Introduction
The prevalence of allergic diseases has increased
rapidly over the past decades, with allergic disorders affecting
>20% of the general population worldwide, including food
allergies (1), allergic
dermatitis (2), allergic rhinitis
(AR) and asthma (3). These
disorders have been classified under type I hypersensitivity.
Cumulative studies have demonstrated that allergic inflammation is
characterized by a T helper 2 (Th2) cell-driven immune response.
IgE binds to the high-affinity receptor of IgE on the surface of
mast cells in order to sensitize them. Re-exposure to the specific
antigens cross-links the IgE/FcεRI complexes on the surface of mast
cells to trigger degranulation and release of inflammatory
mediators (4,5). Skewed Th2-cell proliferation plays a
critical role in the induction of allergic inflammation by
secreting interleukin (IL)-4, IL-5 and IL-13 after activation
(1), while the factors involved
in the maintenance of Th2-cell polarization in allergic conditions
require further elucidation.
The T-cell immunoglobulin and mucin domain (TIM)
family is a recently identified transmembrane molecular group; it
consists of 8 members (TIM1-TIM8) in mice and 3 members (TIM1, TIM3
and TIM4) in humans (6). TIMs are
involved in the regulation of innate and adaptive immune responses,
such as allergy, asthma, autoimmunity and transplant tolerance
(7,8). TIM1, TIM3 and TIM4 have distinct
functions in immune responses and they are expressed by different
immune cells. Previous studies demonstrated that TIM1 was a
susceptibility gene for asthma and allergy, which was
preferentially expressed on activated Th2 cells; by contract, TIM4
is mainly expressed on antigen-presenting cells (APCs) (8). As one of the potent co-stimulatory
signals from APCs, TIM4̸TIM1 interaction facilitates Th2-cell
activation and plays an important role in allergic conditions
(7,9). The crystal structures of TIM1 and
TIM4 include an immunoglobulin variable (IgV) domain (10). TIM1 is co-localized with CD3 on
the T-cell surface, and may be functional as part of the T-cell
receptor (TCR) signaling complex during T-cell activation, possibly
through IL-2-induced T-cell kinase (ITK) and phosphoinositide
3-kinase (PI3K) phosphorylation. Overexpression of TIM1 in T cells
enhances the transcription of the IL-4 promoter (8,11).
However, further experimental research is required to evaluate the
intracellular mechanisms of the TIM1 pathway on CD4 T-cell
responses in allergic conditions.
Silent information regulator 1 (SIRT1), as a class
III histone deacetylase, belongs to one of seven SIRT family
members. It was previously revealed that SIRT members had conserved
sequences, expressed in bacteria, mammalian and humans (12). SIRT1 deacetylates histones and
other non-histone proteins, and is a multifunctional molecule
involved in a variety of pathways, such as cell differentiation,
apoptosis, cell aging, tumor suppression and regulation of
asthmatic inflammation (12,13). Notably, SIRT1 was reported to be
associated with regulation of asthmatic inflammation and Th2-cell
responses by inhibiting gene expression via post-translational
modification of histone proteins (14,15). The expression of SIRT1 in allergic
conditions and the role of SIRT1 in the TIM4/TIM1
interaction-mediated CD4+ T-cell response has not been
clearly determined. The aim of the present study was to investigate
the expression of SIRT1 in mice with AR, and determine whether
SIRT1 levels are increased in the nasal mucosa of mice with AR. In
addition, the effect of the TIM4/TIM1 interaction on SIRT1
expression in splenic CD4+ T cell from mice with AR was
investigated, as was the effect of increased SIRT1 expression on
Th2-cell polarization.
Materials and methods
Reagents
Alum and ovalbumin (OVA), carboxyfluorescein
succinimidyl ester (CFSE) (21888) LY-294,002 (L9908), sirtinol
(S7942) and resveratrol (R5010) were purchased from
Sigma-Aldrich/Merck KGaA (Shanghai, China). The OVA-specific IgE
ELISA kits were obtained from Wuhan EIAab Science (Wuhan, China).
TIM4 shRNA, TIM1 shRNA, anti-mouse p-Akt (monoclonal; 1:200;
sc-293125) and anti-mouse p53 (monoclonal; 1:100, sc-393031) were
purchased from Santa Cruz Biotechnology, Inc. (Guangzhou, China).
Recombinant mouse TIM4 protein was purchased from R&D Systems
(Shanghai, China). Rat allophycocyanin-anti-mouse CD4 (monoclonal;
1:200; cat. no. MCD0405), rat phycoerythrin (PE)-anti-mouse CD11c
(monoclonal; 1:200; cat. no. 12-0114-82), rat
PE-Cy7-conjugated-anti-mouse IL-4 (monoclonal; 1:200; cat. no.
25-7042-82), rat PE-conjugated-anti-mouse interferon-γ (IFN-γ;
monoclonal; 1:200; cat. no. 12-7311-82), anti-mouse CD178 (Fas
ligand) FITC (monoclonal; 1:200; cat. no. 11-5911-82) and Annexin V
kit were obtained from eBioscience, Inc. (Shanghai, China), rat
FITC-conjugated anti-mouse TIM4 (polyclonal; 1:200; EL924747) was
obtained from EterLife (Tianjin China), the caspase-3 fluorometric
assay kit (K105) was obtained from Biovision (Shanghai, China), and
magnetic bead-conjugated antibodies (monoclonal; 100 µl
antibody per 108 total cells; 130-049-201) were
purchased from Miltenyi Biotec (Shanghai, China).
Mice
A total of 48 female BALB/c mice (6–8 weeks old and
weighing 18–22 g) were purchased from the Guangzhou Experimental
Animal Center and were housed under pathogen-free conditions. The
experimental procedures were approved by the Animal Ethics
Committee at the ENT Institute of Shenzhen.
