Introduction
Telomeres protect chromosomal ends from replicative
attrition (1,2). The maintenance of telomere length
and structure is critical to preserving telomere integrity. The
majority of human cancer cells maintain telomere length via the
activation of telomerase, which allows unlimited cell growth
(3). Telomerase reverse
transcriptase (TERT) is a rate-limiting component of telomerase
that is expressed only in telomerase-positive cells (4,5).
Telomere DNA, tandem (TTAGGG)n repeats, is tightly associated with
specialized telomeric protein complexes, termed shelterin (6).
Telomeres are transcribed from the subtelomere
toward the telomere into heterogeneous long non-coding RNA, which
are referred to as telomeric repeat-containing RNA or TERRA
(7,8). TERRA is found in most eukaryotes
(9–11), and its transcription is carried
out primarily by RNA polymerase II (RNAPII) in humans and yeasts
(8,9,11,12). As with other RNAPII-mediated
transcription, transcriptional activity is repressed by telomere
heterochromatin regulation (12,13). Human TERRA has a short half-life
and a cap structure at its 5′-end, and a small fraction of TERRA is
polyadenylated at its 3′-end; polyadenylation is known to increase
TERRA stability (14). TERRA
accumulates in highly proliferating cancer cells, and elevated
TERRA levels are found in various types of human cancer (15). However, whether elevated TERRA
levels result from changes in transcriptional activity or increased
stability of the RNA remains poorly understood.
TERRA is involved in various processes, such as
telomere protection, telomere replication and the epigenetic state
of telomere chromatin (16).
However, the effects of telomere length on TERRA transcription
remain controversial. A correlation between TERRA upregulation and
short telomeres has been found in patients with immunodeficiency,
centromeric instability and facial anomalies syndrome (17). The induction of TERRA
transcription at a specific chromosome leads to telomere shortening
of that chromosome (18). TERRA
mimicking RNA oligonucleotide acts as a telomerase inhibitor
(19). Moreover, cancer cells
exhibit decreased TERRA levels upon telomere elongation mediated by
telomerase overexpression (13).
Taken together, the data from these studies suggest that telomere
length is related to TERRA expression in a negative manner. By
contrast, telomere over-elongation does not cause marked
differences in TERRA abundance in Saccharomyces cerevisiae
(20), which has also been
observed in primary human lung fibroblasts and HeLa cells (21). Moreover, a lack of correlation
between these two parameters has been reported in human cell lines
(22). Thus, further studies are
warranted to explore the functional effects of telomere length on
TERRA expression.
TERRA is well characterized in HeLa, a cervical
cancer cell line, as a model cell line (14). However, TERRA stability and
abundance have not been examined in other cervical cancer cells, at
least to the best of our knowledge. Thus, in this study, we
measured TERRA level and stability, as well as telomere length in 6
human cervical cancer cells and examined whether TERRA abundance is
related to stability and telomere length. We found that TERRA was
more stable in cells with high levels of TERRA, but that telomere
length is unlikely to be associated with TERRA expression in
cervical cancer cells.
Materials and methods
Cell culture
The cells were grown in Dulbecco's modified Eagle's
medium or RPMI-1640 supplemented with 10% fetal bovine serum (both
from Wellgene, Seoul, Korea), 100 µg/ml of streptomycin, and
100 U/ml of penicillin (Gibco, Carlsbad, CA, USA). The SiHa, CaSki,
HeLa, HeLa S3 and SNU-17 cells were purchased from the Korean Cell
Line Bank (KCLB, Seoul, Korea), and C-33A was generously provided
by the Yonsei Cancer Center, Seoul, Korea.
Slot-blot analysis of TERRA
Total RNA was isolated from the cells using
TRIzol® reagent (Invitrogen, Carlsbad, CA, USA) or
Tri-RNA (Favorgen, Koahsiung, Taiwan) according to the
manufacturer's instructions. Total RNA was diluted to 0.02
µg/µl, and 20, 100 or 200 µl of the RNA
dilution was immobilized onto a nylon membrane (Hybond™ N+;
Amersham Biosciences, Piscataway, NJ, USA) for the detection of
TERRA. Similarly, 10, 50 or 100 µl of RNA dilution at a
concentration of 0.004 µg/µl was loaded for the
detection of 18S rRNA. After UV cross-linking (Stratagene, La
Jolla, CA, USA), the filters were hybridized with either a 3′-end
DIG-labeled oligonucleotide d(CCCTAA)4 or a
DIG-random-labeled 18S rDNA probe overnight at 45°C, and detection
was performed using a detection starter kit (Roche, Mannheim,
Germany). The blot hybridized with DIG-d(CCCTAA)4 was
erased in 50% formamide, 5% sodium dodecyl sulfate and 50 mM
Tris-HCl, pH 7.4 at 60°C, for 60 min twice, and reprobed with
DIG-d(TTAGGG)4. DIG-labeled d(CCCTAA)4 and
d(TTAGGG)4, and DIG-random-labeled 18S rDNA probes were
generated using terminal transferase and DIG-High Prime (both from
Roche), respectively. For the TERRA stability assay, total RNA was
isolated from the cells treated with actinomycin D (Act D; Sigma,
St. Louis, MO, USA) at 5 µg/ml for the times indicated in
the figures, and slot blotting was performed as described
above.
