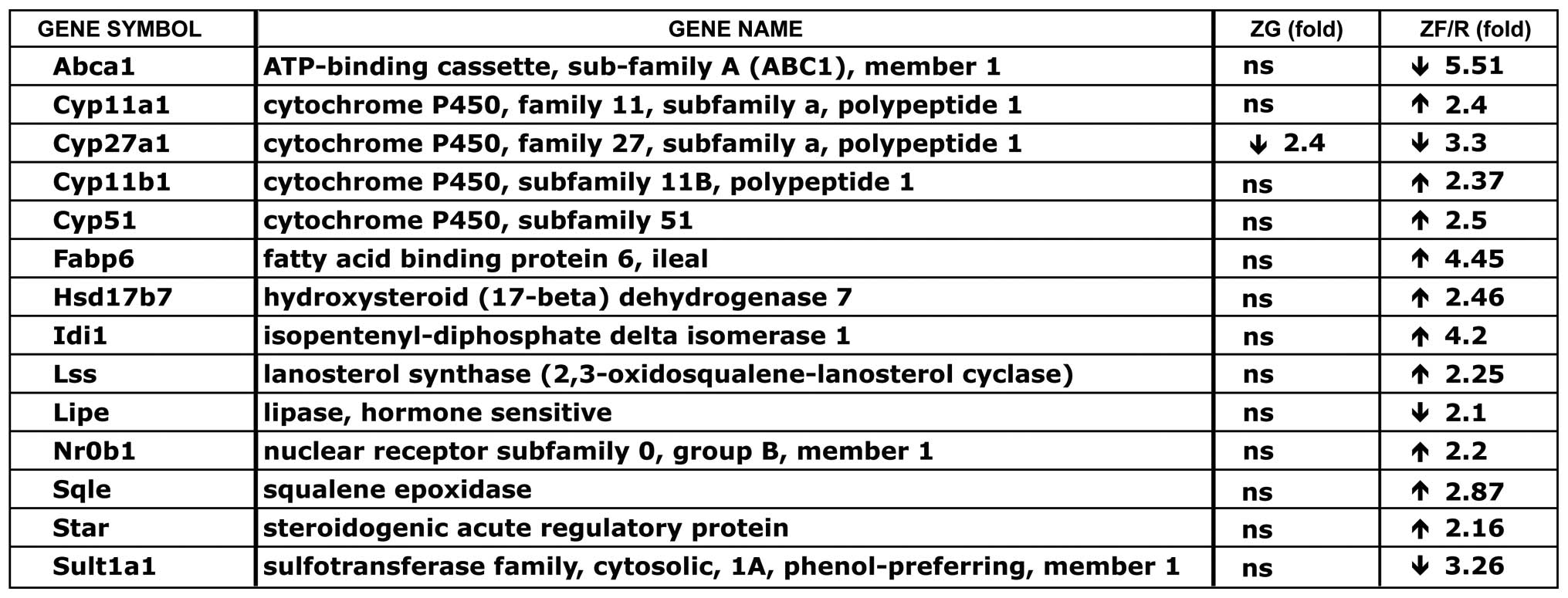
Furthermore, analyses of ZF/R genes represented in
cluster 1, subcluster ‘steroid metabolism process’ (from Table II) revealed numerous genes, the
expression levels of which were different in the ZF/R (Fig. 6). Of these genes, the expression
levels of only 4 of them [ATP-binding cassette, sub-family A
(ABC1), member 1 (Abca1), cytochrome P450, family 27, subfamily A,
polypeptide 1 (Cyp27a1), lipase, hormone-sensitive (Lipe) and
Sult1a1] were lower in the females compared to the males. On the
contrary, the remaining genes exhibited higher expression levels in
the females [Cyp11a1, cytochrome P450, family 11, subfamily B,
polypeptide 1 (Cyp11b1), cytochrome P450, family 51 (Cyp51), fatty
acid binding protein 6, ileal (Fabp6), hydroxysteroid (17-beta)
dehydrogenase 7 (Hsd17b7), isopentenyl-diphosphate delta isomerase
1 (Idi1), lanosterol synthase (2,3-oxidosqualene-lanosterol
cyclase) (Lss), Nr0b1, squalene epoxidase (Sqle) and steroidogenic
acute regulatory protein (Star)].
By means of RT-qPCR, we also validated the
expression levels of selected genes with gender-specific
differences in their expression levels in the ZG and/or ZF/R of
adult male and female rat adrenal glands (Fig. 7). In all cases, the results of
RT-qPCR confirmed the differences revealed by the
Affymetrix® Rat Gene 1.1 ST Array. The expression levels
of Star, Cyp11a1 and Nr0b1 were notably higher in the ZF/R of the
female rats, while no differences were observed in the ZG. No
sex-related differences were observed in the expression levels of
cytochrome P450, family 11, subfamily B, polypeptide 2 (Cyp11b2) in
the ZG, while the expression level Cyp11b1 was notably higher in
the female ZF/R. Lipe mRNA levels were notably higher in the male
ZF/R, while in the ZG its expression was similar in both genders.