Induction of nasal allergic inflammation
in mice
The AR murine model was constructed as previously
described, with slight modifications (16). The mice were sensitized with OVA
(40 µg/kg) diluted in sterile normal saline and aluminum
hydroxide (alum adjuvant, 40 mg/kg), four times by intraperitoneal
(i.p.) injection on days 1, 5, 14 and 21. Intranasal challenge with
OVA (20 µl of 25 mg/ml OVA) diluted with sterile normal
saline was performed daily on days 22–35. The control groups were
treated with normal saline i.p. and nasal challenge. At 24 h after
the last challenge, the mice were sacrificed and samples were
collected from each mouse. The serum OVA-specific IgE was measured
by ELISA. The nasal mucosa was excised for immunohistochemical
analysis, and spleen mononuclear cells were isolated;
CD4+ CD25− T cells and dendritic cells (DCs)
were further isolated from the spleen by magnetic cell sorting with
commercial reagent kits [CD4 (L3T4) MicroBeads, mouse (cat. no.
130-049-201) and CD11c MicroBeads UltraPure, mouse (cat. no.
130-108-338) (both from Miltenyi Biotec)] following the
manufacturer's instructions. The purity of the CD4+ CD25
T cells and DCs was >98%, as determined by flow cytometry.
Animal groups
The 48 mice were randomly divided into control and
AR groups (n=24 each), with 4 subgroups in each category (n=6 per
subgroup). One subgroup from each group was used to examine: i)
Splenic mononuclear cells for the frequency of CD4+
IL-4+ T cells and CD4+ IFN-γ+ T
cells by fluorescence-activated cell sorting (FACS; Fig. 1); ii) splenic DCs for TIM4
expression and CD4+ T-cell proliferation (Fig. 2); iii) nasal mucosa tissues and
splenic CD4+ T cells (Fig.
3); and iv) splenic mononuclear cells and DCs CD4+ T
cells to determine the role of SIRT1 in their proliferation and
measure the Fas ligand (FasL), caspase-3 and p53 expression
(Figs. 4 and 5).
Histology
The nasal mucosa samples were collected and fixed in
4% paraformaldehyde overnight and processed for paraffin embedding.
Sections (4 μm) were prepared and stained with hematoxylin
and eosin for detection of inflammatory cell infiltration.
ELISA
The serum levels of OVA-specific IgE were measured
with purchased reagent kits (Wuhan EIAab Science) following the
manufacturers' instructions.
Gene silencing
The TIM4 gene in splenic DCs and the TIM1 gene in
CD4+ T cells were knocked down by RNA interference with
reagent kits [Lipofectamine™ 3000 Transfection Reagent (L3000015);
Invitrogen, Shanghai, China] following the manufacturer's
instructions. The effect of gene silencing was assessed by western
blotting.
Flow cytometry
Cells were collected from the culture and fixed with
1% formaldehyde and 0.1% Triton x-100 (and permeabilization buffer,
if necessary) for 30 min at 4°C, washed with 1% bovine serum
albumin (BSA)/phosphate-buffered saline (PBS) 3 times, and blocked
for 30 min at 4°C with 1% BSA. Cells were incubated with the
fluorescence-labeled antibodies for 1 h at room temperature. After
washing with PBS, the cells were analyzed with a flow cytometer
(FACSCanto II; BD Biosciences, Franklin Lakes, NJ, USA).
Western blot analysis
Total protein was extracted from the cells with a
protein extraction buffer [M-PER™ Mammalian Protein Extraction
Reagent (cat. no. 78503); Thermo Fisher, Shanghai, China]. The
protein concentration was measured by the bicinchoninic acid
method. After denaturing, the samples were loaded in duplicate onto
a 10% sodium dodecyl sulfate polyacrylamide gel; proteins were
separated by electrophoresis and transferred onto a nitrocellulose
membrane. The membrane was blocked by 5% skimmed milk for 30 min
and incubated with the primary antibodies (50–100 ng/ml) followed
by the secondary antibodies. The immune complex on the membrane was
developed by enhanced luminol-based chemiluminescence and the
results were photographed using the UVP BioSpectrum Imaging system
(BioSpectrum, Upland, CA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Nasal mucosa was removed and total RNA was extracted
using an RNeasy mini kit (Qiagen, Inc., Valencia, CA, USA). A total
of 1 µg RNA was reverse-transcribed into cDNA with the
IScript™ cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) according to the manufacturer's protocol. The resulting
complementary DNA was then subjected to qPCR on the MiniOpticon PCR
system. The primers used in the experiments were as follows: SIRT1:
Forward, ctgttgaccgatggactcct and reverse, gccacagcgtcatatcatcc;
β-actin: Forward, gtgggaatgggtcagaagga and reverse,
tcatcttttcacggttggcc. The amplification protocol was performed as
follows: 1 cycle at 98°C for 1 min followed by 40 cycles at 98°C
for 10 sec, 55°C for 20 sec, and 72°C for 30 sec. The relative
SIRT1 gene expression compared with a housekeeping gene was
analyzed using the comparative quantification cycle method.
Statistical analysis
All values are presented as mean ± standard
deviation of a minimum of three independent experiments. The values
were analyzed by one-way analysis of variance, followed by Tukey's
test for multiple comparisons. P<0.05 was considered to indicate
statistically significant differences.