Reverse
transcription-quantitative-polymerase chain reaction (RT-qPCR)
cDNA was generated using M-MLV reverse transcriptase
(Invitrogen). cDNA synthesis was performed with 1.0 µg of
total RNA in a total volume of 20 µl according to the
manufacturer's instructions (Invitrogen). The reaction was
incubated at 37°C for 50 min and then terminated at 70°C for 15 min
for specific primers or at 25°C for 10 min, 37°C for 50 min, and
70°C for 15 min for random primers (Takara, Kyoto, Japan). The
primers used for reverse transcription are as follows:
5′-AGTCCGCCTAGAAGCATTTG-3′ for β-actin (14), and
5′-CCCTAACCCTAACCCTAACCCTAACCCTAA-3′ for TERRA (14). PCR reactions were performed in 10
µl containing 1X SYBR®-Green PCR master mix
(Roche), 0.2 µM of each forward and reverse primer (Table I) and 1.0 µl of cDNA. The
negative control included nuclease-free water instead of cDNA. PCR
was performed with a LightCycler® 480 Sequence Detection
system (Roche) under the following conditions: denaturation at 95°C
for 5 min, then 45 cycles at 95°C for 15 sec, 60°C for 15 sec, and
72°C for 15 sec. The comparative CT method
(ΔΔCT) was used for quantification and was calculated as
follows: ΔCT = CT (target gene) −
CT (reference gene), and ΔΔCT =
ΔCT (sample) − ΔCT (control). Relative
quantification was derived by 2−ΔΔCq.
 | Table IOligonucleotides used for
RT-qPCR. |
Table I
Oligonucleotides used for
RT-qPCR.
| Transcript | Primers sequences
(5′→3′) | Refs. |
|---|
| 10q-TERRA | F:
GAATCCTGCGCACCGAGAT | (13) |
| R:
CTGCACTTGAACCCTGCAATAC | (13) |
| 13q-TERRA | F:
GCACTTGAACCCTGCAATACAG | (24) |
| R:
CCTGCGCACCGAGATTCT | (24) |
| 15q-TERRA | F:
CAGCGAGATTCTCCCAAGCTAAG | (14) |
| R:
AACCCTAACCACATGAGCAACG | (14) |
| 16p-TERRA | F:
TGCAACCGGGAAAGATTTTATT | (24) |
| R:
GCCTGGCTTTGGGACAACT | (24) |
| 17q-TERRA | F:
AGCTACCTCTCTCAACACCAAGAAG | (24) |
| R:
GTCCATGCATTCTCCATTGATAAG | (24) |
| XqYq-TERRA | F:
CCCCTTGCCTTGGGAGAA | (24) |
| R:
GAAAGCAAAAGCCCCTCTGA | (24) |
| XpYp-TERRA | F:
GCAAAGAGTGAAAGAACGAAGCTT | (14) |
| R:
CCCTCTGAAAGTGGACCAATCA | (14) |
| TERT | F:
CTGGAACCATAGCGTCAGGG | This study |
| R:
ACAGAAACCACGGTCACTCG | This study |
| PinX1 | F:
CACTCCAGAGGAGAACGAAACC | This study |
| R:
CACCGGCTTGGCAAAGTACT | This study |
| TRF1 | F:
TGCTTTCAGTGGCTCTTCTG | (34) |
| R:
ATGGAACCCAGCAACAAGAC | (34) |
| TRF2 | F:
TTGTGGGGTCCTTGGACATA | (34) |
| R:
CCAGTAGAAAACTGGTCAAGGAA | (34) |
| β-actin | F:
TGTACGCCAACACAGTGCTG | (13) |
| R:
GCTGGAAGGTGGACAGCG | (13) |
| snU1 | F:
GGCGAGGCTTATCCATTG | (14) |
| R:
CCCACTACCACAAATTATGC | (14) |
Southern blot analysis of the terminal
restriction fragment
Genomic DNA was isolated using standard
phenol-chloroform extraction following proteinase K (Invitrogen)
treatment, and digested with HinfI overnight at 37°C.
HinfI-digested DNA (5 µg) was fractionated on an 0.8%
agarose gel and transferred onto a nylon membrane using the upward
capillary method. Hybridization was performed with a 3′-end
DIG-labeled d(TTAGGG)4 (Roche) probe at 45°C overnight.
The membrane was washed and hybridization was detected as
recommended by the manufacturer (Roche). Telomere length was
measured as the highest peak of the signal intensity using Image
Lab software (Bio-Rad, Hercules, CA, USA).
Statistical analysis
Statistical analysis was assessed using a Student's
t-test and Spearman's correlation as deemed appropriate. The level
of statistical significance was set at P<0.05.
