Introduction
MicroRNAs (miRNAs or miRs) are a novel class of
small, non-coding RNAs that post-transcriptionally regulate gene
expression in eukaryotic organisms (1). In humans, over 1,900 miRNAs have
been reported (2), and 30% of
human genes may be regulated by these miRNAs (3). To date, it has been demonstrated
that miRNAs are involved in various biological processes, including
cell activation, differentiation and apoptosis (4,5).
In addition, several miRNAs have also been found to be involved in
the regulation of immune cell development and immune signaling
pathways (6). It has been
previously reported that the aberrant expression of miRNAs may play
significant roles in the pathogenesis of several autoimmune
diseases (7-9). For example, in systemic lupus
erythematosus, miR-146 is downregulated; decreased miR-146
expression is associated with prolonged interferon (IFN) signaling,
which results in increased disease activity (10). In multiple sclerosis, miR-326
regulates the differentiatiation of a subset of interleukin
(IL)-17-producing T helper cells (Th17) and levels correlate with
the pathogenesis of the disease (11).
Primary biliary cirrhosis (PBC) is an organ-specific
autoimmune disease that is characterized by the presence of serum
anti-mitochondrial antibodies (AMA) and immune-mediated destruction
of the intrahepatic bile ducts (12,13). Although there has been a
substantial increase in the prevalence of PBC, the pathogenesis of
this disease remains unclear (14). Mounting evidence suggests that
CD4+ T cells play a critical role in immune-mediated
cholangitis in PBC (15).
Traditionally, PBC has been associated with an imbalance of T
helper type 1 (Th1)/T helper type 2 (Th2) cells as demonstrated by
increased IFN-γ levels and decreased IL-10 and IL-4 levels
(16,17). A previous study has detected a
decrease in circulating FoxP3+ regulatory T (Treg) cell
frequency and an increase in Th17 frequency in the peripheral blood
of patients with PBC, providing an opportunity to explore the
mechanisms of PBC (18). However,
the underlying mechanisms that cause the imbalance in
CD4+ T cells in PBC remain to be elucidated.
The aim of the present study was to examine the
expression pattern of miRNAs in the plasma and peripheral blood
mononuclear cells (PBMCs) from patients with PBC, and to analyze
the role of these miRNAs in the development of PBC.
Materials and methods
Study population
All 20 patients (16 females, four males; mean age,
43.2±10.5 years) were recruited from the Infectious Disease
Department of Songjiang Central Hospital (Shanghai, China). The
diagnosis of PBC was based on internationally established criteria
(the consensus of diagnosis and treatment of PBC 2015). Twenty
patients with chronic hepatitis B (CHB) and 20 healthy controls
matched with PBC patients based on gender and age were also
included in this study. The study was approved by the Research
Ethics Committee of SongJiang Central Hospital (Shanghai, China),
and written informed consent was obtained from each patient.
Sample preparation
Blood samples (10 ml) from each participant were
collected in EDTA-treated tubes. Plasma was separated by
centrifugation at 3,000 × g for 10 min, and PBMCs were isolated
through density gradient centrifugation within 1 h (Lymphoprep;
Axis-Shield, Oslo, Norway).
Microarray analysis
An Agilent human miRNA 18.0 micro-array (Agilent
Technologies, Santa Clara, CA, USA) was used to identify miRNAs in
the plasma of three PBC patients and three healthy controls. Total
RNA was extracted and purified using the mirVana™ isolation kit
(Ambion, Austin, TX, USA) according to the manufacturer's
instructions. The miRNA in total RNA was labeled and hybridized
using the miRNA Complete Labeling and Hyb kit (Agilent
Technologies). Slides were then scanned using an Agilent Microarray
Scanner and Feature Extraction software 10.7 (both from Agilent
Technologies). The raw data were normalized using the quantile
algorithm, GeneSpring Software 11.0 (Agilent Technologies).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) verification of miRNA in plasma
and PBMCs
Differentially expressed miRNAs in the plasma and
PBMCs from 60 samples were validated by RT-qPCR (including the
samples used in miRNA array). Total RNA was isolated using an
miRcute miRNA isolation kit (Tiangen Biotech Co., Ltd., Beijing,
China). cDNA was synthesized from total RNA using miRNA-specific
stem-loop RT primers and an miRNA Reverse Transcription kit (Takara
Bio, Otsu, Japan). qPCR was performed using SYBR-Green PCR Master
Mix and an ABI 7500 system (both from Applied Biosystems, Foster
City, CA, USA). All reactions were performed in triplicate. The
mean value of the threshold cycle (Ct) was calculated relative to
U6, an endogenous RNA that served as a control. The relative
expression of each miRNA was calculated using the 2−ΔΔCt
method.
Flow cytometric analysis
The PBMCs were isolated by density gradient
centrifugation and incubated for 4 h at 37°C in 5% CO2
in the presence of 25 ng/ml phorbol myristate acetate, 1
µg/ml ionomycin and 2 µM monensin. The cells were
stained for 30 min with fluorescein isothiocyanate-labeled
anti-human CD4 antibodies (Cat. no. 11-0048-42). The cells were
fixed and permeabilized using Perm/Fix solution. Intracellular
staining was performed with phycoerythrin (PE)-conjugated
anti-human IL-17A (Cat. no. 12-7178-42) or PE-conjugated anti-human
Foxp3 (Cat. no. 12-4777-42) monoclonal antibodies. Isotype controls
were used to confirm antibody specificity. All cells were
resuspended in washing buffer and analyzed by flow cytometry. All
reagents were purchased from eBioscience, Inc. (San Diego, CA,
USA).
Dual staining combining fluorescence in
situ hybridization (FISH) and immunohistochemistry (IHC)
Firstly, the freshly-isolated PBMCs were incubated
with 5 µg/ml anti-CD3, 2 µg/ml anti-CD28, 20 ng/ml
IL-6, 2 ng/ml TGF-β1, 10 µg/ml anti-IFN-γ and anti-IL-4
(eBioscience, Inc.). Subsequently, 25 ng/ml phorbol myristate
acetate, 1 µg/ml ionomycin, and 2 µM monensin were
added and the cells were incubated for another 4 h. Finally, the
cells were collected and seeded on polyane-covered slides.
For detection of miR-92a, the slides were fixed for
15 min at 25°C with paraformaldehyde and washed in
phosphate-buffered saline (PBS) three times. After digestion with
proteinase K for 10 min at 25°C, the slides were washed with PBS
and hydrated in ethanol solutions. Hybridization with the
DIG-labeled miR-92a probe, U6 (positive control), or scrambled
miRNA (negative control) was performed for 1 h at 54°C in
hybridization buffer. Following hybridization, the sections were
washed with 2X SSC, 1X SSC and 0.2X SSC, blocked with 4% horse
serum, and incubated for 12 h at 4°C with alkaline
phosphatase-conjugated Fab-anti-DIG antibody (Cat. no. ab119345;
Abcam, Cambridge, MA, USA) in 1% sheep serum. Staining was
performed by TSA Plus Direct-Cyanine 3 deposition following the
manufacturer's instructions (PerkinElmer, Inc., Waltham, MA,
USA).
