Introduction
Acute lymphocytic leukemia (ALL) is one of the most
common malignant tumors and has the highest morbidity rates among
children, accounting for ~80% of leukemia cases. The incidence rate
of ALL is 5-fold higher than that of acute myeloid leukemia (AML).
The development of medical technology, has led to improvement in
the treatment of ALL. However, 20–30% of children with leukemia
suffer ALL relapse and subsequently have a poor prognosis (1–3).
Clinical studies have shown that the relapse of AML
after treatment is strongly associated with the expression of
homeobox (HOX) genes, whose main role is to control the
proliferation and differentiation of hematopoietic stem and
progenitor cells (4). It has also
been shown that even the development of various types of acute
leukemia such as acute myeloid leukemia, is associated with HOX
gene expression (5,6). HOX genes are divided into four
clusters according to the similarity and chromosomal location of
the human HOX gene sequence. These clusters are HOXA, HOXB, HOXC
and HOXD, which are located on chromosome number VII, XVII, XII and
II, respectively. Each of the HOX contains 9–11 genes. HOXA5
belongs to type 1 of the HOXA gene and is located on
chromosome VII (7p15.2). HOXA encodes a DNA-binding transcription
factor that regulates the expression of genes which control cell
differentiation (6). The abnormal
expression of HOX may affect cell differentiation and maturation in
hematopoietic disorders (6). It
may also decrease hematopoietic ability and result in the
occurrence and development of leukemia (6). Findings by Delval et al
(7) have shown that HOXA1
interacts with B-cell leukemia transcription factor through a HOX
polypeptide. The mutation of the conserved tryptophan and
methionine residues led to loss of its ability to stimulate cell
proliferation, anchorage-independent cell growth and loss of
contact inhibition (7). A study
by Okada et al (8) showed
that HOXA5 methylation plays an important role in leukemic
transformation, which is induced by the CALM-AF10 fusion protein
(8). Bach et al (9) found that the high expression of
HOXA5 may contribute to the occurrence and phenotype of
leukemia.
RNA interference (RNAi) is a type of simple and
effective genetic tool that has been developed in recent years and
is used instead of gene knockout (10,11). RNA interference (RNAi) is the
process of sequence-specific, post-transcriptional gene silencing
in the same direction, initiated by double-stranded RNA (10). RNAi technology is a type of
small-interfering RNA (siRNA) with 21–23 bp that is derived from
double-stranded DNA (dsRNA) by effect of RNase III endonuclease
Dicer (11). It is a highly
efficient gene-blocking technology that blocks the expression of
target genes by mediating specific degradation of complementary
homologous mRNA (12). In the
present study, HOXA5 gene expression in ALL was detected by
clinical tests, and the expression levels of HOXA5 mRNA and protein
were detected by quantitative fluorescent-polymerase chain reaction
(QF-PCR) and western blot analysis. Subsequently, through the
synthesis of HOXA5 targeting-specific siRNA, cationic liposome was
used to transfect Jurkat cells, a human acute T-cell leukemia cell
line. HOXA5-specific siRNA may inhibit the expression of
HOXA5 gene. We detected the expression of HOXA5 mRNA and
protein in Jurkat cells and investigated the effect of HOXA5
gene in cell cycle and apoptosis. In the present study, the results
showed that HOXA5 can be used as a target for gene therapy in
leukemia and provide a new treatment for acute lymphoblastic
leukemia.
Materials and methods
Cell lines and reagents
Human lymphocyte separation medium were obtained
from Tianjin TBD Co. (Tianjin, China), and Jurkat leukemic T cells
from human peripheral blood (Shanghai Institute of Cell Library,
Shanghai, China). RPMI-1640 and fetal bovine serum (FBS) were
purchased from HyClone (Logan, UT, USA) and Lipofectamine™ 2000
from Invitrogen-Life Technologies (Carlsbad, CA, USA). G418 and
CCK-8 were obtained from Beyotime Institute of Biotechnology
(Shanghai, China) and DMSO from Sigma (St. Louis, MO, USA). Annexin
V-PE/7AAD and the cell apoptosis detection kit were obtained from
Nanjing KeyGen Biotech (Nanjing, China) and the TRIzol reagent from
Invitrogen-Life Technologies. The RNA extraction reagent was
purchased from BioFlux, Hangzhou, China. The QF-PCR kit, HOXA5,
glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primer,
restriction endonuclease BamHI, T4 DNA ligase, and gel
purification kit were purchased from Takara (Shiga, Japan). siRNA
sequence targeting HOXA5, and the negative control siRNA sequence
were purchased from Adicon Co. (Shanghai, China). Rabbit anti-human
HOXA5 polyclonal antibodies were purchased from Abcam (Cambridge,
UK) and horseradish peroxidase-labeled goat anti-rabbit secondary
antibodies were purchased from the Beyotime Institute of
Biotechnology. Other reagents were developed and purified in
China.
Cases
Fifty children newly diagnosed with ALL were
enrolled in the study between October 2013 and June 2015.
The patients were divided into three groups: i) the
acute phase group included 25 newly diagnosed cases of ALL. The ALL
cases were confirmed by morphological analysis of bone marrow cells
and MICM typing, had not previously received any treatment and
excluded other neoplastic diseases, such as multiple myeloma tumor
and malignant lymphoma, according to the basic criteria for the
diagnosis of ALL. ii) The ALL remission group comprised 25 cases.
In this group, ALL remission induction therapy administered
achieved complete remission as per CR standards with efficacy
standards of ALL. iii) The control group comprised 20 cases.
Selected bone marrow samples were obtained from children with
immune thrombocytopenia (ITP) (13). The samples were collected with the
consent of the children's parents.
The ALL acute phase group comprised 13 male and 12
female children with a median age of 6.6 years (10 months to 14
years). In the ALL remission group, there were 15 male and 10
female children with a median age of 6.3 years (10 months to 14
years). The control group included 9 male and 11 female children
with a median age of 6.7 years (10 months to 14 years).
Isolation of mononuclear cells from bone
marrow
Prior to diagnosis with ALL, routine biopsy was
performed to obtain bone marrow samples of ~2 ml from each child.
Subsequently, lymphocyte bone marrow mononuclear cell samples were
isolated. Bone marrow cells (2 ml) were diluted by adding an equal
volume of saline solution. Human lymphocyte separation medium (4
ml) was added to the centrifuge tube. Diluted bone marrow fluid was
gently and gradually layered along the wall until it adhered to the
lymphocyte separation medium, and then centrifuged at 599.4 × g for
25 min. The intermediate buffy coat layer was then collected and
placed into a new tube. Four volumes of saline were added, followed
by centrifugation at 599.4 × g for 20 min. The cells were washed
twice with RPMI-1640, which was purchased from Hyclone (Logan, UT,
USA), prior to discarding the supernatant. The cell pellet was then
washed with 10% FBS RPMI-1640, after the dispersion count. The
cells were seeded in a culture flask (Hyclone) at a concentration
of 3×107/ml, and placed in a cell incubator at a
temperature of 37°C, carbon dioxide (CO2) concentrations
of 5 and 30% moisture saturation. The medium was changed after 2–3
days and passaged once.
Detection of HOXA5 mRNA expression levels
in mononuclear cells using QF-PCR
RNA was extracted from the mono nuclear cells of the
bone marrow. The absorbance of the samples was determined by the UV
spectrophotometer A ratio (A260/A280), at a range of 1.8–2.2, by
identification of 1% agarose gel electrophoresis. Amplification of
HOXA5 and GAPDH genes was performed by QF-PCR.
HOXA5 gene was amplified using the primers: upstream,
5′-TTTTGCGGTCGCTATCC-3′, and downstream,
5′-CTGAGATCCATGCCATTGTAG-3′ and the amplified fragment length was
140 bp. For the GAPDH gene, the primers used were: upstream,
5′-ATGCTG GCGCTGAGTACGTC-3′ and downstream,
5′-GGTCATGAGTCCTTCCACGATA-3′ and the amplified fragment length was
262 bp. The conditions used to set up the PCR reaction were: 95°C
denaturation 30 sec, followed by 95°C denaturation for 5 sec, 58°C
annealing for 34 sec, for 40 cycles. The condition for drawing the
dissolution curve was 95°C denaturation for 15 min, 60°C annealing
for 60 sec, and 95°C denaturation for 15 sec. Data were analyzed
using the formula RQ = 2−ΔΔCt while 2−ΔΔCt
was used to represent the relative expression levels of mRNA HOXA5.