Results
Establishment of mouse model with nasal
Th2 type inflammation
A mouse model of AR was developed as previously
reported (16). Compared with the
control group, mice sensitized with OVA and alum (AR group) had
more denuded skin around the nose and a higher number of scratching
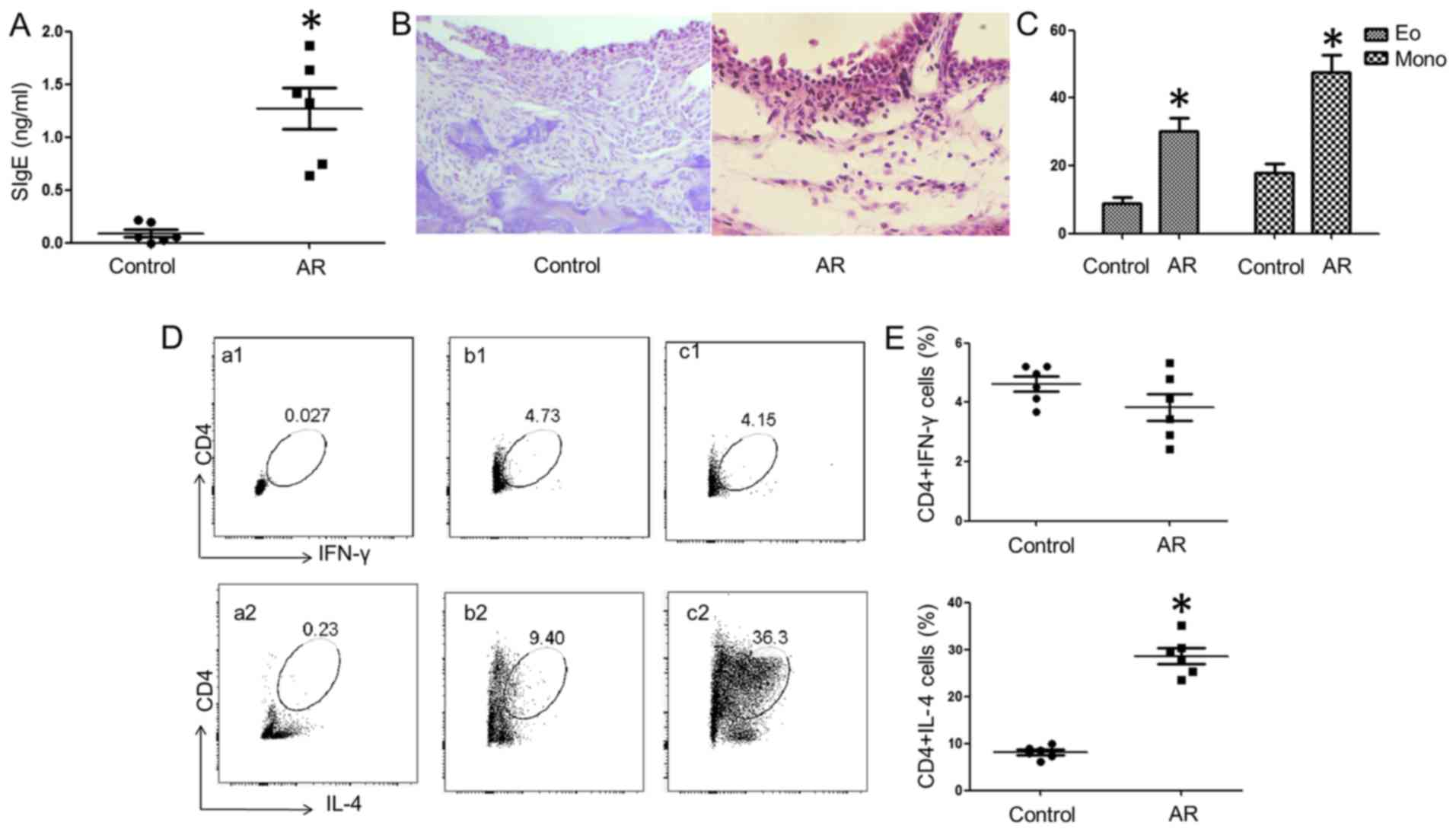
events (data not shown). OVA-specific IgE (SIgE) was detected in
the serum (Fig. 1A). Inflammatory
cell infiltration in the nasal mucosa was observed in the AR group
(Fig. 1B and C). As shown by flow
cytometry, more IL-4+ CD4+ T cells and fewer
IFN-γ+ CD4 T cells were detected in the spleens of the
allergy group compared with the control group (Fig. 1D).
The TIM4̸TIM1 interaction promotes
Th2-cell proliferation
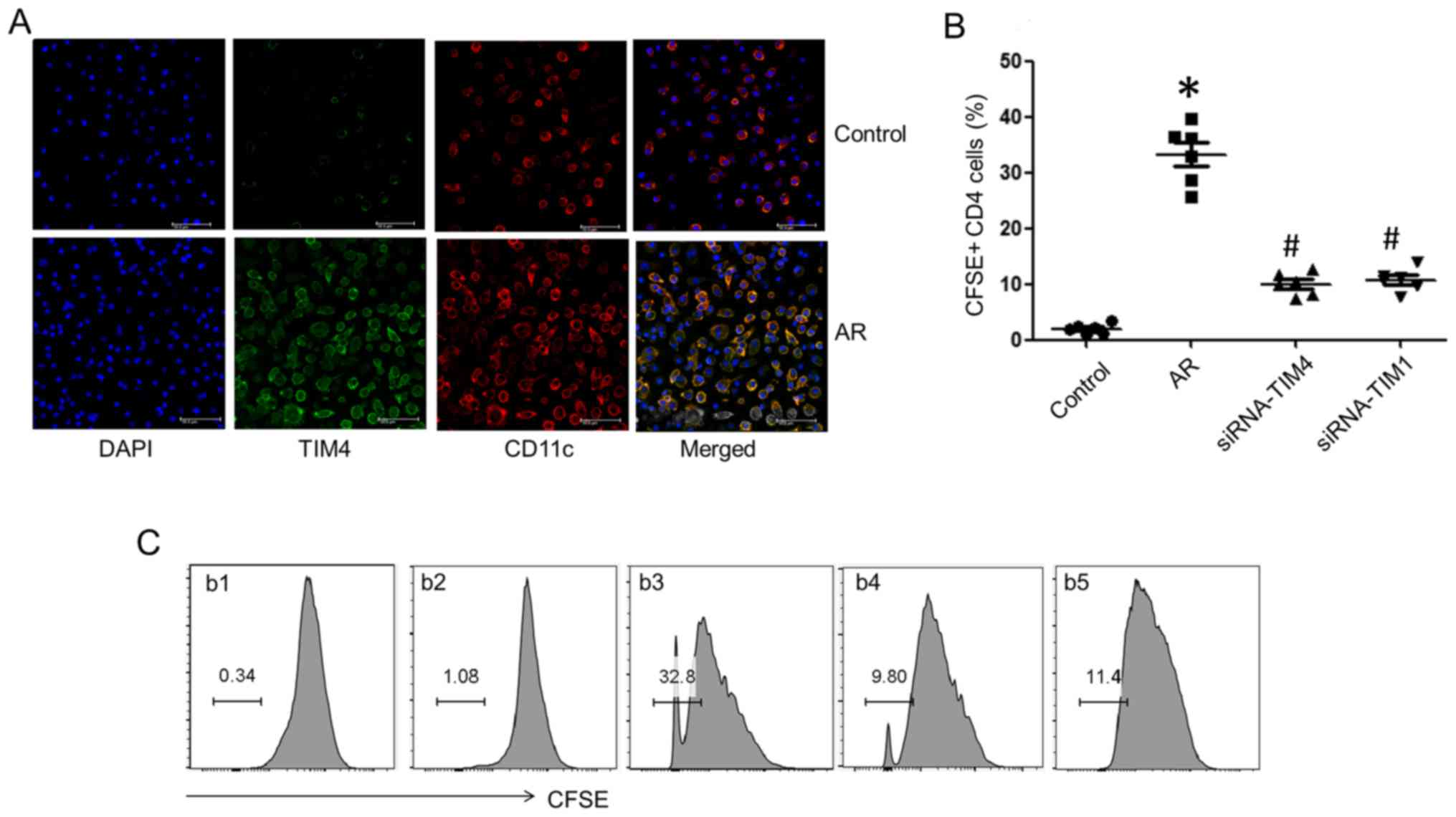
Splenic DCs were isolated from AR mice and control
mice. The DCs were pulsed with OVA (10 ng/ml) in the culture for 3
days. TIM4 expression was detected in DCs by confocal microscopy
(Fig. 2A). DCs from the AR group
exhibited higher expression of TIM4 compared with the control
group. Splenic CD4+ T cells were isolated and
co-cultured with DCs in the presence of OVA (10 ng/ml) for 3 days
and analyzed by flow cytometry. CD4+ T cell
proliferation from the AR group was markedly increased, which was
inhibited by gene silencing of TIM4 in DCs and TIM1 in