Moreover, in both the ZG and ZF/R, the expression levels of
hypocretin (orexin) receptor 2 (Hcrtr2) were notably lower in the
female than in the male adrenal glands.
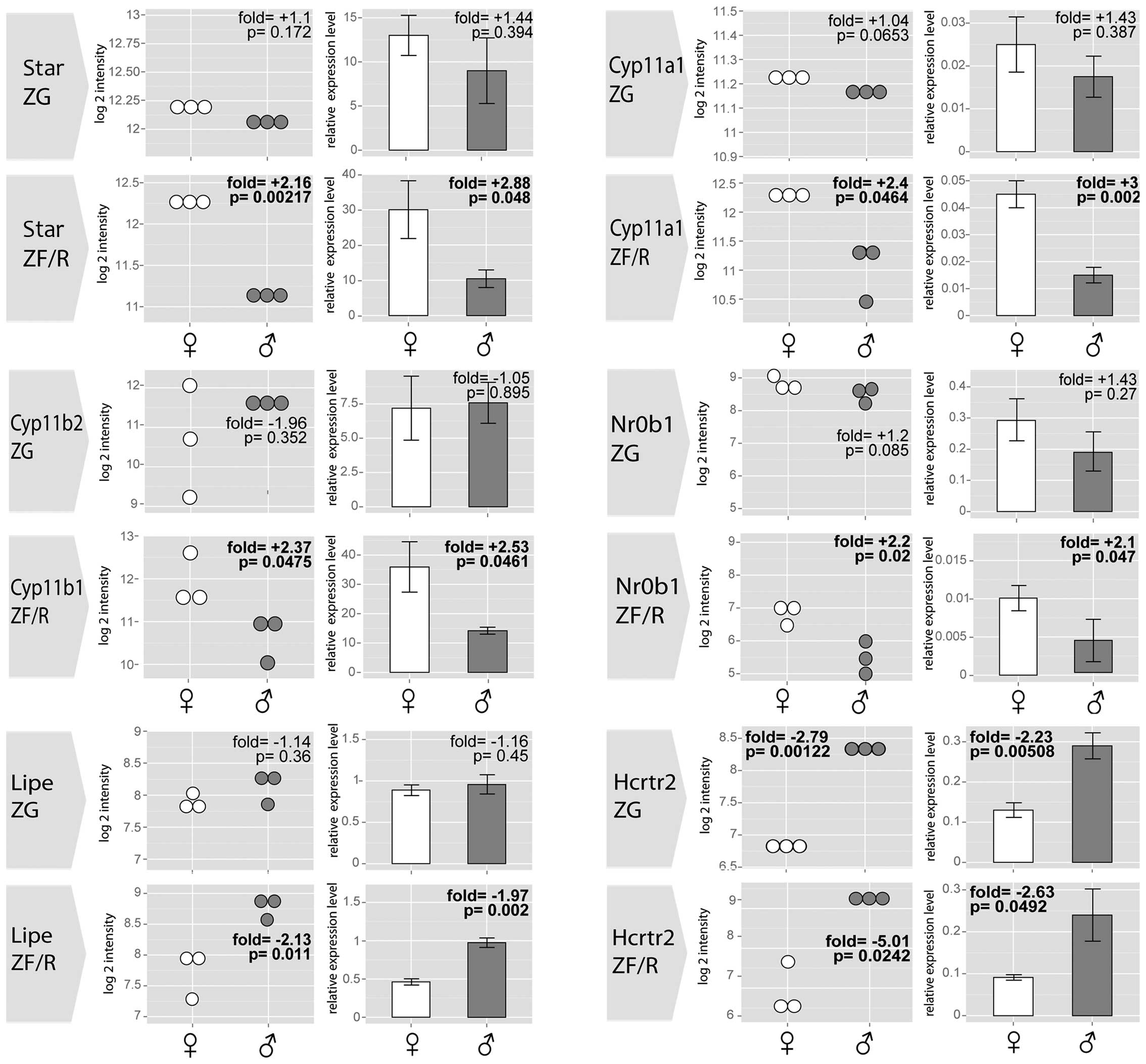 | Figure 7Results of RT-qPCR validation for
selected genes with gender-specific differences in their expression
levels in the ZG and ZF/R of adult male and female rats. Dot plots
present data obtained from Affymetrix® Rat Gene 1.1 ST
Array, bar plots present the means ± SE from RT-qPCR. Eeach group
n=3. Data are in comparison to male adrenal glands. Fold changes
and p-values are shown. Star - steroid acute regulatory protein;
Cyp11a1, cytochrome P450, family 11, subfamily A, polypeptide 1;
Cyp11b2, cytochrome P450, family 11, subfamily B, polypeptide 2;
Cyp11b1, cytochrome P450, family 11, subfamily B, polypeptide 1;
Lipe, hormone sensitive lipase; Nr0b1, nuclear receptor subfamily
0, group B, member 1; Hcrtr2, hypocretin (orexin) receptor 2; ZG,
zona glomerulosa; ZF/R, zona fasciculata/reticularis. |
Discussion
Global gene expression profiling allows for a
simultaneous analysis of thousands of genes in a single sample.