The gray-level ratio with HOXA5 and internal reference gene
GAPDH expressed was used to indicate the relative expression
of HOXA5 mRNA. The experiment was repeated three times.
Detection of HOXA5 protein expression in
bone marrow mononuclear cells using western blot analysis
Bone marrow mononuclear cells were washed with cold
phosphate-buffered saline (PBS) twice, cell lysis was performed and
the cell lysate was collected and stored at −80°C. The protein
concentration in the lysate was determined using the BCA method to
ensure that the same amount of protein was added in each reaction.
Subsequently, 5X SDS [sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE)] polyacrylamide gel electrophoresis
sample buffer (Abcam, Cambridge, UK) was added to this cell lysate
and boiled for 5 min. The proteins were then transferred to PVDF
membranes following SDS-PAGE. Tris-HCl buffer solution (TBS)
sealing liquid with 5% skim milk powder as well as 1 g/l Tween-20
were used to block the solution for 2 h on the table concentrator.
Subsequently, 1X TBST was used to fully rinse the solutions three
times for 5 min. HOXA5 polyclonal antibody and anti-HOXA5 antibody
were used at a dilution of 1:1,000 and incubated overnight at 4°C.
After fully rinsing the primary antibody the following day, goat
anti-rabbit secondary antibody was added at a dilution of 1:1,000.
Following incubation at room temperature for 1 h, the blot was
developed with ECL light developing film in the dark using the
Gel-Pro Analyzer software (Media Cybernetics, Rockville, MD, USA).
The relative ratio of target protein was determined using HOXA5
protein bands of gray value and GAPDH protein bands of gray value.
The gray-level ratio with HOXA5 and the internal reference gene
GAPDH, the relative expression quantity of HOXA5 protein
expressed in the experimental group was carried out. The experiment
was repeated three times.
Short hairpin RNA (shRNA) design,
screening and synthesis
For the Jurkat cell experiments, three specific
sequences of HOXA5 siRNA were chemically designed and synthesized.
To overcome the influence of siRNA, we designed a negative control
sequence (siRNA-NC), which had no homology with any of the human
genes. The three siRNA sequences targeting the HOXA5 gene
are shown in Table I. The
sequences contained restriction sites for the BamHI and
HindIII enzymes. siRNA was synthesized by Adicon Co.
(Jiangsu, China).
 | Table ISequences of the siRNA targeting
HOXA5 gene. |
Table I
Sequences of the siRNA targeting
HOXA5 gene.
| Group | HindIII | Sense | Loop | Antisense | Termination
signal | HindIII |
|---|
| siRNA insert A 1:75
bp | GGATCCCG |
TTATGGAGATCATAGTTCCGT | TTCAAGAGA |
ACGGAACTATGATCTCCATAA | TTTTTT | CCAAAAGCTT |
| siRNA insert B 1:75
bp | GGATCCCG |
TACGGCTACAATGGCATGGAT | TTCAAGAGA |
ATCCATGCCATTGTAGCCGTA | TTTTTT | CCAAAAGCTT |
| siRNA insert C 1:75
bp | GGATCCCG |
TTGCGGTCGCTATCCAAATGG | TTCAAGAGA |
CCATTTGGATAGCGACCGCAA | TTTTTT | CCAAAAGCTT |
Cell culture and transfection
Jurkat cells were cultured in RPMI-1640 medium at
37°C, with a 5% volume fraction of CO2 and 30% saturated
humidity. The medium was supplemented with 10% fetal calf serum,
penicillin and streptomycin at a concentration of 100 IU/ml. The
cells were grown in suspension, the medium was changed after 2–3
days, and the cells were passaged once. Experiments were conducted
with cells in the logarithmic growth phase. To perform
transfection, the cell concentration was adjusted to
3×107/ml in the RPMI-1640 medium with no serum and no
antibiotics. The cells were divided into group A, blank control
group (plus an equal amount of cells and culture media only); group
B, the negative control group (liposomal transfection with negative
control siRNA); and group C, the experimental group (liposomal
transfection with HOXA5 targeting siRNA). The siRNA concentration
for each transfection was 135 ng/μl according to the
Lipofectamine™ 2000 specification, mixed with serum-free medium
without antibiotics. The mixed liquid was transfected into Jurkat
cells. G418 (200 g/ml) was added to the screened cells after 24 h
of transfection. Monoclonal cells were selected after screening for
4 weeks and G418 (200 g/ml) medium was used to expand the culture.
The experiment was repeated three times.
Detection of HOXA5 mRNA expression levels
in Jurkat cells using QF-PCR
Jurkat cells in the logarithmic phase at
3×105 cells/well were transfected in the 6-well culture
plate as the control group. The experimental and negative control
groups were established using the early stably transfected cells
seeded in a 6-well plate. The cells in each group were seeded in 2
wells and total RNA was extracted after 24 h. The conditions used
to set up the PCR reaction and the calculation of the relative
quantity of gene expression were described earlier. The gray-level
ratio with HOXA5 and the internal reference gene GAPDH was
used to indicate the relative expression of HOXA5 mRNA, prior to
calculation of the experimental group HOXA5 mRNA inhibition rate.
The HOXA5 mRNA inhibition rate was calculated as: [1- experimental
group (HOXA5 mRNA relative expression level)/blank control group
(HOXA5 mRNA relative expression level)] ×100%. The experiment was
repeated three times.
Detection of HOXA5 protein expression
levels using western blot analysis
Jurkat cells in the logarithmic phase were
vaccinated in a 6-well culture plate. The cell groups, the density
of inoculation and the transfection steps were similar to those
described earlier. After 24 h of transfection, the cells were
washed with cold PBS twice, cell lysis was performed and the cell
lysate was collected and stored at −80°C. The conditions for the
PCR reaction were performed as described earlier. The gray-level
ratio with HOXA5 and the internal reference gene GAPDH were
determined, the relative expression quantity of HOXA5 protein was
expressed in the experimental group, and the calculation of HOXA5
protein inhibition rate was performed. The HOXA5 protein inhibition
rate was calculated as: [1- experimental group (HOXA5 protein
relative expression)/blank control group (HOXA5 protein relative
expression)] ×100%. The experiment was repeated three times.
Detection of cell cycle
The cells in each group containing 0.5% serum in
RPMI-1640 medium were cultured for 48 h after cell synchronization.
The cells were cultured in complete medium for 48 h and then seeded
at 5×105/well in a 6-well culture plate in a final
volume of 1 ml. The cells were washed twice with ice-cold PBS
solution, followed with ice-cold 70% ethanol and then fixed at 4°C
for 2 h overnight. Subsequently, the cells were washed with PBS to
remove the ethanol. Conventional PI staining was measured using
flow cytometry and FACScan DNA analysis was performed to determine
the content of DNA. The results were analyzed using MultiCycle
software. Experiments were repeated three times.
Determination of the apoptotic rate using
Annexin V-PE/7AAD
Jurkat cells in the logarithmic phase were seeded at
5×105 cells/well in a 6-well culture plate in a final
volume of 1 ml. The cells were transfected with HOXA5-specific
siRNA or the control siRNA. The cells were collected and washed
with cold PBS twice. The cells were then resuspended in 50
μl of binding buffer, followed by 5 μl of 7-AAD.
Staining was performed at a room temperature of 25°C for 5–15 min
in the dark. This was followed by the addition of 450 μl of
binding buffer, 1 μl of Annexin V-PE, at room temperature
for 5–15 min in the dark. The apoptotic rate was determined using
flow cytometry within 1 h. Experiments were repeated three
times.
Statistical analysis
Data were analyzed using SPSS 13.0 statistical
software (SPSS, Inc., Chicago, IL, USA). Measurement data were
presented as means ± SD. ANOVA was used to compare groups, and
multiple pairwise comparisons were made by the Dixon's q-test.
P<0.05 was considered statistically significant.
