Introduction
Primary liver cancer, which consists predominantly
of hepatocellular carcinoma (HCC), is the sixth most common cancer
worldwide and the second most common cause of cancer mortality
(1). HCC accounts for 70 to 90%
of primary liver cancers. There is an increasing understanding of
the molecular mechanisms that induce hepatocarcinogenesis,
including abrogation of cell-cycle checkpoints, and activation of
oncogenic pathways and Notch (2–4).
Further detailed clarification of the molecular pathways involved
in hepatocarcinogenesis could improve the treatment of HCC
patients.
Numb, a membrane-associated protein, asymmetrically
localizes during the division of neuroblasts and subsequently
segregates to one daughter cell at telophase, where it functions as
an intrinsic determinant of cell fate (5–7).
Drosophila and vertebrate Numb genes have a role in
binary cell fate decision in the peripheral and central nervous
system (8–9). Intrinsically inherited Numb
interacts with the cell surface receptor Notch and a
serine-threonine kinase, Numb-associated kinase (Nak), and
functions, at least in part, by antagonizing Notch activity
(10–11). Subversion of Numb is
associated with important human pathologies, including cancer. An
in vivo RNA interference screen in a model of mouse
lymphomagenesis identified Numb as a putative tumor
suppressor, whose ablation can accelerate the onset of lymphomas
(12). In breast cancer there is
a frequent loss of Numb expression, which causes increased
activity of the receptor Notch (13). Furthermore, Numb enters in
a tricomplex with p53 and the E3 ubiquitin ligase, HDM2, thereby
preventing ubiquitination and degradation of p53, and results in
increased p53 protein levels and activity (14). The present study was designed to
assess the role of Numb in liver carcinogenesis.
Materials and methods
Patients and follow-up
Fresh tumor samples and corresponding non-tumor
tissues used in reverse transcription quantitative-polymerase chain
reaction (RT-qPCR) and western blot analyses were randomly
collected from HCC patients who underwent curative resection
between 2006 and 2009 in The Second Xiangya Hospital (Central South
University, Hunan, China). All the specimens were removed and
preserved as described (15).
Tumor specimens used in immunohistochemistry were obtained from 85
HCC patients (62 males and 23 females, 28–72 years of age) who
underwent hepatectomy in the department. All these patients had
hepatitis B virus infection. The median follow-up period of these
patients was 34 months (range, 1–78 months). Previous informed
consent was obtained, and the study protocol was approved by the
Ethics Committee of The Second Xiangya Hospital.
Cell lines
Huh7 and HepG2 cell lines were purchased from the
certified biological resources center China Center for Type Culture
Collection (Wuhan, China). These cell lines were maintained at 37°C
in a humidified incubator under 5% CO2 conditions, as
described previously (2,16).
Immunohistochemistry
A total of 85 samples of HCC and their corresponding
non-tumoral liver tissue were assessed by immunohistochemistry.
Immunostaining was performed as described previously (17). Polyclonal rabbit anti-human Numb
(sc-25668, 1:100 dilution; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) was used to detect the expression of Numb.
Methylation-specific PCR (MSP), RT-qPCR
and western blot analyses
RT-qPCR of 13 HCC samples and related non-tumor
counterparts were analyzed as described previously (17). The primers are exhibited in
Table I. For other analyses, RNA
was collected from cultured cells with the TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA), according to the
manufacturer's instructions.
 | Table IDNA sequences of the primers used in
the study. |
Table I
DNA sequences of the primers used in
the study.
| Primer name | Sequence 5′→3′ |
|---|
| RT-qPCR | |
| β-actin-F |
ATCATGTTTGAGACCTTCAACA |
| β-actin-R |
CATCTCTTGCTCGAAGTCCA |
| Numb-F |
GGCATACAGAGGTTCCTACA |
| Numb-R |
TGCTCCTTTGACCGCTAC |
| BAK-F |
CAGGGCTTAGGACTTGGTTT |
| BAK-R |
TTTTTTCAGGGTGAGGGGAT |
| SKP2-F |
CTTTCTGGGTGTTCTGGATT |
| SKP2-R |
GGAGCAATTAATCTGTAGATGAGG |
| p21-F |
GCAGCGGAACAAGGAGT |
| p21-R |
GGAGAAACGGGAACCAG |
| CDK4-F |
CCCGAAGTTCTTCTGCAGTC |
| CDK4-R |
GTCGGCTTCAGATTTCCAC |
| MSP | |
| Numb
MSPm-F |
TTTCGAAAGTGTTGGGATTATATAC |
| Numb
MSPm-R |
AACTACAATAAACCAAAATCGCG |
| Numb
MSPu-F |
TTGAAAGTGTTGGGATTATATATGT |
| Numb
MSPu-R |
AACTACAATAAACCAAAATCACACC |
Nine HCC samples and the related
non-tumor-counterparts were further used in western blot analysis
as described previously (15).
The following primary antibodies were used in western blot
analysis: Rabbit polyclonal antibodies against Numb,
cyclin-dependent protein kinase 4 (CDK4) (sc-260), S-phase
kinase-associated protein 2 (SKP2) (sc-7164), Bcl-2 homologous
antagonist/killer (BAK) (sc-832), cyclin-dependent kinase inhibitor
1 (p21) (sc-397) and GAPDH (sc-25778) (all from Santa Cruz
Biotechnology, Inc.).
Thirty primary HCC tumors and their paired
non-tumorous tissues were investigated by MSP, as described
previously (18).
Cell proliferation, colony formation,
cell cycle and apoptosis assay
The functional role of Numb in HCC cells was
analyzed with small interfering RNA (siRNA). The Numb siRNA
sequence was UGGAACAUAAACAUCCUUCUUUCUC. The negative control siRNA
(cat. no. 12935-200; Invitrogen Life Technologies) was used as the
negative control for all the experiments. The transfection of siRNA
into HepG2 and Huh7 was performed with Lipofectamine 2000 (cat. no.
11668-019; Invitrogen Life Technologies) according to the
manufacturer's instructions.
