Introduction
The heart is one of the most frequently studied
human organs and is the organ most susceptible to disease.
Congenital heart defects (CHD), the most common of the human birth
defects, occur in nearly 1% of the population worldwide (1,2).
Significant progress in CHD treatment has occurred over the years;
consequently, more patients with CHD live to adulthood, creating a
new and steadily growing patient population. These patients require
long-term expert medical care and healthcare, which is expensive
(3,4). Correspondingly, the global health
burden resulting from CHD has rapidly increased. However, CHD
progresses to degenerative conditions that subsequently afflict CHD
survivors. The developmental process of the heart is complicated,
involving a series of genes and multiple signaling pathways (Notch,
bone morphogenetic protein and transforming growth factor-β)
(5,6); a small mutation in any of these
genes or pathways will result in embryonic heart defects.
MicroRNAs (miRNAs or miRs) are endogenous
18–22-nucleotide RNAs that have important regulatory roles in
animals and plants by targeting mRNAs for cleavage or translational
repression (7,8). Currently, numerous aspects of miRNA
function in animals, including their involvement in cell
proliferation, apoptotic events, differentiation, fat and lipid
metabolism, cancer, diabetes, and other diseases, have been
researched and established (9,10).
It is becoming evident that miRNAs are involved in different
aspects of cardiomyogenesis (11,12), and have recently been demonstrated
to serve as diagnostic biomarkers (13,14). Based on these findings, prenatal
detection of fetal CHD was performed in our previous studies,
identifying that miRNA-375 (miR-375) is significantly upregulated
in maternal serum at 18–22 weeks of gestation with fetal CHD
(15,16), indicating that it may be involved
in the occurrence or development of CHD. Using bioinformatics
analysis, the downstream target genes of miR-375 were predicted,
which predicted mediation of Notch2, a key protein in the Notch
signaling pathway.
Notch signaling is an evolutionarily conserved
pathway that controls cell fate in metazoans through local
cell-cell interactions. In canonical Notch signaling, transmembrane
receptors (Notch1–4) bind with transmembrane ligands [Jagged1,
Jagged2, Delta-like 1 (Dll1), Dll3 and Dll4] through their
extracellular domains on adjacent cells, initiating proteolysis of
the receptors and subsequent release of the signal-transducing
Notch intracellular domain (NICD). NICD subsequently translocates
to the nucleus and associates with the nuclear proteins of the
recombination signal binding protein for the immunoglobulin kappa J
region (RBP-Jk) family [also known as CSL or CBF1/Su (H)/Lag-1] to
assemble a transcription complex, which activates the expression of
the target genes of Notch signaling, such as the HES and
homocysteine-induced endoplasmic reticulum protein families
(17–19). Notch signaling has an essential
role in cardiac cell differentiation (20,21). We hypothesized that miR-375 may
influence heart development through the Notch signaling pathway,
however, the detailed mechanism requires elucidation. The present
study explored the role of miR-375 in cardiogenesis in
vitro, which may occur via the Notch signaling pathway.
Materials and methods
P19 cell culture and induction of
differentiation
P19 cells were obtained from the American Type
Culture Collection (Manassas, VA, USA) and cultivated as aggregates
for 4 days in Minimum Essential Medium α-modification (α-MEM)
containing 10% fetal bovine serum (FBS) (Gibco-BRL; Thermo Fisher
Scientific, Grand Island, NY, USA) and 1% dimethyl sulfoxide (DMSO;
Sigma-Aldrich, St. Louis, MO, USA) in bacteriological dishes in 5%
CO2 at 37°C. After 4 days of aggregation, the cell
clusters were transferred to culture flasks in α-MEM with 10% FBS.
The medium was replaced every 2 days. Cells were harvested at
differentiation days 0, 4, 6 and 10. The cell morphological changes
were observed and images were captured using an inverted microscope
(Nikon, Tochigi, Japan).
Establishment of miR-375-overexpressing
P19 cell lines
Plasmids overexpressing miR-375 and negative control
vectors contained enhancing green fluorescence protein (GFP) and
were constructed by GenePharma (Shanghai, China). Briefly, partial
primary transcript sequences for the mouse miR-375 genes were
amplified from embryonic telencephalon cDNA and cloned into
pcDNA™6.2-GW/EmGFPmiR (Promega, Madison, WI, USA). miRNA
transfection was performed with Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's protocol.
Fluorescence microscopy was used to observe the transfection
efficiency via GFP expression of the miR-375 expression vector and
control vector in P19 cells. miR-375 expression was verified using
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR). All the data were normalized to the internal standard
(U6). Sequences for RT-qPCR primer pairs are listed as follows:
mmu-miR-375 forward, AGCCGTTTGTTCGTT CGGCT and reverse primer,
GTGCAGGGTCCGAGGT; and U6 forward, CGCTTCGGCAGCACATATAC and reverse
primer, TTCACGAATTTGCGTGTCAT.
Cell proliferation assays
Cell counting kit-8 (CCK-8) assay
To analyze the proliferation of the stable cell
lines bearing miR-375 expression plasmids or vector, 2,000
cells/well in 100 µl medium were seeded in 96-well plates
subsequent to obtaining total cell counts using a hemocytometer,
and were cultured in α-MEM supplemented with 10% FBS at 37°C with
5% CO2. Cell viability was monitored at 0, 24, 36, 48
and 72 h. At each time-point, each well was treated with 10 µl
CCK-8 (Dojindo, Kumamoto, Japan) solution, and the optical density
was measured 2 h later with a microplate reader at 450 and 650
nm.
Cell cycle assay
Flow cytometry was used to evaluate the distribution
of cells in different phases of the cell cycle following
transfection, basing the evaluation on the DNA content of propidium
iodide (PI)-stained nuclei (22).
The cells were synchronized by culturing with serum-free α-MEM for
24 h. Subsequently, all the cells were digested with EDTA-free
trypsin (Invitrogen), washed in phosphate-buffered saline (PBS),
and centrifuged at 1,000 rpm for 5 min. The supernatant was
discarded and the pellets were fixed overnight at 4°C in cold 70%
ethanol. Following this, the cells were washed with PBS and
incubated with 100 mg/ml RNase A (Sigma-Aldrich) at 37°C for 1 h,
and incubated at 4°C in the dark for 30 min with 100 µg/ml PI
(Sigma-Aldrich). Disposed cells were analyzed using a BD FACScan
system and CellQuest software (BD Biosciences, San Jose, CA,
USA).