For IL-17A detection, the sections were first
blocked using PBS-BB (PBS containing 0.2% powdered skim milk, 1 %
bovine serum albumin and 0.3% Triton X-100) for 30 min, followed by
incubation for 1 h at room temperature with the mouse anti-human
IL-17A monoclonal antibody (Cat. no. 14-7179-82; eBioscience,
Inc.). After washing, we used the TSA Plus Direct-Green kit
according to the manufacturer's instructions (PerkinElmer, Inc.,
Shelton, CT, USA). Finally, the sections were incubated with
4′,6-diamidino-2-phenylindole for 5 min, covered with coverslips
and analyzed under a laser scanning confocal microscope (TCSSP2;
Leica, Wetzlar, Germany).
Statistical analysis
For microarray analysis, Student's t-tests were used
to differentiate the expression of miRNAs among the patients with
PBC and the healthy controls. For RT-qPCR data, one-way analysis of
variance (ANOVA) was used to determine overall differences between
independent groups. The Spearman's correlation coefficient was used
to evaluate correlations between variables. A p-value <0.05 was
considered to indicate a statistically significant difference.
Results
miRNA expression profile in plasma
obtained from patients with PBC
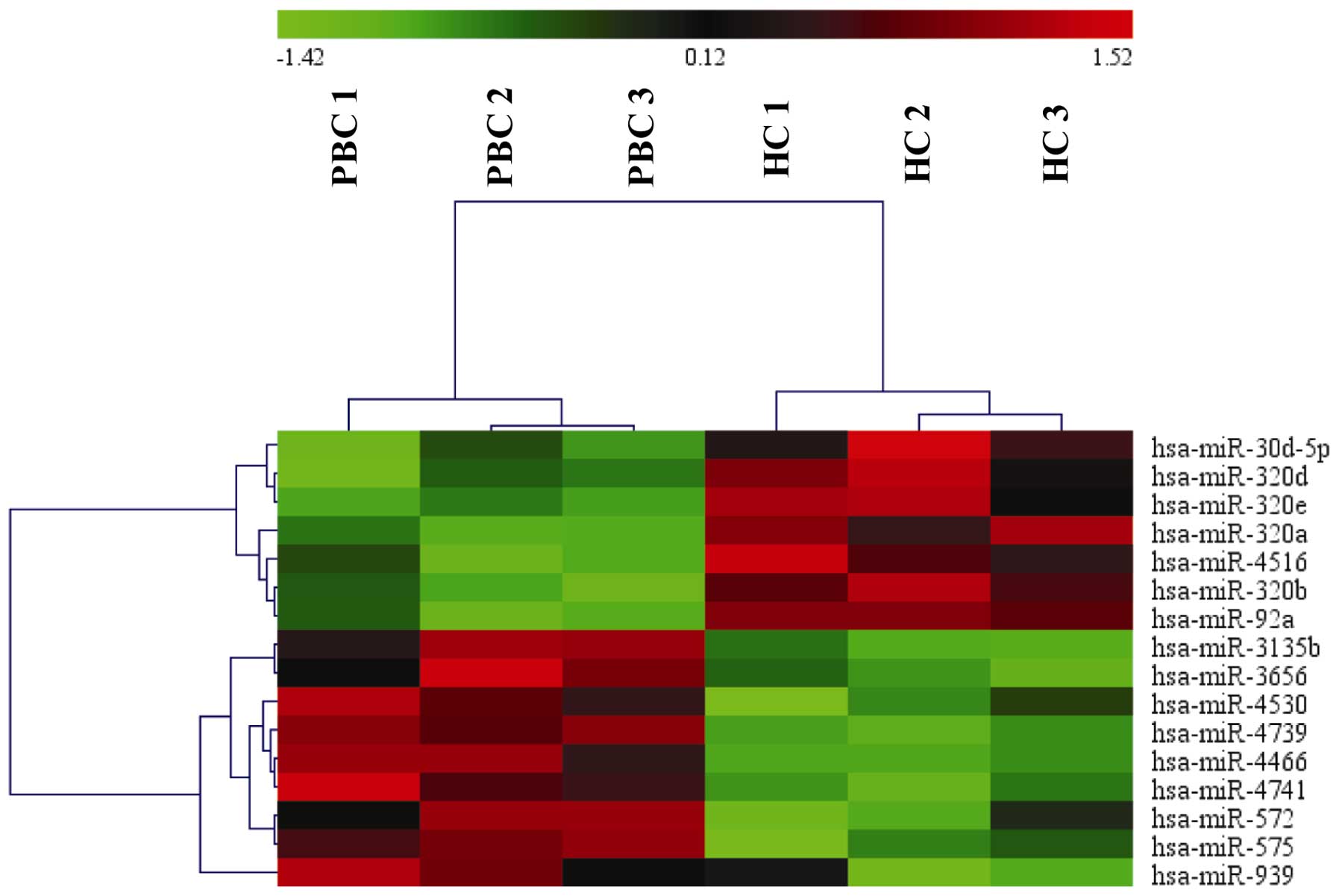
The microarray analysis of plasma from three
patients with PBC and three healthy controls identified 16 miRNAs
that were differentially expressed (Fig. 1), and of these, nine miRNAs were
upregulated and seven miRNAs were downregulated in the patients
with PBC (Table I).
 | Table IDifferentially expressed miRNAs in
plasma from patients with PBC compared with healthy controls. |
Table I
Differentially expressed miRNAs in
plasma from patients with PBC compared with healthy controls.