Results
HOXA5 mRNA expression levels in bone
marrow mononuclear cells
HOXA5 expression was observed in 7 of 20 control
group patients with ITP (positive rate of 28%). HOXA5 was expressed
in 16 of the 25 cases of children with ALL in the acute stage
(positive rate of 64%). In the ALL remission group, HOXA5 was
expressed in 10 of 25 cases (positive rate of 40%). The results of
QF-PCR for the HOXA5 mRNA relative expression analysis in each
group were: ALL acute phase 2−ΔΔCt 0.76±0.05% (F=16.31,
P<0.05); ALL remission 2−ΔΔCt 0.48±0.07%; and control
group (ITP) 2−ΔΔCt 0.47±0.08% (Fig. 1 and Table II).
 | Table IIExpression of HOXA5 mRNA in bone
marrow of each group. |
Table II
Expression of HOXA5 mRNA in bone
marrow of each group.
| Group | n | HOXA5 mRNA positive
rate
[cases (%)] | HOXA5
mRNA
(mean ± SD) |
|---|
| Control group | 20 | 7 (28) | 0.47±0.08 |
| ALL acute
phase | 25 | 16 (64)a | 0.76±0.05a |
| ALL remission
stage | 25 | 10 (40)b | 0.48±0.07b |
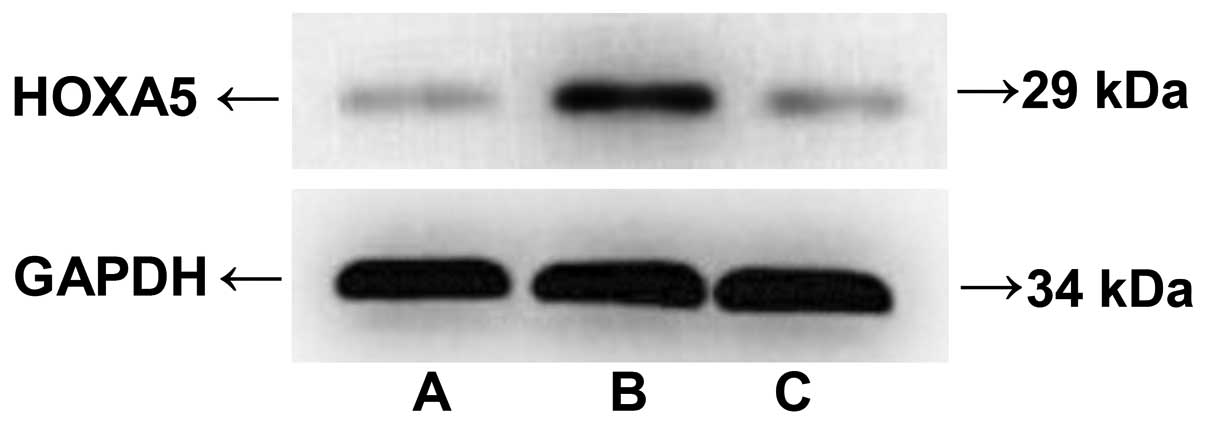
HOXA5 protein expression levels in bone
marrow mononuclear cells
The results of the western blot analysis of HOXA5
protein expression levels in bone marrow mononuclear cells were:
ALL acute phase (0.70±0.02), ALL remission (0.39±0.03), control
group ITP (0.42±0.02) (Fig.
2).
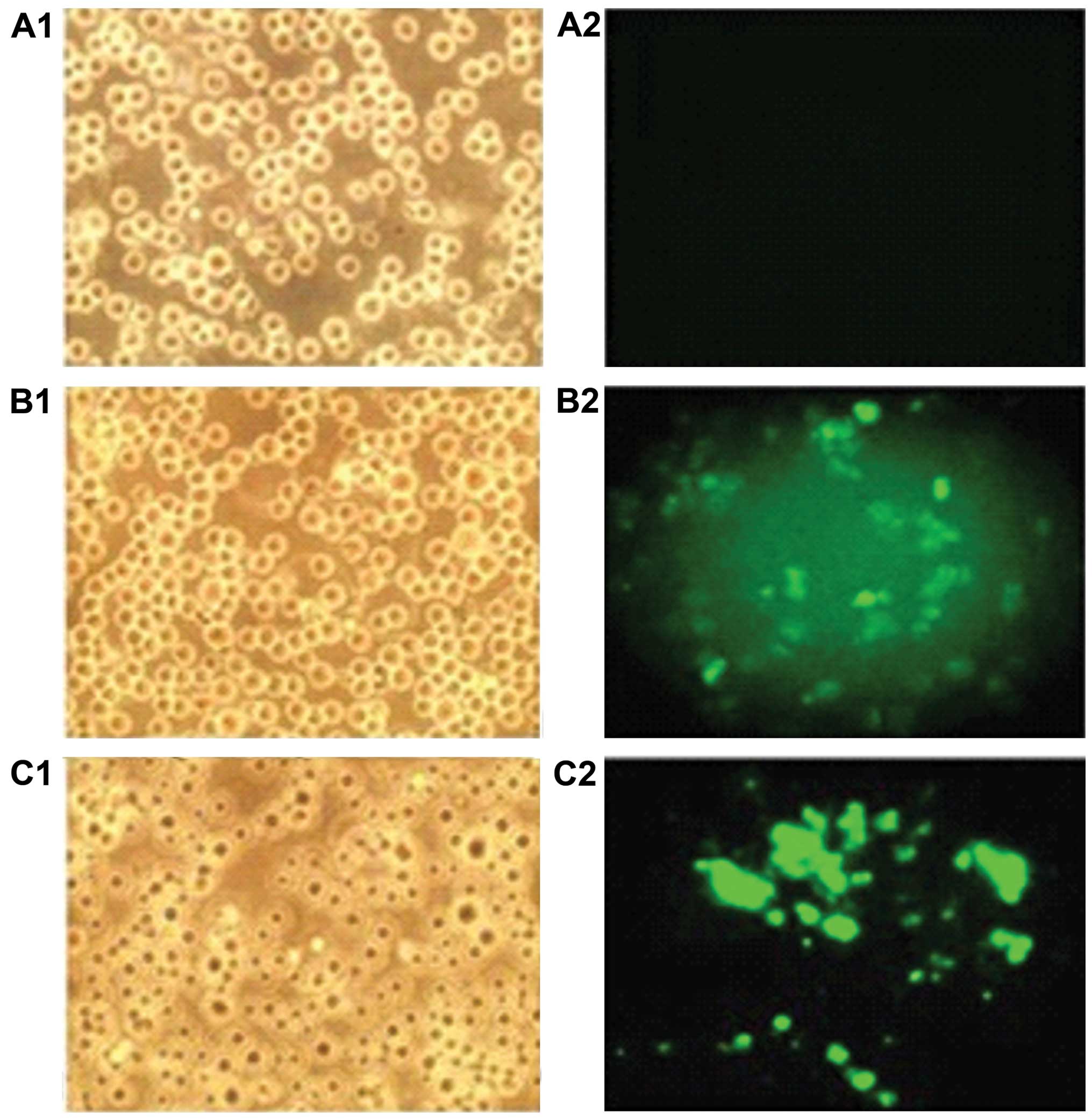
Jurkat cells transfected with recombinant
vector
Green fluorescent protein was expressed in Jurkat
cells transfected with pRNAT-GFP-Neo-siHOXA5C recombinant vector
(Fig. 3). The transfection
efficiency was ~60%.
