Introduction
Glioma is the most common primary central nervous
system tumor, which accounts for approximately 50% of all
neuroepithelial tumors (1). The
5-year survival rate of patients with glioma is generally <50%.
The median survival time is <1 year, and the majority of
patients with this tumor succumb to the disease within 2 years
following diagnosis (2,3), particularly those with glioblastoma
multiforme (GBM), which is a highly malignant tumor with a high
post-operative recurrence rate (4). According to the statistics of WHO in
1998, malignant glioma is the second leading cause of death for all
patients, the second leading cause of death for tumor patients aged
<34 years, and the third leading cause of death for tumor
patients between the ages of 35–54 years (5). Current opinions suggest that the
development and progression of glioma is a complex, multistep
process with multiple genes involved.
The activation of multiple cell signaling pathways
has been demonstrated to be involved in the development and
progression of glioma, among which the vascular endothelial growth
factor (VEGF) and Ras pathways have been widely considered to be
involved (6–8). In recent years, some information has
already been obtained regarding the pathogenesis and treatment of
malignant glioma (9). However,
the mechanisms involved in the invasive growth and infinite
proliferation remain unclear. Further investigations of the
pathogenesis of malignant glioma and search for novel treatment
methods are of great importance for reducing the mortality
associated with malignant glioma.
Over the past decade, the efforts in the development
of more effective, novel gene therapies for the treatment of GBM
have resulted in some pre-clinical information and in many
promising gene therapies. The rat hyperplasia suppressor gene
(rHSG), also known as the rat mitofusin-2 (rMfn2) gene, is a newly
discovered gene. rHSG was found firstly in vascular smooth muscle
cells (VSMCs) in hypertensive rats. The expression of rHSG has been
shown to be significantly downregulated in the VSMCs of
hypertensive rats compared with normal rats (10). It has been demonstrated that rHSG
regulates the apoptosis of VSMCs via the mitochondrial apoptotic
pathway (11). In vitro
and in vivo studies have also demonstrated that the abnormal
cell apoptosis and invasion during tumorigenesis is closely
associated with the activation of extracellular signal-regulated
kinase (Erk) (12–15). Therefore, inhibiting the activity
of Erk may effectively inhibit tumor invasion and may promote cell
apoptosis (16–19). A previous study demonstrated the
evident disruption of the constitutively activated phosphoinositide
3-kinase (PI3K)/Akt signaling pathway in malignant glioma (20). However, to the best of our
knowledge, no study on the effects of rHSG on the
PKCa/mitogen-activated protein kinase (MAPK) and PI3K/Akt pathways
in glioma has been published to date.
Our previous in vitro study demonstrated that
the overexpression of rHSG suppressed the proliferation of rat
glioma cells, based on the findings that the protein expression
level of rHSG was higher in the C6 cells in the group infected with
Adv-rHSG-GFP; cell cycle was arrested at the G0/G1 phase, and rat
glioma cell proliferation was markedly inhibited; the expression of
p27Kip1 and p21Cip1 was increased, while the
expression of PCNA was decreased (21).
In the present study, we investigated the effects of
rHSG on the apoptosis of C6 rat glioma cells and the roles of the
protein kinase C (PKC)α/MAPK and PI3K/Akt pathways.
Materials and methods
Materials
C6 cells (Type Culture Collection of the Chinese
Academy of Sciences, Shanghai, China) and purified Adv-rHSG-GFP
virus-containing solution (titer, 1×1011 pfu/ml) were
preserved in our laboratory. Purified Adv-GFF virus-containing
solution (titer, 1×1011 pfu/ml) was purchased from
Baosai Biological Technology Co., Ltd. (Beijing, China). Fetal
bovine serum (FBS), Dulbecco's modified Eagle's medium (DMEM; high
glucose), trypsin (0.25%), penicillin and streptomycin were
purchased from HyClone (Logan, UT, USA). Insulin-like growth factor
(IGF)-1 was purchased from Sigma-Aldrich (St. Louis, MO, USA).
Mouse-anti-rat rHSG monoclonal antibody (Cat. no. ab56889) was
purchased from Abcam (Cambridge, MA, USA); rabbit-anti-rat
monoclonal antibodies to poly(ADP-ribose) polymerase (PARP; Cat.
no. 9542S), caspase-3 (Cat. no. 9664S), phosphorylated (p-)Akt
(Cat. no. 9271S), Akt (Cat. no. 9272S), p-Erk1/2 (Cat. no. 9101S)
and Erk1/2 (Cat. no. 9102S) were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA). Mouse-anti-rat β-actin
polyclonal (Cat. no. TA-09) and goat-anti-mouse IgG (Cat. no.
ZB-2305) antibodies, the immunohistochemical staining kits and DAB
solution were purchased from Zhongshan Jinqiao Biotechnology Co.,
Inc. (Beijing, China). The Hoechst 33342/PI double staining kits,
comet assay kits, total protein extraction kits, and BCA protein
quantification kits were purchased from Kaiji Biotechnology Co.,
Ltd. (Nanjing, China). ECL luminol solution was purchased from
Pierce Biotechnology, Inc. (Rockford, IL, USA).
Cell culture
C6 cells were thawed, and DMEM culture medium
containing 10% FBS was added, and the cells were then incubated at
37°C, 5% CO2 (v/v) in an incubator with constant
humidity. Following adhesion, DMEM medium containing 0.2% FBS was
used to synchronize the cells for 24 h. Afterwards, adenovirus with
optimal multiplicity of infection (MOI) was used to transfect the
cells (PBS was used for the PBS control group) for 4 h, and the
culture medium was then discarded; then the cells were continuously
cultured with fresh full culture medium until use in the subsequent
experiments.
Transfection of C6 cells with
adenovirus
The C6 cells were divided into 3 groups, as follows:
the PBS control, Adv-rHSG-GFP and Adv-GFP groups. The cells were
seeded into a 6-well plate at a density of 1×105
cells/well, and then Adv-rHSG-GFP or Adv-GFP at an MOI of 0, 10,
50, 80 and 100 was added to transfect the cells. An inverted
fluorescence microscope (Olympus TL-4; Olympus Optical Co., Ltd.,,
Tokyo, Japan) and a flow cytometer (BD FACSCalibur; BD Biosciences,
San Jose, CA, USA) were used to measure the MOI at the optimal
transfection efficiency 24 h later. Green-stained cells were
considered positive cells that had been successfully transfected.
The transfection efficiency was considered high if 90% of the cells
exhibited green fluorescence. The cells were collected at 24, 48,
72 and 96 h post-transfection for use in apoptosis assay.
Measurement of cellular rHSG expression
by immunocytochemistry
The C6 cells were seeded into a 24-well plate
pre-coated with coverslips at a density of 5×104
cells/well and transfection was carried out following culture for
24 h. The cells were collected at different time points (24, 48, 72
and 96 h), and immunocytochemistry was then performed using
immunohistochemical staining kits according to the manufacturer's
instructions. Following visualization with DAB, contrast staining
with hematoxylin, and dehydration with gradient ethanol, the slides
were dried and mounted. The C6 cells with a brown-yellow or dark
brown stained cytoplasm were considered as rHSG-positive. Upright
microscope (Olympus BH2-RFCA) was used to obtain images at a
magnification of ×400. The images were imported to an image
analysis system (Image-Pro Plus 6.0) to calculate the mean optical
density of the positive areas on each slide (IOD/area), which was
used as the index to reflect the positive strength of rHSG.
Determination of IGF-1 concentration and
stimulation of C6 cells
The C6 cells were seeded into 60-mm culture dishes
at a density of 1×106 cells/well, and 10% DMEM medium
containing 0, 1, 5, 8 and 10 ng/ml IGF-1 was used to stimulate the
C6 cells for 15 min following culture for 24 h. Western blot
analysis was applied for analyzing the expression level of p-Akt
and p-Erk1/2 in order to determine the optimal concentration. By
using the same method described above, the cells were stimulated
with IGF-1 at the optimal concentration for 0, 5, 15, 30 and 60 min
to determine the optimal stimulating time. Subsequently, the
optimal concentration and treatment time for stimulation with IGF-1
would be applied to the relevant experiment.