| miRNA | Fold-change | P-values | Regulation | FDR |
|---|
| hsa-miR-30d-5p | 0.39 | 0.003927 | Down | 0.0125 |
| hsa-miR-4516 | 0.18 | 0.011847 | Down | 0.0257 |
| hsa-miR-320a | 0.27 | 0.003692 | Down | 0.0176 |
| hsa-miR-320b | 0.57 | 0.025506 | Down | 0.0349 |
| hsa-miR-320d | 0.48 | 0.017229 | Down | 0.0247 |
| hsa-miR-320e | 0.48 | 0.041163 | Down | 0.0383 |
| hsa-miR-92a | 0.4 | 0.006802 | Down | 0.0098 |
| hsa-miR-4466 | 1.5 | 0.011165 | Up | 0.0147 |
| hsa-miR-3135b | 1.90 | 0.008485 | Up | 0.0245 |
| hsa-miR-4530 | 2.2 | 0.017937 | Up | 0.0314 |
| hsa-miR-4739 | 2.0 | 0.000162 | Up | 0.0021 |
| hsa-miR-4741 | 2.1 | 0.011578 | Up | 0.0359 |
| hsa-miR-572 | 4.2 | 0.02568 | Up | 0.0417 |
| hsa-miR-575 | 3.1 | 0.011811 | Up | 0.0219 |
| hsa-miR-3656 | 1.58 | 0.031537 | Up | 0.03456 |
| hsa-miR-939 | 2.4 | 0.044987 | Up | 0.01432 |
RT-qPCR validation of miRNA microarray
results
RT-qPCR was performed to confirm the differential
expression of miRNAs identified by microarray analysis. In addition
to comparisons with the healthy controls, 20 patients with CHB were
also included in this study. Validation of miRNA expression was
conducted in all samples (20 patients with PBC, 20 patients with
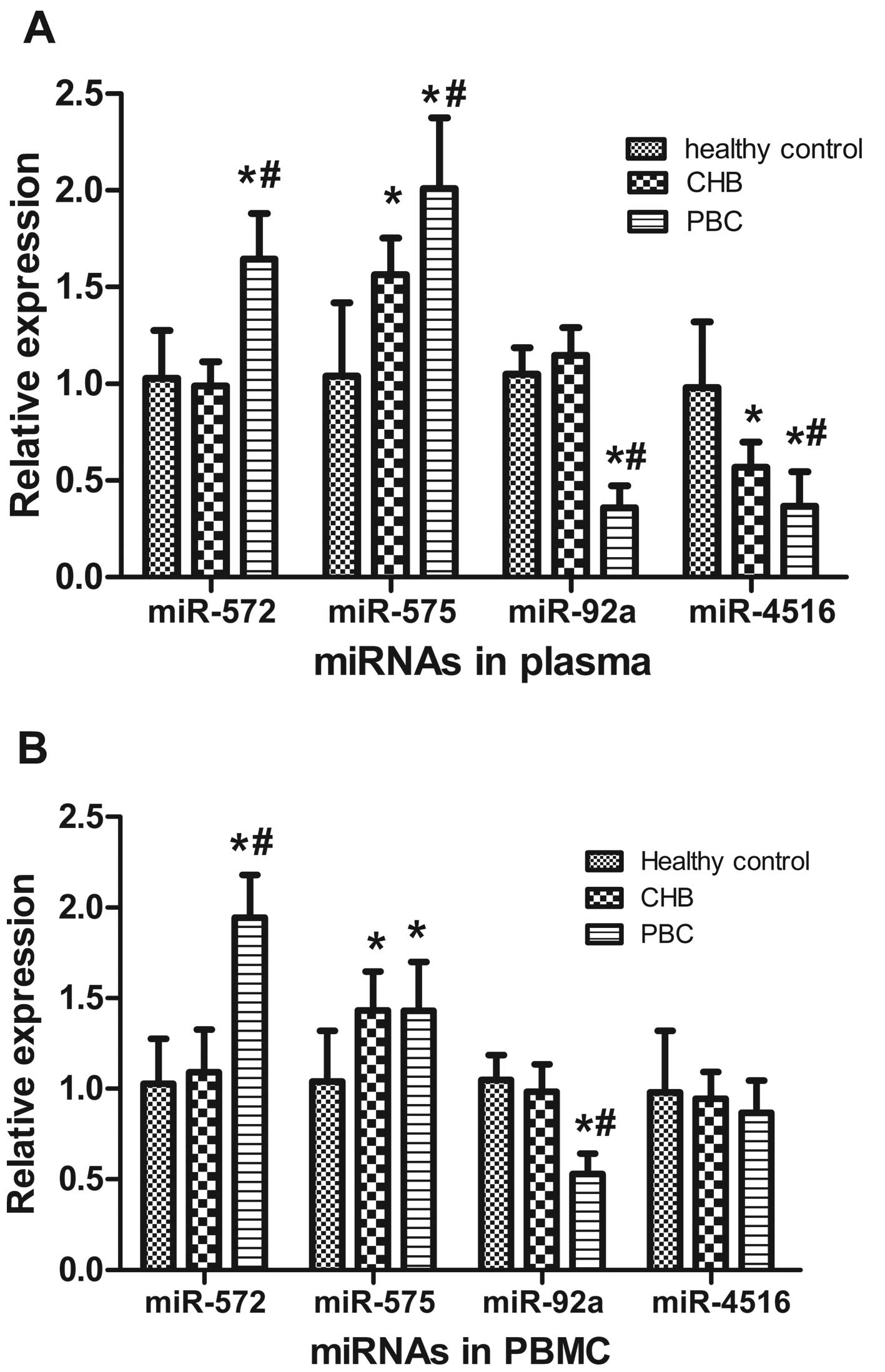
CHB and 20 healthy controls). Among the 16 miRNAs, two miRNAs
(miR-572 and miR-575) were upregulated in the plasma of patients
with PBC compared with the healthy controls and the patients with
CHB. Two miRNAs (miR-92a and miR-4516) were downregulated compared
with the healthy controls and the patients with CHB (Fig. 2A). There were no differences in
the expression of other miRNAs among the patients with PBC or CHB
and the healthy controls (data not shown).
Expression patterns of miRNAs in PBMCs
compared with those in plasma
We also analyzed the expression of the four
validated miRNAs in PBMCs from the patients with PBC or CHB, and
the healthy controls using RT-qPCR. In the PBMCs, miR-572 was
significantly increased in the patients with PBC compared with the
healthy controls and patients with CHB. miR-575 was increased in
the patients with PBC compared with the healthy controls; however,
miR-575 levels did not differ from those in the patients with CHB.
miR-92a was significantly decreased in the patients with PBC
compared with the healthy controls and patients with CHB. miR-4516
expression was unchanged in the PBMCs from the patients with PBC
compared with the healthy controls and patients with CHB, which
differed from the plasma expression pattern (Fig. 2B).