Results and Discussion
Variable TERRA levels in cervical cancer
cells
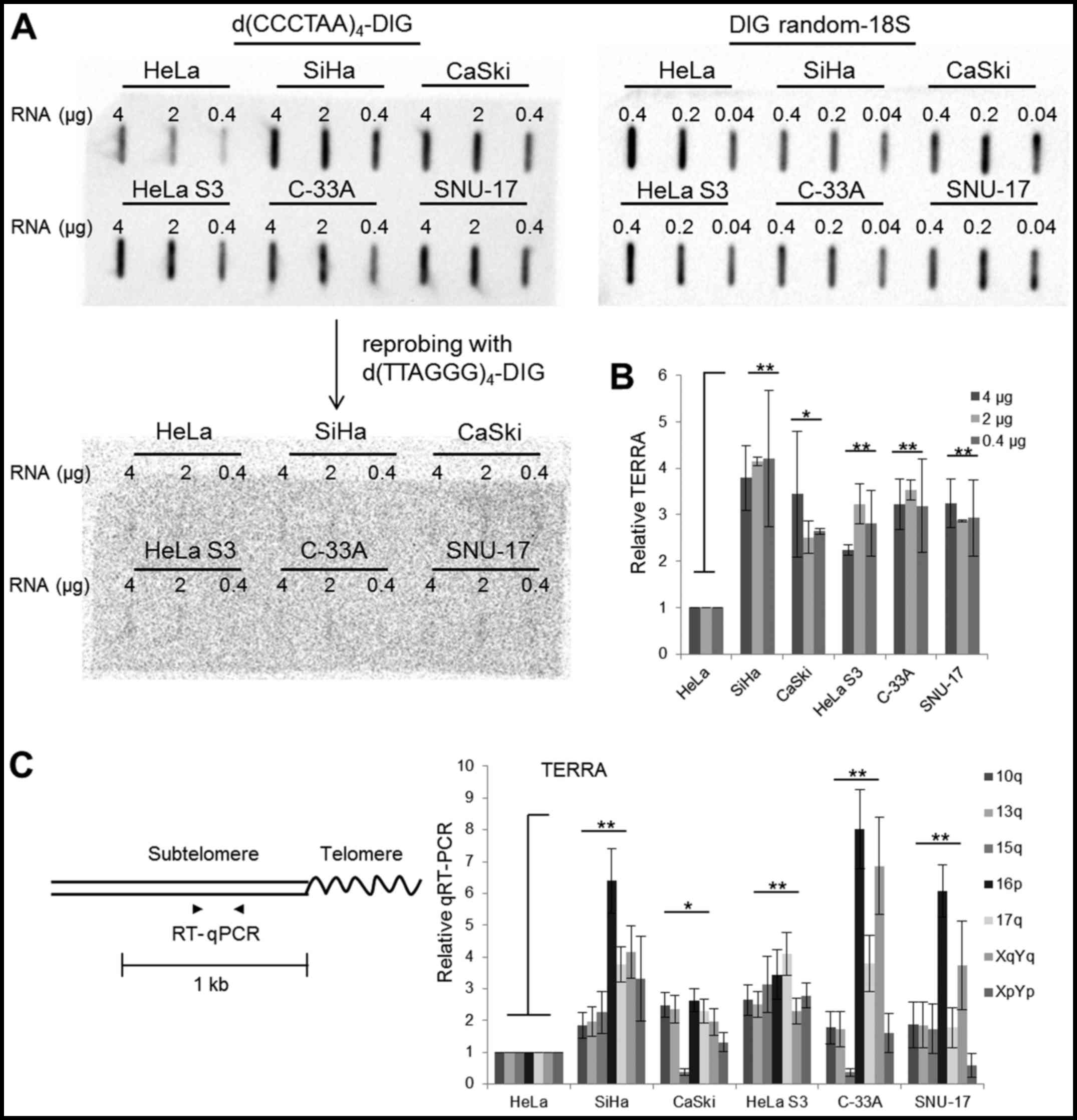
Slot-blot analysis was performed to monitor the
TERRA levels in the 6 cervical cancer cells, HeLa, SiHa, CaSki,
HeLa S3 (a clonal derivative of the parent HeLa line), C-33A and
SNU-17. Total RNA was blotted onto a membrane and hybridization was
followed with a strand-specific TERRA probe,
d(CCCTAA)4-DIG (Fig.
1A). Hybridization with the d(CCCTAA)4-DIG probes
was quantified as a ratio relative to the level measured in HeLa
cells following normalization by 18S rRNA (Fig. 1B). The hybridization signal was
approximately 2- to 4-fold greater in the SiHa, CaSki, HeLa S3,
C-33A and SNU-17 cells than in the HeLa cells (Fig. 1A and 1B). The blot used for TERRA detection
was stripped and reprobed with a DIG-labeled d(TTAGGG)4
probe identical to the TERRA repeat sequences, and this
hybridization resulted in weak signals (Fig. 1A). This indicated that RNAs with
UUAGGG repeats, which are considered to be TERRA, are abundant in
cervical cancer cells, whereas CCCUAA-containing RNA molecules
exist at very low levels.
Slot blotting is a tool which is used to monitor the
UUAGGG repeat content rather than the number of TERRA molecules
(23). Thus, the TERRA level was
also assessed in subtelomeres using chromosome-specific RT-qPCR
with primers matching the different chromosome arms (Fig. 1C and Table I). Note that the primer pairs used
in this study amplify DNA fragments from more than one subtelomere
due to the repetitive nature of subtelomeric DNA (7,13,24–27). TERRA for almost all subtelomeres
tested in this study was more abundant in 5 of the cell lines
compared with the HeLa cells (Fig.
1C). TERRA transcription at specific chromosome ends appeared
to behave differently from that in the independent chromosome ends
in some cells. For example, 16p TERRA was detected at a high level
in the SiHa, C-33A and SNU-17 cells, and 15q TERRA was detected at
a low level in the CaSki and C-33A cells. Overall, the results of
RT-qPCR were consistent with those of slot blotting.
