Introduction
Hepatic fibrosis is a common pathophysiological
outcome of various chronic hepatic diseases, which may be caused by
viral infection, alcohol abuse, drug or chemical toxicity, hepatic
autoimmunity, biliary obstruction, etc. During the process of
hepatic injury, injured hepatocytes degenerate, and stimulate
hepatocyte regeneration and liver inflammation (1). Chronic hepatocyte injury and
degeneration is able to promote the deposition of extracellular
matrix components, including collagens, glycoproteins,
mucopolysaccharide and proteoglycan, thus leading to liver fibrosis
(2). Without optimal control,
hepatic fibrosis can progress to cirrhosis, hepatocellular
carcinoma and liver failure (3).
Cirrhosis and hepatocellular carcinoma are major causes of
morbidity and mortality worldwide (4); however, the molecular mechanisms
underlying hepatic fibrosis remain unclear.
Post-translational modification by protein
ubiquitination can degrade misfolded and unwanted proteins through
proteasomes. This process affects various signaling processes,
including cell signaling, DNA repair, protein trafficking,
chromatin modifications, cell cycle progression and cell death
(5). Arkadia is a RING-type E3
ubiquitin ligase, also known as ring finger protein 11, which can
catalyze degradation of key signaling molecules in the transforming
growth factor (TGF)-β1 signaling pathway (6,7).
TGF-β1 is a profibrotic factor that activates Smad signaling and is
closely associated with the development and progression of hepatic
fibrosis (8–10). Previous studies have reported that
Arkadia can degrade Smad6, Smad7, c-Ski and SnoN, which are
negative regulators of the TGF-β1 signaling (6,11–14). However, little is currently known
regarding the expression levels of Arkadia during the process of
hepatic fibrosis, and the location of Arkadia expression in the
liver.
The present study employed two rat models of hepatic
fibrosis in order to characterize the expression levels of Arkadia
in fibrotic livers from rats, as well as liver samples from
patients with various stages of hepatic fibrosis. The results
indicated that Arkadia was predominantly expressed in the
cytoplasm, whereas expression was reduced in the nucleus of
hepatocytes and cholangiocytes in liver samples from rats with
hepatic fibrosis. The relative mRNA expression levels of Arkadia
were increased in the livers of rats with hepatic fibrosis, whereas
the protein expression levels of Arkadia were gradually reduced in
the livers of rats hepatic fibrosis and in human liver samples from
patients with hepatic fibrosis.
Materials and methods
Human liver samples
Human liver tissues used in the present study were
remnants from biopsies obtained for routine clinical purposes or
were cirrhotic liver samples acquired during hepatectomy and were
collected between September 2014 and July 2015. Each liver biopsy
sample was processed for routine histology. Informed consent was
obtained from each patient prior to participation in the present
study. The experimental protocol was approved by the Ethics
Committee of Beijing Friendship Hospital, Capital Medical
University (Beijing, China).
Animals
Male Wistar rats (weight, 180–220 g) were obtained
from the Laboratory Animal Center of Chinese Academy of Medical
Sciences (Beijing, China). The rats were housed in a specific
pathogen-free facility with controlled temperature (25±1°C) and
humidity (55±5%), under a 12-h light/dark cycle. The rats were
allowed free access to rat chow and water. All animals received
humane care according to the criteria outlined in the ‘Guide for
the Care and Use of Laboratory Animals’ prepared by the National
Academy of Sciences and published by the National Institutes of
Health (NIH publication no. 86–23). The animal study was approved
by the Ethics Committee of Beijing Friendship Hospital, Capital
Medical University.
Animal models of hepatic fibrosis
Carbon tetrachloride (CCl4)
intoxication
The rats were randomly assigned to five groups (n=6
rats/group). The control group was injected intraperitoneally
(i.p.) with olive oil (0.2 ml/100 g body weight) twice per week for
8 consecutive weeks. The hepatic fibrosis groups were injected with
50% CCl4 in olive oil (i.p; Sinopharm Chemical Reagent
Co., Ltd., Shanghai, China) at a dose of 0.15 ml/100 g body weight
twice per week for 2, 4, 6 or 8 weeks. After the final injection,
rats were fasted overnight and anesthetized with pentobarbital
sodium (40 mg/kg). The rats were then sacrificed. Subsequently,
blood and liver samples were collected for further analysis.
Bile duct ligation (BDL)
The rats were randomly divided into five groups (n=6
rats/group), including the sham operation group, and BDL 7, 14, 28
and 42 day groups. The rats were anesthetized with pentobarbital
sodium, and the BDL and sham operations were performed, as
previously described (15,16).
Briefly, a 2-cm midline laparotomy was performed, after which the
common bile duct was identified and ligated twice with 6.0 silk
sutures (17). The first ligature
was made under the junction of the hepatic ducts and the second
ligature was located above the entrance to the pancreatic ducts.
The rats in the sham operation group were subjected to abdominal
incision, and the common bile duct was identified but not ligated.
After the abdomen was sutured, the animals were recovered on a
heating pad. After 24 h, the rats were allowed free access to food
and water. The rats in the sham operation group were sacrificed 42
days post-surgery. The rats in the experimental groups were
sacrificed at 7, 14, 28 or 42 days post-surgery. Liver and blood
samples were obtained for further analysis.
Detection of serum biochemical
indicators
Individual serum samples were prepared by
centrifugation at 1,811 × g for 5 min at 4°C, and the levels of
serum aspartate transaminase (AST), alanine transaminase (ALT),
direct bilirubin (DBIL) and total bilirubin (TBIL) were analyzed
using an automatic biochemistry analyzer (Abbott Laboratories,
Chicago, IL, USA), according to the manufacturer’s protocol.
Histopathological examination
The liver tissues were fixed with 4%
paraformaldehyde for 24 h at room temperature, and embedded in
paraffin. Subsequently, liver tissue sections (4 μm) were
stained with hematoxylin and eosin (H&E) or Masson’s trichrome.
Finally, the sections were dehydrated by increasing concentrations
of ethanol and xylene. In Masson’s staining, the sections were
dewaxed and rehydrated by H2O and distilled water, and
then stained by Harris’s hematoxylin dye for 1 min. After washed
with H2O, the sections were stained with Masson’s acid
magenta dye for 10 min, differentiated by 1% phosphomolybdic acid
aqueous solution for 5 min and washed with Glacial acetic acid for
a few seconds. Images of the sections were captured under a light
microscope (DM6000B; Leica Microsystems GmbH, Mannheim,
Germany).