This powerful tool of molecular biology is widely used, among
others, in studies on basic biology or in the diagnosis of various
diseases. This method has also been used in studies on adrenal
glands. In recent years increasing amounts of data linking gene
expression with adrenal biology have been generated. In this area,
the first reports were focused on gene profiles for steroidogenic
enzymes in adrenocortical diseases, in particular in
aldosterone-producing adenomas and other adrenocortical tumors
(29–34). Gene profiling methods applied to
freshly isolated adrenocortical cells or established cell lines
(Y1, H295R human adrenocortical cells), allows the identification
of numerous genes involved in the regulation of adrenocortical
growth and functioning and provides novel data on intracellular
pathways involved in the regulation of aldosterone and
corticosterone secretion (13,35,36).
Transcriptional profiling has also been used in
in vivo experiments. By means of this method, the circadian
regulation of steroid hormone biosynthesis genes was examined in
the rat adrenal gland (37). In
knockout mice lacking Star, numerous up- and downregulated genes
were identified by Ishii et al (38). Furthermore, recent studies on the
regulation of the renin-angiotensin-aldosterone system (RAAS) in
rat adrenal glands identified transcripts involved in RAAS
activation (39). Transcriptional
profiling has also been applied for the characterization of
enucleation-induced adrenal regeneration in the rat (21–23).
From the above short survey, it appears that in
in vivo experiments, gene profiling methods are used mainly
for investigations of adrenal glands with experimentally modified
function. Not considering studies on the circadian regulation of
steroid hormone biosynthesis genes, to the best of our knowledge,
only one study has been performed on the intact rat adrenal gland
(14). The authors investigated
differentially expressed transcripts in the adrenal ZG and ZF of
the adult male rat. It appeared that in the ZG, 235 transcripts
were upregulated by >2-fold compared to ZF and 231 transcripts
were upregulated in the ZF (14).
We performed a similar analysis of the adrenal glands of adult male
and female rats. In our study, in both genders the number of
differentially expressed transcripts in the adrenal ZG and ZF/R was
notably higher than that previously reported for the ZG and ZF of
male rats (14). The study by
Nishimoto et al (14)
identified 466 such transcripts, while the figures obtained in the
present study amounted to 571 for male and 702 for female rats.
These differences may be due to the various methods applied in two
studies, as there are many methods gerenarally used (40–42). We used Affymetrix
GeneChip® WT Terminal Labeling and the Hybridization
system, while Nishimoto et al applied the Illumina platform;
we sampled fresh tissue (vs. frozen), used the RMA algorithm (vs.
the percentile shift method) and our experiment was performed on
Wistar rats (vs. the Sprague-Dawley strain) [we have also used
Wistar rats in a previous study (43)]. Furthermore, the study by
Nishimoto et al (14)
obtained samples by means of a very precise laser-capture
microdissection method, while our samples contained cells of both
the zona fasciculata and reticularis (ZF/R). This means that our
zona fasciculata cells were contaminated with the zona reticularis
ones. Be as it may, our data also revealed notable differences in
the number of differentially expressed transcripts in the adrenal
glands of adult male and female rats, and in females this number
was significantly higher.
It is well known that in the rat, the structure and
function of the adrenal cortex exhibits sex-dependent differences
(reviewed in refs. 4,5). The genetic background of these
differences is little known, therefore we performed whole
transcriptome analyses on the adrenal glands of mature male and
female rats which allowed as to compare the expression levels of
approximately 27,000 genes by applying microarray technology. In
the adrenal cortex of the examined rats, in both the ZG and ZF/R,
we identified 265 differentially expressed genes (male vs. female)
and this figure is comparable to data previously reported for mice
(269 genes regulated in the female adrenal compared to the male
gland, and analyses were performed on the entire glands with
Agilent array) (16). Our
analysis revealed that the most pronounced sex-related differences
in gene expression in the rat adrenal cortex were present in the
ZF/R. In this compartment, the expression levels of 146
sex-regulated genes were lower in the female than in the male
gland.
To describe the functions of differentially
expressed genes in the male and female rat adrenal cortex, we
performed the clustering of identified transcripts. By using DAVID
tool, in the ZG, 3 annotations clusters were obtained and they were
mainly related to ion transport and the response to endogenous
stimuli. Unexpectedly, the expression levels of these genes were
notably lower in the female rats. In the ZF/R, on the other hand, 5
annotation clusters with various numbers of subclusters were
obtained. Of these, only in annotation cluster 1 (combining genes
regulating steroid biosynthesis and metabolism) the numbers of
upregulated transcripts in the female adrenal glands were higher
than those in the male ZF/R. In the 4 remaining clusters, the
numbers of downregulated transcripts in female adrenal glands were
higher than those of the upregulated transcripts. These findings
suggest the higher steroidogenic activity of the female than the
male ZF/R and are in line with the findings of previous studies
showing higher corticosterone secretion by the adrenal glands of
female rats (4,5). Moreover, these observations are in
accordance with those of previous studies, showing that stimulation
of the highly-specialized function of adrenocortical cells (i.e.,
steroid hormone release) is always coupled with the inhibition or
lowering of their basal biological processes, for instance the
proliferation rate (44–46).