Analysis of apoptosis
Hoechst staining
The apoptosis rate in the transfected cells was
first evaluated by Hoechst staining. Briefly, cells were seeded on
sterile cover glasses in 6-well plates the day before treatment.
Subsequently, the cells were fixed, washed twice with PBS, and
stained with Hoechst 33258 (apoptosis Hoechst staining kit;
Beyotime Institute of Biotechnology, Jiangsu, China) according to
the manufacturer's protocol. The stained cells were examined and
immediately images were captured under a fluorescence microscope
(Nikon).
Flow cytometry
Cells were cultured in FBS-free α-MEM (serum-free)
for 24 h to induce apoptosis, and were harvested with EDTA-free
trypsin, washed in PBS, resuspended in 500 µl binding buffer, and
stained with 5 µl Annexin V-allophycocyanine (Annexin V-APC) and 5
µl 7-amino-actinomycin D (7-AAD) at room temperature for 10 min
(Annexin V-APC/7-AAD Apoptosis Detection kit; KeyGen Biotech,
Jiangsu, China). Disposed cells were analyzed using a BD FACScan
system and CellQuest software (BD Biosciences).
RT-qPCR
Total RNA was isolated from cultured P19 cells using
the TRIzol method (Invitrogen) with an miRNeasy mini kit (Qiagen,
Limburg, The Netherlands). Complementary DNA was synthesized from 1
µg total RNA using an AMV reverse transcriptase kit (Promega).
RT-qPCR using the SYBR-Green method was performed using an ABI 7500
Sequence Detection System (Applied Biosystems, Foster City, CA,
USA) according to the manufacturer's protocols. The PCR conditions
involved a denaturation step (95°C for 10 min), and amplification
and quantification were repeated 40 times (95°C for 15 sec and 60°C
for 1 min, respectively). The relative gene expression levels were
quantified based on the threshold cycle (Ct) and normalized to the
reference gene glyceraldehyde-3-phosphate dehydrogenase
(GAPDH). Table I lists the
primer sequences used.
 | Table ISequences of the primer sets used in
the reverse transcription-quantitative polymerase chain
reaction. |
Table I
Sequences of the primer sets used in
the reverse transcription-quantitative polymerase chain
reaction.
| Gene | Forward primer
(5′-3′) | Reverse primer
(3′-5′) |
|---|
| cTnT |
GGAGTACGAGGAGGAACAGG |
GTCCACTCTCTCTCCATCGG |
| GATA4 |
CCAACTGCCAGACTACCAC |
GGACCAGGCTGTTCCAAGA |
| MEF2C |
CAGCACTGACATGGATAAGG |
CTGCCAGGTGGGATAAGAACG |
| NOTCH2 |
GGTCGCTGTTGTCATCATCC |
TGACACTTGCACGGAGAGAT |
| DLL1 |
CGATGAGTGTGCTAGCAACC |
GCAGTGGTCTTTCAGGTGTG |
| HES1 |
CAGTGCCTTTGAGAAGCAGG |
CAGATAACGGGCAACTTCGG |
| BAX |
CCAGCCCATGATGGTTCTGAT |
CCGGCGAATTGGAGATGAACT |
| BCL-2 |
CAGACATGCACCTACCCAGC |
GTCGCTACCGTCGTGACTTC |
| GAPDH |
CCAACTGCCAGACTACCAC |
GGACCAGGCTGTTCCAAGA |
Antibodies and western blotting
Anti-Notch2 (5732s), anti-Dll1 (2588s), anti-hes
family bHLH transcription factor 1 (Hes1; 11988s), and anti-GAPDH
antibodies were purchased from Cell Signaling Technology (Danvers,
MA, USA). GATA binding protein 4 (GATA4; BS1747), myocyte enhancer
factor 2C (Mef2c; BS6401), and cardiac troponin T (cTnT/TNNT2;
BS6013) were purchased from Bioworld Technology Inc. (St. Louis
Park, MN, USA). The antibodies used were all specific monoclonal
antibodies. GAPDH was used as the internal reference. Lysis buffer
[1% Triton X-100, 50 mmol/l Tris-HCl, 0.2% sodium dodecyl sulfate,
0.2% sodium deoxycholate, 1 mmol/l EDTA (pH 7.4)] was directly
added to the cultured cells, which were subsequently transferred
into tubes and vortexed briefly. The supernatant was collected
after centrifugation at 15,200 × g for 15 min at 4°C. Protein
concentrations were detected using a Bicinchoninic Acid Protein
assay kit (Beyotime Institute of Biotechnology).
Luciferase reporter gene assay
The 3′ untranslated region (3′UTR) sequences of the
Notch2 gene containing the predicted miR-375 binding site or a
mutant seed sequence were termed W.T.Notch2–3′UTR and
M.T.Notch2–3′UTR, respectively. In the luciferase assays, P19 cells
cultured in 96-well plates were transiently transfected with either
the W.T.Notch2–3′UTR or M.T.Notch2–3′UTR vector, along with the
miR-375 overexpression plasmid using Lipofectamine 2000, according
to the manufacturer's protocol. A Dual-Luciferase Reporter assay
system (Promega) was used to analyze the luciferase activity after
48-h transfection.
Statistical analysis
Each experiment was performed at least three times.
All the results are presented as the mean ± standard deviation.
Data were analyzed using t- or t′-test with correction for multiple
comparisons as appropriate. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-375 expression in P19 cells
Plasmids overexpressing miR-375 and the negative
vectors were transfected into P19 cells that were 70–80% confluent.
