Introduction
Osteoporosis is a complex disease associated with
mineral composition and bone strength; this disease affects
millions of individuals worldwide, particularly those with
pathological fractures (1,2).
Estrogen deficiency significantly influences skeletal homeostasis
by inducing oral bone loss, which aggravates with aging (3–5).
Previous studies have indicated that a decline in ovarian estrogen
production during menopause results in the rapid loss of the
trabecular microarchitecture, enhances cortical bone resorption and
increases cortical porosity; these effects promote the development
of osteoporosis and increase the risk of fragility fractures
(6). In fact, estrogen deficiency
alters the regional distribution of tissue mineral density
(7,8), leading to alterations in the
mechanical properties of bone at the tissue level. These
dysregulations are commonly observed in alveolar bone (9–12).
Recent studies have demonstrated that estrogen deficiency-induced
bone loss also occurrs in the mandible (13–16).
Non-coding RNAs, including microRNAs (miRNAs or
miRs) and long non-coding RNAs (lncRNAs), play important roles in
biological processes. miRNAs are involved in cell differentiation,
fate and apoptosis, as well as in the pathogenesis of various
diseases; miRNAs have also been shown to be involved in regulating
bone mass (17). A previous study
reported that several miRNAs were dysregulated in bone tissues in
ovariectomized (OVX) mice (18).
Specifically, miR-133 expression has been shown to be enhanced
during estrogen deficiency, modulating osteogenic differentiation
of mesenchymal stem cells and inducing post-menopausal osteoporosis
(19). The functions of other
miRNAs in regulating bone loss have also been extensively
investigated (20). For example,
miR-705 and miR-3077-5p were found to synergistically mediate the
shift of mesenchymal stem celll lineage commitment to adipocyte in
osteoporosis bone marrow (21).
miR-34a can block osteoporosis by inhibiting osteoclastogenesis and
tgif2 which is pro-osteoclastogenic (22). miR-26a has been reported
effectively to improve the osteogenic differentiation capability of
mesenchymal stem cells isolated from ovariectomized osteoporotic
mice both in vitro and in vivo (23). There is evidence to indicate that
circulating monocytes are directly involved in osteoclastogenesis,
and lncRNAs participate in osteoblast differentiation (24). It was also shown that the
lncRNA-DANCR-induced expression of interleukin (IL)-6 and tumor
necrosis factor (TNF)-α in blood mononuclear cells promoted bone
resorption in post-menopausal women with low bone mineral density
(BMD) (24). The potential
therapeutic and biomarker functions of miRNAs and lncRNAs in
treating bone disorders have received increasing research
attention.
In the present study, we comprehensively analyzed
the expression profiles of miRNAs, mRNAs and lncRNAs in the
mandible of OVX mice. We then constructed a complex regulatory
network to investigate the regulatory mechanisms of miRNAs and
lncRNAs in mandible bone mass in OVX mice with estrogen
deficiency.
Materials and methods
Animals
Female C57BL/6 mice, aged 8 weeks (mean weight of 19
g), were purchased from SLAC Laboratory Animal Company, Ltd.
(Shanghai, China). The mice were anesthetized with an
intraperitoneal injection of chloral hydrate (10%, 4 ml/kg body
weight). Bilateral ovariectomy (performed on 8 mice) or sham
operation (Sham-op, performed on 8 mice) was performed using
standard methods. An incision was made on the midline of the
abdomen, in order to find the uterine body and one side of the
uterine horn. The ovary was at the end of the uterine horn. A
suture was placed around the ovarian artery and vein prior to the
removal of the ovary. After removing the ovary, we tightly closed
the incision with a nylon 4-0 suture. The mice in the sham-operated
group underwent the same procedure, except that the ovarian artery
was not ligated and the ovary was not removed.
All the mice were placed in cages under
standard laboratory conditions and fed standard chow during the
study period
After 12 weeks, all mice were prepared for
subsequent experiments. The procedures performed on the mice were
approved by the Institute of Animal Care and Use Committee of
Tongji University (no. TJLAC-014-016).
Preparation of specimens
The animals were sacrificed 12 weeks after the
surgery. The right mandible and femur were dissected, filled with
4% paraformaldehyde for 2 days at room temperature, and then stored
in 0.5% paraformaldehyde at 4°C. The specimens were used to
evaluate their BMD and micro-structure through micro-computerized
tomography (micro-CT) analysis. Subsequently, the right mandibles
were used for histological analysis. For the left mandibles, the
molars and most of the incisors were first dissected; the entire
mandible was then used for microarray and reverse
transcription-quantitative RT-PCR (RT-qPCR) assays.
Micro-CT and histological analyses
The right mandibles and femurs of the mice (n=3),
without sample preparation or decalcification, were scanned using a
high-resolution micro-CT system (µCT50; Scanco Medical AG,
Bassersdorf, Switzerland). Images of the femur and mandible were
acquired at 70 kV of energy and 114 µA of intensity with a
voxel size of 10 µm, and we then analyzed some indicators
[BMD, bone volume over total volume (BV/TV), trabecular separation
(Tb.Sp) and trabecular number (TB.N)] of bone quality by micro-CT.
Following image acquisition, the mandible and femur samples were
decalcified and embedded in paraffin. Serial sections of 4
µm in thickness were cut and then stained with H&E in
accordance with the manufacturer's instructions (Beyotime,
Shanghai, China).
RNA extraction and array analysis
Mandibles were extracted from sham-operated and OVX
mice. The left mandibles from the sham operation and OVX groups
were harvested using TRIzol reagent (Sigma-Aldrich, St. Louis, MO,
USA) and used to extract total RNA. The expression profiles of the
lncRNAs, miRNAs and mRNAs in the mandible were analyzed. The
microarrays used in this study were the Agilent Mouse lncRNA
(4×180K, design ID: 049801; Agilent Technologies, Santa Clara, CA,
USA) and Affymetrix miRNA 4.0 (Affymetrix, Santa Clara, CA,
USA).
Data normalization
Feature extraction software was applied to extract
the raw data from the array images. GeneSpring software was
employed for basic analysis and normalization of the raw data by
the quantile algorithm. Data symmetry and dispersed degree
distribution were described by box-and-whisker plot. The scatter
plot was used to display the overall distribution of central
tendency.
Differential expression analysis
Probes with at least 100% of the samples in any one
of the two conditions have flags in 'P' and were selected for data
analysis. Differentially expressed miRNAs (DEmiRs) and
differentially expressed genes (DEGs) were identified through fold
changes, and P-values were calculated using the t-test. The
threshold for up- and downregulated genes was set at a fold change
of ≥2.0 and a value of P≤0.05.
Prediction of miRNA target genes
GeneSpring 12.5 software was used to predict target
genes and analyze the function of DEmiRs. The TargetScan
(http://www.targetscan.org/vert_71/),
PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_data.html)
and microRNAorg (http://www.microrna.org/microrna/home.do) databases
were applied for target prediction. The analysis of biological
processes and pathways was performed using the DAVID database
(https://david.ncifcrf.gov/summary.jsp). Screening of
miRNA targets related DElncRNAs was performed by using UCSC Genome
Browser (http://genome.ucsc.edu/).
Gene screening using the neighborhood
scoring algorithm
To systematically identify genes affected by
ovariectomy, we mapped the DEGs into the protein signaling network.
Protein interactions in the OVX mice were obtained from the STRING
9.05 database (https://string-db.org/). The
parameters of STRING were set as at a confidence score of 0.4 and
interaction types of neighborhood, experiment, database and text
mining. Subsequently, a specific protein-protein interaction (PPI)
network using the data from the OVX mice was constructed using
Cytoscape software (www.cytoscape.org/). Generally, disease-related genes
and their adjacent proteins participate in the same disease
pathways or biological processes. The core protein and its neighbor
protein significantly correlate at the expression level. Thus, we
calculated the neighborhood score of signature genes in the
mandible of OVX mice using the following formula:
where i is the node in the network, FC(i) is the fold
change value of genes, and N(i) is the nodes adjacent to
i. Score(i) represents the relevance between node
i and the disease. The neighbor-hood scoring algorithm is
used to calculate the variation degree of nodes under the disease
condition, as well as the influence of the node on neighboring
genes using fold changes in the core node and its neighbor nodes to
identify disease-related genes. When the core node and neighbor
nodes exhibit a high (score >0) or low expression (score <0),
the absolute scores ranked in the top 100 are designated as
significant genes.
RNA extraction and RT-PCR
Mandibles were extracted from the sham-operated and
OVX mice. The mandibles from the sham-operated and OVX groups were
harvested using TRIzol reagent (Sigma-Aldrich) and used to extract
total RNA. Complementary DNA (cDNA) were synthesized using a
PrimeScript RT reagent kit (Takara, Dalian, China) under the
following conditions: 37°C for 15 min, 85°C for 5 sec, and holding
at 4°C. For miRNA expression analysis, miRNA was reverse
transcribed using specific RT primers (RiboBio, Guangzhou, China)
under the following conditions: 42°C for 60 min, 70°C for 10 min,
and holding at 4°C.
RT-qPCR reactions were performed using the FastStart
Universal SYBR-Green (Roche, Basel, Switzerland). RT-qPCR was
performed using the following cycles: 10 min at 95°C, 10 sec at
95°C, 10 sec at 60°C and 10 sec at 72°C for 45 cycles. β-actin and
U6 were used as loading controls for quantitation of mRNA and
miRNAs respectively. The Bulge-Loop miRNA RT-qPCR Primer Set
(RiboBio) was used for RT-qPCR. The primer information of related
mRNAs was: Lrp2bp forward, 5′-ACCCAAAAGCTAGTGTGAAGGC-3′ and
reverse, 5′-GGACTCCAGACTTCCGTTGC-3′; Plin4 forward,
5′-CACTACCAAGTCCGTGCTCAT-3′ and reverse,
5′-CAGACCCTTTAGCCACGTTAAT-3′; RhoQ forward,
5′-GCTAAAGGAATATGCGCCAAAC-3′ and reverse,
5′-TTCTGGGTTAAAGCTGAACACTC-3′; Phka1 forward,
5′-GTTGCCCGTTATTTAGACCGC-3′ and reverse,
5′-AAGTCGCAAGTTGTTTGCACA-3′; Gyk forward,
5′-TGAACCTGAGGATTTGTCAGC-3′ and reverse,
5′-CCATGTGGAGTAACGGATTTCG-3′; Ppp1r3c forward,
5′-CAGGAAGCCAAATCGCAGAGT-3′ and reverse,
5′-TTGGAGTCCGCAAACACGAC-3′; Lrp2bp forward,
5′-ACCCAAAAGCTAGTGTGAAGGC-3′ and reverse,
5′-GGACTCCAGACTTCCGTTGC-3′; Acsl6 forward,
5′-GAACTCAACTACTGGACCTGC-3′ and reverse,
5′-CCGTGGACGTAGATTTGTGC-3′; Fabp5 forward,
5′-CATCACGGTCAAAACCGAGAG-3′ and reverse,
5′-ACTCCACGATCATCTTCCCAT-3′; Mapk10 forward,
5′-CAAGAGGGCTTACCGGGAG-3′ and reverse, 5′-AGGTTGGCGTCCATCAGTTC-3′.
Raw data can then be analyzed with LightCycler® 96 SW1.1
(Roche), generally using the automatic cycle threshold (Ct) setting
for assigning baseline and threshold for Ct determination.