As revealed by DAVID analysis, the differentially
expressed genes were significantly enriched in sets of genes
involved in steroid hormone metabolism and their expression levels
in the ZF/R of adult female rats were significantly higher than
those in male adrenal glands. On the contrary, DAVID analysis did
not reveal this group of transcripts in the ZG. Thus, the group of
genes directly involved in steroid hormone synthesis expresses a
sexually dimorphic pattern in the rat ZF/R, but not ZG. In this
regard, the study by El Wakil et al (16) on sexual dimorphism of gene
expression in the mouse adrenal gland revealed only 2 genes (Nr5a1
and Nr0b1) differentially expressed in the mouse, with higher
expression levels in females. However, in their study none of the
genes directly involved in steroid biosynthesis was found to be
differentially expressed (16).
We confirmed their findings on genes encoding known transcription
factors and in the case of Nr0b1, we demonstrated that this
difference was due to its higher expression in the ZF/R, but not in
the ZG. Furthermore, the present study revealed that in the female
ZF/R of the rat, there were higher expression levels of Cyp11a1,
Cyp11b1, Fabp6, Hsd17b7, Idi1 and Star. These differences may be
dependent on the species (rat vs. mouse) and the method of
sampling, e.g., the separate anlaysis of the ZG and ZF/R in our
study as opposed to the analysis of the entire glands in mice.
In contrast to ‘steroidogenic’ genes, in the female
ZF/R, prevailing numbers of genes linked to cell fraction,
oxidation/reduction processes and response to nutrients, as well as
to extracellular stimuli or steroid hormone stimuli were found to
be downregulated. Also this finding is rather unexpected, it seems
to be scientifically justified (the negative interrelationship
between highly specialized cell function and basic cell
functions).
In present study, the expression levels of selected
genes involved in the regulation of steroid biosynthesis was
validated by RT-qPCR. As is known, cholesterol is the precursor for
the entire adrenal steroidogenesis. Cholesterol may originate from
different sources: i) de novo synthesis in the endoplasmic
reticulum from acetyl CoA, ii) the mobilization of cholesteryl
esters stored in lipid droplets through cholesteryl ester
hydrolase, iii) plasma lipoprotein-derived cholesteryl esters
obtained by either low-density lipoprotein (LDL) receptor-mediated
endocytosis and/or scavenger receptor class B type I
(SR-BI)-mediated selective uptake (47,48). Earlier data have demonstrated that
the concentration of total lipids, total cholesterol, phospholipids
and glycerides is similar in the adrenal glands of adult male and
female rats; however their content, due to larger adrenal glands,
is markedly higher in females (49). In this regard, Lipe is a major
cholesterol hydrolase of the adrenal glands (50,51). The specific activity of this
enzyme in 105,000 g supernatant of adrenal homogenates is higher in
male than in female rats (52).
In the present study, we demonstrated higher Lipe mRNA levels in
the male ZF/R, while in the ZG the expression was similar in both
genders. Thus, in the male ZF/R the higher activity of Lipe is
accompanied by higher expression levels of the Lipe gene. Moreover,
we demonstrated that the expression of the Lipe gene in the rat ZG
was similar in both genders.
It has been well documented that the Star gene
encodes a protein involved in the acute regulation of steroid
hormone synthesis. This protein is responsible for the transport of
cholesterol from the outer to the inner mitochondrial membrane
(53). Earlier
immunohistochemical and immunofluorescence studies have revealed
the presence of Star protein in both the ZG and ZF/R of the rat
adrenal cortex (54–57). In the mouse adrenal glands, Star
mRNA levels (assessed by RT-PCR) have been reported be slightly
higher in males than in females, although the differences were not
statistically significant (58).
In the present study, the mRNA expression levels of Star were
notably higher in the ZF/R of female rats, while no differences
were observed in the ZG. To the best of our knowledge, this is the
first demonstration of specific sex-related differences in Star
gene expression in the rat adrenal cortex.
Cyp11a1 encodes the P450scc enzyme (cholesterol
20–22 desmolase) that catalyzes the first and rate-limiting step of
steroid biosynthesis (59,60).
This enzyme is expressed in all adrenocortical zones (14,61). As previously reported, the rate of
cholesterol transformation into pregnenolone (side chain cleavage
activity) is markedly lower in male than in female rats (2,62,63). Moreover, the levels of adrenodoxin
in decapsulated (i.e., devoid of ZG cells) adrenal glands have been
reported to be lower in male than in female rats (64). In this study, we revealed the
higher expression of the Cyp11a1 gene in the ZF/R of female rats.
Again, as in the case of the Star gene, the mRNA levels of Cyp11a1
were comparable in the ZG of adult male and female rats. Thus, the
previously reported higher rate of pregnenolone synthesis in the
female rat adrenal cortex depends on the higher expression of the
Cyp11a1 gene and the ZF/R compartment of the cortex is responsible
for these differences.