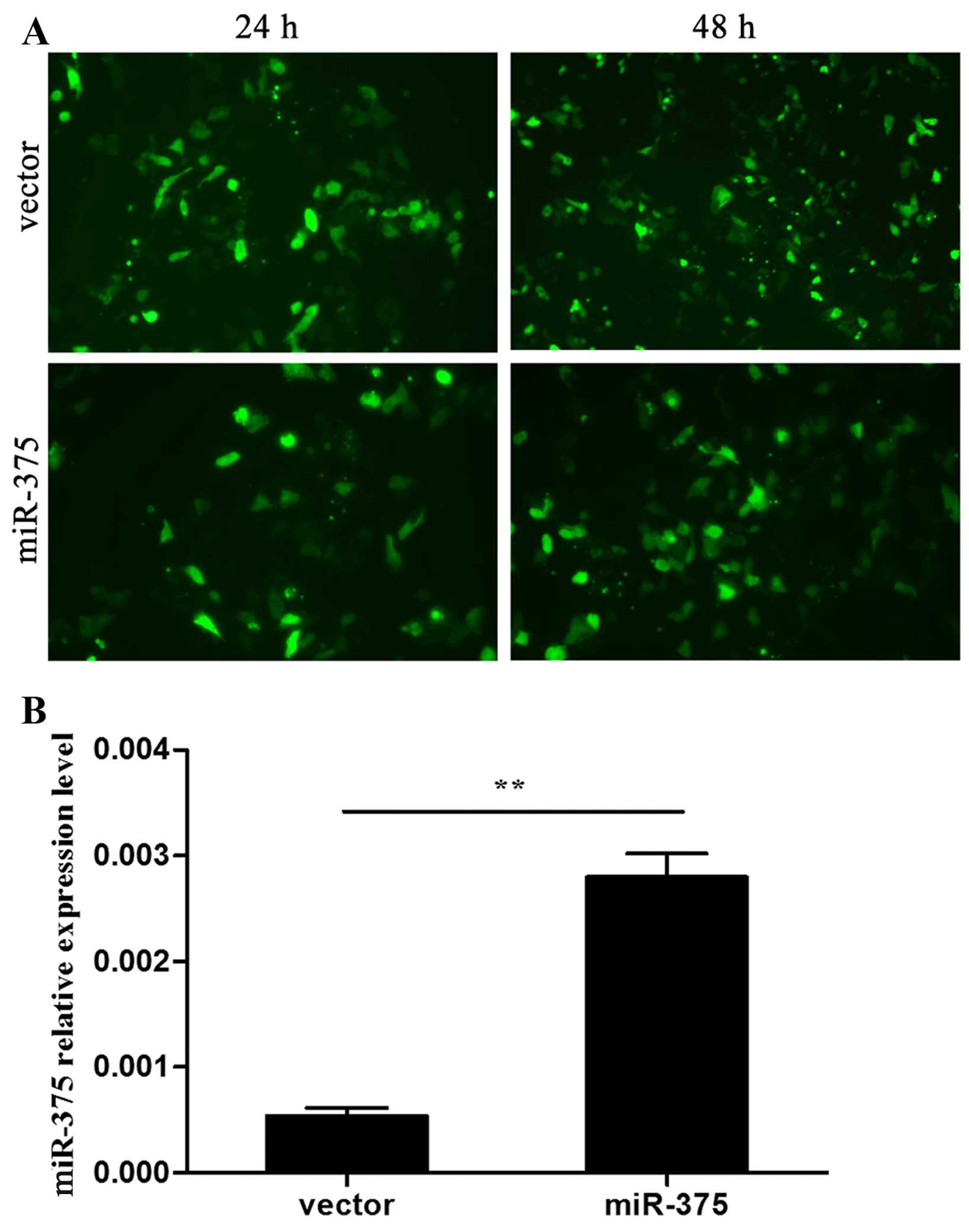
Transfection efficiency was observed after 24 and 48 h via the
expression of GFP under a fluorescence microscope (Fig. 1A). miR-375 expression in the two
groups was confirmed by RT-qPCR (P<0.01) (Fig. 1B). At day 7 during
differentiation, spontaneous contraction of the myocardium-like
cell clusters was observed occasionally. Cluster contraction
increased over the subsequent days, and peaked at around day 12
(data not shown).
miR-375 overexpression inhibits cell
proliferation
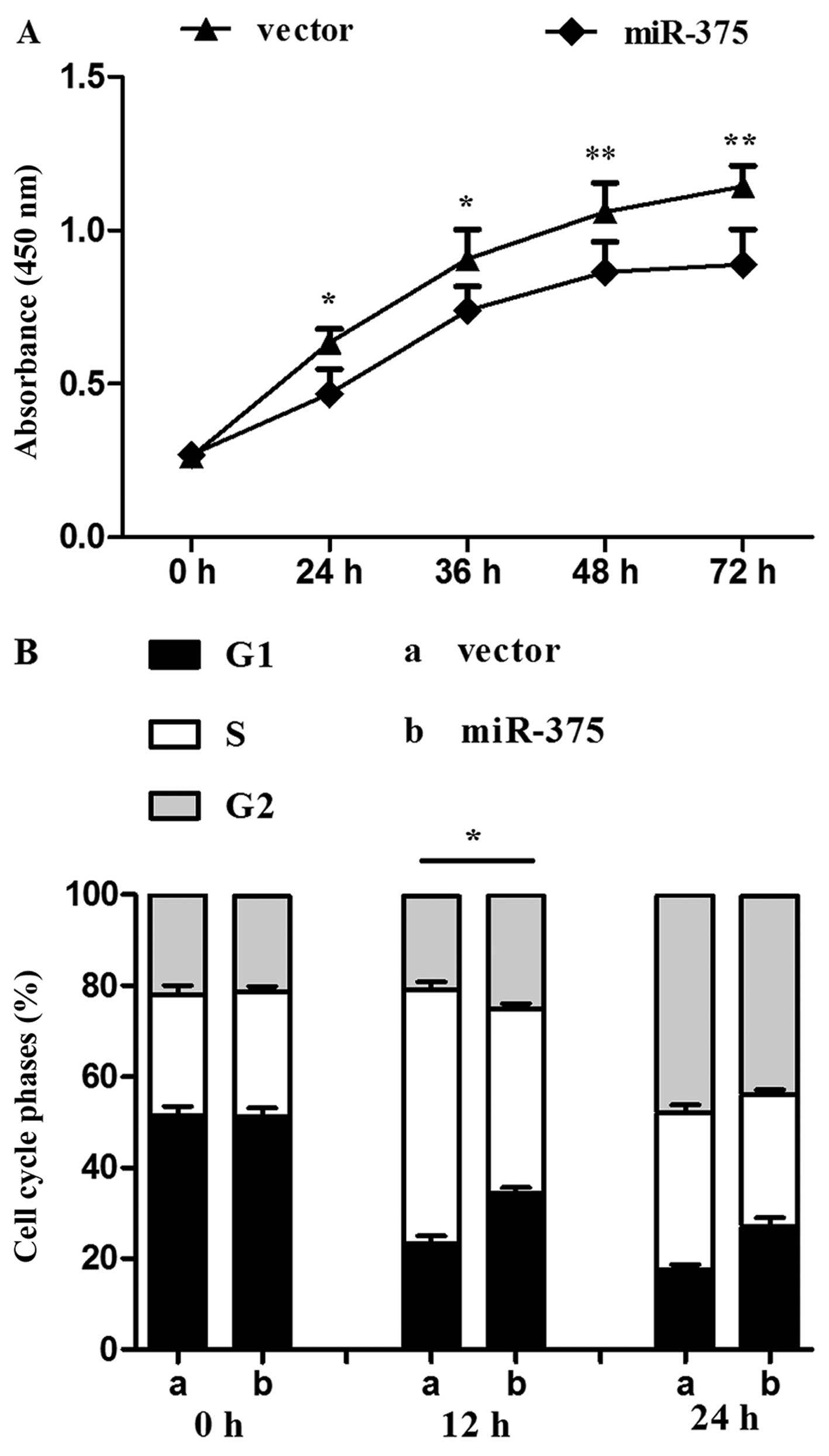
The proliferation rate of P19 cells overexpressing
miR-375 was evaluated using the CCK-8 assay. Compared with the
control group, continuous 72-h monitoring determined that
proliferation was reduced in the miR-375 overexpression group
(P<0.05 and P<0.01) (Fig.