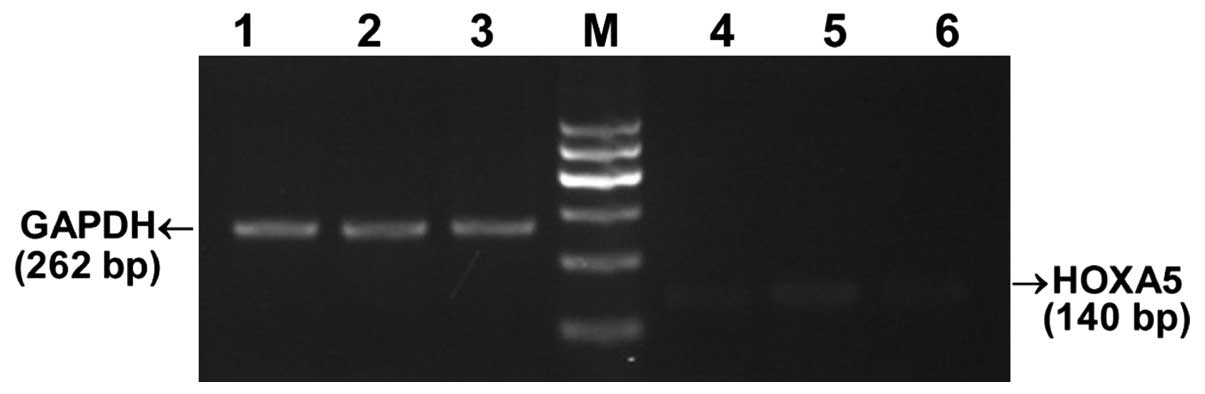
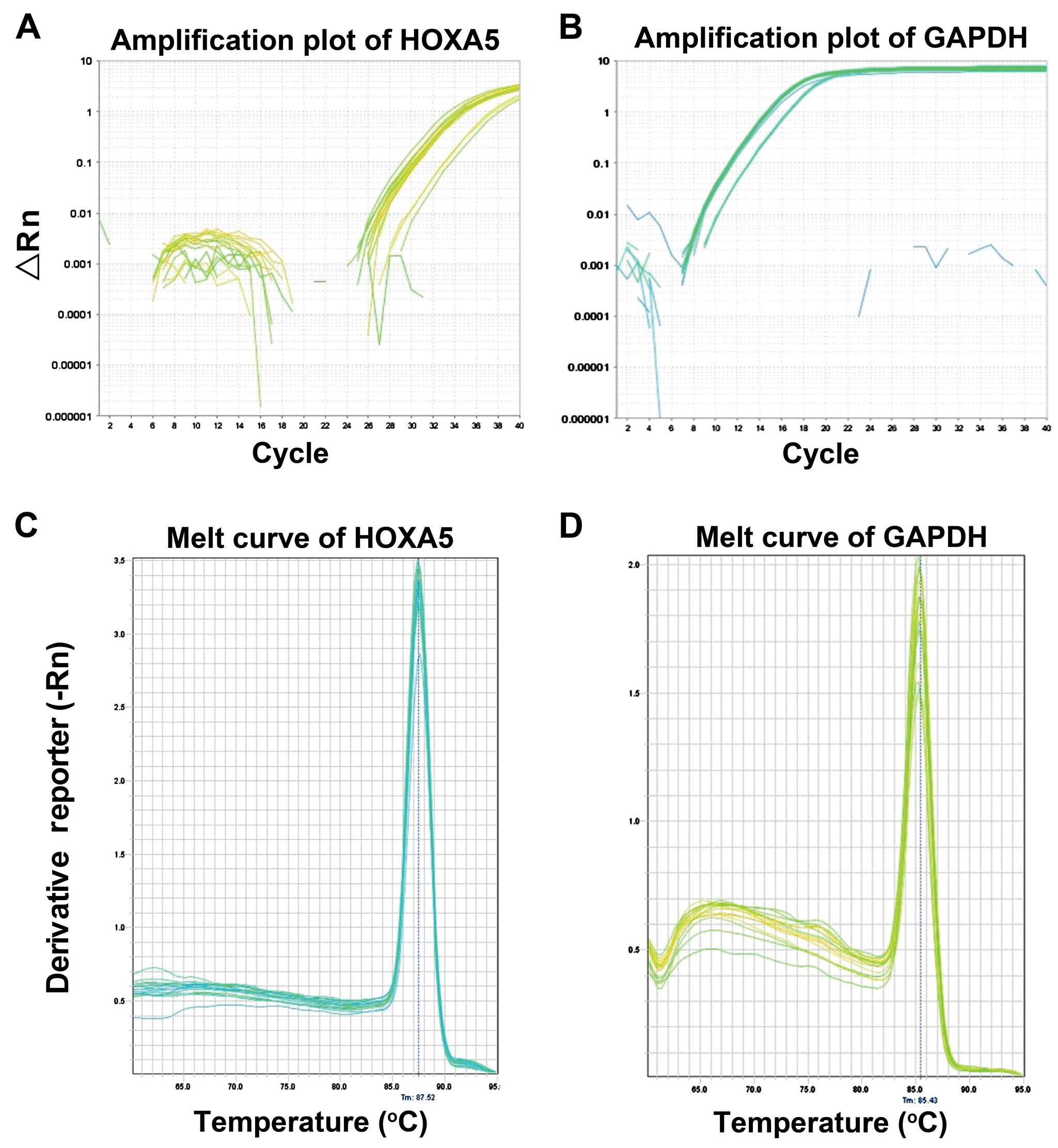
QF-PCR amplification and melting
curve
The experimental amplification curve shown in
Fig. 4A and B is an s-shaped
curve that demonstrates line dynamics. Following QF-PCR reaction at
a temperature of 65–65°C, melting curve analysis was conducted and
the results are shown in Fig. 4C and
D. The homogeneous melting point of the HOXA5 gene and
GAPDH was 84–85°C. The graphs have a single sharp absorption peak
(Fig. 4C and D). No other product
was observed and primer-dimer formation did not occur, indicating
that the design of the primer had a good specificity (Fig. 4).
Effects of the recombinant vector on the
expression of HOXA5 mRNA in Jurkat cells
QF-PCR results for the relative expression quantity
of HOXA5 mRNA were: PRNAT-GFP-Neo-HOXA5A (1.01±0.03%),
PRNAT-GFP-Neo-HOXA5B (0.87±0.02%), PRNAT-GFP-Neo-HOXA5C
(0.39±0.01%), negative control group (1.34±0.06%), and blank
control group (1.29±0.21%) (Fig.
5). The difference between the experimental, negative control
and blank control groups was not statistically significant
(P>0.05). The difference between the experimental, blank control
and negative control groups was statistically significant
(P<0.05), while there was no significant difference between the
negative control and blank control groups (P>0.05). HOXA5 mRNA
inhibition ratios were as follows: PRNAT-GFP-Neo-HOXA5A
(24.62±2.34%), PR NAT- GF P-Neo -HOX A 5B (35. 07±3. 21%) a nd
PRNAT-GFP-Neo-HOXA5C (70.89±6.41%) (Fig. 5). It was evident that of the three
selected siRNAs, PRNAT-GFP-Neo- HOXA5C had the best interfering
interference effects (Fig. 5).
Consequently, PRNAT-GFP-Neo-HOXA5C was selected to conduct the
subsequent experiments.
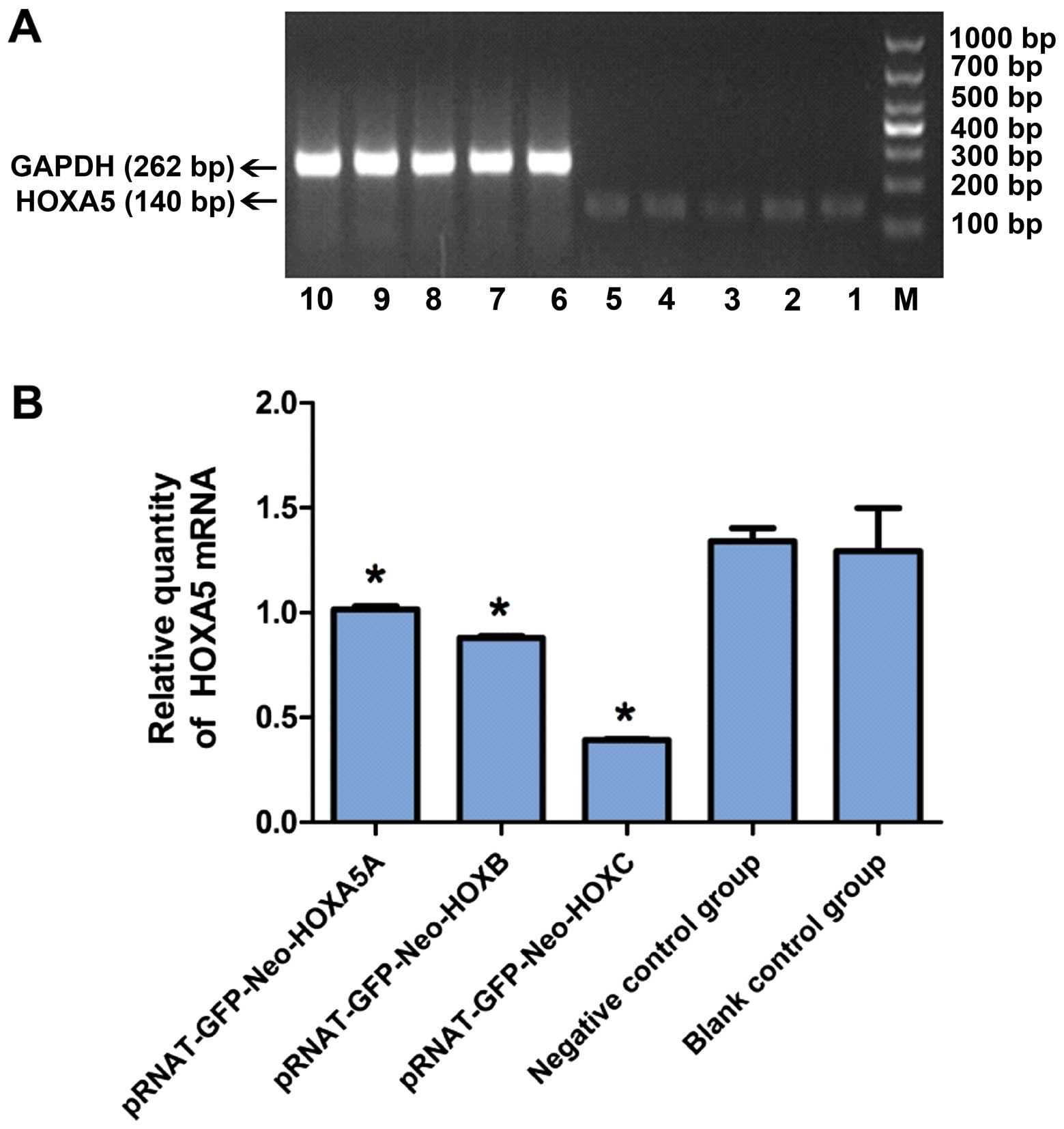 | Figure 5Inhibitory effect of siRNA on the
expression of Jurkat in HOXA5 mRNA cells. (A) Agarose
electrophoresis images of HOXA5; M, marker; Lanes 1,