TERRA stability in cervical cancer
cells
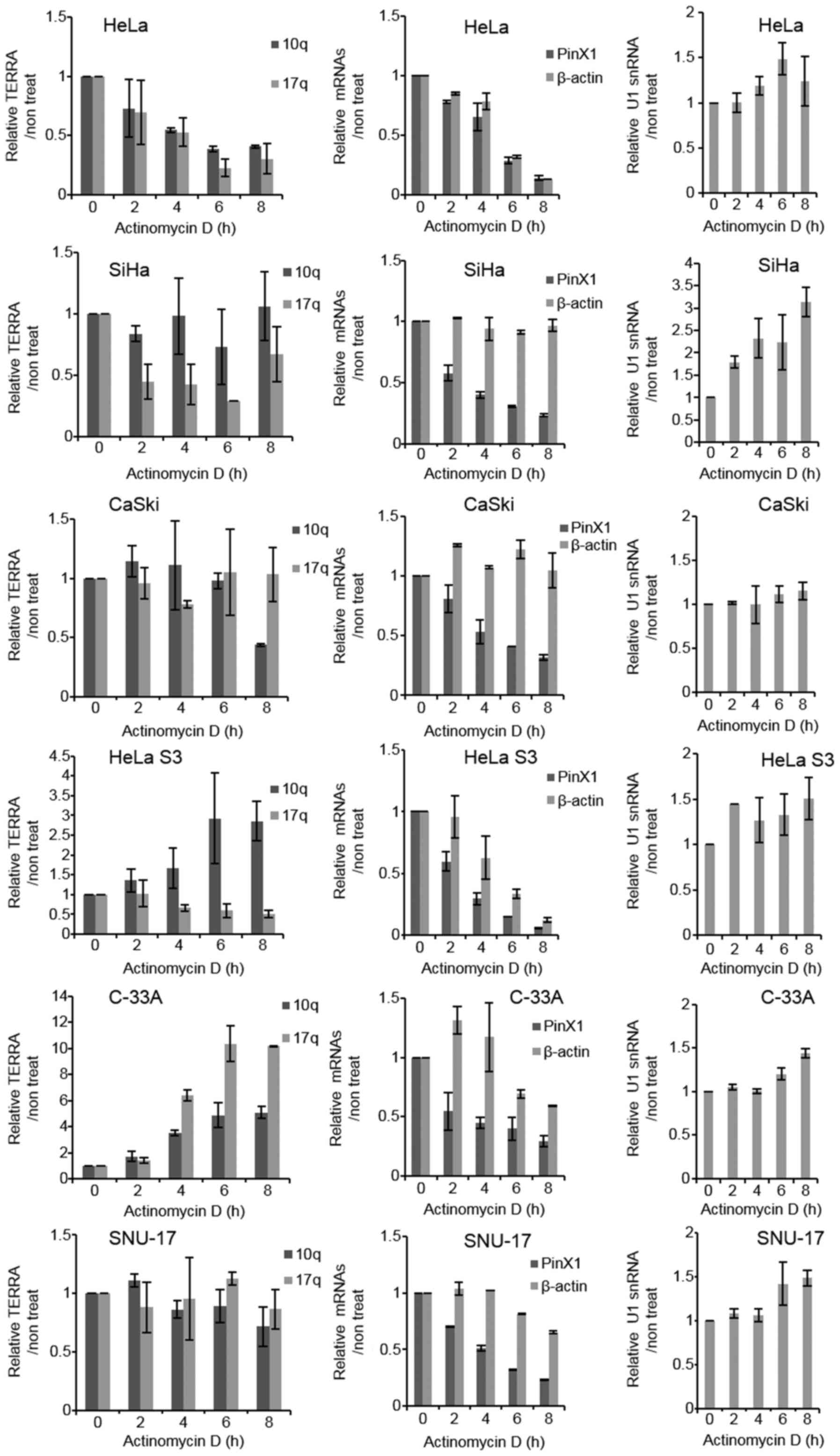
We wondered whether the variable TERRA levels in
cervical cancer cells reflect different degrees of RNA stability.
To assess this possibility, we measured the TERRA half-life by
treating the cells with Act D to block transcription. RT-qPCR was
used to monitor TERRA levels derived from chromosomes 10q and 17q
over time (Fig. 2, left panels).
The cell lines tested in this study were viable under these
conditions. PinX1, which encodes a telomere-binding protein
(6), β-actin and U1 small nuclear
RNA (snRNA) were included in the analysis (Fig. 2). Act D treatment reduced the
PinX1 mRNA levels in all cells to a half-life of 2–3 h in the SiHa,
HeLa S3 and C-33A cells, and 4–5 h in the HeLa, CaSki and SNU-17
cells (Fig. 2, middle panels and
Table II). These results
confirmed that Act D inhibited RNAPII function effectively. The
β-actin transcript was stable in the SiHa, CaSki, C-33A and SNU-17
cells with a half-life of >8 h; the half-life was shorter in the
HeLa and HeLa S3 cells, approximately 5 h (Fig. 2, middle panels and Table II). U1 snRNA is transcribed by
RNAPII and is known to be stable and abundant in human cells
(28,29). This RNA was stably maintained in
all cells tested with a half-life of >8 h (Fig. 2, right panels and Table II). U1 snRNA expression was
slightly increased at later time points in almost all cell lines;
in particular, a gradual increase was observed in the SiHa cells
(Fig. 2, right panels). β-actin
and U1 snRNA were initially included as internal controls; however,
the level of these gene transcripts changed upon Act D treatment in
some cells. Therefore, the RNA levels in the Act D-treated cells
were quantified as a relative value against that of the untreated
cells without normalization (Fig.
2).
 | Table IISummary of the half-life of TERRA,
PinX1, β-actin and U1 snRNA, and telomere length in cervical cancer
cells. |
Table II
Summary of the half-life of TERRA,
PinX1, β-actin and U1 snRNA, and telomere length in cervical cancer
cells.
| Cell line |
t1/2
| Telomere length
mean ± SD (kb) |
|---|
| TERRA | PinX1 | β-actin | U1 snRNA |
|---|
| HeLa | 10q, 4 h
17q, 4 h | 5 h | 5 h | >8 h | 5.6±0.72 |
| SiHa | 10q, >8
h
17q, >2 h | 2 h | >8 h | >8 h | 9.5±0.87 |
| CaSki | 10q, >8
h
17q, >8 h | 4 h | >8 h | >8 h | 5.0±0.63 |
| HeLa S3 | 10q, >8
h
17q, >8 h | 3 h | 5 h | >8 h | 10.4±1.03 |
| C-33A | 10q, >8
h
17q, >8 h | 2 h | >8 h | >8 h | 4.5±1.47 |
| SNU-17 | 10q, >8
h
17q, >8 h | 4 h | >8 h | >8 h | 5.9±0.52 |
Our RT-qPCR experiments revealed that the levels of
TERRA derived from 10q and 17q decreased upon Act D treatment in
HeLa cells (Fig. 2, left panels).