Cell proliferation, colony formation, cell cycle and
apoptosis assays were performed as described previously (16).
Statistical analysis
Statistical analyses were performed with SPSS 16.0
for Windows (SPSS, Inc., Chicago, IL, USA). Cumulative survival
time was calculated by the Kaplan-Meier method and analyzed by the
log-rank test. Univariate and multivariate analyses were based on
the Cox proportional hazards regression model. The χ2
test, Fisher's exact test and Student's t-test were used for
comparison between groups. All the tests were two-tailed. Values of
P<0.05 were considered to indicate a statistically significant
difference.
Results
Numb expression in HCC
Cellular localization of the Numb protein was
assessed by immunohistochemistry in the tumor tissue and their
corresponding non-tumoral liver tissue from 85 samples of HCC. In
general, high expression of the Numb protein was observed in both
tumor tissue (39/85, 46%) and non-tumoral liver tissue (53/85,
62%), and the staining intensity varied widely. In normal liver and
chronic hepatitis samples, Numb expression was low (Fig. 1A-a). The majority of cirrhotic
liver samples showed increased Numb expression, particularly
in regenerative nodules (Fig.
1A–b). High expression of the Numb protein was significantly
more frequent in the samples of cirrhotic liver (49/58, 84%)
compared with the normal liver (0/3, 0%), chronic hepatitis (4/24,
17%) and cancer tissues (39/85, 46%), as shown in Table II. In the cancer tissues, the
histopathological type of HCC was associated with Numb expression;
the expression level decreased with poor cell differentiation. The
majority of well- and moderately differentiated HCCs showed high
Numb expression (36/46, 78%) (Fig.
1A–c), while poorly differentiated HCCs showed a low Numb
expression (36/39, 92%) (Table
III; Fig. 1A–d). The high
expression of Numb was more frequent in liver cirrhosis and
well-differentiated HCCs compared with the normal liver, chronic
hepatitis or poorly differentiated HCCs.
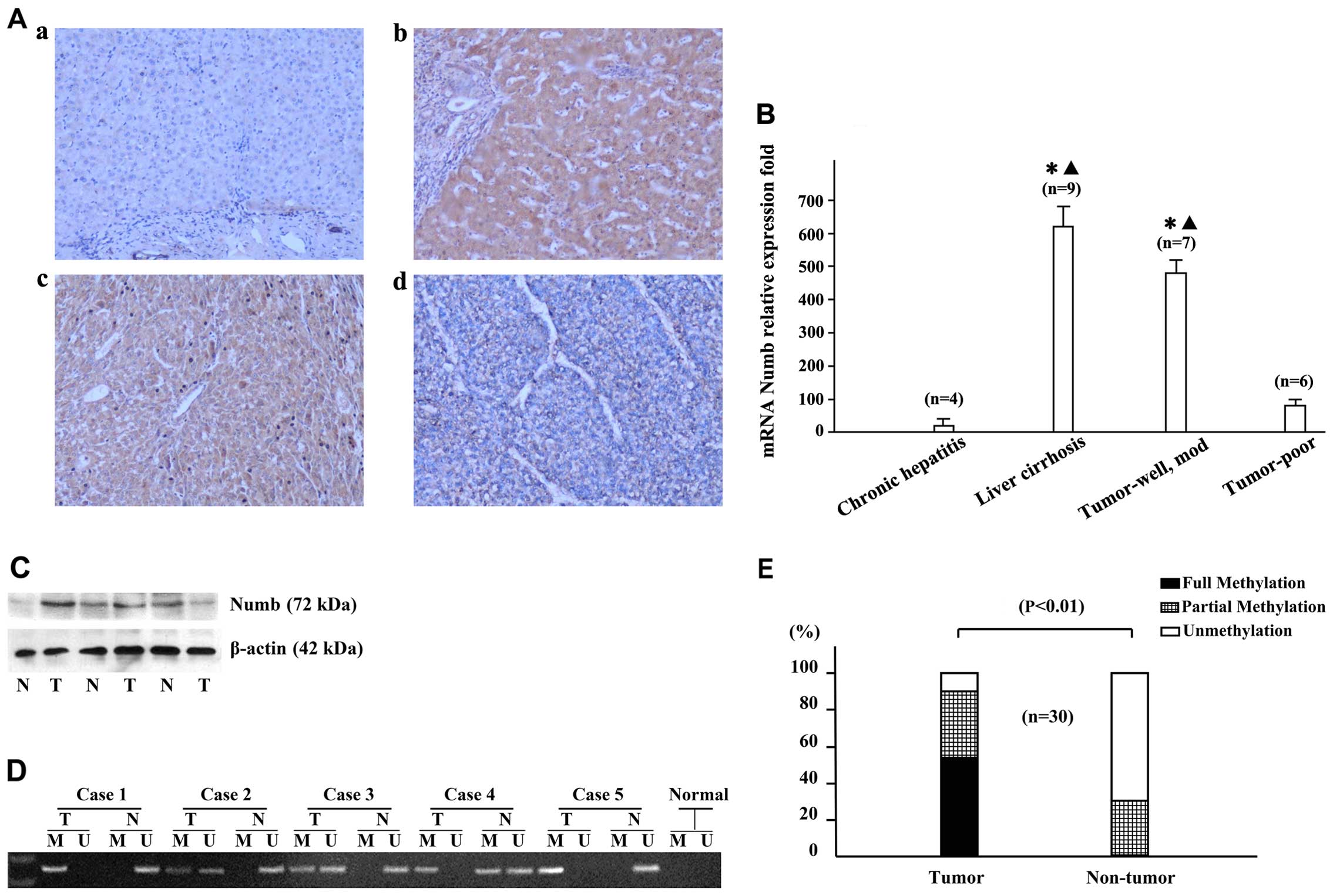 | Figure 1Expression of Numb in samples from
hepatocellular carcinomas (HCC) patients. (A) Expression of Numb in
non-tumorous and tumor liver tissues detected by
immunohistochemistry; (a) chronic hepatitis, (b) liver cirrhosis,
(c) moderately differentiated HCC, (d) poorly differentiated HCC.