CD4+ T cells (Fig. 2B and
C). The results suggest that the TIM4̸TIM1 interaction
facilitates Th2-cell proliferation in AR mice.
The TIM4̸TIM1 interaction modulates SIRT1
expression in splenic CD4+ T cells
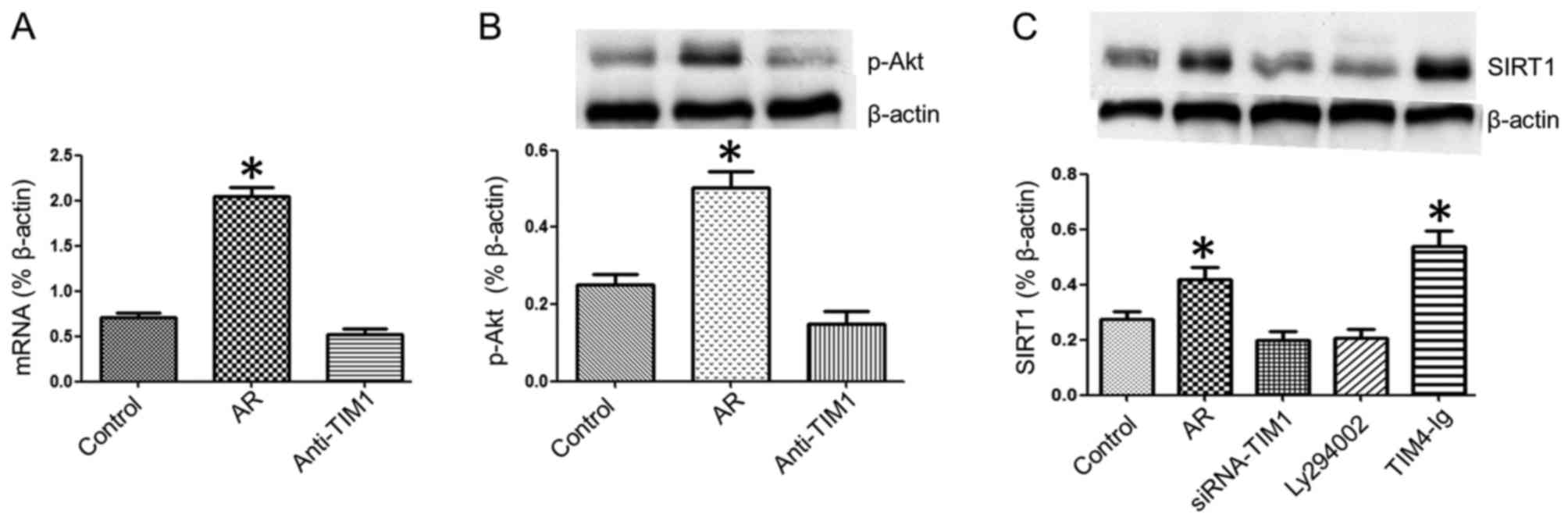
To determine the role of the TIM4̸TIM1 interaction
in the expression of SIRT1 in DCs, samples of splenic DCs were
analyzed by RT-qPCR. It was observed that the SIRT1 mRNA expression
was increased in the nasal mucosa of AR mice, which was blocked by
caudal vein injection of anti-TIM1 blocking antibody (50
µg/mouse) (Fig. 3A). Next,
the effect of the TIM4̸TIM1 interaction on SIRT1 expression was
evaluated in CD4+ T cells. Splenic mononuclear cells in
each group were collected and cultured with OVA (10 ng/ml) for 3
days, CD4+ T cells were isolated by microbeads, and
SIRT1 expression in CD4+ T cells was assessed by western
blot analysis. The results demonstrated that Akt phosphorylation
was increased in splenic CD4+ T cells from the AR group
and it was inhibited by TIM1 gene silencing (Fig. 3B). The SIRT1 expression was higher
in splenic CD4+ T cells from the AR group compared with
the control group, while SIRT1 expression was inhibited by adding
Ly294002 (PI3K/Akt inhibitor) to the culture or via TIM1 gene
silencing, and was enhanced in CD4+ T cells from the AR
group by culturing with TIM4-Ig (Fig.
