Introduction
Duchenne muscular dystrophy is a devastating muscle
wasting disease of early childhood (1) and the most frequently inherited
neuromuscular disorder (2). It
has an X-linked mode of inheritance and is primarily caused by
abnormalities in the Dmd gene, which trigger the almost
complete absence of the membrane cytoskeletal protein dystrophin of
427 kDa (3). The loss of
dystrophin results in a significant reduction of
dystrophin-associated glycoproteins, which in turn impairs the
linkage between the extracellular matrix component laminin and the
actin membrane cytoskeleton (4).
In addition to destabilizing the fibre periphery and thereby making
muscle cells more susceptible to micro-rupturing during
excitation-contraction-relaxation cycles (5), a variety of secondary changes play a
key role in the molecular pathogenesis of X-linked muscular
dystrophy (6). This includes
reactive myofibrosis resulting in tissue scarring and loss of fibre
elasticity, abnormal calcium regulation and increased proteolytic
degradation, impaired cellular signalling and disturbed
excitation-contraction coupling, as well as sterile inflammation of
dystrophin-deficient muscle tissue (7–10).
Therefore, the immune system appears to play a critical role in
X-linked muscular dystrophy (11–14). It is debatable whether the
inflammatory pathology of dystrophic muscles is based on a reactive
and relatively non-specific invasion of immune cells in damaged
muscle fibres or represents a separate mechanism that promotes
fibre degeneration independent of sarcolemmal destabilisation
(15–17). Thus, although dystrophinopathies
are triggered by a single gene defect, the pathophysiological
consequences and adaptive responses of dystrophic muscles are
highly complex.
In the pre-proteomic era, a variety of
muscle-derived protein markers have been established for general
diagnostic purposes. Serum markers with an association to tissue
damage and muscular injury include creatine kinase, pyruvate
kinase, hemopexin, adenylate kinase, carbonic anhydrase and lactate
dehydrogenase (18–22). However, the concentration of some
of these serum markers vary considerably in healthy individuals,
lack a high degree of specificity and/or exhibit inconsistencies in
their concentration changes during acute vs. chronic disease
stages. This potential lack of diagnostic reliability and limited
robustness restricts the clinical usefulness of many general
protein biomarkers of skeletal muscle damage for accurate sample
screening, prognosis or clinical outcome measures. More specific
serum proteins that exhibit elevated levels in Duchenne patients
seem to be the tissue inhibitors of metalloproteinase-1,
transforming growth factor TGF-β1 and the matrix metalloproteinase
MMP-9 (23–26). In contrast to traditional
biochemical studies that usually focus on the characterization of a
restricted number of specific candidate molecules, large-scale
biochemical approaches promise to gain new global insights into the
molecular and cellular mechanisms of muscular dystrophy. Mass
spectrometry-based proteomics is highly suitable to improve our
general understanding of complex changes in dystrophinopathies
(27). Systems biological methods
promise to determine potential pathophysiological hierarchies and
interconnectivities between differing secondary mechanisms within
muscle tissues that are downstream of the pathobiochemical collapse
of the dystrophin-glycoprotein complex (28). Most importantly, the systematic
proteomic identification of new serum biomarkers is crucial for the
improved diagnostic and prognostic evaluation of Duchenne muscular
dystrophy and animal models of dystrophin deficiency (29).
In the field of muscular dystrophy research, the
systematic screening of body fluids from Duchenne patients and
established animal models of dystrophinopathies using proteomic
technology has recently established a variety of new biomarker
candidates, including fibronectin, the coagulation factor XIIIa,
titin, various myosin light chain isoforms, myomesin-3, filamin-C,
lysosomal-associated membrane protein LAMP1 and its accompanying
vesicle proteins, aldolase, phosphoglycerate mutase, enolase,
glycogen phosphorylase, fatty acid-binding protein, myoglobin,
cytochrome c, malate dehydrogenase, fibrinogen, parvalbumin,
electron transfer flavoprotein A, troponin,
Ca2+/calmodulin-dependent protein kinase Camk2b,
metalloproteinase Adamts5, troponin-1, myoglobin and heat-shock
protein HSPA1A (30–39). In analogy to these studies and to
establish new biomarker candidates for the improved evaluation of
animal models of dystrophinopathy, we analysed serum from the
dystrophic mdx-4cv mouse (40). The mdx-4cv model
exhibits substantially less revertant dystrophin-positive fibres
than the conventional mdx mouse (41), which is advantageous for outcome
measurements in experimental treatment studies, such as
exon-skipping therapy (42).
Comparative proteomics of mdx-4cv vs. wild-type serum
established a significantly increased concentration of the
inflammation-inducible plasma marker haptoglobin (43). The elevated levels of this acute
phase response protein agree with skeletal muscle damage and
sterile inflammation in muscular dystrophy. This establishes
haptoglobin as a novel biomarker candidate of the inflammatory
response in dystrophinopathy.
Materials and methods
Materials
Immunodepletion of serum samples was carried out
with Proteome Purify 2 Mouse Serum Protein Immunodepletion Resin
(R&D Systems, Inc., Minneapolis, MN, USA). Ultrapure acrylamide
stock solutions for gel electrophoresis were purchased from
National Diagnostics (Atlanta, GA, USA). Invitrogen (Carlsbad, CA,
USA) supplied Whatman nitrocellulose transfer membranes. The
chemiluminescence substrate and protease inhibitors were obtained
from Roche Diagnostics GmbH (Mannheim, Germany). Superfrost Plus
positively-charged microscope slides were obtained from Menzel
Glaser (Braunschweig, Germany). Sequencing grade modified trypsin
and Lys-C were purchased from Promega Corp. (Madison, WI, USA).
Pierce C18 Spin Columns were obtained from Thermo Fisher Scientific
(Dublin, Ireland). Primary antibodies were purchased from Abcam
(Cambridge, UK) (ab131236 to haptoglobin, ab14225-200 to albumin,
ab52488 to lactate dehydrogenase, ab9033-1 to transferrin, ab15277
to dystrophin and ab11427 to parvalbumin) and Santa Cruz
Biotechnology, Santa Cruz, CA, USA (sc-25607 to myoglobin).
Chemicon International, Inc. (Temecula, CA, USA) provided
peroxidase-conjugated secondary antibodies. Enzyme-linked
immunosorbent assay assays (ELISA) were purchased from Abcam
(ab157714 to haptoglobin and ab157711 to complement C3) and R&D
Systems (FABP-1/L-FABP). Normal goat serum and Cy3-conjugated
antibodies were obtained from Jackson ImmunoResearch Laboratories,
Inc. (West Grove, PA, USA). The embedding medium Fluoromount-G was
obtained from Southern Biotech (Birmingham, AL, USA). A variety of
other general chemicals, including bis-benzimide Hoechst-33342,
were of analytical grade and obtained from Sigma Chemical Company
(Dorset, UK).
Dystrophic mdx-4cv animal model of
Duchenne muscular dystrophy
In order to investigate the serum proteome and
identify potential biomarkers of dystrophinopathy, the
mdx-4cv animal model was employed in the present
study. This mouse model has been generated using N-ethylnitrosourea
to induce a premature stop codon in exon 53 of the Dmd gene
(44). The mdx-4cv
model has 10-fold fewer dystrophin-positive revertant fibres
(41) than the conventional mdx
mouse (45) and thus its serum
proteome may be more representative of the human condition. Mice
were bred in the Bioresource Unit of the University of Bonn under
standard conditions (46) with
the authorization by the District Veterinary Office in Bonn,
Germany. Tissue and organ harvesting was regularly reported to the
District Veterinary Office. Serum and muscle samples from
6-month-old mdx-4cv and age-matched wild-type C57BL6
mice were obtained after sacrificing the animals by cervical
dislocation. Samples were immediately quick-frozen in liquid
nitrogen and stored at −80°C prior to usage. Samples were
transported to Maynooth University on dry ice in accordance with
the Department of Agriculture (animal by-product register number
2016/16 to the Department of Biology, National University of
Ireland, Maynooth).
Immunodepletion of mouse serum
samples
One of the main challenges with the proteomic
profiling of biofluids, particularly for serum and plasma, is the
wide dynamic range of protein expression (47). Since highly abundant proteins such
as albumin and IgG constitute the majority of plasma proteins, and
this seriously interferes with the detection of low-copy-number
proteins by mass spectrometry (48), many profiling studies use
immunodepletion strategies to achieve an enriched pool of
low-abundance proteins. We here used the Proteome Purify 2 Mouse
Serum Protein Immunodepletion Resin to remove the highly abundant
proteins albumin and IgG from the murine serum samples. In summary,
10 µl of serum was added to a micro-centrifuge tube
containing 1 ml of immunodepletion resin. The tubes were placed on
a rotary shaker for 1 h at room temperature. After the incubation
period, the mixture was added to the upper chamber of a Spin-X
filter unit (centrifuge tube with a 0.22-µm cellulose
acetate membrane) and centrifuged for 2 min at 2,000 × g. The
filtrate was collected and used for further analysis.
Sample preparation for mass
spectrometry
Immunodepleted serum samples were acetone
precipitated and the resulting pellets were re-suspended in
label-free solubilisation buffer (6 M urea, 2 M thiourea, 10 mM
Tris, pH 8.0 in LC/MS grade water). Protein concentrations were
determined by the Bradford assay system (49) and sample volumes were equalised
with label-free solubilisation buffer. Samples were reduced for 30
min at 37°C with 10 mM dithiothreitol and alkylated with 25 mM
iodoacetamide in 50 mM ammonium bicarbonate for 20 min in the dark
at room temperature (50). To
limit alkylation of trypsin by any remaining unreacted
iodoacetamide, samples were reduced with a further 10 mM
dithiothreitol for 15 min in the dark at room temperature.
Proteolytic digestion of serum proteins to their constitutive
peptides was carried out in two steps. Firstly samples were
digested with sequencing grade Lys-C at a ratio of 1:100
(protease:protein) for 4 h at 37°C, followed by dilution with four
times the initial sample volume in 50 mM ammonium bicarbonate.
Secondly, samples were digested with sequencing-grade trypsin at a
ratio of 1:25 (protease:protein) overnight at 37°C (51). Samples were acidified with 2%
trifluoroacetic acid (TFA) in 20% acetonitrile (ACN) [3:1 (v/v)
dilution]. Peptide suspensions were purified using Pierce C18 Spin
Columns, dried through vacuum centrifugation and suspended in
loading buffer consisting of 2% ACN and 0.05% TFA in LC-MS grade
water. Samples were vortexed and sonicated to ensure an even
suspension of peptides prior to mass spectrometric analysis.
Label-free liquid chromatography mass
spectrometric analysis
The label-free liquid chromatography mass
spectrometric (LC-MS/MS) analysis of mdx-4cv vs.
wild-type serum samples was carried out using an Ultimate 3000
NanoLC system (Dionex Corporation, Sunnyvale, CA, USA) coupled to a
Q-Exactive mass spectrometer (Thermo Fisher Scientific). Peptide
mixtures (5 µl, corresponding to 1 µg protein) were
loaded by an autosampler onto a C18 trap column (C18 PepMap, 300
µm id × 5 mm, 5 µm particle size, 100 A pore size;
Thermo Fisher Scientific). The trap column was switched on-line
with an analytical Biobasic C18 Picofrit column (C18 PepMap, 75
µm id × 50 cm, 2 µm particle size, 100 A pore size;
Dionex Corporation). Serum-derived peptides were eluted using the
following gradient (solvent A: 80% (v/v) ACN and 0.1% (v/v) formic
acid in LC-MS grade water): 5% solvent A for 120 min, 45% solvent A
for 2.5 min, 90% solvent A for 9 min and 3% solvent A for 43 min
(52). The column flow rate was
set to 0.3 µl/min. Data were acquired with Xcalibur software
(Thermo Fisher Scientific). The Q-Exactive was operated in positive
and data-dependent mode and was externally calibrated. Survey MS
scans were conducted in the 300–1,700 m/z range with a resolution
of 140,000 (m/z 200) and lock mass set to 445.12003.