Magnification, ×100. (B) Reverse transcription-quantitative
polymerase chain reaction (PCR) was used to examine Numb
mRNA expression from 13 paired HCC tissues and their non-tumor
counterparts. The results (mean ± standard deviation) were
normalised for β-actin expression. Statistical analysis was
performed comparing liver cirrhosis, well- and moderately
differentiated tumors, and poorly differentiated tumors with
chronic hepatitis; *P<0.05; and liver cirrhosis, and
well-and moderately differentiated tumors compared with poorly
differentiated tumors; ▲P<0.05. (C) The
representative expression of Numb as assessed by western blot
analysis. Lane 1, chronic hepatitis B; lanes 2 and 4, moderately
differentiated tumors; lanes 3 and 5, liver cirrhosis; lane 6,
poorly differentiated tumor. (D) Methylation of Numb
promoter in HCC tissues was determined by methylation-specific PCR
(MSP). (E) MSP showed that the methylation of Numb was
significantly detected in HCC tissues (T) compared with the
corresponding non-tumorous tissues (N); n=19, P<0.01. M,
methylated DNA; U, unmethylated DNA. |
 | Table IIImmunohistochemical analysis of Numb
in cancer and non-cancer liver tissues. |
Table II
Immunohistochemical analysis of Numb
in cancer and non-cancer liver tissues.
| Pathological
category | No. | Staining intensity
for Numb, no.
|
|---|
| Low | High |
|---|
| Normal liver | 3 | 3 | 0 |
| Chronic
hepatitis | 24 | 20 | 4 |
| Cirrhosis | 58 | 9a,b | 49 |
| HCC | 85 | 46c | 39 |
 | Table IIIAssociation between Numb expression
and clinicopathological parameters in HCC. |
Table III
Association between Numb expression
and clinicopathological parameters in HCC.
| Parameter | n | Expression of Numb,
n
| P-value |
|---|
| Low | High |
|---|
| Age, years | | | | |
| ≥60 | 55 | 24 | 21 | |
| <60 | 30 | 22 | 8 | NS |
| Gender | | | | |
| Male | 62 | 36 | 26 | |
| Female | 23 | 10 | 13 | NS |
| Tumor size, cm | | | | |
| <5 | 12 | 4 | 8 | |
| ≥5 | 73 | 42 | 31 | NS |
| Histological
grade | | | | |
| Well/moderately
differentiated | 46 | 10 | 36 | |
|
Poorly/undifferentiated | 39 | 36 | 3 | 0.001 |
| Portal vein
invasion | | | | |
| Yes | 24 | 16 | 8 | |
| No | 61 | 30 | 31 | 0.022 |
| No. of tumors | | | | |
| Solitary | 58 | 31 | 27 | |
| Multiple | 27 | 15 | 12 | 0.034 |
| Capsular
formation | | | | |
| Yes | 36 | 21 | 15 | |
| No | 49 | 25 | 24 | NS |
RT-qPCR analysis was performed by 13 paired
non-tumor and tumor mRNA extracts. Numb mRNA expression
levels in the tissues of liver cirrhosis (n=9), well- and
moderately differentiated (n=7) and poorly differentiated liver
tumors (n=6) had 41-, 38- and 2-fold increased expression with
respect to chronic hepatitis (n=4) (P<0.05 respectively).
Additionally, decreased expression levels were observed in poorly
differentiated liver tumors compared with those in the tissues of
liver cirrhosis, and well- and moderately differentiated liver
tumors (P<0.05 respectively) (Fig.
1B). In addition, overexpression of Numb was further
confirmed by a western blot analysis in 6 cases (Fig. 1C).
Aberrant methylation of Numb in HCC
The methylation frequency of Numb was
investigated in 30 primary HCC tumors and their paired non-tumorous
tissues by MSP (Fig. 1D).
In the tumor tissues, fully, partially or
unmethylated alleles of Numb were detected in 16/30 (53.3%),
11/30 (36.7%) and 3/30 (10%) cases, respectively, whereas in the
paired non-tumorous tissues, 0/30 (0%), 9/30 (30%) and 21/30 (70%)
cases were identified correspondingly, which supported that
methylation of Numb has a role in hepatocarcinogenesis
(Fig. 1E).
Numb expression and its association with
clinicopathological features and prognosis of the HCC patients
As shown in Table
III, Numb expression in tumor samples was significantly
associated with histological grade (P=0.001), portal vein invasion
(P=0.022) and number of tumors (P=0.034) in the HCC patients.
However, no statistically significant association was identified
between Numb expression and the other clinical
characteristics.
To further evaluate the prognostic value of
Numb for HCC patients, univariate and multivariate analyses
were performed for the clinicopathological characteristics and
expression of Numb. In the univariate analysis, tumor
differentiation, portal vein involvement and number of tumors were
revealed to be associated with disease-free and overall survival of
the HCC patients. No significant prognostic significance was
identified in the other characteristics, including gender, age and
capsular formation of patients for overall or disease-free survival
(Table IV).
 | Table IVUnivariate and multivariate analyses
of factors associated with survival rate and recurrence. |
Table IV
Univariate and multivariate analyses
of factors associated with survival rate and recurrence.