The half-life of TERRA was approximately 4 h in the HeLa cells
(Fig. 2 and Table II), which is similar to the
approximate 3 h value measured using Act D in a northern blot
analysis procedure reported previously (14). Unlike the HeLa cells, the other 5
cell lines did not exhibit a decrease in TERRA levels upon Act D
treatment (Fig. 2, left panels).
The half-life of TERRA was maintained at >8 h in the SiHa, CaSki
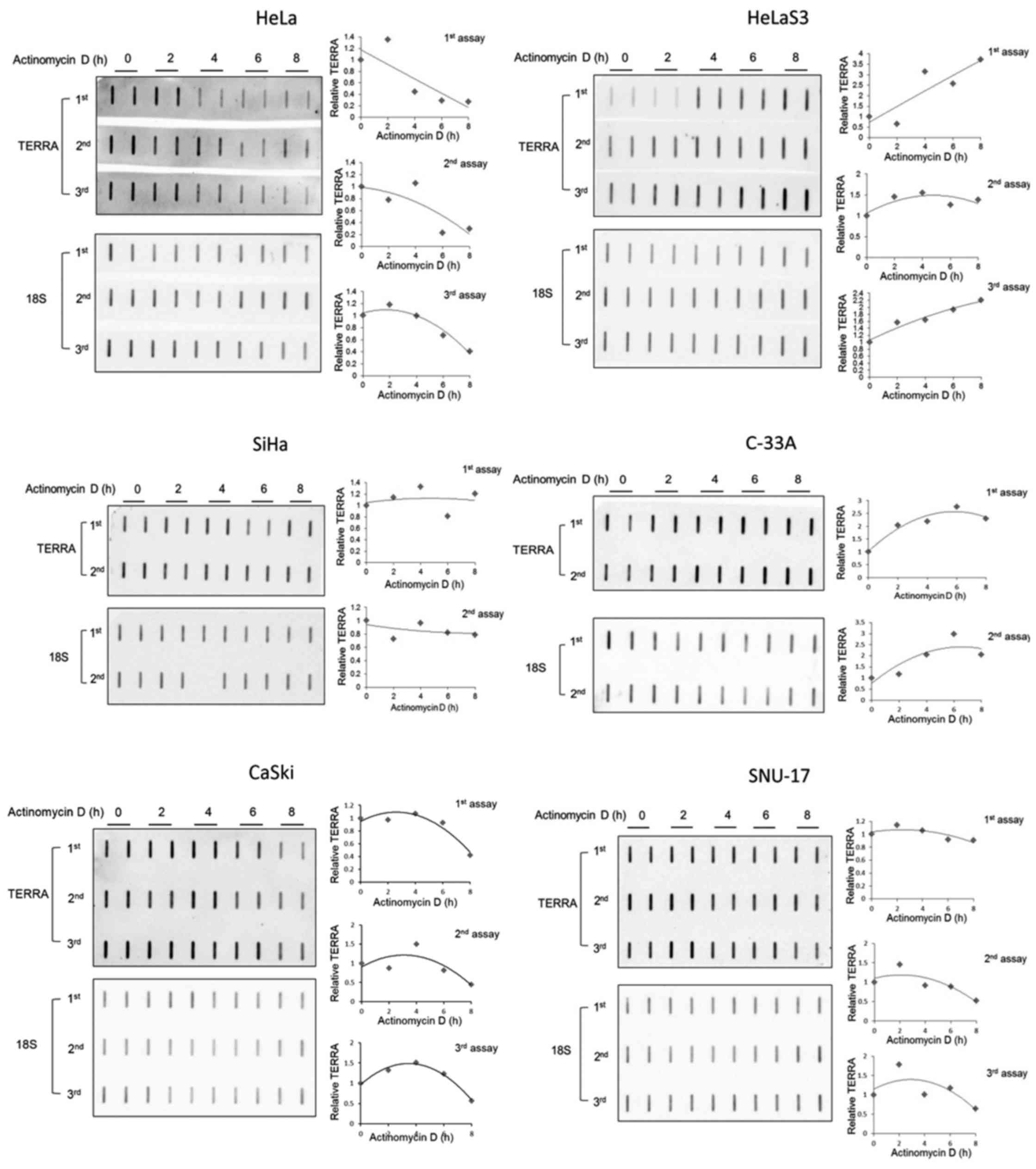
and SNU-17 cells. Similar to the results of RT-qPCR, slot blotting
of the total RNA assessed over time with a TERRA-specific probe
revealed that TERRA was degraded more rapidly in the HeLa cells
than in the SiHa, CaSki and SNU-17 cells (Fig. 3). It has been reported that a
small fraction of TERRA contains a poly(A) tail at the 3′-end in
HeLa (14). This may explain why
TERRA has a short half-life in these cells. Although highly
speculative, the other cervical cancer cells may maintain poly(A)+
TERRA as the predominant form. However, we cannot exclude the
possibility that TERRA is transcribed by other RNA polymerases; for
instance, RNAPI- or III-mediated TERRA transcription was less
sensitive under our experimental condition in the cells, which may
have allowed the persistent synthesis of TERRA.