Measurement of the protein expression of
rHSG, PARP, cleaved caspase-3, p-Akt and p-Erk1/2 by western blot
analysis
The C6 cells were seeded into 60-mm culture dishes
at a density of 1×106 cells/well, and treatments were
then carried out. The cells were collected at different time points
(24, 48, 72 and 96 h). Proteins were extracted from the cells using
the total protein extraction kits according to the manufacturer's
instructions, and the BCA method was used to determine the protein
concentration. The appropriate volume of the protein sample was
added to loading buffer, after boiling at 100°C for 6 min to allow
denaturation. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was
then performed, and the proteins were then transferred to a
polyvinylidene fluoride (PVDF) membrane (Millipore Co., Millipore,
Billerica, MA, USA) and blocked with 5% skim milk powder. Primary
antibody was then added, followed by incubation at 4°C overnight.
The primary antibodies used in the present study include the
antibodies against rHSG (1:400), PARP (1:1,000), caspase-3
(1:1,000), p-Akt (1:500), p-Erk1/2 (1:500) and β-actin (1:1,000).
The membrane was then washed 3 times with TBST, after which the
corresponding secondary antibodies were added. Subsequently, the
chemiluminescence method was used to evaluate the relative protein
level of rHSG.
Measurement of C6 cell apoptosis buy
Hoechst 33342/PI double staining
The C6 cells were seeded into a culture dish with a
glass bottom that is specifically used for a laser scanning
confocal microscope at a density of 1×105 cells/well.
Following treatment, the cells in each group were washed with PBS
once, and then 5 µl of Hoechst 33342 was added and mixed
gently. The cells were then placed in the dark at room temperature
for 10 to 15 min. PBS was then used to wash the cells once. PI
solution (5 µl) was added after washing the cells with PBS
for an additional 2 times, followed by gentle mixing. The cells in
the respective groups were incubated in the dark at room
temperature for a further 10–15 min. A laser scanning confocal
microscope (LSCM; FV1000 IX81; Olympus) was used to observe the
cells at a magnification of ×400 immediately after another wash
with PBS, and bi-channel images were then obtained with Hoechst
(excitation wavelength of 350 nm) and PI staining (excitation
wavelength of 488 nm).
Determination of DNA damage in C6 cells
induced by rHSG by comet assay
The C6 cells were collected at different time points
(24, 48, 72 and 96 h) and were resuspended with PBS to obtain a
density of 1×106 cells/ml. The slides were covered with
3 layers of agarose gel, the upper and lower layers with 0.5%
normal melting point (NMP) agarose and the middle layer with 0.7%
low melting point (LMP) agarose with 104 C6 cells. The
slides with agarose gel were placed in cell lysis solution at 4°C
for 24 h. They were then rinsed with PBS and immersed in fresh
alkaline electrophoresis buffer at room temperature for 1 h to
allow DNA unwinding. Electrophoresis was carried out for 20 min at
25 V, and the slides were then placed in a dish; 0.4 mm/l Tris-HCl
solution (PH 7.5) was added for 10 min. Each slide was stained with
20 µl PI and covered with a coverslip. The slides were
placed in the dark for 10 min to allow staining, and an inverted
fluorescence microscope (Olympus TL-4; Olympus) was then used for
observation and imaging. The cells with comet tails formed by the
migration of DNA from the nuclei were considered to have DNA
damage.
Statistical analysis
SPSS 19.0 statistical software was used for the
statistical analysis. All the experiments were repeated at least 3
times. Quantitative data are expressed as the means ± standard
deviation (SD) (n≥3 experiments); one-way analysis of variance
(ANOVA) was used for comparing the means among different groups. A
value of P<0.05 was considered to indicate a statistically
significant difference.
Results
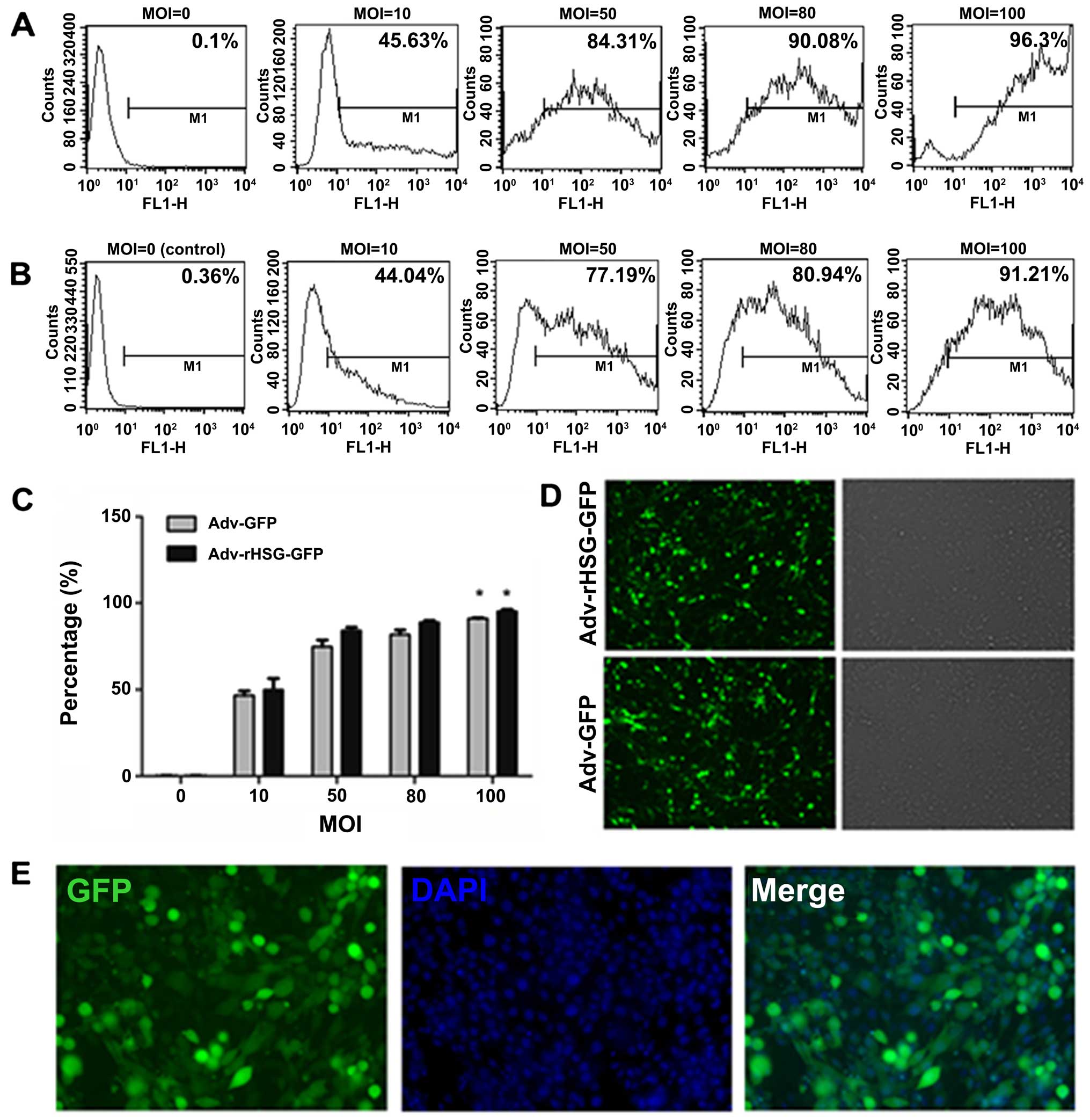
Effecient transfection of C6 cells with
adenovirus
An inverted fluorescence microscope was used to
observe the C6 cells at 24 h post-transfection, and it was found
that the percentage of positive cells with GFP expression increased
with the increasing MOI (Fig. 1C and
D). After obtaining the results using the microscope, the cells
were collected and measured using a flow cytometer. The results
revealed that when the MOI was 100, the transfection efficiency was
>90% (Fig. 1A and B). The C6
cells that were transfected with Adv-rHSG-GFP at an MOI of 100 were
transiently fixed with paraformaldehyde. DAPI was used to stain the
nuclei, and a fluorescence microscope was then used to estimate the
transfection efficiency (Fig.
1E). Therefore, the following experiments were carried out on
the cells transfected with the virus at an MOI of 100.