Statistical analysis
All values were expressed as the means ± standard
deviation. All analyses were conducted using SPSS 20.0 software
(SPSS, Inc., Chicago, IL, USA). The difference in the evaluated
parameters among the groups was tested using the two-tailed
independent sample t-test. A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
Establishment of the mouse model of
osteoporosis induced by estrogen deficiency due to ovariectomy
We dissected the mandible and femur of the mice 12
weeks after the surgery. Micro-CT images and histological sections
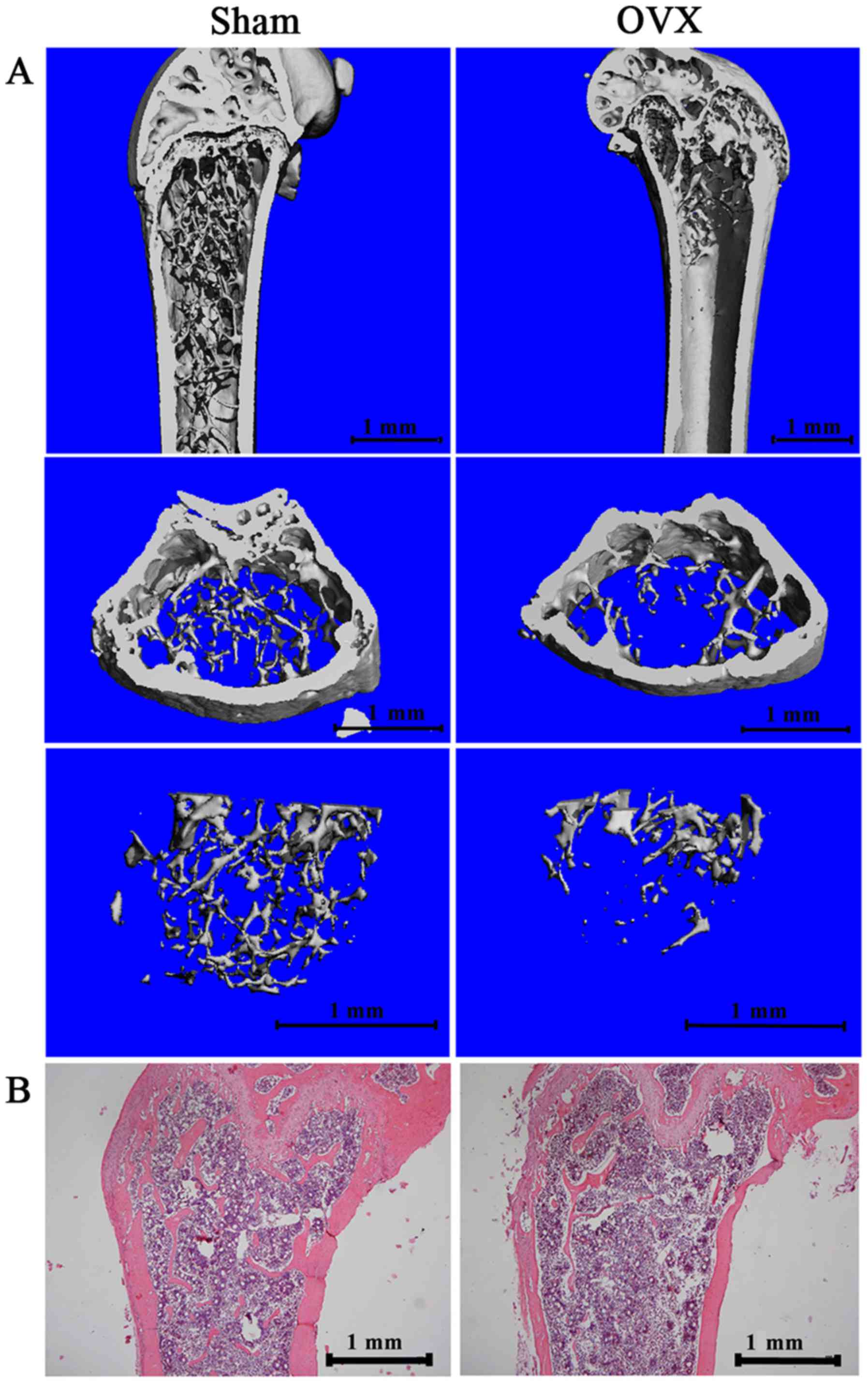
were used to visualize the established osteoporotic model (Fig. 1). The 3D images (Fig. 1A) and results from H&E
staining (Fig. 1B) of the distal
femur of the OVX mice revealed a significantly decreased
subchondral trabecular bone volume compared with the sham-operated
mice. We also analyzed trabecular bone 1 mm under the growth plate
of the distal femur, as previously described (25). Micro-CT data demonstrated a
significant decrease in BMD, BV/TV and TB.N, and an increase in
Tb.Sp in the OVX group compared with the sham-operated group
(Table I).
 | Table IResults from micro-CT analysis data
of the femur between the OVX and sham-operated group. |
Table I
Results from micro-CT analysis data
of the femur between the OVX and sham-operated group.
| Control | Sham | OVX |
|---|
| BMD (mg A/ccm) | 868.7±10.39 | 824.62±3.22a |
| BV/TV(%) | 2.32±0.13 | 1.24±0.04a |
| Tb.Sp (mm) | 0.38±0.05 | 0.63±0.11a |
| Tb.N (1/mm) | 2.81±0.13 | 1.59±0.06a |
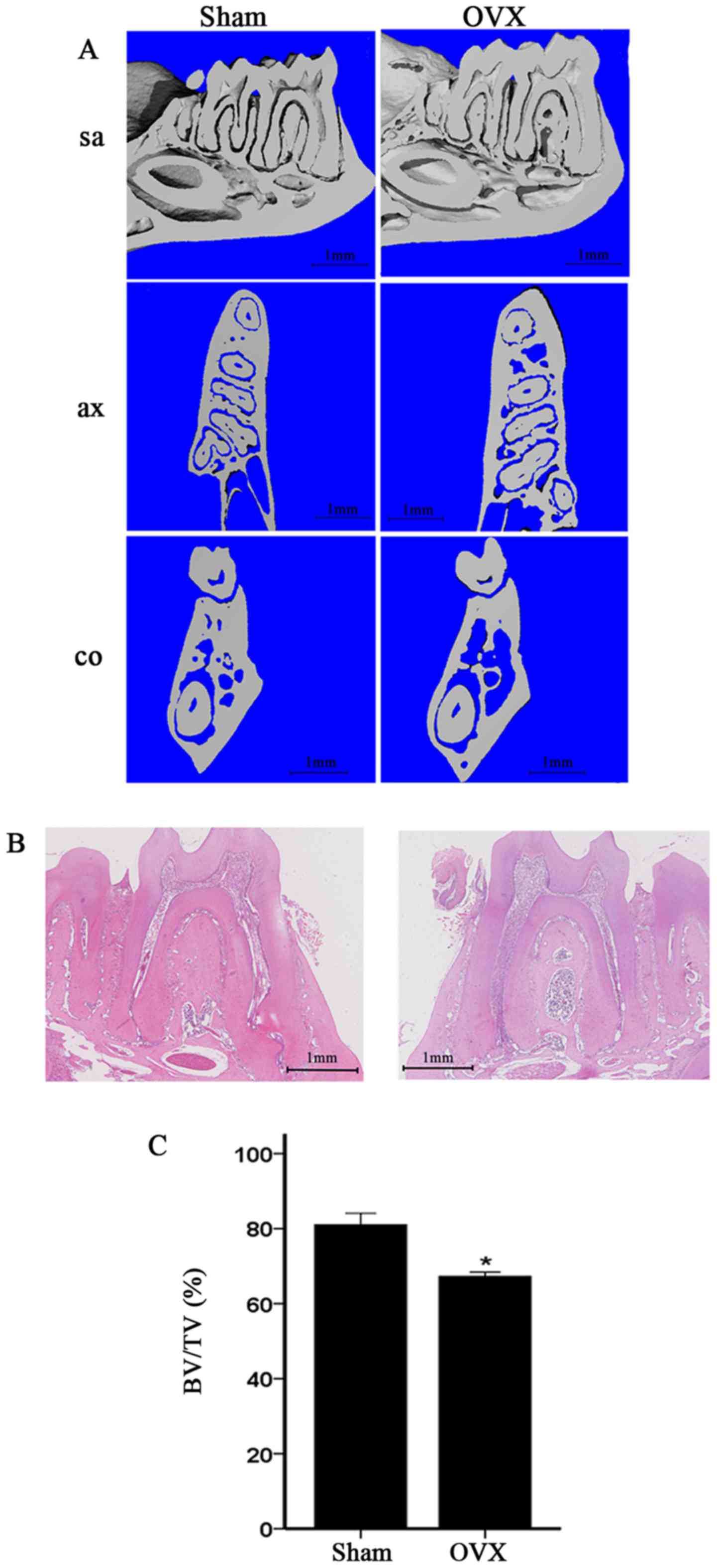
In the present study, the mandibles of the OVX mice
were compared with those of the sham-operated mice. We compared the
alveolar bone of the first molar, particularly the area of the
furcation. The micro-CT images of the OVX mice revealed a
significant decrease in alveolar bone from the coronal sagittal,
and transaxial slices compared with those of the sham-operated mice
(Fig. 2A). Moreover, H&E
staining of the alveolar bone of the first molar in the OVX group
revealed significantly reduced furcation and a relatively scant
marrow space compared with those in the sham-operation group; this
result is consistent with the micro-CT data (Fig. 2B). The alveolar bone of the first
molar was also analyzed. The micro-CT data of the alveolar bone of
the first mandibular molar in the OVX mice demonstrated decreased
BV/TV (Fig. 2C).
Differential expression analysis of
lncRNAs, miRNAs and mRNAs
Differential expression analysis was performed on
the samples from the OVX and sham-operated mice following data
normalization. A total of 2,915 significantly expressed mRNAs were
obtained, including 1,037 upregulated mRNAs and 1,878 downregulated
mRNAs. Moreover, 53 significantly expressed miRNAs were acquired;
of which 18 were upregulated and 35 were downregulated (data not
shown).