| Factors | Overall survival
| Cumulative
recurrence
|
|---|
Univariate
| Multivariate
| Univariate
| Multivariate
|
|---|
| P-value | HR (95% CI) | P-value | P-value | HR (95% CI) | P-value |
|---|
| Age, years | | | | | | |
| <65 | 0.646 | 0.694 | 0.376 | 0.971 | 0.866 | 0.733 |
| ≥65 | | (0.349–2.753) | | | (0.378–1.981) | |
| Gender | | | | | | |
| Male | 0.504 | 0.980 | 0.970 | 0.607 | 0.979 | 0.968 |
| Female | | (0.340–2.647) | | | (0.346–2.772) | |
| Numb
expression | | | | | | |
| Low | <0.001 | 9.303 | <0.001 | <0.001 | 13.600 | <0.001 |
| High | | (2.812–30.779) | | | (3.695–50.056) | |
| Tumor size, cm | | | | | | |
| <3 | 0.681 | 0.925 | 0.598 | 0.856 | 0.950 | 0.906 |
| ≥3 | | (0.396–2.157) | | | (0.405–2.226) | |
| Tumor
differentiation | | | | | | |
| I/II | <0.001 | 0.349 | 0.013 | <0.001 | 0.332 | 0.009 |
| III/IV | | (0.151–0.803) | | | (0.144–0.762) | |
| Tumor number | | | | | | |
| Solitary | 0.001 | 0.565 | 0.207 | 0.010 | 0.848 | 0.719 |
| Multiple | | (0.232–1.373) | | | (0.345–2.084) | |
| Portal vein
invasion | | | | | | |
| No | <0.001 | 0.403 | 0.041 | <0.001 | 0.271 | 0.005 |
| Yes | | (0.168–0.964) | | | (0.110–0.671) | |
| Tumor
encapsulation | | | | | | |
| No | 0.369 | 1.282 | 0.588 | 0.747 | 0.910 | 0.844 |
| Yes | | (0.521–3.156) | | | (0.359–2.311) | |
Individual features that showed significance by
univariate analysis were adopted as covariates in a multivariate
Cox proportional hazards model and subsequently the combined
variables were further analyzed. Numb was shown to be an
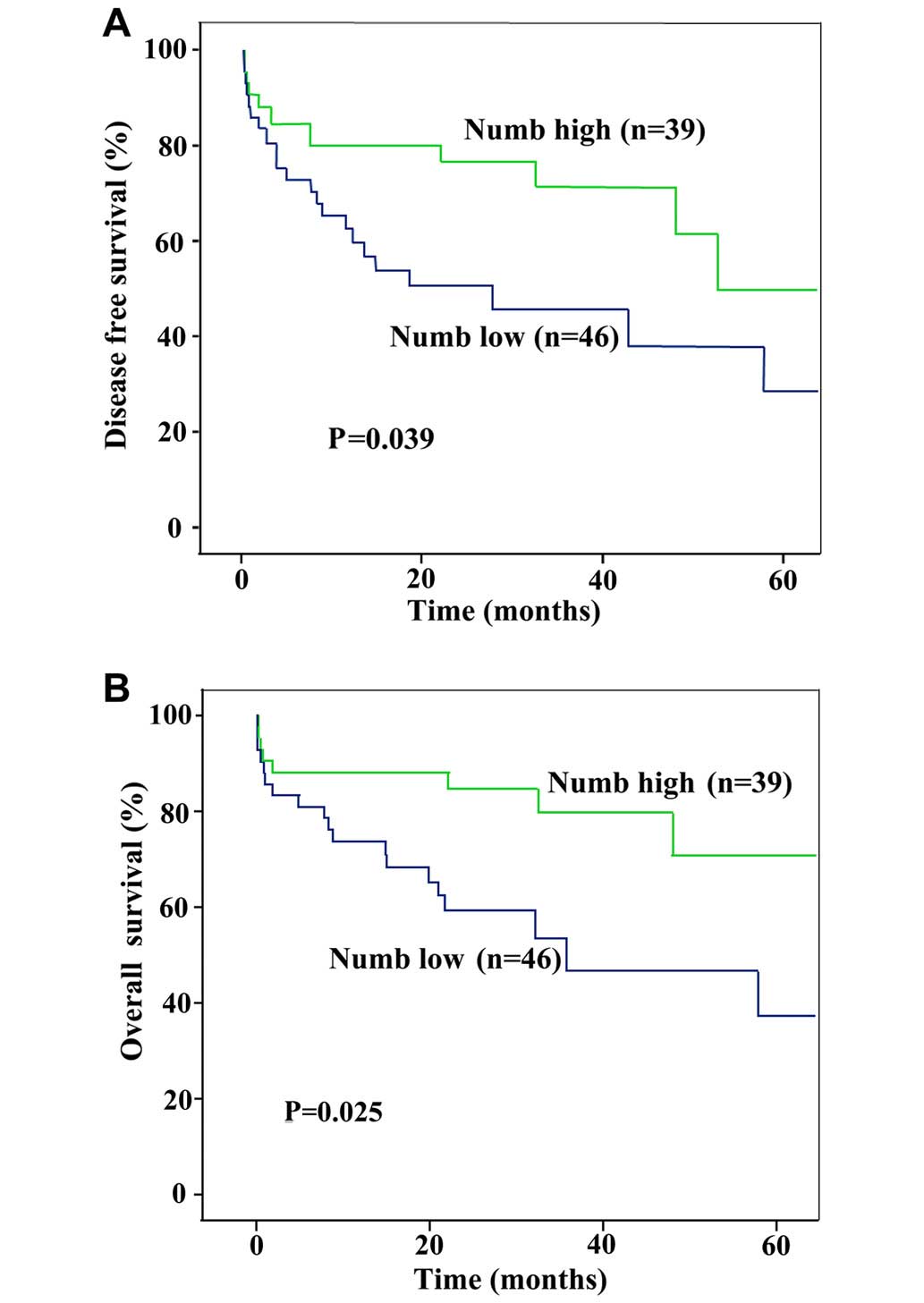
independent prognostic indicator for disease-free (P=0.039) and
overall survival (P=0.025) (Fig. 2A
and B).