Immunohistochemistry
The paraffin-embedded liver sections (4 μm)
were rehydrated, and treated with 3% H2O2 in
methanol to inactivate endogenous peroxidase. The sections were
then incubated with rabbit anti-Arkadia (sc-367716; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C. After
washing, the bound antibodies were detected using horseradish
peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin (Ig)G
(PV-9001; OriGene Technologies, Inc., Beijing, China) at room
temperature for 30 min and visualized using diaminobenzidine (DAB;
ZLI-9017; OriGene Technologies, Inc.), followed by counterstaining
with hematoxylin. The images of the different groups were captured
using a microscope (Leica DM6000B; Leica Microsystems GmbH).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from liver tissues using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and was reverse transcribed (42°C for 15 min,
95°C for 5 min, 4°C for 5 min, only 1 cycle) into cDNA using 3
μl 10X RT buffer, 4 μl MgCl2, 0.5
μl recombinant RNase inhibitor, 2 μl dNTP mixture, 1
μl Oligo dT-Adaptor primer, 0.6 μl AMV reverse
transcriptase (Promega Corp., Madison, WI, USA) and 1 μg RNA
sample. The relative mRNA expression levels of Arkadia in
individual samples were determined by qPCR using the SYBR-Green
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) and
specific primers in an ABI 7500 sequence detection system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling
conditions were as follows: denaturation at 95°C for 10 min, 40
cycles at 95°C for 15 sec, 64°C for 30 sec, 95°C for 15 sec, 60°C
for 1 min, and 95°C for 15 sec, and a final extension step at 60°C
for 15 sec. The primer sequences were as follows: Rat Arkadia,
forward 5′-TCATATTCATGTGCCTCAAACCA-3′, reverse
5′-CCCAGTTCCCAGGCAGTTC-3′; and GAPDH, forward
5′-ACGCATTTGGTCGTATTGGG-3′ and reverse 5′-TGATTTTGGAGGGATCTCGC-3′.
Data were analyzed by 2−ΔΔCq (18).
Western blot analysis
Individual liver tissue samples were homogenized in
radioimmunoprecipitation assay lysis buffer containing a protease
inhibitor cocktail (Roche Diagnostics, Indianapolis, IN, USA).
After centrifugation at 12,000 × g for 15 min at 4°C, protein
concentrations were measured using the bicinchoninic acid protein
assay kit (Pierce; Thermo Fisher Scientific, Inc.). Individual
tissue lysates (30 μg/lane) were separated by 8–12%
SDS-PAGE, and were transferred to polyvinylidene fluoride membranes
(EMD Millipore, Billerica, MA, USA). After blocking with 5%
fat-free dry milk in Tris-buffered saline-0.05% Tween (TBST) for 2
h at room temperature, the membranes were incubated with rabbit
anti-Arkadia (1:500; sc-367716; Santa Cruz Biotechnology, Inc.),
rabbit anti-GAPDH (1:1,000; ab181602; Abcam, Cambridge, UK), rabbit
anti-β-actin antibody (1:1,000; ab8227; Abcam) at 4°C overnight.
Subsequently, the membranes were washed three times with TBST and
incubated with HRP-conjugated goat anti-rabbit IgG (PV-9001;
Zhongshan Goldenbridge Biotechnology Co., Ltd., Beijing, China) for
30 min at room temperature. The blots were visualized using an
enhanced chemiluminescence kit (EMD Millipore). The band density of
target proteins was quantified and normalized to that of GAPDH
using Image Lab version 5.1 software (Bio-Rad Corporation,
Hercules, CA, USA).
Statistical analysis
Data are representative images or are presented as
the means ± standard deviation, unless otherwise specified. These
experiments were repeated for 6 times. All statistical analyses
were conducted using SPSS 20.0 software (IBM Corporation, Armonk,
NY, USA). The difference between two groups was analyzed by
Student’s t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Effects of BDL on liver pathology and
function in rats
To evaluate the effects of BDL on liver pathology
and collagen fiber deposition in rats, the rats were subjected to
sham or BDL surgery and were then sacrificed at various time points
post-surgery. Pathological alterations in the liver tissues were
analyzed by H&E and Masson staining. As shown in Fig. 1A, the sham group displayed normal
lobular architecture with central veins and radiating hepatic
cords. Conversely, small bile duct dilation, portal inflammation
and hepatocyte degeneration, as well as inflammatory infiltration,
were accompanied by obvious fibrosis in the livers of rats in the
BDL 7 day group. Small bile duct dilation around the portal area,
hepatocyte degeneration and inflammatory infiltration, as well as
collagenous fibers were more obvious in the livers of rats in the
BDL 14 day group. In the BDL 28 and 42 day groups, cholestasis
became more severe, epithelial cells were detected in the
interlobular bile ducts, and diffused collagenous fibers and
capillaries extended into the hepatic lobular regions, thus forming
membrane-like structures in the liver lobules; these are hallmarks
of hepatic fibrosis. Masson staining revealed that the liver
sections of the sham group exhibited normal distribution of
collagen fibers (stained blue) around the central veins and portal
tracts, whereas the number of collagenous fibers was gradually
increased after BDL. In the BDL 28 and 42 day groups, marked
collagen fibers were deposited around the central vein and small
bile ducts, which formed membrane-like structures in the hepatic
lobules, a characteristic of pseudolobules.