We also validated the expression levels of 2 genes
coding enzymes of the steroidogenic late pathway in the rat,
Cyp11b2 (aldosterone synthase, responsible for aldosterone
synthesis) and Cyp11b1 (steroid 11beta-hydroxylase, responsible for
corticosterone synthesis). In situ hybridization and
immunohistochemistry revealed the expression of aldosterone
synthase in the rat ZG only (14,61,65). These results are in agreement with
those of a previous study which demonstrated (by RT-PCR) high
expression levels of Cyp11b2 in this zone (66). Our study confirmed these earlier
observations. Moreover, we demonstrated that the expression levels
of the Cyp11b2 gene were similar in the ZG of male and female rats.
These data are in accordance with those of previous studies on the
absence of sex-related differences in aldosterone synthesis in male
and female rats (reviewed in Refs. 4,67).
On the contrary, in the mouse adrenal glands, the mRNA levels of
aldosterone synthase have been shown to be slightly higher in
female than in male adrenal glands (58).
The Cyp11b1 gene encodes steroid 11beta-hydroxylase,
which catalyzes the conversion of deoxycorticosterone to
corticosterone. In the rat adrenal glands this enzyme is localized
in the ZF/R cells (14,61). The activity of steroid
11beta-hydroxylase has been shown to be similar in adult male and
female rats (68,69). However, the level of cytochrome
P-45011β, in the rat adrenal mitochondria has been shown
to be lower in male than in female rats (64). In the present study, we
demonstrated that the Cyp11b1 gene expression levels were notably
higher in the female ZF/R, while no differences were observed in
the ZG of the adrenal cortex.
Nr0b1 encodes the Dax1 protein (dosage-sensitive sex
reversal, adrenal hypoplasia critical region, on chromosome X, gene
1) which is responsible for the development and maintenance of the
steroidogenic axis (70–72). Experimental data have suggested
that the DAX1 protein is a negative regulator of steroidogenesis.
The sexually dimorphic expression of Dax-1 (NR0B1) in the mouse
adrenal cortex has been observed by RT-PCR, western blotting and
immunohistochemistry (73). It
should be mentioned that in the mouse adrenal glands, the mRNA
levels of Dax1 were only slightly higher in female than in male
adrenals (16,58). To the best of our knowledge, for
the first time, we demonstrated that the expression levels of Nr0b1
were notably higher in the ZF/R of female rats, while no
differences were observed in the ZG.
In a series of experiments, the group of Jöhren
et al (74–76) demonstrated a notable sex-dependent
expression of Hcrtr2 in the rat adrenal cortex. They demonstrated
that Hcrtr2 was localized in the ZG and zona reticularis, with
higher expression levels in male adrenal gland. We confirmed these
earlier studies, as in our study, the expression levels of Hcrtr2
in the ZG and ZF/R were notably lower in the female than in the
male adrenal glands. These results confirm the proper preparation
of our tissue samples (ZG and ZF/R) for the analysis presented in
this study.
In conclusion, to the best of our knowledge, the
present study presents the first report of sex-related gene
expression profiles in the adrenal cortex of adult rats. The number
of differentially expressed transcripts in the adrenal ZG and ZF/R
was notably higher in the female than in the male rats (702 vs.
571). The differentially expressed genes were significantly
enriched in sets of genes involved in steroid hormone metabolism
and their expression levels in the ZF/R of adult female rats were
significantly higher than those in the male adrenal glands. In the
female ZF/R, when compared with the male one, the prevailing
numbers of genes linked to cell fraction, oxidation/reduction
processes, response to nutrients and to extracellular stimuli or
steroid hormone stimuli were downregulated.
Acknowledgments
This study was financed from the funds of the
National Science Centre (Poland) allocated on the basis of the
decision number DEC-2013/11/B/NZ4/04746.
References
|
1
|
Bachmann R: Die Nebenniere. Handbuch der
Mikroskopischen Anatomie des Menschen. Bargmann W: 6/5. Springer;
Berlin Göttingen Heidelberg: pp. 1–952. 1954, View Article : Google Scholar
|
|
2
|
Kitay JI: Effects of estrogen and androgen
on the adrenal cortex of the rat. Functions of the Adrenal Cortex.
2. McKerns KW: North Holland Publishing Corpany; Amsterdam: pp.
775–811. 1968
|
|
3
|
Kime D, Vinson GP, Major P and Kilpatrick
R: Adrenal-gonad relationship. Functions of the Adrenal Cortex. 3.
McKerns KW: Academic Press; London: pp. 183–347. 1979
|
|
4
|
Malendowicz LK: Cytophysiology of the
mammalian adrenal cortex as related to sex, gonadectomy and gonadal
hormones. PTPN Press; Poznan: pp. 1–233. 1994
|
|
5
|
Goel N, Workman JL, Lee TT, et al: Sex
differences in the HPA axis. Compr Physiol. 4:1121–1155. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lescoat G, Jego P, Beraud G and Maniey J:
Sex influences on the response of the
hypothalamo-hypophysio-adrenal axis to emotional and systemic
stress in the rat. 164:2106–2113. 1970.(In French).
|
|
7
|
Le Mevel JC, Abitbol S, Beraud G and
Maniey J: Temporal changes in plasma adrenocorticotropin
concentration after repeated neurotropic stress in male and female
rats. Endocrinology. 105:812–817. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lesniewska B, Miskowiak B, Nowak M and
Malendowicz LK: Sex differences in adrenocortical structure and
function. XXVII. The effect of ether stress on ACTH and
corticosterone in intact, gonadectomized, and testosterone- or
estradiol-replaced rats. Res Exp Med (Berl). 190:95–103. 1990.