Antiproliferation of HCC cells by Numb
silencing
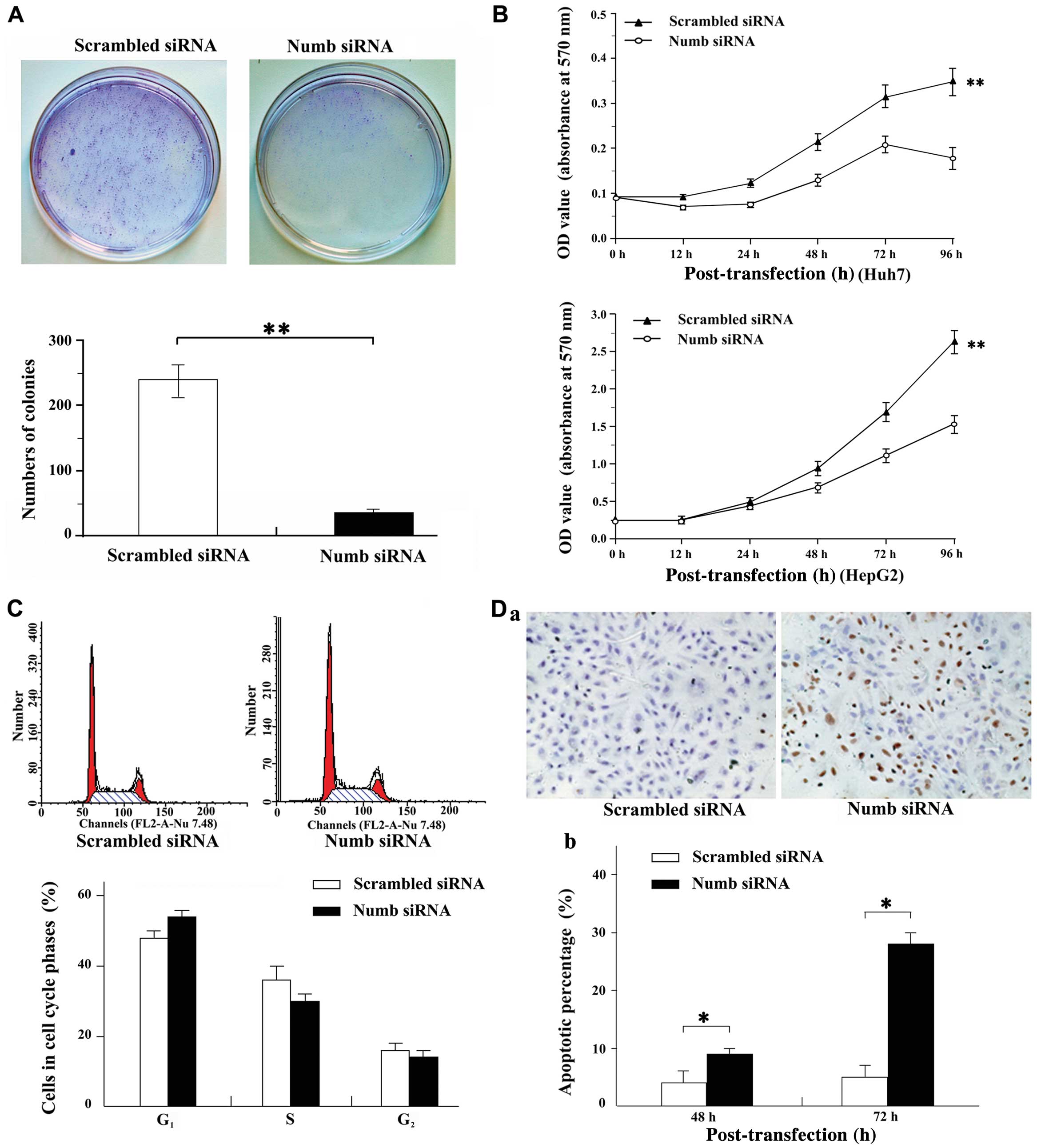
Tumor suppression was assessed by soft agar
formation and cell growth assay.
The soft agar assay showed that the frequency of
colony formation of Huh7 cells was significantly inhibited
(238.67±24.97 vs. 35.00±3.64, P<0.01) in Numb silencing
compared with scramble cells (Fig.
3A).
Numb silencing also significantly inhibited
the cell growth of Huh7 and HepG2 cells by the MTT assay. The
statistical significance (P<0.01) between cultures treated with
Numb siRNA versus scramble is shown in Fig. 3B.
Induction of apoptosis by ectopic
expression of Numb
To explore the molecular mechanism of Numb in
HCC development, the role of Numb in the cell cycle was
investigated with flow cytometry. In the Huh7 cells tested,
treatment with Numb siRNA for 24 h increased the
G0–G1 phase fraction and decreased the S
phase fraction when compared with the scramble-treated cultures. A
representative result is shown in Fig. 3C with the following fraction
values for cultures treated with Numb siRNA versus scramble
(G0–G1: 54.35 vs. 48.91; S phase fraction:
31.22 vs. 35.87; G2-M: 14.44 vs. 15.22,
respectively).
The apoptotic index was compared between Numb
siRNA and scramble cells by TUNEL staining. The TUNEL assay
revealed that apoptotic cells treated by Numb siRNA were
significantly higher compared with those of the scramble control
(48 h: 4.39 vs. 21.94%; 72 h: 4.47 vs. 36.10%, respectively,
P<0.001; Fig. 3D).