Functional enrichment analysis
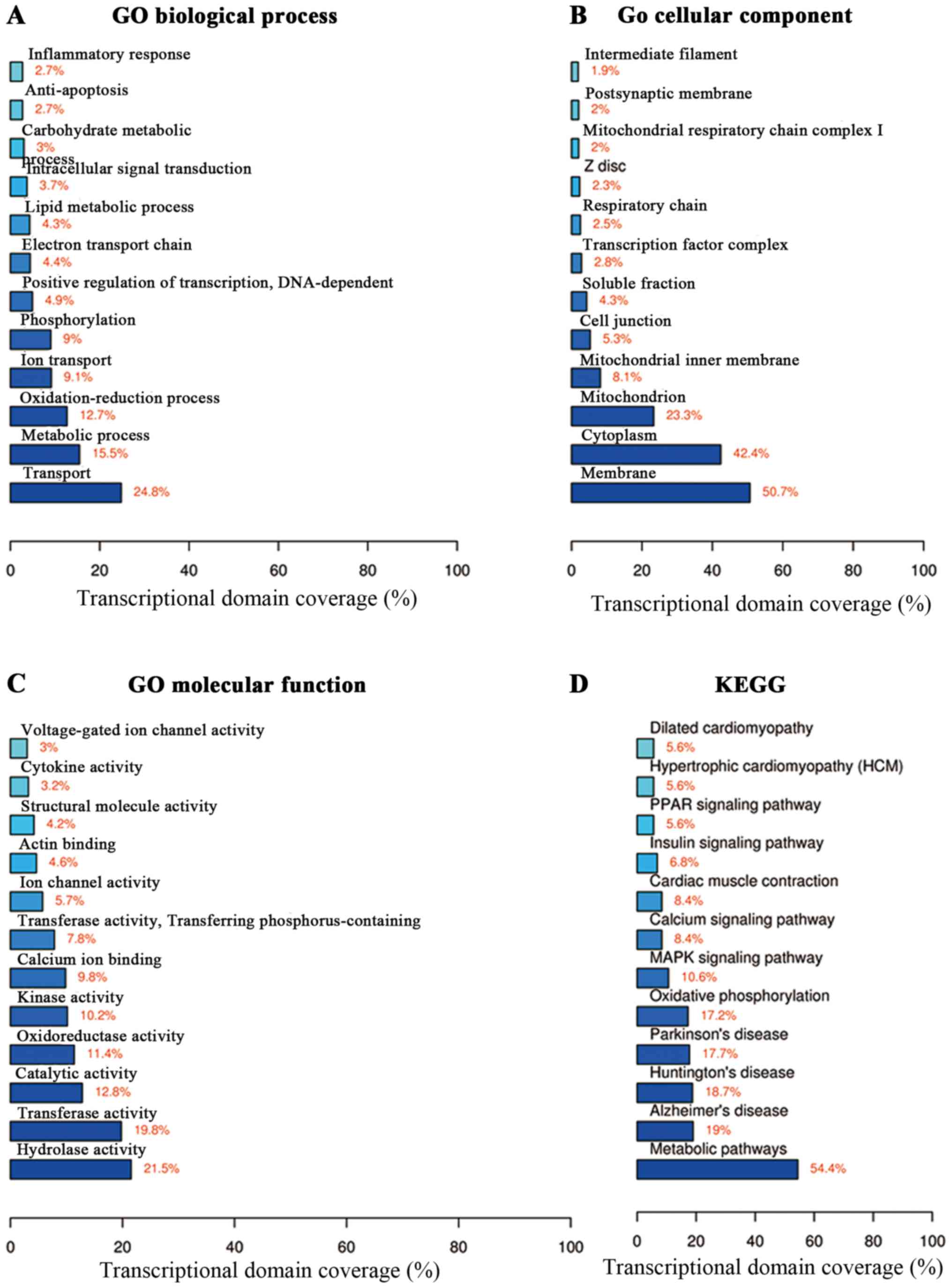
Functional annotation was performed on the DEGs
(2,915 significantly expressed mRNAs, including 1,037 upregulated
and 1,878 downregulated mRNAs) in terms of biological processes,
cell component and molecular function (Fig. 3A–C). In the biological process,
the DEGs regulated transport, metabolic processes and
oxidation-reduction (Fig. 3A).
The DEGs were located in the membrane, cytoplasm and mitochondria
(Fig. 3B). The DEGs also played a
role in hydrolase activity and transferase activity (Fig. 3C). Moreover, the KEGG pathway
analysis was used to identify differential signaling pathways
between the OVX and sham-operated mice. We found that the
differentially expressed mRNAs were involved in pathways such as
the metabolic pathways and mitogen-activated protein kinase (MAPK)
signaling pathway (Fig. 3D).
Furthermore, the coverage scale of metabolic pathways was the
largest, thereby indicating that genes associated with these
pathways were differentially expressed between the OVX and
sham-operated mice.
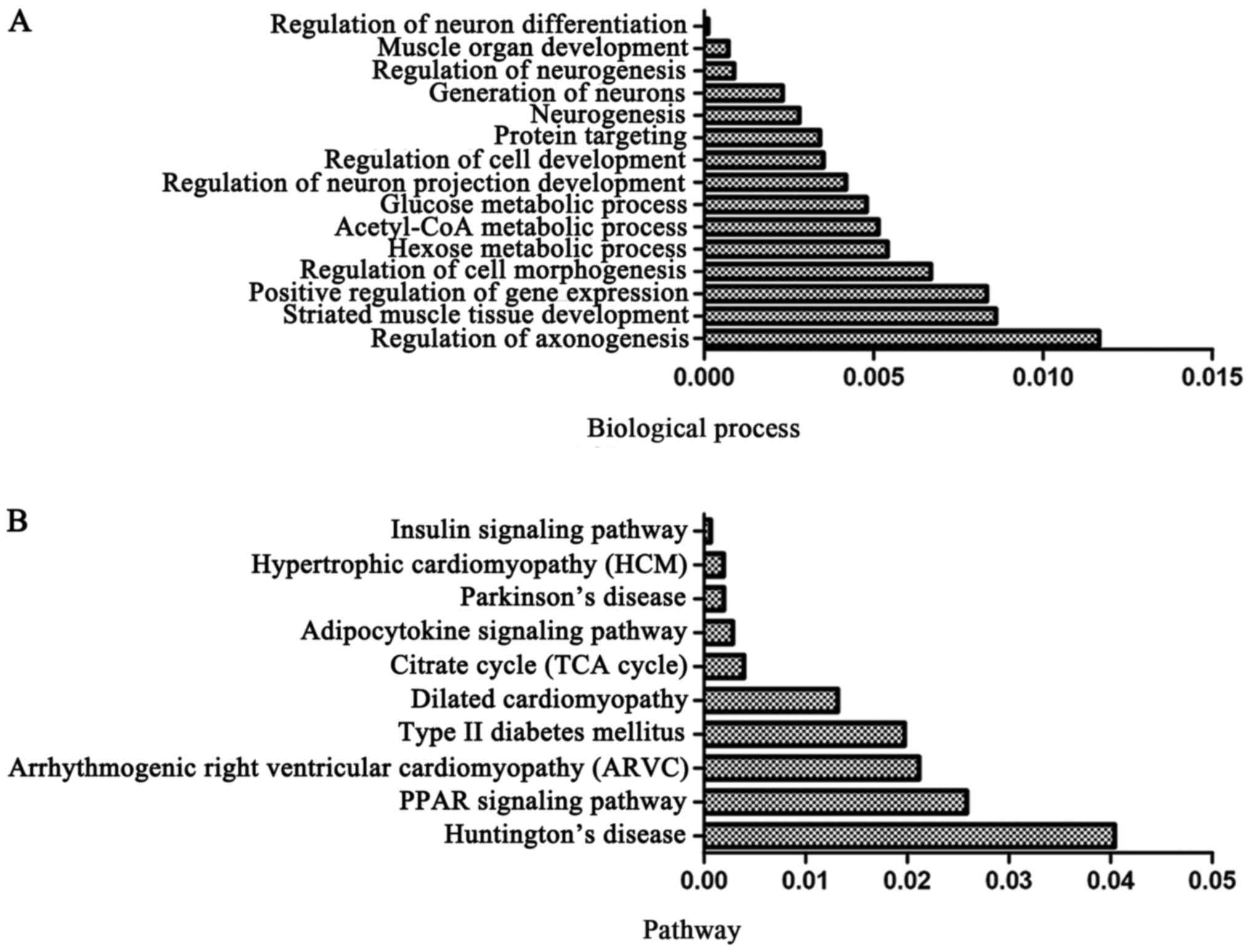
The target genes of DEmiRs may play important roles
in mandible development; therefore, we analyzed the intersection of
DEGs and the putative target genes of DEmiRs and identified this
intersection as DEmiR-targeted DEGs. Functional enrichment analysis
was also applied on the DEmiR-targeted DEGs. The results revealed
that the DEGs targeted by DEmiRs significantly regulated the
biological processes, including regulation of neuronal
differentiation, muscle organ development and regulation of
neurogenesis (Fig. 4A). Moreover,
DEmiR-targeted DEGs were significantly involved in pathways, such
as the peroxisome proliferator-activated receptor (PPAR) and
insulin signaling pathways (26,27) (Fig.
4B).
Specific gene screening by neighborhood
scoring
The DEGs were mapped to the PPI network of mice to
construct differentially expressed PPI network. The nodes in the
network represented the upregulated/downregulated genes, and the
edges indicated the associations among the genes. The network
contained 659 nodes and 3,904 edges, with 246 upregulated DEGs and
300 down-regulated DEGs, as well as 114 non-differential genes
(data not shown). The analysis of topological properties indicated
that the FYN, CREB binding protein (CREBBP), CREBBP, guanine
nucleotide-binding protein subunit beta-2-like 1 (GNB2L1) and
dynein cytoplasmic 1 heavy chain 1 (DYNC1H1) nodes exhibited
relatively high degrees, suggesting their important roles in the
network (data not shown). We subsequently analyzed the significance
of each node in the network using the neighbor-hood scoring
algorithm. The top 100 highest score nodes were obtained. The top 5
and the last 5 genes were extracted for further analysis (Table II). As shown in Table II, the first 5 genes were scored
>0, representing an upregulatory tendency of the genes and their
neighbor genes; conversely, the last 5 genes were scored <0,
indicating a downregulatory tendency. Therefore, these 10 genes
were the most significant in the network and may be potential
biomarkers for clinical analysis and diagnosis.
 | Table IINetwork analysis to screen
biomarkers. |
Table II
Network analysis to screen
biomarkers.