pRNAT-GFP-Neo-HOXA5A; 2, pRNAT-GFP-Neo-HOXA5B; 3, pRNAT-GFP-Neo
-HOXA5C; 4, negative control group: pRNAT-GFP-Neo-siRNAnc; 5, blank
control group; GAPDH; 6–8, experimental group; 9, negative control
group; and 10, blank control group. (B) Statistical results of the
relative quantity of HOXA5 mRNA in (A). Experimental group compared
with the control and negative control groups,
*P<0.05. |
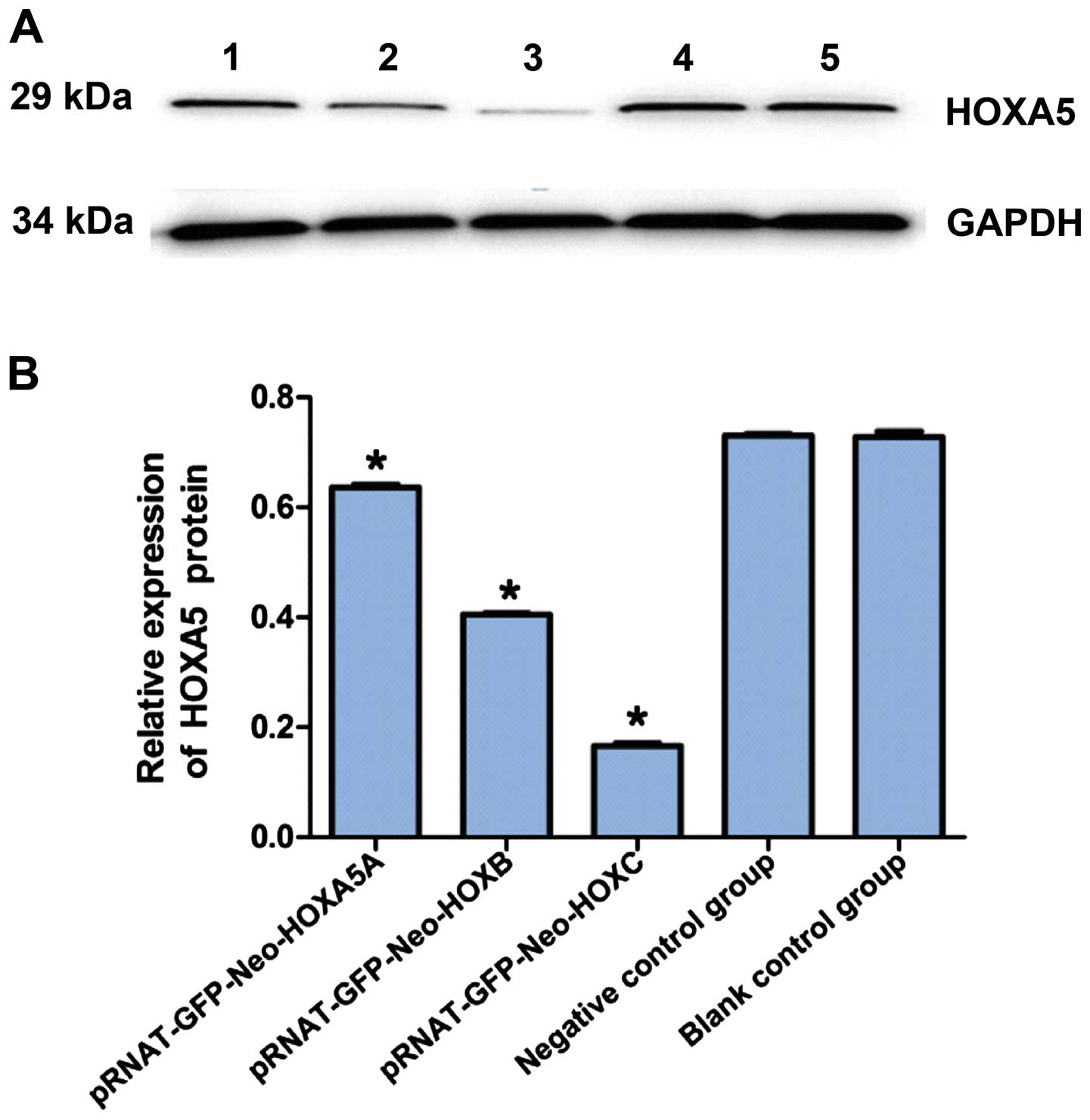
Effects of siRNA on HOXA5 protein
expression levels in Jurkat cells
Western blot analysis was used to examine the
expression of HOXA5 protein. The results revealed that, siRNA
targeting of HOXA5 in Jurkat cells after 24 h decreased the
expression of HOXA5 protein, pRNAT-GFP-Neo-HOXA5A (0.64±0.15),
pRNAT-GFP-Neo-HOXA5B (0.41±0.06), pRNAT-GFP-Neo-HOXA5C (0.17±0.05),
negative control group (0.73±0.12), and the blank control group
(0.73±0.13) (Fig. 6). The
difference between the experimental, blank control and negative
control groups was statistically significant (P<0.05), while
there was no significant difference between the negative control
and blank control groups (P>0.05). The relative HOXA5 protein
expression in the experimental group was significantly lower than
that in the negative control and blank control groups. The HOXA5
protein inhibitory rate was: pRNAT-GFP-Neo-HOXA5A (12.32±3.12%),
pRNAT-GFP-Neo-HOXA5B (43.83±4.13%) and pRNAT-GFP-Neo-HOXA5C
(76.71±5.16%) (Fig. 6). The
expression levels of the pRNAT-GFP-Neo-HOXA5C vector of the HOXA5
protein had a significant inhibitory effect and were shown to be
effective in the interference for subsequent experiments.
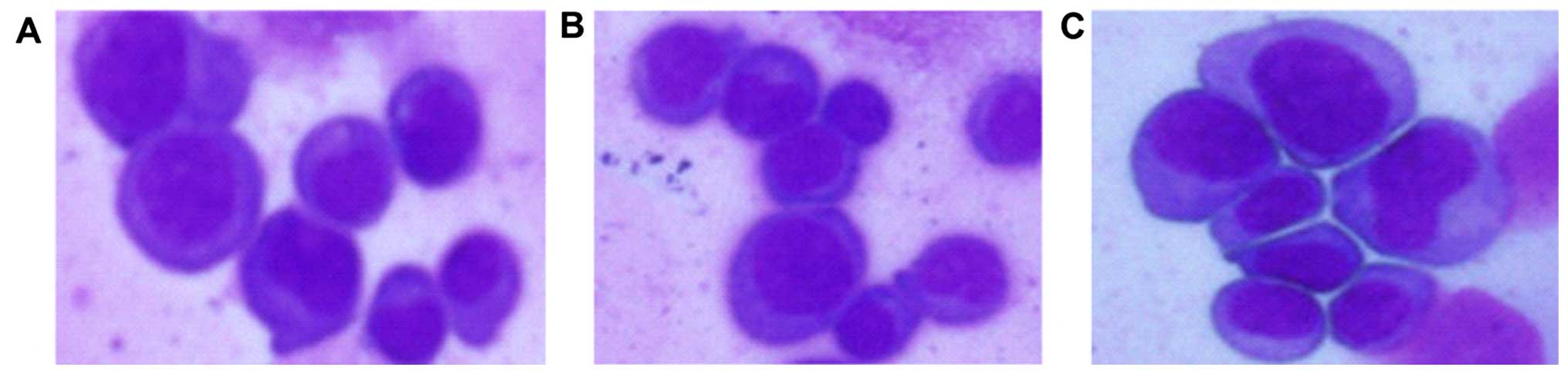
Morphological changes in each group as
revealed by the Wright's stain method
When observed under light microscopy and compared
with the negative control and blank control groups, the
experimental nuclear mass ratio in the experimental group
decreased, and rare nuclear fission and the apoptotic rate
increased (Fig. 7).