2A). miR-375 also influenced the cell cycle: Flow cytometry
revealed a decreased percentage of S phase cells in the miR-375
overexpression group in contrast to the control group (P<0.05)
(Fig. 2B).
miR-375 overexpression promotes
apoptosis
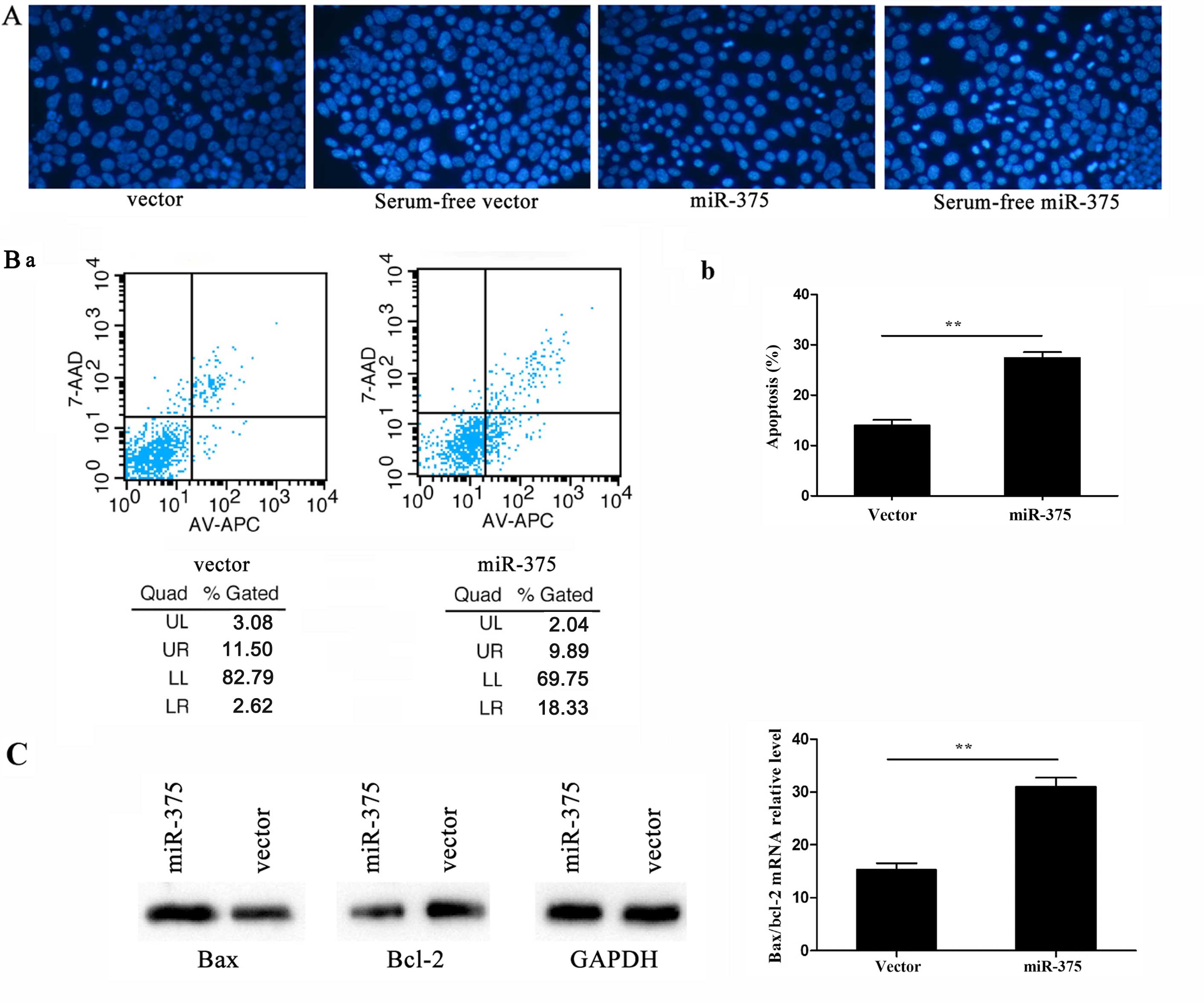
Hoechst staining is a classic, quick and easy method
for distinguishing apoptotic cells from normal cells. As the
chromatin condenses during apoptosis, Hoechst staining would have
rendered it clearly visible in the nuclei of cells overexpressing
miR-375, where it was visible as condensed, brighter spots under
fluorescence microscopy, whereas normal cells appeared more
homogenously stained (Fig. 3A).