| Gene | Neighbor score | Rank |
|---|
| Pla2g4d | 1.3395875 | 1 |
| Lrp2bp | 1.3003 | 2 |
| Rasgrf1 | 1.278100893 | 3 |
| Tmprss11a | 1.275932143 | 4 |
| Cd109 | 1.2653375 | 5 |
| Ppp1r10 | −1.365723214 | −5 |
| Atpaf1 | −1.374365402 | −4 |
| Drd4 | −1.393098215 | −3 |
| Myadml2 | −1.410200893 | −2 |
| Jun | −1.410200893 | −1 |
miRNA-mRNA regulatory network
To enhance the analysis of the DEmiR- and
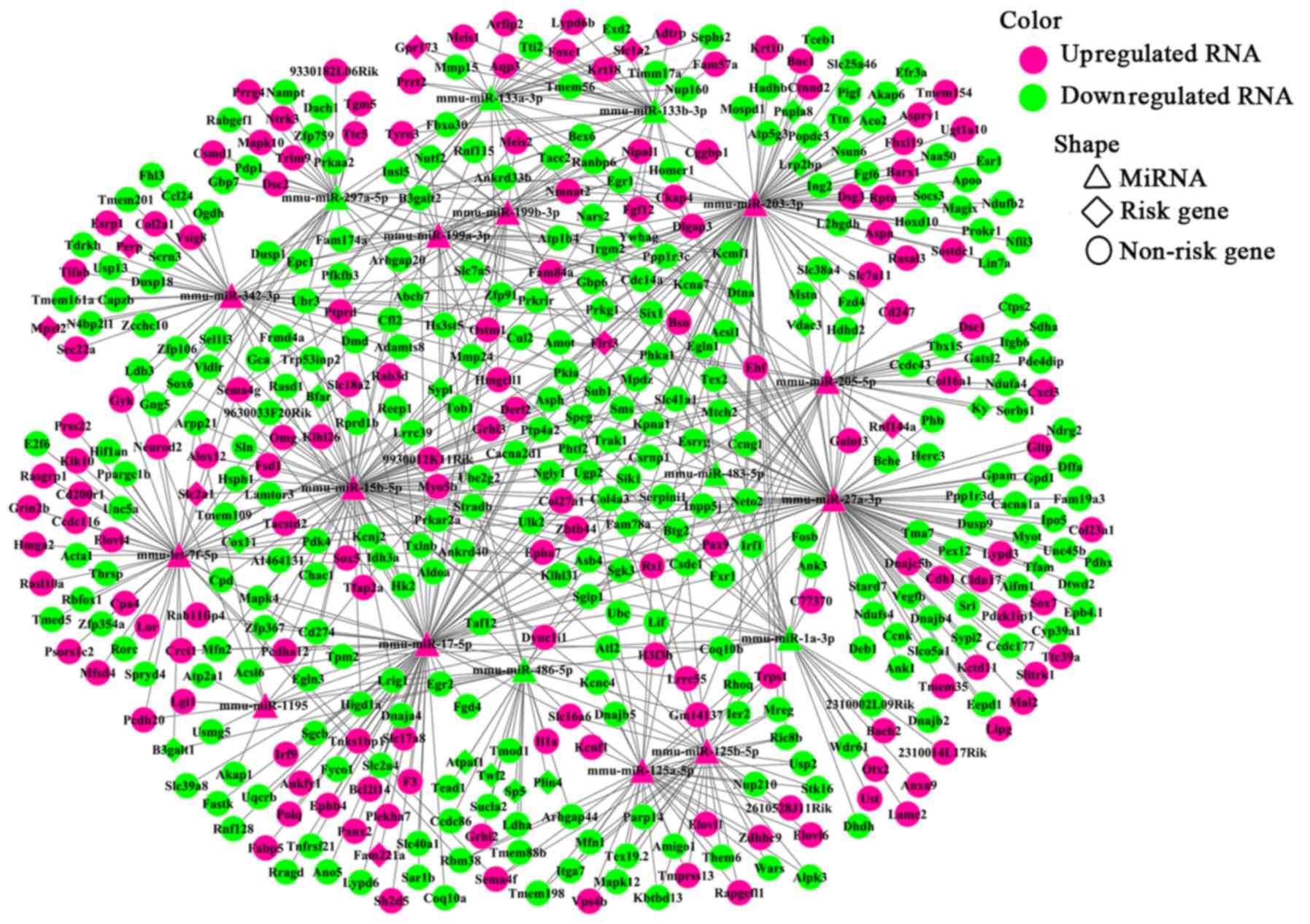
miRNA-targeted DEGs, we extracted 767 pairs of DEmiR-DEGs
comprising 18 DEmiRs and 452 DEGs and constructed an miRNA-mRNA
regulating network (Fig. 5). In
this network, 23 DEmiR-targeted DEGs were significantly altered
(i.e., differentially expressed with relatively high neigh-boring
scores) in the OVX group compared with those in the sham group.
These 23 DEGs corresponded to 12 DEmiRs and presented 36
miRNA-target pairs (Table III).
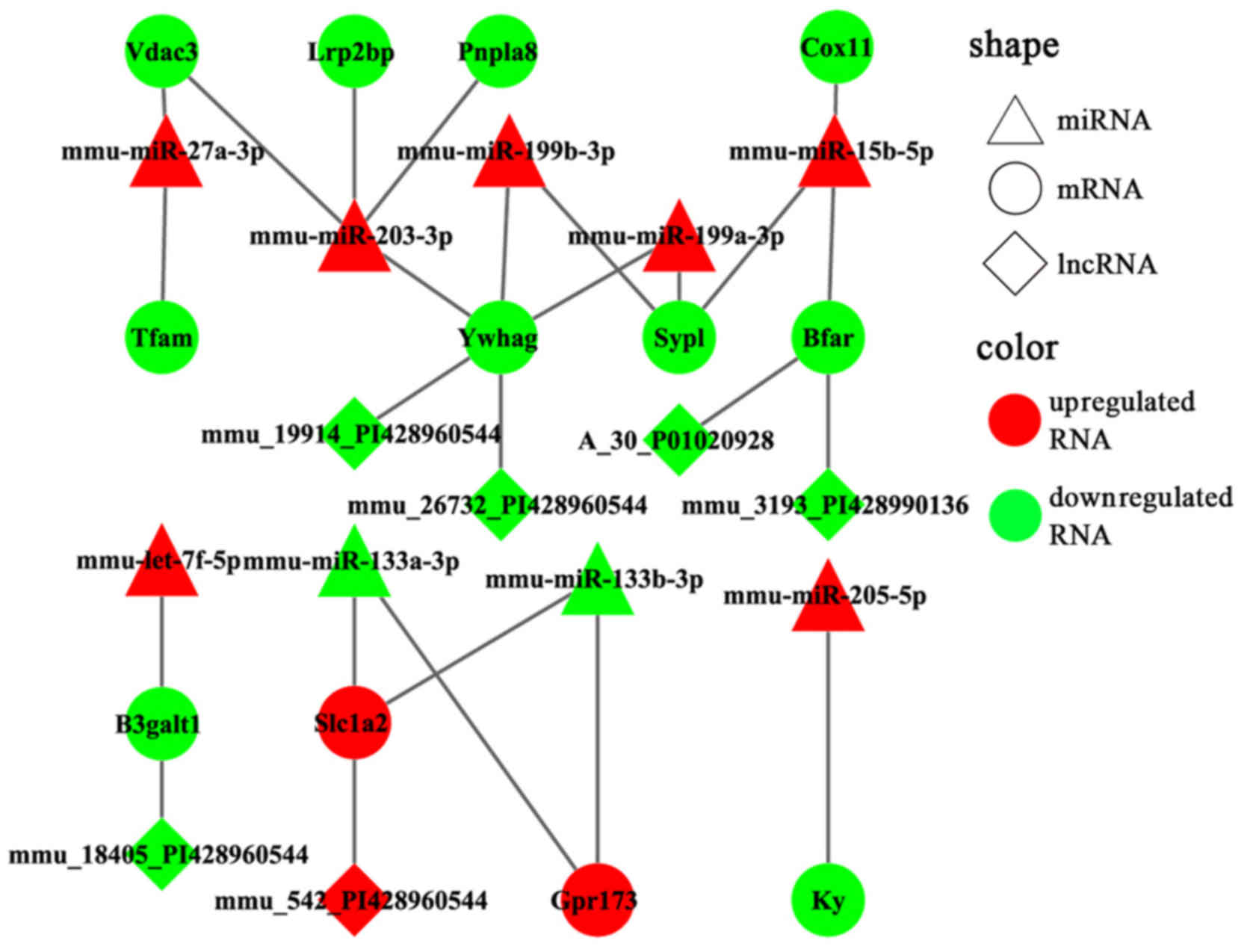
Additionally, the DEmiRs and their own targets exhibited an
opposite expression tendency in 19 of the 36 miRNA-target pairs
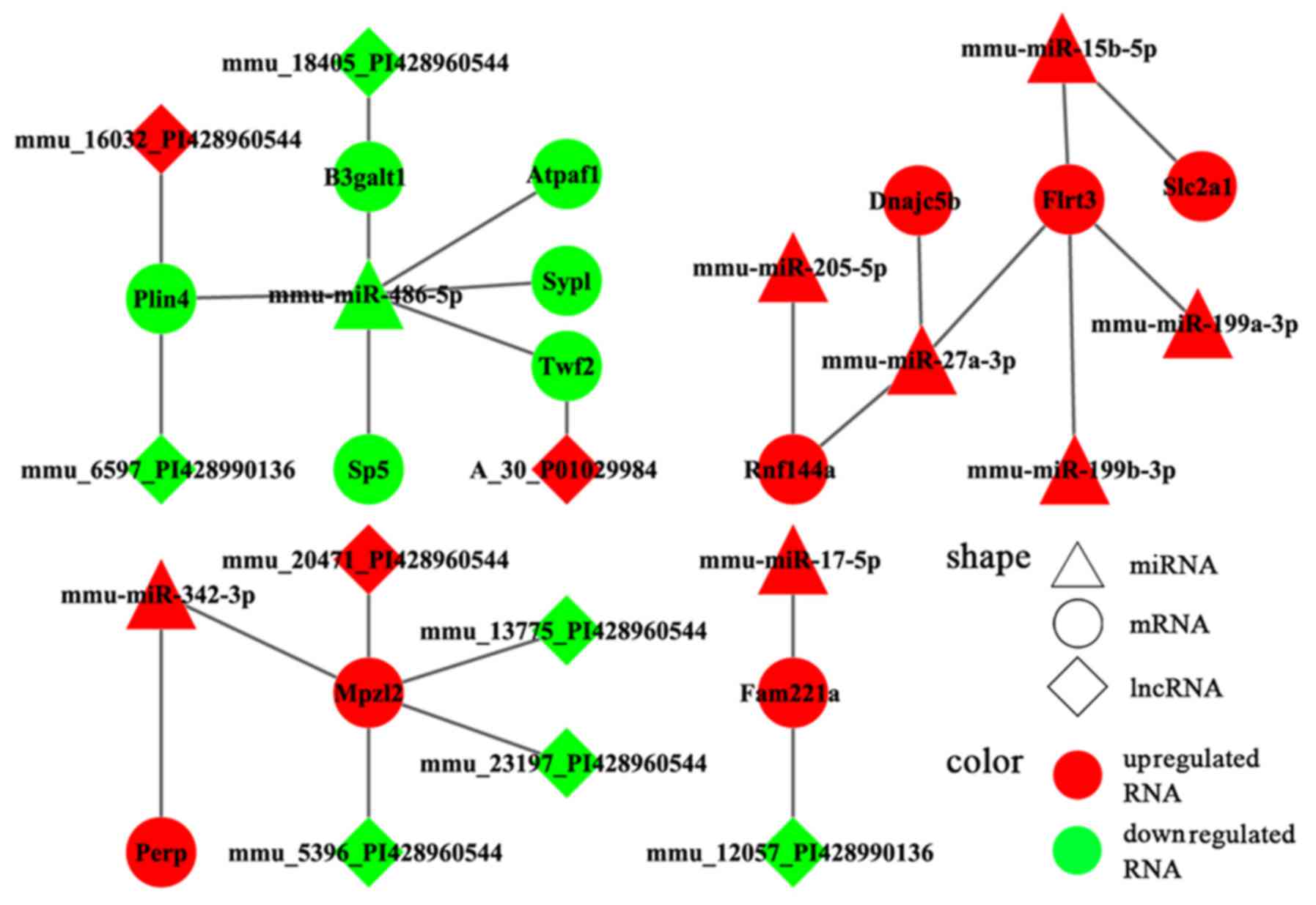
(Fig. 6). Moreover, a consistent
tendency was observed within 17 pairs, 6 of which presented a
downregulatory tendency, and the remaining pairs exhibited an
upregulatory tendency (Fig. 7).