3C). These results indicate that the TIM4̸TIM1 interaction
modulates SIRT1 expression in splenic CD4+ T cells
during allergic inflammation via PI3K/Akt signaling.
SIRT1 facilitates CD4+ T-cell
proliferation
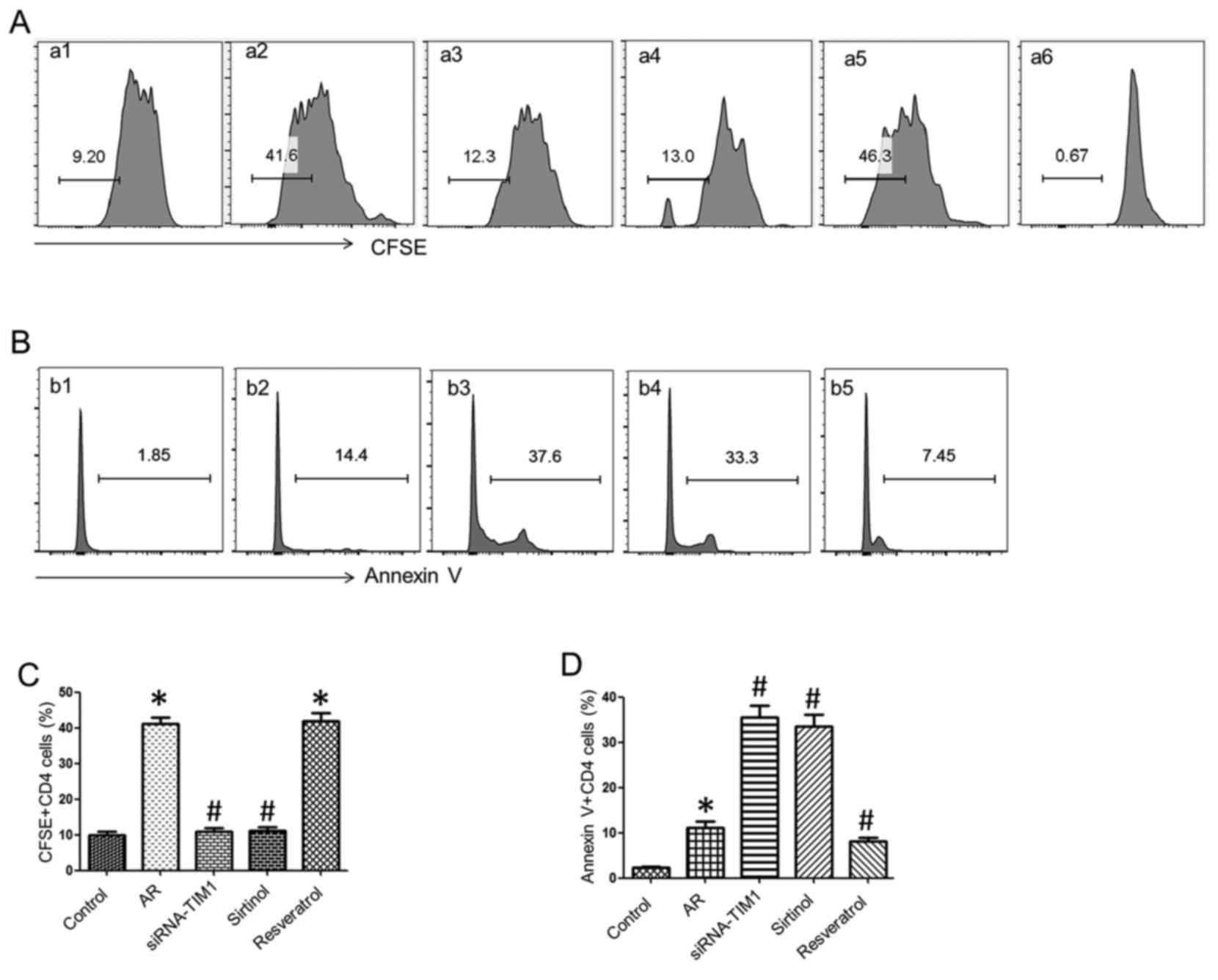
To elucidate the role of SIRT1 in CD4+
T-cell proliferation, splenic DCs (105 cells/well) and
CD4+ T cells (106 cells/well) from the AR or
the control group were co-cultured in the presence of OVA (10
ng/ml) for 3 days. CD4+ T cells were labeled with CFSE
or Annexin V for proliferation and apoptosis assessment by flow
cytometry. The results demonstrated that CD4+ T-cell
proliferation was ~9.2% in the control group, whereas it was
>40% in the AR group, which was markedly reduced by treatment
with siRNA-TIM1 (12.3%) or the addition of sirtinol (13%), and
enhanced by resveratrol stimulation (46.3%). The proportion of
CD4+ T-cell apoptosis in the AR group (14.4%) was higher
compared with that in the control group (1.85%), but the percentage
of apoptosis by treatment with siRNA-TIM1 (37.6%) or sirtinol
(33.3%) increased markedly, and was inhibited by treatment with
resveratrol (7.45%) (Fig. 4).
These findings indicate that SIRT1 regulates CD4+ T-cell
proliferation and apoptosis in allergic inflammation.