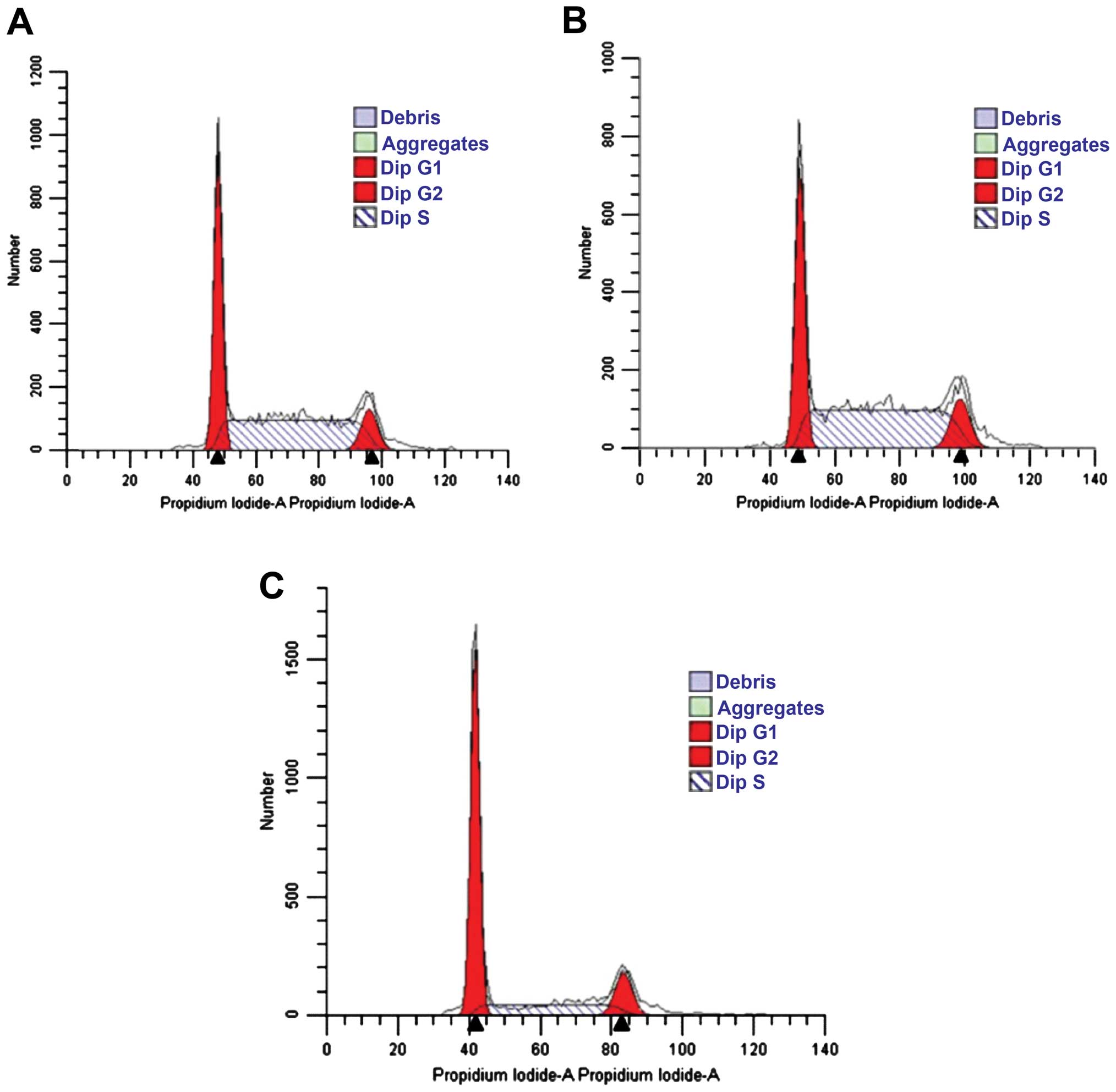
Effects of siRNA on cell cycle of Jurkat
cells
Following the transfection of Jurkat cells with
HOXA5 siRNA for 48 h, the ratio of Jurkat cells in the G0/G1 phase
significantly increased (56.70±6.4 vs. 38.55±6% and 38.69±2.2%),
whereas the ratio of cells in the S phase significantly decreased
(29±5.5 vs. 49.53±8.3% and 48.86±6%) (Fig. 8 and Table III). This difference was
statistically significant (P<0.05). No statistically significant
difference was identified in the distribution of cells in the
control and negative control groups (Fig. 8A and B, respectively, and Table III).
 | Table IIIDistribution of the cell cycle 48 h
after transfection (%, mean ± SD). |
Table III
Distribution of the cell cycle 48 h
after transfection (%, mean ± SD).
| Group | G0/G1 | S | G2/M |
|---|
| Control | 38.69±2.2 | 48.86±6.0 | 11.70±2.8 |
| Negative
control | 38.55±6.0 | 49.53±8.3 | 11.60±3.5 |
Experimental
(pRNAT-GFP-Neo-HOXA5C) | 56.70±6.4a | 29.00±5.5a | 14.29±1.5b |
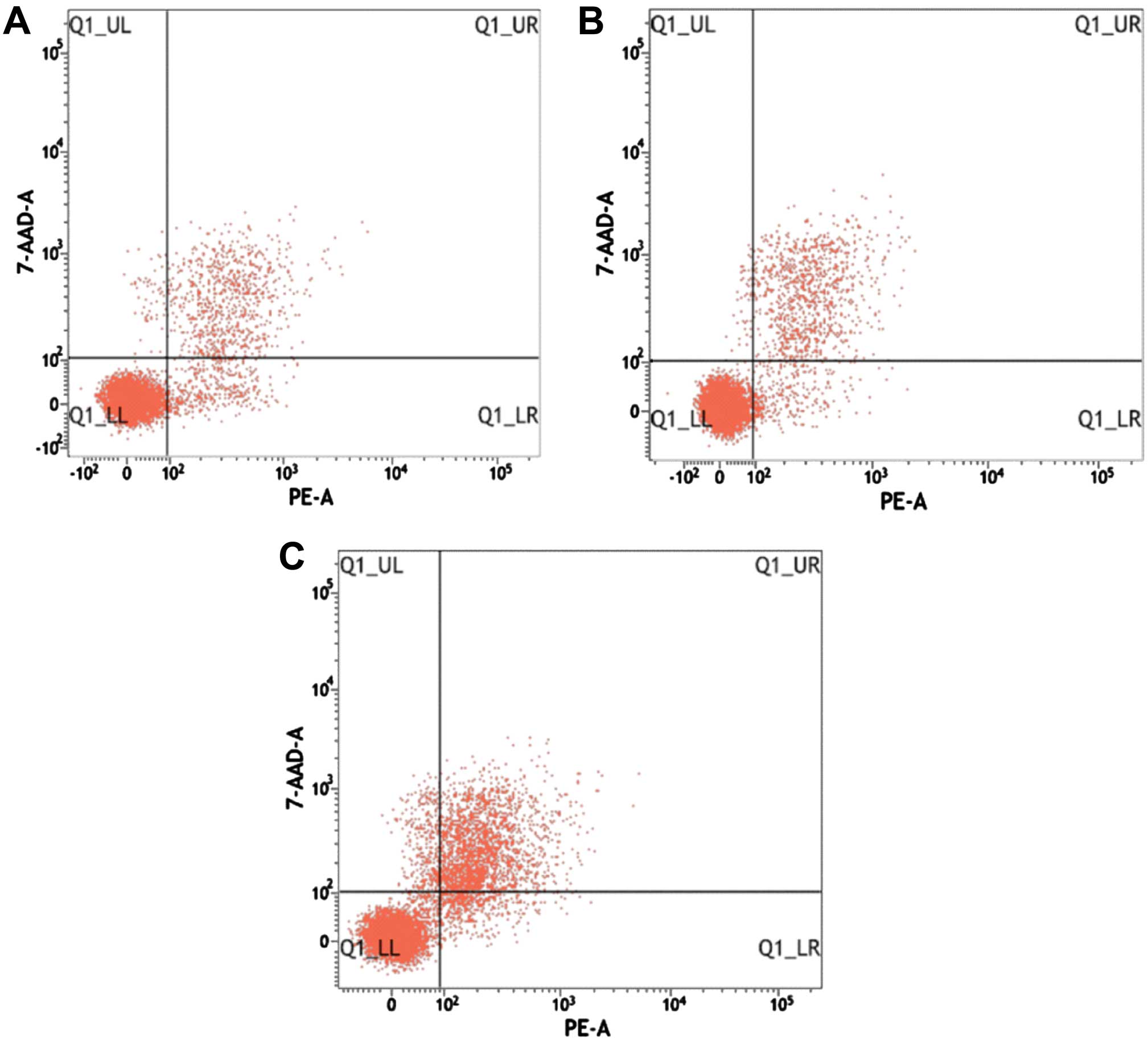
Effects of recombinant vector on
apoptosis in Jurkat cells
After staining with Annexin V-PE and 7-AAD, double
labeling flow cytometry showed that the recombinant vector was
transfected in Jurkat cells after 48 h. The apoptotic cell rate in
the control, negative control and experimental groups was
13.98±1.05, 13.94±0.98 and 24.99±5.16%, respectively. The
difference in the apoptotic rate between the experimental, control
and negative control groups was statistically significant
(P<0.05), whereas the difference between the negative control
and control groups, was not statistically significant (P>0.05)
(Fig. 9 and Table IV).