Considering that some lncRNAs have been suggested to play key roles
in regulating the expression of their neighboring or overlapping
genes in genome-wide analysis, we screened miRNA targets related to
DElncRNAs; screening was performed based on the location
distribution of these targets on mouse chromosomes (28) using UCSC Genome Browser. A total
of 9 differentially expressed lncRNAs were acquired; through the
prediction of lncRNAs, we found that 6 DElncRNAs regulated the
target genes (Tables IV and
V). In the specific pairs, LRP2
binding protein (Lrp2bp) and perilipin 4 (Plin4) exhibited the
largest effect on the surrounding genes (Table III). Lrp2bp was downregulated in
the experimental group and targeted by the upregulated expression
of mmu-miR-203-3p (Fig. 6);
conversely, Plin4 was downregulated in the experimental group and
targeted by downregulated mmu-miR-486-5p and mmu-6597-PI428990136
as well as upregulated mmu-16032-PI428960544 (Fig. 7). These DElncRNAs may function as
competing endogenous RNAs (ceRNAs) and thus affect the development
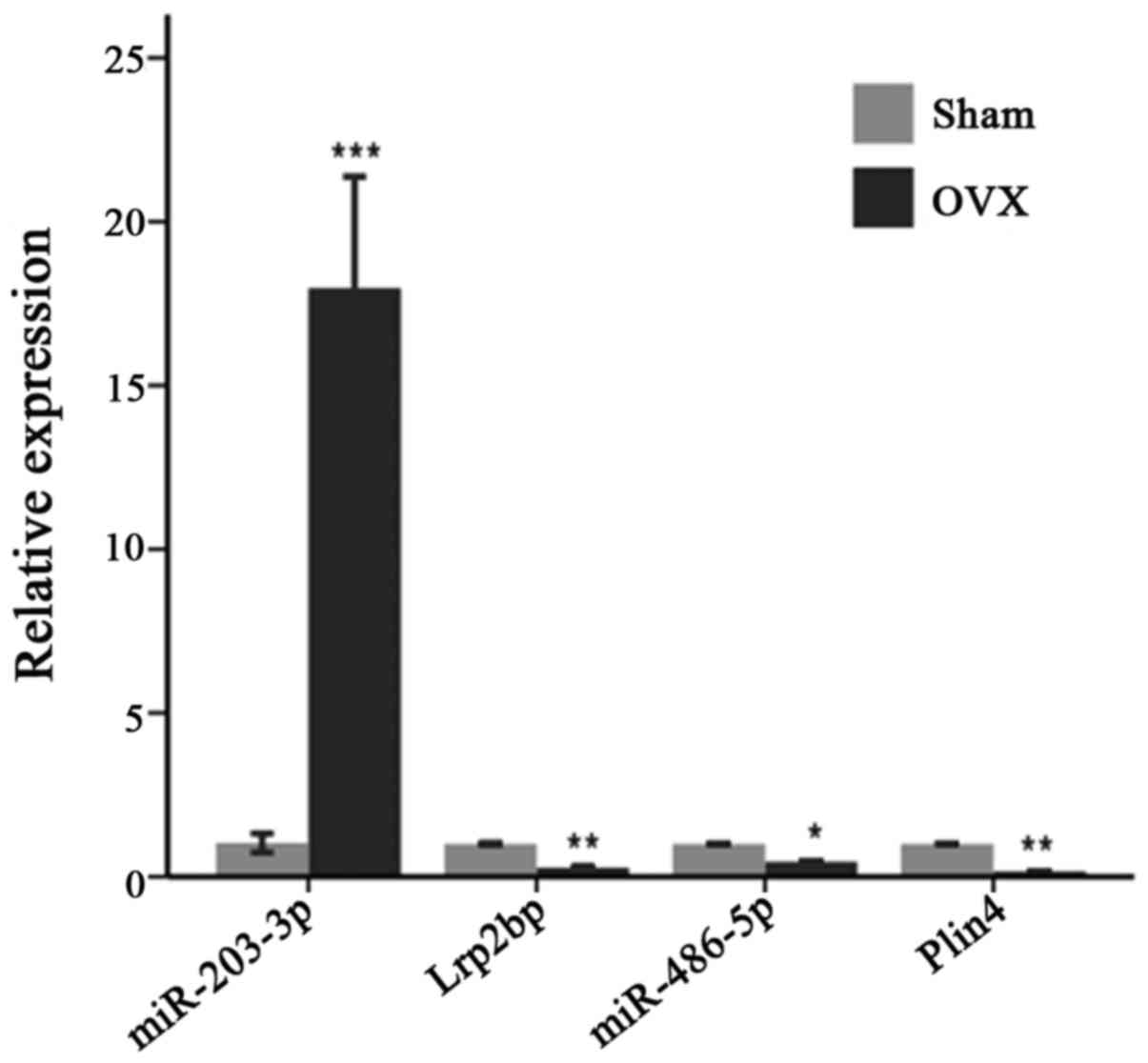
of the mandible. The results of RT-qPCR also validated the presence
of DEmiRs and DEGs in the mandible of the OVX mice (Fig. 8).
 | Table IIIRisk miRNA-target pairs. |
Table III
Risk miRNA-target pairs.
| miRNA | Label_miRNA | mRNA | Label_mRNA | mRNA_score |
|---|
| mmu-miR-203-3p | Up | Lrp2bp | Down | 1.3003 |
| mmu-miR-203-3p | Up | Vdac3 | Down | 1.234167 |
| mmu-miR-27a-3p | Up | Vdac3 | Down | 1.234167 |
| mmu-miR-486-5p | Down | Twf2 | Down | 1.186039 |
|
mmu-miR-133b-3p | Down | Slc1a2 | Up | 1.17075 |
|
mmu-miR-133a-3p | Down | Slc1a2 | Up | 1.17075 |
| mmu-miR-486-5p | Down | Sypl | Down | 1.144257 |
|
mmu-miR-199a-3p | Up | Sypl | Down | 1.144257 |
| mmu-miR-15b-5p | Up | Sypl | Down | 1.144257 |
|
mmu-miR-199b-3p | Up | Sypl | Down | 1.144257 |
|
mmu-miR-199b-3p | Up | Flrt3 | Up | 1.130324 |
| mmu-miR-15b-5p | Up | Flrt3 | Up | 1.130324 |
| mmu-miR-27a-3p | Up | Flrt3 | Up | 1.130324 |
|
mmu-miR-199a-3p | Up | Flrt3 | Up | 1.130324 |
| mmu-miR-342-3p | Up | Mpzl2 | Up | 1.100377 |
|
mmu-miR-199b-3p | Up | Ywhag | Down | 1.093004 |
|
mmu-miR-199a-3p | Up | Ywhag | Down | 1.093004 |
| mmu-miR-203-3p | Up | Ywhag | Down | 1.093004 |
| mmu-miR-205-5p | Up | Ky | Down | 1.073271 |
| mmu-miR-15b-5p | Up | Bfar | Down | 1.060279 |
| mmu-miR-15b-5p | Up | Slc2a1 | Up | 1.055304 |
| mmu-miR-203-3p | Up | Pnpla8 | Down | 1.003898 |
| mmu-miR-342-3p | Up | Perp | Up | 0.800739 |
| mmu-let-7f-5p | Up | B3galt1 | Down | 0.762609 |
| mmu-miR-486-5p | Down | B3galt1 | Down | 0.762609 |
| mmu-miR-15b-5p | Up | Cox11 | Down | −1.18494 |
| mmu-miR-17-5p | Up | Fam221a | Up | −1.19248 |
| mmu-miR-27a-3p | Up | Rnf144a | Up | −1.20161 |
| mmu-miR-205-5p | Up | Rnf144a | Up | −1.20161 |
| mmu-miR-27a-3p | Up | Tfam | Down | −1.21787 |
| mmu-miR-27a-3p | Up | Dnajc5b | Up | −1.2231 |
| mmu-miR-486-5p | Down | Sp5 | Down | −1.33851 |
|
mmu-miR-133b-3p | Down | Gpr173 | Up | −1.36572 |
|
mmu-miR-133a-3p | Down | Gpr173 | Up | −1.36572 |
| mmu-miR-486-5p | Down | Atpaf1 | Down | −1.37437 |
| mmu-miR-486-5p | Down | Plin4 | Down | −1.3931 |
 | Table IVlncRNAs associated with
miRNA-targeted genes with opposite expression trend. |
Table IV
lncRNAs associated with
miRNA-targeted genes with opposite expression trend.
Targets of miRNA
| lncRNA
|
|---|
| mRNA | Label_mRNA | lncRNA | Label_lncRNA | Chr | Start | End | Strand |
|---|
| B3galt1 | Down |
mmu_18405_PI428960544 | Down | chr2 | 67364593 | 67364652 | + |
| Bfar | Down |
mmu_3193_PI428990136 | Down | chr16 | 13672094 | 13674200 | + |
| Bfar | Down | A_30_P01020928 | Down | chr16 | 84706136 | 84706072 | − |
| Slc1a2 | Up |
mmu_542_PI428960544 | Up | chr2 | 103017944 | 103023051 | + |
| Ywhag | Down |
mmu_26732_PI428960544 | Down | chr5 | 15752481 | 15877297 | + |
| Ywhag | Down |
mmu_19914_PI428960544 | Down | chr5 | 15876898 | 15880322 | + |
 | Table VlncRNAs associated with
miRNA-targeted genes with similar expression trend. |
Table V
lncRNAs associated with
miRNA-targeted genes with similar expression trend.