By contrast, flow cytometry assessment of apoptosis revealed higher
apoptotic rates in cells overexpressing miR-375 compared to the
controls (P<0.01; UL, upper left; UR, upper right; LL, lower
left; LR, lower right. The apoptotic rate was calculated by UR+LR
(Fig. 3B-a). In addition, RT-qPCR
detected the apoptosis-related genes Bax and B-cell chronic
lymphocytic leukemia/lymphoma 2 (Bcl-2) following 24-h
culture in serum-free α-MEM for inducing apoptosis. By calculating
the Bax/Bcl-2 ratio, the apoptosis rate was observed to be
increased following miR-375 overexpression; the Bax and Bcl-2
protein levels were consistent with their mRNA expression levels
(P<0.01) (Fig. 3C).
Effect of miR-375 on P19 cell
differentiation
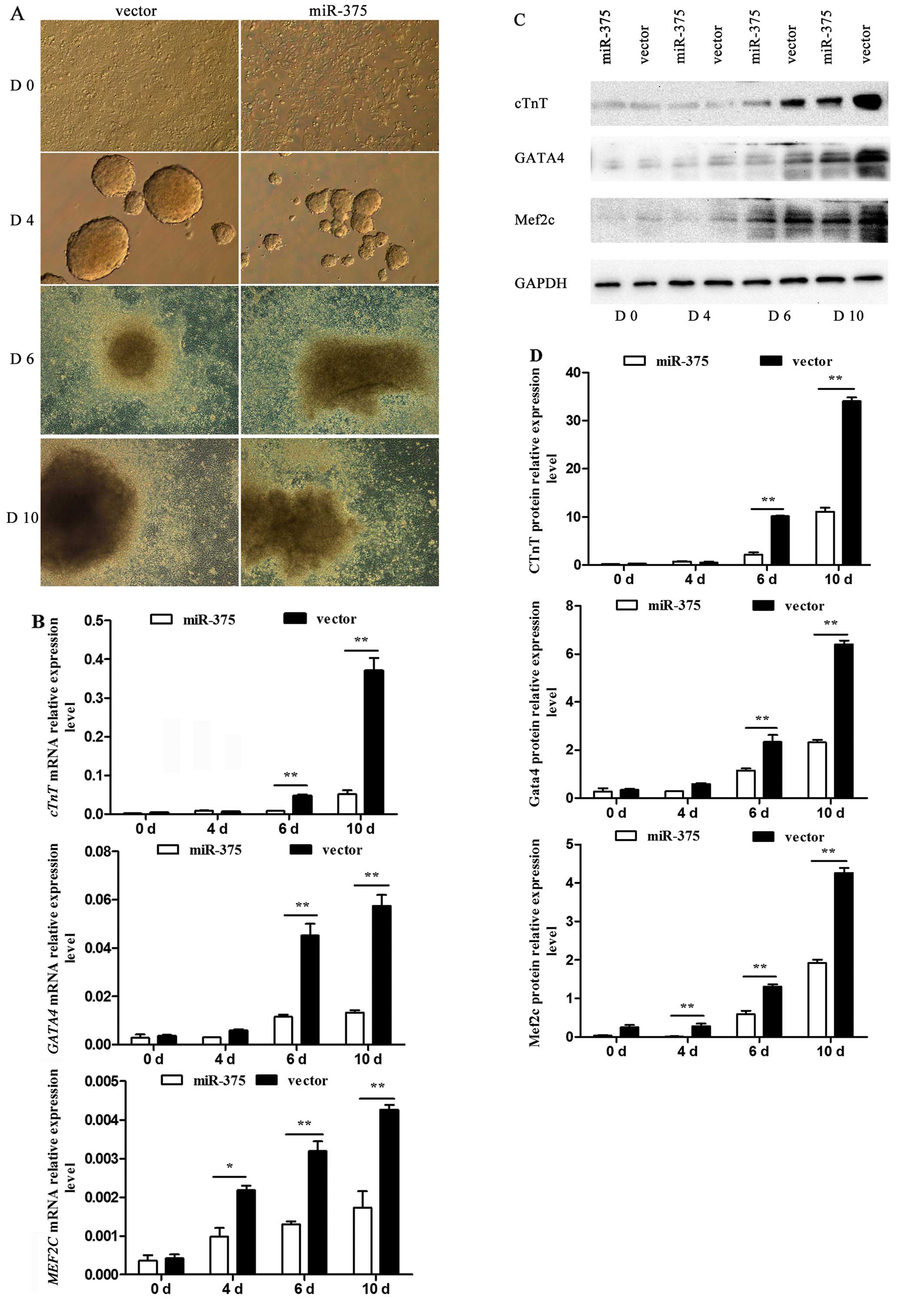
The process by which P19 cells were induced into
cardiomyocytes was observed using an inverted microscope. P19 cells
underwent a series of morphological changes (Fig. 4A). At day 4, the generated cell
bodies in the control group were larger and rounder than those in
the miR-375 overexpression group. Similarly, at day 10, the
condition of cells in the control group was improved compared to
that in the miR-375 overexpression group. At days 0, 4, 6 and 10 of
differentiation, the differential mRNA expression of the
myocardial-specific marker genes cTnT, GATA4 and
MEF2C was determined using RT-qPCR. The corresponding mRNA
levels were barely detectable in the undifferentiated P19 cells,
but their expression was rapidly upregulated following DMSO
induction. However, there was a negative effect on P19 cell
differentiation into cardiomyocytes in the miR-375 overexpression
group compared to the control group (P<0.05 and P<0.01)
(Fig. 4B). The relative protein
levels detected by western blotting verified the trend (P<0.05
and P<0.01) (Fig. 4C and
D).