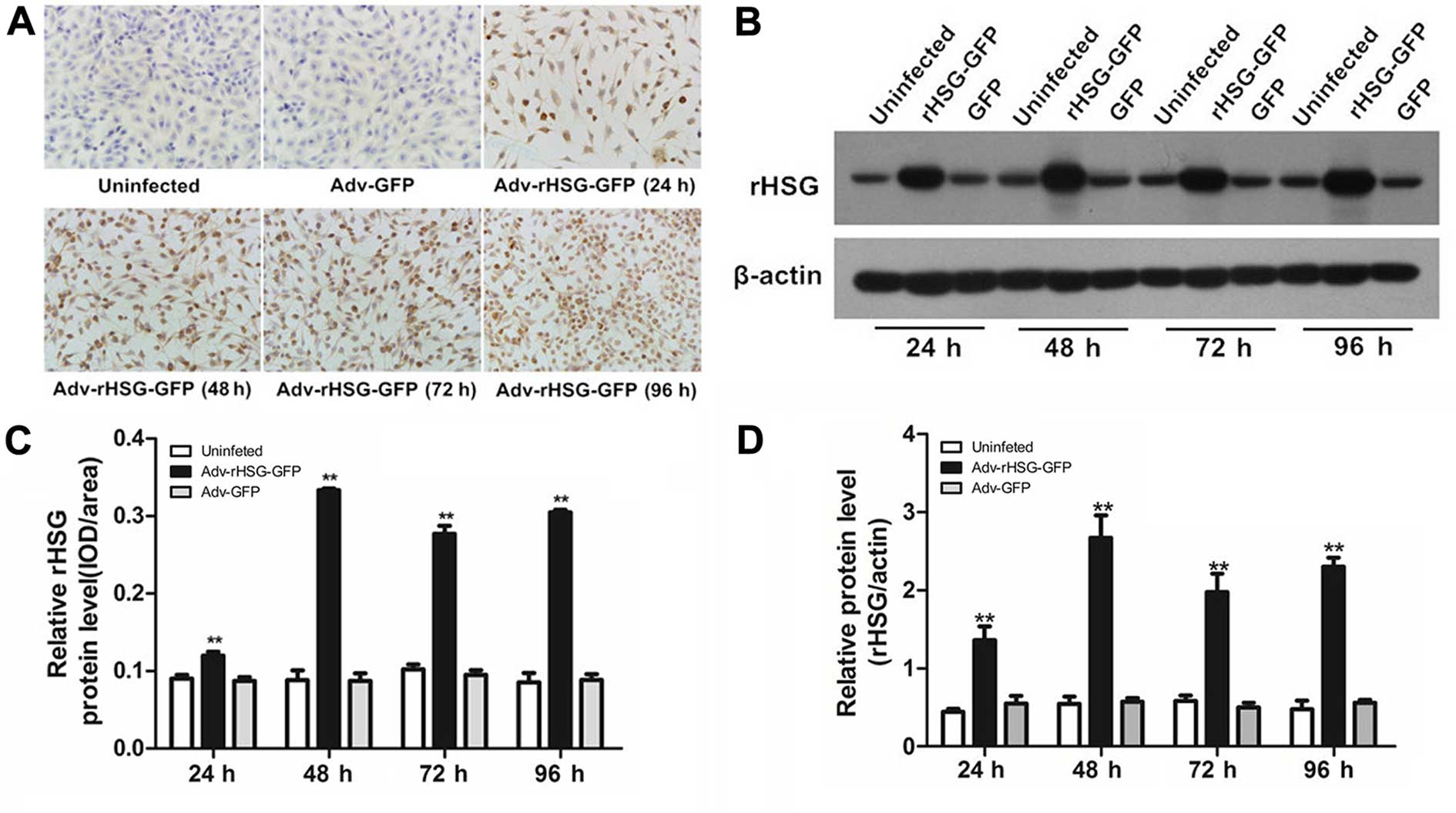
High protein expression level of rHSG in
C6 cells
Immunocytochemistry revealed that rHSG protein was
expressed in the cytoplasm of the C6 cells, which was brown-yellow
in color (Fig. 2A). The protein
expression of rHSG was significantly higher in the Adv-rHSG-GFP
group compared with the control group or the Adv-GFP group
(P<0.01, n=6 experiments; Fig.
2C). The results of western blot analysis indicated the
rHSG-specific band at each time point (Fig. 2B); the expression of rHSG was
significantly higher in the Adv-rHSG-GFP group compared with the
Adv-GFP and control groups (P<0.01, n=3; Fig. 2D). These results demonstrated that
the transfection of C6 cells with Adv-rHSG-GFP resulted in a high
protein expression level of rHSG.
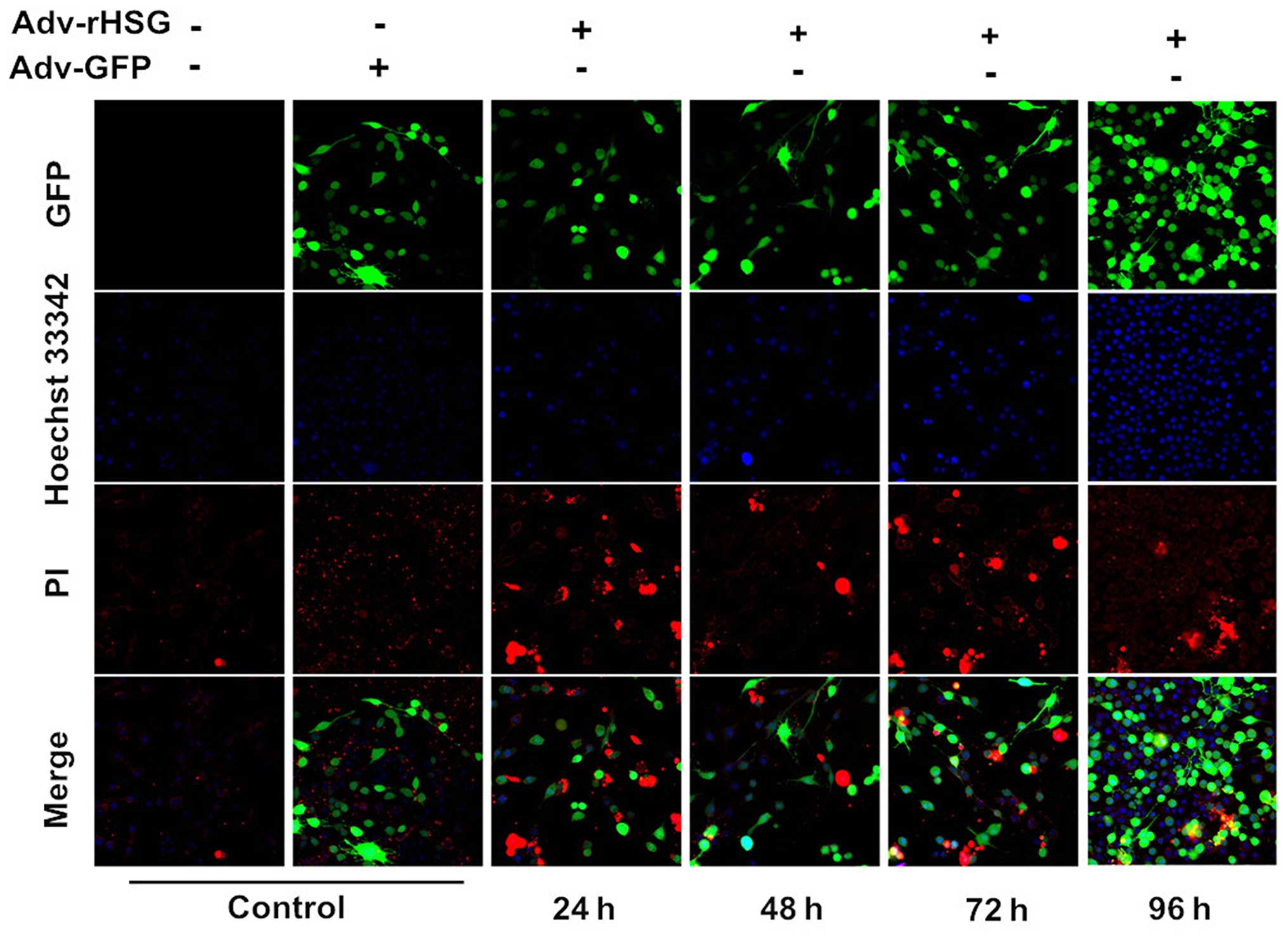
Apoptosis of the C6 cells induced by
rHSG
A laser scanning confocal microscope was used to
observe the cells, and it was found that the C6 cells in the
Adv-rHSG-GFP group exhibited distinct apoptosis-defining
morphological changes at 24–96 h post-transfection. The cells
exhibited chromosome condensation, karyopyknosis and nuclear
fragmentation. In addition, the nuclei of the cells were deeply
stained and presented with a brilliant blue color. The chromatin
gradually aggregated adjacent to the nuclear membrane in the shape
of a crescent moon. However, for the cells in the control and
Adv-GFP groups, the nuclei exhibited homogenously distributed weak
dark-blue fluorescence, and no sign of apoptosis was observed
(Fig. 3).