Targets of miRNA
| lncRNA
|
|---|
| mRNA | Label_mRNA | lncRNA | Label_lncRNA | Chr | Start | End | Strand |
|---|
| B3galt1 | Down |
mmu_18405_PI428960544 | Down | chr2 | 67364593 | 67364652 | + |
| Fam221a | Up |
mmu_12057_PI428990136 | Down | chr6 | 100518251 | 100519726 | − |
| Mpzl2 | Up |
mmu_23197_PI428960544 | Down | chr9 | 79824054 | 79825631 | − |
| Mpzl2 | Up |
mmu_13775_PI428960544 | Down | chr9 | 79596609 | 79597858 | − |
| Mpzl2 | Up |
mmu_20471_PI428960544 | Up | chr9 | 44884261 | 44885520 | + |
| Mpzl2 | Up |
mmu_5396_PI428960544 | Down | chr9 | 79648988 | 79652070 | − |
| Plin4 | Down |
mmu_16032_PI428960544 | Up | chr17 | 56146354 | 56148925 | − |
| Plin4 | Down |
mmu_6597_PI428990136 | Down | chr17 | 56251024 | 56256970 | − |
| Twf2 | Down | A_30_P01029984 | Up | chr9 | 106177744 | 106177800 | + |
Specific miRNA-gene-lncRNA pathway
regulatory network
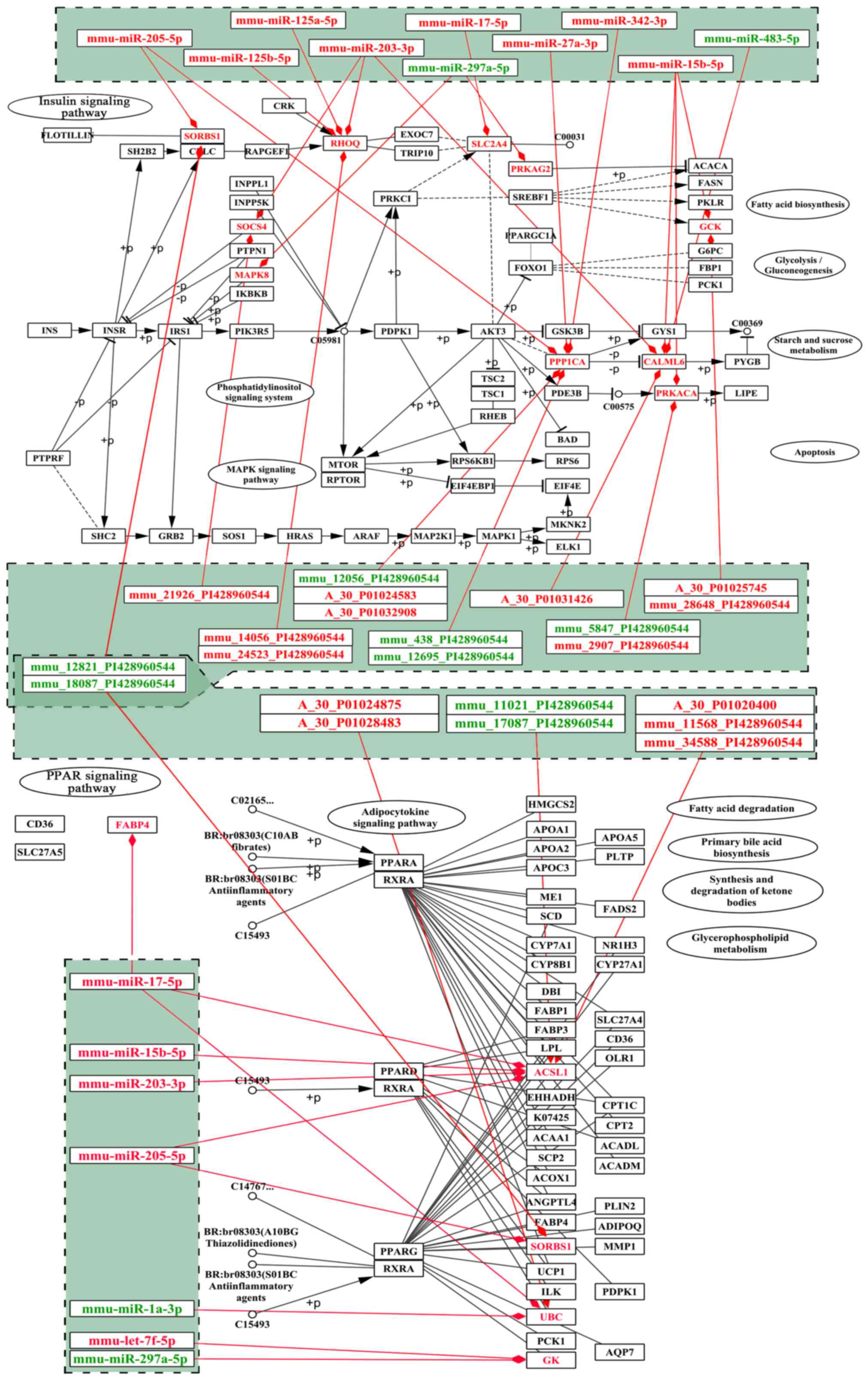
A total of 11 and 6 risk genes were obtained from
the insulin and PPAR signaling pathways, respectively. We extracted
DEmiRs that putatively targeted these risk genes and the DElncRNAs
that correlated with these risk genes. A specific miRNA-gene-lncRNA
pathway of the regulatory network was constructed by integrating
DEmiR-risk genes and DElncRNA-risk genes (Fig. 9). To further analyze the function
of the risk genes, we mapped them into the two signaling
pathways.
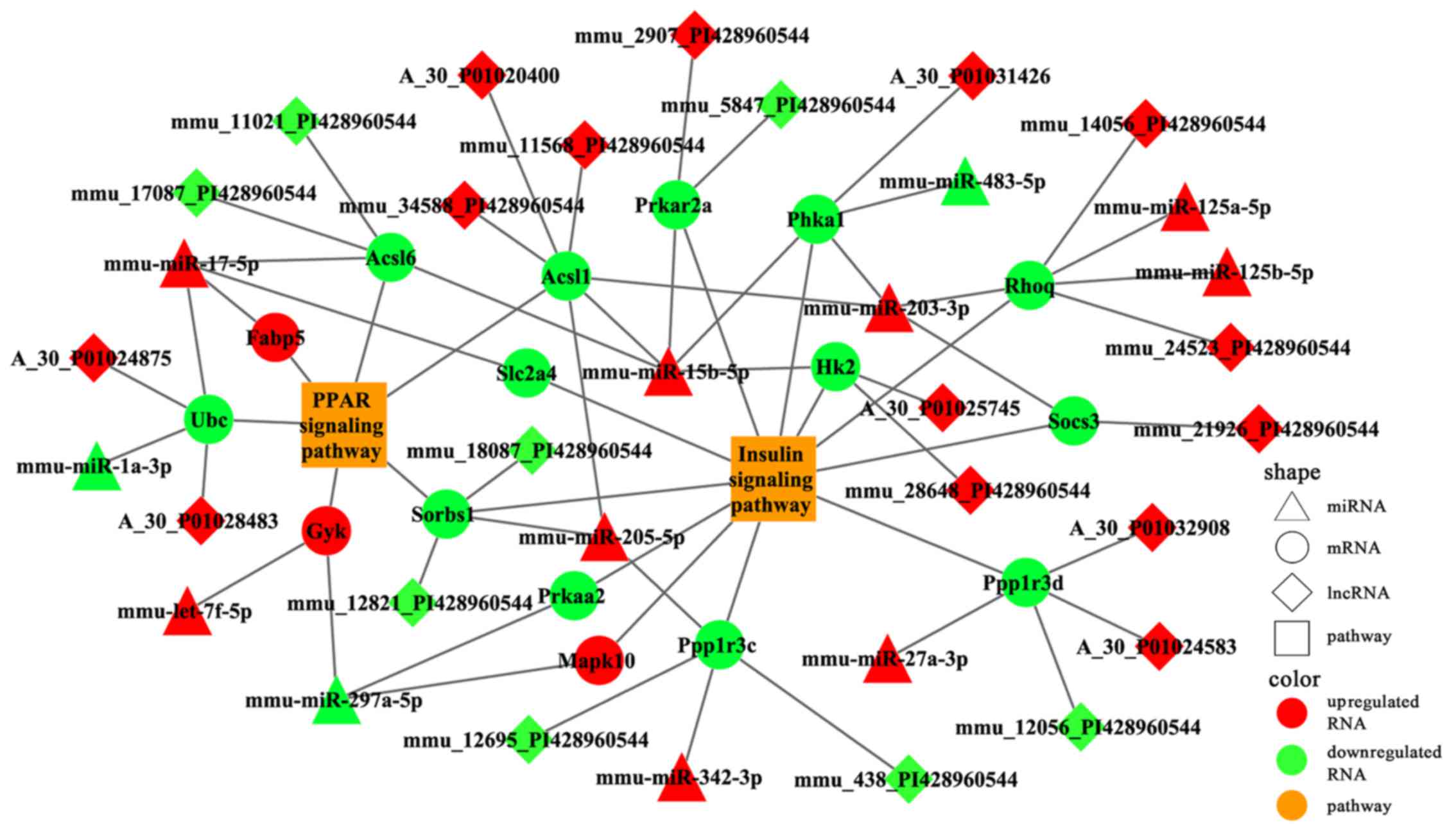
Several differentially expressed lncRNAs were mapped
into the two pathways (Fig. 10).
The results revealed that Sorbs1 participated in both pathways and
was putatively targeted by mmu-miR-205-5p; this process also
possibly involved two lncRNAs (mmu_12821_PI428960544 and
mmu_18087_PI428960544) (Fig.
10). We deduced that these lncRNAs may function as ceRNA to
regulate the signaling pathways associated with osteoporosis in the
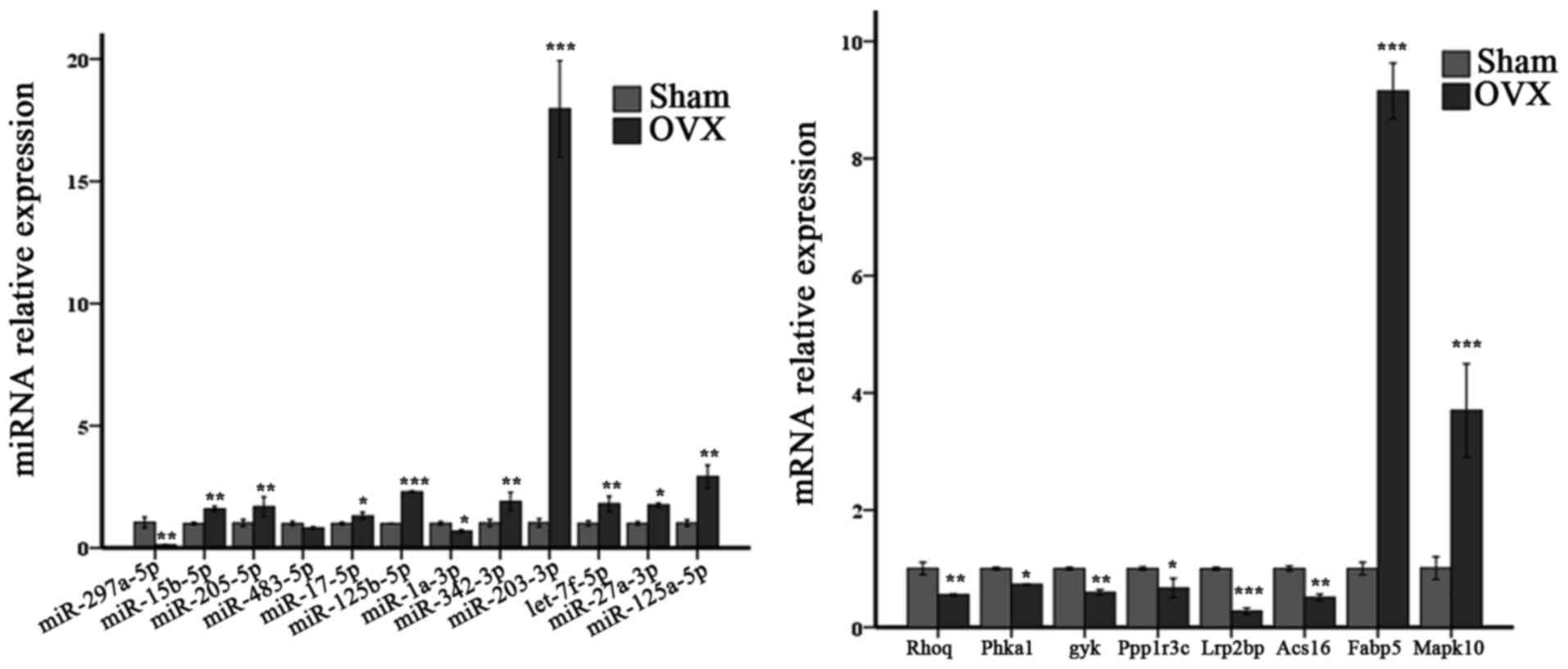
mandible of OVX mice. RT-qPCR analysis was also performed to
validate the relative expression of partial miRNAs and mRNAs in the
two signaling pathways (Fig.