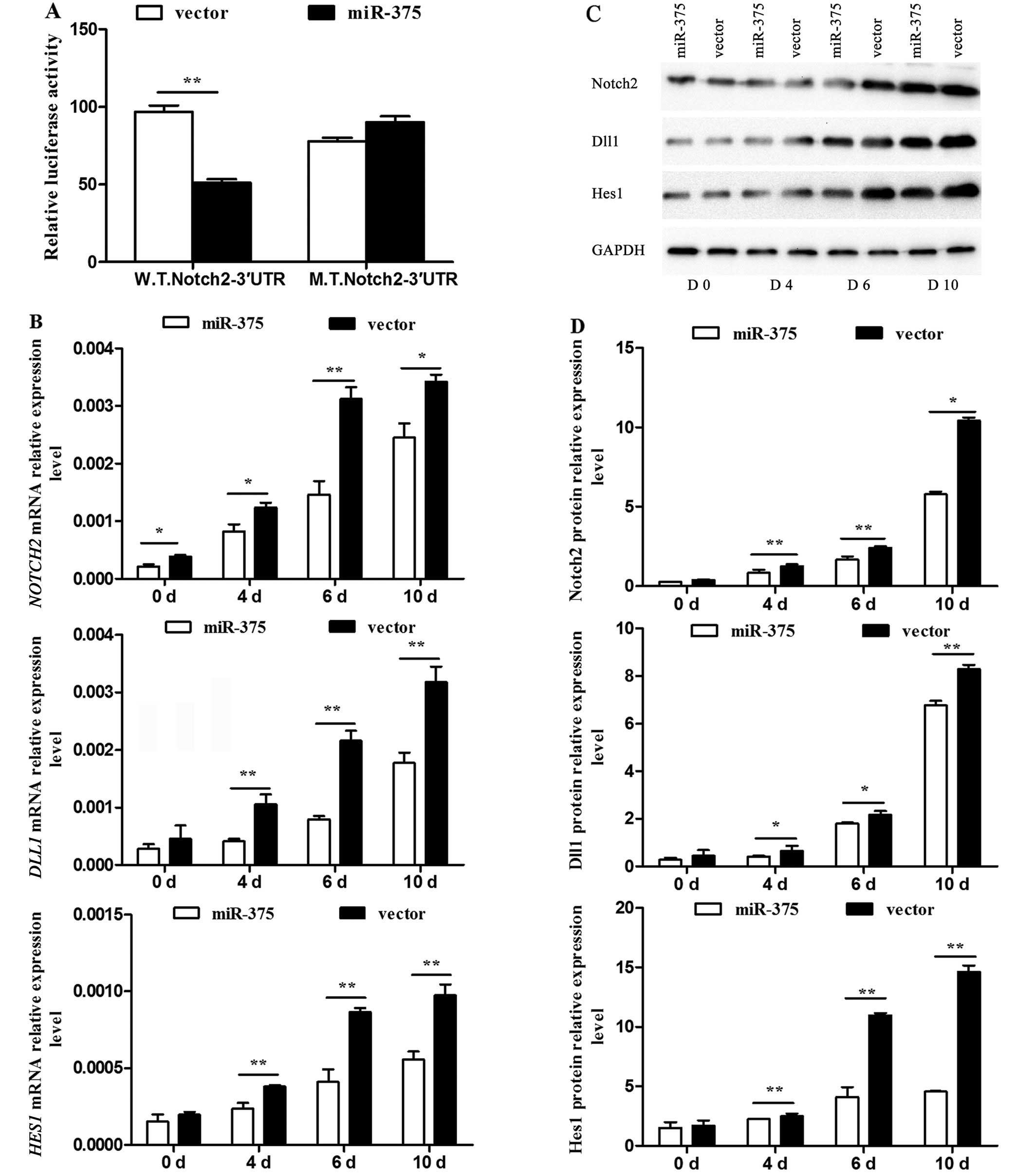
Effect of miR-375 on the Notch signaling
pathway
As NOTCH2 was a predicted downstream target
gene of miR-375, further verification and discussion was necessary.
The 3′UTR of NOTCH2 was confirmed as a target of miR-375. As
expected, miR-375 inhibited the luciferase activity of the
W.T.Notch2–3′UTR reporter efficiently, but not that of the
M.T.Notch2–3′UTR (P<0.01) (Fig.
5A). The mRNA expression levels of the pivotal Notch signaling
pathway regulators NOTCH2, DLL1 and HES1 were
detected by RT-qPCR, and were significantly decreased in the
miR-375 overexpression group at the examined time-points compared
to that of the control group (P<0.05 and P<0.01) (Fig. 5B). The associated protein levels
were detected using western blotting (Fig. 5C) and their relative levels were
calculated (P<0.05 and P<0.01) (Fig. 5D). All data indicated an
inhibitory effect of miR-375 on the Notch signaling pathway.
Discussion
The present study observed that miR-375 promoted
apoptosis and inhibited cell proliferation and differentiation.
Notch2 was also confirmed as a target gene of miR-375, and miR-375
overexpression significantly influenced other vital members of the
Notch signaling pathway. As miR-375 has been identified from the
ventricular septal myocardial tissues from fetuses with ventricular
septal defects, the present findings may shed light on novel
therapeutic methods for CHD.
As the number of adults who have survived CHD is
gradually increasing, CHD is currently becoming a significant
health issue worldwide. Despite various treatments, the high
incidence and serious resultant complications render CHD one of the
leading causes of morbidity and mortality (23,24). Therefore, determining its
pathogenesis and devising useful therapies is urgent. In recent
years, it was determined that miRNAs are responsible for CHD
(25), providing insight into how
they may be exploited to understand and study the mechanism of this
disease, as well as identifying them as promising candidates for
potential novel treatment strategies.
miR-375 regulates insulin secretion, pancreatic
islet development and alveolar epithelial cell transdifferentiation
(26–28). Our previous study determined that
miR-375 is abundantly expressed in the developing heart with
consistent fold changes (29). In
addition, miR-375 is significantly upregulated in maternal serum at
18–22 weeks of gestation with fetal CHD (15). These results demonstrate that
miR-375 is associated with heart development and is involved in the
occurrence of CHD. However, the specific mechanism remains
unknown.
The present results show that miR-375 overexpression
promotes apoptosis and inhibits cell proliferation and
differentiation. The CCK-8 assay and decreased percentage of S
phase cells confirmed the negative effect of miR-375 on cell
proliferation. Simultaneously, Hoechst staining illustrating clear
morphological diversity accompanied by flow cytometry was used to
assess the apoptosis rate, and indicated the promoting effect of
miR-375 on apoptosis. In contrast to necrocytosis, apoptosis is a
process that involves cellular gene expression. Subsequently, the
apoptosis-related genes BAX and BCL-2 were examined,
and the Bax/Bcl-2 ratio in the miR-375 overexpression group was
increased, which did not contradict our previous findings. Bax and
Bcl-2 are members of the Bcl-2 family, which is a pivotal regulator
of apoptosis (30). The Bcl-2
family contains pro-apoptotic (Bax, Bak, Bad and Bcl-xS) and
anti-apoptotic (Bcl-2 and Bcl-xL) members. It is believed that the
BCL-2 gene product inhibits cell death and contributes to
the prolongation of cell survival, while Bax, an important
homologue of Bcl-2, is a promoter of apoptosis (31,32). Evidently, the Bax/Bcl-2 ratio has
a decisive role in cell fate. As aforementioned, the increased
Bax/Bcl-2 ratio and relative protein levels in the present study
demonstrate that miR-375 overexpression promotes apoptosis. Cell
proliferation and apoptosis are basic features of living organisms;
homeostasis of the two is an important biological activity for
maintaining structural stability and environmental functional
balance in multicellular organisms. Therefore, the inhibition of
proliferation and promotion of apoptosis in miR-375 overexpression
P19 cells shows that miR-375 influences P19 cell differentiation
into cardiomyocytes and has a critical role during this
process.