Our RT-qPCR experiments revealed a gradual increase
in TERRA levels, particularly 10q in HeLa S3 and 10q and 17q in
C-33A cells, upon Act D treatment' (Fig. 2, left panels). We wondered whether
this phenomenon would be detected by slot blotting. The results of
slot blotting revealed that the UUAGGG signals increased in these
cells upon Act D treatment (Fig.
3), indicating that the induced RNA detected by RT-qPCR was
derived from telomeric RNA. The mechanism responsible for the
increased TERRA accumulation in the presence of Act D remains
unknown. TERRA may be released from RNA degradation cycles in these
particular cells. It would be interesting to test whether newly
synthesized RNA accounts for the increased TERRA. Collectively,
these finding suggest that TERRA was less abundant and was degraded
more rapidly in the HeLa cells, but was abundant and stable in the
other 5 cell lines.
Telomere length and expression of
telomere genes in cervical cancer cells
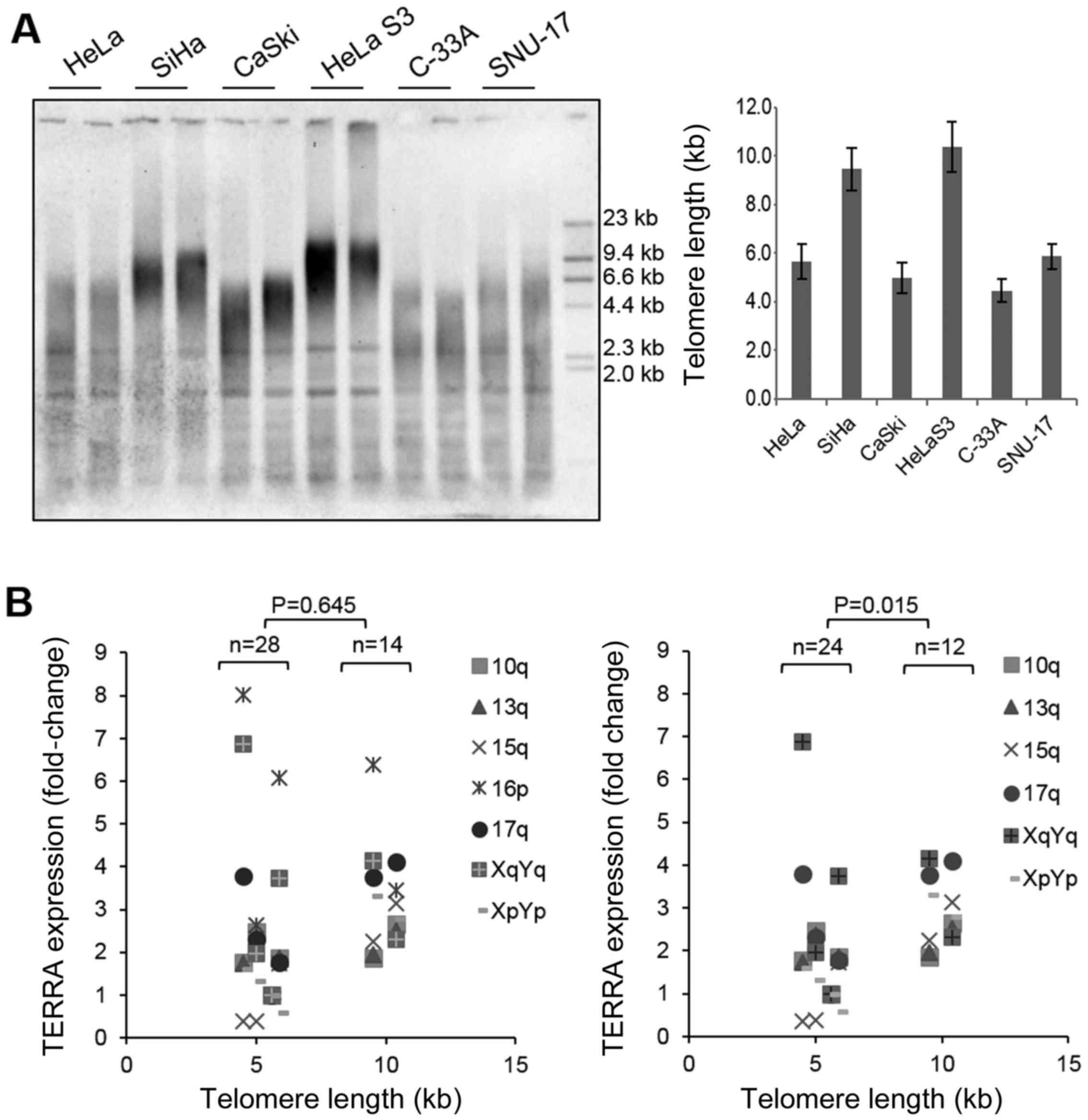
Telomere length was measured using Southern blot
analysis of terminal restriction fragment lengths. The average
telomere length was determined as the highest peak of the signal
intensity. The telomeres were significantly longer in the SiHa and
HeLa S3 cells than in the other 4 cell lines (Fig. 4A and Table II). To determine whether telomere
length is related to TERRA expression in cervical cells, the TERRA
levels measured by RT-qPCR were plotted against telomere length.
Telomere length did not correlate significantly with the TERRA
level (Fig. 4B, P=0.645). If
anything, there was a tendency for cells with long telomeres to
express higher levels of TERRA, and this was somewhat consistent
with the results reported previously (30). This tendency was more evident
after excluding from the analysis TERRA expression at 16p, which
was particularly high compared with the expression at other
chromosome ends (P=0.015) (Fig.
4B, graph on the right). Nonetheless, the number of cell lines
tested in this study may be insufficient to reveal an association.