View Article : Google Scholar
|
|
9
|
Malendowicz LK: Sex differences in
adrenocortical structure and function. I. The effects of
postpubertal gonadectomy and gonadal hormone replacement on nuclear
volume of adrenocortical cells in the rat. Cell Tissue Res.
151:525–536. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Malendowicz LK: Sex differences in
adrenocortical structure and function. II. The effects of
postpubertal gonadectomy and gonadal hormone replacement on the rat
adrenal cortex evaluated by stereology at the light microscope
level. Cell Tissue Res. 151:537–547. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Malendowicz LK, Robba C and Nussdorfer GG:
Sex differences in adrenocortical structure and function. XXII.
Light- and electron-microscopic morphometric studies on the effects
of gonadectomy and gonadal hormone replacement on the rat adrenal
cortex. Cell Tissue Res. 244:141–145. 1986.PubMed/NCBI
|
|
12
|
Nussdorfer GG: Cytophysiology of the
adrenal cortex. Int Rev Cytol. 98:1–405. 1986.PubMed/NCBI
|
|
13
|
Schimmer BP, Cordova M, Cheng H, et al:
Global profiles of gene expression induced by adrenocorticotropin
in Y1 mouse adrenal cells. Endocrinology. 147:2357–2367. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nishimoto K, Rigsby CS, Wang T, et al:
Transcriptome analysis reveals differentially expressed transcripts
in rat adrenal zona glomerulosa and zona fasciculata.
Endocrinology. 153:1755–1763. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nishimoto K, Rainey WE, Bollag WB and Seki
T: Lessons from the gene expression pattern of the rat zona
glomerulosa. Mol Cell Endocrinol. 371:107–113. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
El Wakil A, Mari B, Barhanin J and Lalli
E: Genomic analysis of sexual dimorphism of gene expression in the
mouse adrenal gland. Horm Metab Res. 45:870–873. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dennis G Jr, Sherman BT, Hosack DA, et al:
DAVID: database for annotation, visualization, and integrated
discovery. Genome Biol. 4:P32003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
19
|
Rucinski M, Andreis PG, Ziolkowska A,
Nussdorfer GG and Malendowicz LK: Differential expression and
function of beacon in the rat adrenal cortex and medulla. Int J Mol
Med. 16:35–40. 2005.PubMed/NCBI
|
|
20
|
Spinazzi R, Ziolkowska A, Neri G, et al:
Orexins modulate the growth of cultured rat adrenocortical cells,
acting through type 1 and type 2 receptors coupled to the MAPK
p42/p44- and p38-dependent cascades. Int J Mol Med. 15:847–852.
2005.PubMed/NCBI
|
|
21
|
Tyczewska M, Rucinski M, Trejter M, et al:
Angiogenesis in the course of enucleation-induced adrenal
regeneration - expression of selected genes and proteins involved
in development of capillaries. Peptides. 38:404–413. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tyczewska M, Rucinski M, Ziolkowska A, et
al: Expression of selected genes involved in steroidogenesis in the
course of enucleation-induced rat adrenal regeneration. Int J Mol
Med. 33:613–623. 2014.
|
|
23
|
Tyczewska M, Rucinski M and Ziolkowska A:
Enucleation-induced rat adrenal gland regeneration - expression
profile of selected genes involved in control of adrenocortical
cell proliferation. Int J Endocrinol. 14(130359): 2014
|
|
24
|
Gentleman RC, Carey VJ, Bates DM, et al:
Bioconductor: open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc B. 57:289–300. 1995.
|
|
26
|
Gene Ontology Consortium: The Gene
Ontology project in 2008. Nucleic Acids Res. 36:D440–D444.
2008.
|
|
27
|
Feng G, Shaw P, Rosen ST, et al: Using the
bioconductor GeneAnswers package to interpret gene lists. Methods
Mol Biol. 802:101–112. 2012. View Article : Google Scholar
|
|
28
|
Ashburner M, Ball CA, Blake JA, et al:
Gene ontology: tool for the unification of biology. Nat Genet.