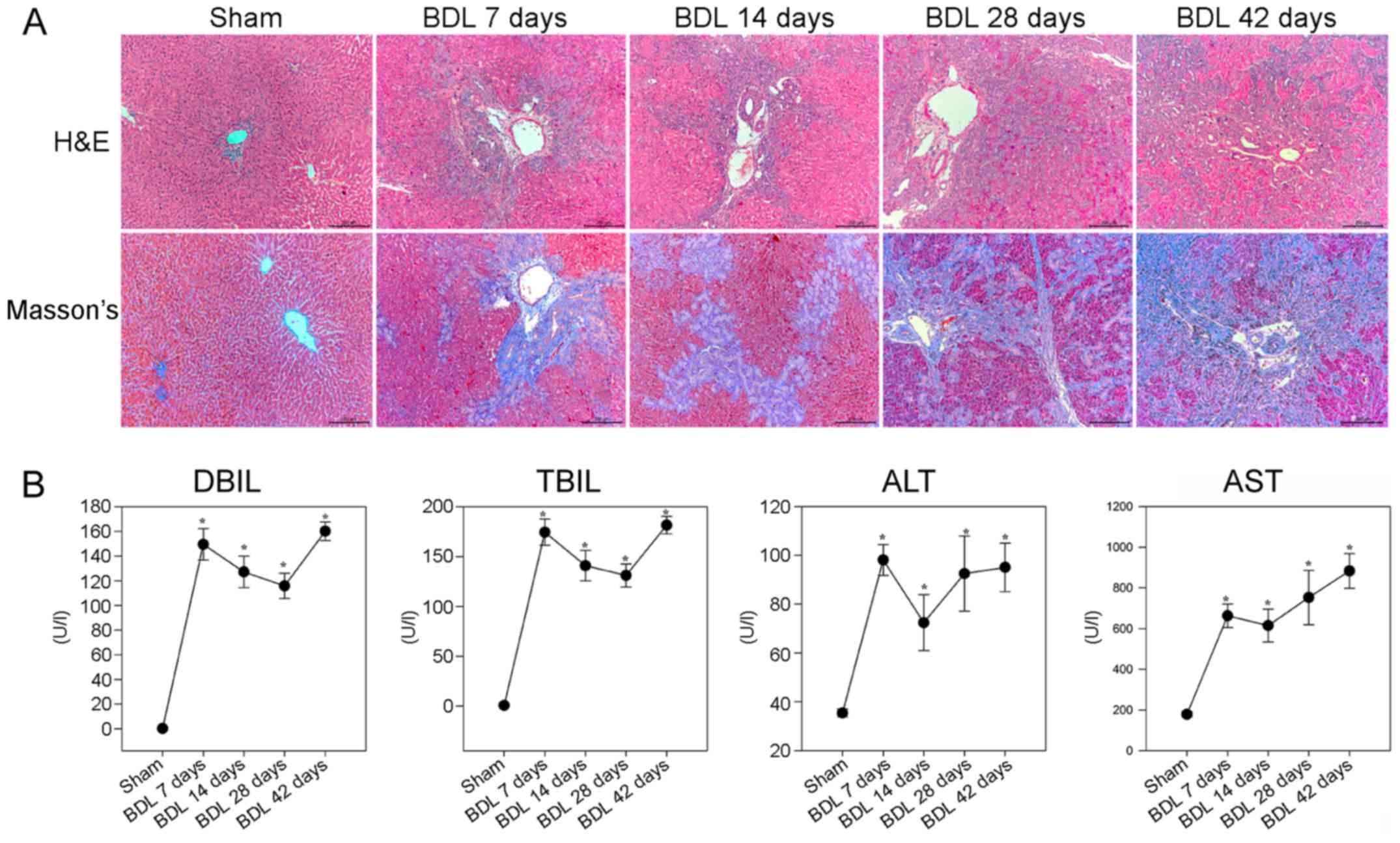 | Figure 1BDL-induced hepatic fibrosis in rats.
Following BDL, rats were sacrificed at various time points
post-surgery and liver sections underwent H&E and Masson
staining. The serum levels of DBIL, TBIL, ALT and AST were
determined in individual rats. (A) Histological examination of the
liver sections (magnification, ×100). Representative images are
presented. (B) Effects of BDL on serum biochemical indicators. Data
are presented as the means ± standard deviation of individual
groups (n=6/group).*P<0.05 vs. the sham operation
group. ALT, alanine transaminase; AST, aspartate transaminase; BDL,
bile duct ligation; DBIL, direct bilirubin; H&E, hematoxylin
and eosin; TBIL, total bilirubin. |
Further analyses indicated that the BDL groups of
rats had significantly higher levels of DBIL, TBIL and ALT, and
gradually increased levels of AST, compared with in the sham group
of rats (P<0.05; Fig. 1B).
These results suggested that BDL may impair liver function, thus
gradually resulting in liver fibrosis in rats.
Effects of CCl4 on liver
pathology and function in rats
To provide additional evidence, a CCl4
model of liver fibrosis was generated in rats. As shown in Fig. 2A, hepatocyte swelling and
steatosis, spotty necrosis in the hepatic lobule and inflammatory
infiltrates around the portal duct areas were detected in the
livers of rats in the CCl4 2 week group. Furthermore,
increased inflammatory infiltrates and fibrocytes surrounding the
hepatic lobules, together with hepatocyte degeneration and focal
necrosis were observed in the livers of rats in the CCl4
4 week group. In addition, disarranged fibrous septa and
pseudolobules were detected in the hepatic lobules of rats in the
CCl4 6 and 8 week groups. Masson staining revealed that
while collagen fibers were detected around the portal tracts and
central veins in the livers of control rats, increased numbers of
collagenous fibers were observed in the livers of
CCl4-injected rats. In addition, collagenous fibers
formed membrane-like structures in the hepatic lobules of rats in
the CCl4 8 week group.