Collectively, there was a lack of correlation between telomere
length and TERRA transcription in the cervical cancer cells. The
functional interaction between these two parameters may depend on
the cell type, as noted previously (21).
The shelterin proteins, telomere repeat-binding
factor 1 (TRF1) and TRF2, are important for recruiting other
shelterin proteins to telomeres and negatively regulate telomere
length (6). PinX1, a
TRF1-interacting protein, inhibits telomerase activity and
regulates telomere length in a negative manner (31). TERT, a catalytic component of
telomerase, acts as a positive regulator of telomere length
(32). It has recently been
reported that TRF1 and TRF2 associate functionally with TERRA
transcription; for instance, the depletion of TRF1 or TRF2 leads to
the accumulation of TERRA (33).
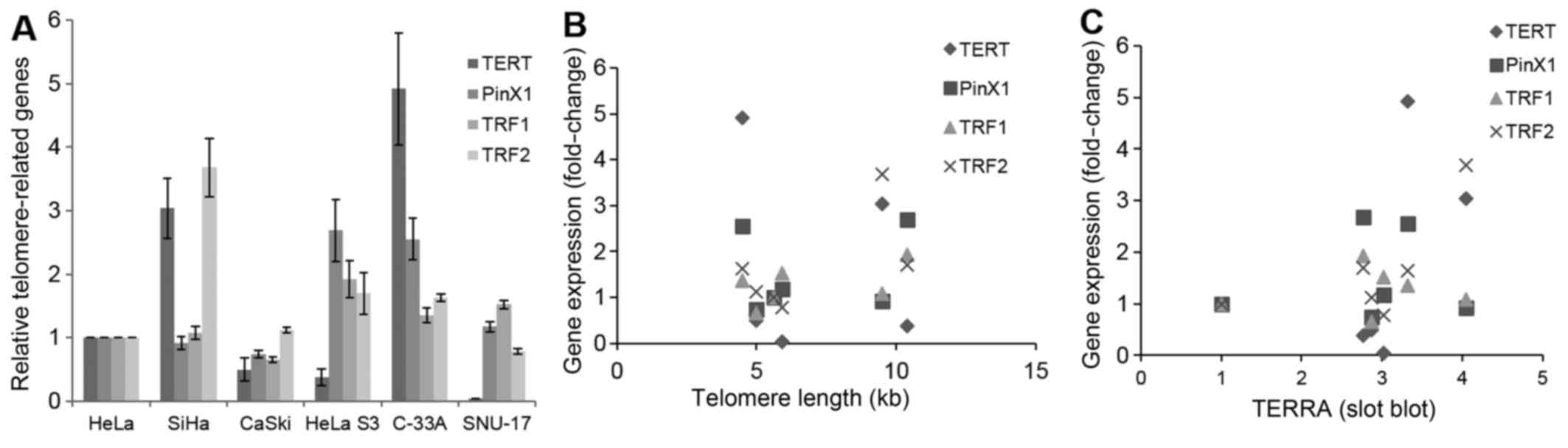
In this study, we measured the mRNA levels of TERT, PinX1, TRF1 and
TRF2 (Fig. 5A), and we examined
whether these are related to telomere length and TERRA level. The
mRNA levels of none of these genes correlated significantly with
telomere length or TERRA level in cervical cancer cells (Fig. 5B and 5C).
In conclusion, TERRA abundance and stability vary
between types of cervical cancer cells. TERRA is degraded rapidly
in HeLa cells, but is maintained stably in other cervical cancer
cell lines. TERRA abundance is associated with the stability of the
RNA in cervical cancer cells; however, there was a lack of
correlation between the TERRA level and telomere length. Additional
studies are warranted to explore the mechanisms that regulate TERRA
steady-state levels.
Acknowledgments
This study was supported by the Basic Science
Research Program through the the National Research Foundation of
Korea (NRF) funded by the Ministry of Education (nos. 2010-0008254,
2014R1A1A2054542 to B.-K.O.) and funded by the Ministry of Science,
ICT and Future Planning (no. 2011-0015638 to B.-K.O.) and by the
Hanyang University (no. 201200000002989 to J.S.C.).
References
|
1
|
Greider CW and Blackburn EH:
Identification of a specific telomere terminal transferase activity
in Tetrahymena extracts. Cell. 43:405–413. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
de Lange T: Shelterin: The protein complex
that shapes and safeguards human telomeres. Genes Dev.
19:2100–2110. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shay JW, Zou Y, Hiyama E and Wright WE:
Telomerase and cancer. Hum Mol Genet. 10:677–685. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bodnar AG, Ouellette M, Frolkis M, Holt
SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S and
Wright WE: Extension of life-span by introduction of telomerase
into normal human cells. Science. 279:349–352. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nakayama J, Tahara H, Tahara E, Saito M,
Ito K, Nakamura H, Nakanishi T, Tahara E, Ide T and Ishikawa F:
Telomerase activation by hTRT in human normal fibroblasts and
hepatocellular carcinomas. Nat Genet. 18:65–68. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Palm W and de Lange T: How shelterin
protects mammalian telomeres. Annu Rev Genet. 42:301–334. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Azzalin CM, Reichenbach P, Khoriauli L,
Giulotto E and Lingner J: Telomeric repeat containing RNA and RNA
surveillance factors at mammalian chromosome ends. Science.
318:798–801. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schoeftner S and Blasco MA:
Developmentally regulated transcription of mammalian telomeres by
DNA-dependent RNA polymerase II. Nat Cell Biol. 10:228–236. 2008.