Collision-induced dissociation fragmentation was carried out with
the 15 most intense ions per scan and at 17,500 resolution. Within
30 sec, a dynamic exclusion window was applied. An isolation window
of 2 m/z and one microscan were used to collect suitable tandem
mass spectra.
Quantitative proteomic profiling by
label-free LC-MS/MS analysis
Quantitative analysis of the raw data generated from
LC-MS/MS was conducted with Progenesis QI for Proteomics software
(version 3.1; Nonlinear Dynamics, Ltd., Newcastle upon Tyne, UK). A
reference run was selected based on the highest number of peptide
ions and the retention times of all the other runs were aligned to
this reference run. This accounted for any drift in LC retention
time, thus giving an adjusted retention time for all runs in the
analysis (53). The data were
filtered using the criteria listed below and were then exported as
a Mascot generic file to Proteome Discoverer 2.0 (Thermo
Scientific); i) peptide features with ANOVA ≤0.05 between
experimental groups, ii) mass peaks with charge states from
+1 to +5 and iii) greater than one isotope
per peptide. The exported MS/MS spectra from Progenesis software
were used for peptide identification using Proteome Discoverer 2.0
against Mascot (version 2.3) and Sequest HT and searched against
the UniProtKB-SwissProt database (taxonomy: Mus musculus).
The following search parameters were used for protein
identification: i) peptide mass tolerance set to 10 ppm, ii) MS/MS
mass tolerance set to 0.02 Da, iii) up to two missed cleavages were
allowed, iv) carbamidomethylation set as a fixed modification and
v) methionine oxidation set as a variable modification (54). For re-importation back into
Progenesis LC-MS software for further analysis, only peptides
classified as high confidence peptides and with ion scores of 40.00
or more (from Mascot) and/or XCorr scores >1.5 for singly
charged ions, >2.0 for doubly charged ions and >3.0 for
triply charged ions (Sequest HT, Thermo Fisher Scientific) were
selected. The following criteria were applied to assign a protein
as properly identified and with differential abundance: i) an ANOVA
score between experimental groups of ≤0.05, and ii) proteins with
≥2 peptides matched (55). The
PANTHER database of protein families (http://pantherdb.org; version 10.0) was used to group
proteins based on their protein class (56).
Immunoblot analysis
Immunoblotting using a panel of select antibodies
was used for the independent verification of the proteomic
findings. Immunoblotting was performed using routine conditions as
previously described in detail (57). Briefly, crude serum samples were
separated by gel electrophoresis using 10% 1D polyacrylamide gels,
followed by wet transfer to nitrocellulose membranes at 100 V for
70 min at 4°C in a Trans-Blot cell from Bio-Rad Laboratories
(Richmond, CA, USA). Transfer efficiency was assessed using Ponceau
reversible stain and membranes were then blocked for 1 h at room
temperature using a milk protein solution [2.5% (w/v) fat-free milk
powder in phosphate buffered saline] (58). Membranes were incubated with
appropriately diluted primary antibodies overnight at 4°C with
gentle agitation. Membranes were then washed with the milk protein
solution twice for 10 min and incubated with peroxidase-conjugated
secondary antibodies for 1.5 h at room temperature with gentle
agitation. Following a series of washing steps with the milk
protein solution and phosphate-buffered saline, antibody-labelled
protein bands were visualised using enhanced chemiluminescence.
Densitometric scanning and statistical analysis of immunoblots were
performed using a HP PSC-2355 scanner and ImageJ software [National
Institutes of Health (NIH), Bethesda, MA, USA] along with Graph-Pad
Prism software (San Diego, CA, USA), in which a P<0.05 was
deemed to be statistically significant.
ELISA
ELISA was employed to independently verify key
findings from the mass spectrometric analysis. Crude serum samples
were screened using quantitative sandwich enzyme immunoassays to
haptoglobin, FABP-1 and complement C3. Appropriately diluted serum
samples (1:400 for FABP-1, 1:1,000 for haptoglobin, and 1:30,000
for complement C3 assay) were added to antibody-coated microtiter
wells and were incubated at room temperature for 20 min (for
complement C and haptoglobin) or 2 h (for FABP-1). After the
incubation period, wells were washed and a peroxidase-labelled
secondary detector antibody was added. After incubation at room
temperature for 20 min in the dark, tetramethylbenzidine chromogen
substrate was added. The reaction was stopped after 10 min (for
complement C and haptoglobin) or 30 min (for FABP-1) and absorbance
was measured at λ=450 nm on a microplate reader (33). The quantity of protein in the test
samples was interpolated from the standard curve and was corrected
for sample dilution. All ELISA assays were carried out with crude
serum from wild-type (n=5) and mdx-4cv (n=8 for
complement C3; n=7 for haptoglobin and FABP-1) mice. All test
samples were assayed in triplicate. The intraplate % CV was
calculated and was found to be less than 10% for all assays.
Statistical ROC (receiver operating characteristics) analyses were
performed using MedCalc for Windows, version 16.8 (MedCalc Software
BVBA, Ostend, Belgium) to determine representaive AUC values for
individual ELISA assays.
Immunofluorescence microscopy
To establish the mutant and dystrophic status of
mdx-4cv mice at the age of 6 months, deficiency in
the dystrophin protein isoform Dp427 and histological skeletal
muscle changes were established by immunofluorescence microscopy
and histological analysis, respectively (59). Dystrophic mdx-4cv
and wild-type gastrocnemius muscles were freshly dissected
and then quick-frozen in liquid nitrogen. Histochemical staining
was carried out with routine haematoxylin and eosin staining.
Cryo-sectioning was employed to generate 10 µm muscle tissue
sections, which were placed on Superfrost Plus positively-charged
microscope slides. Skeletal muscle cryosections were then boiled to
reduce background staining (60)
and permeabilized in 0.1% (v/v) Triton X-100 in phosphate-buffered
saline for 30 min at room temperature (61). Tissue sections were blocked in 20%
(v/v) normal goat serum in phosphate-buffered saline for 30 min and
incubated overnight at 4°C with an appropriately diluted antibody
to dystrophin. Following a careful washing step, skeletal muscle
sections were incubated with Cy3-conjugated anti-IgG antibodies
(1:200) for 30 min at room temperature. Primary antibodies were
omitted for control staining. Antibody-labelled skeletal muscle
sections were embedded in Fluoromount-G medium and viewed under a
Zeiss Axioskop 2 epi-fluorescence microscope equipped with a
digital Zeiss AxioCam HRc camera (Carl Zeiss Jena GmbH, Jena,
Germany).
Results
In order to evaluate whether the dystrophic
phenotype of the mdx-4cv mouse is reflected by an
altered rate of protein release into the circulatory system or
other plasma fluctuations, potential changes in the
mdx-4cv serum proteome were determined by label-free
mass spectrometry. Key findings from the proteomic profiling of
serum samples were verified by comparative immunoblotting and
enzyme-linked immunosorbent assays.
Dystrophic mdx-4cv model of
dystrophinopathy
Prior to the proteomic profiling of serum samples,
the mutant and dystrophic status of the 6-month-old
mdx-4cv animal model of Duchenne muscular dystrophy
was established using immunofluorescence microscopy and
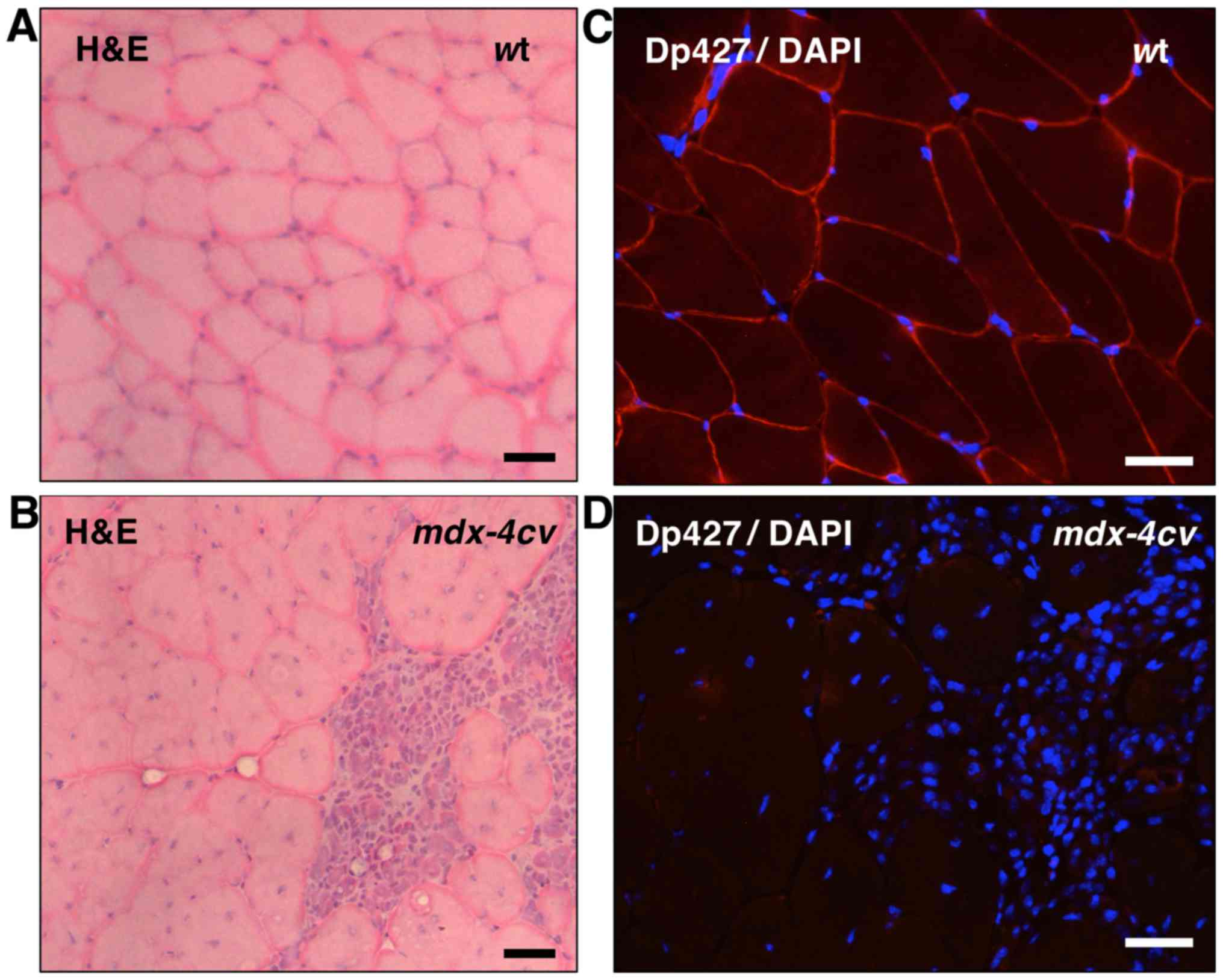
histological assessment. Fig. 1
clearly demonstrates the dystrophic phenotype of the
mdx-4cv gastrocnemius muscle (45). In contrast to the relatively
regular cell shape and peripheral nucleation in wild-type (wt)
mouse muscle (Fig. 1A),
transverse sections of the mdx-4cv mouse muscle
showed central nucleation and the infiltration of immune cells in
areas of fibre damage (Fig. 1B).
The peripheral labelling of dystrophin isoform Dp427 in wt muscle
by immunofluorescence microscopy (Fig. 1C) was in stark contrast to the
lack of dystrophin staining in mdx-4cv mutant muscle
(Fig. 1D).
Pre-fractionation of mdx-4cv serum
Protein depletion, pre-fractionation and
post-fractionation are routinely employed methods in proteomic
studies of body fluids that exhibit a complex proteome with the
presence of a few highly abundant proteins. These protein species
may mask the detection of low-copy-number protein species. Albumin
and IgG molecules are major classes of abundant plasma proteins and
may therefore interfere with the detection of low-abundance
proteins in comparative proteomic studies (48). We therefore employed an
immunodepletion strategy to reduce sample complexity and enrich the
starting material in serum proteins with a relatively low
concentration (62).