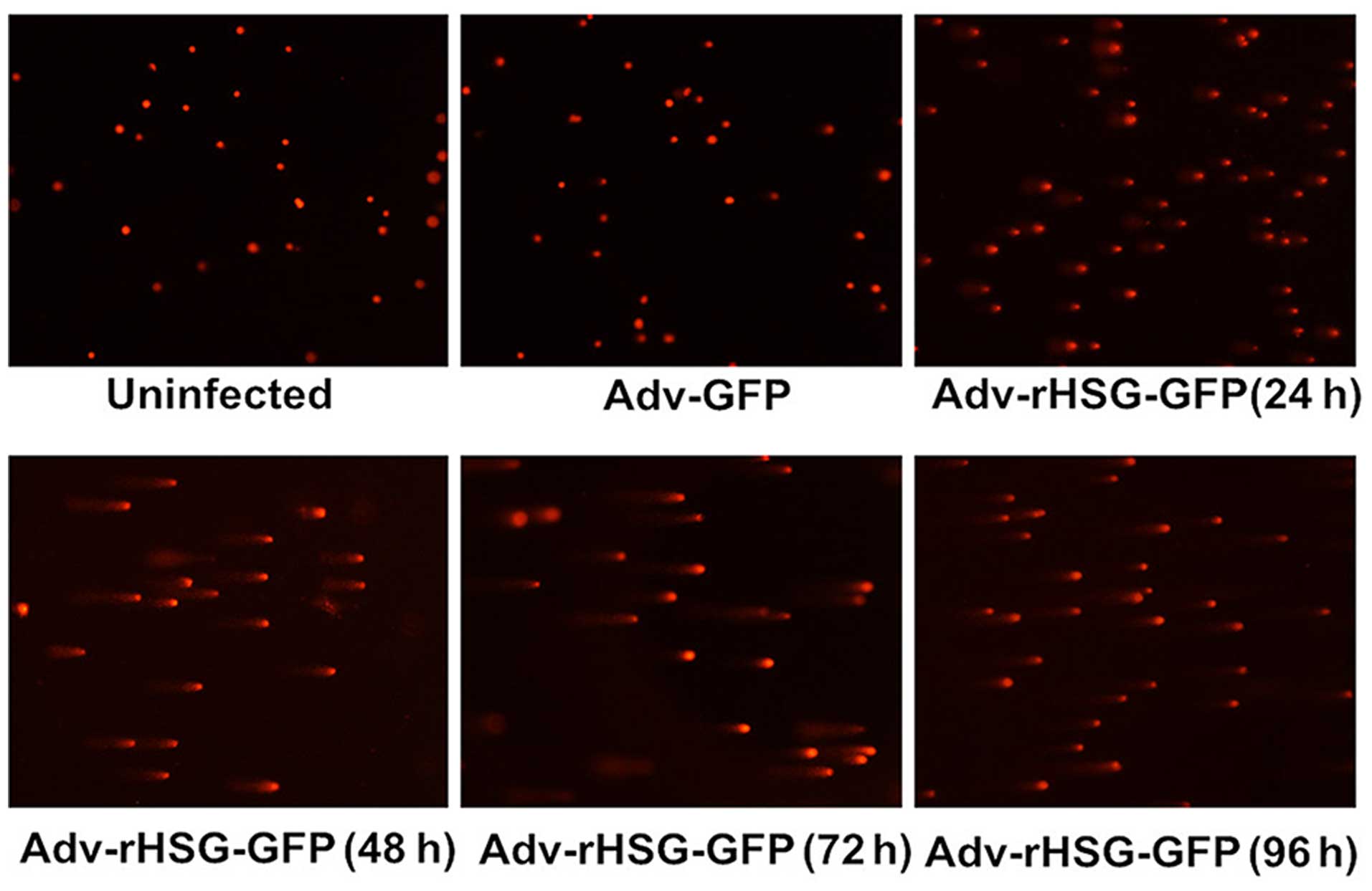
The C6 cells in the Adv-rHSG-GFP group were
collected at 24–96 h post-transfection, and the results of comet
assay indicated that the DNA of the cells had migrated from the
nuclei and formed evident comet tails, suggesting the presence of
DNA damage. However, no evident comet tails were observed in the
control group or the Adv-GFP group (Fig. 4).
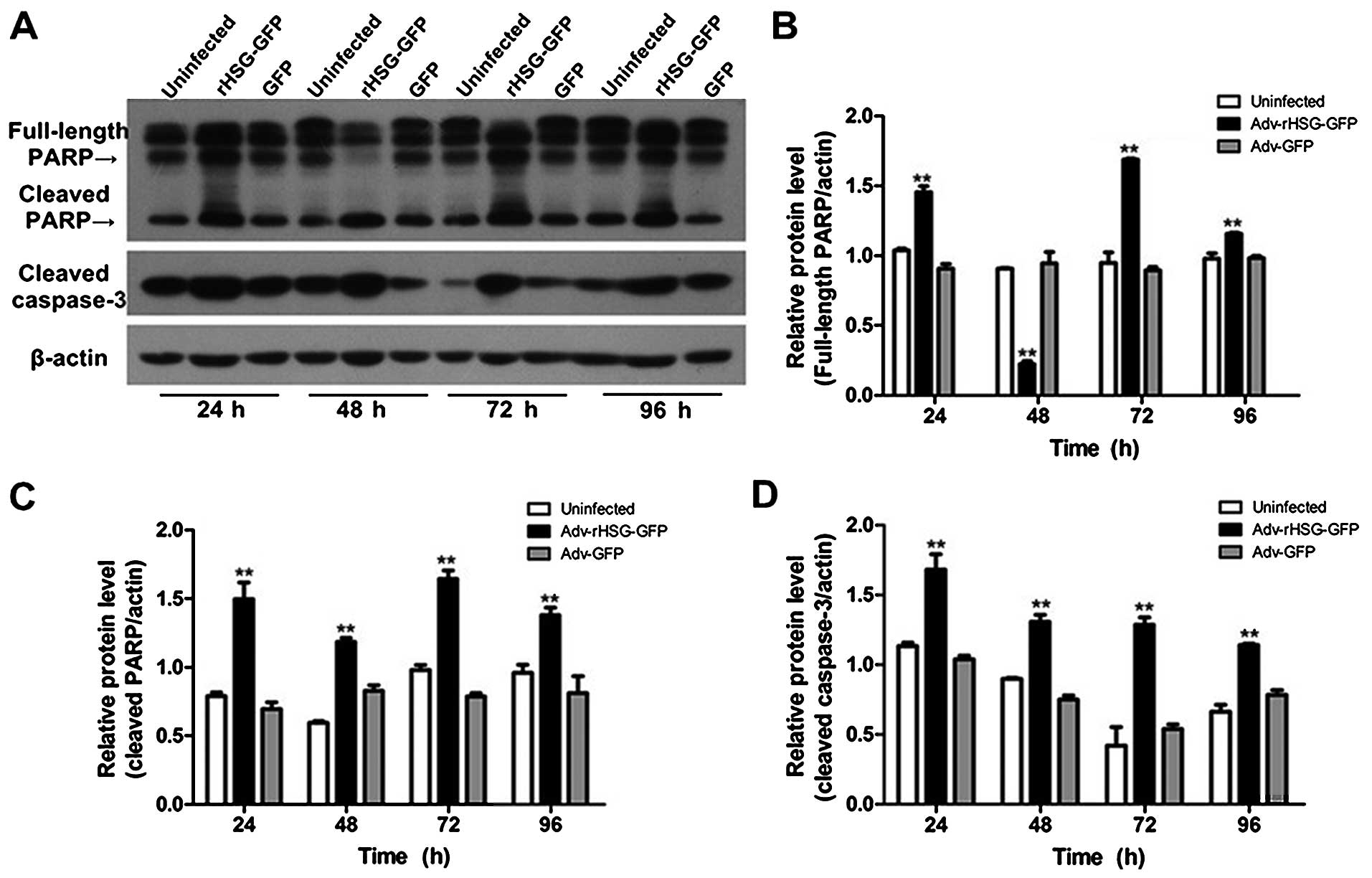
The results of western blot analysis also revealed
that the protein expression of full-length PARP increased
significantly at 24, 72 and 96 h post-transfection in the
Adv-rHSG-GFP group (P<0.01, n=3); however, the expression level
decreased significantly at 48 h (P<0.01, n=3; Fig. 5A and B). The protein expression of
cleaved caspase-3 in the Adv-rHSG-GFP group was significantly
higher than that in the control group or Adv-GFP group (P<0.01,
n=3; Fig. 5C and D); however, no
significant differences were observed between the control group and
Adv-GFP group (P>0.05) (Fig.