View Article : Google Scholar
|
|
9
|
Luke B, Panza A, Redon S, Iglesias N, Li Z
and Lingner J: The Rat1p 5′ to 3′ exonuclease degrades telomeric
repeat-containing RNA and promotes telomere elongation in
Saccharomyces cerevisiae. Mol Cell. 32:465–477. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vrbsky J, Akimcheva S, Watson JM, Turner
TL, Daxinger L, Vyskot B, Aufsatz W and Riha K: siRNA-mediated
methylation of Arabidopsis telomeres. PLoS Genet. 6:e10009862010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bah A, Wischnewski H, Shchepachev V and
Azzalin CM: The telomeric transcriptome of Schizosaccharomyces
pombe. Nucleic Acids Res. 40:2995–3005. 2012. View Article : Google Scholar :
|
|
12
|
Nergadze SG, Farnung BO, Wischnewski H,
Khoriauli L, Vitelli V, Chawla R, Giulotto E and Azzalin CM:
CpG-island promoters drive transcription of human telomeres. RNA.
15:2186–2194. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Arnoult N, Van Beneden A and Decottignies
A: Telomere length regulates TERRA levels through increased
trimethylation of telomeric H3K9 and HP1α. Nat Struct Mol Biol.
19:948–956. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Porro A, Feuerhahn S, Reichenbach P and
Lingner J: Molecular dissection of telomeric repeat-containing RNA
biogenesis unveils the presence of distinct and multiple regulatory
pathways. Mol Cell Biol. 30:4808–4817. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Deng Z, Wang Z, Xiang C, Molczan A, Baubet
V, Conejo-Garcia J, Xu X, Lieberman PM and Dahmane N: Formation of
telomeric repeat-containing RNA (TERRA) foci in highly
proliferating mouse cerebellar neuronal progenitors and
medulloblastoma. J Cell Sci. 125:4383–4394. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Azzalin CM and Lingner J: Telomere
functions grounding on TERRA firma. Trends Cell Biol. 25:29–36.
2015. View Article : Google Scholar
|
|
17
|
Yehezkel S, Segev Y, Viegas-Péquignot E,
Skorecki K and Selig S: Hypomethylation of subtelomeric regions in
ICF syndrome is associated with abnormally short telomeres and
enhanced transcription from telomeric regions. Hum Mol Genet.
17:2776–2789. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pfeiffer V and Lingner J: TERRA promotes
telomere shortening through exonuclease 1-mediated resection of
chromosome ends. PLoS Genet. 8:e10027472012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Redon S, Reichenbach P and Lingner J: The
non-coding RNA TERRA is a natural ligand and direct inhibitor of
human telomerase. Nucleic Acids Res. 38:5797–5806. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iglesias N, Redon S, Pfeiffer V, Dees M,
Lingner J and Luke B: Subtelomeric repetitive elements determine
TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO
Rep. 12:587–593. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Farnung BO, Brun CM, Arora R, Lorenzi LE
and Azzalin CM: Telomerase efficiently elongates highly
transcribing telomeres in human cancer cells. PLoS One.
7:e357142012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smirnova A, Gamba R, Khoriauli L, Vitelli
V, Nergadze SG and Giulotto E: TERRA expression levels do not
correlate with telomere length and radiation sensitivity in human
cancer cell lines. Front Oncol. 3:1152013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Van Beneden A, Arnoult N and Decottignies
A: Telomeric RNA expression: Length matters. Front Oncol.
3:1782013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Deng Z, Wang Z, Stong N, Plasschaert R,
Moczan A, Chen HS, Hu S, Wikramasinghe P, Davuluri RV, Bartolomei
MS, et al: A role for CTCF and cohesin in subtelomere chromatin
organization, TERRA transcription, and telomere end protection.
EMBO J. 31:4165–4178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Riethman HC, Xiang Z, Paul S, Morse E, Hu
XL, Flint J, Chi HC, Grady DL and Moyzis RK: Integration of
telomere sequences with the draft human genome sequence. Nature.
409:948–951. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Riethman H, Ambrosini A, Castaneda C,
Finklestein J, Hu XL, Mudunuri U, Paul S and Wei J: Mapping and
initial analysis of human subtelomeric sequence assemblies. Genome
Res. 14:18–28. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ambrosini A, Paul S, Hu S and Riethman H:
Human subtelomeric duplicon structure and organization. Genome
Biol. 8:R1512007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ro-Choi TS, Raj NB, Pike LM and Busch H:
Effects of alpha-amanitin, cycloheximide, and thioacetamide on low
molecular weight nuclear RNA. Biochemistry. 15:3823–3828. 1976.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Noonberg SB, Scott GK and Benz CC:
Evidence of post-transcriptional regulation of U6 small nuclear
RNA. J Biol Chem. 271:10477–10481. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vitelli V, Falvo P, Khoriauli L, Smirnova
A, Gamba R, Santagostino M, Nergadze SG and Giulotto E: More on the
lack of correlation between terra expression and telomere length.
Front Oncol. 3:2452013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou XZ and Lu KP: The
Pin2/TRF1-interacting protein PinX1 is a potent telomerase
inhibitor. Cell. 107:347–359. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Autexier C and Lue NF: The structure and
function of telomerase reverse transcriptase. Annu Rev Biochem.
75:493–517. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Porro A, Feuerhahn S, Delafontaine J,
Riethman H, Rougemont J and Lingner J: Functional characterization
of the TERRA transcriptome at damaged telomeres. Nat Commun.
5:53792014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Scheibe M, Arnoult N, Kappei D, Buchholz
F, Decottignies A, Butter F and Mann M: Quantitative interaction
screen of telomeric repeat-containing RNA reveals novel TERRA
regulators. Genome Res. 23:2149–2157. 2013. View Article : Google Scholar : PubMed/NCBI
|