In general, immunodepletion is a widely employed and
internationally accepted approach for studying complex biofluids
(63). This protein depletion
technique decisively reduces the problem of dynamic range in both
LC-MS/MS and gel-based biomarker studies (64–68). However, albumin and other major
plasma proteins have been shown to bind a diverse range of minor
peptides and proteins and thus the albumin- and IgG-depleted serum
proteome may be slightly altered in relation to the composition of
select protein species (69–71), in what is generally referred to as
the 'carrier-and-cargo' effect of the albuminome.
Thus, although albumin/IgG depletion can result in a
concomitant decrease in certain low-molecular-mass proteins due to
their biological interactions with albumin or IgG molecules, the
analytical advantages of reducing sample complexity outweigh this
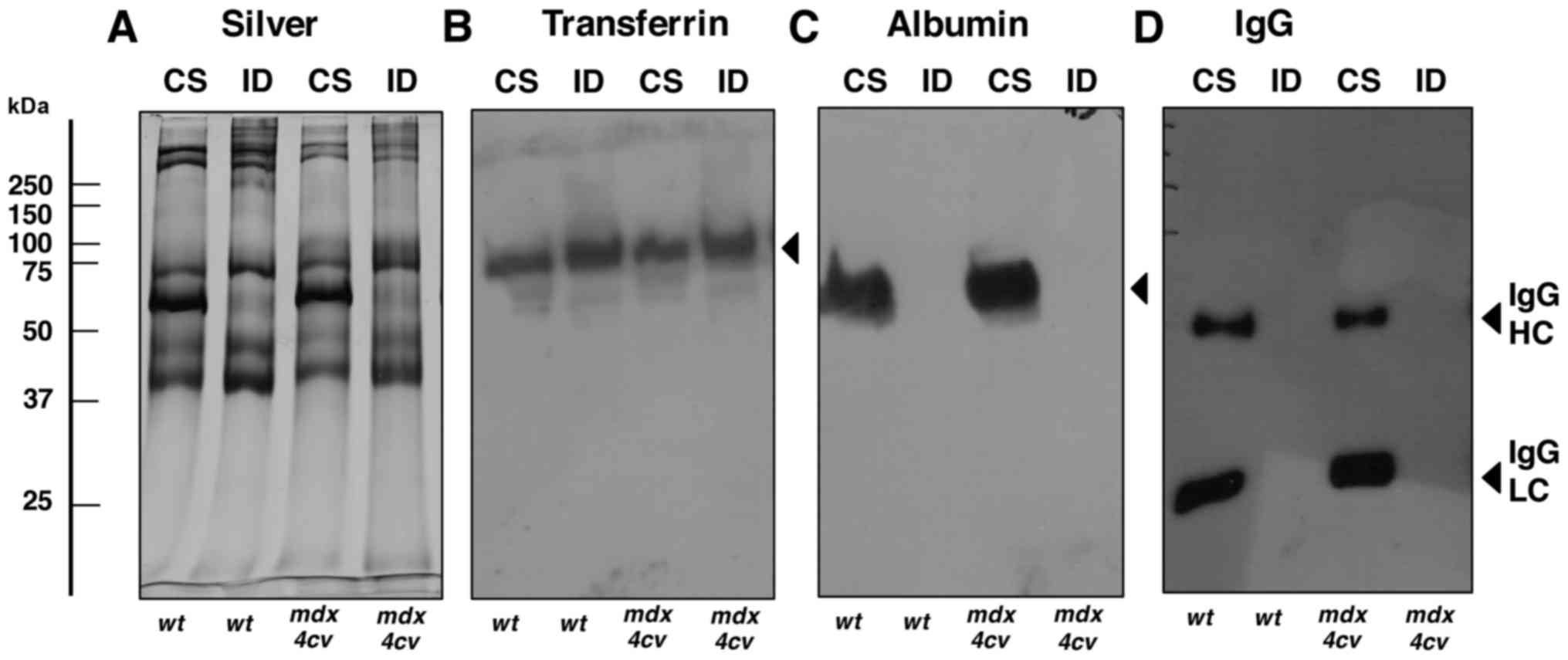
issue (62). Fig. 2 illustrates the successful
depletion of albumin and IgG from the crude serum samples derived
from dystrophic mdx-4cv mice and wild-type animals. A
representative silver-stained gel confirmed the alteration in the
overall protein composition of crude serum vs. immuno-depleted
serum (Fig. 2A). In contrast to
transferrin, for which the abundance was not highly altered
following pre-fractionation (Fig.
2B), the amounts of albumin, IgG heavy chain and IgG light
chain were markedly decreased in the immuno-depleted fractions
(Fig. 2C and D).
Proteomic profiling of 6-month-old
mdx-4cv serum
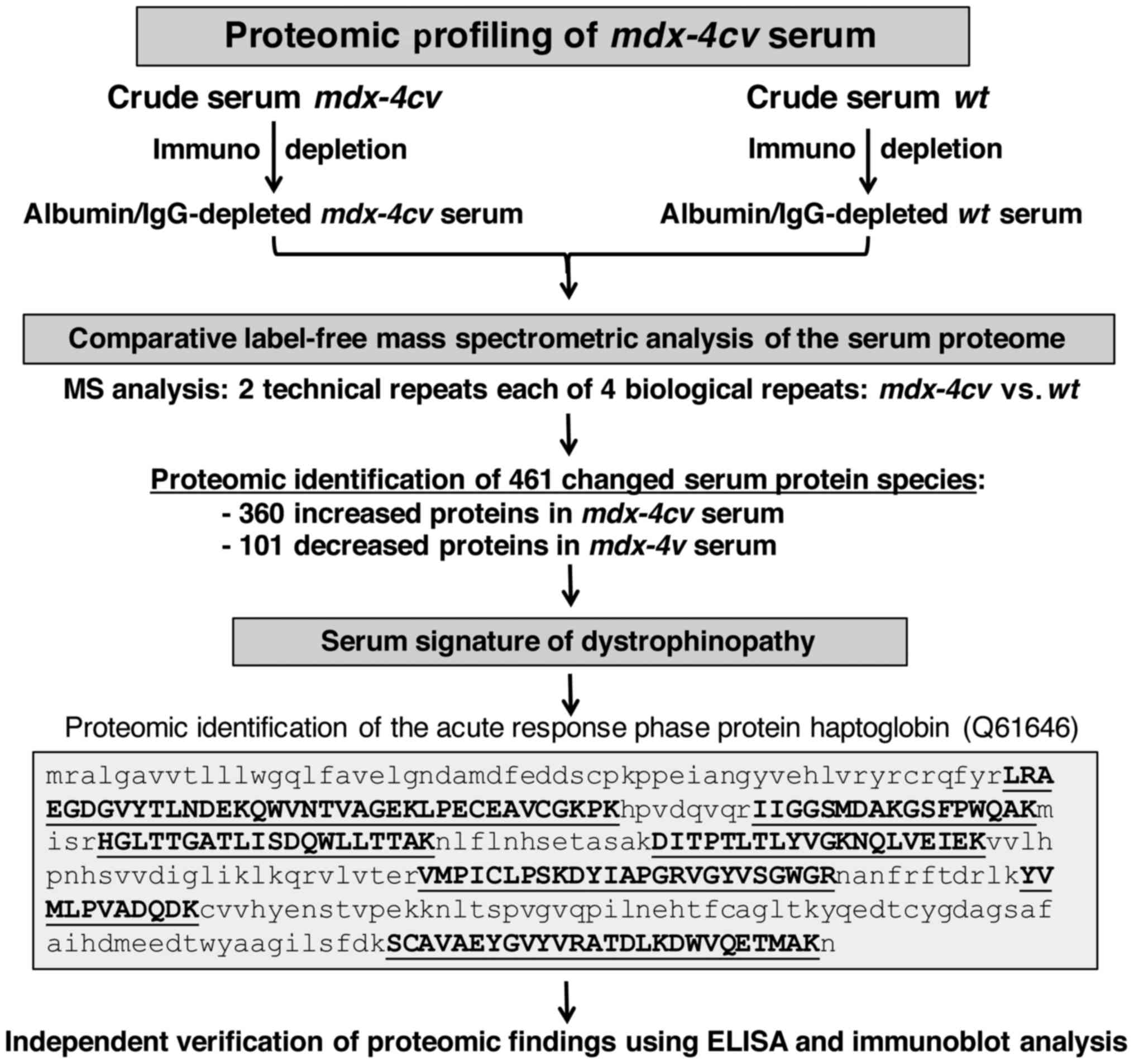
The proteomic profiling approach used in the present
study is summarized in the flow chart of Fig. 3. Following pre-fractionation, the
albumin/IgG-depleted serum from wild-type vs. dystrophic
mdx-4cv mice was analysed by comparative label-free
mass spectrometry to establish the serum signature of
dystrophinopathy. The proteomic survey of 2 technical repeats each
of 4 biological samples identified 461 changed serum protein
species consisting of 360 increased and 101 decreased proteins in
mdx-4cv serum. As an illustrative example, the
crucial proteomic identification and mass spectrometric fingerprint
of the acute response phase protein haptoglobin (Q61646) is shown
in Fig. 3. The independent
verification of key proteomic findings was carried out by
immunoblotting and enzyme-linked immunosorbent assays, as outlined
below. Identified proteins with a high degree of increase vs.
decrease are listed in Tables
ITable II–III, respectively. The tables list
information in relation to accession number, protein name, number
of unique peptides, confidence score and fold change.
 | Table IList of increased proteins in
6-month-old mdx-4cv mouse serum vs. age-matched
wild-type mouse serum as determined by label-free mass
spectrometry. |
Table I
List of increased proteins in
6-month-old mdx-4cv mouse serum vs. age-matched
wild-type mouse serum as determined by label-free mass
spectrometry.
| Accession no. | Protein name | Unique
peptides | Confidence
score | Fold change |
|---|
| P70441 | Na(+)/H(+) exchange
regulatory cofactor NHE-RF1 | 2 | 128.77 | 384.8 |
| Q9D5J6 |
Sedoheptulokinase | 3 | 181.59 | 213.1 |
| Q19LI2 |
α-1B-glycoprotein | 3 | 201.11 | 199.9 |
| Q99020 | Heterogeneous
nuclear ribonucleoprotein A/B | 2 | 120.87 | 131.4 |
| P09405 | Nucleolin | 2 | 66.85 | 104.3 |
| Q62446 | Peptidyl-prolyl
cis-trans isomerase FKBP3 | 2 | 69.16 | 89.1 |
| Q8BG05 | Heterogeneous
nuclear ribonucleoprotein A3 | 2 | 132.17 | 72.8 |
| Q08331 | Calretinin | 5 | 174.3 | 59.9 |
| P07361 | α-1-acid
glycoprotein 2 | 2 | 101.47 | 56.8 |
| P00493 |
Hypoxanthine-guanine
phosphoribosyltransferase | 2 | 73.54 | 53.3 |
| Q61646 | Haptoglobin | 14 | 826.52 | 50.3 |
| Q9D0R2 | Threonine-tRNA
ligase, cytoplasmic | 5 | 187.79 | 48.4 |
| P48428 | Tubulin-specific
chaperone A | 2 | 59.15 | 46.8 |
| Q99KP3 | Lambda-crystallin
homolog | 3 | 173.73 | 43.3 |
| A2ABU4 | Myomesin-3 | 9 | 588.37 | 38.6 |
| Q99KC8 | von Willebrand
factor A domain-containing protein 5A | 2 | 41.66 | 37.2 |
| P05366 | Serum amyloid A-1
protein | 2 | 135.98 | 36.9 |
| Q8VC12 | Urocanate
hydratase | 10 | 751.33 | 36.9 |
| P47199 | Quinone
oxidoreductase | 4 | 206.42 | 36.8 |
| Q91X52 | L-xylulose
reductase | 2 | 99.38 | 32.8 |
| Q8VCX1 | 3-oxo-5-β-steroid
4-dehydrogenase | 5 | 177.09 | 31.2 |
| Q9WTP6 | Adenylate kinase 2,
mitochondrial | 2 | 67.03 | 31.2 |
| A3KMP2 | Tetratricopeptide
repeat protein 38 | 4 | 177.89 | 30.6 |
| Q5SX40 | Myosin-1 | 62 | 4670.12 | 30.3 |
| A2ASS6 | Titin | 88 | 4357.28 | 30.2 |
| P99027 | 60S acidic
ribosomal protein P2 | 2 | 162.5 | 30.1 |
| Q8VCT4 | Carboxylesterase
1D | 5 | 114.1 | 26.8 |
| P53026 | 60S ribosomal
protein L10a | 2 | 97.35 | 26.5 |
| P68040 | Guanine
nucleotide-binding protein subunit β-2-like 1 | 2 | 48.55 | 25.6 |
| Q9QXE0 | 2-Hydroxyacyl-CoA
lyase 1 | 4 | 252.9 | 24.5 |
 | Table IIList of previously established
increased serum marker proteins that were also identified in
6-month-old mdx-4cv mouse serum vs. age-matched
wild-type mouse serum by label-free mass spectrometry. |
Table II
List of previously established
increased serum marker proteins that were also identified in
6-month-old mdx-4cv mouse serum vs. age-matched
wild-type mouse serum by label-free mass spectrometry.