5).
rHSG promotes C6 cell apoptosis through
the PI3K/Akt and Ras-Raf-MAPK pathways
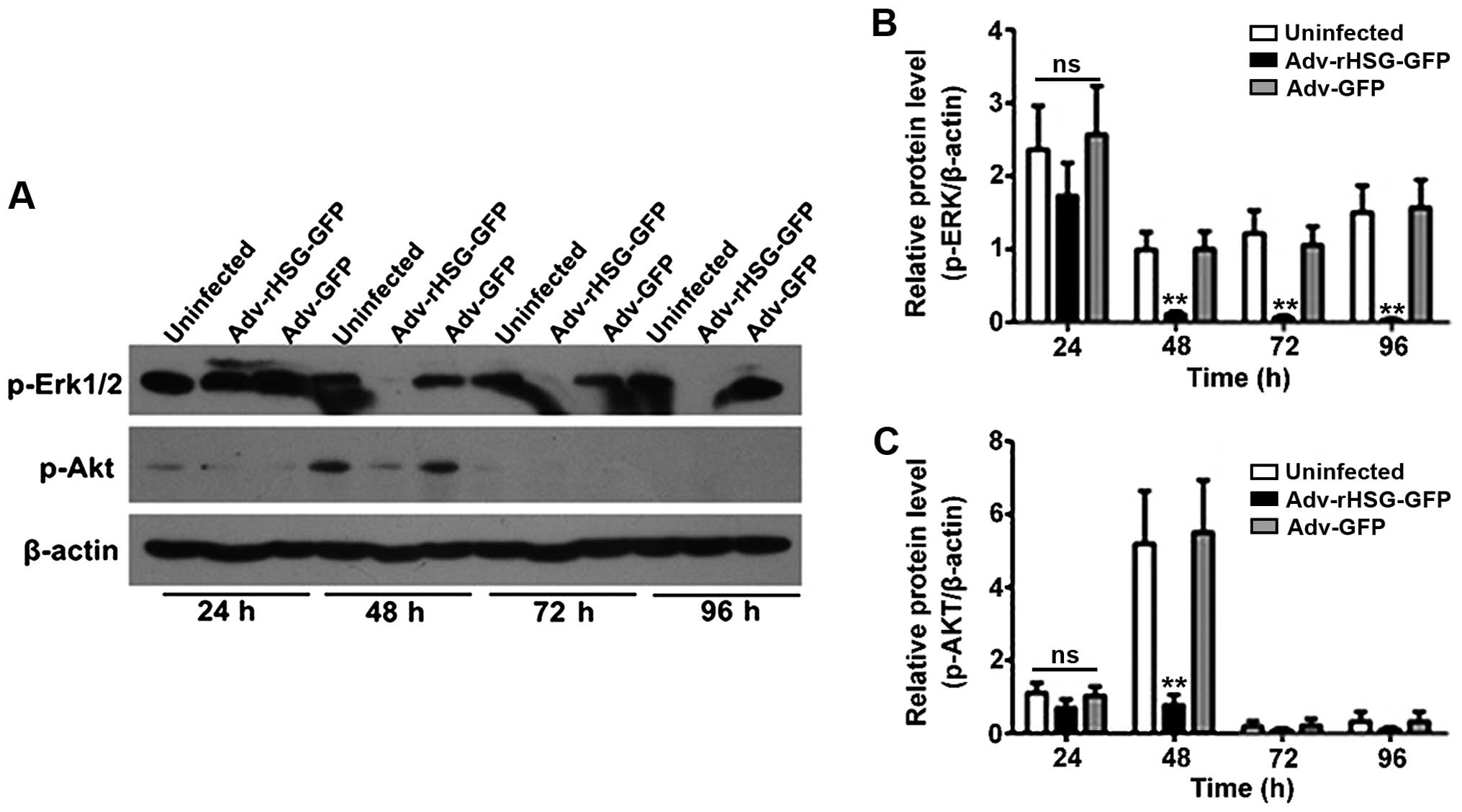
The results of western blot analysis revealed that
the expression of p-Erk1/2 began to decrease at 24 h
post-transfection in the Adv-rHSG-GFP group, although the
difference was not statistically significant as compared to the
control or Adv-GFP group (P>0.05; Fig. 6A and B). By contrast, the decrease
was statistically significant from the time point of 48 h
(P<0.01, n=3; Fig. 6A and B).
The expression of p-Akt was also evident at 48 h, but not at any of
the other time points. In addition, the expression of p-Akt was
significantly lower at 48 h in the Adv-rHSG-GFP group than in the
control or Adv-GFP group (P<0.01, n=3; Fig. 6A and C); no significant difference
was observed between the control and Adv-GFP groups (P>0.05;
Fig. 6).