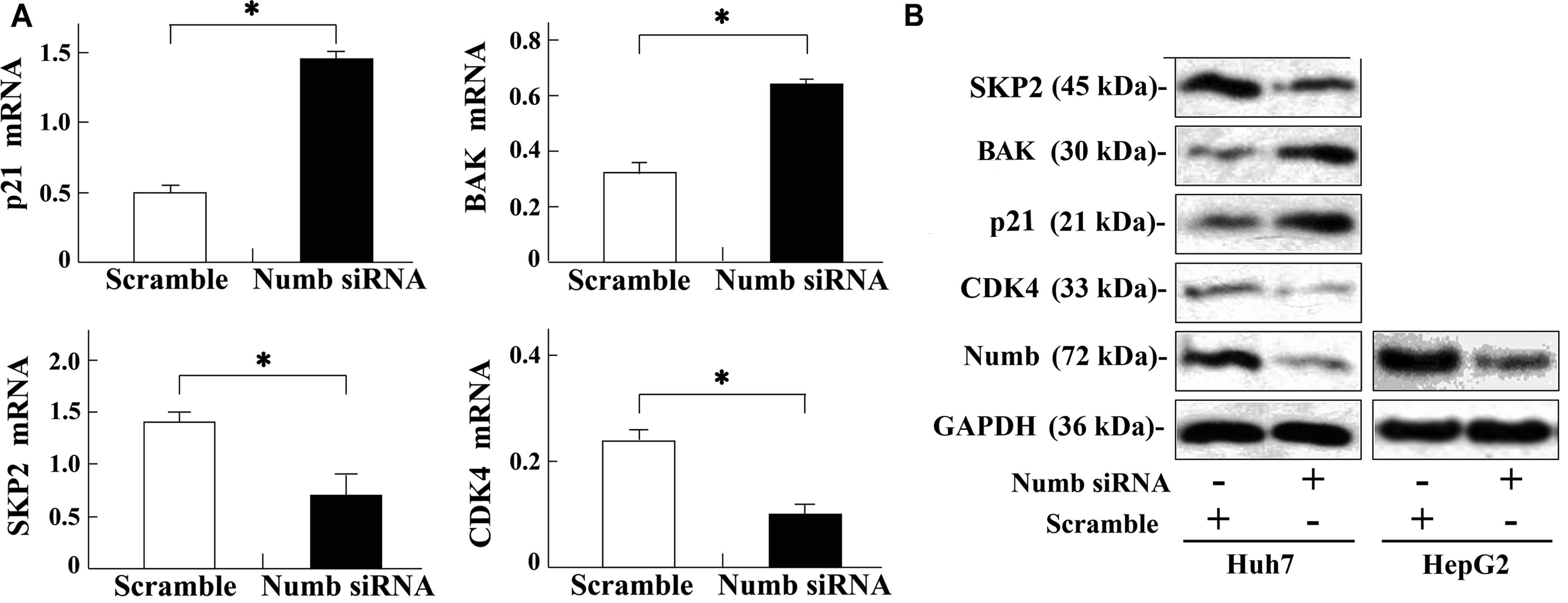
To further elucidate the molecular basis of cell
cycle and apoptosis induced by Numb silencing, the relevant
molecules were screened by RT-qPCR and confirmed by western blot
analysis. RT-qPCR analysis revealed that CDK4 and
SKP2 were downregulated whereas BAK and p21
were upregulated in the Huh7 cell line following Numb siRNA
treatment (P<0.05, respectively) (Fig. 4A). Western blot analysis further
confirmed these results (Fig.
4B).
Discussion
In the present study, Numb was overexpressed
in patients with HCC. A decreased expression of Numb in
human HCC was an independent predictor of poor prognosis.
Numb ablation by siRNA in vitro could inhibit tumor
cell proliferation by downregulating CDK4 and SKP2
and upregulating p21 expression, and enhancing the apoptotic
potential by upregulating BAK.
The present results showed that the Numb gene
was involved in hepatocarcinogenesis and its decreased expression
was associated with a poor prognosis of HCC patients. In the
non-tumor liver tissue, Numb was overexpressed in the
majority of cirrhotic nodules and liver tissue infected with
hepatitis B virus compared to the normal liver tissue. In addition,
Numb was overexpressed in well- and moderately
differentiated tumor tissues, which was similar to our previous
reports on Vanilloid receptor-1, CB1 and CB2 receptors (17,19). Additionally, downregulation of
Numb was associated with aggressive cancer phenotypes,
including portal vein invasion and dedifferentiated histology
(Table III). Univariate and
multivariate analyses showed that a low expression of Numb
was significantly associated with disease relapse and poor survival
of HCC. These data indicate that Numb may serve as an
independent predictor for the prognosis of HCC patients.
Numb is an evolutionary conserved protein
that has been indicated in tumorigenesis with roles in controlling
stem/progenitor cell development (20). In addition, a number of cellular
processes, such as cell adhesion and migration controlled by
Numb, are also involved in mammalian tumorigenesis, such as
in chronic myelogenous leukaemia, breast cancer, non-small cell
lung cancer and salivary gland carcinomas (13,14,21–24). In chronic myelogenous leukaemia,
which progress from a slow-growing chronic phase to an aggressive
blast crisis phase, high levels of Numb expression were
observed in the chronic phase whereas low levels of Numb
expression were observed in the blast crisis phase. In breast
cancer, reduced expression of Numb correlating with
decreased p53 levels and increased chemo-resistance was found to
result in an aggressive tumor phenotype as illustrated by poor
prognostic outcome for these patients (14,25). Concordant with these findings, a
defect of Numb expression in HCC was associated with
aggressive phenotypes of the tumor and poor outcome of the patients
highlighted its involvement in hepatocarcinogenesis.
It has been well documented that DNA methylation of
CpG islands located near gene promoters affects the transcription
of specific genes (26,27). Epigenetic inactivation of genes by
promotor methylation has been recognized as an important and
alternative mechanism in carcinogenesis. In HCC, aberrant promoter
methylation of p16 (INK4) is considered a main cause
resulting in its function loss (28,29). In the present study, methylated
alleles of Numb could be detected in 12/19 (63.2%) of tumor
tissues, indicating the involvement of methylation in
hepatocarcinogenesis.
On the basis of those findings above, the role of
Numb was further explored in the proliferation of HCC cells.
Compared with scramble RNA-treated HCC cells, Numb siRNA
silencing resulted in the inhibition of proliferation and colony
formation in soft agar, cell-cycle arrest and induction of
apoptosis. These results suggest that Numb has an important
role in the proliferative process of HCC cells.
Our previous molecular studies identified that
Numb suppression could cause dysregulation of CDK4, SKP2,
p21 and BAK, suggesting its roles in several signaling pathways.
Numb is involved in p53, Notch and Hedgehog pathways in
several tumors (14,30–32). The potential roles of cyclin-cdk
complexes, such as CDK and cyclin kinase inhibitor families
modulating in the G0–G1 phase of the cell
cycle, have been addressed in human hepatocarcinogenesis (33,34). Progress has further shown that
SKP2, which maintains and preserves the distinct phases during a
cell cycle through protein degradation, has emerged in human
hepatocarcinogenesis (35,36).