| Accession no. | Protein name | Unique
peptides | Confidence
score | Fold change |
|---|
| Q05816 | Fatty acid-binding
protein, epidermal | 8 | 385.47 | 18.3 |
| P13412 | Troponin I, fast
skeletal muscle | 3 | 131.21 | 18.2 |
| P62897 | Cytochrome c,
somatic | 5 | 543.08 | 15.2 |
| P05063 |
Fructose-bisphosphate aldolase C | 9 | 679.32 | 14.8 |
| P05977 | Myosin light chain
1/3, skeletal muscle isoform | 9 | 561.43 | 12.0 |
| P52480 | Pyruvate kinase
PKM | 46 | 4564.3 | 10.4 |
| Q9ET01 | Glycogen
phosphorylase, liver form | 36 | 2184.82 | 9.7 |
| P12710 | Fatty acid-binding
protein, liver | 11 | 928.62 | 8.4 |
| Q04447 | Creatine kinase
B-type | 17 | 1492.20 | 8.2 |
| P11499 | Heat shock protein
HSP 90-β | 4 | 202.62 | 7.6 |
| Q9WUB3 | Glycogen
phosphorylase, muscle form | 48 | 4290.78 | 7.0 |
| Q91Y97 |
Fructose-bisphosphate aldolase B | 20 | 1745.78 | 6.4 |
| P17183 | γ-enolase | 6 | 437.74 | 5.7 |
| P08249 | Malate
dehydrogenase, mitochondrial | 6 | 274.29 | 5.7 |
| P20801 | Troponin C,
skeletal muscle | 3 | 192.78 | 5.6 |
| Q9D0F9 |
Phosphoglucomutase-1 | 22 | 1318.77 | 4.0 |
| Q61316 | Heat shock 70 kDa
protein 4 | 20 | 1146.72 | 3.9 |
| P53657 | Pyruvate kinase
PKLR | 3 | 119.98 | 3.7 |
| Q6P8J7 | Creatine kinase
S-type, mitochondrial | 10 | 596.62 | 3.7 |
| P14602 | Heat shock protein
β-1 | 3 | 150.33 | 3.3 |
| P14152 | Malate
dehydrogenase, cytoplasmic | 13 | 1108.02 | 3.3 |
| P05064 |
Fructose-bisphosphate aldolase A | 30 | 3015.5 | 3.1 |
| P16125 | L-lactate
dehydrogenase B chain | 12 | 1000.89 | 2.9 |
| P07901 | Heat shock protein
HSP 90-α | 4 | 172.93 | 2.8 |
| P17182 | α-enolase | 25 | 1951.59 | 2.7 |
| P32848 | Parvalbumin α | 16 | 1096.30 | 2.6 |
| P63017 | Heat shock cognate
71 kDa protein | 18 | 1479.96 | 2.5 |
| P16015 | Carbonic anhydrase
3 | 15 | 830.57 | 2.4 |
| P07310 | Creatine kinase
M-type | 30 | 2895.13 | 2.4 |
| P11276 | Fibronectin | 6 | 298.37 | 2.3 |
| P04117 | Fatty acid-binding
protein, adipocyte | 9 | 800.82 | 2.2 |
| Q9R0Y5 | Adenylate kinase
isoenzyme 1 | 3 | 247.3 | 2.2 |
| O70250 | Phosphoglucomutase
2 | 10 | 714.82 | 2.2 |
| P21550 | β-enolase | 23 | 2090.28 | 2.1 |
| P11404 | Fatty acid-binding
protein, heart | 5 | 281.57 | 1.6 |
| Q91X72 | Hemopexin | 32 | 2684.29 | 1.5 |
| O88783 | Coagulation factor
V | 6 | 223.63 | 1.3 |
 | Table IIIList of decreased proteins in
6-month-old mdx-4cv mouse serum vs. age-matched
wild-type mouse serum exhibiting a fold-change above 2 as
determined by label-free mass spectrometry. |
Table III
List of decreased proteins in
6-month-old mdx-4cv mouse serum vs. age-matched
wild-type mouse serum exhibiting a fold-change above 2 as
determined by label-free mass spectrometry.
| Accession no. | Protein name | Unique
peptides | Confidence
score | Fold change |
|---|
| P09470 |
Angiotensin-converting enzyme | 2 | 97.76 | 3.5 |
| O35930 | Platelet
glycoprotein Ib α chain | 11 | 865.35 | 3.4 |
| Q07456 | Protein AMBP | 4 | 362.51 | 3.1 |
| P28665 |
Murinoglobulin-1 | 116 | 12973.81 | 2.9 |
| B5X0G2 | Major urinary
protein 17 | 5 | 231.6 | 2.8 |
| Q00898 | α-1-antitrypsin
1-5 | 14 | 1024.28 | 2.7 |
| P05208 | Chymotrypsin-like
elastase family member 2A | 3 | 230.49 | 2.7 |
| O08742 | Platelet
glycoprotein V | 5 | 601.56 | 2.6 |
| Q03311 | Cholinesterase | 7 | 362.36 | 2.6 |
| P01942 | Hemoglobin subunit
α | 7 | 448.81 | 2.6 |
| P42703 | Leukemia inhibitory
factor receptor | 28 | 2444.33 | 2.5 |
| Q62351 | Transferrin
receptor protein 1 | 5 | 278.54 | 2.5 |
| Q9DBB9 | Carboxypeptidase N
subunit 2 | 15 | 1509.87 | 2.4 |
| Q8QZR3 | Pyrethroid
hydrolase Ces2a | 2 | 114.66 | 2.4 |
| Q8BPB5 | EGF-containing
fibulin-like extracellular matrix protein 1 | 4 | 182.66 | 2.4 |
| O88968 |
Transcobalamin-2 | 9 | 577.32 | 2.4 |
| Q61704 | Inter-α-trypsin
inhibitor heavy chain H3 | 7 | 627.52 | 2.3 |
| Q03734 | Serine protease
inhibitor A3M | 9 | 788.27 | 2.3 |
| E9Q414 | Apolipoprotein
B-100 | 35 | 1947.84 | 2.3 |
| Q07235 | Glia-derived
nexin | 5 | 234.21 | 2.3 |
The most markedly elevated levels included
Na+/H+ exchange regulatory cofactor NHE-RF1,
sedoheptulokinase, α-1B-glycoprotein, heterogeneous nuclear
ribonucleoprotein A/B and A3, nucleolin, peptidyl-prolyl
cis-trans isomerase FKBP3, calretinin, α-1-acid
glycoprotein 2, hypoxanthine-guanine phosphoribosyl-transferase,
haptoglobin, cytoplasmic threonine-tRNA ligase, tubulin-specific
chaperone A, lambda-crystallin homolog, myomesin-3, von Willebrand
factor A domain-containing protein 5A, serum amyloid A-1 protein,
urocanate hydratase, quinone oxidoreductase, L-xylulose reductase,
3-oxo-5-β-steroid 4-dehydrogenase, mitochondrial adenylate kinase
2, tetratricopeptide repeat protein 38, myosin-1 and titin. A very
interesting finding was the identification of increased
haptoglobin, which represents a typical acute phase plasma marker
protein of tissue damage and the inflammatory response. The
elevated concentration of haptoglobin was further investigated by
immuno-labelling approaches, as outlined below. Besides serum
haptoglobin, elevated levels of another acute phase plasma protein
were established for amyloid A-1 protein (Table I).
Importantly, a variety of common serum markers of
dystrophinopathy were verified by mass spectrometry (Tables I and II), including increased amounts of
creatine kinase isoforms CK-B, CK-S and CK-M, pyruvate kinase
isoforms PKM and PKLR, hemopexin, adenylate kinase isoforms AK1 and
AK2, carbonic anhydrase isoform CA3 and lactate dehydrogenase,
which have been previously established by non-proteomic techniques
(18–22). Elevated levels of previously
identified proteomic serum markers were also confirmed (Tables I and II), such as fibronectin, coagulation
factor V, titin, myosin light chain 1/3, fructose-bisphosphate
aldolase (isoforms A–C), phosphoglucomutase (isoforms 1 and 2),
enolase (isoforms α, β and γ), glycogen phosphorylase (muscle and
liver isoforms), fatty acid-binding protein (adipocyte, epidermal,
liver and heart isoforms), cytochrome c, malate
dehydrogenase (cytoplasmic and mitochondrial isoform), parvalbumin,
troponin (muscle isoforms TnI and TnC) and various heat-shock
proteins (HSP90-β, HSP90-α, HSP70, HSP β-1, HSP cognate 71)
(30–39). These findings demonstrate the
reproducibility of the findings from a large number of biochemical
and proteomic surveys of serum and plasma preparations from
dystrophic patients and animal models of Duchenne muscular
dystrophy.
The degree of decreased serum proteins was markedly
less pronounced as compared to the increased protein species.