In addition, the effects of miR-375 were examined on
P19 cell differentiation into cardiomyocytes. Murine P19 embryonal
carcinoma cells, which can be induced by DMSO to differentiate into
spontaneously beating cardiomyocytes in vitro (33,34), are the classic model for
investigating heart development and are a valuable means of
studying cardiomyocyte differentiation. Over the years, several
influencing factors associated with cardiac disease have been
explored (35–37), which is an essential part of the
present study. The RT-qPCR and western blotting data also
demonstrate the reliability of this cell model. The relative
expression levels of cTnT, GATA4 and Mef2c mRNA and protein
exhibited an increase during differentiation.
Evolutionarily, the embryonic heart undergoes a
series of complicated morphogenetic and differentiation processes
prior to forming the mature cardiac structures during development.
Notably, these processes are controlled by a conserved network of
genes and signaling pathways (38–40), including the Notch signaling
pathway. During mammalian cardiogenesis, Notch signaling is
involved in the development of the aortic valve, ventricles,
atrioventricular canal and outflow tract (41). Mutations in the Notch signaling
components affect heart development and are eventually involved in
several types of CHD (42).
Jagged1 and Notch2 are involved in Alagille syndrome, an autosomal
dominant genetic disorder that results in pulmonary artery stenosis
and tetralogy of Fallot (43,44). Notch1 is associated with aortic
valve disease (45). Notably
Notch signaling facilitates repair following myocardial injury by
promoting myocardial regeneration, protecting the myocardium from
ischemia, inducing angiogenesis, and inhibiting the transformation
of cardiac fibroblasts to myofibroblasts (46). The present study used luciferase
reporter gene assays to verify the downstream target genes
predicted by bioinformatics analysis, and identified that miR-375
combined with the 3′UTR of NOTCH2, a key gene in the Notch
signaling pathway. Furthermore, NOTCH2, DLL1 and
HES1 mRNA levels were increased significantly during P19
cell differentiation in the control group, whereas this was not the
case in the miR-375 overexpression group; the same trend was
observed for the associated protein levels. The present study
demonstrates that the Notch signaling pathway is involved in P19
cell differentiation and that miR-375 overexpression leads to
abnormal expression of downstream Notch signaling pathway
genes.
In conclusion, miR-375 overexpression promotes cell
apoptosis and inhibits proliferation and differentiation through
the Notch signaling pathway, which may provide potential targets
for therapeutic intervention in CHD.
Acknowledgments
The present study was supported by grants from the
Jiangsu Province Natural Science Foundation of China (no.
BK20130076) and the Natural Science Foundation of China (no.
81501262).
References
|
1
|
Hoffman JI and Kaplan S: The incidence of
congenital heart disease. J Am Coll Cardiol. 39:1890–1900. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Srivastava D: Making or breaking the
heart: From lineage determination to morphogenesis. Cell.
126:1037–1048. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Somerville J: Grown-up congenital heart
disease - medical demands look back, look forward 2000. Thorac
Cardiovasc Surg. 49:21–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van der Linde D, Konings EE, Slager MA,
Witsenburg M, Helbing WA, Takkenberg JJ and Roos-Hesselink JW:
Birth prevalence of congenital heart disease worldwide: A
systematic review and meta-analysis. J Am Coll Cardiol.
58:2241–2247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garside VC, Chang AC, Karsan A and
Hoodless PA: Co-ordinating Notch, BMP, and TGF-β signaling during
heart valve development. Cell Mol Life Sci. 70:2899–2917. 2013.
View Article : Google Scholar
|
|
6
|
High FA and Epstein JA: The multifaceted
role of Notch in cardiac development and disease. Nat Rev Genet.
9:49–61. 2008. View
Article : Google Scholar
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gangaraju VK and Lin H: MicroRNAs: Key
regulators of stem cells. Nat Rev Mol Cell Biol. 10:116–125. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Feng Y and Yu X: Cardinal roles of miRNA
in cardiac development and disease. Sci China Life Sci.
54:1113–1120. 2011. View Article : Google Scholar
|
|
12
|
Thum T, Catalucci D and Bauersachs J:
MicroRNAs: Novel regulators in cardiac development and disease.
Cardiovasc Res. 79:562–570. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kasinski AL and Slack FJ: Epigenetics and
genetics MicroRNAs en route to the clinic: Progress in validating
and targeting microRNAs for cancer therapy. Nat Rev Cancer.
11:849–864. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Callegari E, Gramantieri L, Domenicali M,
D'Abundo L, Sabbioni S and Negrini M: MicroRNAs in liver cancer: A
model for investigating pathogenesis and novel therapeutic
approaches. Cell Death Differ. 22:46–57. 2015. View Article : Google Scholar
|
|
15
|
Zhu S, Cao L, Zhu J, Kong L, Jin J, Qian
L, Zhu C, Hu X, Li M and Guo X: et al Identification of maternal
serum microRNAs as novel non-invasive biomarkers for prenatal
detection of fetal congenital heart defects. Clin Chim Acta.
424:66–72. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu Z, Han S, Hu P, Zhu C, Wang X, Qian L
and Guo X: Potential role of maternal serum microRNAs as a
biomarker for fetal congenital heart defects. Med Hypotheses.
76:424–426. 2011. View Article : Google Scholar
|
|
17
|
Andersson ER, Sandberg R and Lendahl U:
Notch signaling: Simplicity in design, versatility in function.
Development. 138:3593–3612. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kopan R and Ilagan MXG: The canonical
Notch signaling pathway: Unfolding the activation mechanism. Cell.