 | Table IVRestructuring carrier effects on
Jurkat cell apoptosis (%, mean ± SD). |
Table IV
Restructuring carrier effects on
Jurkat cell apoptosis (%, mean ± SD).
| Group | Flow rate of
apoptosis (%) |
|---|
| Control | 13.98±1.05 |
| Negative
control | 13.94±0.98 |
| Experimental
(pRNAT-GFP-Neo-HOXA5C) | 24.99±5.16a |
Discussion
Leukemia is a malignant hyperplastic disease of the
hematopoietic system, which ranks first among tumor diseases in
children (14). Homeobox genes
encode transcription factors that are members of the Hox gene
family and participate in hematopoietic stem/progenitor cell (HSPC)
proliferation, differentiation and maturation (15). They are a type of regulatory gene
that controls embryonic and cell differentiation and is closely
associated with the incidence of leukemia (15–18). Normal mature tissues express
HOX genes, which are silent, or expressed in the embryonic
state during organization, leading to tumor development (19). HOX genes are important in the
regulation of the hematopoietic proliferation and differentiation,
as well as the abnormal expression of HOX genes, leading to
the occurrence of leukemia (20).
The head end (HOXA1-HOXA5) HOX gene, a positive marker of
AML of mixed leukemia genes [mixed lineage leukemia (MLL)] is often
characterized by abnormal protein expression (21). It has been suggested that MLL
protein fusion is achieved by disordering the transcription of
HOX genes (21). As a
member of the family of HOX genes, HOXA5 is expressed in
many organs and regulates gene expression, cell differentiation and
the morphogenesis of body function (22). HOXA5 is a key regulator of the
haematopoietic stem cell (HSC) cycle, and the inappropriate
expression of HOXA5 in lineage-committed progenitor cells leads to
aberrant erythropoiesis (22).
Its structure and dysfunction is closely associated with the
occurrence of leukemia. Kim et al (23) performed pyrosequencing to quantify
the methylation level of the HOXA5 gene in the bone marrow
samples obtained from 50 patients with AML and 19 normal controls.
The results showed that the survival rate of AML patients with
stage 3A cancer correlated with HOXA5 methylation (23).
Under certain conditions, changing the expression
level of the HOXA gene may promote or inhibit the occurrence
and development of a tumor (24).
Although there are many methods of inhibiting gene expression, RNAi
is the most commonly used. RNAi technology is a simple and
effective alternative knockout genetic tool that has been developed
in recent years (25,26). Moore et al (27) found that a significant expression
of HOXA5 mediated by retrovirus causes myeloid differentiation but
not erythroid differentiation of hematopoietic stem cells and
progenitor cells. The findings of Liu et al (28) indicated that miR-196a is
significantly upregulated in non-small cell lung cancer (NSCLC)
tissues and regulates NSCLC cell proliferation, migration and
invasion, partially via the downregulation of HOXA5. Thus, miR-196a
is a potential therapeutic target for NSCLC intervention. The study
of Wang et al (29)
suggested that specific siRNA of CXCR4 effectively downregulates
the expression of the CXCR4 gene and induces cell cycle
arrest and apoptosis of Jurkat cells, while inhibiting cell
proliferation. Other studies have shown that Jurkat cells of human
leukemia cell line is an ideal RNAi experimental cell model
(30,31). Therefore, the application of RNAi
technology to downregulate HOXA5 expression may inhibit the
proliferation and apoptosis of leukemia cells. A study by Zhang
et al (32) showed that
shRNA targeting of silent HOXA10 gene mediated by lentiviral
vector, can effectively inhibit the proliferation and promote the
apoptosis of U937 cells. Fan et al (33) study showed that RNAi technology
combined with a small dose of Ara-C effectively inhibits the
proliferation and induces the apoptosis of K562 cells.
The specific detection of QF-PCR and western blot
analysis in the present study indicated that HOXA5 gene was
expressed at high levels in ALL patients. The expression of ALL
mRNA (0.76±0.05%) and protein (0.70±0.020%) in the acute phase was
significantly higher than that in the remission stage and control
groups. The experimental group (pRNAT-GFP-Neo-siHOXA5C) affected by
siRNA showed lower mRNA (0.39±0.01%) and protein (0.17±0.05%)
levels compared to the negative control and blank control groups.
The results showed that pRNAT-GFP-Neo-siRNAHOXA5C HOXA5 carrier
effectively silences gene expression and inhibits Jurkat cell
proliferation. The cell cycle detected through flow cytometry
showed that, compared with the negative control and blank control
groups, the proportion of G0/G1 cells increased and the proportion
of S phase cells decreased. Annexin V-PE/7-AAD double staining is
an ideal method for detection of the apoptotic rate (34). In the present study, the
experimental group (pRNAT-GFP-Neo-siHOXA5C) under the influence of
siRNA showed a flow apoptotic rate of 24.99±5.16, which was higher
as compared to that of the negative control group (13.94±0.98) and
the blank control group (13.98±1.05). Following transfection, mRNA
in the pRNAT-GFP-Neo siHOXA5C group was effectively reduced, and
the apoptotic rate was significantly increased compared with the
other groups. Another study has shown that the overexpression of
HOXA5 inhibits apoptosis (34) by
inhibiting its target genes. Flow cytometry showed that, in this
group, siRNA carrier inhibited the ability of HOXA5 to promote
Jurkat cell apoptotic rate, which was 24.99±5.16%. Compared with
the negative control and blank control groups, the mRNA and protein
expression of HOXA5 in Jurkat cells in the experimental group
(pRNAT-GFP-Neo-siHOXA5C) was significantly reduced, the cell cycle
was suppressed, and the apoptotic rate increased. The evidence
showed that the construction of pRNAT-GFP-Neo-siHOXA5 in this
experiment was successful.
The aforementioned results show that HOXA5
gene is highly expressed in ALL and closely associated with the
occurrence of ALL in children. Eukaryotic expression carrier
targeting HOXA5 constructed in the present study can effectively
reduce the expression of HOXA5 in Jurkat leukemia cells and inhibit
its proliferative ability by silencing the HOXA5 gene.
Therefore, this eukaryotic expression carrier has the potential to
become an effective gene therapy to treat leukemia.
Acknowledgments
We would like to thank the Science and Technology
Bureau of Sichuan Province for its financial support (grant no.
201410).