Moderately reduced levels were shown to include
angiotensin-converting enzyme, platelet glycoprotein Ib α chain,
protein AMBP, murinoglobulin-1, major urinary protein 17,
α-1-antitrypsin 1–5, chymotrypsin-like elastase family member 2A,
platelet glycoprotein V, cholinesterase, hemoglobin subunit α,
leukemia inhibitory factor receptor, transferrin receptor protein 1
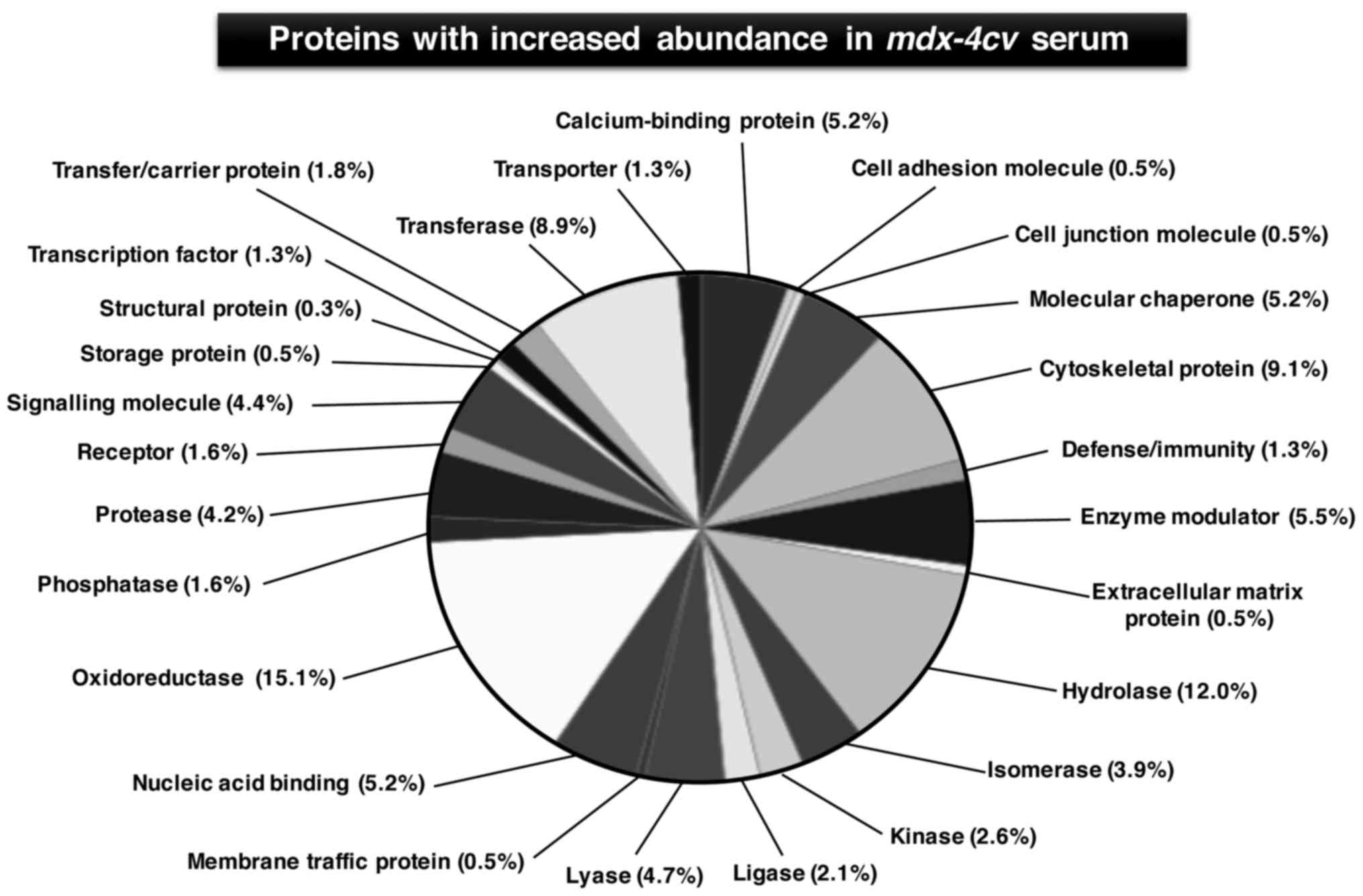
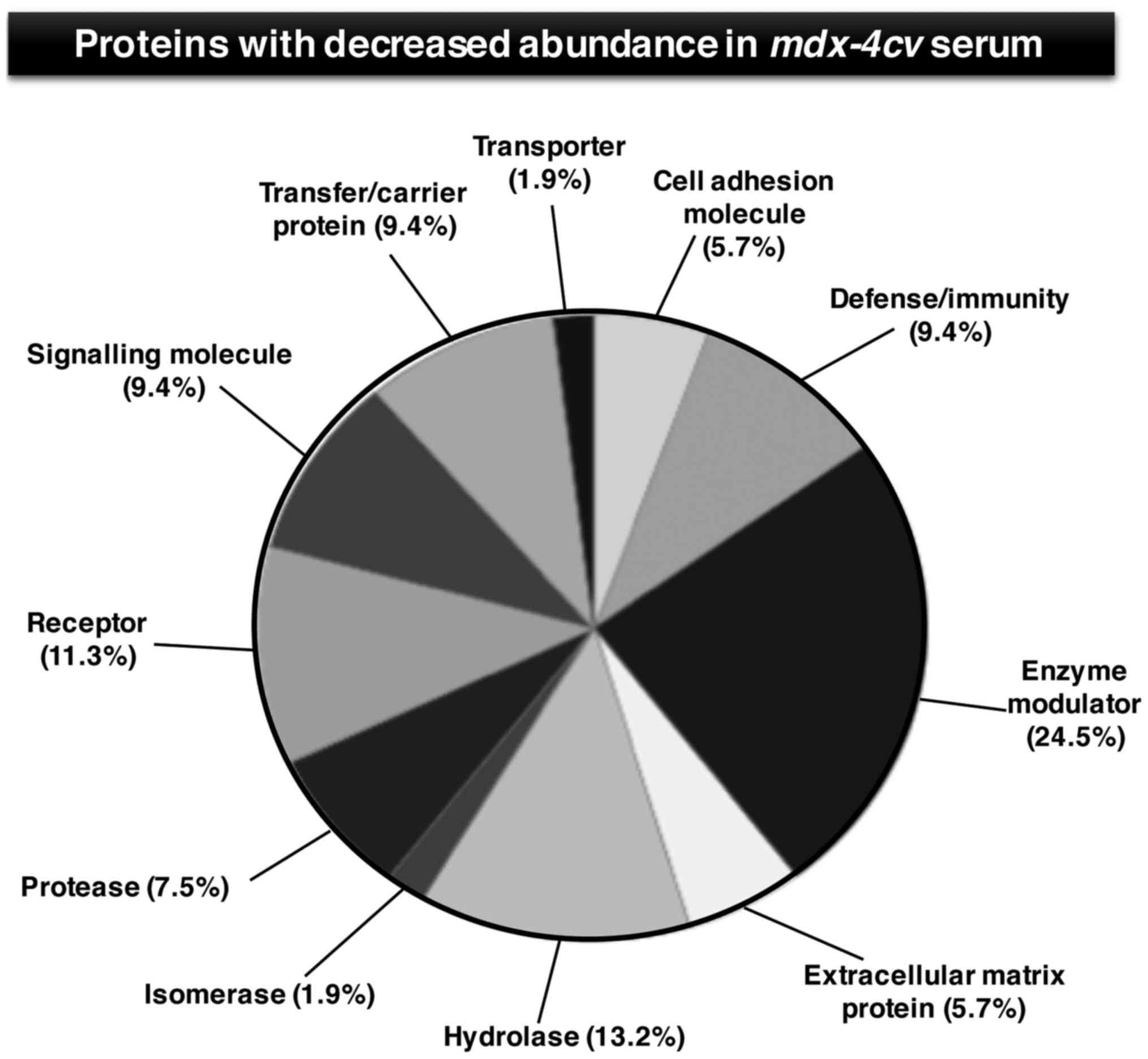
and carboxypeptidase N subunit 2 (Table III). Figs. 4 and 5 show the bioinformatic PANTHER analysis
of protein families (56) that
were established to exhibit an altered concentration in
mdx-4cv mouse serum.
Verification of proteomic profiling
findings
Following the identification of a large number of
altered serum proteins in the mdx-4cv model of
dystrophinopathy, we focused our study on the verification of a few
key proteomic findings with a special emphasis on haptoglobin.
Proteomic results were confirmed by immunoblotting and
enzyme-linked immunosorbent assays with specific antibodies to a
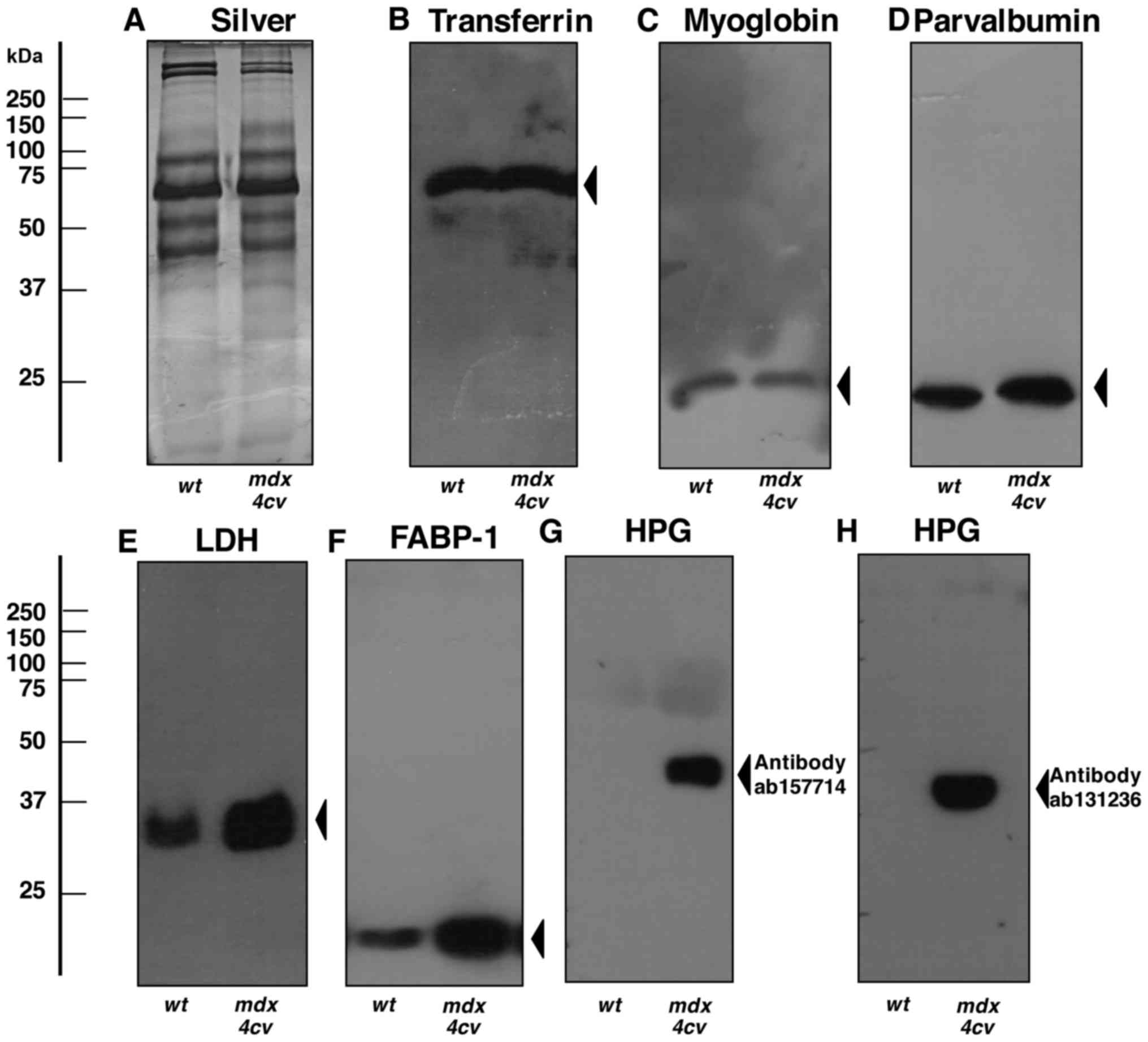
select number of serum proteins (Figs. 6Figure 7–8). The immunoblot analysis of crude
serum showed no significant changes in the abundance of serum
transferrin, myoglobin and parvalbumin (Fig. 6B–D). In contrast, lactate
dehydrogenase and the FABP-1 isoform of fatty acid binding protein
exhibited elevated levels in crude mdx-4cv serum
(Figs. 6E and F). Most
importantly, the highly increased concentration of serum
haptoglobin in dystrophic mice, as shown by mass spectrometric
analysis (Table I) was confirmed
by immunoblotting with two different antibodies to this acute phase
plasma protein (Fig. 6G and H).
The statistical evaluation of immunoblotting is summarized in
Fig. 7 and establishes the
significant elvation of lactate dehydrogenase, fatty acid binding
protein FABP-1 and haptoglobin in crude mdx-4cv
serum.