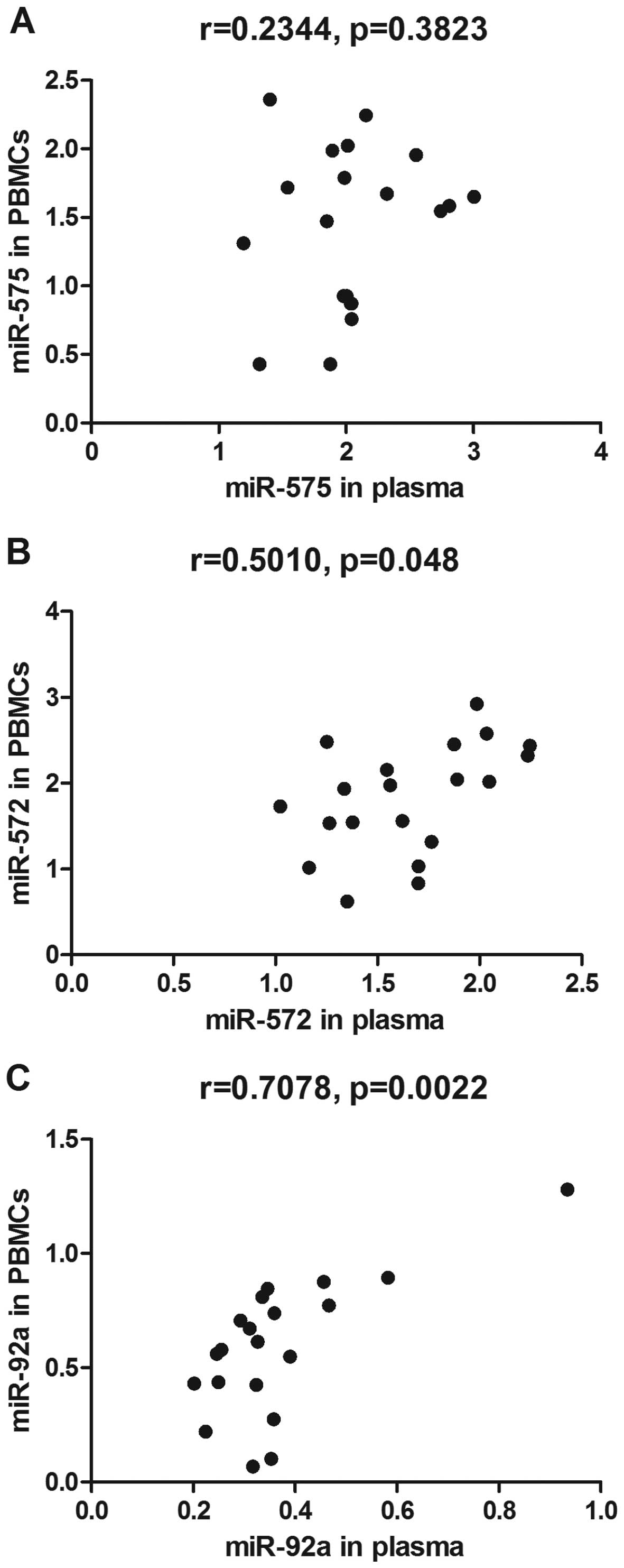
To determine whether there was a correlation between
differentially expressed miRNAs in PBMCs and in plasma, Spearman's
correlation analyses were performed. The results showed that
expression of miR-572 and miR-92a in the PBMCs positively
correlated with their expression in the plasma (Fig. 3B and C). No significant
correlations were observed between the expression of miR-575 in the
plasma and in the PBMCs (Fig.
3A).
Imbalanced T cell subsets in patients
with PBC
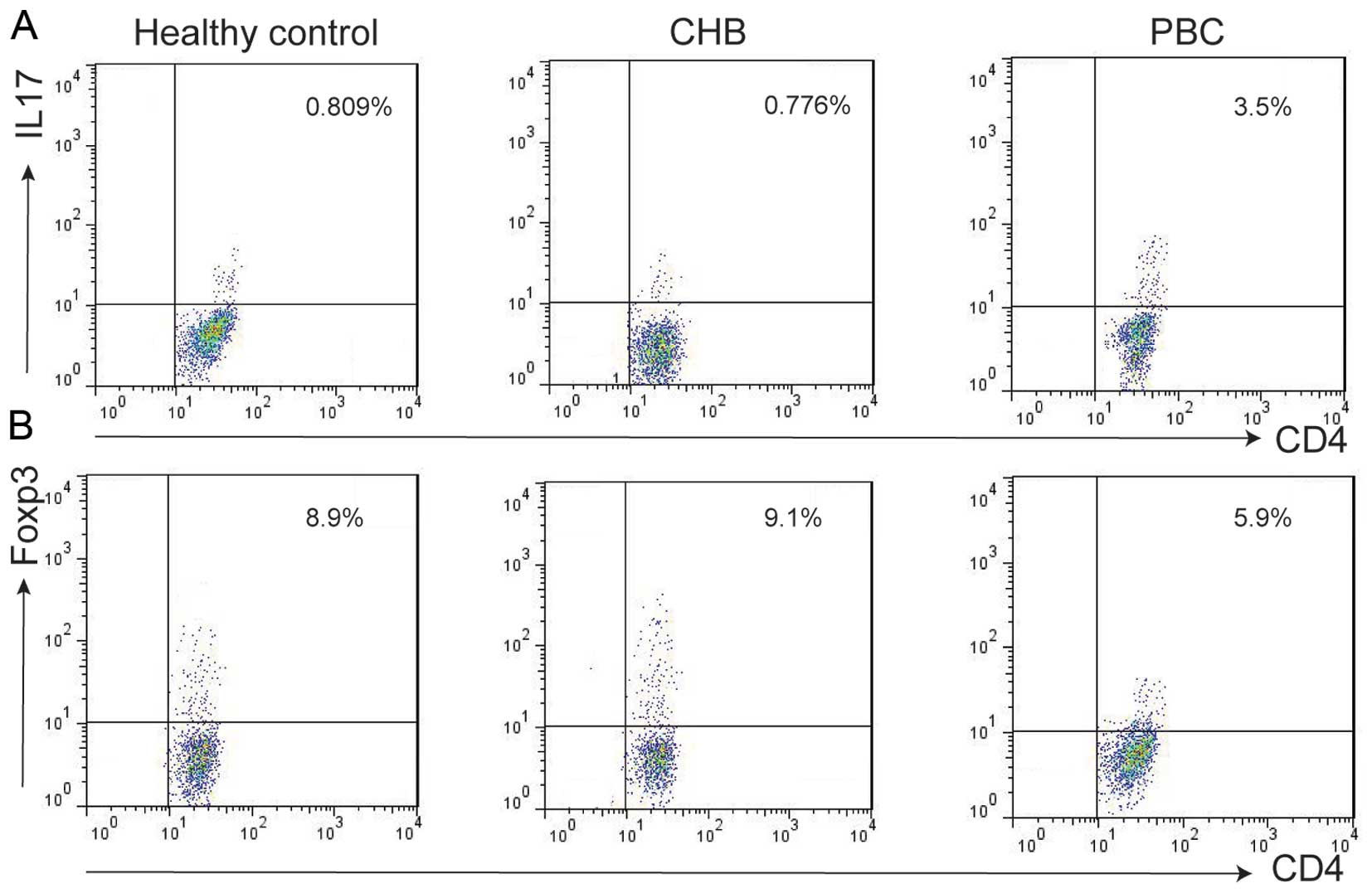
We examined the subset population of T cells from
PBMCs isolated from patients with PBC or CHB, and healthy controls
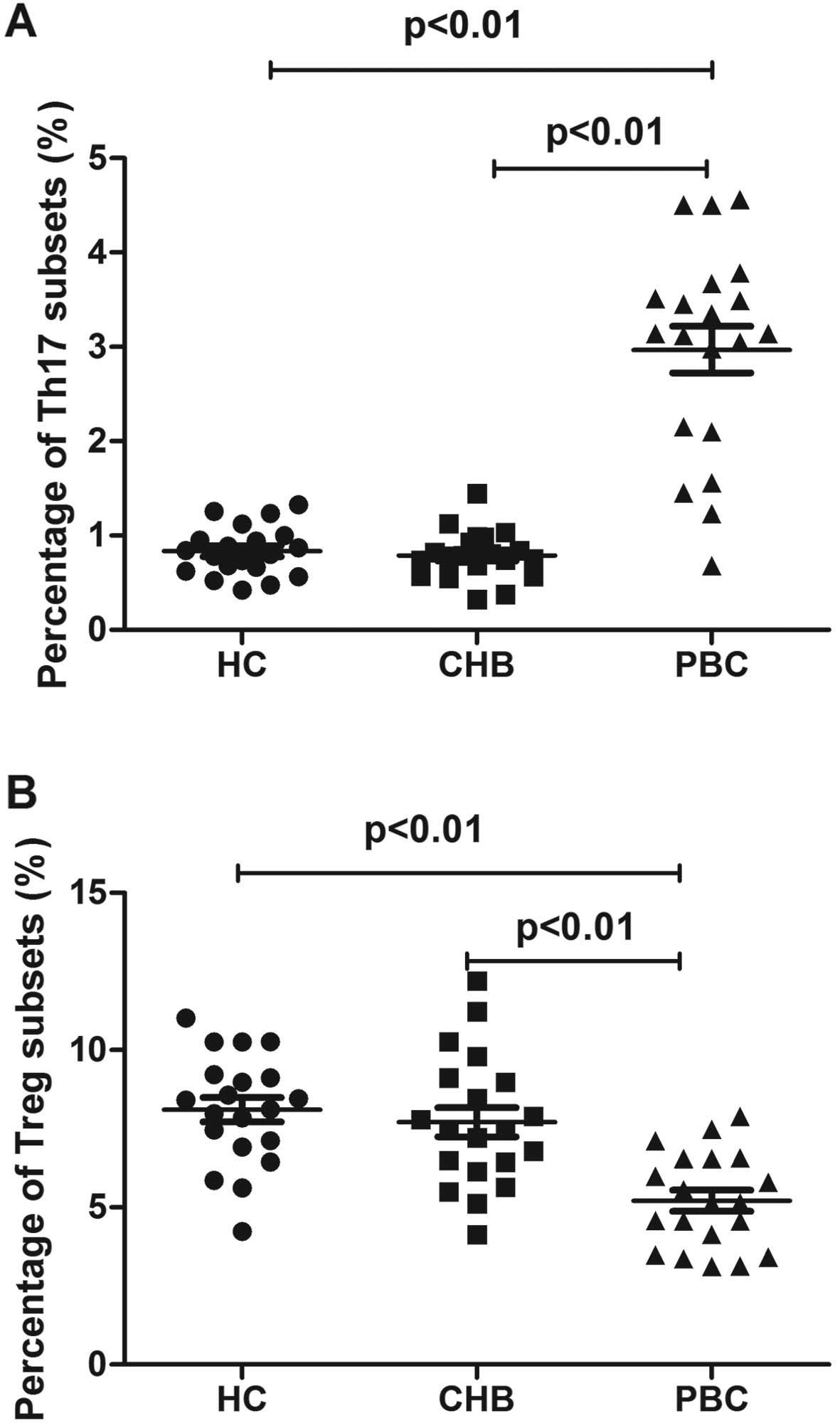
using flow cytometry. Th17 cell populations were increased in
patients with PBC (2.9±1.1%) compared with those in patients with