11).
Discussion
Reduced ovarian production of estrogen during
menopause results in rapid bone loss and increased cortical
porosity (29). Estrogen
deficiency increases the risk of fragility fractures in the hip,
spine and wrist. A number of studies have reported that estrogen
deficiency is associated with tooth loss (30,31), periodontal diseases (32,33) and decreased BMD of the mandible
(34,35). However, studies have shown that
estrogen deficiency-induced bone loss in the mandible is not as
great as that in the long bone (36,37). The BMD and BV/TV of the mandible
decrease less significantly than the long bone in response to OVX.
The specific mechanism of this phenomenon remains unclear. The
irregular shape of the mandible and the increased tooth mastication
in OVX mice, which have been reported to eat approximately 10% more
than the sham-operated controls (38), may possibly be important factors
that alleviate bone loss in the mandible. The embryological
difference between the mandible and long bone may also be the cause
for the difference in the sensitivity of the two skeletal sites to
estrogen deficiency (36).
Therefore, some studies have suggested that mandibular cortical
width can be used to predict osteoporosis (39,40); however, effective means with which
to directly identify mandibular osteoporosis have yet to
determined. We hypothesized that the specific molecular mechanisms
of the mandible and long bone in response to estrogen deficiency
are different. Hence, changes in biomarkers in the mandible of
patients with osteoporosis may contribute to the diagnosis and
treatment of mandibular osteoporosis.
With further studies on miRNAs, which regulate the
expression of genes at the post-transcriptional level, researchers
consider that miRNAs could be the next generation therapeutic
targets in human diseases (41).
Considering the key role of miRNAs in bone metabolism (42), miRNAs which subtly repress gene
expression are anticipated to be highly efficacious in the
treatment of bone diseases. Screening out the candidate miRNAs of
post-menopausal osteoporosis and completing the target validation
of these miRNAs, can provide a basis for the diagnosis and
treatment of post-menopausal osteoporosis.
In the present study, we established a model of
post-menopausal osteoporosis by the successful excision of ovaries
in the mice. Alveolar bone loss significantly increased 3 months
following ovariectomy in the mice. Theereafter, we systematically
analyzed the distinct expression profiles of miRNAs, mRNAs and
lncRNAs, as well as their complex regulatory networks associated
with estrogen deficiency-induced osteoporosis in the mandible of
OVX mice. DEmiRs between the OVX and sham-operated groups were
mainly involved in metabolic systems, such as PPAR and insulin
signaling pathways; hence, these DEmiRs play a crucial role in
osteoporosis in the mandible of OVX mice. In the constructed
DEmiR-DEG regulatory network, Lrp2bp and Plin4 exhibited the most
significant effect on the network putatively regulated by
mmu-miR-203-3p and mmu-miR-486-5p, respectively, and were thus
associated with mandible development. Moreover, the regulatory
network of the complex DEmiR-risk gene-lncRNA pathway was
significantly associated with osteoporosis in the mandible of OVX
mice. These findings may provide insight into the molecular
mechanisms of post-menopausal osteoporosis in the mandible for the
development of therapeutic strategies.
The core DEG Lrp2bp was putatively targeted by
upregulated mmu-miR-203-3p, which may play an important role in
estrogen deficiency-induced osteoporosis in the mandible. Further
analysis through RT-qPCR validated the dysregulation of Lrp2bp and
mmu-miR-203-3p in the mandible of OVX mice. Therefore, we deduced
that upregulated mmu-miR-203-3p was partially targeted to inhibit
Lrp2bp, which contributed to post-menopausal osteoporosis in the
mandible. Similarly, a recent study indicated that mmu-miR-203 was
induced by TNF-α to downregulate lysyl oxidase, thereby inhibiting
osteoblast differentiation and finally inducing osteopenia
(43). Another core gene, namely,
DEG Plin4, was putatively targeted by downregulated mmu-miR-486-5p
in the complex regulatory network; this core may also participate
in mandible osteoporosis. Plin4 is a member of the PATS family of
genes and is involved in the lipolysis of intracellular lipid
deposits. Cusano et al (44) reported a significant association
between a single nucleotide polymorphism (SNP) in Plin4 and height,
but not with bone traits in adult Caucasian participants of the
Framingham Osteoporosis Study. However, a gender-specific
association between one polymorphism of Plin1 and BMD was reported
in a Japanese population (45).
These contradicting results may be due to the different ethnicities
of the participants used in these studies. Nevertheless, the
precise role of Plin4 in estrogen deficiency-induced osteoporosis
in the mandible must be further investigated. The present findings
present potential therapeutic targets for the treatment of
post-menopausal osteoporosis in the mandible.
DEmiR targets were found to be significantly
associated with PPAR and insulin signaling pathways, which affect
the development of the mandible (46,47). Previous studies have reported a
close association between lipid metabolism and bone remodeling, as
well as agents inducing adipogenesis that inhibit osteoblast
differentiation, thereby promoting bone loss. PPARγ plays a
critical role in adipocyte differentiation (48) and acts as a molecular switch
between osteogenic and adipogenic lineage commitment (49). In a previous study, the
overexpression of PPARγ in osteoblasts using collagen type 1
promoter decreased bone mass gain in males and accelerated bone
loss in female OVX mice (50).
Moreover, thiazolidinediones, inducers of PPARγ, and the subsequent
induction of mesenchymal stem cells into adipocytes are associated
with bone loss and osteoporosis. By contrast, PPARγ agonist has
been shown to suppress inflammatory periodontal bone loss by
inhibiting osteoclastogenesis (51). The insulin-like growth factor
(Igf) family, as well as other growth factor families (Hh, Wnt,
Tgf-Bmp, Mapk-Fgf and Notch), which implement biological function
via the insulin signaling pathway, may contribute to the merging of
the mandibular arch (52).
Variations in the promoter region of the IGF-1 gene have been shown
to be associated with BMD and the risk of osteoporosis in a Chinese
post-menopausal population (53,54). Furthermore, we noted that some
DEGs involved in the two signaling pathways were not only
differentially expressed, but were also targeted by DEmiRNAs;
Sorbs1 correlated with the two signaling pathways. The present
results indicated that upregulated mmu-miR-205-5p putatively
targeted the Sorbs1 gene, implicating both PPAR and insulin
signaling pathways. miR-205 expression is enhanced by STAT3
activation, whereas CHOP expression is inhibited in osteoblasts;
these findings may provide a basis for elucidating the mechanisms
underlying the pathogenesis of associated diseases, including
osteoporosis (55). We also
validated the upregulation of miR-205-5p in the mandible of OVX
mice through qRT-qPCR analysis; the present study may provide
insight into the complex molecular mechanisms of estrogen
deficiency-induced osteoporosis in the mandible.
Six differentially expressed lncRNAs were acquired
in the present study. These lncRNAs may act as miRNA sponges, i.e.,
as ceRNAs, to decrease the amount of miRNAs available to target
mRNAs (56). Moreover, two
lncRNAs (mmu_12821_PI428960544 and mmu_18087_PI428960544) may be
associated with the regulation of mmu-miR-205-5p and its putatively
targeted Sorbs1 gene. We deduced that these lncRNAs may function as
ceRNAs to regulate the signaling pathways in osteoporosis in the
mandible of OVX mice. The differentially expressed
lncRNA-mmu-16032-PI428960544 and mmu-6597-PI428990136, as well as
Plin4 and mmu-miR-486-5p may intervene in mandible development via
forming ceRNAs. This hypothesis may highlight a potential method
for the treatment of osteoporosis; however, this requires
validation in future studies.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81470716) and
Science and Technology Committee Foundation of Shanghai (grant no.
14411967200).
References
|
1
|
Simsek G, Uzun H, Karter Y, Aydin S and
Yigit G: Effects of osteoporotic cytokines in ovary-intact and
ovariectomised rats with induced hyperthyroidism; is skeletal
responsiveness to thyroid hormone altered in estrogen deficiency?
Tohoku J Exp Med. 201:81–89. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao L, Mao Z, Schneider LS and Brinton
RD: Estrogen receptor β-selective phytoestrogenic formulation
prevents physical and neurological changes in a preclinical model
of human menopause. Menopause. 18:1131–1142. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mawatari T, Miura H, Higaki H, Kurata K,
Moro-oka T, Murakami T and Iwamoto Y: Quantitative analysis of
three-dimensional complexity and connectivity changes in trabecular
microarchitecture in relation to aging, menopause, and
inflammation. J Orthop Sci. 4:431–438. 1999. View Article : Google Scholar
|
|
4
|
Liang H, Ma Y, Pun S, Stimpel M and Jee
WS: Aging- and ovariectomy-related skeletal changes in
spontaneously hypertensive rats. Anat Rec. 249:173–180. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao Q, Liu X, Zhang L, Shen X, Qi J, Wang
J, Qian N and Deng L: Bone selective protective effect of a novel
bone-seeking estrogen on trabecular bone in ovariectomized rats.
Calcif Tissue Int. 93:172–183. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Manolagas SC, O'Brien CA and Almeida M:
The role of estrogen and androgen receptors in bone health and
disease. Nat Rev Endocrinol. 9:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wronski TJ, Dann LM, Scott KS and Cintrón
M: Long-term effects of ovariectomy and aging on the rat skeleton.
Calcif Tissue Int. 45:360–366. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sharma D, Ciani C, Marin PA, Levy JD, Doty
SB and Fritton SP: Alterations in the osteocyte lacunar-canalicular
microenvironment due to estrogen deficiency. Bone. 51:488–497.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu T, Yan M, Wang Y, Wang Z, Xie L, Tang
C, Zhang G and Yu J: Estrogen deficiency reduces the dentinogenic
capacity of rat lower incisors. J Mol Histol. 45:11–19. 2014.
View Article : Google Scholar
|
|
10
|
Liu X, Zhang R, Zhou Y, Yang Y, Si H, Li X
and Liu L: The effect of Astragalus extractive on alveolar bone
rebuilding progress of tooth extracted socket of ovariectomied
rats. Afr J Tradit Complement Altern Med. 11:91–98. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang R, Liu ZG, Li C, Hu SJ, Liu L, Wang
JP and Mei QB: Du-Zhong (Eucommia ulmoides Oliv.) cortex extract
prevent OVX-induced osteoporosis in rats. Bone. 45:553–559. 2009.
View Article : Google Scholar
|
|
12
|
Tewari D, Khan MP, Sagar N, China SP,
Singh AK, Kheruka SC, Barai S, Tewari MC, Nagar GK, Vishwakarma AL,
et al: Ovariectomized rats with established osteopenia have
diminished mesenchymal stem cells in the bone marrow and impaired
homing, osteoinduction and bone regeneration at the fracture site.
Stem Cell Rev. 11:309–321. 2015. View Article : Google Scholar
|
|
13
|
Bhatnagar S, Krishnamurthy V and Pagare
SS: Diagnostic efficacy of panoramic radiography in detection of
osteoporosis in post-menopausal women with low bone mineral
density. J Clin Imaging Sci. 3:232013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aguilera-Barreirode LA, Dáva los-Vázquez
KF, Jiménez-Méndez C, Jiménez-Mendoza D, Olivarez-Padrón LA and
Rodríguez-García ME: The relationship of nutritional status, body
and mandibular bone mineral density, tooth loss and fracture risk
(FRAX) in pre-and postmenopausal women with periodontitis. Nutr
Hosp. 29:1419–1426. 2014.In Spanish.
|
|
15
|
Nackaerts O, Jacobs R, Devlin H, Pavitt S,
Bleyen E, Yan B, Borghs H, Lindh C, Karayianni K, van der Stelt P,
et al: Osteoporosis detection using intraoral densitometry.