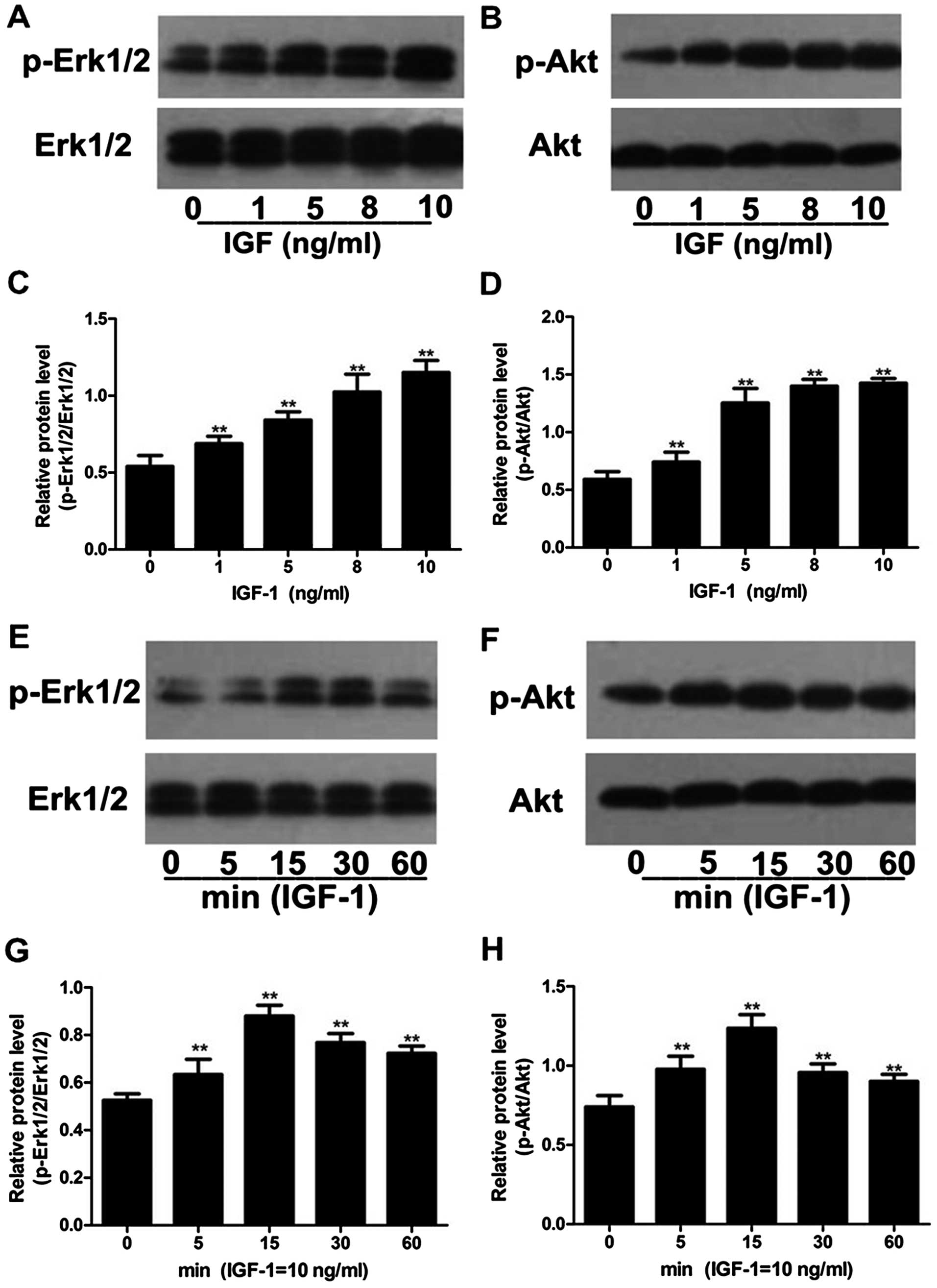
The expression levels of p-Akt and p-Erk1/2
increased after the C6 cells were stimulated with IGF-1, while the
total expression levels of Akt and Erk1/2 were not significantly
altered. The expression levels of p-Akt and p-Erk1/2 increased
slightly when 1 ng/ml of IGF-1 was used, and increased
significantly when 5, 8 and 10 ng/ml of IGF-1 were used (P<0.01,
n=3; Fig. 7A–D). The expression
levels of p-Akt and p-Erk1/2 were the highest when 10 ng/ml of
IGF-1 was used; thus, 10 ng/ml of IGF-1 was selected as the optimal
concentration for the subsequent evaluation of p-Akt and p-Erk1/2
protein expression in the C6 cells. After the C6 cells were treated
with the optimal concentration of IGF-1 for 0, 5, 15, 30 and 60
min, we found that the expression levels of p-Akt and p-Erk1/2 were
the highest at the time point of 15 min (Fig. 7E–H). Therefore, 15 min was
selected as the optimal time point for the following experiments to
measure the expression levels of p-Akt and p-Erk1/2 in the C6 cells
stimulated with IGF-1.
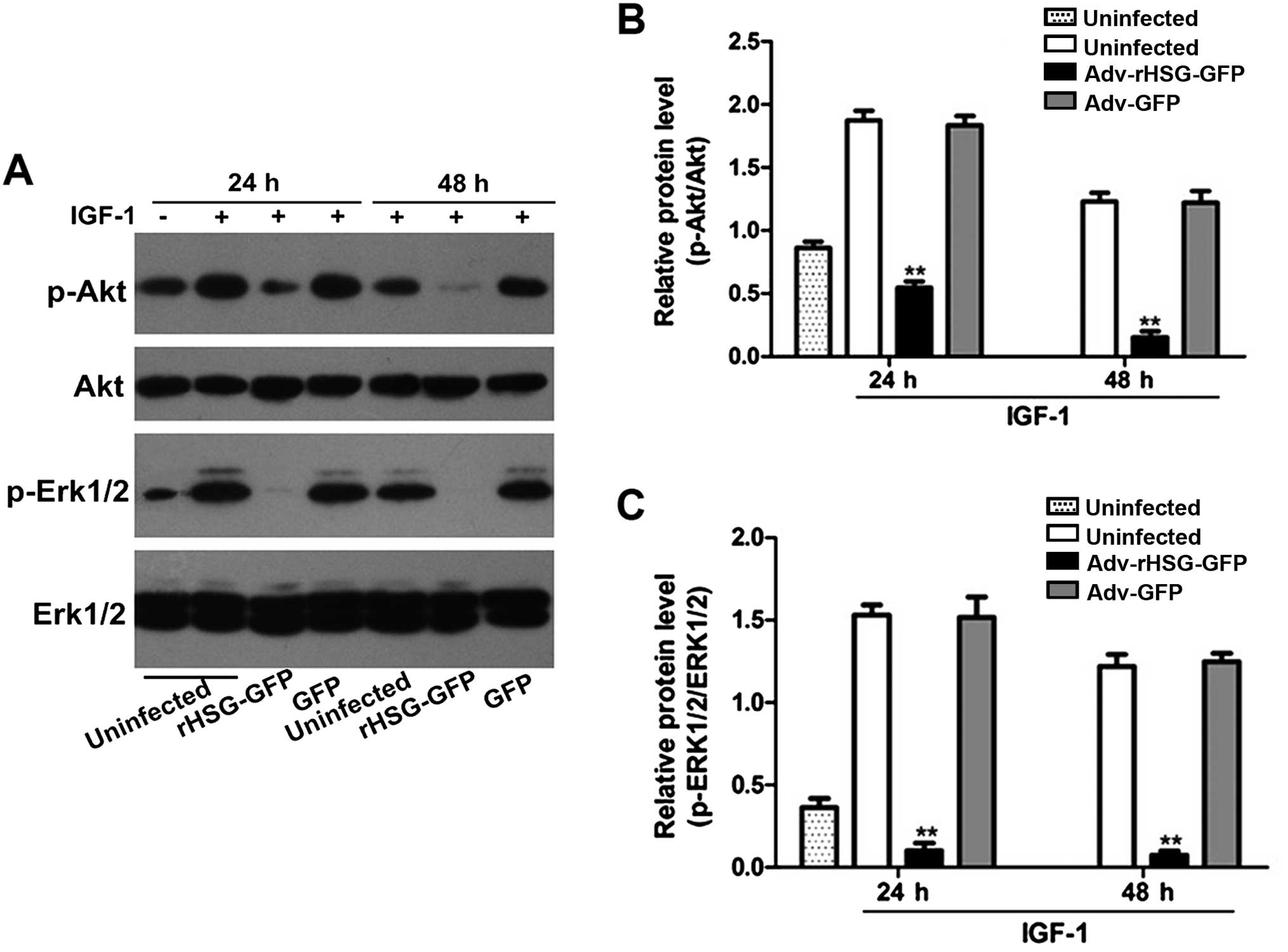
The expression levels of p-Akt and p-Erk1/2 in the
C6 cells stimulated with IGF-1 were significantly higher than the
non-stimulated C6 cells (P<0.01, n=3). By contrast, the
expression levels of p-Akt and p-Erk1/2 did not differ
significantly between the control and Adv-GFP groups, but decreased
significantly with time in the Adv-rHSG-GFP group (P<0.01, n=3).
In addition, the expression levels of these two proteins were also
significantly lower at each time point post-tranfsection compared
with the control group (P<0.01, n=3; Fig. 8).
Discussion
rHSG has now been recognized as a hyperplasia
suppressor gene. It has been demonstrated that transfection with
rHSG promotes the apoptosis of some malignant cell lines, including
hepatocellular carcinoma (HCC) cells and the bladder cancer cell
lines, T24 and 5637, suggesting that rHSG has substantial
pro-apoptotic functions (22,23). However, the effects of rHSG on
glioma cell apoptosis have not been reported to date, at least to
the best of our knowledge. The most important findings of the
present study are that the overexpression of rHSG promoted the
apoptosis of C6 cells, which were confirmed by a series of
experimental findings. Firstly, a recombined adenovirus system was
used to induce the highly efficient expression of rHSG in the C6
cells. Subsequently, several experiments including the observation
of the cell nuclei morphological cahnges, DNA damage, and the
measurement of the expression of apoptosis-related factors
demonstrated that the rHSG gene significantly promoted the
apoptosis of C6 cells. Secondly, the measurement of the expression
of p-Akt and p-Erk1/2, two important factors in the Ras-Raf-MAPK
and PI3K/Akt pathways, revealed that rHSG significantly inhibited
the expression of these two factors. Finally and most importantly,
stimulating the cells with IGF-1, a factor that stimulates the
Ras-Raf-MAPK and PI3K/Akt pathways, confirmed the association
between rHSG and these two pathways, and further provided evidence
of the role of the Ras-Raf-MAPK and PI3K/Akt pathways in the
apoptosis of C6 cells. These findings suggest that rHSG is of great
importance in identifying and treating many malignant cells,
including glioma cells, as well as in apoptosis in cell
proliferative disorders.