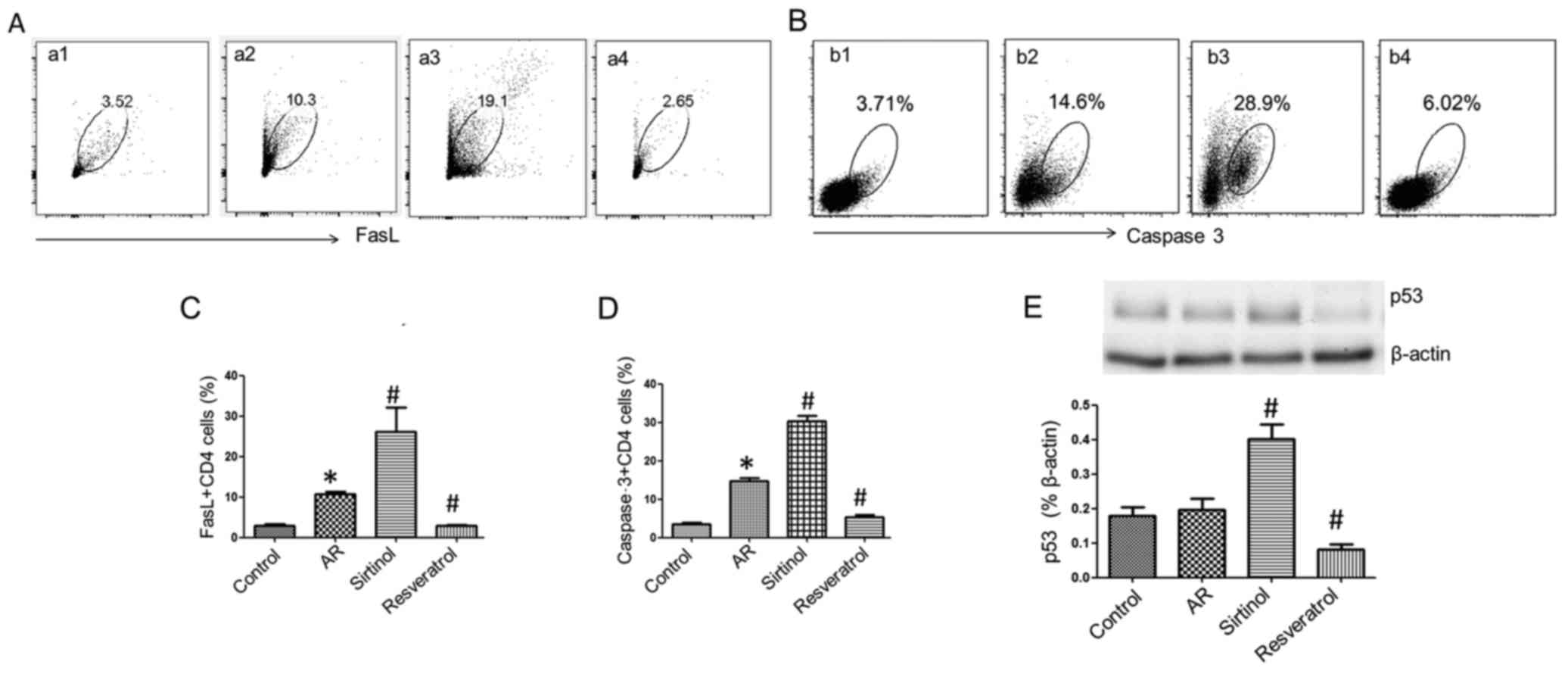
SIRT1 regulates FasL, caspase-3 and p53
expression in CD4+ T cells
To understand the underlying mechanisms, the
expression of FasL, caspase-3 and p53 was analyzed in
CD4+ T cells. Splenic cells from the AR and control
groups were cultured with OVA (10 ng/ml) for 3 days. Compared with
the AR group, the addition of sirtinol (100 µM) to the
culture significantly increased FasL (Fig. 5), caspase-3 and p53 expression in
CD4+ T cells, whereas treatment with resveratrol (100
µM) decreased the expression of FasL, caspase-3 and p53 in
CD4+ T cells to the levels of the control group. These
results suggest that SIRT1 downregulates the expression of FasL,
caspase-3 and p53 in CD4+ T cells.
Discussion
Skewed Th2-cell proliferation plays a critical role
in the induction of allergic inflammation, while the factors that
initiate and maintain Th2-cell polarization in allergic diseases
remain unclear. The TIM4̸TIM1 interaction, as one of the
co-stimulatory signals, is involved in the pathogenesis of allergic
conditions (7–9). Using an AR mouse model, the present
study demonstrated that TIM4 expression in splenic DCs was
increased in AR mice, and the TIM4̸TIM1 interaction promoted the
expression of SIRT1 in CD4+ T cells during allergic
inflammation via promoting PI3K/Akt phosphorylation. SIRT1
facilitated CD4+ T-cell proliferation through
downregulating FasL, caspase-3 and p53 expression in AR mice. TIM1
is associated with the Th2 pattern of inflammation and Th2 cytokine
expression; activated T cells expressed higher levels of TIM1
(17). TIM4, as a type I cell
surface glycoprotein, is specifically expressed by APCs, such as
DCs. Blockade of TIM4 on DCs repressed Th2-cell differentiation and
impeded IL-4 signal transducer and activator of transcription 6
signaling (18). Consistent with
these results, we observed that blockade of TIM1 or TIM4 inhibited
CD4+ T-cell proliferation in AR mice. This result is
supported by other studies reporting that antagonists of TIM1
blocked allergic inflammation in mouse models of asthma (19,20) and allergic gut inflammation
(9).
TIM1 is preferentially expressed on Th2 cells and
may be upregulated following TCR stimulation (21). Stimulation of TIM4 is required by
TIM1-induced T-cell proliferation (22). Previous data, as well as the
present findings, indicated that TIM4/TIM1 interaction is involved
in the pathogenesis of allergic diseases, while the mechanisms
underlying the TIM4̸TIM1 interaction in the maintenance of allergic
conditions, and the precise intracellular downstream signaling by
TIM1 and TIM4 engagement, remain obscure (8). TIM1 is co-localized with CD3 on the
surface of T cells, and may function as part of the TCR signaling
during T-cell activation, possibly through IL-2-induced ITK and
PI3K phosphorylation (8,11,23). SIRT1 plays an important role in
cell differentiation, apoptosis and tumor suppression via
post-translational modification of histone proteins, and is
involved in allergic inflammation (14,15). Published data demonstrated that
the PI3K̸Akt signaling pathway is required for the regulation of
SIRT1 expression (24,25). We also observed that SIRT1 mRNA
expression was increased in the nasal mucosa of AR mice. The
results indicate that the TIM4̸TIM1 interaction modulates SIRT1
expression in splenic CD4+ T cells during allergic
inflammation via PI3K/Akt phosphorylation.