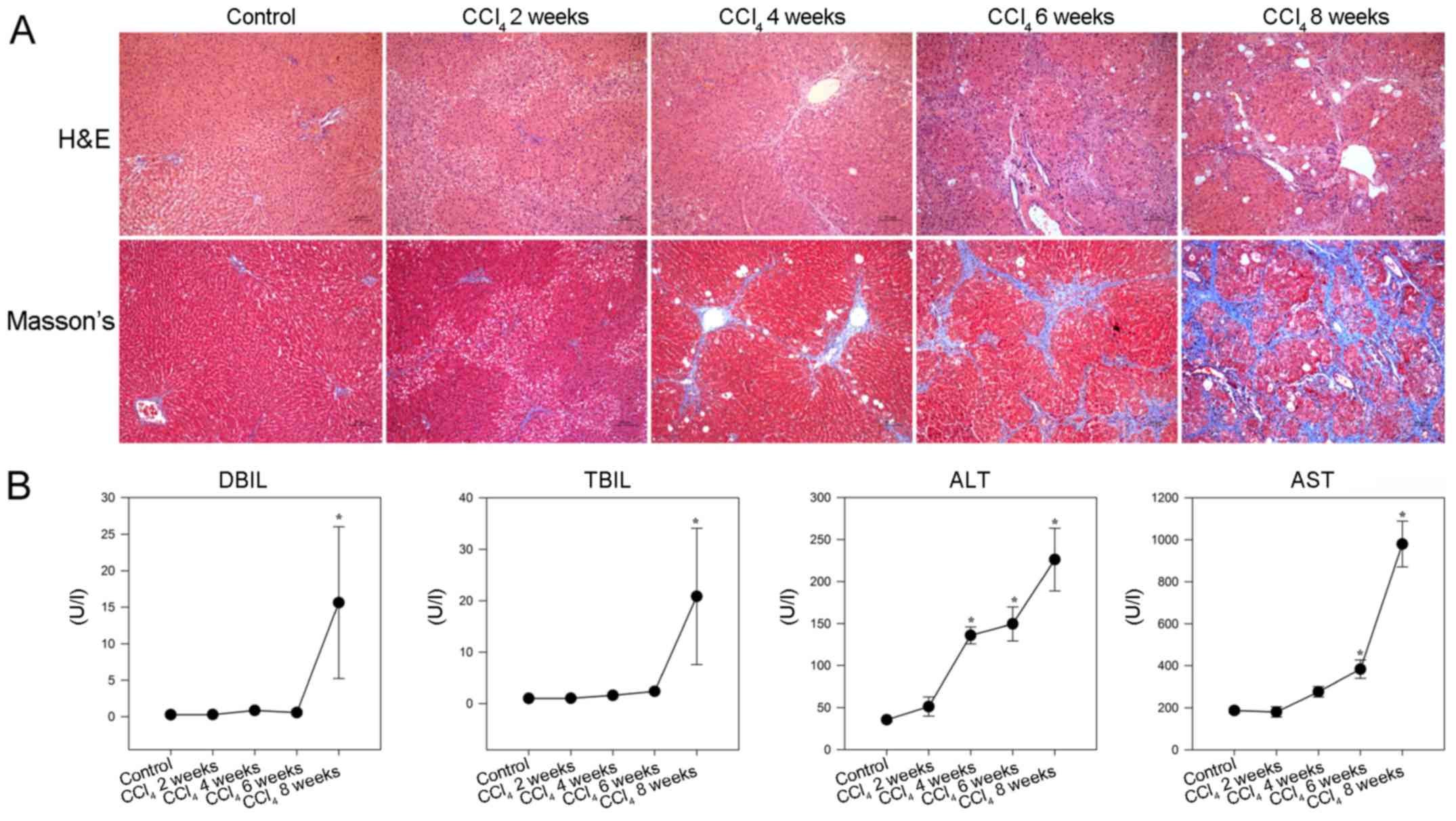 | Figure 2CCl4-induced hepatic
fibrosis in rats. Following CCl4 injection, rats were
sacrificed at the indicated time points and liver sections
underwent H&E and Masson staining. The serum levels of DBIL,
TBIL, ALT and AST were detected in individual rats. (A)
Histological examination of liver sections (magnification, ×100).
Representative images are presented. (B) Effects of CCl4
administration on serum biochemical indicators. Data are presented
as the means ± standard deviation of individual groups
(n=6/group).*P<0.05 vs. the control group. ALT,
alanine transaminase; AST, aspartate transaminase; CCl4,
carbon tetrachloride; DBIL, direct bilirubin; H&E, hematoxylin
and eosin; TBIL, total bilirubin. |
Consequently, significantly increased levels of
serum DBIL and TBIL, as well as gradually elevated ALT and AST,
were detected in the CCl4 -injected rats compared with
in the control rats (P<0.05; Fig.
2B). These results indicated that injection with
CCl4 impaired liver function and induced hepatic
fibrosis in rats.
Expression of Arkadia in rats with
BDL-induced hepatic fibrosis
To evaluate Arkadia expression in rats with
BDL-induced hepatic fibrosis, the expression levels of Arkadia were
detected in fibrotic and normal liver tissues by
immunohistochemical staining. The results demonstrated that Arkadia
was predominantly expressed in the cytoplasm, and was weakly
expressed in the nucleus of hepatocytes and cholangiocytes
(Fig. 3A). There were no notable
differences between sham and BDL rat Arkadia immunohistochemistry
results. Using qPCR, the mRNA expression levels of Arkadia were
revealed to be significantly higher in the livers of the BDL 7 day
group compared with in the control group (P<0.01), after which
they gradually decreased; the expression levels in the livers of
the BDL 28 day group were similar to those in the control group
(Fig. 3B). However, the mRNA
expression levels of Arkadia were significantly elevated in the
liver samples of the BDL 42 day group compared with in the control
group (P<0.01). Western blot analysis indicated that increased
levels of Arkadia expression were detected in the livers of the
control rats, whereas the protein expression levels of Arkadia were
gradually reduced in the fibrotic livers of rats (P<0.01;
Fig. 3C and D). These results
indicated that the levels of Arkadia mRNA transcripts were
increased and the protein expression levels of Arkadia were
decreased in hepatocytes and cholangiocytes from the fibrotic
livers of rats.