Previous studies have demonstrated that PARP is a
substrate of caspase-3; therefore the activation of caspase-3
induces cell apoptosis, and thus PARP is also known as 'death
substrate' (24–26). PARP is associated with the repair
of DNA and the integrity of genes. Caspase-3 recognizes the DVCD
sequences in PARP following activation and thus cleaves PARP, which
disrupts the normal functions of PARP. Altered PARP induces the
activity of endonuclease that is negatively regulated by PARP, and
thus finally degrades the DNA between nucleosomes and consequently
induces cell apoptosis (27–32). The findings of the present study
demonstrating that the transfection of C6 cells with rHSG induced
cell apoptosis also confirmed these theories. However, we also
observed that the expression of cleaved PARP increased during this
process. The expression of full-length PARP was also increased at
24, 72 and 96 h post-transfection, suggesting that rHSG induces the
activation of two pathways with opposite effects, namely one
pro-apoptotic pathway and one anti-apoptotic pathway. We
hypothesized that when rHSG induced DNA damage, in order to
survive, the cells produced PARP for repairing the DNA. rHSG also
activated the pro-apoptotic pathway, which activated the critical
downstream factor, caspase-3, to cleave corresponding PARP, and
thus the expression of cleaved PARP increased significantly. In the
present study, we found that the expression of cleaved PARP
increased considerably, while the expression of full-length PARP
decreased significantly at 48 h post-transfection compared to the
control and Adv-GFP groups, and this is the time point at which the
rHSG protein level was the highest. These findings suggested that
the DNA repairing effects of PARP at this time point were
effectively inhibited, and the C6 cells were mainly in the state of
apoptosis induced by rHSG.
In order to investigate further the effects of rHSG
on the pathways of anti-/pro-apoptosis in the cells, we measured
the levels of p-Akt and p-Erk1/2, which are the two critical
factors in the Ras-Raf-MAPK and PI3K/Akt pathways. It has been
demonstrated that inhibition of the MAPK pathway is also a factor
that causes cell apoptosis; it has been shown that in pancreatic
cancer cells and epithelial cells, the increased expression of
p-Erk1/2 induced the phosphorylation of BAD protein, and thus
inhibited cell apoptosis (33,34). The PI3K/Akt pathway is a classic
pathway that inhibits cell apoptosis and maintains cell survival.
PI3K interacts with phosphoinositide-dependent protein kinase (PDK)
following activation, which activates Akt and thus increases the
transcription of the downstream survival-related genes to maintain
cell survival, or induce the phosphorylation of BAD protein to
inhibit cell apoptosis (35). The
findings of the present study demonstrated that the protein
expression levels of Erk1/2 and Akt decreased significantly at 48 h
post-rHSG transfection, at which time point the rHSG level was the
highest, but no significant decrease was observed at 24 h
post-transfection. Therefore, we introduced IGF-1, a factor that
stimulates the PI3K/Akt and MAPK pathways, to stimulate the cells.
The stimulating effects of IGF-1 were firstly identified on the
PI3K/Akt pathway (36,37), and further studies also found that
IGF-1 activates the MAPK pathway as well (38,39). During the activation of these
pathways, IGF plays an important role in anti-apoptosis (40,41). In the present study, Adv-rHSG-GFP
was used to transfect the C6 cells, after which IGF-1 was used to
activate the Erk1/2 and PI3K/Akt pathways in these cells. The
expression levels of p-Akt and p-Erk1/2 increased significantly to
protect the cells from apoptosis. However, the expression levels of
p-Akt and p-Erk1/2 were significantly lower than the level in the
control group at the corresponding time point of 24 and 48 h
post-transfection. These expression levels decreased further with
time. These findings indicated that the combined use of rHSG and
IGF-1 significantly promoted the pro-apoptotic effects of rHSG on
C6 cells. We hypothesized that the exogenous cell protective
factor, IGF-1, decreased the repair and protective effects of the
cells, and thus enhanced the pro-apoptotic effects of rHSG on the
C6 cells. These findings suggested that the activation of the
Erk1/2 and Akt pathways also protected the cells and inhibied the
apoptosis induced by rHSG.
In conclusion, these findings suggest that the
overexpression of rHSG promotes the apoptosis of rat glioma cells,
and the molecular mechanisms involve the PI3K/Akt and Ras-Raf-MAPK
pathways. The Ras-Raf-MAPK pathway may play a dominant role in the
effects, suggesting that rHSG may be a potential target for the
treatment of gliomas.
The present study provided additional evidence on
the effects of rHSG on rat glioma (42), which is also important for further
studies on glioma. Changes in caspase-3 expression were also
observed in the present study. However, further studies are
required to investigate further the molecular mechanisms involved
in the pro-apoptotic effects of rHSG on glioma cells, particularly
the other two pathways with caspase-3 involvement.
Acknowledgments
This study was supported by the the Ningxia Hui
Autonomous Region Education Department Foundation (no. NGY2011054),
and the Natural Science Foundation Key Project of the Ningxia Hui
Autonomous Region (no. NZ13129).
References
|
1
|
Fuller GN and Scheithauer BW: The 2007
Revised World Health Organization (WHO) classification of tumours
of the central nervous system: newly codified entities. Brain
Pathol. 17:304–307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reifenberger G, Hentschel B, Felsberg J,
Schackert G, Simon M, Schnell O, Westphal M, Wick W, Pietsch T,
Loeffler M, et al German Glioma Network: Predictive impact of MGMT
promoter methylation in glioblastoma of the elderly. Int J Cancer.
131:1342–1350. 2012. View Article : Google Scholar
|
|
3
|
Grossman SA, Ye X, Piantadosi S, Desideri
S, Nabors LB and Rosenfeld M: Survival of patients with newly
diagnosed glioblastoma treated with radiation and temozolomide in
research studies in the United States. Clin Cancer Res.
16:2443–2449. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chinese Glioma Cooperative Group (CGCG);
Chinese Glioma Atlas (CGGA): Chinese Glioma Molecular Guidelines.
Chin J Neurosurg. 30:435–444. 2014.
|
|
6
|
Candolfi M, Kroeger KM, Muhammad AK, Yagiz
K, Farrokhi C, Pechnick RN, Lowenstein PR and Castro MG: Gene
therapy for brain cancer: combination therapies provide enhanced
efficacy and safety. Curr Gene Ther. 9:409–421. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nakada M, Niska JA, Tran NL, McDonough WS
and Berens ME: EphB2/R-Ras signaling regulates glioma cell
adhesion, growth, and invasion. Am J Pathol. 167:565–576. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jeon BN, Yoo JY, Choi WI, Lee CE, Yoon HG
and Hur MW: Proto-oncogene FBI-1 (Pokemon/ZBTB7A) represses
transcription of the tumor suppressor Rb gene via binding
competition with Sp1 and recruitment of co-repressors. J Biol Chem.
283:33199–33210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Westphal M and Lamszus K: The neurobiology
of gliomas: from cell biology to the development of therapeutic
approaches. Nat Rev Neurosci. 12:495–508. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen KH, Guo X, Ma D, Guo Y, Li Q, Yang D,
Li P, Qiu X, Wen S, Xiao RP and Tang J: Dysregulation of HSG
triggers vascular proliferative disorders. Nat Cell Biol.
6:872–883. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo X, Chen KH, Guo Y, Liao H, Tang J and
Xiao RP: Mitofusin 2 triggers vascular smooth muscle cell apoptosis
via mitochondrial death pathway. Circ Res. 101:1113–1122. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu M, Chen Q, Li D, Li X, Li X, Huang C,
Tang Y, Zhou Y, Wang D, Tang K, et al: LRRC4 inhibits human
glioblastoma cells proliferation, invasion, and proMMP-2 activation
by reducing SDF-1 alpha/CXCR4-mediated ERK1/2 and Akt signaling
pathways. J Cell Biochem. 103:245–255. 2008. View Article : Google Scholar
|
|
13
|
Wu M, Huang C, Li X, Li X, Gan K, Chen Q,
Tang Y, Tang K, Shen S and Li G: LRRC4 inhibits glioblastoma cell
proliferation, migration, and angiogenesis by downregulating
pleiotropic cytokine expression and responses. J Cell Physiol.