25:25–29. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bassett MH, Mayhew B, Rehman K, et al:
Expression profiles for steroidogenic enzymes in adrenocortical
disease. J Clin Endocrinol Metab. 90:5446–5455. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Assie G, Auzan C, Gasc JM, et al:
Steroidogenesis in aldosterone-producing adenoma revisited by
transcriptome analysis. J Clin Endocrinol Metab. 90:6638–6649.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lenzini L, Seccia TM, Aldighieri E, et al:
Heterogeneity of aldosterone-producing adenomas revealed by a whole
transcriptome analysis. Hypertension. 50:1106–1113. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Giordano TJ, Kuick R, Else T, et al:
Molecular classification and prognostication of adrenocortical
tumors by transcriptome profiling. Clin Cancer Res. 15:668–676.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang T, Satoh F, Morimoto R, et al: Gene
expression profiles in aldosterone-producing adenomas and adjacent
adrenal glands. Eur J Endocrinol. 164:613–619. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wilmot RH, Vezzosi D, Rizk-Rabin M, et al:
Identification of gene expression profiles associated with cortisol
secretion in adrenocortical adenomas. J Clin Endocrinol Metab.
98:E1109–E1121. 2013. View Article : Google Scholar
|
|
35
|
Romero DG, Plonczynski MW, Welsh BL, et
al: Gene expression profile in rat adrenal zona glomerulosa cells
stimulated with aldosterone secretagogues. Physiol Genomics.
32:117–127. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Romero DG, Gomez-Sanchez EP and
Gomez-Sanchez CE: Angiotensin II-regulated transcription regulatory
genes in adrenal steroidogenesis. Physiol Genomics. 42A:259–266.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oster H, Damerow S, Hut RA and Eichele G:
Transcriptional profiling in the adrenal gland reveals circadian
regulation of hormone biosynthesis genes and nucleosome assembly
genes. J Biol Rhythms. 21:350–361. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ishii T, Mitsui T, Suzuki S, et al: A
genome-wide expression profile of adrenocortical cells in knockout
mice lacking steroidogenic acute regulatory protein. Endocrinology.
153:2714–2723. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nishimoto K, Harris RB, Rainey WE and Seki
T: Sodium deficiency regulates rat adrenal zona glomerulosa gene
expression. Endocrinology. 155:1363–1372. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tan PK, Downey TJ, Spitznagel EL, et al:
Evaluation of gene expression measurements from commercial
microarray platforms. Nucleic Acids Res. 31:5676–5684. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Barnes M, Freudenberg J, Thompson S, et
al: Experimental comparison and cross-validation of the Affymetrix
and Illumina gene expression analysis platforms. Nucleic Acids Res.
33:5914–5923. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Russ J and Futschik ME: Comparison and
consolidation of microarray data sets of human tissue expression.
BMC Genomics. 11(305): 2010
|
|
43
|
Malendowicz LK: Sex differences in
adrenocortical structure and function. XXIV. Comparative
morphometric studies on adrenal cortex of intact mature male and
female rats of different strains. Cell Tissue Res. 249:443–449.
1987.PubMed/NCBI
|
|
44
|
Ramachandran J and Suyama AT: Inhibition
of replication of normal adrenocortical cells in culture by
adrenocorticotropin. Proc Natl Acad Sci USA. 72:113–117. 1975.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rybak SM and Ramachandran J: Primary
culture of normal rat adrenocortical cells. I. Culture conditions
for optimal growth and function. In Vitro. 17:599–604. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dallman MF: Control of adrenocortical
growth in vivo. Endocr Res. 10:213–242. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hu J, Zhang Z, Shen WJ and Azhar S:
Cellular cholesterol delivery, intracellular processing and
utilization for biosynthesis of steroid hormones. Nutr Metab.
7(47): 2010
|
|
48
|
Gallo-Payet N and Battista MC:
Steroidogenesis-adrenal cell signal transduction. Compr Physiol.
4:889–964. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Malendowicz LK and Mlynarczyk W: Sex
differences in adrenocortical structure and function. X. Lipid and
corticosterone in the rat adrenal as affected by gonadectomy and
testosterone or estradiol replacement. Endokrinologie. 79:292–300.
1982.PubMed/NCBI
|
|
50
|
Boyd GS and Trzeciak WH: Cholesterol
metabolism in the adrenal cortex: studies on the mode of action of
ACTH. Ann NY Acad Sci. 212:361–377. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kraemer FB, Shen WJ, Harada K, et al:
Hormone-sensitive lipase is required for high-density lipoprotein
cholesteryl ester-supported adrenal steroidogenesis. Mol
Endocrinol. 18:549–557. 2004. View Article : Google Scholar
|
|
52
|
Trzeciak WH and Malendowicz LK: Sex
differences in adrenocortical structure and function. VII. Adrenal
sterol ester hydrolase activity in the rat and its dependence on
gonadal hormones. Horm Metab Res. 13:519–522. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Stocco DM: StAR protein and the regulation
of steroid hormone biosynthesis. Annu Rev Physiol. 63:193–213.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kim YC, Ariyoshi N, Artemenko I, et al:
Control of cholesterol access to cytochrome P450scc in rat adrenal
cells mediated by regulation of the steroidogenic acute regulatory
protein. Steroids. 62:10–20. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lo YC, Brett L, Kenyon CJ, et al: StAR
protein is expressed in both medulla and cortex of the bovine and
rat adrenal gland. Endocr Res. 24:559–563. 1998. View Article : Google Scholar
|
|
56
|
Lehoux JG, Hales DB, Fleury A, et al: The
in vivo effects of adrenocorticotropin and sodium restriction on
the formation of the different species of steroidogenic acute
regulatory protein in rat adrenal. Endocrinology. 140:5154–5164.