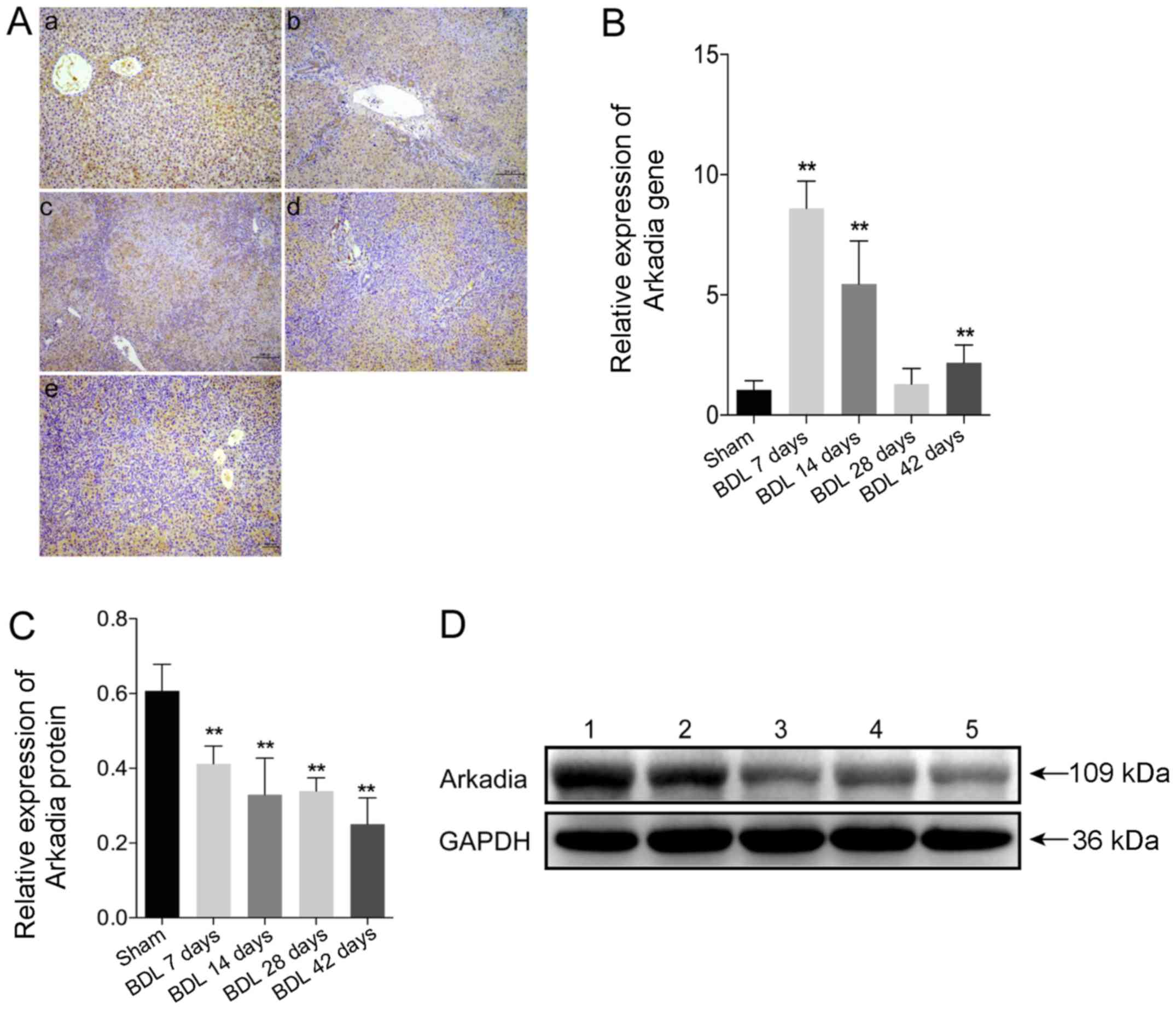 | Figure 3Arkadia expression in rats with
BDL-induced hepatic fibrosis. Following sacrifice, the expression
levels of Arkadia were detected in the liver samples using RT-qPCR,
western blot analysis and immunohistochemistry. Data are presented
as representative images or are expressed as the means ± standard
deviation of individual groups (n=6/group). (A) Immunohistochemical
staining of Arkadia expression in liver sections (original
magnification, ×100). (a) Sham; (b) BDL 7 days; (c) BDL 14 days;
(d) BDL 28 days and (e) BDL 42 days. (B) RT-qPCR analysis of
Arkadia mRNA transcript levels in the liver samples. (C) Relative
protein expression levels of Arkadia. (D) Western blot analysis of
Arkadia protein. Lane 1, Sham; lane 2, BDL 7 days; lane 3, BDL 14
days; lane 4, BDL 28 days and lane 5, BDL 42 days.
**P<0.01 vs. the sham group. BDL, bile duct ligation;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction. |
Expression of Arkadia in rats with
CCl4-induced hepatic fibrosis
The present study used immunohistochemistry to
investigate the expression of Arkadia in CCl4-induced
hepatic fibrosis. Strong Arkadia staining was detected in the
cytoplasm, whereas weak staining was detected in the nucleus of
hepatocytes in the control rats. Conversely, the number of
hepatocytes positive for Arkadia staining was reduced in the liver
samples from rats with hepatic fibrosis (Fig. 4A). Compared with in the control
group, the mRNA expression levels of Arkadia were significantly
increased in the fibrotic liver samples from rats in the
CCl4 6 and 8 week groups (P<0.01; Fig. 4B). Furthermore, western blot
analysis revealed that the protein expression levels of Arkadia in
the liver samples from the rats in the CCl4 6 and 8week
groups were significantly reduced compared with in the control
group (P<0.05; Fig. 4C and
D).