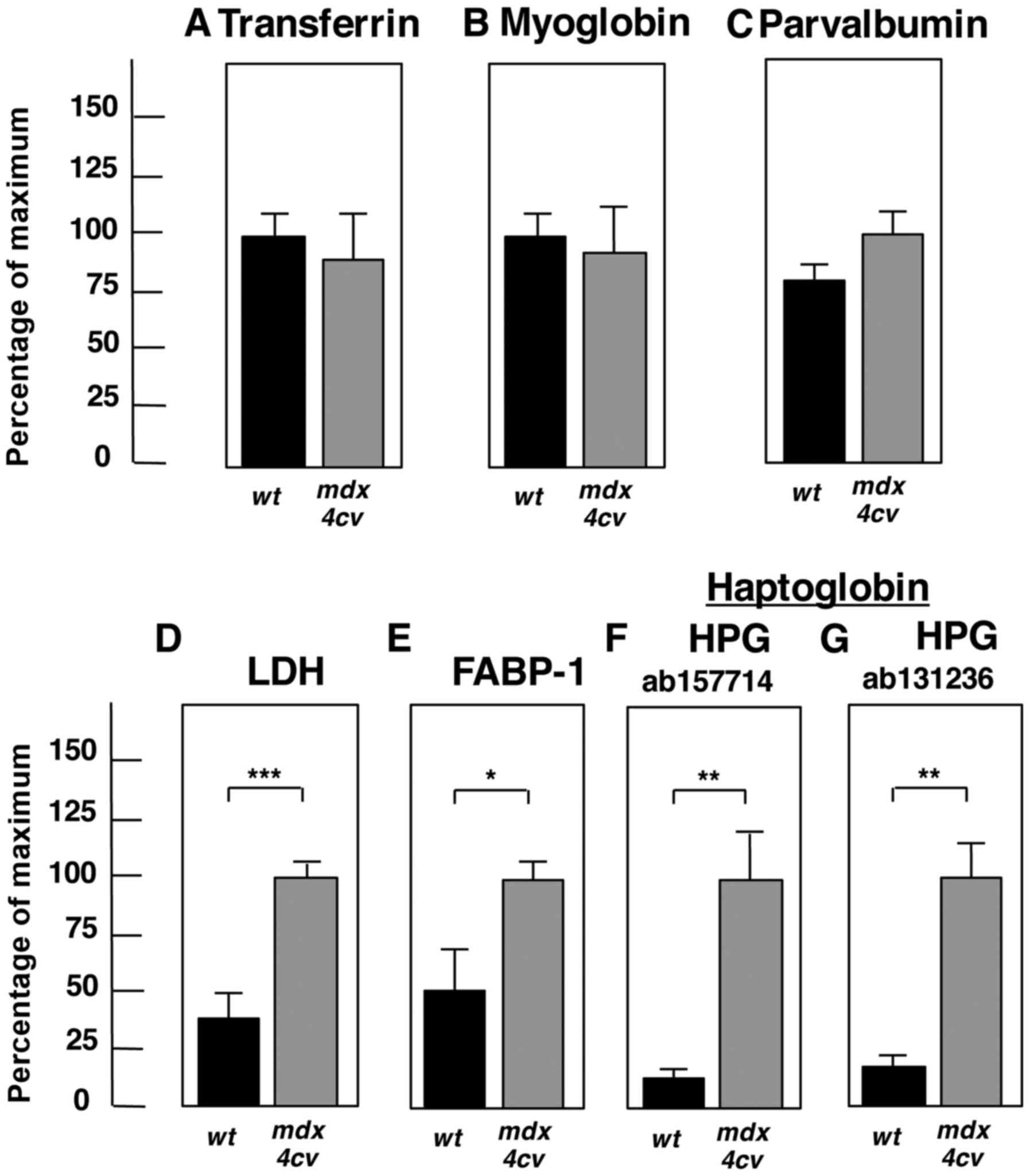 | Figure 7Significant changes of proteins in
the mdx-4cv mouse serum. Shown is the graphical
presentation of the statistical evaluation of the immunoblots
depicted in Fig. 6. The
comparative blotting between crude serum samples from 6-month-old
wild-type (wt) vs. mdx-4cv mice was statistically
evaluated using an unpaired Student's t-test (n=4;
*P<0.05, **P<0.01,
***P<0.001). Shown is the analysis of (A)
transferrin, (B) myoglobin, (C) parvalbumin, (D) LDH, (E) FABP-1,
and (F and G) HPG. LDH, lactate dehydrogenase; FABP, fatty acid
binding protein; HPG, haptoglobin. |
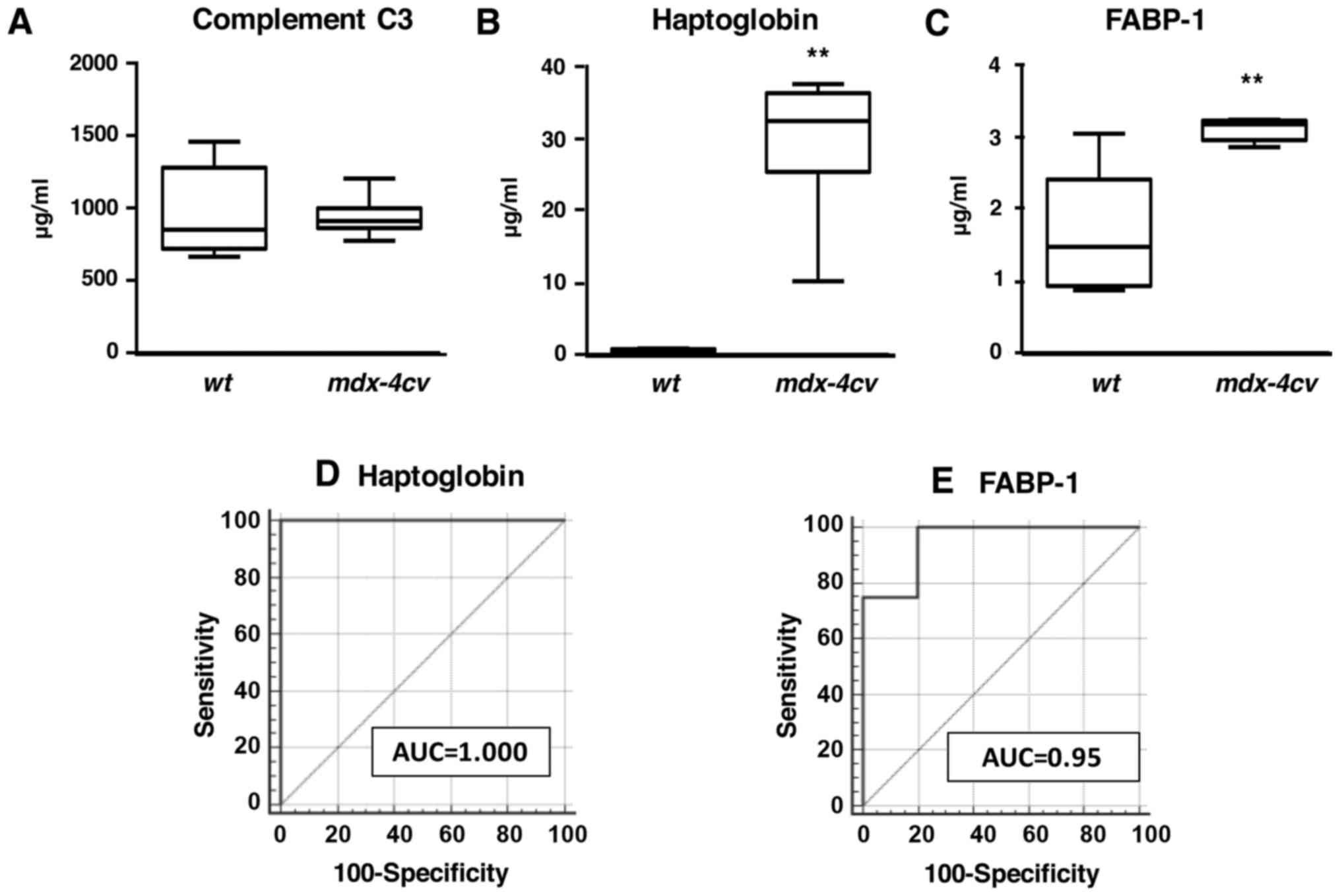
In analogy to immunoblot analyses, enzyme-linked
immunosorbent assays with antibodies to haptoglobin and FABP-1 also
showed increased levels of these two proteins in crude serum from
dystrophic animals (Figs. 8B and
C). Statistical ROC analyses demonstrated that these findings
were highly significant (Figs. 8D and
E). In contrast, values for complement C3 were found to be
comparable between wild-type and mdx-4cv serum
(Fig. 8A).
Discussion
A key step in the molecular pathogenesis of X-linked
muscular dystrophy is mediated by the disintegration of the
dystrophin-glycoprotein complex (4) resulting in sarcolemmal
destabilisation, impaired cellular signalling, abnormal ion fluxes
and increased proteolysis (6).
This primary mechanism of fibre damage is accompanied by adaptive
responses, such as myofibrosis (10), and secondary changes related to an
inflammatory pathology (16). In
addition to massive proteome-wide changes within skeletal muscle
fibres (46,50–52,60,72–74), the deficiency in dystrophin
isoform Dp427 also causes substantial alterations in the rate of
protein release from muscle tissues into the circulatory system and
other plasma fluctuations (30–39). This includes actively secreted
proteins and passively released components from mechanically
damaged fibres and other types of tissue (29). In this respect, one of the major
findings of the proteomic serum study presented here is the
identification of elevated haptoglobin levels in
mdx-4cv serum by mass spectrometry, immunoblotting
and enzyme-linked immunosorbent assays.
This acute phase plasma protein is a tissue damage
marker and is often increased in serum in relation to cellular
degeneration and inflammation (75). Since there is an urgent need to
establish new biomarker candidates of X-linked muscular dystrophy
(76), the proteomic
identification of elevated haptoglobin levels in dystrophinopathy
is an encouraging result. Haptoglobin, in conjunction with other
altered serum proteins, may represent a novel diagnostic,
prognostic and/or therapy-monitoring biomarker candidate for the
improved evaluation of sterile inflammation in the
mdx-4cv animal model of dystrophinopathy. Haptoglobin
is a major acute phase plasma protein that acts as a high-affinity
hemoglobin-binding protein and protective anti-oxidant (77). The liver is the main site of
haptoglobin synthesis, but this protein is also produced in other
organs (43). The increased serum
haptoglobin levels established here by mass spectrometry agree with
a pre-proteomic analysis by two-dimensional gel electrophoresis and
densitometric scanning by John and Purdom (78). A recent serum proteomic profiling
study by Rouillon et al (37) with a focus on the characterization
of the myofibrillar structural protein myomesin isoform MYOM3 also
identified haptoglobin as a potential biomarker candidate for
monitoring the outcome of therapeutic interventions in Duchenne
muscular dystrophy.
The cellular disturbances and multi-system pathology
of dystrophinopathy are closely associated with highly complex and
interconnecting damage pathways, including modifying factors such
as abnormal Ca2+ flux, increased proteolytic
degradation, elevated extracellular ATP levels and altered cytokine
signalling (6,8). In addition to promoting hyperactive
extracellular matrix remodelling and resulting fibrotic scarring,
these processes stimulate harmful inflammatory responses (79). Macrophages are intrinsically
involved in both the natural repair of acutely injured skeletal
muscle fibres and the pathological exacerbation of chronically
damaged contractile cells (80).
Inflammatory monocyte-derived macrophages appear to be closely
associated with disturbed chemokine levels and are key cell types
that promote changes in dystrophin-deficient and degenerating
skeletal muscles (13,81). Sterile tissue injury that occurs
chronically in muscular dystrophy initiates a local inflammatory
response. This in turn triggers the mobilization of the acute phase
reaction causing a massive increase in the level of acute phase
plasma proteins in the circulatory system.
As a biomarker of dystrophinopathy, the advantages
of serum haptoglobin are manifold as compared to muscle-associated
proteins, since the application of diagnostic or prognostic
biomarkers associated with the skeletal muscle interior would
require the undesirable usage of invasive methodology (62). In contrast, the systematic mining
of the biofluid proteome promises to identify novel marker
molecules that can be assayed in a minimally invasive or
non-invasive manner and may also result in the proteomic
identification of new therapeutic targets (82–84). However, it is important to
emphasise that the field of predictive and diagnostic medicine has
had only limited success in the translation from biomarker
discovery to clinical utility, especially in relation to biomarker
specificity (85). This would
also be a critical issue in the future application of haptoglobin
as a biomarker, since this serum protein was recently identified as
a novel marker molecule, which is potentially suitable for
predicting a variety of human diseases including colorectal cancer
hepatic metastasis (86).
In the case of muscular dystrophy, one can assume
that damaged and leaky skeletal muscle tissue has an enhanced
tendency to actively release or passively shed proteins,
metabolites and other biomolecules into their external environment.