References
|
1
|
Ceppi F, Antillon F, Pacheco C, Sullivan
CE, Lam CG, Howard SC and Conter V: Supportive medical care for
children with acute lymphoblastic leukemia in low- and
middle-income countries. Expert Rev Hematol. May 26–2015.Epub ahead
of print. View Article : Google Scholar
|
|
2
|
Lo-Coco F, Fouad TM and Ramadan SM: Acute
leukemia in women. Womens Health (Lond Engl). 6:239–249. 2010.
View Article : Google Scholar
|
|
3
|
Pui CH: Recent research advances in
childhood acute lymphoblastic leukemia. J Formos Med Assoc.
109:777–787. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Strathdee G, Holyoake TL, Sim A, Parker A,
Oscier DG, Melo JV, Meyer S, Eden T, Dickinson AM, Mountford JC,
Jorgensen HG, Soutar R and Brown R: Inactivation of HOXA genes by
hypermethylation in myeloid and lymphoid malignancy is frequent and
associated with poor prognosis. Clin Cancer Res. 13:5048–5055.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
De Braekeleer E, Douet-Guilbert N, Basinko
A, Le Bris MJ, Morel F and De Braekeleer M: Hox gene dysregulation
in acute myeloid leukemia. Future Oncol. 10:475–495. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu TT and Liu W: Studies on the
relationship between HOX genes and leukemia. J Pediatr Hematol
Oncol. 18:1421452013.
|
|
7
|
Delval S, Taminiau A, Lamy J, Lallemand C,
Gilles C, Noël A and Rezsohazy R: The Pbx interaction motif of
Hoxa1 is essential for its oncogenic activity. PLoS One.
6:e252472011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okada Y, Jiang Q, Lemieux M, Jeannotte L,
Su L and Zhang Y: Leukaemic transformation by CALMAF10 involves
upregulation of HOXA5 by hDOT1L. Nat Cell Biol. 8:1017–1024. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bach C, Buhl S, Mueller D, García-Cuéllar
MP, Maethner E and Slany RK: Leukemogenic transformation by HOXA
cluster genes. Blood. 115:2910–2918. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boutter J, Huang Y, Marovca B, Vonderheit
A, Grotzer MA, Eckert C, Cario G, Wollscheid B, Horvath P,
Bornhauser BC and Bourquin JP: Image-based RNA interference
screening reveals an individual dependence of acute lymphoblastic
leukemia on stromal cysteine support. Oncotarget. 5:11501–11512.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Landry B, Valencia-Serna J, Gul-Uludag H,
Jiang X, Janowska-Wieczorek A, Brandwein J and Uludag H: Progress
in RNAi-mediated molecular therapy of acute and chromic myeloid
leukemia. Mol Ther Nucleic Acids. 4:e2402015. View Article : Google Scholar
|
|
12
|
Olivieri D, Sykora MM, Sachidanandam R,
Mechtler K and Brennecke J: An in vivo RNAi assay identifies major
genetic and cellular requirements for primary piRNA biogenesis in
Drosophila. EMBO J. 29:3301–3317. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
China Medical Sciences Branch of
Hematology Group: Editorial Committee Member of Chinese Journal of
Pediatrics. Diagnosis and treatment for children with acute
lymphoblastic leukemia (Third Amendment Bill). Zhonghua Er Ke Za
Zhi. 44:392–395. 2006.In Chinese.
|
|
14
|
Qian X and Wen-jun L: Platelet changes in
acute leukemia. Cell Biochem Biophys. 67:1473–1479. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang Q and Liu WJ: Relationship between
the HOX gene family and the acute myeloid leukemia-review. Zhongguo
Shi Yan Xue Ye Xue Za Zhi. 21:1340–1344. 2013.In Chinese.
PubMed/NCBI
|
|
16
|
Wen-jun L, Qu-lian G, Hong-ying C, Yan Z
and Mei-Xian H: Studies on HOXB4 expression during differentiation
of human cytomegalovirus-infected hematopoietic stem cells into
lymphocyte and erythrocyte progenitor cells. Cell Biochem Biophys.
63:133–141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu WJ, Huang MX, Guo QL, Chen JH and Shi
H: Effect of human cytomegalovirus infection on the expression of
Hoxb2 and Hoxb4 genes in the developmental process of cord blood
erythroid progenitors. Mol Med Rep. 4:1307–1311. 2011.PubMed/NCBI
|
|
18
|
Liu WJ1, Jiang NJ, Guo QL and Xu Q: ATRA
and As2O3 regulate differentiation of human
hematopoietic stem cells into granulocyte progenitor via alteration
of Hoxb8 expression. Eur Rev Med Pharmacol Sci. 19:1055–1062.
2015.
|
|
19
|
Shah N and Sukumar S: The Hox genes and
their roles in oncogenesis. Nat Rev Cancer. 10:361–371. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang N and Liu W: Role of HOX gene in
occurrence of leukemia and study progress. J Appl Clin Pediatr.
27:215–217. 2012.In Chinese.
|
|
21
|
Marschalek R: Mechanisms of leukemogenesis
by MLL fusion proteins. Br J Haematol. 152:141–154. 2011.
View Article : Google Scholar
|
|
22
|
Yang D, Zhang X, Dong Y, Liu X, Wang T,
Wang X, Geng Y, Fang S, Zheng Y, Chen X, et al: Enforced expression
of Hoxa5 in haematopoietic stem cells leads to aberrant
erythropoiesis in vivo. Cell Cycle. 14:612–620. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim SY, Hwang SH, Song EJ, Shin HJ, Jung
JS and Lee EY: Level of HOXA5 hypermethylation in acute myeloid
leukemia is associated with short-term outcome. Korean J Lab Med.
30:469–473. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang ML, Nie FQ, Sun M, Xia R, Xie M, Lu
KH and Li W: HOXA5 indicates poor prognosis and suppresses cell
proliferation by regulating p21 expression in non small cell lung
cancer. Tumour Biol. 36:3521–3531. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fujita Y, Kuwano K and Ochiya T:
Development of small RNA delivery systems for lung cancer therapy.
Int J Mol Sci. 16:5254–5270. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Teng Z and Liu W: The research progress of
RNA interference targeting leukemia HOXA genes. Chin J Pract
Pediatr. 30:396–399. 2015.
|
|
27
|
Moore MA, Dorn DC, Schuringa JJ, Chung KY
and Morrone G: Constitutive activation of Flt3 and STAT5A enhances
self-renewal and alters differentiation of hematopoietic stem
cells. Exp Hematol. 35(Suppl 1): 105–116. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu XH, Lu KH, Wang KM, Sun M, Zhang EB,
Yang JS, Yin DD, Liu ZL, Zhou J, Liu ZJ, et al: MicroRNA-196a
promotes non-small cell lung cancer cell proliferation and invasion
through targeting HOXA5. BMC Cancer. 12:348–360. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Liu XR, Tan YF and Yin XC: Effects
of CXCR4 silence induced by RNA interference on cell cycle
distribution and apoptosis of Jurkat cells. Zhongguo Shi Yan Xue Ye
Xue Za Zhi. 18:625–628. 2010.In Chinese. PubMed/NCBI
|
|
30
|
Crnkovic-Mertens I, Hoppe-Seyler F and
Butz K: Induction of apoptosis in tumor cells by siRNA-mediated
silencing of the livin/ML-IAP/KIAP gene. Oncogene. 22:8330–8336.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu H, Lin H, Zhu Y, Gu C, Ye Z and Zhang
M: Establishment of Jurkat Cell Lines with Knockdown of BIRC7 Gene.
J Sun Yat-Sen Univ. 29:139–143. 2008.In Chinese.
|
|
32
|
Zhang YJ, Jia XH, Li JC and Xu YH: Effect
of HOXA10 gene silenced by shRNA on proliferation and apoptosis of
U937 cell line. Zhongguo Dang Dai Er Ke Za Zhi. 14:785–791. 2012.In
Chinese. PubMed/NCBI
|
|
33
|
Fan W, Jia X, Li J, Tang S and Zhu S:
Effects of RNA interference and low dose cytarabine on
proliferation and apoptosis of K562 cells. J Appl Clin Pediatr.
27:1177–1180. 2012.
|
|
34
|
Chen H, Chung S and Sukumar S:
HOXA5-induced apoptosis in breast cancer cells is mediated by
caspases 2 and 8. Mol Cell Biol. 24:924–935. 2004. View Article : Google Scholar : PubMed/NCBI
|