The present data showed that Numb silencing resulted in
upregulation of p21 and downregulation of CDK4 and
SKP2 in HCC cells, suggesting involvement of Numb in
the G1-S phase of a cell cycle. In addition, BAK is a
well-known cell death initiator in the apoptotic signaling cascade,
and its roles in hepatocarcinogensis or for HCC treatment have also
been documented (37,38). Different agents induce apoptosis
in HCC cells by stimulating BAK expression, suggesting its
pro-apoptotic effect (39,40).
The present findings that upregulation of BAK was caused by
Numb silencing highlights the role of Numb in
apoptosis induction. Interactions among these molecules as targeted
by Numb warrant further detailed investigation.
Based on the findings of the present study, it can
be concluded that downregulation of Numb may be a predictor
of HCC prognosis, and Numb has an important role in the
proliferation of HCC cells in vitro via interaction with
CDK4, p21, SKP2 and BAK. Numb may be a potential therapeutic
target for HCC patients.
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
BAK
|
Bcl-2 homologous antagonist/killer
|
|
CDK4
|
cyclin-dependent protein kinase 4
|
|
SKP2
|
S-phase kinase-associated protein
2
|
|
p21
|
cyclin-dependent kinase inhibitor
1
|
|
MSP
|
methylation-specific polymerase chain
reaction
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu X, Yamamoto H, Liu G, Ito Y, Ngan CY,
Kondo M, Nagano H, Dono K, Sekimoto M and Monden M: CDC25A
inhibition suppresses the growth and invasion of human
hepatocellular carcinoma cells. Int J Mol Med. 21:145–152.
2008.PubMed/NCBI
|
|
3
|
Martínez-López N, Varela-Rey M,
Fernández-Ramos D, Woodhoo A, Vázquez-Chantada M, Embade N,
Espinosa-Hevia L, Bustamante FJ, Parada LA, Rodriguez MS, et al:
Activation of LKB1-Akt pathway independent of phosphoinositide
3-kinase plays a critical role in the proliferation of
hepatocellular carcinoma from nonalcoholic steatohepatitis.
Hepatology. 52:1621–1631. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu J, Yun X, Jiang J, Wei Y, Wu Y, Zhang
W, Liu Y, Wang W, Wen Y and Gu J: Hepatitis B virus X protein
blunts senescence-like growth arrest of human hepatocellular
carcinoma by reducing Notch1 cleavage. Hepatology. 52:142–154.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rhyu MS, Jan LY and Jan YN: Asymmetric
distribution of numb protein during division of the sensory organ
precursor cell confers distinct fates to daughter cells. Cell.
76:477–491. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Spana EP, Kopczynski C, Goodman CS and Doe
CQ: Asymmetric localization of numb autonomously determines sibling
neuron identity in the Drosophila CNS. Development. 121:3489–3494.
1995.PubMed/NCBI
|
|
7
|
Vervoort M, Dambly-Chaudière C and Ghysen
A: Cell fate determination in Drosophila. Curr Opin Neurobiol.
7:21–28. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park M, Yaich LE and Bodmer R: Mesodermal
cell fate decisions in Drosophila are under the control of the
lineage genes numb, Notch, and sanpodo. Mech Dev. 75:117–126. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Spana EP and Doe CQ: Numb antagonizes
Notch signaling to specify sibling neuron cell fates. Neuron.
17:21–26. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo M, Jan LY and Jan YN: Control of
daughter cell fates during asymmetric division: Interaction of Numb
and Notch. Neuron. 17:27–41. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chien CT, Wang S, Rothenberg M, Jan LY and
Jan YN: Numb-associated kinase interacts with the phosphotyrosine
binding domain of Numb and antagonizes the function of Numb in
vivo. Mol Cell Biol. 18:598–607. 1998.PubMed/NCBI
|
|
12
|
Bric A, Miething C, Bialucha CU, Scuoppo
C, Zender L, Krasnitz A, Xuan Z, Zuber J, Wigler M, Hicks J, et al:
Functional identification of tumor-suppressor genes through an in
vivo RNA interference screen in a mouse lymphoma model. Cancer
Cell. 16:324–335. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pece S, Serresi M, Santolini E, Capra M,
Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G and Di
Fiore PP: Loss of negative regulation by Numb over Notch is
relevant to human breast carcinogenesis. J Cell Biol. 167:215–221.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Colaluca IN, Tosoni D, Nuciforo P,
Senic-Matuglia F, Galimberti V, Viale G, Pece S and Di Fiore PP:
NUMB controls p53 tumour suppressor activity. Nature. 451:76–80.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu G, Xie C, Sun F, Xu X, Yang Y, Zhang
T, Deng Y, Wang D, Huang Z, Yang L, et al: Clinical significance of
transient receptor potential vanilloid 2 expression in human
hepatocellular carcinoma. Cancer Genet Cytogenet. 197:54–59. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xie C, Liu G, Liu J, Huang Z, Wang F, Lei
X, Wu X, Huang S, Zhong D and Xu X: Anti-proliferative effects of
anandamide in human hepatocellular carcinoma cells. Oncol Lett.