CHB (0.7±0.2%) and the healthy controls (0.8±0.2%) (Figs. 4A and 5A). The Treg cell population was
decreased in the patients with PBC (5.2±1.5%) vs. the patients with
CHB (7.7±2.1%) and healthy controls (8.1±1.7%) (Figs. 4B and 5B).
Correlation between miRNAs and T cell
subset frequencies
We analyzed the relationship between differentially
expressed miRNAs and the Th cell subset imbalance in the patients
with PBC using Spearman's correlation analyses. These analyses
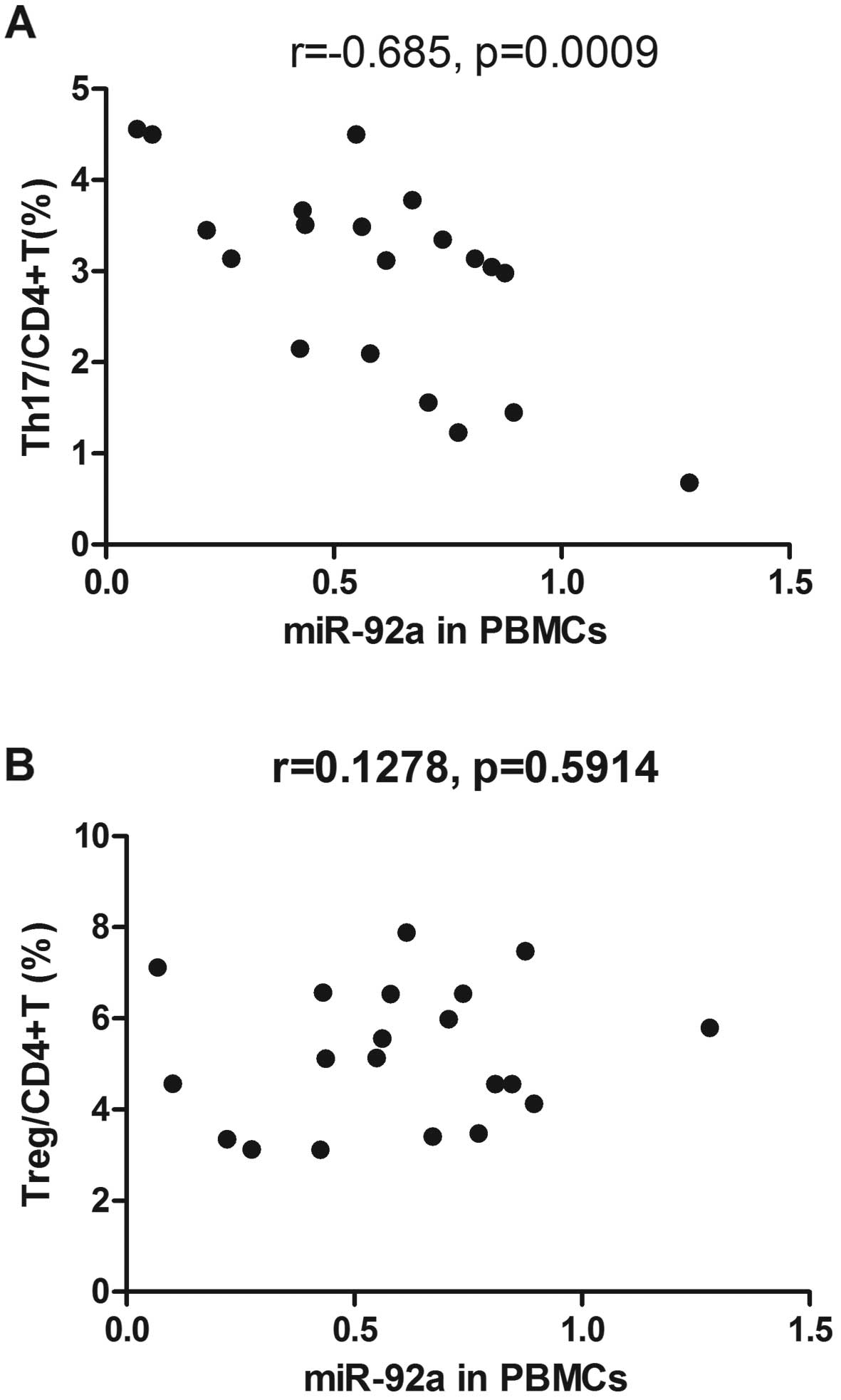
revealed that miR-92a expression in the PBMCs inversely correlated
with the Th17 cell population in the patients with PBC (Fig. 6A); however, there was no
correlation with the frequency of Treg cells (Fig. 6B). There was no correlation
between the expression of other miRNAs and the frequencies of Th
cell subsets (data not shown).