214:65–74. 2008. View Article : Google Scholar
|
|
14
|
Kunapuli P, Kasyapa CS, Hawthorn L and
Cowell JK: LGI1, a putative tumor metastasis suppressor gene,
controls in vitro invasiveness and expression of matrix
metalloproteinases in glioma cells through the ERK1/2 pathway. J
Biol Chem. 279:23151–23157. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bhaskara VK, Sundaram C and Babu PP: pERK,
pAkt and pBad: a possible role in cell proliferation and sustained
cellular survival during tumorigenesis and tumor progression in ENU
induced transplacental glioma rat model. Neurochem Res.
31:1163–1170. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cuevas P, Diaz-González D, Carceller F and
Dujovny M: Dual blockade of mitogen-activated protein kinases ERK-1
(p42) and ERK-2 (p44) and cyclic AMP response element binding
protein (CREB) by neomycin inhibits glioma cell proliferation.
Neurol Res. 25:13–16. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Belsey MJ, Davies AR, Witchel HJ and
Kozlowski RZ: Inhibition of ERK and JNK decreases both
osmosensitive taurine release and cell proliferation in glioma
cells. Neurochem Res. 32:1940–1949. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Betti M, Minelli A, Canonico B, Castaldo
P, Magi S, Aisa MC, Piroddi M, Di Tomaso V and Galli F:
Antiproliferative effects of tocopherols (vitamin E) on murine
glioma C6 cells: homologue-specific control of PKC/ERK and cyclin
signaling. Free Radic Biol Med. 41:464–472. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McDaid HM, Lopez-Barcons L, Grossman A,
Lia M, Keller S, Pérez-Soler R and Horwitz SB: Enhancement of the
therapeutic efficacy of taxol by the mitogen-activated protein
kinase kinase inhibitor CI-1040 in nude mice bearing human
heterotransplants. Cancer Res. 65:2854–2860. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Knobbe CB, Trampe-Kieslich A and
Reifenberger G: Genetic alteration and expression of the
phosphoinositol-3-kinase/Akt pathway genes PIK3CA and PIKE in human
glioblastomas. Neuropathol Appl Neurobiol. 31:486–490. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao P, Jiang S, Guo H, Yang Q, Fang Z,
Zhao W and Shen B: Expression and mechanism of rat hyperplasia
suppressor gene suppressing growth of rat C6 glioma cells. J Third
Mil Med Univ. 36:381–385. 2014.
|
|
22
|
Jin B, Fu G, Pan H, Cheng X, Zhou L, Lv J,
Chen G and Zheng S: Anti-tumour efficacy of mitofusin-2 in urinary
bladder carcinoma. Med Oncol. 28(Suppl 1): S373–S380. 2011.
View Article : Google Scholar
|
|
23
|
Wang W, Lu J, Zhu F, Wei J, Jia C, Zhang
Y, Zhou L, Xie H and Zheng S: Pro-apoptotic and anti-proliferative
effects of mitofusin-2 via Bax signaling in hepatocellular
carcinoma cells. Med Oncol. 29:70–76. 2012. View Article : Google Scholar
|
|
24
|
Gagné JP, Moreel X, Gagné P, Labelle Y,
Droit A, Chevalier-Paré M, Bourassa S, McDonald D, Hendzel MJ,
Prigent C and Poirier GG: Proteomic investigation of
phosphorylation sites in poly(ADP-ribose) polymerase-1 and
poly(ADP-ribose) glycohydrolase. J Proteome Res. 8:1014–1029. 2009.
View Article : Google Scholar
|
|
25
|
Reed AM, Fishel ML and Kelley MR:
Small-molecule inhibitors of proteins involved in base excision
repair potentiate the anti-tumorigenic effect of existing
chemotherapeutics and irradiation. Future Oncol. 5:713–726. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Atorino L, Di Meglio S, Farina B, Jones R
and Quesada P: Rat germinal cells require PARP for repair of DNA
damage induced by gamma-irradiation and H2O2
treatment. Eur J Cell Biol. 80:222–229. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thornberry NA and Lazebnik Y: Caspases:
enemies within. Science. 281:1312–1316. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kato J, Kuwabara Y, Mitani M, Shinoda N,
Sato A, Toyama T, Mitsui A, Nishiwaki T, Moriyama S, Kudo J and
Fujii Y: Expression of survivin in esophageal cancer: correlation
with the prognosis and response to chemotherapy. Int J Cancer.
95:92–95. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Altieri DC: Survivin, versatile modulation
of cell division and apoptosis in cancer. Oncogene. 22:8581–8589.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Beardsmore DM, Verbeke CS, Davies CL,
Guillou PJ and Clark GW: Apoptotic and proliferative indexes in
esophageal cancer: predictors of response to neoadjuvant therapy
[corrected]. J Gastrointest Surg. 7:77–86. 2003. View Article : Google Scholar
|
|
31
|
Kelly RJ, Lopez-Chavez A, Citrin D, Janik
JE and Morris JC: Impacting tumor cell-fate by targeting the
inhibitor of apoptosis protein survivin. Mol Cancer. 10:352011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tamm I, Wang Y, Sausville E, Scudiero DA,
Vigna N, Oltersdorf T and Reed JC: IAP-family protein survivin
inhibits caspase activity and apoptosis induced by Fas (CD95), Bax,
caspases, and anticancer drugs. Cancer Res. 58:5315–5320.
1998.PubMed/NCBI
|
|
33
|
Boucher MJ, Morisset J, Vachon PH, Reed
JC, Lainé J and Rivard N: MEK/ERK signaling pathway regulates the
expression of Bcl-2, Bcl-X(L), and Mcl-1 and promotes survival of
human pancreatic cancer cells. J Cell Biochem. 79:355–369. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Buder-Hoffmann S, Palmer C, Vacek P,
Taatjes D and Mossman B: Different accumulation of activated
extracellular signal-regulated kinases (ERK 1/2) and role in
cell-cycle alterations by epidermal growth factor, hydrogen
peroxide, or asbestos in pulmonary epithelial cells. Am J Respir
Cell Mol Biol. 24:405–413. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Neri LM, Borgatti P, Capitani S and
Martelli AM: The nuclear phosphoinositide 3-kinase/AKT pathway: a
new second messenger system. Biochim Biophys Acta. 1584:73–80.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ueki K, Fruman DA, Brachmann SM, Tseng YH,
Cantley LC and Kahn CR: Molecular balance between the regulatory
and catalytic subunits of phosphoinositide 3-kinase regulates cell
signaling and survival. Mol Cell Biol. 22:965–977. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wymann MP and Pirola L: Structure and
function of phosphoinositide 3-kinases. Biochim Biophys Acta.
1436:127–150. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Khokhlatchev AV, Canagarajah B, Wilsbacher
J, Robinson M, Atkinson M, Goldsmith E and Cobb MH: Phosphorylation
of the MAP kinase ERK2 promotes its homodimerization and nuclear
translocation. Cell. 93:605–615. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Párrizas M, Saltiel AR and LeRoith D:
Insulin-like growth factor 1 inhibits apoptosis using the
phosphatidylinositol 3′-kinase and mitogen-activated protein kinase
pathways. J Biol Chem. 272:154–161. 1997. View Article : Google Scholar
|
|
40
|
Kenchappa P, Yadav A, Singh G, Nandana S
and Banerjee K: Rescue of TNFalpha-inhibited neuronal cells by
IGF-1 involves Akt and c-Jun N-terminal kinases. J Neurosci Res.
76:466–474. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bencomo E, Pérez R, Arteaga MF, Acosta E,
Peña O, Lopez L, Avila J and Palumbo A: Apoptosis of cultured
granulosa-lutein cells is reduced by insulin-like growth factor I
and may correlate with embryo fragmentation and pregnancy rate.
Fertil Steril. 85:474–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gao P, Wang Z, Zhang B, Zou Y, Guo H, Liu
H, Yang Q, Fang Z, Jiang S, Shen B, et al: Suppression of C6
gliomas via application of rat hyperplasia gene. Int J Biol
Markers. 29:e411–e422. 2014. View Article : Google Scholar : PubMed/NCBI
|