1999.PubMed/NCBI
|
|
57
|
Rucinski M, Tortorella C, Ziolkowska A, et
al: Steroidogenic acute regulatory protein gene expression,
steroid-hormone secretion and proliferative activity of
adrenocortical cells in the presence of proteasome inhibitors: in
vivo studies on the regenerating rat adrenal cortex. Int J Mol Med.
21:593–597. 2008.PubMed/NCBI
|
|
58
|
Bastida CM, Cremades A, Castells MT, et
al: Sexual dimorphism of ornithine decarboxylase in the mouse
adrenal: influence of polyamine deprivation on catecholamine and
corticoid levels. Am J Physiol. 292:E1010–E1017. 2007.
|
|
59
|
Simpson ER, Mason JI, John ME, et al:
Regulation of the biosynthesis of steroidogenic enzymes. J Steroid
Biochem. 27:801–805. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Miller WL: Role of mitochondria in
steroidogenesis. Endocr Dev. 20:1–19. 2011.
|
|
61
|
Ishimura K and Fujita H: Light and
electron microscopic immunohistochemistry of the localization of
adrenal steroidogenic enzymes. Microsc Res Tech. 36:445–453. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Colby HD and Kitay JI: Effects of gonadal
hormones on adrenal steroid metabolism in vitro. Steroids.
20:143–157. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Malendowicz LK: Sex differences in
adrenocortical structure and function. III. The effects of
postpubertal gonadectomy and gonadal hormone replacement on adrenal
cholesterol side chain cleavage activity and on steroids
biosynthesis by rat adrenal homogenates. Endokrinologie.
75:311–323. 1976.
|
|
64
|
Brownie AC, Bhasker CR and Waterman MR:
Levels of adrenodoxin, NADPH-cytochrome P-450 reductase and
cytochromes P-45011 beta, P-45021, P-450scc, in adrenal zona
fasciculata-reticularis tissue from androgen-treated rats. Mol Cell
Endocrinol. 55:15–20. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Malee MP and Mellon SH: Zone-specific
regulation of two messenger RNAs for P450c11 in the adrenals of
pregnant and nonpregnant rats. Proc Natl Acad Sci USA.
88:4731–4735. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Oaks MK and Raff H: Differentiation of the
expression of aldosterone synthase and 11 beta-hydroxylase mRNA in
the rat adrenal cortex by reverse transcriptase-polymerase chain
reaction. J Steroid Biochem Mol Biol. 54:193–199. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Solyom J, Ludwig E, Regoly-Merei J and
Vajda A: The effect of potassium ions in vitro upon steroid
biosynthesis by rat adrenals. A study on the effect of different
salt intake and sex difference. Endokrinologie. 59:249–259.
1972.PubMed/NCBI
|
|
68
|
Ruhmann-Wennhold A and Nelson DH:
Testosterone inhibition of estradiol-induced stimulation of adrenal
11-beta- and 18-hydroxylation. Proc Soc Exp Biol Med. 133:493–496.
1970. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Goldman AS, Gustafsson JA and Stenberg A:
Neonatal imprinting of enzyme levels in rat adrenals. Acta
Endocrinol (Copenh). 76:719–728. 1974.
|
|
70
|
Beuschlein F, Keegan CE, Bavers DL, et al:
SF-1, DAX-1, and acd: molecular determinants of adrenocortical
growth and steroidogenesis. Endocr Res. 28:597–607. 2002.
View Article : Google Scholar
|
|
71
|
Iyer AK and McCabe ER: Molecular
mechanisms of DAX1 action. Mol Genet Metab. 83:60–73. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Manna PR, Dyson MT, Jo Y and Stocco DM:
Role of dosage-sensitive sex reversal, adrenal hypoplasia
congenita, critical region on the X chromosome, gene 1 in protein
kinase A- and protein kinase C-mediated regulation of the
steroidogenic acute regulatory protein expression in mouse Leydig
tumor cells: mechanism of action. Endocrinology. 150:187–199. 2009.
View Article : Google Scholar :
|
|
73
|
Mukai T, Kusaka M, Kawabe K, et al:
Sexually dimorphic expression of Dax-1 in the adrenal cortex. Genes
Cells. 7:717–729. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Jöhren O, Neidert SJ, Kummer M, Dendorfer
A and Dominiak P: Prepro-orexin and orexin receptor mRNAs are
differentially expressed in peripheral tissues of male and female
rats. Endocrinology. 142:3324–3331. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Jöhren O, Bruggemann N, Dendorfer A and
Dominiak P: Gonadal steroids differentially regulate the messenger
ribonucleic acid expression of pituitary orexin type 1 receptors
and adrenal orexin type 2 receptors. Endocrinology. 144:1219–1225.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Jöhren O, Bruggemann N and Dominiak P:
Orexins (hypocretins) and adrenal function. Horm Metab Res.
36:370–375. 2004. View Article : Google Scholar : PubMed/NCBI
|