Dentomaxillofac Rad. 37:282–287. 2008. View Article : Google Scholar
|
|
16
|
Barngkgei I, Al Haffar I and Khattab R:
Osteoporosis prediction from the mandible using cone-beam computed
tomography. Imaging Sci Dent. 44:263–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen J, Qiu M, Dou C, Cao Z and Dong S:
MicroRNAs in bone balance and osteoporosis. Drug Dev Res.
76:235–245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
An JH, Ohn JH, Song JA, Yang JY, Park H,
Choi HJ, Kim SW, Kim SY, Park WY and Shin CS: Changes of microRNA
profile and microRNA-mRNA regulatory network in bones of
ovariectomized mice. J Bone Miner Res. 29:644–56. 2014. View Article : Google Scholar
|
|
19
|
Lv H, Sun Y and Zhang Y: MiR-133 is
involved in estrogen deficiency-induced osteoporosis through
modulating osteogenic differentiation of mesenchymal stem cells.
Med Sci Monit. 21:1527–1534. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun M, Zhou X, Chen L, Huang S, Leung V,
Wu N, Pan H, Zhen W, Lu W and Peng S: The regulatory roles of
microRNAs in bone remodeling and perspectives as biomarkers in
osteoporosis. Biomed Res Int. 2016:16524172016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liao L, Yang X, Su X, Hu C, Zhu X, Yang N,
Chen X, Shi S, Shi S and Jin Y: Redundant miR-3077-5p and miR-705
mediate the shift of mesenchymal stem cell lineage commitment to
adipocyte in osteoporosis bone marrow. Cell Death Dis. 4:e6002013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krzeszinski JY, Wei W, Huynh H, Jin Z,
Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, et
al: miR-34a blocks osteoporosis and bone metastasis by inhibiting
osteoclastogenesis and Tgif2. Nature. 512:431–435. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Fan L, Hu J, Zhang L, Liao L, Liu S,
Wu D, Yang P, Shen L, Chen J and Jin Y: MiR-26a rescues bone
regeneration deficiency of mesenchymal stem cells derived from
osteoporotic mice. Mol Ther. 23:1349–1357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tong X, Gu PC, Xu SZ and Lin XJ: Long
non-coding RNA-DANCR in human circulating monocytes: A potential
biomarker associated with postmenopausal osteoporosis. Biosci
Biotechnol Biochem. 79:732–737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kang DM, Yoon KH, Kim JY, Oh JM, Lee M,
Jung ST, Juhng SK and Lee YH: CT imaging biomarker for evaluation
of emodin as a potential drug on LPS-mediated osteoporosis mice.
Acad Radiol. 21:457–462. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao J, Ou G, Yang N, Ding K, Kream BE,
Hamrick MW, Isales CM and Shi XM: Impact of targeted PPARγ
disruption on bone remodeling. Mol Cell Endocrinol. 410:27–34.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Q, Liu H, Yu X, Wang Y, Yang C and Xu
H: Analysis of the role of insulin signaling in bone turnover
induced by fluoride. Biol Trace Elem Res. 171:380–390. 2016.
View Article : Google Scholar
|
|
28
|
Aydin A, Kenar H, Atmaca H, Alici T, Gacar
G, Müezzinoğlu ÜS and Karaöz E: The short- and long- term effects
of estrogen deficiency on apoptosis in musculoskeletal tissues: an
experimental animal model study. Arch Iran Med. 16:271–276.
2013.PubMed/NCBI
|
|
29
|
Väänänen HK and Härkönen PL: Estrogen and
bone metabolism. Maturitas. 23(Suppl): S65–S69. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Darcey J, Horner K, Walsh T, Southern H,
Marjanovic EJ and Devlin H: Tooth loss and osteoporosis: To assess
the association between osteoporosis status and tooth number. Br
Dent J. 214:E102013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jang KM, Cho KH, Lee SH, Han SB, Han KD
and Kim YH: Tooth loss and bone mineral density in postmenopausal
South Korean women: The 2008–2010 Korea National Health and
Nutrition Examination Survey. Maturitas. 82:360–364. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Numoto Y, Mori T, Maeda S, Tomoyasu Y,
Higuchi H, Egusa M and Miyawaki T: Low bone mass is a risk factor
in periodontal disease-related tooth loss in patients with
intellectual disability. Open Dent J. 7:157–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guiglia R, Di Fede O, Lo Russo L, Sprini
D, Rini GB and Campisi G: Osteoporosis, jawbones and periodontal
disease. Med Oral Patol Oral Cir Bucal. 18:e93–e99. 2013.
View Article : Google Scholar :
|
|
34
|
Khojastehpour L, Mogharrabi S,
Dabbaghmanesh MH and Iraji Nasrabadi N: Comparison of the
mandibular bone densitometry measurement between normal, osteopenic
and osteoporotic postmenopausal women. J Dent (Tehran). 10:203–209.
2013.
|
|
35
|
Ejiri S, Tanaka M, Watanabe N, Anwar RB,
Yamashita E, Yamada K and Ikegame M: Estrogen deficiency and its
effect on the jaw bones. J Bone Miner Metab. 26:409–415. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mavropoulos A, Rizzoli R and Ammann P:
Different responsiveness of alveolar and tibial bone to bone loss
stimuli. J Bone Miner Res. 22:403–410. 2007. View Article : Google Scholar
|
|
37
|
Zhang Y, Li Y, Gao Q, Shao B, Xiao J, Zhou
H, Niu Q, Shen M, Liu B, Hu K and Kong L: The variation of
cancellous bones at lumbar vertebra, femoral neck, mandibular angle
and rib in ovariectomized sheep. Arch Oral Biol. 59:663–669. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Elovic RP, Hipp JA and Hayes WC:
Ovariectomy decreases the bone area fraction of the rat mandible.
Calcif Tissue Int. 56:305–310. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nagi R: Relationship between femur bone
mineral density, body mass index and dental panoramic mandibular
cortical width in diagnosis of elderly postmenopausal women with
osteoporosis. J Clin Diagn Res. 8:ZC36–ZC40. 2014.PubMed/NCBI
|
|
40
|
Hekmatin E, Ahmadi SS, Ataiekhorasgani M,
Feizianfard M, Jafaripozve S and Jafaripozve N: Prediction of
lumbar spine bone mineral density from the mandibular cortical
width in postmenopausal women. J Res Med Sci. 18:951–955. 2013.
|
|
41
|
Srinivasan S, Selvan ST, Archunan G,
Gulyas B and Padmanabhan P: MicroRNAs - the next generation
therapeutic targets in human diseases. Theranostics. 3:930–942.
2013. View Article : Google Scholar
|
|
42
|
Zhao X, Xu D, Li Y, Zhang J, Liu T, Ji Y,
Wang J, Zhou G and Xie X: MicroRNAs regulate bone metabolism. J
Bone Miner Metab. 32:221–231. 2014. View Article : Google Scholar
|
|
43
|
Khosravi R, Sodek KL, Xu WP, Bais MV,
Saxena D, Faibish M and Trackman PC: A novel function for lysyl
oxidase in pluripotent mesenchymal cell proliferation and relevance
to inflammation-associated osteopenia. PLoS One. 9:e1006692014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cusano NE, Kiel DP, Demissie S, Karasik D,
Cupples LA, Corella D, Gao Q, Richardson K, Yiannakouris N and
Ordovas JM: A Polymorphism in a gene encoding Perilipin 4 is
associated with height but not with bone measures in individuals
from the Framingham Osteoporosis Study. Calcif Tissue Int.
90:96–107. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yamada Y, Ando F and Shimokata H:
Association of polymorphisms in forkhead box C2 and perilipin genes
with bone mineral density in community-dwelling Japanese
individuals. Int J Mol Med. 18:119–127. 2006.PubMed/NCBI
|
|
46
|
Surve VV, Andersson N, Alatalo S,
Lehto-Axtelius D, Halleen J, Väänänen K and Håkanson R: Does
combined gastrectomy and ovariectomy induce greater osteopenia in
young female rats than gastrectomy alone? Calcif Tissue Int.
69:274–280. 2001. View Article : Google Scholar
|
|
47
|
Reinwald S, Mayer LP, Hoyer PB, Turner CH,
Barnes S and Weaver CM: A longitudinal study of the effect of
genistein on bone in two different murine models of diminished
estrogen-producing capacity. J Osteoporos. 2010: View Article : Google Scholar : 2009.
|
|
48
|
Tontonoz P, Hu E and Spiegelman BM:
Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a
lipid-activated transcription factor. Cell. 79:1147–1156. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lecka-Czernik B, Gubrij I, Moerman EJ,
Kajkenova O, Lipschitz DA, Manolagas SC and Jilka RL: Inhibition of
Osf2/Cbfa1 expression and terminal osteoblast differentiation by
PPARgamma2. J Cell Biochem. 74:357–371. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cho SW, Yang JY, Her SJ, Choi HJ, Jung JY,
Sun HJ, An JH, Cho HY, Kim SW, Park KS, et al: Osteoblast-targeted
overexpression of PPARγ inhibited bone mass gain in male mice and
accelerated ovariectomy-induced bone loss in female mice. J Bone
Miner Res. 26:1939–1952. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hassumi MY, Silva-Filho VJ, Campos-Júnior
JC, Vieira SM, Cunha FQ, Alves PM, Alves JB, Kawai T, Gonçalves RB
and Napimoga MH: PPAR-gamma agonist rosiglitazone prevents
inflammatory periodontal bone loss by inhibiting
osteoclastogenesis. Int Immunopharmacol. 9:1150–1158. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Fujita K, Taya Y, Shimazu Y, Aoba T and
Soeno Y: Molecular signaling at the fusion stage of the mouse
mandibular arch: Involvement of insulin-like growth factor family.
Int J Dev Biol. 57:399–406. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li F, Xing WH, Yang XJ, Jiang HY and Xia
H: Influence of polymorphisms in insulin-like growth factor-1 on
the risk of osteoporosis in a Chinese postmenopausal female
population. Int J Clin Exp Pathol. 8:5727–5732. 2015.PubMed/NCBI
|
|
54
|
Yun-Kai L, Hui W, Xin-Wei Z, Liang G and
Jin-Liang Z: The polymorphism of Insulin-like growth factor-I
(IGF-I) is related to osteoporosis and bone mineral density in
postmenopausal population. Pak J Med Sci. 30:131–135.
2014.PubMed/NCBI
|
|
55
|
Zhuang J, Gao R, Wu H, Wu X and Pan F:
Signal transducer and activator of transcription 3 regulates
CCAAT-enhancer-binding homologous protein expression in osteoblasts
through upregulation of microRNA-205. Exp Ther Med. 10:295–299.
2015.PubMed/NCBI
|
|
56
|
Paci P, Colombo T and Farina L:
Computational analysis identifies a sponge interaction network
between long non-coding RNAs and messenger RNAs in human breast
cancer. BMC Syst Biol. 8:832014. View Article : Google Scholar : PubMed/NCBI
|