These factors also trigger secondary changes in other tissue and
organ systems in the body and thereby cause considerable plasma
fluctuations. The range and density of released protein species
relates relatively well to the degree of cellular damage (29). In addition to the effects on acute
phase plasma protein concentration, this proteomic study has shown
that highly complex proteome-wide alterations occur in the serum in
a dystrophic organism. As established by the systems bioinformatic
analysis of the proteomic mdx-4cv serum data
(56), affected protein families
include cytoskeletal proteins, hydrolases, oxidoreductases,
transferases, nucleic acid binding proteins, molecular chaperones,
proteases, signalling molecules and ion-handling proteins. These
proteins with an altered serum concentration are involved in many
complex cellular activities, such as energy metabolism,
physiological regulation, fibre adaptations, intercellular and
intracellular signalling, the stress response, fibre transitions
and the excitation-contraction-relaxation cycle. The proteomic
findings emphasize the pathobiochemical complexity of the
dystrophic phenotype and suggest that future therapeutic approaches
have to address a variety of downstream changes from the primary
abnormality in the membrane cytoskeleton (3).
In conclusion, the identification of robust and
unambiguous biomarkers of high biomedical and clinical value is an
important aim of the systematic assessment of pathological
specimens related to X-linked muscular dystrophy. Here, we
identified elevated levels of the acute phase plasma protein
haptoglobin in the serum from dystrophic mdx-4cv mice
as a new potential indicator molecule of the inflammatory phenotype
of dystrophinoapthy. Since antibody-based tests have confirmed the
significantly increased concentration of serum haptoglobin, this
protein represents a solid biomarker candidate. Haptoglobin is
easily assessable, exhibits a high diagnostic potential and has
suitable physicochemical properties. Hence, this serum protein is
an ideal candidate molecule for the future establishment of a
sensitive and cost-effective test system to evaluate inflammatory
processes related to skeletal muscle damage in muscular
dystrophy.
Acknowledgments
The present research was supported by a Hume
scholarship from Maynooth University and project grants from the
Deutsche Duchenne Stiftung aktion benni & co e.V. the Health
Research Board and Muscular Dystrophy Ireland. The Q-Exactive
quantitative mass spectrometer was funded under the Research
Infrastructure Call 2012 by the Science Foundation of Ireland
(SFI-12/RI/2346/3).
References
|
1
|
Flanigan KM: Duchenne and Becker muscular
dystrophies. Neurol Clin. 32:671–688. viii2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mah JK, Korngut L, Dykeman J, Day L,
Pringsheim T and Jette N: A systematic review and meta-analysis on
the epidemiology of Duchenne and Becker muscular dystrophy.
Neuromuscul Disord. 24:482–491. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guiraud S, Aartsma-Rus A, Vieira NM,
Davies KE, van Ommen GJ and Kunkel LM: The pathogenesis and therapy
of muscular dystrophies. Annu Rev Genomics Hum Genet. 16:281–308.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohlendieck K: Towards an understanding of
the dystrophin-glycoprotein complex: Linkage between the
extracellular matrix and the membrane cytoskeleton in muscle
fibers. Eur J Cell Biol. 69:1–10. 1996.PubMed/NCBI
|
|
5
|
Allen DG, Zhang BT and Whitehead NP:
Stretch-induced membrane damage in muscle: Comparison of wild-type
and mdx mice. Adv Exp Med Biol. 682:297–313. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Allen DG, Whitehead NP and Froehner SC:
Absence of dystrophin disrupts skeletal muscle signaling: Roles of
Ca2+, reactive oxygen species, and nitric oxide in the
development of muscular dystrophy. Physiol Rev. 96:253–305. 2016.
View Article : Google Scholar
|
|
7
|
Hopf FW, Turner PR and Steinhardt RA:
Calcium misregulation and the pathogenesis of muscular dystrophy.
Subcell Biochem. 45:429–464. 2007. View Article : Google Scholar
|
|
8
|
Shin J, Tajrishi MM, Ogura Y and Kumar A:
Wasting mechanisms in muscular dystrophy. Int J Biochem Cell Biol.
45:2266–2279. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mázala DA, Grange RW and Chin ER: The role
of proteases in excitation-contraction coupling failure in muscular
dystrophy. Am J Physiol Cell Physiol. 308:C33–C40. 2015. View Article : Google Scholar :
|
|
10
|
Holland A, Murphy S, Dowling P and
Ohlendieck K: Pathoproteomic profiling of the skeletal muscle
matrisome in dystrophinopathy associated myofibrosis. Proteomics.
16:345–366. 2016. View Article : Google Scholar
|
|
11
|
Serra F, Quarta M, Canato M, Toniolo L, De
Arcangelis V, Trotta A, Spath L, Monaco L, Reggiani C and Naro F:
Inflammation in muscular dystrophy and the beneficial effects of
non-steroidal anti-inflammatory drugs. Muscle Nerve. 46:773–784.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
De Paepe B and De Bleecker JL: Cytokines
and chemokines as regulators of skeletal muscle inflammation:
Presenting the case of Duchenne muscular dystrophy. Mediators
Inflamm. 540370:2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mojumdar K, Liang F, Giordano C, Lemaire
C, Danialou G, Okazaki T, Bourdon J, Rafei M, Galipeau J, Divangahi
M and Petrof BJ: Inflammatory monocytes promote progression of
Duchenne muscular dystrophy and can be therapeutically targeted via
CCR2. EMBO Mol Med. 6:1476–1492. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Villalta SA, Rosenberg AS and Bluestone
JA: The immune system in Duchenne muscular dystrophy: Friend or
foe. Rare Dis. 3:e10109662015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tidball JG: Mechanisms of muscle injury,
repair, and regeneration. Compr Physiol. 1:2029–2062.
2011.PubMed/NCBI
|
|
16
|
Rosenberg AS, Puig M, Nagaraju K, Hoffman
EP, Villalta SA, Rao VA, Wakefield LM and Woodcock J:
Immune-mediated pathology in Duchenne muscular dystrophy. Sci
Transl Med. 7:299rv42015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Villalta SA, Rosenthal W, Martinez L, Kaur
A, Sparwasser T, Tidball JG, Margeta M, Spencer MJ and Bluestone
JA: Regulatory T cells suppress muscle inflammation and injury in
muscular dystrophy. Sci Transl Med. 6:258ra1422014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Proud CM: 50 years ago in the Journal of
Pediatrics: The use of serum creatine phosphokinase and other serum
enzymes in the diagnosis of progressive muscular dystrophy. J
Pediatr. 163:16562013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Percy ME, Andrews DF and Thompson MW:
Duchenne muscular dystrophy carrier detection using logistic
discrimination: Serum creatine kinase, hemopexin, pyruvate kinase,
and lactate dehydrogenase in combination. Am J Med Genet. 13:27–38.
1982. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Carter ND, Heath R, Jeffery S, Jackson MJ,
Newham DJ and Edwards RH: Carbonic anhydrase III in Duchenne
muscular dystrophy. Clin Chim Acta. 133:201–208. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fröhlich T, Reitter B, Scheffner D,
Schirmer RH and Untucht-Grau R: Muscle adenylate kinase in Duchenne
muscular dystrophy. Biochim Biophys Acta. 883:598–603. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Percy ME, Chang LS, Murphy EG, Oss I,
Verellen-Dumoulin C and Thompson MW: Serum creatine kinase and
pyruvate kinase in Duchenne muscular dystrophy carrier detection.
Muscle Nerve. 2:329–339. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
D'Amore PA, Brown RH Jr, Ku PT, Hoffman
EP, Watanabe H, Arahata K, Ishihara T and Folkman J: Elevated basic
fibroblast growth factor in the serum of patients with Duchenne
muscular dystrophy. Ann Neurol. 35:362–365. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bernasconi P, Torchiana E, Confalonieri P,
Brugnoni R, Barresi R, Mora M, Cornelio F, Morandi L and Mantegazza
R: Expression of transforming growth factor-beta 1 in dystrophic
patient muscles correlates with fibrosis. Pathogenetic role of a
fibrogenic cytokine. J Clin Invest. 96:1137–1144. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun G, Haginoya K, Chiba Y, Uematsu M,
Hino-Fukuyo N, Tanaka S, Onuma A, Iinuma K and Tsuchiya S: Elevated
plasma levels of tissue inhibitors of metalloproteinase-1 and their
overexpression in muscle in human and mouse muscular dystrophy. J
Neurol Sci. 297:19–28. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nadarajah VD, van Putten M, Chaouch A,
Garrood P, Straub V, Lochmüller H, Ginjaar HB, Aartsma-Rus AM, van
Ommen GJ, den Dunnen JT and 't Hoen PA: Serum matrix
metalloproteinase-9 (MMP-9) as a biomarker for monitoring disease
progression in Duchenne muscular dystrophy (DMD). Neuromuscul
Disord. 21:569–578. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Holland A, Carberry S and Ohlendieck K:
Proteomics of the dystrophin-glycoprotein complex and
dystrophinopathy. Curr Protein Pept Sci. 14:680–697. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fuller HR, Graham LC, Llavero Hurtado M
and Wishart TM: Understanding the molecular consequences of
inherited muscular dystrophies: Advancements through proteomic
experimentation. Expert Rev Proteomics. 13:659–671. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hathout Y, Seol H, Han MH, Zhang A, Brown
KJ and Hoffman EP: Clinical utility of serum biomarkers in Duchenne
muscular dystrophy. Clin Proteomics. 13:92016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Alagaratnam S, Mertens BJ, Dalebout JC,
Deelder AM, van Ommen GJ, den Dunnen JT and 't Hoen PA: Serum
protein profiling in mice: Identification of Factor XIIIa as a
potential biomarker for muscular dystrophy. Proteomics.
8:1552–1563. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Duguez S, Duddy W, Johnston H, Lainé J, Le
Bihan MC, Brown KJ, Bigot A, Hathout Y, Butler-Browne G and
Partridge T: Dystrophin deficiency leads to disturbance of
LAMP1-vesicle-associated protein secretion. Cell Mol Life Sci.
70:2159–2174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hathout Y, Marathi RL, Rayavarapu S, Zhang
A, Brown KJ, Seol H, Gordish-Dressman H, Cirak S, Bello L, Nagaraju
K, et al: Discovery of serum protein biomarkers in the mdx mouse
model and cross-species comparison to Duchenne muscular dystrophy
patients. Hum Mol Genet. 23:6458–6469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cynthia Martin F, Hiller M, Spitali P,
Oonk S, Dalebout H, Palmblad M, Chaouch A, Guglieri M, Straub V,
Lochmüller H, et al: Fibronectin is a serum biomarker for Duchenne
muscular dystrophy. Proteomics Clin Appl. 8:269–278. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ayoglu B, Chaouch A, Lochmüller H,
Politano L, Bertini E, Spitali P, Hiller M, Niks EH, Gualandi F,
Pontén F, et al: Affinity proteomics within rare diseases: A
BIO-NMD study for blood biomarkers of muscular dystrophies. EMBO
Mol Med. 6:918–936. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rouillon J, Zocevic A, Leger T, Garcia C,
Camadro JM, Udd B, Wong B, Servais L, Voit T and Svinartchouk F:
Proteomics profiling of urine reveals specific titin fragments as
biomarkers of Duchenne muscular dystrophy. Neuromuscul Disord.
24:563–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Coenen-Stass AM, McClorey G, Manzano R,
Betts CA, Blain A, Saleh AF, Gait MJ, Lochmüller H, Wood MJ and
Roberts TC: Identification of novel, therapy-responsive protein
biomarkers in a mouse model of Duchenne muscular dystrophy by
aptamer-based serum proteomics. Sci Rep. 5:170142015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rouillon J, Poupiot J, Zocevic A, Amor F,
Léger T, Garcia C, Camadro JM, Wong B, Pinilla R, Cosette J, et al:
Serum proteomic profiling reveals fragments of MYOM3 as potential
biomarkers for monitoring the outcome of therapeutic interventions
in muscular dystrophies. Hum Mol Genet. 24:4916–4932. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hathout Y, Brody E, Clemens PR, Cripe L,
DeLisle RK, Furlong P, Gordish-Dressman H, Hache L, Henricson E,
Hoffman EP, et al: Large-scale serum protein biomarker discovery in
Duchenne muscular dystrophy. Proc Natl Acad Sci USA. 112:7153–7158.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Oonk S, Spitali P, Hiller M, Switzar L,
Dalebout H, Calissano M, Lochmüller H, Aartsma-Rus A, 't Hoen PA
and van der Burgt YE: Comparative mass spectrometric and
immunoassay-based proteome analysis in serum of Duchenne muscular
dystrophy patients. Proteomics Clin Appl. 10:290–299. 2016.