Co-expression of miR-92a and IL-17A in
PBMCs isolated from patients with PBC
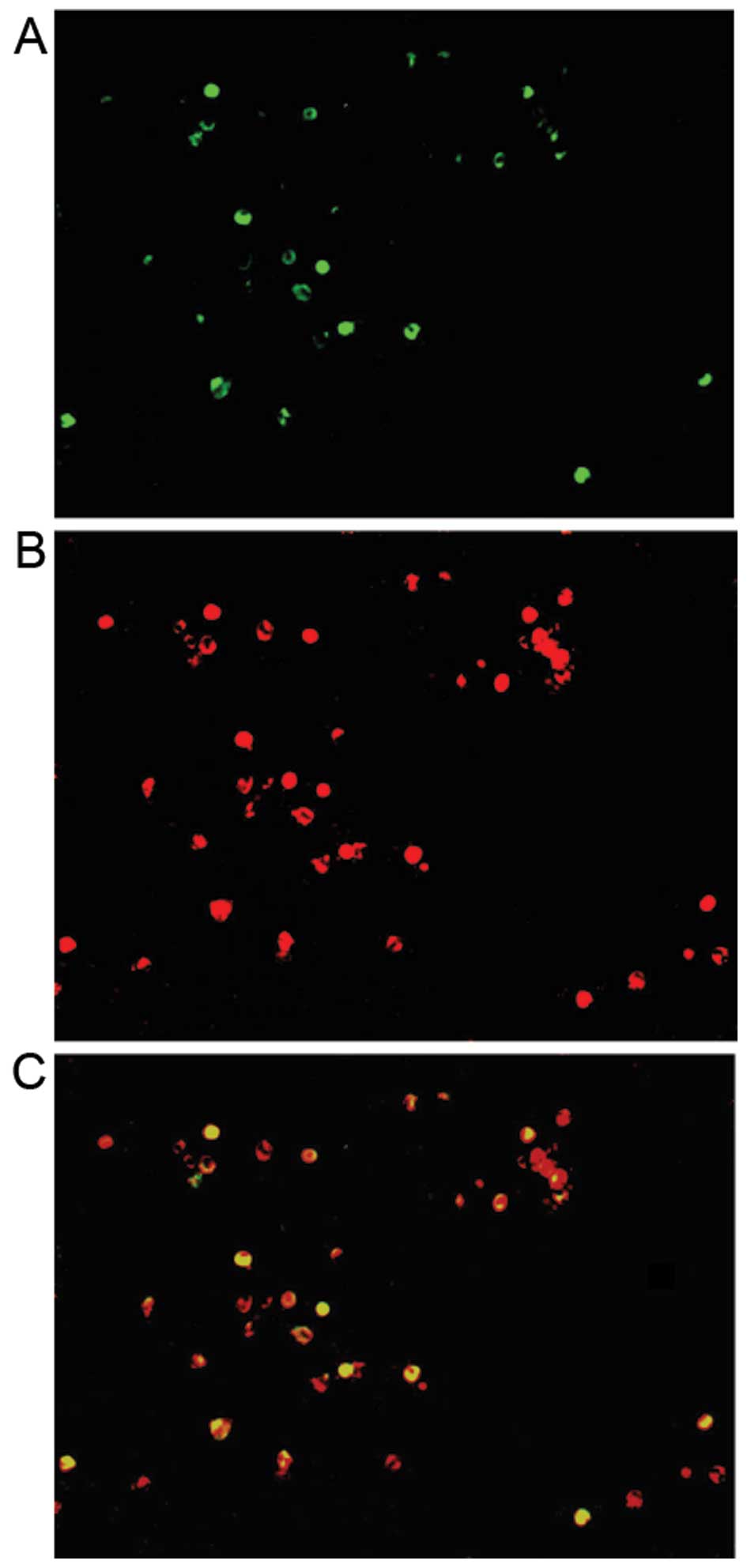
To determine whether miR-92a is involved in Th17
cell differentiation, we evaluated the expression of miR-92a in
PBMCs using sequential miR-92a FISH and IL-17A IHC. Double staining
revealed that both miR-92a and IL-17A were observed in a subset of
PBMCs from patients with PBC (Fig.
7C). Additionally, all IL-17A-positive cells (Fig. 7B) expressed miR-92a (Fig. 7A).
Discussion
Changes in miRNA expression have been reported in
several human diseases, including hepatocellular carcinoma (HCC)
and lung cancer (19–21). However, there is limited
information regarding the expression of miRNAs in PBC (22). In the present study, microarray
analysis was performed in order to screen the miRNA expression
profile in the plasma of patients with PBC. We identified 16 miRNAs
that were differentially expressed. To determine whether these
differentially expressed miRNAs are involved in the development of
PBC, we confirmed their expression in PBMCs and plasma from
patients with PBC or CHB as well as healthy controls using RT-qPCR.
Our results showed that miR-92a and miR-4516 were downregulated in
the plasma from patients with PBC compared with their expression in
healthy controls and patients with CHB, whereas miR-572 and miR-575
were upregulated in the plasma from patients with PBC. However, the
expression of other miRNAs was not significantly altered in the
plasma from patients with PBC. In PBMCs, miR-572 expression was
significantly increased in patients with PBC compared with that in
the healthy controls and patients with CHB. miR-575 was increased
in the patients with PBC compared with healthy controls; however,
there was no difference in expression compared with that in the
patients with CHB. miR-92a was significantly decreased in the
patients with PBC compared with the healthy controls and patients
with CHB. miR-4516 expression was unchanged in the PBMCs from
patients with PBC compared with the healthy controls and the
patients with CHB, which differed from the expression pattern in
plasma. In order to determine whether differentially expressed
miRNAs in plasma were derived from the immune system, correlation
analyses of miRNA expression in the plasma and PBMCs were then
performed. The results demonstrated that the expression of miR-572
and miR-92a in PBMCs positively correlated with the expression in
the plasma. However, there was no correlation between miR-575
levels in the plasma and the PBMCs. We hypothesized that miR-575
upregulation may be derived from the overactivity of immune cells
as well as from hepatocyte injury.