SIRT1 is one of the deacetylases associated with
asthma, and Kim et al reported that SIRT1 expression was
increased in an OVA-induced murine allergic airway model, which was
correlated with increased levels of IL-4, IL-5 and IL-13 and
inflammatory cell infiltration in lung tissues (26). SIRT1 promotes adaptive Th2-cell
responses by repressing peroxisome proliferation-activated
receptor-γ activity in DCs in an induced allergic airway mouse
model (15). Our data
demonstrated the role of SIRT1 in CD4+ T-cell
proliferation. The findings indicated that SIRT1 enhances
CD4+ T-cell proliferation and inhibits their apoptosis
in allergic inflammation. As a multifunctional molecule, SIRT1 is
involved in a variety of molecular pathways, such as cell
differentiation, cell aging and anti-inflammation. Our results
revealed a novel functional aspect of SIRT1 that promotes adaptive
Th2-cell responses. Other studies reported that SIRT1 plays a role
in maintaining T-cell balance and exerts anti-inflammatory effects
by inhibiting proinflammatory transcription factors (12). Lung SIRT1 expression decreased,
while serum SIRT1 increased, in the setting of asthma (13). SIRT1 expression was reduced in the
peripheral blood mononuclear cells of patients with severe asthma,
and the inhibition of SIRT1 promotes a Th2-like phenotype in T
cells and IL-4 gene expression via acetylation of GATA-3, but there
was no correlation between IL-5 transcripts and SIRT1 activity.
These inconsistent results may be due to the fact that the decrease
in SIRT1 appears to be associated with oxidative stress in patients
with severe asthma (14).
SIRT1 localizes in the nucleus as well as the
cytoplasm and, thus, may interact with both nuclear and cytosolic
proteins, and deacetylates histones and various transcription
factors, such as p53 and FOXO (27). It was previously reported that
SIRT1 may be a potential oncogene, which prevents apoptosis and
senescence by interacting with and targeting p53 for deacetylation
and decreasing p53-dependent transcriptional activity (12). It also suppresses FasL expression
in activated T cells to interfere with activation-induced cell
death (AICD) (28). Caspase-3
plays an important role in the induction of cell apoptosis
(29). Our previous study
suggested that Fas/FasL, p53 and caspase-3 are involved in the
course of CD4+ T-cell apoptosis and AICD (30). In the present study, to elucidate
the mechanisms underlying SIRT1 regulation of CD4+
T-cell proliferation in allergic inflammation, the expression of
FasL, caspase-3 and p53 was determined in CD4+ T cells.
The results suggested that SIRT1 downregulates FasL, caspase-3 and
p53 expression in CD4+ T cells.
In conclusion, the present study demonstrated that
the TIM4̸TIM1 interaction promotes PI3K/Akt phosphorylation in
CD4+ T cells, resulting in increased SIRT1 expression;
SIRT1 then facilitates CD4+ T-cell proliferation through
downregulating FasL, caspase-3 and p53 expression in AR mice. These
results suggest that the TIM4/TIM1 interaction modulates Th2-cell
inflammation through enhancing SIRT1 expression.
Acknowledgments
The present study was supported by grants from the
Natural Science Foundation of China (no. 81571790), the Innovation
of Science and Technology Commission of Shenzhen Municipality (nos.
JCYJ20140411150916749, JCYJ20160429091935720, ZDSYS201506050935272
and YLWS20140609111127924), the Medical Science and Technology
Research Fund of Guangdong province (A2016272; no. 2014A030313781),
and the Health Committee Foundation of Shenzhen (nos. 201401097 and
201401096). The study was also supported by a grant from the
Innovation of Science and Technology Commission of Shenzhen
Municipality (no. JCYJ20150403091931195).
Abbreviations:
|
Th2
|
T helper 2
|
|
TIM
|
T-cell immunoglobulin and mucin
domain
|
|
SIRT1
|
silent information regulator 1
|
|
CD
|
cluster of differentiation
|
|
IgE
|
immunoglobulin E
|
|
APCs
|
antigen-presenting cells
|
|
TCR
|
T-cell receptor
|
|
ITK
|
IL-2-induced T-cell kinase
|
|
PI3K
|
phosphoinositide 3-kinase
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
AR
|
allergic rhinitis
|
|
OVA
|
ovalbumin
|
|
SIgE
|
OVA-specific IgE
|
|
DC
|
dendritic cells
|
|
CFSE
|
carboxyfluorescein succinimidyl
ester
|
|
BSA
|
bovine serum albumin
|
References
|
1
|
Sicherer SH and Sampson HA: Food allergy:
Epidemiology, pathogenesis, diagnosis, and treatment. J Allergy
Clin Immunol. 133:291–307; quiz 308. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heratizadeh A: Atopic dermatitis: New
evidence on the role of allergic inflammation. Curr Opin Allergy
Clin Immunol. 16:458–464. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rosati MG and Peters AT: Relationships
among allergic rhinitis, asthma, and chronic rhinosinusitis. Am J
Rhinol Allergy. 30:44–47. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eifan AO and Durham SR: Pathogenesis of
rhinitis. Clin Exp Allergy. 46:1139–1151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oettgen HC and Burton OT: IgE receptor
signaling in food allergy pathogenesis. Curr Opin Immunol.
36:109–114. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hiraishi Y, Nambu A, Shibui A, Nakanishi
W, Yamaguchi S, Morita H, Iikura M, McKenzie AN, Matsumoto K, Sudo
K, et al: TIM-3 is not essential for development of airway
inflammation induced by house dust mite antigens. Allergol Int.
65:459–465. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Z, Ju Z and Frieri M: The T-cell
immunoglobulin and mucin domain (Tim) gene family in asthma,
allergy, and autoimmunity. Allergy Asthma Proc. 34:e21–e26. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Freeman GJ, Casasnovas JM, Umetsu DT and
DeKruyff RH: TIM genes: A family of cell surface phosphatidylserine
receptors that regulate innate and adaptive immunity. Immunol Rev.