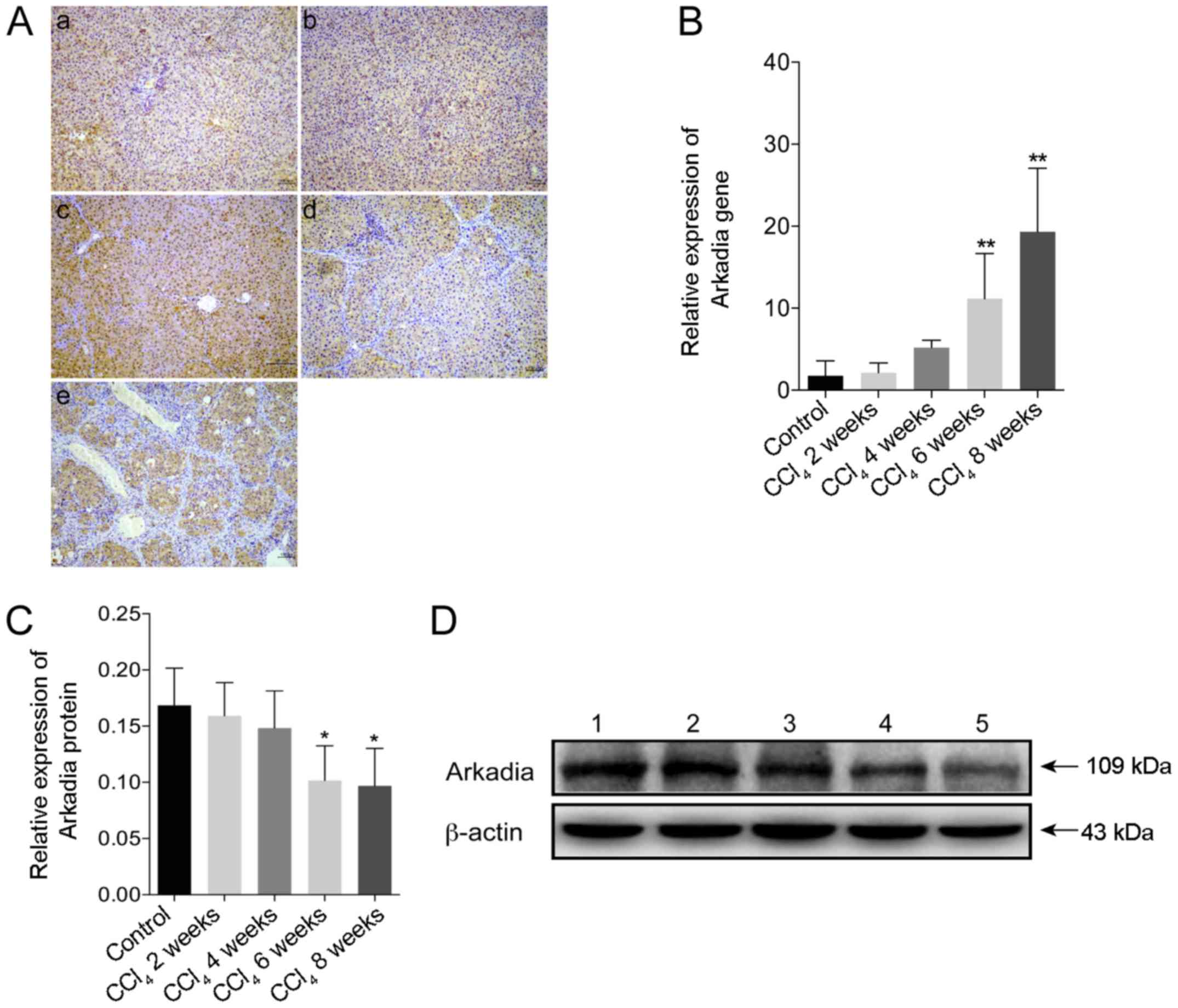 | Figure 4Arkadia expression in rats with
CCl4-induced hepatic fibrosis. Following sacrifice, the
expression levels of Arkadia in the liver samples were determined
by RT-qPCR, western blot analysis and immunohistochemistry. Results
are presented as representative images or are expressed as the
means ± standard deviation of individual groups (n=6/group). (A)
Immunohistochemical staining of Arkadia expression in the liver
sections (original magnification, ×100). (a) control; (b)
CCl4 2 weeks; (c) CCl4 4 weeks; (d)
CCl4 6 weeks and (e) CCl4 8 weeks. (B)
RT-qPCR analysis of Arkadia mRNA transcript levels in the liver
samples. (C) Relative protein expression levels of Arkadia. (D)
Western blot analysis of Arkadia protein. Lane 1, control; lane 2,
CCl4 2 weeks; lane 3, CCl4 4 weeks; lane 4,
CCl4 6 weeks and lane 5, CCl4 8 weeks.
*P<0.05, **P<0.01 vs. the control
group. CCl4, carbon tetrachloride; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction. |
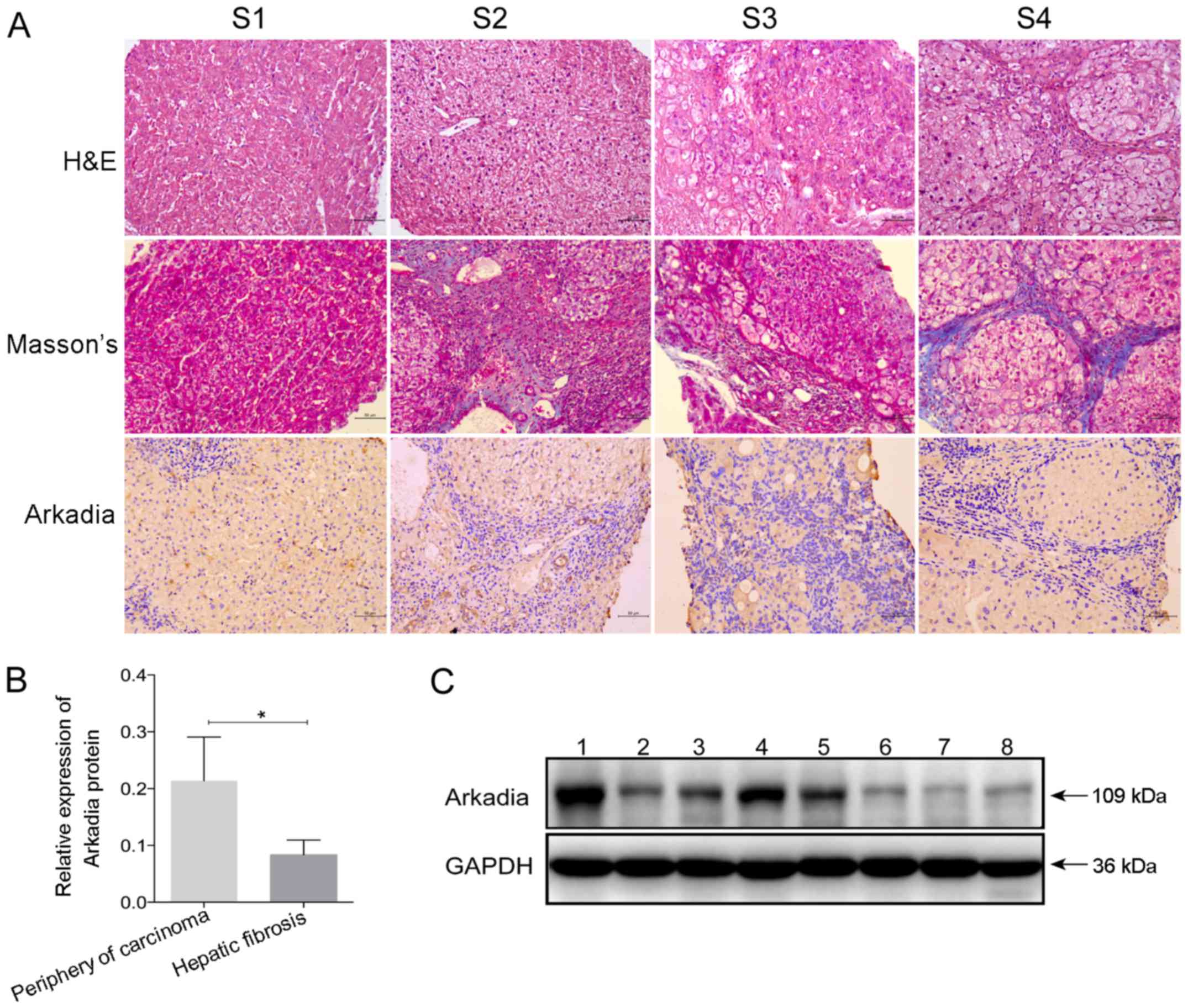
Expression of Arkadia in patients with
hepatic fibrosis
To examine the expression of Arkadia in the fibrotic
livers of human patients, the remnants of human liver tissues were
collected from biopsies conducted for routine clinical purposes,
and fibrotic liver samples were collected during surgery. The
characteristics of the study population are presented in Tables I and II. According to the region and extent
of collagen deposition, and the severity of liver structural
damage, patients were classified into stages S1–S4 of hepatic
fibrosis (Fig. 5A). Hepatocyte
swelling, cytoplasm rarefaction, spotty necrosis in the hepatic
lobule and inflammatory cell infiltration into the portal duct
areas were detected in the liver sample from the patient with S1
stage hepatic fibrosis. At S2 stage, numerous inflammatory cells
had infiltrated into the portal areas, and a few fibrous septa
formed by collagen fibers were located in the portal area; however,
the structure of the liver lobule retained normal architecture. The
liver lobules at S3 stage were disordered, alongside deposition of
fibrous septa, inflammatory cell infiltration and hepatocyte
degeneration. Eventually, pseudolobules were formed in the livers
at S4 stage, and hepatic fibrosis progressed to cirrhosis.