137:216–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iso T, Kedes L and Hamamori Y: HES and
HERP families: Multiple effectors of the Notch signaling pathway. J
Cell Physiol. 194:237–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang ACY, Fu Y, Garside VC, Niessen K,
Chang L, Fuller M, Setiadi A, Smrz J, Kyle A and Minchinton A: et
al Notch initiates the endothelial-to-mesenchymal transition in the
atrioventricular canal through autocrine activation of soluble
guanylyl cyclase. Dev Cell. 21:288–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chau MDL, Tuft R, Fogarty K and Bao ZZ:
Notch signaling plays a key role in cardiac cell differentiation.
Mech Dev. 123:626–640. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bishayee K, Ghosh S, Mukherjee A,
Sadhukhan R, Mondal J and Khuda-Bukhsh AR: Quercetin induces
cytochrome-c release and ROS accumulation to promote apoptosis and
arrest the cell cycle in G2/M, in cervical carcinoma: Signal
cascade and drug-DNA interaction. Cell Prolif. 46:153–163. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Verheugt CL, Uiterwaal CS, Grobbee DE and
Mulder BJ: Long-term prognosis of congenital heart defects: A
systematic review. Int J Cardiol. 131:25–32. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zomer AC, Vaartjes I, van der Velde ET, de
Jong HM, Konings TC, Wagenaar LJ, Heesen WF, Eerens F, Baur LH and
Grobbee DE: et al Heart failure admissions in adults with
congenital heart disease; risk factors and prognosis. Int J
Cardiol. 168:2487–2493. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sayed AS, Xia K, Salma U, Yang T and Peng
J: Diagnosis, prognosis and therapeutic role of circulating miRNAs
in cardiovascular diseases. Heart Lung Circ. 23:503–510. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
El Ouaamari A, Baroukh N, Martens GA,
Lebrun P, Pipeleers D and van Obberghen E: miR-375 targets
3′-phosphoinositide-dependent protein kinase-1 and regulates
glucose-induced biological responses in pancreatic beta-cells.
Diabetes. 57:2708–2717. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kloosterman WP, Lagendijk AK, Ketting RF,
Moulton JD and Plasterk RH: Targeted inhibition of miRNA maturation
with morpholinos reveals a role for miR-375 in pancreatic islet
development. PLoS Biol. 5:e2032007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang H, Mishra A, Chintagari NR, Gou D
and Liu L: Micro-RNA-375 inhibits lung surfactant secretion by
altering cytoskeleton reorganization. IUBMB Life. 62:78–83.
2010.
|
|
29
|
Cao L, Kong LP, Yu ZB, Han SP, Bai YF, Zhu
J, Hu X, Zhu C, Zhu S and Guo XR: microRNA expression profiling of
the developing mouse heart. Int J Mol Med. 30:1095–1104.
2012.PubMed/NCBI
|
|
30
|
Adams JM and Cory S: The Bcl-2 protein
family: Arbiters of cell survival. Science. 281:1322–1326. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Golestani Eimani B, Sanati MH, Houshmand
M, Ataei M, Akbarian F and Shakhssalim N: Expression and prognostic
significance of bcl-2 and bax in the progression and clinical
outcome of transitional bladder cell carcinoma. Cell J. 15:356–363.
2014.PubMed/NCBI
|
|
32
|
Oltvai ZN, Milliman CL and Korsmeyer SJ:
Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that
accelerates programmed cell death. Cell. 74:609–619. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
van der Heyden MA and Defize LH: Twenty
one years of P19 cells: What an embryonal carcinoma cell line
taught us about cardiomyocyte differentiation. Cardiovasc Res.
58:292–302. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
van der Heyden MA, van Kempen MJ, Tsuji Y,
Rook MB, Jongsma HJ and Opthof T: P19 embryonal carcinoma cells: A
suitable model system for cardiac electrophysiological
differentiation at the molecular and functional level. Cardiovasc
Res. 58:410–422. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qin DN, Qian L, Hu DL, Yu ZB, Han SP, Zhu
C, Wang X and Hu X: Effects of miR-19b overexpression on
proliferation, differentiation, apoptosis and Wnt/β-catenin
signaling pathway in P19 cell model of cardiac differentiation in
vitro. Cell Biochem Biophys. 66:709–722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhu C, Hu DL, Liu YQ, Zhang QJ, Chen FK,
Kong XQ, Cao KJ, Zhang JS and Qian LM: Fabp3 inhibits proliferation
and promotes apoptosis of embryonic myocardial cells. Cell Biochem
Biophys. 60:259–266. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu S, Hu X, Han S, Yu Z, Peng Y, Zhu J,
Liu X, Qian L, Zhu C and Li M: et al Differential expression
profile of long non-coding RNAs during differentiation of
cardiomyocytes. Int J Med Sci. 11:500–507. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yuan S, Zaidi S and Brueckner M:
Congenital heart disease: Emerging themes linking genetics and
development. Curr Opin Genet Dev. 23:352–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Staudt D and Stainier D: Uncovering the
molecular and cellular mechanisms of heart development using the
zebrafish. Annu Rev Genet. 46:397–418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Olson EN: Gene regulatory networks in the
evolution and development of the heart. Science. 313:1922–1927.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Penton AL, Leonard LD and Spinner NB:
Notch signaling in human development and disease. Semin Cell Dev
Biol. 23:450–457. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
de la Pompa JL: Notch signaling in cardiac
development and disease. Pediatr Cardiol. 30:643–650. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
McDaniell R, Warthen DM, Sanchez-Lara PA,
Pai A, Krantz ID, Piccoli DA and Spinner NB: NOTCH2 mutations cause
Alagille syndrome, a heterogeneous disorder of the notch signaling
pathway. Am J Hum Genet. 79:169–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kamath BM, Spinner NB, Emerick KM, Chudley
AE, Booth C, Piccoli DA and Krantz ID: Vascular anomalies in
Alagille syndrome: A significant cause of morbidity and mortality.
Circulation. 109:1354–1358. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Garg V, Muth AN, Ransom JF, Schluterman
MK, Barnes R, King IN, Grossfeld PD and Srivastava D: Mutations in
NOTCH1 cause aortic valve disease. Nature. 437:270–274. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou XL and Liu JC: Role of Notch
signaling in the mammalian heart. Braz J Med Biol Res. 47:1–10.
2014. View Article : Google Scholar :
|