235:172–189. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng BS, Chen X, He SH, Zheng PY, Foster
J, Xing Z, Bienenstock J and Yang PC: Disruption of T-cell
immunoglobulin and mucin domain molecule (TIM)-1/TIM4 interaction
as a therapeutic strategy in a dendritic cell-induced peanut
allergy model. J Allergy Clin Immunol. 122:55–61. e1–7. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Santiago C, Ballesteros A, Tami C,
Martínez-Muñoz L, Kaplan GG and Casasnovas JM: Structures of T Cell
immunoglobulin mucin receptors 1 and 2 reveal mechanisms for
regulation of immune responses by the TIM receptor family.
Immunity. 26:299–310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Binne LL, Scott ML and Rennert PD: Human
TIM-1 associates with the TCR complex and up-regulates T cell
activation signals. J Immunol. 178:4342–4350. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Haigis MC and Sinclair DA: Mammalian
sirtuins: Biological insights and disease relevance. Annu Rev
Pathol. 5:253–295. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Li D, Ma G, Li W, Wu J, Lai T,
Huang D, Zhao X, Lv Q, Chen M, et al: Increases in peripheral
SIRT1: A new biological characteristic of asthma. Respirology.
20:1066–1072. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Colley T, Mercado N, Kunori Y, Brightling
C, Bhavsar PK, Barnes PJ and Ito K: Defective sirtuin-1 increases
IL-4 expression through acetylation of GATA-3 in patients with
severe asthma. J Allergy Clin Immunol. 137:1595–1597.e7. 2016.
View Article : Google Scholar
|
|
15
|
Legutko A1, Marichal T, Fiévez L, Bedoret
D, Mayer A, de Vries H, Klotz L, Drion PV, Heirman C, Cataldo D, et
al: Sirtuin 1 promotes Th2 responses and airway allergy by
repressing peroxisome proliferator-activated receptor-gamma
activity in dendritic cells. J Immunol. 187:4517–4529. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim YH, Yang TY, Park CS, Ahn SH, Son BK,
Kim JH, Lim DH and Jang TY: Anti-IL-33 antibody has a therapeutic
effect in a murine model of allergic rhinitis. Allergy. 67:183–190.
2012. View Article : Google Scholar
|
|
17
|
de Souza AJ, Oriss TB, O'malley KJ, Ray A
and Kane LP: T cell Ig and mucin 1 (TIM-1) is expressed on in
vivo-activated T cells and provides a costimulatory signal for T
cell activation. Proc Natl Acad Sci USA. 102:17113–17118. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li J, Zhao X, Liu X and Liu H: Disruption
of TIM-4 in dendritic cell ameliorates hepatic warm IR injury
through the induction of regulatory T cells. Mol Immunol.
66:117–125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sonar SS, Hsu YM, Conrad ML, Majeau GR,
Kilic A, Garber E, Gao Y, Nwankwo C, Willer G, Dudda JC, et al:
Antagonism of TIM-1 blocks the development of disease in a
humanized mouse model of allergic asthma. J Clin Invest.
120:2767–2781. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim HY, Chang YJ, Chuang YT, Lee HH,
Kasahara DI, Martin T, Hsu JT, Savage PB, Shore SA, Freeman GJ, et
al: T-cell immunoglobulin and mucin domain 1 deficiency eliminates
airway hyperreactivity triggered by the recognition of airway cell
death. J Allergy Clin Immunol. 132:414–425.e6. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yeung MY, McGrath M and Najafian N: The
emerging role of the TIM molecules in transplantation. Am J
Transplant. 11:2012–2019. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mariat C, Degauque N, Balasubramanian S,
Kenny J, DeKruyff RH, Umetsu DT, Kuchroo V, Zheng XX and Strom TB:
Tim-1 signaling substitutes for conventional signal 1 and requires
costimulation to induce T cell proliferation. J Immunol.
182:1379–1385. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
de Souza AJ, Oak JS, Jordanhazy R,
DeKruyff RH, Fruman DA and Kane LP: T cell Ig and mucin
domain-1-mediated T cell activation requires recruitment and
activation of phosphoinositide 3-kinase. J Immunol. 180:6518–6526.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Koga T, Suico MA, Shimasaki S, Watanabe E,
Kai Y, Koyama K, Omachi K, Morino-Koga S, Sato T, Shuto T, et al:
Endoplasmic reticulum (ER) stress induces sirtuin 1 (SIRT1)
expression via the PI3K-Akt-GSK3β signaling pathway and promotes
hepato-cellular injury. J Biol Chem. 290:30366–30374. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ming GF, Tang YJ, Hu K, Chen Y, Huang WH
and Xiao J: Visfatin attenuates the ox-LDL-induced senescence of
endothelial progenitor cells by upregulating SIRT1 expression
through the PI3K/Akt/ERK pathway. Int J Mol Med. 38:643–649. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim SR, Lee KS, Park SJ, Min KH, Choe YH,
Moon H, Yoo WH, Chae HJ, Han MK and Lee YC: Involvement of sirtuin
1 in airway inflammation and hyperresponsiveness of allergic airway
disease. J Allergy Clin Immunol. 125:449–460.e14. 2010. View Article : Google Scholar
|
|
27
|
Tanno M, Sakamoto J, Miura T, Shimamoto K
and Horio Y: Nucleocytoplasmic shuttling of the
NAD+-dependent histone deacetylase SIRT1. J Biol Chem.
282:6823–6832. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arakaki R, Yamada A, Kudo Y, Hayashi Y and
Ishimaru N: Mechanism of activation-induced cell death of T cells
and regulation of FasL expression. Crit Rev Immunol. 34:301–314.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Porter AG and Jänicke RU: Emerging roles
of caspase-3 in apoptosis. Cell Death Differ. 6:99–104. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang L, Xu LZ, Liu ZQ, Yang G, Geng XR, Mo
LH, Liu ZG, Zheng PY and Yang PC: Interleukin-13 interferes with
activation-induced T-cell apoptosis by repressing p53 expression.
Cell Mol Immunol. 13:669–677. 2016. View Article : Google Scholar :
|