View Article : Google Scholar
|
|
40
|
Im WB, Phelps SF, Copen EH, Adams EG,
Slightom JL and Chamberlain JS: Differential expression of
dystrophin isoforms in strains of mdx mice with different
mutations. Hum Mol Genet. 5:1149–1153. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Danko I, Chapman V and Wolff JA: The
frequency of revertants in mdx mouse genetic models for Duchenne
muscular dystrophy. Pediatr Res. 32:128–131. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mitrpant C, Fletcher S, Iversen PL and
Wilton SD: By-passing the nonsense mutation in the 4 CV mouse model
of muscular dystrophy by induced exon skipping. J Gene Med.
11:46–56. 2009. View Article : Google Scholar
|
|
43
|
Wang Y, Kinzie E, Berger FG, Lim SK and
Baumann H: Haptoglobin, an inflammation-inducible plasma protein.
Redox Rep. 6:379–385. 2001. View Article : Google Scholar
|
|
44
|
Chapman VM, Miller DR, Armstrong D and
Caskey CT: Recovery of induced mutations for X chromosome-linked
muscular dystrophy in mice. Proc Natl Acad Sci USA. 86:1292–1296.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Partridge TA: The mdx mouse model as a
surrogate for Duchenne muscular dystrophy. FEBS J. 280:4177–4186.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Carberry S, Zweyer M, Swandulla D and
Ohlendieck K: Comparative proteomic analysis of the
contractile-protein-depleted fraction from normal versus dystrophic
skeletal muscle. Anal Biochem. 446:108–115. 2014. View Article : Google Scholar
|
|
47
|
Hortin GL and Sviridov D: The dynamic
range problem in the analysis of the plasma proteome. J Proteomics.
73:629–636. 2010. View Article : Google Scholar
|
|
48
|
Anderson L and Anderson NG: The human
plasma proteome: History, character, and diagnostic prospects. Mol
Cell Proteomics. 1:845–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Holland A, Henry M, Meleady P, Winkler CK,
Krautwald M, Brinkmeier H and Ohlendieck K: Comparative label-free
mass spectrometric analysis of mildly versus severely affected mdx
mouse skeletal muscles identifies annexin, lamin, and vimentin as
universal dystrophic markers. Molecules. 20:11317–11344. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Holland A, Dowling P, Meleady P, Henry M,
Zweyer M, Mundegar RR, Swandulla D and Ohlendieck K: Label-free
mass spectrometric analysis of the mdx-4cv diaphragm identifies the
matricellular protein periostin as a potential factor involved in
dystrophinopathy-related fibrosis. Proteomics. 15:2318–2331. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Murphy S, Zweyer M, Mundegar RR, Henry M,
Meleady P, Swandulla D and Ohlendieck K: Concurrent label-free mass
spectrometric analysis of dystrophin isoform Dp427 and the
myofibrosis marker collagen in crude extracts from mdx-4cv skeletal
muscles. Proteomes. 3:298–327. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Di Luca A, Henry M, Meleady P and O'Connor
R: Label-free LC-MS analysis of HER2+ breast cancer cell
line response to HER2 inhibitor treatment. Daru. 23:402015.
View Article : Google Scholar
|
|
54
|
Linge A, Maurya P, Friedrich K, Baretton
GB, Kelly S, Henry M, Clynes M, Larkin A and Meleady P:
Identification and functional validation of RAD23B as a potential
protein in human breast cancer progression. J Proteome Res.
13:3212–3222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Murphy S, Dowling P, Zweyer M, Mundegar
RR, Henry M, Meleady P, Swandulla D and Ohlendieck K: Proteomic
analysis of dystrophin deficiency and associated changes in the
aged mdx-4cv heart model of dystrophinopathy-related
cardiomyopathy. J Proteomics. 145:24–36. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mi H, Muruganujan A and Thomas PD: PANTHER
in 2013: Modeling the evolution of gene function, and other gene
attributes, in the context of phylogenetic trees. Nucleic Acids
Res. 41(D1): D377–D386. 2013. View Article : Google Scholar :
|
|
57
|
Staunton L, Zweyer M, Swandulla D and
Ohlendieck K: Mass spectrometry-based proteomic analysis of
middle-aged vs. aged vastus lateralis reveals increased levels of
carbonic anhydrase isoform 3 in senescent human skeletal muscle.
Int J Mol Med. 30:723–733. 2012.PubMed/NCBI
|
|
58
|
Holland A, Dowling P, Zweyer M, Swandulla
D, Henry M, Clynes M and Ohlendieck K: Proteomic profiling of
cardiomyopathic tissue from the aged mdx model of Duchenne muscular
dystrophy reveals a drastic decrease in laminin, nidogen and
annexin. Proteomics. 13:2312–2323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lewis C, Jockusch H and Ohlendieck K:
Proteomic profiling of the dystrophin-deficient MDX heart reveals
drastically altered levels of key metabolic and contractile
proteins. J Biomed Biotechnol. 2010:6485012010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Murphy S, Henry M, Meleady P, Zweyer M,
Mundegar RR, Swandulla D and Ohlendieck K: Simultaneous
pathoproteomic evaluation of the dystrophin-glycoprotein complex
and secondary changes in the mdx-4cv mouse model of Duchenne
muscular dystrophy. Biology (Basel). 4:397–423. 2015.
|
|
61
|
Murphy S, Zweyer M, Henry M, Meleady P,
Mundegar RR, Swandulla D and Ohlendieck K: Label-free mass
spectrometric analysis reveals complex changes in the brain
proteome from the mdx-4cv mouse model of Duchenne muscular
dystrophy. Clin Proteomics. 12:272015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dowling P, Holland A and Ohlendieck K:
Mass spectrometry-based identification of muscle-associated and
muscle-derived proteomic biomarkers of dystrophinopathies. J
Neuromuscul Dis. 1:15–40. 2014.PubMed/NCBI
|
|
63
|
Gianazza E, Miller I, Palazzolo L,
Parravicini C and Eberini I: With or without you - Proteomics with
or without major plasma/serum proteins. J Proteomics. 140:62–80.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Omenn GS, States DJ, Adamski M, Blackwell
TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes
JS, et al: Overview of the HUPO Plasma Proteome Project: Results
from the pilot phase with 35 collaborating laboratories and
multiple analytical groups, generating a core dataset of 3020
proteins and a publicly-available database. Proteomics.
5:3226–3245. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Pietrowska M, Marczak L, Polanska J,
Behrendt K, Nowicka E, Walaszczyk A, Chmura A, Deja R, Stobiecki M,
Polanski A, et al: Mass spectrometry-based serum proteome pattern
analysis in molecular diagnostics of early stage breast cancer. J
Transl Med. 7:602009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Smith MP, Wood SL, Zougman A, Ho JT, Peng
J, Jackson D, Cairns DA, Lewington AJ, Selby PJ and Banks RE: A
systematic analysis of the effects of increasing degrees of serum
immunodepletion in terms of depth of coverage and other key aspects
in top-down and bottom-up proteomic analyses. Proteomics.
11:2222–2235. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Dowling P, Hayes C, Ting KR, Hameed A,
Meiller J, Mitsiades C, Anderson KC, Clynes M, Clarke C, Richardson
P and O'Gorman P: Identification of proteins found to be
significantly altered when comparing the serum proteome from
Multiple Myeloma patients with varying degrees of bone disease. BMC
Genomics. 15:9042014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Araújo JE, Jorge S, Teixeira E, Costa F,
Ramos A, Lodeiro C, Santos HM and Capelo JL: A cost-effective
method to get insight into the peritoneal dialysate effluent
proteome. J Proteomics. 145:207–213. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Gundry RL, Fu Q, Jelinek CA, Van Eyk JE
and Cotter RJ: Investigation of an albumin-enriched fraction of
human serum and its albuminome. Proteomics Clin Appl. 1:73–88.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Gundry RL, White MY, Nogee J, Tchernyshyov
I and Van Eyk JE: Assessment of albumin removal from an
immunoaffinity spin column: Critical implications for proteomic
examination of the albuminome and albumin-depleted samples.
Proteomics. 9:2021–2028. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tirumalai RS, Chan KC, Prieto DA, Issaq
HJ, Conrads TP and Veenstra TD: Characterization of the low
molecular weight human serum proteome. Mol Cell Proteomics.
2:1096–1103. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Rayavarapu S, Coley W, Cakir E, Jahnke V,
Takeda S, Aoki Y, Grodish-Dressman H, Jaiswal JK, Hoffman EP, Brown
KJ, et al: Identification of disease specific pathways using in
vivo SILAC proteomics in dystrophin deficient mdx mouse. Mol Cell
Proteomics. 12:1061–1073. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Roberts TC, Johansson HJ, McClorey G,
Godfrey C, Blomberg KE, Coursindel T, Gait MJ, Smith CI, Lehtiö J,
El Andaloussi S and Wood MJ: Multi-level omics analysis in a murine
model of dystrophin loss and therapeutic restoration. Hum Mol
Genet. 24:6756–6768. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Turk R, Hsiao JJ, Smits MM, Ng BH,
Pospisil TC, Jones KS, Campbell KP and Wright ME: Molecular
signatures of membrane protein complexes underlying muscular
dystrophy. Mol Cell Proteomics. 15:2169–2185. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Schrödl W, Büchler R, Wendler S, Reinhold
P, Muckova P, Reindl J and Rhode H: Acute phase proteins as
promising biomarkers: Perspectives and limitations for human and
veterinary medicine. Proteomics Clin Appl. 10:1077–1092. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ohlendieck K: Proteomic identification of
biomarkers of skeletal muscle disorders. Biomarkers Med. 7:169–186.
2013. View Article : Google Scholar
|
|
77
|
Levy AP, Asleh R, Blum S, Levy NS,
Miller-Lotan R, Kalet-Litman S, Anbinder Y, Lache O, Nakhoul FM,
Asaf, et al: Haptoglobin: Basic and clinical aspects. Antioxid
Redox Signal. 12:293–304. 2010. View Article : Google Scholar
|
|
78
|
John HA and Purdom IF: Elevated plasma
levels of haptoglobin in Duchenne muscular dystrophy:
Electrophoretic variants in patients with a severe form of the
disease. Electrophoresis. 10:489–493. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Górecki DC: Dystrophin: The dead calm of a
dogma. Rare Dis. 4:e11537772016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Kharraz Y, Guerra J, Mann CJ, Serrano AL
and Muñoz-Cánoves P: Macrophage plasticity and the role of
inflammation in skeletal muscle repair. Mediators Inflamm.
2013:4914972013. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Sinadinos A, Young CN, Al-Khalidi R, Teti
A, Kalinski P, Mohamad S, Floriot L, Henry T, Tozzi G, Jiang T, et
al: 2R X7 purinoceptor: A therapeutic target for ameliorating the
symptoms of duchenne muscular dystrophy. PLoS Med. 12:e10018882015.
View Article : Google Scholar
|
|
82
|
Veenstra TD, Conrads TP, Hood BL, Avellino
AM, Ellenbogen RG and Morrison RS: Biomarkers: Mining the biofluid
proteome. Mol Cell Proteomics. 4:409–418. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Savino R, Paduano S, Preianò M and
Terracciano R: The proteomics big challenge for biomarkers and new
drug-targets discovery. Int J Mol Sci. 13:13926–13948. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Stastna M and Van Eyk JE: Secreted
proteins as a fundamental source for biomarker discovery.
Proteomics. 12:722–735. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Drucker E and Krapfenbauer K: Pitfalls and
limitations in translation from biomarker discovery to clinical
utility in predictive and personalised medicine. EPMA J. 4:72013.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sun L, Hu S, Yu L, Guo C, Sun L, Yang Z,
Qi J and Ran Y: Serum haptoglobin as a novel molecular biomarker
predicting colorectal cancer hepatic metastasis. Int J Cancer.
138:2724–2731. 2016. View Article : Google Scholar : PubMed/NCBI
|