 | Table ICharacteristics of the study
population used for immunohistochemical staining. |
Table I
Characteristics of the study
population used for immunohistochemical staining.
| Characteristic | S1 stage | S2 stage | S3 stage | S4 stage |
|---|
| Age (years) | 21 | 24 | 32 | 38 |
| Sex | Male | Female | Male | Female |
| Etiology | HBV infection | HBV infection | HBV infection | HBV infection |
| Treatment | No | No | No | No |
 | Table IICharacteristics of the study
population used for western blot analysis (n=8). |
Table II
Characteristics of the study
population used for western blot analysis (n=8).
| Characteristic | Total (n=8) | Periphery of
carcinoma (n=5) | Hepatic fibrosis
(n=3) |
|---|
| Age (years) | 39.38±17.51 | 50.6±9.79 | 20.67±6.43 |
| Sex
(male:female) | 7:1 | 4:1 | 3:0 |
| Etiology | HBV infection | HBV infection | HBV infection |
| Degree of
fibrosis | – | – | S4 |
Further immunohistochemical staining indicated that
Arkadia was predominantly expressed in the cytoplasm, whereas
expression was reduced in the nucleus of hepatocytes and
cholangiocytes in the livers of human patients with various stages
of fibrosis (Fig. 5A). There was
no marked difference in the positive staining rate among the groups
of patients with different stages of hepatic fibrosis (S1–S4
stages). Western blot analysis revealed that the protein expression
levels of Arkadia were significantly lower in the fibrotic liver
samples compared with samples from the periphery of hepatocellular
carcinoma tissues (P<0.05; Fig. 5B
and C). The samples from the periphery of hepatocellular
carcinoma were considered the control.
Discussion
The present study investigated Arkadia expression in
the livers of rats with hepatic fibrosis. The results indicated
that Arkadia was highly expressed in the cytoplasm, whereas it was
weakly expressed in the nucleus of cholangiocytes and hepatocytes
from rat liver samples. Furthermore, reduced levels of Arkadia mRNA
transcripts, but increased protein expression levels of Arkadia
were detected in the livers of control rats. The mRNA expression
levels of Arkadia were increased in the livers of rats with
BDL-induced hepatic fibrosis, particularly at 7 days post-surgery.
Subsequently, the mRNA expression levels of Arkadia were gradually
reduced; however, they remained significantly higher compared with
in the sham group, with the exception of the insignificant
difference between the BDL 28 day group and sham group. Conversely,
the relative protein expression levels of Arkadia were
significantly reduced in the liver samples of rats with BDL-induced
hepatic fibrosis compared with in the sham group; the levels were
gradually reduced in the livers of rats with hepatic fibrosis
throughout the observation period. In the CCl4 -induced
rat model of hepatic fibrosis, gradually increased levels of
Arkadia mRNA transcripts and decreased levels of protein expression
were detected in the livers of the CCl4-treated groups
of rats. Similarly, significantly reduced levels of Arkadia protein
were detected in the liver samples from patients with various
stages of hepatic fibrosis. These findings suggested that Arkadia
mRNA and protein expression may be inversely associated in fibrotic
livers during the development and progression of hepatic
fibrosis.
There are numerous potential reasons regarding the
absence of a positive correlation between the mRNA and protein
expression levels of Arkadia, and these may not be mutually
exclusive (19,20). Firstly, there are numerous complex
and varied post-transcriptional mechanisms that regulate the
translation of mRNA into protein, which are not yet sufficiently
well defined to be able to compute protein concentrations from mRNA
transcripts. A previous study reported that some genes exhibited
minimal variation in their mRNA expression throughout the cell
cycle and were likely to have little or no correlation with final
protein level; the cells may control the open reading frames at the
translational and/or post-translational level, with the mRNA levels
being somewhat independent of final protein concentration (19). Furthermore, while a positive
correlation does exist between mRNA and protein abundance, mRNA
abundance is not a predictor of protein abundance in multicellular
organisms. Secondly, proteins may differ in their in vivo
half lives, as a result of varied protein synthesis and degradation
(20). Cells can control the
rates of degradation and synthesis for specific proteins, and there
is significant heterogeneity, even within proteins that have
similar functions (21,22). Alternatively, it is possible that
the ubiquitin proteasome system may degrade the Arkadia protein in
the progression of hepatic fibrosis. We aim to further investigate
how hepatic fibrosis regulates Arkadia expression in the liver.