Immune cells, particularly CD4+ T cells,
play an important role in immune-mediated cholangitis in PBC
(15). Traditionally, based on
their cytokine production profile, CD4+ T cells are
divided into two subsets: Th1 and Th2. Th1 cells, characterized by
the production of IFN-γ, are responsible for immunity against
intracellular pathogens, whereas Th2 cells, characterized by IL-4,
IL-5 and IL-13 secretion, play important roles in clearing
extracellular pathogens and mediating allergic responses (23). Two additional subsets, Th17 and
Treg, have been classified (24,25). Th17 cells belong to the
pro-inflammatory Th cell subset, which induce tissue inflammation
through IL-17A secretion, rather than IFN-γ or IL-4. Treg cells
directly contact or secrete suppressive cytokines that suppress
inflammation (26,27). Each subset plays a unique role,
and the dysregulation of subset differentiation has been associated
with disease (28,29). An imbalance between Th17/Treg
cells has been reported in the progression of atherosclerosis
(30). In the present study, in
addition to observing the altered expression of miRNAs in PBMCs, we
also confirmed the frequency of T cell subsets in patients with
PBC. Our results showed that Th17 cells were upregulated and Treg
cells were down-regulated in patients with PBC, which is in
agreement with previous findings (18).
Mounting evidence suggests that miRNAs are important
modulators of Th cell differentiation and effector function. The
first study regarding miRNA-mediated regulation of Th cell
differentiation involved Dicer-deficient CD4+ T
cells, which exhibited increased differentiation into the Th1 cell
subset, with increased production of IFN-γ (31). Functional screens in
DGCR8-deficient CD4+ T cells showed that the high
expression of miR-29a in naive T cells inhibited Th1 cell
differentiation and IFN-γ expression (32). Several other miRNAs were also
reported to regulate the differentiation and function of Th1 cells
(33-35). In addition to Th1 cell regulation,
the miRNA-mediated regulation of other CD4+ T cells has
also been studied. In miR-155 knockout (miR-155-KO
or miR-155−/−) mice, both Th1 and Th17 cells were
defective (36). Furthermore, the
proportion and absolute number of Treg cells were also smaller in
miR-155-deficient mice (37). In
summary, these studies indicated that miRNAs are essential for the
differentiation of Th cell subsets. In the present study, we
demonstrated an imbalance of Th17/Treg cells in patients with PBC,
with an increased peripheral Th17 population and simultaneously
decreased Treg population in the same subjects. To identify whether
miR-572 and miR-92a were involved in the differentiation of Th
cells in patients with PBC, we performed correlation analyses in
order to examine the expression of miRNAs and the frequency of Th
cells. We found that miR-92a expression in the PBMCs inversely
correlated with the Th17 cell population in patients with PBC;
however, there was no correlation between miR-92a expression and
the frequency of Treg cells. There was no correlation between the
expression of other miRNAs and the frequencies of Th cell
subsets.
Th17 cells are a new subset of Th cells which have
been implicated in the etiology and pathology of many autoimmune
diseases, including psoriasis, multiple sclerosis, colitis,
rheumatoid arthritis (RA) and asthma (38). Previous research has reported that
miRNAs expressed in IL-17-producing T cells may regulate the
differentiation of Th17 cells. In patients with RA, miR-146a was
co-expressed with IL-17 in PBMCs isolated from patients with
early-stage disease, and was associated with Th17 differentiation
(39). In addition to direct
regulation by miRNAs, Th17 cell differentiation may be indirectly
affected by miRNAs that target genes in immune cells other than
CD4+ T cells. For example, miR-155 regulates Th17 cell
differentiation by modulating Th17 cell-polarizing cytokine
secretion by dendritic cells (40). To determine whether miR-92a plays
a T cell-intrinsic role in Th17 cell differentiation or an indirect
role in the regulation of other genes in immune cells, we performed
sequential miR-92a FISH and IL-17A IHC using TSA Plus
Direct-Cyanine 3 and TSA Plus Direct-Green. We found that miR-92a
was co-expressed with IL-17A in the PBMCs, suggesting that miR-92a
may directly regulate Th17 cells in patients with PBC. However,
more studies are warranted in order to clarify whether alternative
transcripts are regulated by miR-92a during the pathogenesis of
PBC.
Taken together, these findings demonstrate that
miR-92a was downregulated in patients with PBC. There was an
inverse correlation between miR-92a expression and the Th17 cell
population, and miR-92a was co-expressed with IL-17A in PBMCs
isolated from patients with PBC. Our study suggests that miR-92a
may be involved in the imbalance of Th cell subsets, particularly
the upregulation of Th17 cells, which may play an important role in
the development of PBC. However, due to limitations in the number
of subjects and methods used in the present study, the association
between miR-92a expression and Th cell differentiation in patients
with PBC was not completely resolved. Further studies are warranted
to elucidate the precise relationship between miR-92a and Th cell
differentiation, as well as the mechanisms responsible for the
miR-92a-mediated regulation of Th cell differentiation in patients
with PBC.
Acknowledgments
The present study was supported by the Health Bureau
of Shanghai. The funders had no role in study design, data
collection and analysis, decision to publish, or preparation of the
manuscript.
Abbreviations:
|
PBC
|
primary biliary cirrhosis
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
PBMCs
|
peripheral blood mononuclear cells
|
|
CHB
|
chronic hepatitis B
|
|
FISH
|
fluorescence in situ
hybridization
|
|
AMA
|
anti-mitochondrial antibodies
|
|
IFN
|
interferon
|
|
PE
|
phycoerythrin
|
|
PBS
|
phosphate-buffered saline
|
|
ANOVA
|
analysis of variance
|
|
HCC
|
hepatocellular carcinoma
|
|
Th1
|
T helper type 1
|
|
Th2
|
T helper type 2
|
|
Th17
|
subset of IL-17-producing T helper
cells
|
|
Treg
|
regulatory T cells
|
References
|
1
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
miRBase: the microRNA database. http://www.mirbase.org/.
Accessed, 2013.
|
|
3
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lindsay MA: microRNAs and the immune
response. Trends Immunol. 29:343–351. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maes OC, An J, Sarojini H and Wang E:
Murine microRNAs implicated in liver functions and aging process.
Mech Ageing Dev. 129:534–541. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pauley KM, Cha S and Chan EK: MicroRNA in
autoimmunity and autoimmune diseases. J Autoimmun. 32:189–194.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Furer V, Greenberg JD, Attur M, Abramson
SB and Pillinger MH: The role of microRNA in rheumatoid arthritis
and other autoimmune diseases. Clin Immunol. 136:1–15. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iborra M, Bernuzzi F, Invernizzi P and
Danese S: MicroRNAs in autoimmunity and inflammatory bowel disease:
Crucial regulators in immune response. Autoimmun Rev. 11:305–314.
2012. View Article : Google Scholar
|
|
9
|
Pauley KM, Satoh M, Chan AL, Bubb MR,
Reeves WH and Chan EK: Upregulated miR-146a expression in
peripheral blood mononuclear cells from rheumatoid arthritis
patients. Arthritis Res Ther. 10:R1012008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y,
Huang X, Zhou H, de Vries N, Tak PP, et al: MicroRNA-146A
contributes to abnormal activation of the type I interferon pathway
in human lupus by targeting the key signaling proteins. Arthritis
Rheum. 60:1065–1075. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Du C, Liu C, Kang J, Zhao G, Ye Z, Huang
S, Li Z, Wu Z and Pei G: MicroRNA miR-326 regulates TH-17
differentiation and is associated with the pathogenesis of multiple
sclerosis. Nat Immunol. 10:1252–1259. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nakanuma Y and Ohta G: Histometric and
serial section observations of the intrahepatic bile ducts in
primary biliary cirrhosis. Gastroenterology. 76:1326–1332.