4:403–407. 2012.PubMed/NCBI
|
|
17
|
Miao X, Liu G, Xu X, Xie C, Sun F, Yang Y,
Zhang T, Hua S, Fan W, Li Q, et al: High expression of vanilloid
receptor-1 is associated with better prognosis of patients with
hepatocellular carcinoma. Cancer Genet Cytogenet. 186:25–32. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fu L, Qin YR, Xie D, Hu L, Kwong DL,
Srivastava G, Tsao SW and Guan XY: Characterization of a novel
tumor-suppressor gene PLC delta 1 at 3p22 in esophageal squamous
cell carcinoma. Cancer Res. 67:10720–10726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu X, Liu Y, Huang S, Liu G, Xie C, Zhou
J, Fan W, Li Q, Wang Q, Zhong D, et al: Overexpression of
cannabinoid receptors CB1 and CB2 correlates with improved
prognosis of patients with hepatocellular carcinoma. Cancer Genet
Cytogenet. 171:31–38. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Neumüller RA and Knoblich JA: Dividing
cellular asymmetry: Asymmetric cell division and its implications
for stem cells and cancer. Genes Dev. 23:2675–2699. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao C, Chen A, Jamieson CH, Fereshteh M,
Abrahamsson A, Blum J, Kwon HY, Kim J, Chute JP, Rizzieri D, et al:
Hedgehog signalling is essential for maintenance of cancer stem
cells in myeloid leukaemia. Nature. 458:776–779. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ito T, Kwon HY, Zimdahl B, Congdon KL,
Blum J, Lento WE, Zhao C, Lagoo A, Gerrard G, Foroni L, et al:
Regulation of myeloid leukaemia by the cell-fate determinant
Musashi. Nature. 466:765–768. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Westhoff B, Colaluca IN, D'Ario G,
Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G,
Viale G, et al: Alterations of the Notch pathway in lung cancer.
Proc Natl Acad Sci USA. 106:22293–22298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maiorano E, Favia G, Pece S, Resta L,
Maisonneuve P, Di Fiore PP, Capodiferro S, Urbani U and Viale G:
Prognostic implications of NUMB immunoreactivity in salivary gland
carcinomas. Int J Immunopathol Pharmacol. 20:779–789. 2007.
|
|
25
|
Rennstam K, McMichael N, Berglund P,
Honeth G, Hegardt C, Rydén L, Luts L, Bendahl PO and Hedenfalk I:
Numb protein expression correlates with a basal-like phenotype and
cancer stem cell markers in primary breast cancer. Breast Cancer
Res Treat. 122:315–324. 2010. View Article : Google Scholar
|
|
26
|
Sakai T, Toguchida J, Ohtani N, Yandell
DW, Rapaport JM and Dryja TP: Allele-specific hypermethylation of
the retinoblastoma tumor-suppressor gene. Am J Hum Genet.
48:880–888. 1991.PubMed/NCBI
|
|
27
|
Merlo A, Herman JG, Mao L, Lee DJ,
Gabrielson E, Burger PC, Baylin SB and Sidransky D: 5′CpG island
methylation is associated with transcriptional silencing of the
tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med.
1:686–692. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Matsuda Y, Ichida T, Matsuzawa J, Sugimura
K and Asakura H: p16(INK4) is inactivated by extensive CpG
methylation in human hepatocellular carcinoma. Gastroenterology.
116:394–400. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li X, Hui AM, Sun L, Hasegawa K, Torzilli
G, Minagawa M, Takayama T and Makuuchi M: p16INK4A hypermethylation
is associated with hepatitis virus infection, age, and gender in
hepatocellular carcinoma. Clin Cancer Res. 10:7484–7489. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chapman G, Liu L, Sahlgren C, Dahlqvist C
and Lendahl U: High levels of Notch signaling down-regulate Numb
and Numblike. J Cell Biol. 175:535–540. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McGill MA, Dho SE, Weinmaster G and
McGlade CJ: Numb regulates post-endocytic trafficking and
degradation of Notch1. J Biol Chem. 284:26427–26438. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Di Marcotullio L, Ferretti E, Greco A, De
Smaele E, Po A, Sico MA, Alimandi M, Giannini G, Maroder M,
Screpanti I, et al: Numb is a suppressor of Hedgehog signalling and
targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol.
8:1415–1423. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Greenbaum LE: Cell cycle regulation and
hepatocarcinogenesis. Cancer Biol Ther. 3:1200–1207. 2004.
View Article : Google Scholar
|
|
34
|
Hui AM, Makuuchi M and Li X: Cell cycle
regulators and human hepatocarcinogenesis. Hepatogastroenterology.
45:1635–1642. 1998.PubMed/NCBI
|
|
35
|
Calvisi DF, Ladu S, Pinna F, Frau M,
Tomasi ML, Sini M, Simile MM, Bonelli P, Muroni MR, Seddaiu MA, et
al: SKP2 and CKS1 promote degradation of cell cycle regulators and
are associated with hepatocellular carcinoma prognosis.
Gastroenterology. 137:1816–26.e10. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ho C, Wang C, Mattu S, Destefanis G, Ladu
S, Delogu S, Armbruster J, Fan L, Lee SA, Jiang L, et al: AKT
(v-akt murine thymoma viral oncogene homolog 1) and N-Ras
(neuroblastoma ras viral oncogene homolog) coactivation in the
mouse liver promotes rapid carcinogenesis by way of mTOR (mammalian
target of rapamycin complex 1), FOXM1 (forkhead box M1)/SKP2, and
c-Myc pathways. Hepatology. 55:833–845. 2012. View Article : Google Scholar :
|
|
37
|
Cory S and Adams JM: The Bcl2 family:
Regulators of the cellular life-or-death switch. Nat Rev Cancer.
2:647–656. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Griffiths GJ, Dubrez L, Morgan CP, Jones
NA, Whitehouse J, Corfe BM, Dive C and Hickman JA: Cell
damage-induced conformational changes of the pro-apoptotic protein
Bak in vivo precede the onset of apoptosis. J Cell Biol.
144:903–914. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hu R, Zhai Q, Liu W and Liu X: An insight
into the mechanism of cytotoxicity of ricin to hepatoma cell: Roles
of Bcl-2 family proteins, caspases, Ca(2+)-dependent proteases and
protein kinase C. J Cell Biochem. 81:583–593. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang QF, Chen JC, Hsieh SJ, Cheng CC and
Hsu SL: Regulation of Bcl-2 family molecules and activation of
caspase cascade involved in gypenosides-induced apoptosis in human
hepatoma cells. Cancer Lett. 183:169–178. 2002. View Article : Google Scholar : PubMed/NCBI
|