In conclusion, these findings suggested that
decreased Arkadia protein expression may be associated with the
progression of hepatic fibrosis. Although several studies have
reported that Arkadia positively contributes to renal tubular
epithelial to mesenchymal transition through the degradation of
Smad7 (23), the mechanistic
roles of Arkadia are rarely reported in hepatic fibrosis.
Therefore, the current findings will encourage us to further
investigate how Arkadia affects the progression of hepatic
fibrosis.
The present study hypothesized that Arkadia may
participate in hepatic fibrosis. Using two hepatic fibrosis models
induced by BDL and CCl4 intoxication, and human liver
samples from patient with hepatic fibrosis, the results identified
the expression and localization of Arkadia in hepatic fibrosis. The
results indicated that Arkadia protein expression was significantly
decreased, whereas the levels of Arkadia mRNA transcripts were
elevated in fibrotic livers.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81300334).
References
|
1
|
Puche JE, Saiman Y and Friedman SL:
Hepatic stellate cells and liver fibrosis. Compr Physiol.
3:1473–1492. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brenner DA, Waterboer T, Choi SK,
Lindquist JN, Stefanovic B, Burchardt E, Yamauchi M, Gillan A and
Rippe RA: New aspects of hepatic fibrosis. J Hepatol. (32)(Suppl):
32–38. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moreira RK: Hepatic stellate cells and
liver fibrosis. Arch Pathol Lab Med. 131:1728–1734. 2007.PubMed/NCBI
|
|
4
|
Lim YS and Kim WR: The global impact of
hepatic fibrosis and end-stage liver disease. Clin Liver Dis.
12:733–746. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Richburg JH, Myers JL and Bratton SB: The
role of E3 ligases in the ubiquitin-dependent regulation of
spermatogenesis. Semin Cell Dev Biol. 30:27–35. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nagano Y, Mavrakis KJ, Lee KL, Fujii T,
Koinuma D, Sase H, Yuki K, Isogaya K, Saitoh M, Imamura T, et al:
Arkadia induces degradation of SnoN and c-Ski to enhance
transforming growth factor-beta signaling. J Biol Chem.
282:20492–20501. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mavrakis KJ, Andrew RL, Lee KL,
Petropoulou C, Dixon JE, Navaratnam N, Norris DP and Episkopou V:
Arkadia enhances Nodal/TGF-beta signaling by coupling
phospho-Smad2/3 activity and turnover. PLoS Biol. 5:e672007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ling H, Roux E, Hempel D, Tao J, Smith M,
Lonning S, Zuk A, Arbeeny C and Ledbetter S: Transforming growth
factor β neutralization ameliorates pre-existing hepatic fibrosis
and reduces cholangiocarcinoma in thioacetamide-treated rats. PLoS
One. 8:e544992013. View Article : Google Scholar
|
|
9
|
Massagué J and Wotton D: Transcriptional
control by the TGF-beta/Smad signaling system. EMBO J.
19:1745–1754. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu F, Liu C, Zhou D and Zhang L:
TGF-β/SMAD pathway and its regulation in hepatic fibrosis. J
Histochem Cytochem. 64:157–167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Koinuma D, Shinozaki M, Komuro A, Goto K,
Saitoh M, Hanyu A, Ebina M, Nukiwa T, Miyazawa K, Imamura and
Miyazono K: Arkadia amplifies TGF-beta superfamily signalling
through degradation of Smad7. EMBO J. 22:6458–6470. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Levy L, Howell M, Das D, Harkin S,
Episkopou V and Hill CS: Arkadia activates Smad3/Smad4-dependent
transcription by triggering signal-induced SnoN degradation. Mol
Cell Biol. 27:6068–6083. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Le Scolan E, Zhu Q, Wang L, Bandyopadhyay
A, Javelaud D, Mauviel A, Sun L and Luo K: Transforming growth
factor-beta suppresses the ability of Ski to inhibit tumor
metastasis by inducing its degradation. Cancer Res. 68:3277–3285.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsubakihara Y, Hikita A, Yamamoto S,
Matsushita S, Matsushita N, Oshima Y, Miyazawa K and Imamura T:
Arkadia enhances BMP signalling through ubiquitylation and
degradation of Smad6. J Biochem. 158:61–71. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Aghaei I, Shabani M, Doustar N, Nazeri M
and Dehpour A: Peroxisome proliferator-activated receptor-γ
activation attenuates motor and cognition impairments induced by
bile duct ligation in a rat model of hepatic cirrhosis. Pharmacol
Biochem Behav. 120:133–139. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Javadi-Paydar M, Ghiassy B, Ebadian S,
Rahimi N, Norouzi A and Dehpour AR: Nitric oxide mediates the
beneficial effect of chronic naltrexone on cholestasis-induced
memory impairment in male rats. Behav Pharmacol. 24:195–206. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Uchinami H, Seki E, Brenner DA and
D’Armiento J: Loss of MMP 13 attenuates murine hepatic injury and
fibrosis during cholestasis. Hepatology. 44:420–429. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Greenbaum D, Colangelo C, Williams K and
Gerstein M: Comparing protein abundance and mRNA expression levels
on a genomic scale. Genome Biol. 4:1172003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Khositseth S, Pisitkun T, Slentz DH, Wang
G, Hoffert JD, Knepper MA and Yu MJ: Quantitative protein and mRNA
profiling shows selective post-transcriptional control of protein
expression by vasopressin in kidney cells. Mol Cell Proteomics.
10:M110.0040362011. View Article : Google Scholar :
|
|
21
|
Lian Z, Kluger Y, Greenbaum DS, Tuck D,
Gerstein M, Berliner N, Weissman SM and Newburger PE: Genomic and
proteomic analysis of the myeloid differentiation program: Global
analysis of gene expression during induced differentiation in the
MPRO cell line. Blood. 100:3209–3220. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gerner C, Vejda S, Gelbmann D, Bayer E,
Gotzmann J, Schulte-Hermann R and Mikulits W: Concomitant
determination of absolute values of cellular protein amounts,
synthesis rates, and turnover rates by quantitative proteome
profiling. Mol Cell Proteomics. 1:528–537. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu FY, Li XZ, Peng YM, Liu H and Liu YH:
Arkadia regulates TGF-beta signaling during renal tubular
epithelial to mesenchymal cell transition. Kidney Int. 73:588–594.
2008. View Article : Google Scholar
|