1979.PubMed/NCBI
|
|
13
|
Kaplan MM and Gershwin ME: Primary biliary
cirrhosis. N Engl J Med. 353:1261–1273. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lindor K: Ursodeoxycholic acid for the
treatment of primary biliary cirrhosis. N Engl J Med.
357:1524–1529. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jones DE: Pathogenesis of primary biliary
cirrhosis. Postgrad Med J. 84:23–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nagano T, Yamamoto K, Matsumoto S, Okamoto
R, Tagashira M, Ibuki N, Matsumura S, Yabushita K, Okano N and
Tsuji T: Cytokine profile in the liver of primary biliary
cirrhosis. J Clin Immunol. 19:422–427. 1999. View Article : Google Scholar
|
|
17
|
Selmi C, Ichiki Y, Invernizzi P, Podda M
and Gershwin ME: The enigma of primary biliary cirrhosis. Clin Rev
Allergy Immunol. 28:73–81. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rong G, Zhou Y, Xiong Y, Zhou L, Geng H,
Jiang T, Zhu Y, Lu H, Zhang S, Wang P, et al: Imbalance between T
helper type 17 and T regulatory cells in patients with primary
biliary cirrhosis: the serum cytokine profile and peripheral cell
population. Clin Exp Immunol. 156:217–225. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang J, Zhao H, Xin Y and Fan L:
MicroRNA-198 inhibits proliferation and induces apoptosis of lung
cancer cells via targeting FGFR1. J Cell Biochem. 115:987–995.
2014. View Article : Google Scholar
|
|
20
|
Hu QY, Jiang H, Su J and Jia YQ: MicroRNAs
as biomarkers for hepatocellular carcinoma: a diagnostic
meta-analysis. Clin Lab. 59:1113–1120. 2013.PubMed/NCBI
|
|
21
|
Spoerl D, Duroux-Richard I, Louis-Plence P
and Jorgensen C: The role of miR-155 in regulatory T cells and
rheumatoid arthritis. Clin Immunol. 148:56–65. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qian C, Wang HZ, Fan HJ, Gu ML, Ren CL,
Deng AM and Zhong RQ: MicroRNA profiling in T cells of peripheral
blood mononuclear cell from patients with primary biliary
cirrhosis. Zhonghua Yi Xue Za Zhi. 92:2265–2267. 2012.In Chinese.
PubMed/NCBI
|
|
23
|
Liew FY: T(H)1 and T(H)2 cells: a
historical perspective. Nat Rev Immunol. 2:55–60. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Costantino CM, Baecher-Allan CM and Hafler
DA: Human regulatory T cells and autoimmunity. Eur J Immunol.
38:921–924. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ouyang W, Kolls JK and Zheng Y: The
biological functions of T helper 17 cell effector cytokines in
inflammation. Immunity. 28:454–467. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Smith E, Prasad KM, Butcher M, Dobrian A,
Kolls JK, Ley K and Galkina E: Blockade of interleukin-17A results
in reduced atherosclerosis in apolipoprotein E-deficient mice.
Circulation. 121:1746–1755. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Taleb S, Tedgui A and Mallat Z: Adaptive T
cell immune responses and atherogenesis. Curr Opin Pharmacol.
10:197–202. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Becker H, Langrock A and Federlin K:
Imbalance of CD4+ lymphocyte subsets in patients with
mixed connective tissue disease. Clin Exp Immunol. 88:91–95. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen DY, Lan JL, Lin FJ, Hsieh TY and Wen
MC: Predominance of Th1 cytokine in peripheral blood and
pathological tissues of patients with active untreated adult onset
Still's disease. Ann Rheum Dis. 63:1300–1306. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xie JJ, Wang J, Tang TT, Chen J, Gao XL,
Yuan J, Zhou ZH, Liao MY, Yao R, Yu X, et al: The Th17/Treg
functional imbalance during atherogenesis in ApoE(−/−)
mice. Cytokine. 49:185–193. 2010. View Article : Google Scholar
|
|
31
|
Chong MM, Rasmussen JP, Rudensky AY and
Littman DR: The RNAseIII enzyme Drosha is critical in T cells for
preventing lethal inflammatory disease. J Exp Med. 205:2005–2017.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Steiner DF, Thomas MF, Hu JK, Yang Z,
Babiarz JE, Allen CD, Matloubian M, Blelloch R and Ansel KM:
MicroRNA-29 regulates T-box transcription factors and interferon-γ
production in helper T cells. Immunity. 35:169–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang L, Boldin MP, Yu Y, Liu CS, Ea CK,
Ramakrishnan P, Taganov KD, Zhao JL and Baltimore D: miR-146a
controls the resolution of T cell responses in mice. J Exp Med.
209:1655–1670. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huffaker TB, Hu R, Runtsch MC, Bake E,
Chen X, Zhao J, Round JL, Baltimore D and O'Connell RM: Epistasis
between microRNAs 155 and 146a during T cell-mediated antitumor
immunity. Cell Reports. 2:1697–1709. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Loeb GB, Khan AA, Canner D, Hiatt JB,
Shendure J, Darnell RB, Leslie CS and Rudensky AY:
Transcriptome-wide miR-155 binding map reveals widespread
noncanonical microRNA targeting. Mol Cell. 48:760–770. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
O'Connell RM, Kahn D, Gibson WS, Round JL,
Scholz RL, Chaudhuri AA, Kahn ME, Rao DS and Baltimore D:
MicroRNA-155 promotes autoimmune inflammation by enhancing
inflammatory T cell development. Immunity. 33:607–619. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu LF, Thai TH, Calado DP, Chaudhry A,
Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K and
Rudensky AY: Foxp3-dependent microRNA155 confers competitive
fitness to regulatory T cells by targeting SOCS1 protein. Immunity.
30:80–91. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Weaver CT, Elson CO, Fouser LA and Kolls
JK: The Th17 pathway and inflammatory diseases of the intestines,
lungs, and skin. Annu Rev Pathol. 8:477–512. 2013. View Article : Google Scholar
|
|
39
|
Niimoto T, Nakasa T, Ishikawa M, Okuhara
A, Izumi B, Deie M, Suzuki O, Adachi N and Ochi M: MicroRNA-146a
expresses in interleukin-17 producing T cells in rheumatoid
arthritis patients. BMC Musculoskelet Disord. 11:209–221. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Murugaiyan G, Beynon V, Mittal A, Joller N
and Weiner HL: Silencing microRNA-155 ameliorates experimental
autoimmune encephalomyelitis. J Immunol. 187:2213–2221. 2011.
View Article : Google Scholar : PubMed/NCBI
|